Abstract
Purpose
To develop 1H MR spectroscopy that provides distinction of GABA signal at 3T in vivo.
Methods
Triple-refocusing was tailored at 3T, with numerical simulations and phantom validation, for distinction of the GABA 2.29-ppm resonance from the neighboring glutamate (Glu) resonance. The optimization was performed on the inter-RF pulse time delays and the duration and carrier frequency of a non-slice selective RF pulse. The optimized triple refocusing was tested in multiple regions in six healthy subjects, including hippocampus. The in-vivo spectra were analyzed with LCModel using in-house basis spectra. After normalization of the metabolite signal estimates to water, the metabolite concentrations were quantified with reference to medial-occipital creatine at 8 mM.
Results
A triple-refocusing scheme with optimized inter-RF pulse time delays (TE=74ms) was obtained for GABA detection. With optimized duration (14 ms) and carrier frequency (4.5 ppm) of the non-slice selective RF pulse, the triple refocusing gave rise to distinction between the GABA 2.29-ppm and Glu 2.35-ppm signals. The GABA 2.29-ppm signal was clearly discernible in spectra in vivo (voxel size 4 – 12 mL; scan times 4.3 – 17 min). In total 24 spectra from six gray or white matter dominant regions, the GABA concentration was measured to be 0.62 – 1.15 mM (CRLB of 8 – 14%), and the Glu level 5.8 – 11.2 mM (CRLB of 3 – 6%).
Conclusion
The optimized triple refocusing provided distinction between GABA and Glu signals and permitted direct co-detection of these metabolites in the human brain at 3T in vivo.
Keywords: γ-Aminobutyric acid (GABA), 1H MRS, 3T, Triple refocusing, Human brain, Gray matter, White matter
INTRODUCTION
γ-Aminobutyric acid (GABA) and glutamate (Glu) are the major inhibitory and excitatory neurotransmitters in a mammalian central nervous system, respectively. Perturbations in the neurotransmitter activities associated with GABAergic and glutamatergic neurotransmission have been implicated in several neuropsychiatric disorders (1,2). The capability to measure GABA and Glu in the human brain in vivo is therefore of considerable therapeutic potential for brain disorders. Precise measurement of GABA by short-TE 1H MRS at 3T is challenging due to the extensive overlap of the low-concentration metabolite signals with neighboring abundant resonances. GABA has six J-coupled protons of the 2CH2, 3CH2, and 4CH2 groups, resonating at 2.29, 1.89, 3.01 ppm respectively (3). The C3- and C4-proton signals are strongly obscured by the large singlets of N-acetylaspartate (NAA) and total creatine (tCr), whilst the C2-proton resonance at 2.29 ppm is extensively overlapped with the abundant Glu C4-proton resonance (2.35 ppm).
The signal overlap is usually overcome by means of some form of spectral editing that utilizes the J coupling evolution of the GABA spin system. One example is J-difference editing, in which the GABA 3.01 ppm resonance is difference edited via its coupling to the 1.89 ppm resonance. This editing has several limitations. First, the macromolecule (MM) species that has J coupled resonances at approximately 3.0 and 1.7 ppm can be partially co-edited and interfere with GABA measurement. The co-edited signal of the MM species (bound lysine) that has short T2 diminishes rapidly (4,5), but mobile lysine, whose T2 may be as long as metabolite T2 (6), can bring about a co-edited signal at 3.01 ppm as large as the edited GABA signal even when the editing 180° radio-frequency (RF) pulse is highly selective (e.g., 26 ms long DANTE) (7). Although the co-editing of mobile lysine can be effectively minimized by means of symmetric excitation when the editing sequence can house a highly selective long editing RF pulse (8), mitigation of lysine co-editing is not straightforward in a widely-used form of J-difference editing (MEGA) at 3T (9), in which two editing 180° RF pulses are inserted within a point-resolved spectroscopy (PRESS) sequence with TE of 68 – 80 ms (10). Second, homocarnosine (HC) is co-edited in editing of GABA 3.01 ppm resonance, which occurs due to the close proximity of the C4- and C3-proton resonances of the HC GABA moiety to those of free GABA (3). Given that the homocarnosine level was measured to be approximately 50% of the free GABA level in the human brain biopsies (11), contamination of the HC GABA moiety signal is very concerning because HC is entirely co-edited in GABA editing. Third, complete cancelation of the tCr CH3 singlet cannot be taken for granted in vivo. For GABA and tCr concentrations of 1 and 8 mM respectively, for which the unedited GABA to tCr CH3 signal ratio is only about 4%, the MEGA-edited GABA signal at 3.01 ppm may be always smaller than 2% of the unedited tCr CH3 signal because of the chemical shift displacement effects. Given the prevalence of frequency drifts due to subject motions and potential instability of shimming currents, the spectral location and lineshape of the tCr CH3 singlet could be varied during the relatively long GABA MRS scans. Acceptable suppression of the tCr signal (for example, suppression ratio of 500 for residual tCr signal as small as 10% of the edited GABA signal) may be achievable only by rigorous retrospective data processing following data acquisitions with cardiac triggering (12). Lastly, frequency drifts may incur variations in the rotation angle of the GABA 1.89 ppm spins and consequently the difference-edited GABA signal strength with time during the scan, which can lead to erroneous estimation of GABA. The frequency drift artifacts can be mitigated using editing 180° RF pulses with large bandwidth, but use of these pulses will increase MM co-editing. Taken together, measurement of GABA using MEGA, which is widely used at 3T, is prone to several types of measurement errors and thus may not be ideally suitable for studying brain diseases in which alterations in GABA level are moderate, suggesting development of an alternative, robust MRS technique without considerable complications in data processing and signal contamination at 3T, a field strength which is widely used in clinical studies.
Given the presence of complexities with detection of the GABA 3.01 ppm resonance, one may consider detection of the GABA C2-proton resonance (2.29 ppm) for measuring GABA as an alternative. The Glu C4-proton resonance at 2.35 ppm, which is a major obstacle, is relatively distant from the GABA 2.29 ppm resonance. The Glu C4 protons are strongly coupled to the C3 protons while the GABA C2 protons are weakly coupled to the C3 protons at 3T. The signal intensity and pattern of the Glu 2.35 ppm resonance are therefore much more sensitive to the MRS sequence parameters such as inter-RF pulse time delays, compared with the GABA 2.29 ppm resonance. In the case of STEAM (stimulated-echo acquisition mode) with optimized echo and mixing times (TE=168 ms and TM=28 ms) (13), the single-quantum coherence evolution during the echo time periods, together with the zero-quantum filtering effects of the sequence, can lead to drastic reduction of the Glu C4-proton multiplet, thereby making a GABA signal discernible on the shoulder of the suppressed Glu signal. One more example of the J evolution difference between Glu and GABA is optimized-TE PRESS (point-resolved spectroscopy) at 7T (14), in which a PRESS TE of 91 ms (TE1 = 31 ms and TE2 = 61 ms) gave rise to narrowing of the Glu and GABA multiplets between 2.2 and 2.4 ppm and thus the GABA and Glu signals were completely discriminated without considerable suppression of the signals, thereby making it possible to co-detect GABA and Glu with precision. The significance of these two prior studies is that the GABA estimation may not contain considerable contaminations from lysine and HC because the lysine resonances are all far from 2.29 ppm (15) and the C2-proton resonance of the HC GABA moiety is relatively distant (~0.1 ppm) from the GABA C2-proton resonance (16). In addition, direct measurement of the target signal of GABA without need of subtraction between spectra is a great plus.
