Abstract
Receptor tyrosine kinase-mediated growth factor signaling is essential for proper formation and development of the neural crest. The many ligands and receptors implicated in these processes signal through relatively few downstream pathways, frequently converging on the MAPK and PI3K pathways. Despite decades of study, there is still considerable uncertainty about where and when these signaling pathways are required and how they elicit particular responses. This review summarizes our current understanding of growth factor-induced MAPK and PI3K signaling in the neural crest.
Keywords: Receptor Tyrosine Kinases, ERK, AKT, cell signaling
1. Introduction
Ever since Wilhelm His initially described the ‘intermediate cord’, biologists have been fascinated by this population of cells, later dubbed the neural crest (NC). Considering the NC’s eventual dissemination throughout the body and the variety of cell types it can form, it is not hard to understand why. This group of cells, sometimes dubbed the fourth germ layer, is specified shortly after gastrulation at the border between neural and non-neural ectoderm. These cells then undergo an epithelial-to-mesenchymal transition and subsequent migration throughout the developing embryo where their descendants proliferate and subsequently differentiate into a panoply of cell types and tissues. These cell types include the bones, connective tissue, and pericytes in the face, melanocytes throughout the skin, smooth muscle in the outflow tract of the heart, and numerous neural derivatives such as the facial nerves, dorsal root ganglia, peripheral sensory, sympathetic, and parasympathetic neurons and their associated Schwann cells, the adrenal medulla, and the enteric nervous system, which innervates the digestive tract (Mayor and Theveneau, 2013; Simões-Costa and Bronner, 2015). For a more thorough review of the NC and multipotency, see reviews from LeDouarin and Dupin (LeDouarin, 2018; Dupin, 2018) in this issue. A natural question relates to the signals that direct neural crest specification, survival, growth, migration, and ultimate differentiation. One family of signaling molecules particularly relevant to all of these steps are growth factor ligands and the receptor tyrosine kinases (RTKs) they activate (Fantauzzo and Soriano, 2015). The link between NC and RTKs is in fact quite old. Mouse fanciers across the globe have prized mice with unique coat patterns for centuries, and classic alleles such as Dominant Spotting (W-locus) and Steel (Sl-locus) are caused by mutations in the genes Kit and Kitl, respectively, that encode the RTK KIT and its ligand Stem Cell Factor (SCF) (Chabot et al., 1988; Copeland et al., 1990; Flanagan and Leder, 1990; Geissler et al., 1988; Silver, 1995; Silvers, 1979; Tokuda, 1935; Zsebo et al., 1990). The importance of RTKs in NC during development was dramatically illustrated by Hans Grüneberg’s studies of the Patch mouse in the 1960s (Grueneberg and Truslove, 1960), which turned out to have defects in two RTK genes, Pdgfra (Smith et al., 1991; Stephenson et al., 1991) and Kit (Wehrle-Haller et al., 1996).
Neural crest RTK signaling is relevant to human disease. Mutations in RTK genes, their effectors, and the signaling pathways they activate cause both congenital malformations and neoplasms attributed to defects in NC, collectively known as neurocristopathies (Bolande, 1974). These include congenital malformations such as craniofrontonasal dysplasia (EFNB1), craniosynostosis (FGFR1, FGFR2, FGFR3), Hirschsprung disease (RET, GDNF), piebaldism (KIT), and a group of syndromes known as the neuro-cardio-facio-cutaneous syndromes that involve NC and non-NC cells (MAPK1, MAPK3, PTPN11, KRAS, HRAS, SOS1, BRAF, RAF1, MEK1, MEK2) as well as neoplasms such as melanoma (KIT, BRAF) (Bolande, 1997; Etchevers et al., 2006; Samuels et al., 2009; Watt and Trainor, 2013).
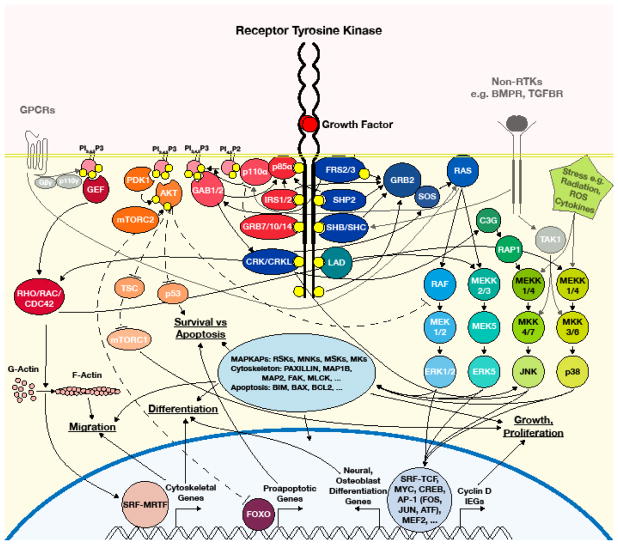
RTKs are a diverse group of membrane-spanning receptor proteins that engage multiple intracellular signaling cascades. Typically, receptors dimerize in response to ligand-binding. Dimerization stimulates the receptors’ intrinsic tyrosine kinase activity that in turn activates a number of downstream signaling cascades. This is achieved through autophosphorylation of the cytoplasmic domain of the RTK itself, thus creating docking sites for signaling proteins, and through direct phosphorylation of signaling effectors. RTK signaling has been shown to regulate a broad suite of cellular responses including proliferation, differentiation, migration, and apoptosis. In the human, there are at least 58 RTKs, organized into twenty subfamilies, which are activated by an even greater number of ligands (Lemmon and Schlessinger, 2010). So far, twelve of these families have been clearly implicated in NC development (Fantauzzo and Soriano, 2015). Despite the hundreds of unique receptor-ligand complexes RTKs have been shown to form, they converge on a comparatively limited number of intracellular signaling pathways. These include the mitogen-activated protein kinases (MAPK) such as the extracellular-signal regulated kinases (ERK) 1/2, p38, the c-JUN N-terminal kinases (JNK), and ERK5, as well as by phosphatidylinositol 3-kinase (PI3K), phospholipase-c gamma (PLCγ), protein kinase C (PKC), Src-family tyrosine kinases, and activation of several Signal Transducer and Activator of Transcription (STAT) proteins.
There are multiple explanations for the superficial disconnect between the number of receptor-ligand pairs and the limited number of signaling pathways they activate. First, RTKs and their ligands are not ubiquitously expressed, so the large number of genes allows for temporal, spatial, and cell-type specificity of signaling activity. Second, not every receptor activates every potential downstream pathway. Thus, specific receptor dimers or receptor-ligand complexes may activate unique combinations of downstream signaling pathways that, when activated together, elicit a unique cellular response (Lemmon and Schlessinger, 2010; Vasudevan et al., 2015). Finally, even receptors that activate the same subset of signaling pathways do not necessarily activate these pathways to the same degree or with the same temporal dynamics, which in itself can lead to specific gene expression and fate decisions (Amit et al., 2007; J.-Y. Chen et al., 2012; Marshall, 1995; Purvis and Lahav, 2013; Toettcher et al., 2013; Wilson et al., 2017). The importance of proper signaling strength and duration for development is attested by the striking phenotypes in mouse mutants for positive and negative feedback proteins, the focus of a recent review (Neben et al., 2017). The mechanisms by which RTKs achieve specificity have been described (Lemmon and Schlessinger, 2010; Tan and S. K. Kim, 1999; Vasudevan and Soriano, 2016; Volinsky and Kholodenko, 2013). Nevertheless, there is considerable overlap in expression of RTKs and members of the signaling pathways they engage in the neural crest, making their study a challenging objective (Fantauzzo and Soriano, 2015). Complicating matters, these signaling cascades are not exclusively activated by RTKs, but can also be activated, directly or indirectly, downstream of other receptors such as transforming growth factor beta (TGF-β) receptors, bone morphogenetic protein (BMP) receptors, and G-Protein Coupled Receptors (GPCRs) or in response to environmental stress (Cargnello and Roux, 2011; Fruman et al., 2017; Rahman et al., 2015).
One approach to demystify the question of how signaling regulates development is to study the problem at the level of the receptor and its ligands. Expression analysis coupled to gain-of-function (GoF) and loss-of-function (LoF) studies of ligands and receptors is an essential component in advancing our knowledge of RTK signaling and development. The observed effect (or lack thereof) of manipulating a particular receptor or ligand subsequently raises two issues. First, which downstream pathway(s) is responsible for the observed effect? Because RTKs engage multiple downstream signaling cascades, attributing mutant phenotypes to a particular pathway(s) is rarely easy. This can be tested in vitro using chemical agonists and antagonists, although their specificity may be an issue. It can be even more challenging to relate an effect to a specific upstream RTK in vivo without further genetic manipulation, for instance by making mutations in the RTKs that prevent docking of particular signaling effectors (Tallquist et al, 2003; Klinghoffer et al. 2002; Brewer et al. 2015; (Jain et al., 2006; Jijiwa et al., 2004; Kimura et al., 2004; Maina et al., 2001; Wong et al., 2005). Second, to what extent is the system buffered by redundancy through other RTKs or non-RTK mechanisms that stimulate the same downstream pathways? Cells in vivo often simultaneously express multiple RTKs of the same and different families (for instance we observe cranial neural crest cells (cNCCs) expressing PDGFRα, PDGFRβ, FGFR1, and FGFR2) and loss of multiple receptors can enhance a mutant phenotype (Fantauzzo and Soriano, 2016; Klinghoffer et al., 2002; Park et al., 2006). It is therefore likely that cells integrate multiple receptor signaling inputs at the level of the downstream signaling cascade (Kholodenko et al., 2010). Thus, studying development at the level of the intracellular signaling pathway is an equally important task.
This review focuses on MAPK and PI3K signaling and their role in neural crest development, using mainly genetic data from the mouse, but also from the chick, frog, and zebrafish. Both MAPK and PI3K are commonly activated by RTKs and there is extensive published research linking them to NC. In some cases these pathways have not been studied by conditional genetic approaches, so we discuss global knockouts and whether there is a likely NC defect. We also review how MAPK and PI3K signaling regulate the transcription factor Serum Response Factor (SRF), which is critical to induce rapid expression of immediate early genes (IEGs), one of the classic functions of growth factor signaling. For a discussion of RTK signaling generally or the role of the specific RTK families themselves in neural crest development, we direct the reader to these reviews (Fantauzzo and Soriano, 2015; Lemmon and Schlessinger, 2010).
