Abstract
In multiple myeloma, next-generation sequencing (NGS) has expanded our knowledge of genomic lesions, and highlighted a dynamic and heterogeneous composition of the tumor. Here we used NGS to characterize the genomic landscape of 418 multiple myeloma cases at diagnosis and correlate this with prognosis and classification. Translocations and copy number abnormalities (CNAs) had a preponderant contribution over gene mutations in defining the genotype and prognosis of each case. Known and novel independent prognostic markers were identified in our cohort of proteasome inhibitor and immunomodulatory drug-treated patients with long follow-up, including events with context-specific prognostic value, such as deletions of the PRDM1 gene. Taking advantage of the comprehensive genomic annotation of each case, we used innovative statistical approaches to identify potential novel myeloma subgroups. We observed clusters of patients stratified based on the overall number of mutations and number/type of CNAs, with distinct effects on survival, suggesting that extended genotype of multiple myeloma at diagnosis may lead to improved disease classification and prognostication.
Key points
Next-generation sequencing allows analysis of the integrated spectrum of gene mutations, aneuploidies and IGH translocations in multiple myeloma.
Karyotypic events have a stronger impact on prognosis than mutations, but extended genotyping shows novel prognostic categories.
Introduction
Multiple myeloma (MM) derives from the neoplastic transformation and proliferation of a post-germinal center B-cell. Karyotypic events are the main drivers of early stages of transformation [1–3], and most MM cases are either hyperdiploid (HDMM) or harbor translocations of the immunoglobulin heavy chain (IGH) locus [4]. In addition, several additional recurrent translocations and copy number abnormalities (CNAs) can be found [5], although at diagnosis, their analysis is usually limited to events that have an established prognostic role and may guide treatment: del17p, t(4;14) and t(14;16) [6, 7]. Gene mutations are thought to be secondary events associated with tumor progression rather than initiation [2, 8]. Several next-generation sequencing (NGS) studies to date have expanded our knowledge on the spectrum of gene mutations in MM [8–13]. However, such studies have also highlighted substantial heterogeneity and low recurrence rates as compared to other hematological malignancies. Furthermore, the integrated spectrum of gene mutations, rearrangements and CNAs of each case is composed of alterations that may belong to different subclones, often in dynamic and differential evolution during the various stages of disease [14]. NGS technologies are ideally suited to return information on those driver events, whose analysis and correlation with clinical and laboratory features of the patient can impact prognosis and disease classification. Initial efforts based on whole exome sequencing (WES) have shown that integration of certain genomic lesions into standard risk models can improve prognostication in MM [13]. However, the full potential of NGS studies has not been exploited so far. Custom target pulldown (TPD) has significant advantages over WES in that it allows analysis of a limited fraction of the genome, thus reducing the cost and complexity of downstream analysis per sample. The success of this methodology in detecting gene mutations at diagnosis or relapse has been illustrated in myeloid malignancies [15–18] and MM [19–21]. We have previously shown that TPD panels can be designed to interrogate the integrated spectrum of gene mutations, CNAs and IGH translocations in MM, thus providing a potential one-stop platform for prognostication of newly diagnosed MM cases [22]. Here we applied our custom TPD to a large cohort of multiple myeloma samples at diagnosis to understand MM genomic and clinical interrelationships. We report our unbiased analysis of a large set of MM driver events, leading to the identification of novel prognostic factors and significant interactions between genomic events, and suggesting that analysis of the extended myeloma genotype in larger cohorts can identify novel subgroups that are biologically and clinically distinct.
Methods
Sequencing and identification of mutations
In total 10 ng of genomic DNA from CD138+ bone marrow cells were subjected to whole-genome amplification. Patient-level tags were added and samples pooled before custom target enrichment and sequencing, which was performed on Illumina HiSeq2000 machines on a paired-end 75 bp protocol. Reads were aligned with BWA-mem and variants called with in-house algorithms [23, 24] using a previously described pipeline to filter out probable artifacts and germline variants, and to rank somatic variants based on their likely oncogenic potential [10, 15].
CNAs, IGH rearrangements and VAF adjustment
Coverage data were used to identify regions of aneuploidies, after normalization to diploid samples. This was performed at the whole-chromosome level, and from there down to cytogenetic bands and gene loci. Hyperdiploid samples were defined by a gain in at least two of the following chromosomes: 3, 5, 7, 9, 11, 15, 19, 21. Translocations were called as previously described [22]. Gene-level CNA information was also used to adjust the variant allelic frequency (VAF) of each variant, to estimate the number of cells bearing a given variant, as previously described [16].
Statistical analysis
Pairwise association studies were performed using Fisher test corrected for multiple hypotheses testing. For survival analysis, both progression-free (PFS) and overall survival (OS) were used as end-points, and log-rank tests were used for univariate hypothesis tests after correction for multiple hypothesis testing. For multivariate survival analyses, sparse Cox regression was performed on the full set of driver events. Further information on methodology can be found in the online supplement.
Results
Patients and sequencing metrics of the study
We used TPD to sequence unmatched genomic DNA from 373 MM patients at diagnosis, and we added 45 patients from a previously published WES study [10] for a total of 418 (Table 1). Mean age was 56.6 years. Most patients (76.3%) received bortezomib-based induction treatment followed by autologous hemopoietic stem cell transplant. First-line bortezomib-treated patients had a younger median age (55.6 vs 59.8 years, p = 1 × 10−5, Student's t-test). Median follow-up was 5.4 years. At the time of analysis, 85.9% of patients relapsed, and 48.3% died.
Table 1.