While a PRESS sequence can offer excellent separation between the GABA and Glu signals at 7T (14), which is due to the effects of the subecho time optimization on top of the enhanced spectral resolution of J coupled resonances at the high field, it is unlikely that at 3T, distinction of the GABA 2.29 ppm signal from the Glu 2.35 ppm signal without sacrificing the Glu signal strength is straightforward by tailoring the PRESS subecho times. A triple refocusing sequence (90° - 180° - 180° - 180°) has additional inter-RF pulse time delays and thus provides a platform for amplifying the coherence evolution difference between spin resonances, as shown in our recent studies (17,18). In this study we explore triple-refocusing for detection of the GABA 2.29 ppm resonance at 3T. For given RF envelopes, all possible components of the sequence, which included the four inter-RF pulse time delays and the duration and carrier frequency of a non-slice selective 180° RF pulse, were tailored, using density-matrix simulations, to achieve acceptable signal separation between the GABA C2-proton and Glu C4-proton resonances without considerable reduction of the signal strengths. Following validation of the GABA-optimized triple refocusing in a phantom solution, we tested the method in healthy subjects. Preliminary data from multiple brain regions, which are known to be implicated in many neuropsychiatric disorders, are presented.
METHODS
Density-matrix simulations were performed at 3T to optimize the triple-refocusing sequence parameters for distinction between the GABA and Glu signals between 2.2 and 2.4 ppm. The triple refocusing sequence, with three 180° RF pulses following a 90° excitation RF pulse, had four inter-RF pulse time delays, τ1, τ2, τ3, and τ4 (Figure 1a). The second 180° was non-slice selective (NS180) while three other RF pulses were slice selective. Volume localization was obtained with a 9.8-ms 90° pulse with bandwidth (BW) of 4.2 kHz at half amplitude and two 13.2-ms 180° pulses with BW of 1.3 kHz (19), at an RF field strength (B1) of 13.5 μT. The time evolution of the density operator was calculated by solving the Liouville-von Neumann equation for the Hamiltonian that included Zeeman, chemical shift, scalar coupling terms and shaped RF and gradient pulses, using a product operator based transformation matrix algorithm, as described in a prior study (Supplementary Information) (20). The spatial resolution of slice selection was set to 1% with respect to the slice thickness, namely, 0.01 = sample length/number of pixels/slice thickness, where the sample length was twofold greater than the slice thicknesses with number of pixels (isochromats) of 200. The carrier frequency of the slice selective RF pulses was set to 2.5 ppm. The 3D localized spectra of Glu and GABA were numerically calculated for nine NS180 durations (14 – 30 ms, with 2 ms increments) and for TE (= τ1+τ2+τ3+τ4) between 70 and 100 ms. The carrier frequency of NS180 was varied between 1 and 4.5 ppm with 0.5 ppm increments, ensuring 180° rotation of resonances between 0.5 and 5 ppm by the NS180 (17 = kHz BW × ms duration), whose frequency profile is shown in our prior study (18). With 1 ms increments for each of τ values that refocus the chemical shift evolution (i.e., τ1+τ3 = τ2+τ4), the total number of spectra was approximately 120,000 for each of GABA and Glu. With broadening of the spectra to singlet linewidth of 4 Hz and use of GABA and Glu T2 of 180 ms (21), a sequence parameter set was searched for using the following criteria: 1) GABA C2-proton peak-to-peak amplitude greater than 95% of the zero-TE GABA C2-proton signal amplitude, 2) Glu C4-proton peak-to-peak amplitude greater than 60% of the zero-TE Glu C4-proton signal amplitude, and 3) small variation of Glu signal between 2.25 and 2.32 ppm. The zero-TE signals of GABA and Glu were obtained by scaling 90°-acquire calculated signals for the localized volume. Published chemical shift and J coupling constants were used for the simulations (3). The computer simulation was programmed with Matlab (The MathWorks, Inc., Natick, MA). Glutamine (Gln) was not included in the simulation for sequence optimization because the Gln resonances, which are fairly distant from the GABA C2-proton resonance, may not interfere with GABA measurement.
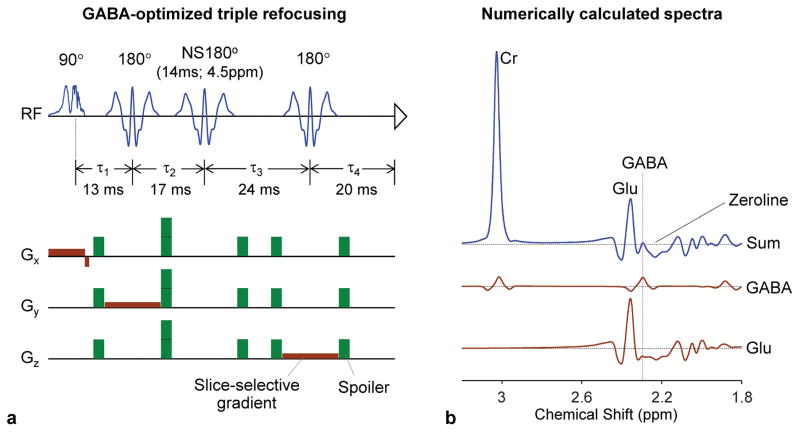
Figure 1.
(a) Schematic diagram of the GABA-optimized triple-refocusing sequence. The 90° pulse (9.8 ms; BW 4.2 kHz) and the first and third 180° pulses (13.2 ms; BW 1.3 kHz) were slice selective while the second 180° was non-slice selective (NS180; 14 ms long; BW 1.2 kHz). With the four inter-RF pulse time delays (τ1, τ2, τ3, τ4) of (13, 17, 24, 20) ms, the total TE was 74 ms. Slice selective gradients are shown in brown and spoiling gradients in green (each spoiler with strength 16 mT/m and duration 2.8 ms). The 14 ms NS180 pulse was tuned to 4.5 ppm. (b) Numerically calculated triple-refocused spectra of GABA, Glu and Cr are presented at the Cr FWHM of 4 Hz (similar to the in vivo mean singlet linewidth of the present study). The concentration ratio of GABA:Glu:Cr = 1:10:5. A vertical line is drawn at the GABA C2-proton resonance (2.29 ppm).
1H MR experiments were conducted in a whole-body 3T scanner (Philips Medical Systems, Best, The Netherlands), equipped with a whole-body coil for RF transmission and a 32-channel phased-array head coil for reception. In-vitro tests of the GABA-optimized triple-refocusing sequence were conducted on an aqueous solution (pH = 7.4) of GABA (3 mM), Glu (30 mM) and Cr (15 mM). Data were acquired, with TR 9 s and TE 74 ms, from a 2×2×2 cm3 voxel at the center of the phantom sphere (6 cm diameter). In addition, in-vitro experiments were conducted on a phantom with GABA (3 mM), Glu (30 mM), N-acetylaspartate (NAA) (30 mM) and glycine (Gly) (36 mM) for comparison of GABA editing by the triple refocusing and MEGA (TE 74 ms for both).