2. MAPK Pathways
There are four canonical MAPK pathways in mammals: the ERK1/2, JNK, p38, and ERK5 pathways. All four pathways consist of a three-tiered kinase cascade including an upstream MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK), and the MAPK itself. MAPKs are proline-directed Ser/Thr kinases that phosphorylate hundreds of substrates in the cytoplasm and nucleus. MAPKs are accordingly implicated in numerous cellular behaviors such as proliferation, survival, apoptosis, migration, and differentiation, as well as in human disease (E. K. Kim and Choi, 2010). There are also three non-canonical MAPK pathways, ERK 3/4, ERK7/8, and NLK, that are outside the scope of this review (Cargnello and Roux, 2011). Most RTKs seem to activate MAPK signaling to some degree, though the intensity, duration, and importance of this activity varies considerably by RTK (McKay and D. K. Morrison, 2007). The four canonical MAPK cascades share a consensus phosphorylation site (P-X-S/T-P), but show only partial overlap in their known substrates and elicit particular responses through the activation of unique ensembles of effectors, spatial and dynamic regulation, feedback mechanisms, and scaffolding proteins (Cargnello and Roux, 2011; Casar and Crespo, 2016; Keshet and Seger, 2010; Sheridan et al., 2008; Wortzel and Seger, 2011).
2a. ERK1/2 Pathway
The ERK1/2 MAPK pathway is activated in response to a broad range of stimuli including growth factor-induced RTK signaling, GPCRs, non-canonical TGF-β and BMP signaling, and cellular stress (Cargnello and Roux, 2011). ERK1/2 signaling is strongly associated with the FGF pathway, but is also activated in response to NC-expressed PDGF, EPH, ERBB, RET, MET, KIT, and TRK family RTKs (Fantauzzo and Soriano, 2015). Receptors activate the cascade by recruitment of adapter molecules such as FRS2/3 (Kouhara et al., 1997), SHB (Cross et al., 2002), SHC (Pelicci et al., 1992), SHP2 (Kazlauskas et al., 1993; Lechleider et al., 1993), and CRK/CRKL (CRK) (Brewer et al., 2016; Lemmon and Schlessinger, 2010). These molecules recruit the guanine nucleotide exchange factor (GEF) Son-of-Sevenless (SOS) through the adapter protein GRB2, which is itself also sometimes recruited to RTKs directly (N. Li et al., 1993). SOS mediates activation of the membrane-associated GTP-binding protein RAS. GTP-bound RAS activates the ERK1/2 signaling cascade. This consists of the MAPKKKs A-RAF, B-RAF, and RAF-1/C-RAF; the MAPKKs MEK1 and MEK2; and the MAPKs ERK1 and ERK2 (Figure 1A). Once phosphorylated by MEK, ERK1/2 can then act on targets in the cytoplasm, such as a broad group of Ser/Thr kinases known as the MAPK-activated protein kinases (MAPKAPs) (Cargnello and Roux, 2011), and in the nucleus, primarily transcription factors such as ELK1, FOS, MYC, and MEF2 (Plotnikov et al., 2011). ERK1/2 activity has been linked in various contexts to the processes of proliferation, survival, migration and fate choice (Cargnello and Roux, 2011; Wortzel and Seger, 2011).
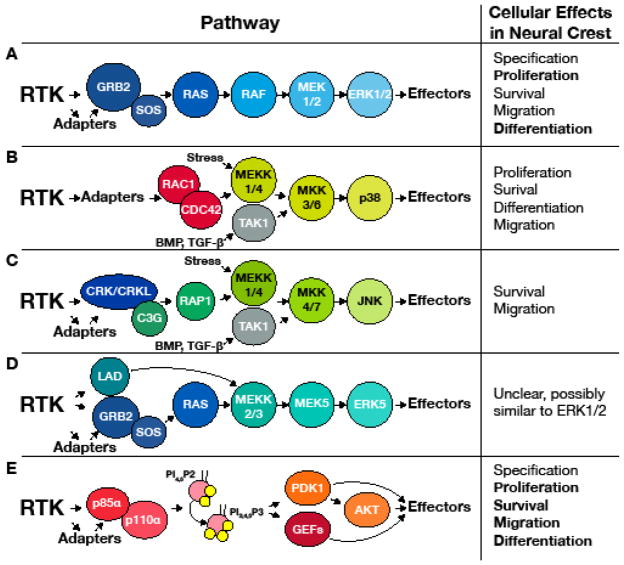
Figure 1. RTK signaling pathways in isolation.
The ERK1/2 (A), p38 (B), JNK (C) ERK5 (D), and PI3K (E) pathways and their known or likely means of activation downstream of RTKs are shown at left. For simplicity only crucial adapter proteins are shown and specific effector proteins and crosstalk between pathways have been omitted. See the text for further clarification. To the right are demonstrated (bold) and suggested (regular) cellular downstream behaviors in NC.
ERK1/2 and Neural Crest Developmental Phenotypes
The ERK1/2 pathway has long been of interest in regard to both development and disease, which has resulted in a wealth of mouse null and conditional alleles available to study the pathway’s in vivo functions. The mouse models discussed in the review and their phenotypes are summarized in Table 1. These models highlight the neural crest as particularly sensitive to ERK1/2 perturbation. ERK1 and ERK2 are encoded by the genes Mapk3 and Mapk1, respectively. Mapk3−/− mice are viable and fertile (Pages et al., 1999), with a background-dependent hyperactivity phenotype (Mazzucchelli et al., 2002), whereas Mapk1−/− mice die shortly after implantation with a defect in extraembryonic tissue (Hatano et al., 2003; Saba-El-Leil et al., 2003). Similarly, mice lacking one of the two kinases upstream of ERK1/2, MEK2, are viable and fertile, while mice lacking MEK1 die around E10.5 with placental and vascular defects (Belanger et al., 2003; Giroux et al., 1999).
Table 1.
Summary of MAPK and PI3K mutant phenotypes discussed in the text. For simplicity, growth factor receptor mutations have been omitted.
| Genotype | NC Defects | Lethality/Other Defects | Reference |
|---|---|---|---|
| ERK1/2 Pathway LoF | |||
| Mapk1−/− (Erk2) | NA | Peri-implantation lethal. Extraembryonic tissue defects. | (Hatano et al., 2003; Saba-El-Leil et al., 2003) |
| Mapk3−/− (Erk1) | NA | Viable and fertile. Background-dependent hyperactivity. | (Mazzucchelli et al., 2002; Pages et al., 1999) |
| Map2k1−/− (Mek1) | NA | Mid-gestation lethal. Placental defects. | (Giroux et al., 1999) |
| Map2k2−/− (Mek2) | NA | Viable and fertile. | (Belanger et al., 2003) |
| Mapk1flox/flox; Wnt1-Cre | Micrognathia, hypoplastic maxilla, cleft palate, DORV, PTA, VSD, osteoblast differentiation defects. | Perinatal lethal. | (Newbern et al., 2008; Parada et al., 2015) |
| Mapk1flox/flox; Mapk3−/−; Wnt1-Cre | Above + aglossia, missing external ear and thymus, misplaced thyroid. Hypoplastic PAs at E10.5. No NCC- derived Schwann cells. DRG degenerate. Lacrimal aplasia. | Lethal E18-E19. | (Garg et al., 2017; Newbern et al., 2008; 2011) |
| Mek1flox/flox; Mek2−/−; Wnt1-Cre | Similar to above. | Similar to above. | (Garg et al., 2017; Newbern et al., 2008; 2011) |
| Brafflox/flox; Raf1flox/flox; Wnt1-Cre | Similar craniofacial and cardiovascular phenotypes as above. | Similar to above. | (Newbern et al., 2008) |
| Mapk1flox/flox; Mapk3−/−; Dhh-Cre | PNS not analyzed. Hypo- and unmyelinated PNS axons. Tremor, hindlimb paresis. | Lethal by P28. | (Newbern et al., 2011) |
| Mapk1Δ/flox; Sox2-Cre | Agnathia, aglossia, hypoplastic maxilla, VSD, PSA. | Lethal by E16.5. Short forelimbs and missing hindlimbs. | (Frémin et al., 2015) |
| Mapk1Δ/flox; Mapk3+/−; Sox2-Cre | NA | Small size, lethal E10.5. | (Frémin et al., 2015) |
| Ptpn11flox/flox (Shp2); Wnt1-Cre | Reduced or absent NCC-derived craniofacial bones and cartilage, PTA, variable VSD. Osteoblast differentiation defects. Lacrimal aplasia. | Lethal between E15.5 and birth. | (Garg et al., 2017; Nakamura et al., 2009a) |
| ERK1/2 Pathway GoF | |||
| Tg(CAG-cat,- Ptpn11Q79R); Wnt1-Cre | Variable cleft lip/palate. Domed skull, hypertelorism, small size. Osteoblast differentiation defects. | Lethal P1–P5 if cleft lip/palate present. | (Nakamura et al., 2009b) |
| ROSA26-Map2k1DD; Dhh-Cre and ROSA26- Map2k1DD; Egr2-Cre | Continuous myelin growth in PNS Schwann cells. | NA | (Sheean et al., 2014) |
| Tg(P0-Raf1/ER); Activated in adult Schwann cells | Schwann cell dedifferentiation, increased proliferation, muscle weakening. | NA | (Napoli et al., 2012) |
| ERK1/2 Pathway Effectors | |||
| Rps6ka−/− (Rsk2) | Delayed mineralization of craniofacial bones. Wide cranial sutures at birth. Dental defects. | Viable. Reduced long bone length and decreased mineral density. | (Laugel-Haushalter et al., 2014; X. Yang et al., 2004) |
| Ets1−/− | Ectopic NCC-derived cartilage in heart, VSD. Rare surviving mouse had white belly spot. Increased apoptosis of melanoblasts. | Perinatal lethal on C57Bl/6 background. | (Gao et al., 2010; Saldana-Caboverde et al., 2015) |
| Srfflox/flox; Wnt1-Cre | Facial cleft, micrognathia, thymic and thyroid aplasia, PTA, VSD, DRG innervation defects. | Background dependent lethality between E13.5 and late gestation. | (Newbern et al., 2008; Vasudevan and Soriano, 2014; Wickramasinghe et al., 2008) |
| P38 LoF | |||
| Mapk14−/− (p38α) | NA | Lethal by E12.5. Anemia, liver, heart, and placenta defects. | (Adams et al., 2000; Allen et al., 2000; Mudgett et al., 2000; Tamura et al., 2000) |
| Mapk14flox/flox; Mapk11−/−; Sox2-Cre (p38α/β) | NA | Lethal by E16.5. Exencephaly, small liver, thin myocardium, VSD. | (del Barco Barrantes et al., 2011) |
| Mapk12/13−/− (p38γ/δ) | NA | Viable and fertile. | (Sabio et al., 2005) |
| Tak1flox/flox; Wnt1-Cre | Cleft palate. | Perinatal lethal. | (Song et al., 2013; Yumoto et al., 2013) |
| JNK LoF | |||
| Mapk8−/− (Jnk1), Mapk9−/− (Jnk2), Mapk10−/− (Jnk3), Mapk8/10−/−, Mapk9/10−/− | NA | Viable and fertile. | (Kuan et al., 1999) |
| Mapk8/9−/− | Normal. | Lethal by E12.5. Exencephaly and neural apoptosis defects. | (Kuan et al., 1999; Sabapathy et al., 1999) |
| Mkk4−/− | NA | Lethal by E14. | (Di Yang et al., 1997; Ganiatsas et al., 1998; Nishina et al., 1999) |
| Mkk7−/− | NA | Viable and fertile. | (Schramek et al., 2011) |
| JNK Effectors | |||