Overall clinical features of cases included in the study
| Variable | Baseline distribution in cohort |
|---|---|
| Sample | |
| Sample size for sequencing | 418 |
| Sample size for outcome data | 418 |
| Median (range) follow-up | 5.4 (0.1–11.5) years |
| Demographics | |
| Sex | |
| Male | 243 (58.1%) |
| Female | 171 (40.9%) |
| NA | 4 (1%) |
| Age, mean ± SD | 56.6 ± 8.4 years |
| Biochemical | |
| β2 microglobulin, mean ± SD | 7.8 ± 9.6 (mg/L) |
| MM staging | |
| ISS I | 150 (35.9%) |
| ISS II | 102 (24.4%) |
| ISS III | 86 (20.6 %) |
| NA | 80 (19.1%) |
| Treatment | |
| VD-HDM | 266 (63.6%) |
| VTD-HDM | 53 (12.7%) |
| VAD-HDM | 67 (16.0%) |
| MPV/KMP | 5 (1.2%) |
| MPT | 10 (2.4%) |
| RD | 5 (1.2%) |
| SMM | 1 (0.2%) |
| NA | 11 (2.6%) |
| Outcome | |
| Relapse | 359/418 (85.9%) |
| Death | 202/418 (48.3%) |
We sequenced an average of 1.46 Gb per sample to a target region depth of 118.9× (Supplementary Figure 1). The average on-target efficiency was 34.3%, including the IGH locus that shows extensive deletions in plasma cells, leading to underestimation of the actual on-target efficiency [22]. Although coverage was lower than our previous WES study (Supplementary Figure 2A), the distribution of gene mutations did not show any significant differences (Supplementary Figure 2B), suggesting that the overall performance of our TPD was comparable to the previous WES study.
Analysis of gene mutations in multiple myeloma
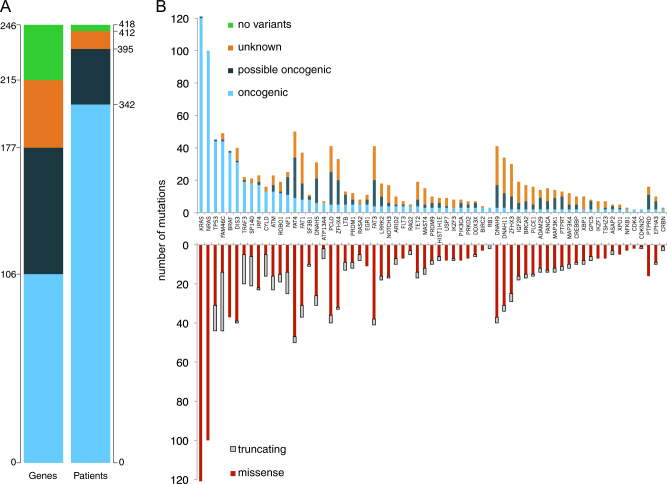
After excluding artifacts and likely germline single-nucleotide polyorphisms, we identified 2269 high-confidence, likely somatic variants in 215 of the 246 genes included in the design in 412 out of the 418 patients analyzed (Supplementary Table S1). We then compared the expected pattern of mutations in each given gene derived from the literature—typically inactivating mutations for tumor suppressor genes and hot-spot mutations for oncogenes—to the pattern observed in our series to triage each variant into “oncogenic”, “possible oncogenic”, or “unknown” classes (see Supplemental Methods for further details). Looking at oncogenic mutations only, we found 695 variants in 106 genes in 342 patients (Fig. 1a). Numbers increased to 1250 variants in 177 genes in 395 patients with the addition of possible oncogenic variants. At least one oncogenic or possible oncogenic variant was found in 94.5% of patients, with a median of two per patient. The list of most commonly mutated genes closely recapitulated previously published data (Fig. 1b, top) [9–13]. Clustered missense mutations were prevalent in known driver oncogenes such as KRAS, NRAS and BRAF, whereas an excess of truncating mutations was found in known tumor suppressors such as TP53, FAM46C, SP140 (Fig. 1b, bottom). Most oncogenic mutations (63%) were accounted for by nine of the top driver genes previously identified, i.e., KRAS, NRAS, TP53, FAM46C, BRAF, DIS3, TRAF3, SP140, IRF4 (Fig. 1b). A mutation in at least one of these nine genes was found in 64% of patients. Conversely, many genes showed a large excess of variants of unknown or possible oncogenic potential. Among these, large genes like FAT1, FAT3, FAT4, DNAH9, DNAH11, PCLO, whose role in myeloma pathogenesis remains unclear [10, 13, 25]. Last, we found sporadic oncogenic mutations with potential clinical impact in CRBN and IKZF1, previously associated with resistance to immunomodulatory drug [19, 26] in <1% of patients each. Our mixed confirmation/discovery effort did therefore not identify novel genes mutated at a significant recurrence rate in MM, but at the same time we showed how the long tail of uncommonly mutated genes contributed a significant fraction of the heterogeneous genomic landscape of MM. Given the possible inclusion of artifacts and/or germline variants in mutations of “unknown” class, we only considered variants of oncogenic or possible oncogenic classes as driver mutations for subsequent analysis.
Fig. 1.
Absolute number of mutations in the study: a Left: absolute numbers of genes with at least one mutation of the specified class found in the study. Right: Number of patients carrying at least one mutation of the specified class. Oncogenic variants: light blue. Possible oncogenic variants: dark blue. Variants of unknown significance: orange. No variants identified: green. b Top: stacked bar chart of mutations in the study, limited to genes with >2 oncogenic mutations, broken down by mutation class. Bottom: For the same genes, missense variants are in red, truncating variants are in gray
Overall, 56% of patients displayed activation of the RAS/MAPK pathway, as clustered missense mutations of KRAS, NRAS, BRAF, and truncating mutations of NF1 and RASA2. The NF-kB pathway was recurrently hit as well, with mutations—mostly inactivating—of TRAF3,CYLD and LTB in 12% of patients. Last, mutations or deletions of genes broadly implicated in the DNA damage response (TP53, ATM, ATR, BRCA2) were observed in 22% of patients. Although the mutational spectrum was heterogeneous and most genes showed low recurrence rates, frequent involvement of these pathways confirms their functional relevance in MM pathogenesis.