In-vivo MR scans were carried out in 6 healthy subjects (3 males and 3 females, age 27±5 years). The protocol was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center. Written informed consent was obtained from patients prior to MR scans. Triple-refocused MR spectra were obtained from the medial occipital and left parietal regions from all 6 subjects, of whom 3 subjects had additional MRS acquisitions in medial frontal, left frontal, posterior cingulate, and left hippocampus. MRS scan parameters included TR 2 s, TE 74 ms, sweep width 2.5 kHz, and 1024 sampling points. The number of signal averages (NSA) ranged from 128 to 512 averages, depending on the voxel size (3.9 – 12.2 mL). The carrier frequencies of the slice selective RF pulses were set at 2.5 ppm to minimize the chemical shift misregistration of resonances between 1 and 4 ppm and were real-time adjusted for B0 drifts in each excitation using a vendor-supplied tool (frequency stabilization). Water suppression was obtained with a vendor-supplied four-pulse variable-flip-angle scheme (22). Triple-refocused unsuppressed water was acquired from each voxel for eddy current compensation and multi-channel combination. Unsuppressed water was additionally obtained with short-TE (13 ms) MRS and TR 20 s for quantification of metabolites concentrations. Up to second order shimming was carried out using a vendor-supplied shimming tool. Proper flip angles of the RF pulses were obtained with MRS voxel localized B1 calibration followed by confirming equal strengths of triple refocused and PRESS water signals at TE 74 ms. 3D sagittal T1-weighted images were acquired with 1×1×1 mm3 resolution.
Following the combination of the 32-channel using a vendor-supplied algorithm, the data were zero filled to 4096 data points prior to Fourier transformation. Spectral fitting was performed with LCModel software (Ver 6.2F) (23), using numerically-calculated basis spectra of 17 metabolites, which included GABA, Glu, Gln, NAA, HC (homocarnosine), mI (myo-inositol), Gly (glycine), Tau (taurine), sI (scyllo-inositol), Asp (aspartate), PE (phosphoethanolamine), Lac (lactate), tCr (creatine + phosphocreatine), NAAG (N-acetylaspartylglutamate), tCho (glycerophosphocholine + phosphocholine), Glc (glucose), and GSH (glutathione). The spectral fitting was performed between 0.5 and 4.0 ppm. The Cramer-Rao lower bound (CRLB) was returned as a percentage standard deviation (SD) by LCModel. The contents of gray matter (GM), white matter (WM) and cerebrospinal fluids (CSF) within the MRS voxels were obtained from segmentation of the T1-weighted images using Statistical Parametric Mapping software (SPM5) (24). The metabolite signal estimates from LCModel were normalized to the short-TE water in GM and WM, and subsequently the metabolite concentrations were calculated by setting the mean tCr estimate of the medial occipital brain at 8 mM (25,26), similarly as in our prior study (14). To obtain the metabolite concentrations in pure GM and WM, the metabolite estimates from the voxels were fitted with a linear function of fractional GM content, fGM = GM/(GM+WM). Student’s t-test was performed for comparison of metabolite estimates between groups. Data are presented as mean ± SD.
RESULTS
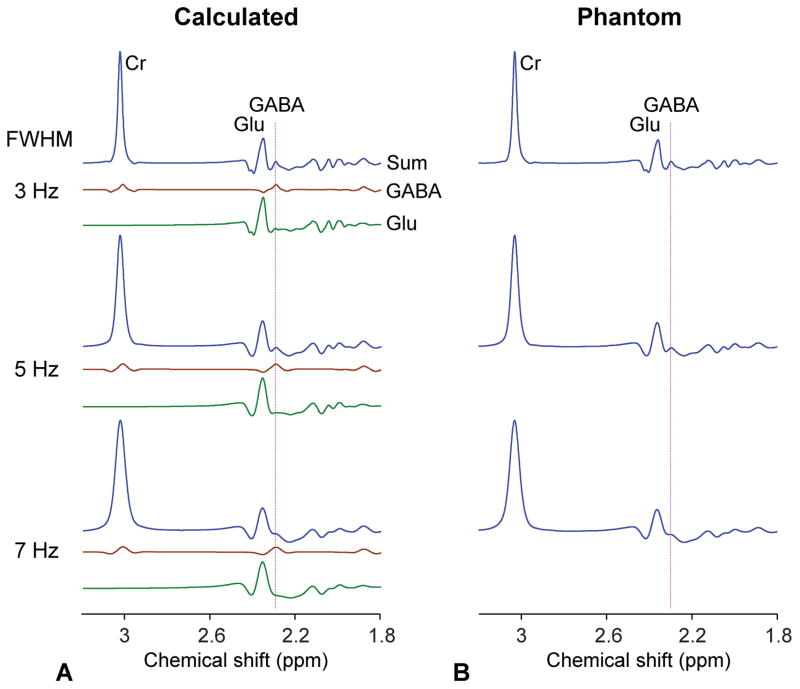
For co-detection of GABA and Glu signals, triple-refocusing sequence parameters were optimized as NS180 duration of 14 ms and carrier frequency of 4.5 ppm and the inter-RF pulse durations (τ1, τ2, τ3, τ4) = (13, 17, 24, 20) ms (TE = 74 ms) (Figure 1a). A numerical simulation showed that, for these triple-refocusing sequence parameters, the GABA signal at 2.29 ppm was clearly separated from the Glu 2.35 ppm signal, when broadened to singlet (Cr) linewidth of 4 Hz, (Figure 1b). The upfield sideband of the Glu multiplet was fairly flat between 2.25 and 2.32 ppm, making the GABA 2.29 ppm signal clearly discernible in the sum spectrum. Figure 2a presents numerically-calculated spectra of GABA, Glu and Cr at a concentration ratio of 1:10:5 for Cr-CH3 singlet linewidths (FWHM) of 3, 5 and 7 Hz. The spectral resolution of GABA with respect to Glu decreased progressively as FWHM increased. The GABA signal was well distinguishable from Glu for FWHM up to 7 Hz. The optimized triple refocusing was tested in an aqueous solution of GABA, Glu and Cr at a concentration ratio of 1:10:5 (Figure 2b). The signal strength and pattern of the calculated sum spectra were in excellent agreement with the phantom data. The FWHM dependence of GABA resolution agreed well between calculation and experiment. Spectral analysis of the phantom data reproduced the prepared concentration ratio within 2% errors incorporating phantom T2 values of the chemical compounds (650, 650 and 1200 ms for GABA, Glu and Cr-CH3 respectively).
Figure 2.
(a) Numerically-calculated triple-refocusing spectra of GABA, Glu, and Cr are presented for Cr FWHM of 3 – 7 Hz. The concentration ratio GABA:Glu:Cr equals 1:10:5. (b) Spectra from an aqueous solution with GABA (3 mM), Glu (30 mM), and Cr (15 mM), obtained using the GABA-optimized triple refocusing, are presented for various Cr FWHM, similarly as in calculated spectra. Each of the calculated and phantom spectra was normalized to its Cr peak amplitude. A vertical line is drawn at 2.29 ppm.