| Jun−/− | Lack of NCCs in cardiac OFT. | Lethal E13. Apoptosis of hepatoblasts and hematopoietic cells. | (Eferl et al., 1999) |
| JunAA/AA | NA | Viable and fertile. Small size, resistant to epileptic seizures and neuronal apoptosis. | (Behrens et al., 1999) |
| ERK5 LoF | |||
| Mapk7−/− (Erk5) | Disorganized cranial mesenchyme, small PAs | Lethal at E10.5. Cardiovascular defects. | (Regan et al., 2002) |
| Mek5−/− | NA | Similar to above. | (Wang et al., 2004) |
| Mapk7flox/flox; Wnt1-Cre | Micrognathia, truncated ear, reduced size. | Viable and fertile. | (Newbern et al., 2011) |
| PI3K LoF | |||
| Pik3ca−/− (p110α) or Pik3caD933A/D933A | NA | Lethal at E10.5. Hemorrhaging, subepidermal blebs, angiogenesis defects. | (Bi et al., 1999; Graupera et al., 2008) |
| Pik3cb−/− (p110β) | NA | Lethal at E3.5. | (Bi et al., 2002) |
| Pik3cbK805R/K805R | NA | Subviable. | (Ciraolo et al., 2008) |
| Pik3cd−/− (p110δ) | NA | Viable with immune defects. | (Jou et al., 2002; Okkenhaug et al., 2002) |
| Pik3r1−/− or Pik3r2−/− | NA | Viable and fertile. | (Brachmann et al., 2005) |
| Pik3r1/2−/− | Facial cleft. | Lethal between E10.5-E12.5. Hemorrhaging, subepidermal blebs, wavy neural tube. | (Brachmann et al., 2005) |
| PI3K GoF | |||
| Ptenflox/flox; Wnt1-Cre | Overgrowth of craniofacial structures. Increased proliferation, enhanced osteoblast differentiation. | Perinatal lethal. | (T. Yang et al., 2017) |
| Ptenflox/flox; Dct-Cre (melanocytes) | Temporary overgrowth of dermal melanocytes. Resistance to graying. | Most mice die within two months of life due to neurologic defects. Small size. | (Inoue-Narita et al., 2008) |
| PI3K Effectors | |||
| Pdk1−/− | Absent PAs and DRG. | Lethal E9.5. No somites or heart, generally delayed. | (Lawlor et al., 2002) |
| Akt1−/− | NA | Viable and fertile. Growth retardation. | (Chen et al., 2001) |
| Akt1−/−, Akt1/2−/− | NA | Perinatal lethal. Dwarfism, skin, muscle, and bone defects. | (Peng et al., 2003) |
| Akt2−/−, Akt3−/−, Akt2/3−/− | NA | Viable and fertile. | (Dummler et al., 2006) |
| Akt1/3−/− | NA | Lethal between E11-E12. Morphologically normal at E10.5. | (Z. Z. Yang et al., 2005) |
| Mkl2−/− (also regulated by other pathways) | Cardiac OFT smooth muscle defect. | Perinatal lethal. | (Li et al., 2005) |
| Rac1flox/flox; Wnt1-Cre | Cleft lip and palate, PTA, ectodermal detachment, reduced proliferation. Possible differentiation defects. Reduced DRG. | Lethal between E12-E13.5. | (Fuchs et al., 2009; Thomas et al., 2010) |
| Cdc42flox/flox; Wnt1-Cre | Cleft lip and palate, shortened snout, PTA, VSD, possible migration defects, reduced proliferation, small DRG. | Lethal between E14.5- birth. | (Fuchs et al., 2009; Liu et al., 2013) |
| Tg(RockDN); Wnt1-Cre | Facial cleft, hypoplastic PAs, shortened snout, OFT defects, cytoskeletal disorganization, increased apoptosis. | Lethality between midgestation and E18.5. | (Phillips et al., 2012) |
Fremin et al. deleted Mapk1 throughout the embryo proper using the epiblast-specific Sox2-Cre in order to bypass the placental defect. Embryos survived to E16.5, lacked a mandible and tongue, had a shortened maxilla, and had cardiovascular defects such as VSD and PSA. Embryos were also smaller than controls, had shortened forelimbs, and lacked hindlimbs (Frémin et al., 2015). These phenotypes correlate with domains of ERK1/2 activity detected in E9.5 and E10.5 mouse embryos by whole-mount immunohistochemistry for double-phosphorylated ERK1/2 (ppERK1/2), where high levels of ERK activity were observed in the pharyngeal arches, frontonasal prominence, and limb buds, as well as the heart, forebrain, midbrain-hindbrain boundary, liver primordia, and tailbud (Corson et al., 2003; Frémin et al., 2015). Loss of one copy of Mapk3 in addition to both copies of Mapk1 led to growth arrest at E10.5. Intriguingly, mice completely lacking both Mapk1 and Mapk3 in the embryo proper can be rescued by a ubiquitously expressed Mapk3 transgene (Frémin et al., 2015). Together the results argue that ERK1 and ERK2 are functionally redundant, act in a dose-dependent manner during development, and that NCCs (and limb) are particularly sensitive to ERK1/2 depletion. This study, however, could not distinguish between NCC autonomous and non-autonomous roles for ERK1/2.
Newbern et al. took a complementary approach and combined null and conditional alleles to interrogate the ERK1/2 pathway requirement in the neural crest (Newbern et al., 2008). The investigation was prompted by the discovery that human patients with craniofacial and cardiovascular defects similar to DiGeorge Syndrome carried a deletion that spanned MAPK1. In an extensive mouse genetic analysis, Newbern et al. used the Wnt1-Cre driver, active in early NCCs (discussed further below), to analyze Mapk1flox/flox; Mapk3−/−; Wnt1-Cre embryos, Mek1flox/flox; Mek2−/−; Wnt1-Cre embryos, and Brafflox/flox; Raf1flox/flox; Wnt1-Cre embryos, thereby interrogating the ERK1/2 cascade at all three levels. Conditional loss of Mapk1 alone resulted in perinatal lethality. Embryos showed significant craniofacial and cardiovascular defects including cleft palate, a shortened maxilla, a drastically shortened mandible, and variably penetrant cardiac outflow tract defects such as double outlet right ventricle (DORV), persistent truncus arteriosus (PTA), and ventricular septal defect (VSD). Global disruption of Mapk3 in addition to conditional loss of Mapk1 enhanced the severity of the craniofacial phenotypes, the penetrance of cardiovascular defects, and produced novel defects such as absence of the tongue, absence of the external ear, failure of thymus development, and a misplaced thyroid gland. Double mutant mice also showed hypoplastic pharyngeal arches at E10.5, suggesting NC migration, proliferation, or survival defects. Brafflox/flox; Raf1flox/flox; Wnt1-Cre and Mek1flox/flox; Mek2−/−; Wnt1-Cre mice largely phenocopied the Mapk1/3 mice, emphasizing the linear nature of this cascade (Newbern et al., 2008).
NC-specific loss of Mapk1/3 or Mek1/2 also affected the peripheral nervous system (PNS). These mice were completely devoid of NC-derived Schwann cells (Newbern et al., 2011). The dorsal root ganglia (DRG) were specified correctly and motor and sensory nerves initially projected axons peripherally, but the DRG later underwent cell death and peripheral nerve projections degenerated, both apparently caused by the lack of Schwann cells. Conditional deletion of these genes in Schwann Precursor Cells slightly later in development (E12-13) using Dhh-Cre did not affect Schwann cell number but inhibited myelination and these mice died by four weeks of age (Newbern et al., 2011).
ERK1/2 activity has also been manipulated in the NC through the phosphatase SHP2, which promotes RTK-mediated ERK1/2 activation through recruitment of GRB2 (Araki et al., 2003; Tajan et al., 2015), and can also regulate the PI3K pathway and dephosphorylate activated RTKs (Bennett et al., 1994; Edouard et al., 2010; Tajan et al., 2015). Deletion of Ptpn11, the gene encoding SHP2, in NC using Wnt1-Cre causes reduced ppERK1/2 levels in NCCs, embryonic lethality between E15.5 and birth, and developmental defects similar to the ERK1/2 pathway mutants discussed above, including a hypoplastic maxilla, vastly reduced mandible and tongue, smaller ears, misplaced eyes, PTA and VSD (Nakamura et al., 2009a). Conversely, expression of a Q79R GoF Shp2 mutant allele in NC leads to increased ppERK1/2 levels (Nakamura et al., 2009b). A subset of these mice was born with a cleft lip and palate and quickly succumbed, while the rest survived but exhibited growth retardation, a domed skull, a shortened but correctly-patterned mandible, and premature fusion of the frontal bones. These phenotypes were ameliorated by injecting pregnant dams with the MEK1/2 inhibitor U01216, indicating that the observed phenotypes are in fact due to ERK1/2 hyperactivation and demonstrating the potential for therapeutic modulation of growth factor signaling in utero (Nakamura et al., 2009a). NC-specific loss of SHP-2 also caused a failure in induction of the lacrimal gland from the ectoderm overlying the NC-derived peri-ocular mesenchyme (Garg et al., 2017). A similar phenotype was seen in Mapk1flox/flox; Mapk3−/−; Wnt1-Cre embryos, Mek1flox/flox; Mek2−/−; Wnt1-Cre embryos, and Fgfr1flox/flox; Fgfr2flox/flox; Wnt1-Cre embryos, showing that FGFR-SHP2-ERK1/2 signaling is required in the NC for lacrimal gland induction (Garg et al., 2017).
Together, these studies show that precise ERK1/2 signaling is required for proper NC development. This is seen in multiple NC populations including the cranial and cardiac crest, as well as trunk NC-derived Schwann cells. These studies did not evaluate melanocytes or enteric crest, but both require RTKs capable of inducing ERK1/2 (e.g. KIT and RET, respectively (Fantauzzo and Soriano, 2015)) so defects in these populations would not be surprising.