Identification of copy number changes and IGH translocations allows analysis of the integrated genomic landscape of myeloma
With most MM patients displaying prognostically relevant structural abnormalities by karyotype or fluorescence in situ hybridization (FISH) analysis, we sought to derive this information from our NGS data [22]. Recurrent IGH translocations were identified in 135 out of 418 patients. FISH validation for t(4;14) on 119 of them showed our approach had 91% sensitivity and 98% specificity (Supplementary Table S2). We identified 57% of patients as HDMM and 32% as IGH-translocated (Fig. 2a). 9% of patients harbored both an hyperdiploid karyotype and an IGH translocation, and t(4:14) was the most prevalent in this subgroup [27]. We then identified segmental chromosomal aneuploidies with prognostic significance in 61% of patients, at rates comparable to previous studies based on SNP arrays [28]. Furthermore, we identified gene-level gains and losses. Many gene amplifications were concordant with whole-chromosome trisomies in hyperdiploid cases, such as CCND1 in chromosome 11, prompting exclusion of these events from further analysis. The most frequent gene deletions instead consisted of known tumor suppressors, and for TP53, CYLD, SNX7 these coexisted so frequently with larger segmental chromosomal deletions that we aggregated loss of the gene locus and loss of the chromosome segment as one variable for further analysis. After these adjustments, genuine gene-level copy number alterations were found in 51% of patients.
Fig. 2.
The genomic landscape of multiple myeloma: a Pie chart of the breakdown of samples in the study, classified based on the presumed founder cytogenetic abnormalities: hyperdiploidy and IGH translocations. HDMM hyperdiploid multiple myeloma, NA not available for analysis, None no cytogenetic event identified. IGH_Tx translocation involving the IGH region. b Bar chart of the prevalence of each class of alteration (copy number abnormalities -CNA-, karyotype, gene-level copy number abnormalities, gene mutations) in samples in the study. c Stacked bar chart of the most frequent genomic events of any class, broken down by cytogenetic group
Considering recurrent translocations and aneuploidies, deletions of tumor suppressor genes, amplification of oncogenes, and mutations pertaining to “oncogenic” or “possible oncogenic” classes, at least one such driver event was present in >99% of patients (Supplementary Figure 3A). Overall, a median of 6 events were present in each patient (Fig. 2b). KRAS and NRAS were the only point mutations present in the 15 most frequent driver events, highlighting how karyotypic events dominate the integrated genomic landscape of MM (Fig. 2c). In addition, we interestingly found frequent deletions in genes required for plasma cell development such as XBP1 and PRDM1 (Fig. 2c) that also show recurrent mutations, often truncating, suggesting they may represent novel tumor suppressors in MM (Supplementary Table S1). Hyperdiploid cases showed a small but significantly higher average number of mutations than cases with IGH translocations (2.75 vs 2.5, chi-square test p = 0.002) (Supplementary Figure 3B), suggesting that while additional genetic events are required for disease progression, the evolutionary trajectory of each myeloma case may vary based on the initiating event.
Predictive and prognostic value of individual genomic events
We asked whether genomic features were correlated with clinico-laboratoristic features. Using linear models, we found only two associations. 17p deletions presented a modest but significant (p = 0.001, likelihood ratio test) correlation with increasing age. Unlike myeloid disorders [15, 16] and clonal hemopoiesis [29–33], here TP53 mutations were not correlated with age (Supplementary Figure 4A, B), suggesting a different evolutionary trajectory. Furthermore, 1q amplification was significantly correlated with higher beta-2 microglobulin levels (p = 0.003, Supplementary Figure 4C). Among disease-independent predictors of survival, older age was associated with poorer survival as previously reported (Cox p = 0,019, Supplementary Figure 4D) [34]. First line proteasome inhibitor (PI) treatment was associated with improved PFS, and with a trend towards improved overall survival (OS, Supplementary Figure 4E), whose impact was more difficult to evaluate given the availability of PI-based salvage treatment in the non-PI induction cohort. Finally, we found no significant association between genomic lesions and survival for PI-treated patients, including no significant benefit from bortezomib in t(4;14) [7]. The low frequency of most driver events, and the likely modest size of the effect of a given treatment on a driver event, warrant larger cohorts are analyzed to answer this question in a comprehensive way.
We performed a univariate Cox analysis to infer the prognostic value of each genomic feature for PFS and OS (Supplementary Table S3). We found 12 variants significantly associated with poorer PFS and/or OS (Supplementary Figure 5). Interestingly, most gene mutations were not relevant for prognosis. The only mutated gene with a clear prognostic impact on both PFS and OS was TP53, while DNAH11 mutations conferred worse OS only (Log-rank test, p = 6.9 × 10−4 and 7.3 × 10−3 for OS, respectively). Looking at gene-level gains and losses, we found 5 events conferring shorter OS, including losses of TP53/17p, CYLD/16q, FAT1, and amplifications of MYC and NRAS. We could confirm the negative impact on survival of t(4;14) [4] and that of regions of recurrent CNAs [28] (that were selected for their prognostic impact in the first place). Despite this selection bias, the lack of prognostic value of most gene mutations and their overall lower level of recurrence compared to cytogenetic changes underscores the relevance of karyotypic events in shaping the clinical behavior of MM.