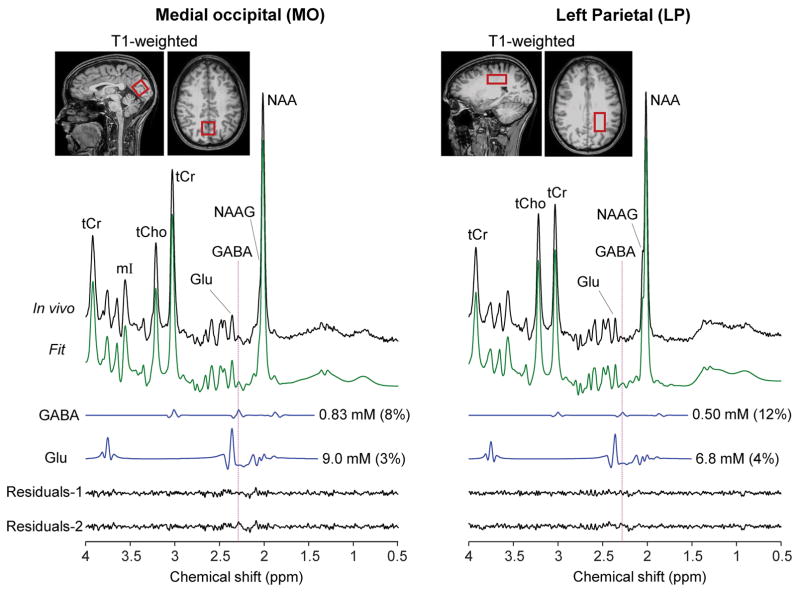
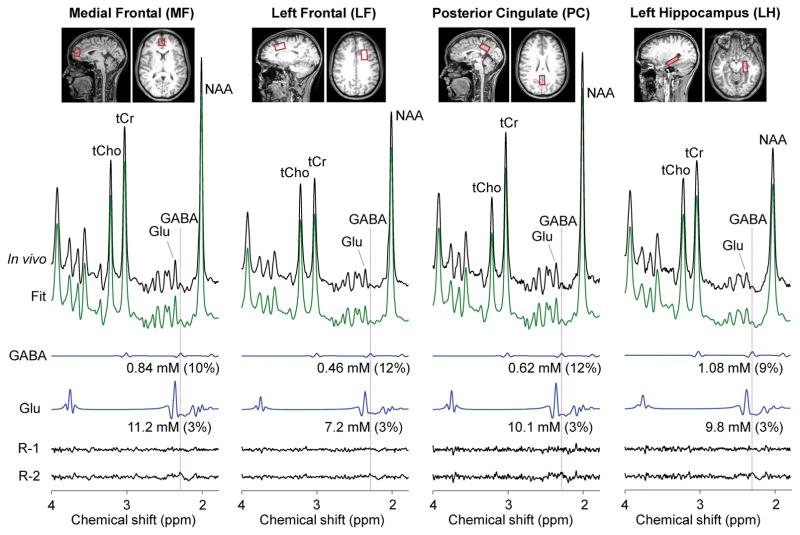
The GABA-optimized TE 74 ms triple refocusing was tested in six healthy volunteers. Representative in-vivo spectra from medial occipital (MO) and left parietal (LP) brain of a healthy volunteer are shown in Figure 3, together with the voxel positioning and spectral analysis results. The spectral patterns were well reproduced by the spectral fit between 0.5 and 4.0 ppm. A fairly large signal was clearly discernible at 2.29 ppm in the spectrum from the MO voxel, whilst the spectrum from the LP region showed a relatively small signal at 2.29 ppm. The GABA concentration estimate was higher by approximately twofold in MO than in LP region. The GABA CRLB was 8% and 12% for the MO and LP regions, respectively. The Glu signal at 2.35 ppm was also greater in MO than in LP region. The Glu CRLB was 3 – 4% in the spectra. To further validate the GABA detection, spectral fitting was undertaken with and without GABA signal in the basis set. In the fitting with GABA, the in vivo spectrum was well reproduced by the fit, leading to noise-level residuals in the proximity of 2.29 ppm (Residuals-1). When GABA was excluded from the fitting, residuals were clearly discernible at ~2.29 ppm (Residuals-2), indicating that the signal at 2.29 ppm was primarily attributed to GABA. In addition, we tested the GABA-optimized triple refocusing in medial frontal (MF), left frontal (LF), posterior cingulate (PC), and left hippocampus (LH) regions (Figure 4). The GABA signal at 2.29 ppm was also readily discernible in spectra from these brain regions. The spectral fittings with and without GABA in the basis set resulted in notable differences in the residuals in the proximity of the GABA 2.29 ppm resonance.
Figure 3.
In-vivo triple-refocused spectra from the medial occipital (MO) and left parietal (LP) regions of a healthy subject, normalized to the GM+WM water signals, are presented together with spectral fitting outputs and voxel positioning. The GABA and Glu signals, returned by LCModel spectral fitting, are displayed with the concentration estimates and CRLB in brackets. The voxel size was 23×23×23 and 35×23×15 mm3 for MO and LP, respectively (NSA = 128 for both). Residual-1 and Residuals-2 were obtained from spectral fittings with and without GABA in the basis set, respectively. Vertical lines are drawn at 2.29 ppm.
Figure 4.
In-vivo triple-refocused spectra from four brain regions of a healthy subject are presented similarly as in Figure 3. For medial frontal (MF), left frontal (LF), posterior cingulate (PC), and left hippocampus (LH), the voxel size was 18×23×28, 32×18×20, 26×18×15, and 30×13×10 mm3, with NSA of 128, 128, 320 and 512, respectively. R-1 and R-2 denote residuals from with-GABA and without-GABA fittings, respectively.
Data were acquired from the MO and LP regions in six subjects, of whom three had additional scans in MF, LF, PC and LH regions. For each brain region, the overall spectral pattern was similar between subjects. The residuals at 2.29 ppm from without-GABA fitting were overall larger in spectra from GM-rich regions than in spectra from WM-rich regions. For the total 24 spectra from the six subjects, the mean signal-to-noise ratios (SNR) of NAA (2.01 ppm) and tCr (3.03 ppm) were 224±41 and 147±30, while the mean FWHMs of NAA and tCr were 4.4±0.2 and 4.6±0.3 Hz, respectively. Here, SNR was the ratio of the peak amplitude with respect to the SD of the residuals between 0.5 and 1 ppm and the FWHM was obtained from the LCModel-returned singlets.
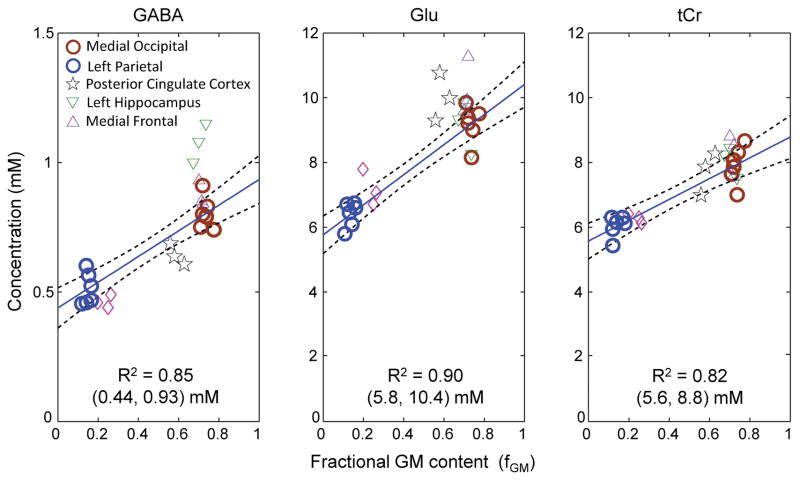
For the six subjects, the mean fractions of GM, WM, and CSF were estimated to be 0.61±0.03, 0.23±0.02, and 0.16±0.04 for MO, and 0.16±0.05, 0.81±0.08, and 0.03±0.04 for LP, respectively. The fractional GM content, i.e., fGM = GM/(GM+WM), was then calculated as 0.73±0.02 and 0.14±0.02 for the MO and LP voxels, respectively (Table 1). The GABA concentration was significantly higher in MO than in LP (0.80±0.06 vs. 0.51±0.07 mM; p = 2×10−5). We performed linear regression of these MO and LP GABA estimates with respect to fGM (Figure 5). The GABA levels in pure GM and pure WM (i.e., Y-axis intercepts at fGM = 1 and 0) were estimated as 0.93 and 0.44 mM respectively, suggesting that GABA is approximately 2 fold higher in pure GM than in pure WM. Glu was well measurable in all data with high precision (CRLB 3 – 4%), and significantly higher in MO than in LP (9.2±0.6 vs. 6.4±0.4 mM, p < 0.001), resulting in the pure-GM and pure-WM concentrations at 10.4 and 5.8 mM respectively. The tCr level in MO was also significantly higher than that of LP (p < 0.001).