ERK1/2 in Neural Crest Specification
Neural crest cells are induced in the border region between neural and non-neural ectoderm. Work primarily in chick, frog, and zebrafish has shown that a balance of WNT, FGF, and BMP signals are important for specifying the NC. Several recent reviews address NC specification in more detail (Milet and Monsoro-Burq, 2012; Pegoraro and Monsoro-Burq, 2012; Sauka-Spengler and Bronner-Fraser, 2008; Stuhlmiller and García-Castro, 2012a). Because FGF is a strong inducer of ERK1/2 signaling in frog, zebrafish, and mouse (Christen and Slack, 1999; Corson et al., 2003; Curran and Grainger, 2000; Shinya et al., 2001), this pathway seemed a good candidate for regulating neural crest induction. In chick, FGF-dependent ppERK1/2 is detected at the presumptive neural plate border region prior to NC induction, with higher levels in the more medial neural plate region and lower levels in the more lateral non-neural ectoderm. Inhibition of ERK1/2 activity via overexpression of an ERK1/2 phosphatase, DUSP6, reduced expression of the NC marker PAX7 in its endogenous domain, but upregulated it ectopically in the neural plate, where ppERK1/2 levels were initially higher. This result was confirmed by culturing explants from different lateral regions with a MEK1/2 inhibitor (Stuhlmiller and García-Castro, 2012b).
Xenopus embryos expressing a dominant negative FGFR4 showed reduced NC gene expression at mid-neurula stages. Animal cap explants initially had high ERK1/2 activity, but transitioned to a low ERK1/2, high PI3K state as they differentiated. Both signaling pathways were inhibited by dominant negative FGFR4 expression. However, caps directed to a NC fate (via Pax3 and Zic3 expression) show persistent high ERK1/2 and low PI3K activity, inhibiting MEK or stimulating PI3K signaling in caps interferes with their adoption of a NC fate, and MEK-inhibited animal caps show reduced NC gene expression. This suggests ERK1/2 downstream of FGFR4 is important for multipotent animal cap cells to adopt a NC identity (Geary and LaBonne, 2018).
The mechanism(s) by which ERK1/2 signaling promotes NC induction remains unresolved. Studies from chick demonstrate FGF can act directly on presumptive NC (Stuhlmiller and García-Castro, 2012b). Work in cultured cells and Xenopus indicate it may function by antagonizing BMP activity through inhibitory phosphorylation of SMAD1 by ERK1/2 (Kretzschmar et al., 1997; Pera et al., 2003; Sater et al., 2003). However, knock-in mice expressing a mutant form of SMAD1 in which the linker MAPK phospho-sites are mutated do not have NC defects, so this pathway may not be conserved in the mouse, MAPK may regulate SMAD1 at other sites, other kinases may act in a redundant fashion at other sites, and/or defects in SMAD1 may be compensated for by the related SMAD5 and SMAD8 proteins (Aubin et al., 2004). Indeed, regulation of SMAD1 by MAPK is likely to be complex as there is evidence that other kinases can phosphorylate these linker residues and MAPK may phosphorylate SMAD1 outside the linker region (Eivers et al., 2009; Sapkota et al., 2007).
Data in Xenopus shows that FGF signals to underlying mesoderm, which induces NC-promoting Wnt signals (Monsoro-Burq et al., 2003). These mechanisms are not mutually exclusive nor do they preclude further roles for ERK1/2 activity. Verifying these mechanisms in the mouse has been challenging due to a combination of genetic redundancy, methodological approaches, and possible species differences (Barriga et al., 2015). Of particular utility would be a Cre driver that is active in the neural plate border region or E7.0-E8.0 ectoderm, allowing for genetic manipulation prior to NC induction while circumventing implantation and gastrulation defects. The widely used Wnt1-Cre may only turn on after neural crest specification has already begun (Barriga et al., 2015). Wnt1-Cre mediated reporter activity is first detected around E8.0 (6 somites) in the midbrain and soon expands to other regions of NC, as well as portions of the neuroepithelium (G. Chen et al., 2017), roughly coinciding with the appearance of NC markers, but 0.5–1.0 days after the neural plate border region from which NC arises is specified (Stuhlmiller and García-Castro, 2012a). Indeed, Chen et al. noted that some SOX9-positive NC cells at the level of the hindbrain escape reporter labeling, suggesting Wnt1-Cre is expressed as NC emerges, but may miss the earliest patterning steps and first-specified cells (G. Chen et al., 2017). It would also be informative to assess NC specification in previously generated Mapk3Δ/flox; Mapk1+/−; Sox2-Cre embryos (Frémin et al., 2015), although as discussed above this experiment would not distinguish between autonomous versus non-autonomous requirements for ERK1/2 as Sox2-Cre deletes throughout the epiblast.
ERK1/2 and Neural Crest Proliferation
The ERK1/2 pathway is well known for its ability to drive proliferation (Chambard et al., 2007). This is perhaps best exemplified in the number of activating mutations found in this pathway in human cancer (Dhillon et al., 2007). Conversely, mouse embryonic fibroblasts (MEFs) completely devoid of ERK1/2 fail to proliferate (Pages et al., 1993; Voisin et al., 2010). ERK1/2 stimulates cell-cycle progression by directly phosphorylating and activating growth-promoting transcription factors including TCFs, other Ets-family transcription factors, JUN, and MYC. ERK1/2 also activates MAPKAP families such as the Ribosomal Protein S6 Kinases (RSKs) and Mitogen and Stress-Activated Kinases (MSKs). Phosphorylated RSKs translocate to the nucleus and phosphorylate transcriptional targets including SRF, FOS, and the cAMP Response Binding Protein (CREB). MSKs target CREB, Activating Transcription Factor 1 (ATF1) which can form part of the Activating Protein 1 (AP-1) complex, and histones (Cargnello and Roux, 2011). Together, this ensemble directs rapid induction of IEGs and cell cycle genes, such as CyclinD1, in response to growth factors (Albanese et al., 1995; Lavoie et al., 1996; O’Donnell et al., 2012).
It might seem obvious that NC proliferation relies on ERK1/2 pathway activity, but in vivo evidence is mixed. It is clear that ERK1/2 activity is necessary for growth-factor induced proliferation in a number of NC-derived cell lines. Classic experiments in rat PC12 pheochromocytoma cells showed that a short pulse of ERK1/2 activity in response to Epidermal Growth Factor (EGF) could stimulate proliferation (Marshall, 1995). Intriguingly, extended ERK1/2 activity in response to Nerve Growth Factor (NGF) stimulated differentiation and neurite outgrowth, highlighting the importance of signaling dynamics in encoding different ERK1/2 responses and suggesting ERK1/2 could be an important node regulating a balance between proliferation and differentiation (Marshall, 1995). Dynamic encoding of RTK signaling has been described in several non-NC cell lines, as well (Francavilla et al., 2013; Freed et al., 2017). As shown in our own laboratory, FGF stimulates proliferation of NC-derived mouse embryonic palatal mesenchyme cells (MEPMs) and expression of several growth-associated genes is MEK-dependent (Vasudevan et al., 2015). Some in vivo GoF studies further support a role for ERK1/2 driven proliferation in NC, while others do not. Activation of RAF in adult Schwann cells stimulated their partial dedifferentiation, reduced myelin gene expression, and increased proliferation (Napoli et al., 2012). On the other hand, expressing a constitutively active MEK in Schwann cells beginning at E16 with Egr2Cre or E12-E13 with Dhh-Cre activated ERK1/2 and caused hypermyelination and increased translation, but the study did not note differences in proliferation or cell number (Sheean et al., 2014). Whether this discrepancy is caused by activation of RAF versus MEK, the timing of induction, expression differences, or a combination of these is unclear. Wnt1-Cre-driven expression of an activated allele of Shp2 causes upregulated ppERK1/2 but no change in proliferation in the frontal bone (Nakamura et al., 2009b). This is consistent with the observation that hyperactive ERK1/2 activity can block the cell cycle in the absence of additional signaling perturbations (Chambard et al., 2007; Pumiglia and Decker, 1997).
LoF studies are even less clear. Several reports in which ERK1/2 pathway components were deleted in NC failed to detect any defect in proliferation, but rather found striking defects in NC differentiation, discussed in more detail below (Nakamura et al., 2009a; Parada et al., 2015). Mosaic loss of X-linked Ephrin-B1 in female heterozygous mice caused reduced proliferation in the anterior palatal shelf mesenchyme at E13.5 and E14.5. Stimulation of Ephrin-B1 in cultured MEPMs activated ERK1/2 but not p38, JNK, or AKT, and induced MEK-dependent proliferation and IEG expression, suggesting the in vivo proliferation defect was due to decreased ERK1/2 activity (Bush and Soriano, 2010). Crucially, NC proliferation has not been assayed in vivo in cells lacking all ERK1/2 activity, so proliferation may be supported by low or transient levels of ERK1/2 signaling. Clonal analysis of ERK1/2 null NCCs could resolve this discrepancy.
ERK1/2 and Neural Crest Survival
The ERK1/2 pathway can act as a survival signal by promoting turnover of the pro-apoptotic protein BIM and antagonizing apoptotic MAPK signaling such as the p38 pathway (Lu and S. Xu, 2006). Stimulation of PC12 cells with NGF results in ERK1/2-dependent phosphorylation (and subsequent turnover) of BIM, promoting survival (Ley et al., 2005). In vivo, however, ERK1/2 LoF studies found no defect in NC proliferation or apoptosis, though this observation is subject to the same caveat that ERK1/2 activity was not completely disrupted (Nakamura et al., 2009a; Parada et al., 2015). Loss of SHP2 in NC reduced ppERK1/2 levels, but one study found no increase in apoptosis and another found only a slight increase (Garg et al., 2017). Mutation in Fgf8, expressed in the pharyngeal arch epithelium and forebrain, results in a striking increase in NC apoptosis in the underlying mesenchyme without significant changes in proliferation (Abu-Issa et al., 2002; Frank et al., 2002; Griffin et al., 2013; Storm et al., 2006; Trumpp et al., 1999). This argues that ERK1/2, strongly activated by FGF receptors in this region of the mouse embryo (Corson et al., 2003), could be an important survival signal for NC. However, loss of Fgf8 must suppress ERK1/2 activity to a greater extent than conditional deletion of Mapk1 or Shp2, or the apoptosis in Fgf8 mutants must be dependent on additional pathways besides ERK1/2. Finally, a missense mutation in the ERK1/2 substrate Ets1 underlies the mouse allele variable spotting and Ets1−/− mice that survive postnatally have a white belly spot due to increased apoptosis in melanoblasts (Gao et al., 2010; Saldana-Caboverde et al., 2015), though whether this process is ERK1/2-dependent is unknown. ERK1/2 may therefore promote NC survival, perhaps in combination with other pathways.