Independent prognostic value of driver genomic lesions and their interaction
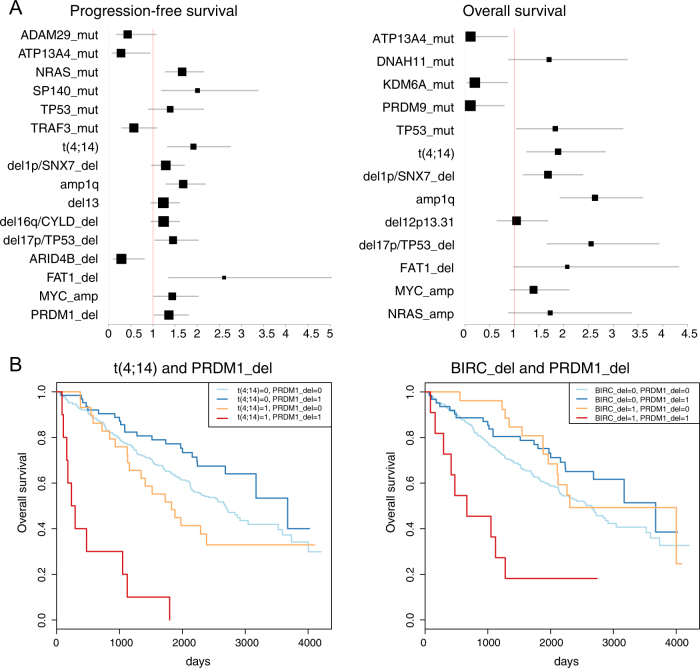
We next performed a multivariate sparse Cox regression on the full set of genomic variables. For PFS, 18 features had non-zero coefficients, with 9 of them being significant in a second multivariate Cox regression restricted to the first shortlist of 18 variables. Among these, mutations in ATP13A4 and deletions in ARID4B had a favorable impact, whereas mutations in SP140 and NRAS, t(4;14), amp(1q), del(17p13) and deletions of FAT1 and PRDM1 had a negative impact (Fig. 3a, left). For the OS regression, 13 features had non-zero coefficients with 7 of them being significant in the restricted analysis: mutations in ATP13A4, KDM6A and PRDM9 had a favorable impact, while t(4;14), amp(1q), del(17p13), del(1p) had a negative impact (Fig. 3a, right). Finally, because our cohort was heterogeneous in demographics and treatment received, we used multivariate linear regression models to exclude a confounding effect of clinical features on the prognostic value of each of these genomic variables, confirming they can be considered as independent prognostic events (Supplementary Figure 6).
Fig. 3.
Independent variants affecting survival: a Forest plot of variables implicated in progression-free survival (left) and overall survival (right). Hazard ratio on the X axis, values <1 confer better prognosis, values >1 confer worse prognosis. For each variable, the confidence interval is a horizontal gray bar and the hazard ratio—referenced to the X-axis—is represented by a black box. b Kaplan–Meier survival curves for two significant instances of interaction found in the study: prognosis for overall survival is significantly worse in case of coexistence of PRDM1 deletions and t(4;14) (left) or PRDM1 deletions and BIRC2/3 deletions (right) than with either variable alone
Subsequently, we systematically looked for significant interactions between driver events, i.e. combinations where the prognostic impact on OS of one is significantly altered if another is co-occurring. Despite the large set of variables, we found only two significant interactions: cases bearing both t(4;14) and PRDM1 deletion had a dismal median OS of 265 days (HR 9.05, p = 3 × 10−4 for interaction, Fig. 3b, left); BIRC2/3 deletion and PRDM1 deletion conferred a median OS of 666 days (HR 6.3, p = 0.04) (Fig. 3b, right). When analyzed in isolation, PRDM1 deletions had no effect (Supplementary Table S3), implying that the prognostic value of driver oncogenic events in MM may differ based on their genomic context.
We also looked for combinations of variables that could be relevant for OS, even if not significant for interaction, across the whole dataset. We first analyzed the association of TP53 mutations and chr17p deletions, two events that are known to co-occur [10], and the prognosis was slightly worse for the combination (20/418 patients), in a trend suggestive of an additive effect (Supplementary Figure 7A). However, the shortest OS was that of t(4;14) and TP53 mutations (Supplementary Figure 7B), conferring a dismal 228 day median overall survival to patients bearing both events. HDMM patients bearing an IGH translocation showed a worse OS compared to those without (Supplementary Figure 7C). On the same lines, OS was negatively affected by the total number of driver oncogenic events, irrespective of their nature, in a stepwise decline (Supplementary Figure 7D), mainly supported by karyotypic events (Supplementary Figures 7E-F). This reinforces the notion that myeloma evolution toward more aggressive disease is driven by acquisition of additional driver events.