Table 1.
The concentration estimates (mM) and CRLBs (%) of GABA, Glu and tCr are tabulated for the 6 brain regions studied, together with the voxel size, number of signal averages (NSA), and mean fractional GM content (fGM), GM/(GM+WM). The metabolite concentration was estimated with reference to the mean tCr of the medial occipital region at 8 mM, following normalization of metabolite signal estimates to the GM+WM water signal. Data are mean ± SD (n = 6 for medial occipital and left parietal, and n = 3 for other brain regions). Asterisks (*) represent the significance of the metabolite level difference from the estimates in the medial occipital region (* p ≤ 0.05 and **p ≤ 0.001).
| Brain region | Voxel size | NSA | fGM | Concentration (mM) (CRLB %) | ||
|---|---|---|---|---|---|---|
| GABA | Glu | tCr | ||||
| Medial Occipital (MO) | 23×23×23 mm3 (12.2 mL) | 128 | 0.73±0.02 | 0.80±0.06 (8.8±0.8) | 9.20±0.60 (3.2±0.4) | 8 (±0.57) (1±0) |
| Left Parietal (LP) | 35×23×15 mm3 (12.1 mL) | 128 | 0.14±0.02 | 0.51±0.07** (11.7±2.0) | 6.44±0.44** (3.8±0.4) | 6.05±0.43** (1±0) |
| Medial Frontal (MF) | 18×23×28 mm3 (11.6 mL) | 128 | 0.70±0.02 | 0.87±0.06 (9.7±0.6) | 10.30±0.90 (3.0±0.0) | 8.30±0.66 (1±0) |
| Posterior Cingulate (PC) | 26×18×15 mm3 (7.0 mL) | 320 | 0.57±0.04 | 0.66±0.04** (12.0±1.5) | 10.12±0.74 (3.0±0.0) | 7.81±0.66 (1±0) |
| Left Hippocampus (LH) | 30×13×10 mm3 (3.9 mL) | 512 | 0.70±0.03 | 1.08±0.08** (10.7±2.1) | 9.15±0.81 (4.7±1.2) | 8.08±0.48 (1±0) |
| Left Frontal (LF) | 32×18×20 mm3 (11.5 mL) | 128 | 0.24±0.03 | 0.46±0.03** (12.3±1.5) | 7.19±0.55** (3.3±0.6) | 6.28±0.02** (1±0) |
Figure 5.
Linear regression of the GABA, Glu and tCr estimates vs. fractional GM contents fGM (= GM/(GM+WM)). The regression was performed on the medial occipital and left parietal brain data (thick circles in blue and brown). For each metabolite, the Y-axis intercepts at fGM = 0 and 1 are shown in a bracket, below the coefficient of determination (R2). Dashed lines indicate 95% confidence intervals of the linear fits.
Significant difference in GABA between GM-rich and WM-rich regions was also observed in frontal brain (i.e., 0.87 and 0.46 mM in MF and LF respectively; p = 0.02) (Table 1). For the PC voxel, which showed approximately equal contents of GM and WM (i.e., fGM = 0.57), the GABA estimate was significantly lower compared with other GM-dominant regions (Figure 5). Of note, the GABA level in LH was significantly higher than those in MO (p = 0.008) and MF (p = 0.02) while the GM contents of the LH, MF and MO regions were about the same (fGM = 0.7 – 0.73). For Glu, the concentration was significantly different between GM-rich and WM-rich regions similarly as in GABA (Figure 5), but its distribution throughout the brain (i.e., multiple brain regions) was somewhat different than GABA; namely, the Glu estimate was not significantly different between MO, LH, MF and PC (Table 1). The tCr concentrations at the multiple locations was largely governed by the GM and WM contents in the voxels.
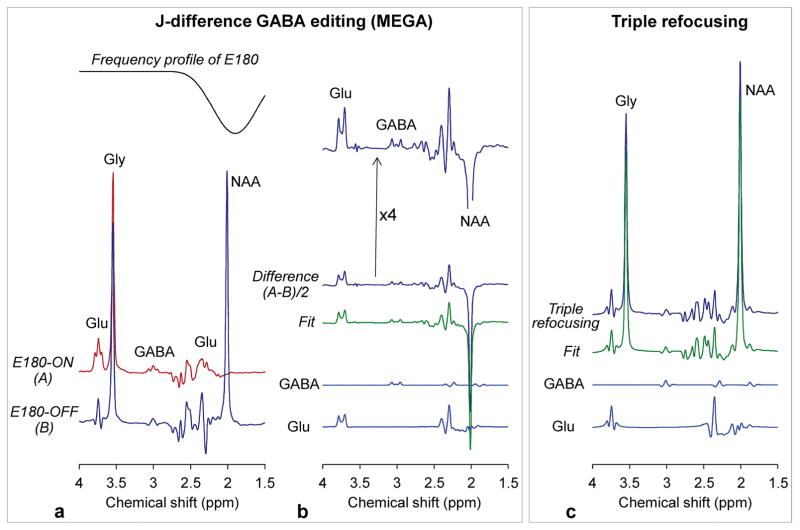
Lastly, in-vitro comparison of MEGA and triple refocusing was performed (TE 74 ms for both) (Figure 6). In MEGA, GABA and Glu were edited using an editing 180° RF pulse (E180) with large bandwidth (88 Hz; 13 ms Gaussian envelope truncated at 10%), implemented within a PRESS sequence with 6.9 ms slice selective 180° RF pulses (BW 1.3 kHz), whose envelope can be found in a prior paper [Choi et al. NMR Biomed 2013;26:1242–1250]. Due to the effect of the large E180 bandwidth, the NAA singlet (2.01 ppm) and Glu C3-proton resonances (2.04 and 2.12 ppm) were largely dephased in the E180-ON scan (Figure 6a). The edited GABA (3.01 ppm) signal was as small as 23% relative to the coedited Glu (3.75 ppm) signal for their concentration ratio of 1/10. The GABA and Glu signals were 1.6% and 6.8% with respect to that of Gly, whose concentration was prepared to produce a tCr signal strength in in-vivo situations (i.e., [GABA]:[Glu]:[tCr] = 1:10:8). LCModel analyses of the MEGA and triple-refocusing data showed good agreement, reproducing the prepared concentration ratio, similarly as in the earlier triple-refocusing in-vitro data analysis.
Figure 6.
In-vitro comparison of MEGA and triple refocusing in an aqueous solution with GABA (3 mM), Glu (30 mM), NAA (30 mM) and Gly (36 mM). (a) Shown below the inversion profile of the editing 180° RF pulse (E180) (13 ms Gaussian pulse truncated at 10%; Bandwidth 88 Hz) are MEGA E180-ON and E180-OFF subspectra. (b) A difference spectrum is shown together with an LCModel fit and the signals of GABA and Glu. A fourfold-magnified difference spectrum is shown at the top. (c) A triple-refocused spectrum is shown together with an LCModel fit and the GABA and Glu signals. For both MEGA and triple refocusing, spectra are scaled equal (except the magnified spectrum). Spectra were broadened to Gly singlet FWHM of 4 Hz.