ERK1/2 and Neural Crest Migration
ERK1/2 may regulate cell migration, though its role in this process remains controversial. ERK1/2 can bind the focal adhesion protein PAXILLIN, which can facilitate ERK1/2 activation by simultaneously associating with MEK1/2. ERK1/2 can also phosphorylate PAXILLIN and the cytoskeletal regulatory proteins Focal Adhesion Kinase (FAK) and Myosin Light Chain Kinase (MLCK), and can repress integrin activation (Huang, 2004). Waves of ERK1/2 were recently shown to orient collective cell migration in epithelial cells (Aoki et al., 2017), and elevated ERK downstream of EGFR oriented border cell migration in Drosophila (Bianco et al., 2007). It was demonstrated that ‘trailblazing’ cells at the migration front in chick NC migration have a distinct gene expression profile and are enriched for Fgfr2 relative to the cells migrating behind them (McLennan et al., 2015), though a connection between FGFRs, ERK1/2, and migration-specific gene expression has not been demonstrated. FGF2 and FGF8 have chemotactic activity for mouse mesencephalic neural crest (Kubota and Ito, 2000). It was found in chick that FGF8 could act as a chemoattractant for cardiac crest and this activity was partially blocked by MEK1/2 inhibition, though PI3K inhibition had a stronger effect (Sato et al., 2011). Goto et al. utilized transgenic mice expressing FRET-based biosensors to assess signaling activity in migrating enteric neural crest. The study noted some ERK1/2 activity in these cells but there was no enrichment of activity in cells at the leading edge of migration and MEK1/2 inhibition did not affect migration in this system (Goto et al., 2013). However, a separate study cultured E11.0 small intestine fragments with GDNF-expressing cells, and NCC outgrowth was sensitive to MEK1/2 inhibition (Natarajan et al., 2002). Newbern et al. noted hypoplastic pharyngeal arches in compound ERK1/2 conditional mutants and suggested migration defects as one possible explanation, though this was not directly assessed (Newbern et al., 2008). Fate mapping NCCs in Shp2 conditional LoF or GoF mutants did not indicate a widespread NC migration defect, though there was a reduced number of NC cells in the cardiac OFT in the LoF model (Garg et al., 2017; Nakamura et al., 2009b; 2009a). The transcription factor ETS1 is a known ERK1/2 substrate and Ets1−/− mice have deficient migration of cardiac NCCs and melanocytes as assessed by Wnt1-Cre-mediated fate-mapping, although cell migration was not directly imaged or evaluated in culture and the importance of ERK1/2 upstream of ETS1 was not tested in regard to migration (Gao et al., 2010).
Several mutant RTK models draw a link between ERK1/2 and migration. Homozygous null mutations in Erbb2 or Erbb3 or in their ligand Nrg1 results in migration defects of the NC-derived sympathetic neurons with no defects in apoptosis (Britsch et al., 1998). Conditional loss of Erbb3 or its downstream effector Shp2 in Schwann cells causes myelination defects that can be rescued by expressing an activated Mek allele (Sheean et al., 2014). If ERBB2/3 functions similarly in the two cell types, it would be logical that ERK1/2 controls migration downstream of ERBB2/3 in sympathetic neurons, though it remains possible that ERBB2/3 is utilizing different pathways in E10 neurons compared to later Schwann cells. Epha4−/−; Twist+/− mice show craniosynostosis of the coronal suture, staining indicated reduced ppERK1/2 levels, and the primary cellular defect was ectopic migration of osteogenic cells that would normally contribute to the frontal and parietal bones into the coronal suture (Ting et al., 2009). Finally, mice with homozygous Y567F and Y569F mutations in Kit, which disrupts binding site for SFKs, SHP-1, and SHP-2, completely lack melanocytes in adult mice. Mutant KIT protein no longer activates ERK1/2 in response to the KIT ligand SCF, while AKT activation is normal (Kimura et al., 2004). Cellular behavior was not analyzed in this model, however KIT null mice initially specify melanoblasts, which then fail to migrate and are subsequently lost (Bernex et al., 1996). Therefore ERK1/2 may regulate NCC migration in some situations but this effect seems to be highly context-dependent. Precisely when, where, and how this occurs needs to be studied in more detail.
ERK1/2 and Neural Crest Differentiation
ERK1/2 signaling regulates NC differentiation in a variety of contexts. As discussed above, an extended pulse of ERK1/2 activity causes differentiation of neural crest-derived PC12 pheochromocytoma cells into autonomic-nerve like cells (Marshall, 1995). This is reminiscent of the pathway’s role in mouse embryonic stem cells, where ERK1/2 inhibition is required to maintain pluripotency (Nichols et al., 2009; Stuhlmiller and García-Castro, 2012a; Yamanaka et al., 2010). On the other hand, FGF treatment of MEPMs led to proliferation at the expense of differentiation down the osteoblast lineage (Vasudevan et al., 2015). Inhibition of MEK promoted differentiation in these cells and reduced expression of genes associated with proliferation. Supporting this, Fgfr1flox/flox; Wnt1-Cre conditional mutant mice showed expanded alkaline phosphatase activity in the face, indicating ectopic or precocious osteoblast differentiation, though the involvement of other pathways downstream of FGFR1 cannot be ruled out (Vasudevan et al., 2015). Parada et al. generated Mapk1flox/flox; Wnt1-Cre mice that had less severe phenotypes than similar mice analyzed by Newbern et al., due to differences in genetic background (Newbern et al., 2008; Parada et al., 2015). These Mapk1flox/flox; Wnt1-Cre mice exhibited cleft palate, maxillary hypoplasia, a small and asymmetric mandible, micrognathia, tongue defects, and perinatal lethality. This study found no defects in NC proliferation or apoptosis, but rather found specific defects in the ability of NC to differentiate down the osteoblast lineage. Consistent with this observation, mice conditionally lacking the phosphatase SHP2 in NC had vastly reduced ppERK1/2 staining and a noted defect in osteoblast differentiation in the frontal bone with no defect in proliferation or apoptosis in the pharyngeal arches or outflow tract. The same study showed that MEK1/2 inhibition reduced NCC expression of differentiation markers TUJ1 and SMA in a cardiac NCC explant assay (Nakamura et al., 2009a). A separate report observed a slight increase in apoptosis in the pharyngeal arches of Shp2flox/flox; Wnt1-Cre embryos, without significant defects in NC specification or migration, but primarily noted a failure to induce Alx genes necessary for lacrimal gland induction (Garg et al., 2017). Curiously, expression of a GoF Shp2 allele in NC increased ppERK1/2 but also resulted in osteoblast differentiation defects in the frontal bone, as assessed by osteopontin expression, without defects in proliferation or survival (Nakamura et al., 2009b). Loss of the ERK1/2 substrate Ets1 leads to ectopic NC-derived cartilage in the heart, and culture of hearts in the presence of a MEK1/2 inhibitor is sufficient to induce ectopic cartilage (Gao et al., 2010).
Mice mutant for Rps6ka, the gene encoding the ERK1/2 substrate RSK2, have craniofacial abnormalities and decreased bone mass (Laugel-Haushalter et al., 2014; X. Yang et al., 2004). RSK2 mutations in humans cause Coffin-Lowry Syndrome, an inherited disorder distinguished by mental retardation and skeletal abnormalities. Mice deficient for the RSK2 substrate ATF4 show delayed mineralization, like Rsk2 mice, and Rsk2 and Atf4 interact genetically, suggesting an ERK1/2-RSK2-ATF4 signaling axis as one mechanism by which ERK1/2 signaling promotes osteoblast differentiation (X. Yang et al., 2004).
Altogether these studies argue for a critical role for ERK1/2 in the regulation of NC differentiation. Just what that effect is, however, depends on the context. ERK1/2 signaling can promote proliferation and/or antagonize differentiation (in cultured cells, Fgfr1 conditional mutants, and Ets1 mutant hearts), promote differentiation with little effect on proliferation (in the various ERK1/2 LoF mouse models), and inhibit differentiation when overactive (in SHP2 GoF experiments). The inner workings of these pathways are not well understood, but the answer may once again lie within the PC12 paradigm that emphasizes the importance of quantitative differences in signaling, rather than a simple on/off model (Marshall, 1995). Proper differentiation may require high or sustained, but still physiological levels of signaling, whereas proliferation may be supported by low or transient levels of signaling. It is also important to remember that many of these genetic experiments may perturb additional signaling pathways beyond ERK1/2 that also regulate the process under investigation. An ability to better quantify the effects of genetic manipulations on signaling, and identifying which other pathways may be affected, will be important for squaring these disparate observations.
Other Canonical MAPK Pathways
2b. P38
The p38 MAPK pathway is primarily known as a stress-activated protein kinase pathway. It is robustly activated downstream of environmental stress, such as ultraviolet light or hydrogen peroxide, and inflammatory cytokines, but can also be activated by various growth factors (Cuenda and Rousseau, 2007; Katz et al., 2007; Raingeaud et al., 1995; Rice et al., 2002; Tangkijvanich et al., 2002). The mechanism linking RTKs to p38 is unclear, but may proceed via RAC1 and CDC42 (Bagrodia et al., 1995; S. Zhang et al., 1995) (Figure 1B). There are four p38 isoforms in mammals: α, β, γ, and δ encoded by the Mapk14, Mapk11, Mapk12, and Mapk13 genes, respectively. All four kinases have been knocked out, but only p38α has a developmental phenotype (Cuenda and Rousseau, 2007). Embryos lacking p38α die around E12.5 exhibiting anemia, liver, heart, and placental defects (Adams et al., 2000; Allen et al., 2000; Mudgett et al., 2000; Tamura et al., 2000). The placental failure is the cause of the other phenotypes, however, as p38α mutant embryos can be rescued by tetraploid complementation (Adams et al., 2000) or deletion of Mapk14 specifically in the embryo proper using Sox2-Cre (Hui et al., 2007). Such embryos survive to term but subsequently succumb with apparent lung defects. Although p38β deficient mice are viable, Mapk14flox/flox; Mapk11−/−; Sox2-Cre mice die before E16.5 and exhibit exencephaly, a small liver, thin myocardium, and VSD (del Barco Barrantes et al., 2011). While defects in neural crest development were not specifically assayed, craniofacial structures appeared to form normally in these embryos at E13.5. Defects in cardiac crest can lead to VSD, however these mice had numerous myocardial abnormalities, making myocardial dysfunction the most parsimonious explanation. Mapk12−/−; Mapk13−/− double mutants are viable (Sabio et al., 2005).