Clonal and subclonal driver mutations
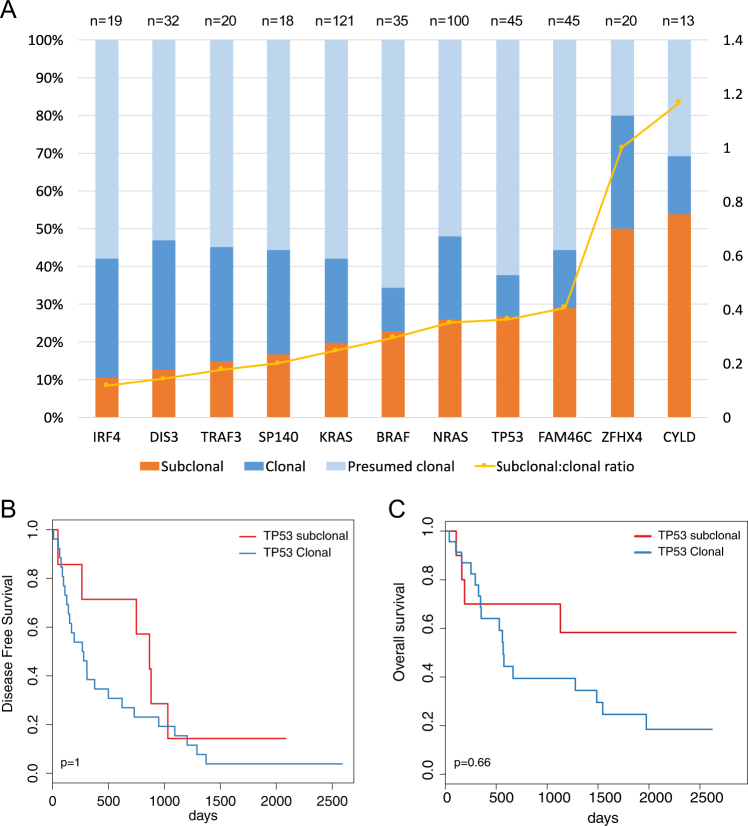
Adjusted VAF (aVAF) can be used to infer the fraction of cells carrying each mutation, so to time its order of acquisition [15, 16]. Subclonal mutations in known driver genes such as KRAS, NRAS, TP53 were identified in 19.8%, 26% and 26.7% of cases, respectively (examples in Supplementary Figure 8A). When ranked based on the fraction of subclonal occurrences, all genes had evidence of both clonal and subclonal mutations, but CYLD and ZFHX4 had a somewhat higher percentage of subclonal variants, suggesting these mutations may preferentially be acquired later in myeloma evolution (Fig. 4a). TP53 was also more frequently subclonal, but was nevertheless clonal in 11% of myeloma cases. Conversely, IRF4 and DIS3 mutations were most likely to be clonal and therefore acquired earlier in evolution. We then looked at recurrent precedencies in pairwise analysis of mutated genes, and again found no recurrent pattern suggesting that mutations of driver genes, in our sample size, do not follow preferential evolutionary trajectories (Supplementary Table S4). Interestingly, we found that multiple mutated alleles of driver gene mutations -up to 5 for TP53- could be found at the subclonal level in a significant fraction of patients (Supplementary Figure 8B), again highlighting the heterogeneous subclonal structure of MM due to convergent evolution [35, 36].
Fig. 4.
Subclonal mutations: a Stacked bar chart of the clonality status of the top 11 mutated genes in our cohort: bars refer to the left Y-axis and plotted is the proportion of variants that are subclonal (orange), clonal (dark blue) and presumed clonal (light blue)—i.e., cases where confidence intervals of aVAF are all overlapping and low tumor purity could lead to over-estimation of clonality. The yellow line refers to the right Y-axis and represents, for each gene, the ratio between subclonal and clonal variants, i.e., higher values correspond to genes with more subclonal variants. (b)Kaplan–Meier plot of progression-free survival in patients with subclonal (red) or clonal (blue) TP53 mutations with p-values from univariate analysis (log-rank test) adjusted for multiple hypotheses testing. c Kaplan–Meier plot of overall survival in patients with subclonal (red) or clonal (blue) TP53 mutations with p-values from univariate analysis (log-rank test) adjusted for multiple hypotheses testing
The impact of clonal status on the prognostic value of gene mutations is unclear for most cancers and has never been studied in myeloma. Interestingly, we found that the clonality status of mutations did not influence survival, with the exception of a trend towards improved OS for TP53 subclonal mutations (Fig. 4b, c, Supplementary Figure 9).
Patterns of co-occurring and mutually exclusive events
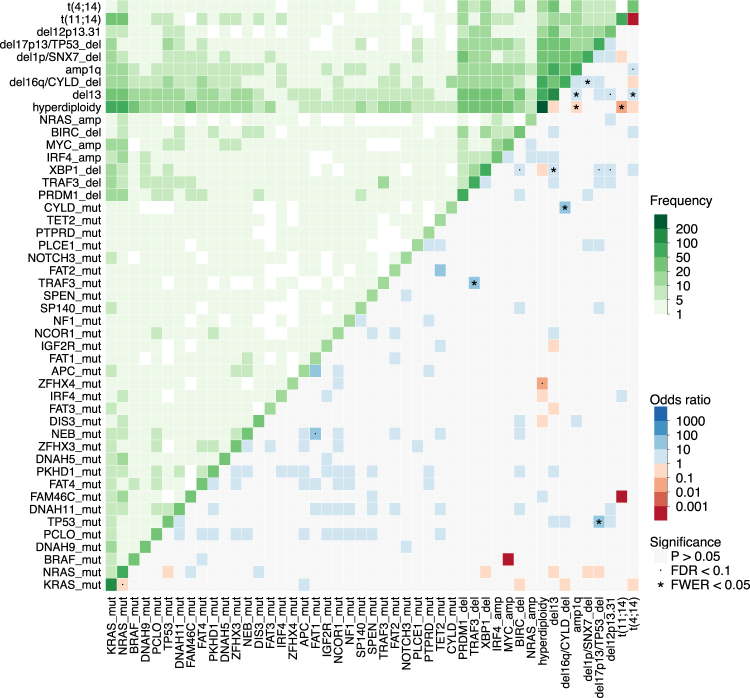
Patterns of co-occurrence of driver events were systematically analyzed to look for potential cooperativity in pairwise associations and functional redundancy for mutually exclusive variables. In the list of 197 driver events we identified 843 significant interactions at p < 0.05 (Fisher Test), 15 at false-discovery rate <0.1 and 9 at family-wise error rate <0.05 out of a total 19306 possible interactions (Supplementary Figure 10, most frequent events in Fig. 5). Among, significant pairwise interactions, some were previously described, such as the positive correlation between chr13 and known poor prognostic factors such as t(4;14), amp1q, del12p13.31, del17p13, TP53 mutations. KRAS and NRAS mutations were mutually exclusive between each other but not with BRAF mutations. We found a general pattern where mutations in tumor suppressor genes co-occurred with deletion of the wild-type allele in a typical double-hit manner for TP53, CYLD, and TRAF3 mutations, supporting that these bi-allelic events not only underlie relapse but may be present at diagnosis as well [37]. Hyperdiploid and IGH-translocated cases showed significant differences in the spectrum of associated genomic events. Chr13 deletions, chr1q amplifications, DIS3 and ZFHX4 mutations were less frequent in HDMM, which in turn was enriched for FAM46C mutations. XBP1 deletions clustered with deletions of NF-kB pathway genes TRAF3 and BIRC2/3, and with chromosomal events such as chr12p, chr13, chr17p deletions and t(4;14). Finally, t(11;14) and t(4;14) showed a diverse spectrum of associated driver lesions, with t(4;14) cases in particular being associated with chr13 deletions and chr1q amplifications.