DISCUSSION AND CONCLUSION
The present study reports a novel triple-refocusing scheme for in vivo measurement of GABA at 3T. Taking advantage of the high variability of strongly-coupled spin signals with respect to changes in the inter-RF pulse time delays as well as the non-selective 180° RF pulse duration and carrier frequency, the Glu C4-proton signal was effectively manipulated to accomplish distinction between the GABA 2.29 ppm and Glu 2.35 ppm multiplets and thereby co-detection of GABA and Glu with precision. Of note, the TE of the optimized triple refocusing was not very long (74 ms) and thus the T2 signal loss was moderate. Using this new GABA MRS, we evaluated regional variation of GABA in medial occipital and left parietal regions in healthy human brain. We further tested the method in other clinically relevant brain regions such as frontal brain, hippocampus, and posterior cingulate. Frontal brain MRS may be relatively vulnerable to subject motion artifacts, and thus precise measurement of low-concentration metabolites such GABA could be challenging. Measurement of GABA in hippocampus, which is also a big challenge due to the small volume of interest in the temporal region, is especially of high significance given the implications of hippocampus in neuropsychiatric disorders. Our triple-refocused data that were acquired from these technically-challenging brain regions in relatively short time frames showed clearly discernible GABA signals at 2.29 ppm, completely separated from the Glu C4-proton resonance, without complicated data processing. This may be the major accomplishment of the present study.
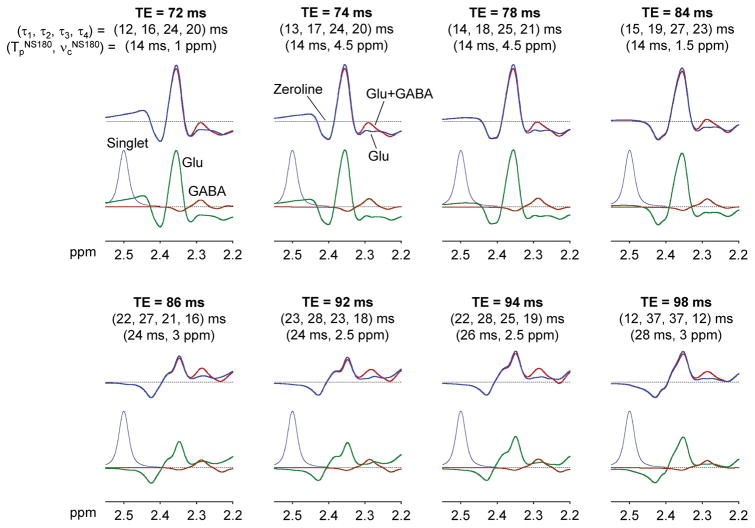
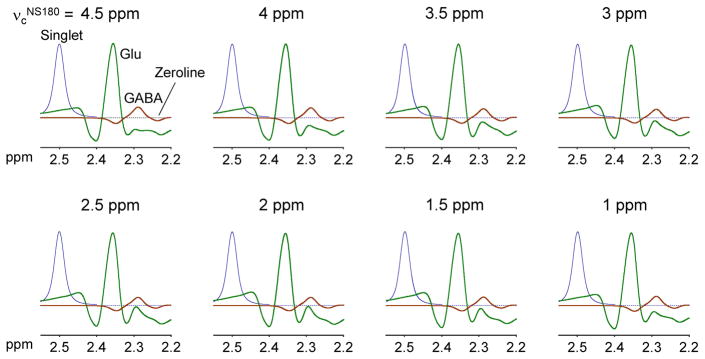
The triple refocusing optimization was carried out for TE ≤ 100 ms to avoid extensive T2 signal loss of the low-concentration metabolite GABA. It appears that small variation of the Glu signal between 2.25 and 2.32 ppm can be achieved at many TEs between 70 and 100 ms, making the GABA C2-proton signal discernible with respect to the Glu signal (Figure 7). Computer simulations showed that the GABA signal variation in the relatively small TE range (30 ms) was minimal (within 20% with respect to the largest signal at TE 74 ms), which is because the coherence evolution of weakly-coupled spins is governed by TE. In contrast, the case of Glu, whose spins are strongly coupled, was very different. For singlet FWHM of 4 Hz, the Glu signal amplitude was decreased to approximately 60% at TE ≥ 86 ms when compared with those at TE ≤ 84 ms. Unlike the case of weakly-coupled spins, the J evolution of strongly-coupled spins is sensitive to many sequence parameters of triple refocusing, which is largely because the J-coupling Hamiltonian of strongly-coupled spins has in-plane operator terms (i.e., IxSx and IySy for J-coupled spins I and S) and these terms do not commute with other Hamiltonians. The Glu signal intensity and pattern can be substantially altered with small changes in inter-RF pulse time delays for an arbitrary constant TE. The Glu signal is also affected by the NS180 duration and carrier frequency. Specifically, the upfield side of the Glu C4-proton multiplet, which is critical for distinction of the GABA 2.29 ppm signal from Glu, was sensitive to the NS180 carrier frequency while the GABA signal was virtually independent of the carrier frequency in the simulation results (Figure 8).
Figure 7.
Numerically-calculated triple-refocused spectra of GABA and Glu (concentration ratio 1:10) are shown for TE 70 – 100 ms, together with inter-RF pulse time delays (τ1, τ2, τ3, τ4) and the NS180 duration (TpNS180) and carrier frequency (νcNS180). Each of the eight-TE sequence parameter sets was chosen for minimum variation of the Glu signal between 2.25 and 2.32 ppm for each of TE = 70 – 72, 74 – 76, 78 – 80, 82 – 84, 86 – 88, 90 – 92, 94 – 96, and 98 – 100 ms. For each TE, upper panel shows spectra of GABA+Glu (in red) and Glu (in blue) and lower panel shows spectra of GABA and Glu and a triple-refocused artificial singlet (2.5 ppm, 4 protons). Signals were broadened to singlet FWHM 4 Hz. T2 relaxation effects were not included in the simulation.
Figure 8.
Calculated triple-refocused spectra of GABA and Glu (concentration ratio 1:10) are shown for NS180 carrier frequency (νcNS180) of 1 – 4.5 ppm (0.5 ppm increments), together with triple-refocused artificial singlet (2.5 ppm, 4 protons). Other sequence parameters were the same as those of the GABA-optimized triple refocusing, i.e., (τ1, τ2, τ3, τ4) = (13, 17, 24, 20) ms and NS180 duration = 14 ms. Signals were broadened to singlet FWHM 4 Hz. T2 relaxation effects were not included in the simulation.
GABA may be reliably measurable with MEGA in a contamination-free situation (as shown in our in-vitro tests), but the in-vivo performance of MEGA is different due to contamination of MM and HC. In MEGA editing of GABA in brain, the C2-proton resonances (~3.75 ppm) of Glu, Gln and GSH are coedited due to the effect of the editing 180° RF pulse (E180) on their C3-proton resonances (2.04 – 2.16 ppm). Since this coediting and MM coediting are all affected by the E180 bandwidth, the MM contamination can be qualitatively assessed by comparing the MEGA-edited 3 ppm signal with the coedited 3.75 ppm signal. Computer simulations indicate that, when the E180 bandwidth is 98 Hz (as in our phantom test), the edited GABA to coedited 3.75 ppm signal ratio may be approximately 20% at maximum for [GABA]:[Glu]:[Gln]:[GSH] of 1:10:3:1. The signal ratio somewhat increases with decreasing E180 bandwidth, but it may not be larger than 40% even when the E180 bandwidth is as small as 38 Hz (e.g., 30 ms Gaussian envelope truncated at 10%). It follows that, when a MEGA-edited signal at ~3 ppm is not much smaller compared with the coedited 3.75 ppm signal (27–29), the edited signal at 3 ppm may contain considerable MM contamination, requiring careful data interpretation.