These results argue that the p38 MAPK pathway is not essential for NC specification or early development. However, p38 has been linked to mechanisms that may be relevant at later stages. First, p38 regulates tracheal smooth muscle cell migration downstream of PDGF in tissue culture (Hedges et al., 1999). Because the neural crest is the source of pericytes and vascular smooth muscle in the head region of the embryo (Etchevers et al., 2001) and it is known that recruitment and stabilization of these cells around blood vessels is dependent on PDGF signaling (Levéen et al., 1994; Soriano, 1994), p38 may be an important effector in this process if both populations of smooth muscle function similarly. Second, p38 has been shown in fibroblasts to phosphorylate FGFR1 in order to regulate its transport to the nucleus and subsequent regulation of transcription (Sørensen et al., 2008). FGFR1 is important for neural crest development (Brewer et al., 2015; Trokovic et al., 2003; C. Wang et al., 2013), however it is currently unknown whether this particular mechanism is at play in NCCs or whether the signal driving p38 activity in this context is FGF itself or another pathway. p38 may regulate NC cell death, as PC12 cells overexpressing the RTK TRKA underwent apoptosis following NGF treatment and this was associated with a marked increase in p38 signaling (Yan et al., 2002). P38 can phosphorylate and activate the transcription factors DLX5 and SP7 (Osterix), promoting osteoblast differentiation in response to BMP2 (Ortuño et al., 2010; Ulsamer et al., 2008; X. Wang et al., 2007), and Dlx5−/− mice have craniofacial bone defects (Acampora et al., 1999). Finally, NCC deletion of the non-canonical TGF-β pathway effector Tak1, a p38 and JNK MAPKKK, leads to cleft palate (Song et al., 2013; Yumoto et al., 2013). These mice have strikingly reduced p38 signaling and additional defects in both JNK and SMAD activity. Conversely, hyperactive p38 signaling in Tgfbr2flox/flox; Wnt1-Cre craniofacial mesenchyme is associated with reduced proliferation (Iwata et al., 2012). p38 is therefore likely to be an important pathway in NCCs, though it would be helpful to disrupt p38 directly in NC as the signaling changes and phenotypes discussed above are currently associated with mutations in more promiscuous upstream components. Furthermore, p38 is implicated primarily downstream of BMP and TGF-β rather than growth factor-mediated RTK signaling.
2c. JNK
The JNK pathway, like the p38 pathway, is a stress-associated MAPK pathway and can be activated by inflammatory cytokines, pathogens, UV radiation, oxidative stress, TGF-β signaling, as well as some GPCRs and WNTs (Zeke et al., 2016). JNK can also be activated downstream of RTKs through the CRK family of adapter proteins, which can engage JNK via the GEF C3G and small GTPase RAP1 (Larsson et al., 1999; Tanaka et al., 1997) (Figure 1C). CRK has also been reported to bind JNK, MKK4, and other signaling components in the JNK pathway, so the primary path(s) between CRK and JNK downstream of RTKs remains obscure (Girardin and Yaniv, 2001; Zeke et al., 2016). JNK is an important regulator of proliferation, apoptosis, and cell migration. Activated JNK regulates transcription, most famously by phosphorylating JUN, from which JNK derives its name, and through other TFs such as MYC and TWIST1 (Zeke et al., 2016). It also has cytosolic targets, many of which are cytoskeletal proteins including PAXILLIN, β-CATENIN, and the microtubule associated proteins MAP1B, MAP2, and DCX, underlying its role in migration (Huang, 2004). There are three JNK isoforms in mammals, JNK1, JNK2, and JNK3 encoded by the Mapk8, Mapk9, and Mapk10 genes, respectively. Mice deficient in any single JNK isoform, JNK1 and JNK3 together, or JNK2 and JNK3 together are viable (Kuan et al., 1999). Mice lacking JNK1 and JNK2 die by E12.5 and exhibit hindbrain exencephaly with reduced hindbrain apoptosis, but increased apoptosis in the forebrain (Kuan et al., 1999; Sabapathy et al., 1999). No difference in BrdU incorporation was noted in the neuroepithelium or cranial mesenchyme. The pharyngeal arches appear to form normally in these embryos and HNK-1/NCAM staining of neural crest cells in E10.5 embryos was indistinguishable from control littermates (Kuan et al., 1999). Additionally, a subset of Mapk8−/−; Mapk9+/− embryos also display exencephaly, while surviving up to E15.5, but neural crest derivatives were not specifically examined (Sabapathy et al., 1999). JNKs have two upstream MAPK kinases, MKK4 and MKK7, that have also been mutated. Mkk4−/− mice die at E12.5 with liver defects (Di Yang et al., 1997; Ganiatsas et al., 1998; Nishina et al., 1999), while Mkk7−/− mice are viable (Schramek et al., 2011). The mice described to date argue against an essential role for JNK signaling in NC specification, growth, or survival, with the important caveat that Jnk triple mutants and Mkk4/7 double mutants have not been analyzed either globally or specifically in NCCs using conditional mutagenesis. To the contrary, a recent study showed that morpholino-mediated depletion of mRNA encoding the tight junction protein MarvelD3 in Xenopus caused NC induction defects associated with elevated JNK signaling. Inhibiting JNK rescued the phenotype, suggesting suppression of JNK may be an important step in NC specification (Vacca et al., 2018).
Like p38, there are several studies indicating a later role for JNK in NCCs, particularly regarding cell migration. Cardiac crest migration was inhibited following treatment with retinoic acid in a neural tube explant assay, an effect mediated by inhibition of JNK activity (J. Li et al., 2001). Intriguingly, Jun mutants lack NCCs in the cardiac outflow tract (Eferl et al., 1999). Mice harboring mutations in two JNK phosphorylation sites in c-JUN are viable, so JNK activity may not be necessary for c-JUN-dependent NCC migration (Behrens et al., 1999), though JNK also phosphorylates c-JUN at additional sites (Hibi et al., 1993). Mice homozygous for a S697A point mutation that disrupts a PKA phosphorylation site in the RTK RET have enteric NC migration defects and this mutation inhibits JNK phosphorylation, but not ERK1/2, p38, SRC, or AKT phosphorylation following stimulation with the RET ligand Glial Derived Neurotrophic Factor (GDNF) (Asai et al., 2006). Chemical inhibition of the JNK pathway negatively affects enteric NC migration, though this study did not find increased JNK activity following treatment with GDNF, so JNK may instead function as a permissive motility signal (Goto et al., 2013). Another substrate of JNK is TWIST1, a transcription factor that regulates epithelial to mesenchymal transition and is important in neural crest emergence and migration (Vincentz et al., 2008; 2013). JNK phosphorylation stabilizes TWIST1 in culture (Hong et al., 2011), however whether JNK affects TWIST1 in a meaningful way in NCCs is unknown. These data suggest JNK could be important for NCC migration in multiple contexts, but in vivo NC defects due to Jnk mutation has not yet been demonstrated and whether JNK is required downstream of specific growth factors in NCCs is unclear.
2d. ERK5
ERK5, also known as Big MAPK (it is twice the size of the other canonical MAPKs due to an extended C-terminus), is encoded by the Mapk7 gene. Like the other MAPKs, it can be activated by multiple stimuli such as cellular stress and growth factors, notably EGF (Kato et al., 1998) and NGF (Nishimoto and Nishida, 2006; Van T Hoang et al., 2017; Watson et al., 2001). Biochemical studies showed ERK5 can be activated by RAS in PC12 cells (Kamakura et al., 1999), although this view has been challenged (Fukuhara et al., 2000; Obara and Nakahata, 2010). ERK5 may instead be activated through the adapter LAD (Sun et al., 2003). It has a single upstream MAPKK, MEK5, which is in turn activated by the MAPKKKs MEKK2/3 (Figure 1D). Uniquely among MAPKs, the ERK5 C-terminus contains a MEF2-interacting domain and a transcriptional activation domain. Mice lacking either ERK5 or MEK5 succumb around E10.5 with cardiovascular defects (Regan et al., 2002; X. Wang et al., 2004). ERK5 mutants were also noted for having defects in cephalic mesenchyme and hypoplastic pharyngeal arches, possibly indicating a NC defect (Regan et al., 2002). and can regulate transcription from SREs by phosphorylating the ternary complex factor ELK4/SAP1 in HeLa cells. Newbern et al. deleted ERK5 in NC using the Wnt1-Cre driver and found this yields viable, fertile mice with reduced size, a shortened mandible, and truncated ear (Newbern et al., 2011). Previous studies had implicated ERK5 PNS development (Finegan et al., 2009), however the PNS was normal in these conditional mutants (Newbern et al., 2011). Thus, ERK5 supports cNCC development, though its impact on development is relatively minor and the underlying cellular defects have not been studied in NC.
3. PI3K Pathway
The PI3K pathway functions through the conversion of phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3) by the PI3K enzyme (Figure 1E). PIP3 recruits and activates the 3-phosphoinositide-dependent protein kinase-1 (PDPK1/PDK1) and recruits the PDK1 substrate protein kinase B/Ak thymoma viral proto-oncogene (PKB/AKT) (Fruman et al., 2017; Vanhaesebroeck et al., 2012). PDK1 phosphorylates AKT at T308 (Alessi et al., 1997; Stokoe et al., 1997), partially activating it and priming it for full activation by phosphorylation at S473 by the mammalian target of rapamycin complex 2 (mTorc2) (Sarbassov et al., 2005) or the DNA-dependent protein kinase (DNA-PK) (Bozulic et al., 2008; Manning and Toker, 2017). AKT is a Ser/Thr kinase which regulates cell survival, proliferation, migration, and differentiation via the mTorc1 pathway, p53 pathway, Forkhead box O (FOXO) transcription factors, Rho-family GTPases, and numerous other substrates (Manning and Toker, 2017). AKT activation is traditionally viewed as the principal output of growth factor-induced PI3K signaling, though more attention has recently been paid to AKT-independent roles of PI3K-PDK1 signaling in development (Grego-Bessa et al., 2016). PIP3 can also activate Rho-family GTPases through PIP3-binding GEFs (Campa et al., 2015; Goicoechea et al., 2014).
PI3K enzymes are divided into three functional classes, Class I, II, and III. Class I is further subdivided into Class IA enzymes, which mainly function downstream of RTKs and will be discussed further, and Class IB enzymes, which primarily act downstream of GPCRs. For information on Classes IB, II and III we direct the reader to these recent reviews (Jean and Kiger, 2014; Thorpe et al., 2015). The PI3K enzyme is composed of a regulatory and a catalytic subunit. In the mouse, there are three Class IA regulatory genes, Pik3r1, Pik3r2, and Pik3r3 encoding p85α/p55α/p50α, p85β, and p55γ, respectively. The Pik3r1 gene encodes multiple subunits via alternative splicing. There are also three Class IA catalytic genes, Pik3ca, Pik3cb, and Pik3cd encoding p110α, p110β, and p110δ, respectively. Class IA regulatory subunits bind to and inhibit Class IA catalytic subunits in the absence of stimulation. Following stimulation with growth factors, regulatory subunits can interface with RTKs through their phospho-tyrosine binding SH2 domains by directly binding the phosphorylated receptor or a phosphorylated adapter protein. Binding has the dual purposes of relieving inhibition of the catalytic subunit and positioning it close to its substrate PIP2 at membranes (Fruman et al., 2017). Catalytic subunits can also be stimulated by the small G-proteins RAS (in the cases of p110α and p110δ) and RAC (in the case of p110β) (Fritsch et al., 2013). PI3K signaling is restricted both in space and time through the degradation of PIP3 by lipid phosphatases, most notably Phosphatase and Tensin homologue (PTEN), which removes the 3′ phosphate added by PI3K (Maehama and Dixon, 1998).