Fig. 5.
Pairwise association between variables: Heatmap showing pairwise analysis of occurrence of the most frequent genomic events in MM. The same variable is plotted in the X and Y axis, and the intensity of color in the leading diagonal indicated the frequency of the variable in the dataset. In the upper triangle, intensity of green indicates the frequency of co-occurrence of any two variables. In the lower triangle, associations are colored by odds ratio: non-significant ones are in gray, while significant ones are in blue if co-occurring, and red if mutually exclusive (p-value < 0.05, fisher test corrected for multiple hypothesis testing). Events with false-discovery rate <0.1 are marked with a dot and events with family-wise error rate <0.05 are marked with a star. While only 47 variables are shown, statistical significance is computed on the full dataset of 192 variables shown in Supplementary Figure 10
We then extended this analysis to uncover preferential trajectories of cancer evolution highlighting different associations of hotspot mutations within genes (Supplementary Figure 11). We found a tendency for hotspot mutations in KRAS, NRAS, BRAF, IRF4 to show a differential pattern of co-occurrence, but none with a significant false-discovery rate.
Genomic structure suggests new myeloma subtypes with clinical impact
The identification of preferential patterns of associations or mutual exclusivity of driver lesions, prompted us to explore whether new myeloma subtypes could be identified based on their extended genotype using a previously described Bayesian model [16]. We could reliably identify four clusters (Fig. 6a) of MM cases with good posterior probability of class assignment (Supplementary Figure 12Ai–Aiii), albeit lower than acute myeloid leukemia, which carry a simpler genomic structure [16]. Aside from a small cluster 1 (3 patients) where no driver events could be identified, most hyperdiploid and IGH-translocated cases clustered together in the large cluster 3 (291 patients, 69.6%). We then identified two clusters, 2 and 4, composed of 58 (13.9%) and 66 (15.8%) patients, respectively. Both clusters were characterized by a significantly lower number of mutations but showed opposing features otherwise. Cluster 2 was enriched for IGH translocations, had the highest number of CNAs, was enriched for amp(1q), del(13), del(17p), deletions of BIRC2/3 and XBP1 and carried more TP53 mutations (Fig. 6b, Supplementary Figure 12B). On the contrary, cluster 4 was mostly composed of hyperdiploid cases and showed fewest CNAs and mutations overall (Fig. 6b, Supplementary Figure 12B). There was no association between treatment (PI, ASCT) and subgroups, while cluster 4 was characterized by a slightly lower age (Supplementary Figure 12C).
Fig. 6.
Clustering of myeloma samples based on genomic landscape: a Bayesian Dirichlet clustering process of the 418 MM cases (in columns) based on genomic variables (in rows, top panel), with verticals black lines showing separation between identified clusters. A zoomed in view for the karyotypic abnormalities is provided in the bottom panel. Cluster 1 is composed of three patients where no driver event could be identified. b For each of the four clusters, the histogram of the distribution of gene mutations (top) and that of the CNAs (bottom) are provided. c Progression-free (left) and overall survival (right) Kaplan–Meier analysis of the four clusters
Prognosis was different for both PFS (Fig. 6c, left) and OS (Fig. 6c, right) across the clusters. Cluster 2 showed the worse median OS—1973 days—whereas cluster 4 was associated with longer survival (median OS not reached), again showing concordance between the number of CNAs and prognosis.
Discussion
We provide the first large-scale TPD effort in MM to comprehensively describe its driver genomic landscape and how this impacts on clinical behavior. Compared to WES, we reduced the amount of input DNA, the complexity of downstream analysis and forwent the need for a matched sample, to move this technology one step closer to the clinic. Our approach was robust and conservative, and even larger studies may allow re-classification of events discarded in our list as non-driver.
Gene mutations were frequent in our cohort; however, we found a preponderant contribution of the few highly recurrently mutated genes that were already described. Nevertheless in individual cases, rare gene mutations may impact treatment decisions. For example, CRBN, IKZF1 or IKZF3 mutations (totaling a combined 1.6% of patients in our cohort) would predict resistance to immunomodulatory drug [26], whereas XBP1 deletions could predict bortezomib resistance [38], suggesting future clinical-grade targeted platforms must include rare driver events that have predictive value for treatment.