The in-vivo GABA estimates of the present study appear to be overall lower than prior GABA measurements by MEGA editing at 3T. Many studies measured GABA in the medial occipital brain and reported the estimate with reference to tCr at 8 mM, similarly as in this study. Compared with these prior MEGA studies, our GABA estimate in medial occipital brain (0.8 mM, ~12 mL voxel) is notably lower. A recent study, which aimed to find regional differences of GABA at 3T, reported 2.7 mM of GABA in the medial occipital cortex (30). Several other 3T studies using MEGA reported medial-occipital GABA/tCr of 0.15 – 0.2 (31–33), which corresponds to 1.2 – 1.6 mM when the tCr level is set to 8 mM. Discrepancy between our estimation and prior MEGA GABA measurements at 3T are also seen in other brain regions. Frontal-brain GABA was measured as 1.4 – 2.1 mM in prior studies at 3T (30,34), higher than our estimate (0.87 mM). For posterior cingulate, our estimate of 0.66 mM in a 7 mL voxel is approximately 50% relative to a prior MEGA study in an 18 mL voxel (35). For hippocampus, our GABA estimate from a 3.9 mL voxel is about 60% of a MEGA measurement of GABA in a much larger voxel (11.3 mL) with most likely lower GM content. In contrast, our GABA estimates are in acceptable agreement with some prior studies. A 7T MEGA study in occipital cortex reported GABA level as 0.75 mM after removing MM contamination (36). In a double-quantum filtered GABA imaging study (37), in which the MM contamination was relatively small (i.e., < 20% of the edited GABA signal), the GABA/tCr estimate in regions with fGM of ~70% (similar to our medial-occipital voxel fGM) was approximately 0.12. Another GABA double-quantum filtering study reported frontal-brain GABA as 0.8 mM after correction for homocarnosine contamination (16), which is close to the GABA estimate of the current study (0.87 mM). Of note, our GABA estimations in medial occipital and frontal brain are in excellent agreement with two prior studies (13,14) that were focused on detection of the GABA 2.29 ppm resonance using optimized-TE MRS. Taken together, the MEGA edited signal at ~3 ppm may contain signals from other compounds such as MM and HC and thus likely result in overestimation of GABA, depending on the sequence design. Approaches that are focused on distinction of the GABA 2.29 ppm resonance appear to give relatively consistent estimates of GABA among the methods, which suggests that the methodology may confer a relatively robust tool for evaluating the clinically-important, low-concentration metabolite GABA.
Our data indicate presence of regional variation of GABA in the healthy brain. Although the mean fGM of the hippocampal voxel was similar to those of the voxels in medial occipital and frontal brain, the GABA level in hippocampus was significantly higher compared with other GM dominant regions (MO, MF and PC). This suggests that the regional variability of GABA is not solely dependent on the GM and WM contents. The regional variation in the GABA pool size may be in part due to distinct functions attributed to the specific brain regions. Medial occipital and frontal regions, which are broadly categorized as cortical subtypes, have different functional roles between them (38–40). As a consequence the GABAergic and/or glutamatergic activities may be distinct and thus may lead to variation in the metabolic pool size. Early studies reported that there are distinct GABAergic interneurons in hippocampus (21 subtypes) and brain cortex (41,42). Similarly, posterior cingulate, which is cytoarchitectonically characterized as a cortical region, showed significantly lower GABA level compared with the medial occipital and frontal cortical regions. Our posterior cingulate voxels were composed of approximately equal fractions of GM and WM (fGM ~ 57%) unlike the voxels in medial occipital and frontal brain. The relatively low fGM and differential functional property may incur low GABA level in posterior cingulate compared to other cortical regions. The GABA levels in the WM dominant regions (left parietal and left frontal) showed lower GABA levels compared with the GM dominant regions, which may be because WM rich regions are composed of nerve fibers that connect neurons, whilst GM rich regions have neuronal core parts which may be more metabolically active and regulated by the functional properties of the brain regions.
The Glu estimates of the present study are in agreement with many prior studies (25,26,43,44). The Glu concentration was estimated to be higher by approximately twofold in pure GM than in pure WM (10.4 vs. 5.8 mM). The Glu level was not significantly different between the medial occipital, medial frontal, and hippocampus regions, unlike GABA. Prior immunohistological studies suggested that excitatory pyramidal neurons account for approximately 85% of all cortical neurons and are relatively homogeneous (45,46). Owing to the uniform and dominant distribution of excitatory pyramidal neurons, the Glu level may be similar across these GM dominant cortical and hippocampal regions. However, the interneurons, which are mainly GABAergic and comprise 16 – 30% of cortical neurons, display synaptic and physiological variability (47–49). The diversity and variability of the GABAergic interneurons across the brain may explain the GABA pool size variations seen in the present study.
In conclusion, we have developed a novel triple-refocusing scheme that provides distinction of the GABA 2.29 ppm resonance from the neighboring Glu signal. With well-preserved Glu signal, the optimized triple refocusing permits precise co-detection of GABA and Glu in the human brain at 3T. Importantly, the method performs well in multiple brain locations, including temporal brain such as hippocampus. Linear regression of GABA estimates with respect to fractional GM content showed approximately twofold higher concentration of GABA in pure GM than in pure WM. With minimal contaminations from other compounds, the proposed triple refocusing may provide an effective tool for detecting potential small alterations in GABA and Glu levels in brain diseases at 3T, a field strength which is widely used in clinical studies. Further work will be required to determine the sensitivity to alterations in GABA level in patient populations and to accomplish multi-voxel imaging of GABA using the triple refocusing.
Acknowledgments
This work was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA184584 and by a Cancer Prevention Research Institute of Texas grant RP130427. We thank Ms. Jeannie Baxter and Janet Jerrow for subject recruitment and Dr. Ivan Dimitrov for technical assistance.