PI3K and Neural Crest Developmental Defects
Like the ERK1/2 pathway, mouse mutants exist for most of the PI3K signaling cascade. An important point to remember is that disruption of Class IA PI3K genes particularly affects PI3K signaling downstream of RTKs, whereas mutation of downstream pathway components, effectors, or feedback enzymes affects non-RTK mediated PI3K signaling as well.
Mutant mice have been generated for all three Class IA catalytic genes. Global loss of p110α, or knock-in of a catalytically dead p110α, results in embryonic lethality around E10.5 (Bi et al., 1999; Graupera et al., 2008; Lelievre et al., 2005). Embryos develop to E9.5 with no organogenesis defects, but are smaller, have widespread hemorrhaging and occasionally exhibit blood-filled subepidermal blebs around the neural tube. Some of these defects are autonomous to endothelium as conditional deletion in endothelial cells results in lethality by E12.5 with angiogenesis defects (Graupera et al., 2008). Embryos null for p110β die around E3.5 and cells cultured from these blastocysts fail to proliferate (Bi et al., 2002). However, homozygous knock-in mice expressing catalytically inactivated p110β are recovered at birth at sub-Mendelian ratios indicating the early lethality is not due to loss of p110β enzymatic activity and PIP3 generation, but may instead be due to a non-enzymatic role for p110β in RTK and GPCR trafficking (Ciraolo et al., 2008). Further complicating matters, despite associating with Class IA regulatory subunits, p110β was initially found to be activated more strongly downstream of GPCRs (Guillermet-Guibert et al., 2008). It was then demonstrated that it could be stimulated synergistically by both RTKs and GPCRs, illustrating the potential for complex interactions among signaling pathways and receptors (Houslay et al., 2016). p110δ is primarily expressed in immune cells and accordingly p110δ null mice are viable with immune system defects (Jou et al., 2002; Okkenhaug et al., 2002).
Mutants in Class IA regulatory subunits more clearly demonstrate a role for growth factor-induced PI3K signaling in NC. Mutations that disrupt Pik3r1 or Pik3r2 alone are viable, but Pik3r1−/−; Pik3r2−/− double mutants die between E10.5 and E12.5 (Brachmann et al., 2005). Like p110α mutants, there is widespread hemorrhaging and subepidermal blebs. Some mice also exhibit a wavy neural tube and those that survive to E11.5 and E12.5 show a facial cleft. This is strikingly similar to the phenotype of Pdgfra mutant mice (Soriano, 1997). Indeed, point mutations in PDGFRα that disrupt its interaction with PI3K indicate PDGFRα signaling is primarily mediated through the PI3K pathway (He and Soriano, 2013; Klinghoffer et al., 2002). Furthermore, conditional mutagenesis indicates that the facial cleft and midfacial blebbing, but not the trunk blebbing or wavy neural tube, are autonomous to the NC (Tallquist and Soriano, 2003). PDGFRα can form heterodimers with PDGFRβ to support its activity in NC and this signaling is also primarily through the PI3K pathway (Fantauzzo and Soriano, 2016; Klinghoffer et al., 2002). Together these results indicate that Pik3r1 and Pik3r2 exhibit redundancy and are required downstream of PDGF in the NC to regulate craniofacial morphogenesis. Mice lacking Pik3r3 have not been described.
Mutations in the downstream effectors PDK1 and the three mouse AKT isoforms exist but have not been especially informative regarding NC. PDK1 mutants die around E9.5 and lack pharyngeal arches and dorsal root ganglia at that time, though the embryos also lack somites and a heart and are developmentally delayed, so whether there is a specific defect in NC or the embryos simply experience growth retardation and/or toxicity is unclear (Lawlor et al., 2002). Akt2−/− and Akt3−/− single mutants and Akt2−/−; Akt3−/− double mutants are viable (Dummler et al., 2006), Akt1−/− mutants are viable but exhibit growth retardation (W. S. Chen et al., 2001), Akt1−/−; Akt2−/− mutants are born with reduced size, skeletal muscle atrophy, a delay in bone development, and die within a few hours of birth likely from respiratory failure (Peng et al., 2003), and Akt1−/−; Akt3−/− mutants die between E11 and E12 with unclear defects, though they appear morphologically normal at E10.5 (Z. Z. Yang et al., 2005). None of the Class IA PI3K enzymes and few PI3K downstream effectors and regulators (PTEN being the exception, see below) have been studied conditionally in NC. It would be interesting to know if NC conditional compound AKT mutants phenocopy the facial cleft seen in Pik3r1−/−; Pik3r2−/− mutants or if the phenotype is AKT-independent.
PI3K and Neural Crest Specification
Studying PI3K in mouse NC specification is hampered by the same drawbacks as those outlined above for ERK1/2. There is, however, some evidence from both mouse and other organisms that PI3K may be important in NC specification. A recent screen for chemicals that inhibit the NC reporter Crestin:EGFP in zebrafish identified a molecule, CAPE, which reduced reporter and NC gene expression as well as NCC migration. The study noted reduced AKT phosphorylation following CAPE treatment and the phenotype was rescued by constitutively active AKT expression whereas constitutively active PTEN caused similar defects to CAPE treatment (Ciarlo et al., 2017). The target of CAPE is unknown as is the signal driving PI3K/AKT activity. A recent study in Xenopus found that late stages of NC specification and subsequent NCC migration were disrupted following PFKFB4 depletion and these effects were due to reduced AKT activity. PFKFB4 was induced by the gene regulatory network governing NC specification, but whether the AKT activity it supports is dependent on a particular growth factor is unknown, as is the precise mechanism by which PFKFB4 promotes AKT activity (Figueiredo et al., 2017). In mouse, p110α mutants, various AKT compound mutants, Pik3r1−/−; Pik3r2−/− double mutants, and PdgfraPI3K/PI3K; PdgfrbPI3K/PI3K all form at least some NC, so RTK-stimulated PI3K activity may not be strictly required to induce NC, though genetic redundancy precludes an unambiguous ruling on the matter. Furthermore, NC induction in these models has not been studied quantitatively or in great detail, so patterning could be negatively affected even if not absolutely inhibited. The missing pharyngeal arches and dorsal root ganglia in Pdk1−/− mice could reflect a NC induction defect, but this possibility has not been specifically investigated (Lawlor et al., 2002).
PI3K in Neural Crest Proliferation
PI3K is known to promote growth through AKT and mTorc1. Indeed, activating mutations in many PI3K components and inactivating mutations in Pten are common in many cancers (Fruman et al., 2017). This connection between PI3K and the cell cycle is unsurprisingly borne out in NCCs during development. Pdgfraflox/flox; Wnt1-Cre mice show reduced proliferation in the medionasal prominence (MNP) (He and Soriano, 2013). At least some of this activity is PI3K dependent because PdgfraPI3K/PI3K mice also show reduced proliferation in this region (He and Soriano, 2013). Expression of a constitutively active allele of Pdgfra in the Meox2-Cre (epiblast), Wnt1-Cre2 (NC), or Col2a1-Cre (chondrocyte) lineages led to cranial suture malformations and ectopic cartilage in this region (He and Soriano, 2017). The opposite phenotype was seen in PdgfraPI3K/PI3K embryos. Indeed, PDGFRα activation drove increased AKT and PLCγ activity. BrdU assays indicated an increase in proliferation in this region, though additional effects on migration and differentiation cannot be ruled out (He and Soriano, 2017). Furthermore, loss of Pten in NCCs caused enhanced PI3K-AKT signaling and subsequent overgrowth of tissue (T. Yang et al., 2017). GDNF-stimulated proliferation of enteric neuroblasts is blocked by a PI3K inhibitor (Focke et al., 2001). PI3K is therefore an important driver of NC proliferation.
PI3K and Neural Crest Survival
PI3K signaling can promote cell survival through two well-characterized mechanisms. One is the phosphorylation and subsequent inhibition of the pro-apoptotic FOXO transcription factors. In enteric NC, a GDNF-RET-PI3K-AKT signaling pathway was shown to promote survival by antagonizing FOXO1 and FOXO3A (Srinivasan et al., 2005). FOXO4 and FOXO6 are expressed in cranial NC in Xenopus where they are antagonized by AKT, and depletion of FOXO4 causes craniofacial abnormalities (Schuff et al., 2010). Foxo4−/− mice have no detectable phenotype, although this could be due to redundancy of Foxo genes (Hosaka et al., 2004). AKT can also promote survival through antagonizing the p53 pathway. A proteomics study from our own laboratory assayed for PDGFRα-induced AKT substrates in MEPMs and identified several negative regulators of p53 (Fantauzzo and Soriano, 2014). Consistent with this observation, p53 mRNA levels are reduced following PDGF-AA treatment, loss of one copy of Tp53 rescues a subset of PdgfraPI3K/PI3K skeletal phenotypes including a strong reduction in the incidence of cleft palate and significant rescue of vertebral malformations (Fantauzzo and Soriano, 2014), and Pdgfra−/− mice have increased apoptosis in cranial mesenchyme (Soriano, 1997).
PI3K and Neural Crest Migration
PI3K activity is critically important for NC migration and numerous studies support a role for growth factor-induced PI3K signaling in this process. Motility of cardiac crest towards Fgf8 (Sato et al., 2011) and enteric neurons towards GDNF (Goto et al., 2013) is sensitive to PI3K inhibition. Supporting this, mouse embryos expressing a Ret Y1062F mutation that disrupts a binding site for multiple adapter proteins had a reduced or missing ENS (Jijiwa et al., 2004; Wong et al., 2005). This mutation affected both ERK1/2 and PI3K signaling, but a later study partially rescued ERK1/2 activity by deleting the negative regulator Sprouty2, while PI3K activity, as assessed by pAKT levels, remained suppressed. The ENS in these double mutant Sprouty2−/−; RetY1062F/Y1062F embryos better colonized the stomach compared to RetY1062F/Y1062F single mutants, but failed to rescue along the length of the intestine, suggesting the phenotype is primarily PI3K dependent (Miyamoto et al., 2011). It remains to be determined whether the primary defect in these embryos is migration, as suggested by the inhibitor study (Goto et al., 2013), or proliferation.