Confirming previous reports, the prognostic yield of gene mutations was rather low in our large cohort as compared to chromosomal events. PFS and OS were both impacted only by TP53 mutations and by rare mutations in ATP13A4, an ion transporter previously associated with developmental disorders [39] but whose role in MM was never reported before. We did not confirm a prognostic role of genes such as IRF4, EGR1 and ZFXH4 [13], possibly because of front-line PI treatment and longer follow-up of our cohort (63 months here vs 25 in ref. [13]). We identified subclonal mutations for all recurrent driver genes, showing that lesions classically associated with disease progression can spontaneously arise before diagnosis. As these lesions can be positively selected by treatment, sensitive methods must be employed for their identification at diagnosis to achieve an accurate prognostication. Some events had a strong, context-dependent, prognostic value, such as deletions of PRDM1 coupled with either t(4;14) or BIRC2/3 deletions. These interactions may be infrequent but are highly instructive and clinically relevant, supporting the use of extended gene panels in prognostic studies. However, to identify more such instances, much larger sample sizes will be needed given the heterogeneous genome of MM and the expected modest effect of most interactions. In the future, global risk calculators that incorporate extended clinical, demographic, laboratoristic and genomic variables will be able to address these points [40]. BIRC2/3 genes deletions have been described in myeloma [41, 42], whereas PRDM1 has only recently been identified as a tumor suppressor based on recurrent truncating mutations [11]. Truncations and deletions in PRDM1 were nevertheless previously described in lymphomas [43, 44], where they are associated with poor prognosis [45]. Deletions of the 6q21 chromosome band containing the PRDM1 locus are known to be recurrent in MM [46]. The targeted nature of our study, however, did not allow us to characterize the size of such deletions and thus we cannot exclude that other tumor suppressors deleted in 6q contributed to this observation.
Overall, driver events had an additive effect on prognosis, showing that progressive accumulation of abnormalities correlates with clinical aggressiveness, almost independently of the nature of such events. Therefore, our findings extend previous initial observations on modulation of prognosis by additive negative prognostic factors in FISH [47], and that of aneuploidies in the prognosis of HDMM or IGH translocations [48, 49].
Current MM classification based on IGH translocations or hyperdiploidy is reliable and biologically relevant, as it is based on known early driver events. Here, we report on the first attempt toward a genomic classification of myeloma using innovative clustering algorithms based on the extended genotype of each patient. Allowing for the heterogeneity of the disease, we could identify disease subgroups characterized by a different spectrum of translocations, CNAs and gene mutations, as well as different contribution by each class of events. A cluster of hyperdiploid cases characterized by the fewest gene mutations and CNAs showed a relatively good prognosis, whereas non-hyperdiploid cases carrying multiple segmental chromosomal aneuploidies and fewer gene mutations carried a worse prognosis. Again, specific gene mutations did not contribute much to the clustering. Similar to our analysis, a report integrating MM expression profiles with mutational data to identify subgroups with biological and prognostic values showed little contribution of gene mutations to the clustering [50]. However, knowledge of the mutational status of a large number of genes allowed us to perform a prognostically relevant clustering of cases based on the overall mutational burden, supporting the value of analysis of infrequently mutated genes. Lastly, a recent study also reported on the dissection of hyperdiploid cases with different prognosis based on specific additional cytogenetic lesions, supporting the validity of extended genotyping for prognostication of MM [51].
The increasing availability of novel drugs and the better understanding of pathways involved in MM progression prompts the need for rationalized treatment approaches and mandates that high-risk disease is diagnosed with accuracy [52,53]. Probably, future genomic studies will provide not only improved prognostication, but also predictive factors of response and actionable mutations that will help in treatment choices. The methodology and results described here represent an important advance that can accelerate the introduction of genomics in the clinical approach to MM.
Electronic supplementary material
Acknowledgements
This work was supported by a grant from NIH PO1-155258 (NCM, PJC, KCA, HAL, and SM), RO1-124929 (NCM) P50-100007 (KCA, NCM) and by the Wellcome Trust (grant reference 077012/Z/05/Z). NB was funded by a MFAG Grant No. 17658 from the Associazione Italiana Ricerca sul Cancro (AIRC). FM was supported by the Associazione Italiana Leucemie e Linfomi (AIL) and by the Società Italiana di Ematologia Sperimentale (SIES). PJC is personally funded through a Wellcome Trust senior clinical research fellowship. KCA is an American Cancer Society clinical research professor.
Author contributions
NB designed the study, collected and analyzed the data and wrote the paper; GB, MM, SG, YL, CdP, FM, TB, MG analyzed data. Y-TT, EK, MD, PM, SM provided samples. VS, LM, SO’M, KR, JWT, APB, CC processed samples and raw sequencing data. PC, KCA, PJC, EP analyzed data and critically revised the manuscript. HA-L and NCM conceived the project, provided samples, interpreted data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41375-018-0037-9) contains supplementary material, which is available to authorized users.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 3.Corre J, Munshi N, Avet-Loiseau H. Genetics of multiple myeloma: another heterogeneity level? Blood. 2015;125:1870–6. doi: 10.1182/blood-2014-10-567370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca R, Leif Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zingone A, Kuehl WM. Pathogenesis of monoclonal gammopathy of undetermined significance and progression to multiple myeloma. Semin Hematol. 2011;48:4–12. doi: 10.1053/j.seminhematol.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosiñol L, et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J Clin Oncol. 2015;33:2863–9. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avet-Loiseau H, LeLeu X, Roussel M, Moreau P, Guerin-Charbonnel C, Caillot D, et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p) J Clin Oncol. 2010;28:4630–4. doi: 10.1200/JCO.2010.28.3945. [DOI] [PubMed] [Google Scholar]