References
- 1.Sanacora G, Gueorguieva R, Epperson CN, Wu YT, Appel M, Rothman DL, Krystal JH, Mason GF. Subtype-specific alterations of gamma-aminobutyric acid and glutamate in patients with major depression. Arch Gen Psychiatry. 2004;61:705–713. doi: 10.1001/archpsyc.61.7.705. [DOI] [PubMed] [Google Scholar]
- 2.van Elst LT, Valerius G, Buchert M, Thiel T, Rusch N, Bubl E, Hennig J, Ebert D, Olbrich HM. Increased prefrontal and hippocampal glutamate concentration in schizophrenia: evidence from a magnetic resonance spectroscopy study. Biol Psychiatry. 2005;58:724–730. doi: 10.1016/j.biopsych.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 3.Govind V. 1H-NMR Chemical Shifts and Coupling Constants for Brain Metabolites. eMagRes. 2016;5:1347–1362. doi: 10.1002/9780470034590.emrstm1530. [DOI] [Google Scholar]
- 4.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32:294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 5.Kreis R, Slotboom J, Hofmann L, Boesch C. Integrated data acquisition and processing to determine metabolite contents, relaxation times, and macromolecule baseline in single examinations of individual subjects. Magn Reson Med. 2005;54:761–768. doi: 10.1002/mrm.20673. [DOI] [PubMed] [Google Scholar]
- 6.Choi C, Bhardwaj PP, Kalra S, Casault CA, Yasmin US, Allen PS, Coupland NJ. Measurement of GABA and contaminants in gray and white matter in human brain in vivo. Magn Reson Med. 2007;58:27–33. doi: 10.1002/mrm.21275. [DOI] [PubMed] [Google Scholar]
- 7.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci USA. 1993;90:5662–5666. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45:517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Mullins PG, McGonigle DJ, O’Gorman RL, Puts NA, Vidyasagar R, Evans CJ, Edden RA Cardiff Symposium on MRSoG. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. NeuroImage. 2014;86:43–52. doi: 10.1016/j.neuroimage.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lajtha A. Handbook of Neurochemistry: Volume 1. Chemical and Cellular Architecture. New York and London: Plenum Press; 1982. [Google Scholar]
- 12.Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW. Efficient spectral editing at 7 T: GABA detection with MEGA-sLASER. Magn Reson Med. 2012;68:1018–1025. doi: 10.1002/mrm.24131. [DOI] [PubMed] [Google Scholar]
- 13.Hanstock CC, Coupland NJ, Allen PS. GABA X2 multiplet measured pre- and post-administration of vigabatrin in human brain. Magn Reson Med. 2002;48:617–623. doi: 10.1002/mrm.10265. [DOI] [PubMed] [Google Scholar]
- 14.Ganji SK, An Z, Banerjee A, Madan A, Hulsey KM, Choi C. Measurement of regional variation of GABA in the human brain by optimized point-resolved spectroscopy at 7 T in vivo. NMR Biomed. 2014;27:1167–1175. doi: 10.1002/nbm.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chazin WJ, Rance M, Wright PE. Complete assignment of lysine resonances in 1H NMR spectra of proteins as probes of surface structure and dynamics. FEBS Lett. 1987;222:109–114. doi: 10.1016/0014-5793(87)80201-6. [DOI] [PubMed] [Google Scholar]
- 16.Choi C, Coupland NJ, Hanstock CC, Ogilvie CJ, Higgins AC, Gheorghiu D, Allen PS. Brain gamma-aminobutyric acid measurement by proton double-quantum filtering with selective J rewinding. Magn Reson Med. 2005;54:272–279. doi: 10.1002/mrm.20563. [DOI] [PubMed] [Google Scholar]
- 17.An Z, Ganji SK, Tiwari V, Pinho MC, Patel T, Barnett S, Pan E, Mickey BE, Maher EA, Choi C. Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo. Magn Reson Med. 2017;78:40–48. doi: 10.1002/mrm.26347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari V, An Z, Ganji SK, Baxter J, Patel TR, Pan E, Mickey BE, Maher EA, Pinho MC, Choi C. Measurement of glycine in healthy and tumorous brain by triple-refocusing MRS at 3 T in vivo. NMR Biomed. 2017;30:e3747. doi: 10.1002/nbm.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi C, Ganji S, Hulsey K, Madan A, Kovacs Z, Dimitrov I, Zhang S, Pichumani K, Mendelsohn D, Mickey B, Malloy C, Bachoo R, Deberardinis R, Maher E. A comparative study of short- and long-TE 1H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR Biomed. 2013;26:1242–1250. doi: 10.1002/nbm.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi C, Ganji SK, Deberardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganji SK, Banerjee A, Patel AM, Zhao YD, Dimitrov IE, Browning JD, Brown ES, Maher EA, Choi C. T2 measurement of J-coupled metabolites in the human brain at 3T. NMR Biomed. 2012;25:523–529. doi: 10.1002/nbm.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994;104:1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 23.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 24.Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD. Statistical Parametric Mapping: The Analysis of Functional Brain Images. London: Academic Press; 2006. [Google Scholar]
- 25.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61:1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 26.Tkac I, Oz G, Adriany G, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: metabolite quantification at 4T vs. 7T. Magn Reson Med. 2009;62:868–879. doi: 10.1002/mrm.22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasler G, Neumeister A, van der Veen JW, Tumonis T, Bain EE, Shen J, Drevets WC, Charney DS. Normal prefrontal gamma-aminobutyric acid levels in remitted depressed subjects determined by proton magnetic resonance spectroscopy. Biol Psychiatry. 2005;58:969–973. doi: 10.1016/j.biopsych.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 28.Simpson HB, Shungu DC, Bender J, Jr, Mao X, Xu X, Slifstein M, Kegeles LS. Investigation of cortical glutamate-glutamine and gamma-aminobutyric acid in obsessive-compulsive disorder by proton magnetic resonance spectroscopy. Neuropsychopharmacology. 2012;37:2684–2692. doi: 10.1038/npp.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumner P, Edden RA, Bompas A, Evans CJ, Singh KD. More GABA, less distraction: a neurochemical predictor of motor decision speed. Nat Neurosci. 2010;13:825–827. doi: 10.1038/nn.2559. [DOI] [PubMed] [Google Scholar]
- 30.Grewal M, Dabas A, Saharan S, Barker PB, Edden RA, Mandal PK. GABA quantitation using MEGA-PRESS: Regional and hemispheric differences. J Magn Reson Imaging: JMRI. 2016;44:1619–1623. doi: 10.1002/jmri.25324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhagwagar Z, Wylezinska M, Jezzard P, Evans J, Boorman E, PMM, PJC Low GABA concentrations in occipital cortex and anterior cingulate cortex in medication-free, recovered depressed patients. Int J Neuropsychopharmacol. 2008;11:255–260. doi: 10.1017/S1461145707007924. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya PK, Phillips MD, Stone LA, Lowe MJ. In vivo magnetic resonance spectroscopy measurement of gray-matter and white-matter gamma-aminobutyric acid concentration in sensorimotor cortex using a motion-controlled MEGA point-resolved spectroscopy sequence. Magn Reson Imaging. 2011;29:374–379. doi: 10.1016/j.mri.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging: JMRI. 2014;40:1445–1452. doi: 10.1002/jmri.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray DE, Durazzo TC, Schmidt TP, Abe C, Guydish J, Meyerhoff DJ. Frontal Metabolite Concentration Deficits in Opiate Dependence Relate to Substance Use, Cognition, and Self-Regulation. J Addict Res Ther. 2016:7. doi: 10.4172/2155-6105.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao G, Edden RAE, Gao F, Li H, Gong T, Chen W, Liu X, Wang G, Zhao B. Reduced GABA levels correlate with cognitive impairment in patients with relapsing-remitting multiple sclerosis. Eur Radiol. 2017 doi: 10.1007/s00330-017-5064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47:1009–1012. doi: 10.1002/mrm.10146. [DOI] [PubMed] [Google Scholar]
- 37.Choi IY, Lee SP, Merkle H, Shen J. In vivo detection of gray and white matter differences in GABA concentration in the human brain. NeuroImage. 2006;33:85–93. doi: 10.1016/j.neuroimage.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Bedny M, Pascual-Leone A, Dodell-Feder D, Fedorenko E, Saxe R. Language processing in the occipital cortex of congenitally blind adults. Proc Natl Acad Sci USA. 2011;108:4429–4434. doi: 10.1073/pnas.1014818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76:1057–1070. doi: 10.1016/j.neuron.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JS, Kanjlia S, Merabet LB, Bedny M. Development of the Visual Word Form Area Requires Visual Experience: Evidence from Blind Braille Readers. J Neurosci. 2017;37:11495–11504. doi: 10.1523/JNEUROSCI.0997-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fishell G, Rudy B. Mechanisms of inhibition within the telencephalon: “where the wild things are”. Annu Rev Neurosci. 2011;34:535–567. doi: 10.1146/annurev-neuro-061010-113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan R, Cunningham C, Chen A, Vigneron D, Hurd R, Nelson S, Pelletier D. TE-averaged two-dimensional proton spectroscopic imaging of glutamate at 3 T. NeuroImage. 2006;30:1171–1178. doi: 10.1016/j.neuroimage.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 44.Pan JW, Avdievich N, Hetherington HP. J-refocused coherence transfer spectroscopic imaging at 7 T in human brain. Magn Reson Med. 2010;64:1237–1246. doi: 10.1002/mrm.22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 46.Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 47.Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 49.Hevner RF. Layer-specific markers as probes for neuron type identity in human neocortex and malformations of cortical development. J Neuropathol Exp Neurol. 2007;66:101–109. doi: 10.1097/nen.0b013e3180301c06. [DOI] [PubMed] [Google Scholar]