- 8.Walker BA, Walker BA, Wardell CP, Wardell CP, Melchor L, Melchor L, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–90. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–86. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 13.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–20. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14:100–13. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- 15.Papaemmanuil E, Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolli N, Manes N, McKerrell T, Chi J, Park N, Gundem G, et al. Characterization of gene mutations and copy number changes in acute myeloid leukemia using a rapid target enrichment protocol. Haematologica. 2015;100:214–22. doi: 10.3324/haematol.2014.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortüm KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128:1226–33. doi: 10.1182/blood-2016-02-698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortüm KM, Langer C, Monge J, Bruins L, Egan JB, Zhu YX, et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M(3) P) in -17p high risk disease. Br J Haematol. 2015;168:507–10. doi: 10.1111/bjh.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortuem KM, Braggio E, Bruins L, Barrio S, Shi CS, Zhu YX, et al. Panel sequencing for clinically oriented variant screening and copy number detection in 142 untreated multiple myeloma patients. Blood Cancer J. 2016;6:e397. doi: 10.1038/bcj.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolli N, Li Y, Sathiaseelan V, Raine K, Jones D, Ganly P, et al. A DNA target-enrichment approach to detect mutations, copy number changes and immunoglobulin translocations in multiple myeloma. Blood Cancer J. 2016;6:e467. doi: 10.1038/bcj.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones D, Raine KM, Davies H, Tarpey PS, Butler AP, Teague JW, et al. cgpCaVEManWrapper: simple execution of CaVEMan in order to detect somatic single nucleotide variants in NGS Data. Curr Protoc Bioinf. 2016;56:15.10. 1–15.10.18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raine KM, Hinton J, Butler AP, Teague JW, Davies H, Tarpey P, et al. cgpPindel: identifying somatically acquired insertion and deletion events from paired end sequencing. Curr Protoc Bioinf. 2015;52:15.7.1–12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohr JG, Lohr JG, Stojanov P, Stojanov P, Lawrence MS, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan JB, Kortuem KM, Kurdoglu A, Izatt T, Aldrich J, Reiman R, et al. Extramedullary myeloma whole genome sequencing reveals novel mutations in Cereblon, proteasome subunit G2 and the glucocorticoid receptor in multi drug resistant disease. Br J Haematol. 2013;161:748–51. doi: 10.1111/bjh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca R, Debes-Marun CS, Picken EB, Dewald GW, Bryant SC, Winkler JM, et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood. 2003;102:2562–7. doi: 10.1182/blood-2003-02-0493. [DOI] [PubMed] [Google Scholar]
- 28.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–45. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig H, Bolejack V, Crowley J, Bladé J, Miguel JS, Kyle RA, et al. Survival and years of life lost in different age cohorts of patients with multiple myeloma. J Clin Oncol. 2010;28:1599–605. doi: 10.1200/JCO.2009.25.2114. [DOI] [PubMed] [Google Scholar]
- 35.Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF, et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28:1705–15. doi: 10.1038/leu.2014.13. [DOI] [PubMed] [Google Scholar]
- 36.Raab MS, Lehners N, Xu J, Ho AD, Schirmacher P, Goldschmidt H, et al. Spatially divergent clonal evolution in multiple myeloma: overcoming resistance to BRAF inhibition. Blood. 2016;127:2155–7. doi: 10.1182/blood-2015-12-686782. [DOI] [PubMed] [Google Scholar]
- 37.Weinhold N, Ashby C, Rasche L, Chavan SS, Stein C, Stephens OW, et al. Clonal selection and double hit events involving tumor suppressor genes underlie relapse from chemotherapy: myeloma as a model. Blood. 2016;128:1735–44. doi: 10.1182/blood-2016-06-723007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano GP, Besse L, Li N, Kraus M, Besse A, Meeuwenoord N, et al. Proteasome inhibitor-adapted myeloma cells are largely independent from proteasome activity and show complex proteomic changes, in particular in redox and energy metabolism. Leukemia. 2016;30:2198–207. doi: 10.1038/leu.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwasnicka-Crawford DA, Carson AR, Roberts W, Summers AM, Rehnström K, Järvelä I, et al. Characterization of a novel cation transporter ATPase gene (ATP13A4) interrupted by 3q25-q29 inversion in an individual with language delay. Genomics. 2005;86:182–94. doi: 10.1016/j.ygeno.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Gerstung M, Papaemmanuil E, Martincorena I, Bullinger L, Gaidzik VI, Paschka P, et al. Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet. 2017;49:332–40. doi: 10.1038/ng.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–44. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–7. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 45.Xia Y, Xu-Monette ZY, Tzankov A, Li X, Manyam GC, Murty V, et al. Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of activated B-cell-like diffuse large B-cell lymphoma. Leukemia. 2016;31:625–36. doi: 10.1038/leu.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cigudosa JC, Rao PH, Calasanz MJ, Odero MD, Michaeli J, Jhanwar SC, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–10. [PubMed] [Google Scholar]
- 47.Boyd KD, Ross FM, Chiecchio L, Dagrada GP, Konn ZJ, Tapper WJ, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26:349–55. doi: 10.1038/leu.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlyn C, Melchor L, Murison A, Wardell CP, Brioli A, Boyle EM, et al. Coexistent hyperdiploidy does not abrogate poor prognosis in myeloma with adverse cytogenetics and may precede IGH translocations. Blood. 2015;125:831–40. doi: 10.1182/blood-2014-07-584268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chretien ML, Corre J, Lauwers-Cances V, Magrangeas F, Cleynen A, Yon E, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015;126:2713–9. doi: 10.1182/blood-2015-06-650242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laganà A, Perumal D, Melnekoff D, Readhead B, Kidd BA, Leshchenko V, et al. Integrative network analysis identifies novel drivers of pathogenesis and progression in newly diagnosed multiple myeloma. Leukemia. 2017 doi: 10.1038/leu.2017.197. [DOI] [PubMed] [Google Scholar]
- 51.Shah V, Sherborne AL, Walker BA, Johnson DC, Boyle EM, Ellis S, et al. Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia. 2017;32:102–10. doi: 10.1038/leu.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Usmani SZ, Rodriguez-Otero P, Bhutani M, Mateos MV, Miguel JS. Defining and treating high-risk multiple myeloma. Leukemia. 2015;29:2119–25. doi: 10.1038/leu.2015.209. [DOI] [PubMed] [Google Scholar]
- 53.Maura F, Petljak M, Lionetti M, Cifola I, Liang W, Pinatel E. et al. Biological and prognostic impact of APOBEC-induced mutations in the spectrum of plasma cell dyscrasias and multiple myeloma cell lines. Leukemia. 2017. ePub 6 December 2017. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.