In Xenopus, NC cells migrate toward PDGF-AA and PDGFRα was shown to mediate cell dispersion and contact inhibition of locomotion through regulation of N-Cadherin. This effect was sensitive to inhibition of PDGFRα, PI3K, and AKT (Bahm et al., 2017). In mouse, Pdgfraflox/flox; Wnt1-Cre conditional mutants have lineage tracing defects in the cranial crest, with fewer cells reaching the frontonasal prominence and pharyngeal arches (He and Soriano, 2013). Neural crest explant assays of Pdgfra conditional mutants showed reduced migration out of the explant compared to controls. Cells that did emerge had a smaller size, fewer focal adhesions, and disorganized lamellipodia (He and Soriano, 2013). PDGF signaling is associated with robust PI3K activity, however lineage tracing in PdgfraPI3K/PI3K; Rosa26RLacZ/+; Wnt1-Cre embryos did not reveal a migration defect and MEPMs cultured from these embryos responded normally to PDGF-AA in transwell assays (He and Soriano, 2013). Perhaps PDGFRA regulates migration via a different signaling pathway than PI3K in vivo. He et al. further implicated RAC1 downstream of PDGFRA (He and Soriano, 2013) and RAC1 can be activated by RTKs independent of PI3K (Feng et al., 2011; Schiller, 2006). An alternative explanation is that PdgfraPI3K/PI3K NCCs can still activate PI3K through heterodimer formation with PDGFRB (Fantauzzo and Soriano, 2016). Supporting the involvement of Rho-family GTPases in NC development, conditional NC mutants of Rac1 (Fuchs et al., 2009; Thomas et al., 2010), Cdc42 (Fuchs et al., 2009; Y. Liu et al., 2013), or mice with NC-specific expression of a dominant negative Rho Kinase (ROCK) (Phillips et al., 2012) all display pronounced facial clefts. RTK-driven PI3K signaling can activate these GTPases, but so can PI3K independent mechanisms also active in NC (Fort and Theveneau, 2014; Schiller, 2006). It is currently unknown to what extent these GTPases are dependent on PDGFRα, other RTKs, PI3K, and other stimuli altogether for their overall activity. Inhibitor studies clearly demonstrate the importance of PI3K in NCC migration and the necessity of PI3K activity to induce motility following treatment with growth factors, but genetic studies have yet to conclusively show the source of this PI3K activity originates at a specific RTK. Directly visualizing migration in Pik3r1/2−/− (Brachmann et al., 2005) or PdgfraPI3K/PI3K; Pdgfrbflox/flox; Wnt1-Cre (Fantauzzo and Soriano, 2016) mutant embryos and conducting genetic interaction studies between RTKs, small GTPases and/or PI3K components would be enlightening.
PI3K and Neural Crest Differentiation
There are also several reports pointing to PI3K as a regulator of NCC differentiation. One recent study ablated the negative regulator Pten in NC, resulting in enhanced proliferation as well as osteoblast differentiation in cephalic NCCs (T. Yang et al., 2017). This result is consistent with previous work in our laboratory that PDGF-AA treatment of MEPMs promotes osteoblast differentiation in a PI3K-dependent manner (Vasudevan et al., 2015). It is also likely that PI3K plays a role in smooth muscle differentiation through the positive regulation of SRF-MRTF complexes, which will be discussed in more detail below. Overexpression of FGF8 in the anterior palate caused increased cartilage formation at the expense of bone as well as increased proliferation (J. Xu et al., 2018). Surprisingly, ppERK1/2 levels were unchanged but p85α protein levels were reduced, suggesting FGF-signaling may antagonize PI3K signaling to prevent bone differentiation (J. Xu et al., 2018).
4. Serum Response Factor: A shared MAPK and PI3K Effector
An interesting example of how MAPK and PI3K signaling can elicit some of their effects in NC through a common effector is found in Serum Response Factor (SRF). This transcription factor cooperates with MAPK-activated Ternary Complex Factors (TCFs) such as ELK1 to induce expression of genes regulated by Serum Response Elements (SREs). Alternatively, it can form a complex with Myocardin or the two Myocardin Related Transcription Factors (MRTFs), MRTF-A and MRTF-B (D. Wang et al., 2001; Z. Wang et al., 2004; Zaromytidou et al., 2006). SRF-MRTF complexes are regulated by PI3K, RHO-actin, and ERK1/2 signaling, and are important for regulating genes involved in the cytoskeleton, migration, and muscle differentiation (Panayiotou et al., 2016; Posern and Treisman, 2006).
Newbern et al., investigated NC-specific conditional mutants of Srf in conjunction with their study of the ERK1/2 pathway in NC. Interestingly, these mice partially phenocopied ERK1/2 conditional mutants. They displayed mandibular hypoplasia, outflow tract defects, a midline cleft, and defects of the thymus and thyroid gland, but had less severe maxillary defects and retained their ear and tongue, arguing that the effects of ERK1/2 may be partially mediated through SRF (Newbern et al., 2008). In DRG neurons, SRF was required for axon growth, branching, and target innervation but not survival (Wickramasinghe et al., 2008). Restoring expression in NCCs rescues the phenotype, indicating the defect is autonomous to NC (J. Li et al., 2005). SRF is also linked more directly to RTKs. The craniofacial phenotype of Srfflox/flox; Wnt1-Cre mice is strikingly similar to Pdgfraflox/flox; Wnt1-Cre mice and both genes interact genetically (Vasudevan and Soriano, 2014).
Unsurprisingly, given their similar phenotypes, SRF has been shown to regulate many of the same cellular behaviors as the upstream signaling pathways that regulate it. Deletion of Srf in NC causes a reduction in proliferation in the frontonasal region (Vasudevan and Soriano, 2014). Cultured primary frontonasal prominence cells from Srf conditional mutants failed to migrate towards a source of PDGF-AA, unlike their control littermate counterparts (Vasudevan and Soriano, 2014). This is consistent with SRF’s well-characterized role in cell migration and cytoskeletal gene expression (Franco et al., 2013; Schwartz et al., 2014). Mkl2−/− embryos, which lack SRF’s partner MRTF-B, underwent initial NC specification and migration, but cardiac NC then failed to differentiate into smooth muscle, causing cardiac outflow tract defects.
SRF seems to be deployed differentially via distinct signaling inputs. Our lab recently found that whereas both FGF and PDGF could cause SRF-TCF complex formation in MEPMs in an ERK1/2-dependent fashion, only PDGF stimulated SRF-MRTF complex formation, in a PI3K-dependent fashion (Vasudevan and Soriano, 2014). A detailed study of MRTF-A activation further showed that both ERK1/2 and actin signaling were in fact required for full MRTF activation in NIH3T3 cells, illustrating how multiple pathways can combine to differentially regulate outcomes through a single protein (Panayiotou et al., 2016).
5. The Agony and Ecstasy of Crosstalk
The importance of achieving the proper level and duration of a particular signaling pathway is attested by the striking example of the PC12 cells (Marshall, 1995), the fact that GoF and LoF mutants in the same pathway both generate severe defects, and the importance of feedback genes in development and disease (Neben et al., 2017). In addition to feedback of one pathway onto itself, however, many pathways also cross regulate each other. Indeed, the point has been made that activation of signaling pathways by RTKs might better be described as activation of signaling networks (Jordan et al., 2000; Manning and Toker, 2017; Raju et al., 2014) (Figure 2). This is especially important to keep in mind and test for, as it can fill in crucial gaps connecting pathway perturbations with phenotypes. A defect that seems due to loss of ERK1/2 activity could be more directly attributable to a consequent hyperactivation of PI3K/AKT signaling, or a combination of both. Indeed, cross-regulation between the ERK1/2 and PI3K pathways has long been recognized (Mendoza et al., 2011). In MEPMs, we see increased AKT activation in response to MEK1/2 inhibition and increased ERK1/2 phosphorylation in response to PI3K inhibition (Vasudevan et al., 2015). Consistent with this, FGF1 induces differentiation of MEPMs, normally a PDGFAA-PI3K driven process, when MEK1/2 is also inhibited. A similar phenomenon was noted in zebrafish, where FGF did not normally stimulate AKT activity, but did in the presence of a MEK1/2 inhibitor (Ciarlo et al., 2017). There is also crosstalk between MAPK pathways, likely mediated partially by dual specificity phosphatases (DUSPs), negative feedback genes that are specific to particular MAPKs (Owens and Keyse, 2007). MEK1/2 inhibition leads to an ectopic induction of JNK activity in response to either FGF1 or PDGFAA in MEPMs (Vasudevan et al., 2015). It is therefore possible that a phenotype such as apoptosis thought to be due to reduced ERK1/2 or PI3K activity may instead be caused by non-physiological JNK or p38 signaling. These possibilities will be exciting to explore as our understanding of growth factor signaling in NC advances.
Figure 2. RTK signaling pathways acting in concert.
A generic RTK is shown at center in an activated state. Signaling proteins that activate the MAPK pathways (e.g. GRB2-SOS) and PI3K pathway (e.g. p85α) can be recruited directly to the RTK or via signaling effectors that act as intermediates (e.g. FRS2/3, SHB/SHC, CRK/CRKL, SHP2, GRB7/10/14, IRS1/2, etc.). Many of these proteins can recruit one another and receptor activation thereby creates multimeric signaling complexes. The exact composition of effectors and activated pathways will depend on the RTK, ligand, and cell type involved. MAPK signaling regulates cytoplasmic targets such as MAPKAPs, cytoskeletal proteins, and apoptosis/survival proteins, and nuclear targets, mainly transcription factors. Some MAPKAPS will translocate to the nucleus and regulate TFs, potentiating the response. PI3K activity can activate Rho-family GTPases that regulate the cytoskeleton and cytoskeleton-regulated TFs (e.g. MKL1/2), and activate AKT signaling, which in turn regulates p53, FOXO, and mTORC1 activity. MAPK and PI3K can regulate each other and some of this crosstalk is shown (e.g. AKT inhibition of RAF). MAPK and PI3K can also be activated by other receptors or cellular stress and these sources are shown in a lighter tone. Last, RTKs can activate additional signaling pathways not covered in this review (PLCγ, PKC, Src Family Kinases, and STATs). While many of these pathways can interact with MAPK and PI3K pathways, they are omitted for clarity. Effectors and pathways more associated with MAPK signaling are shown in blue tones and green tones, while those more tightly associated with PI3K signaling are shown in red and orange themes, though the high degree of interconnectedness makes such distinctions somewhat contrived.
6. Future Questions and Approaches
Considerable headway has been made towards understanding where and when growth factors and the signaling networks that they activate are deployed during NC development. Genetic studies have made clear the importance of RTK signaling for NC development and generated a long list of RTK families and signaling proteins known to be involved. Nevertheless, detailed knowledge of how specific growth factors modulate signaling in vivo and how these signaling changes are decoded by NCCs to generate specific behaviors remains lacking. Indeed, as simple on-off signaling models are discarded in favor of network-oriented, dynamic, and quantitative ones, the problems of studying this in vivo only grow. Thankfully, several new approaches are allowing developmental biologists to interrogate signaling with methods once strictly in the domain of cell biologists.
To characterize pathway activity, fluorescent signaling sensors for MAPK and PI3K signaling represent an exciting opportunity to assay pathway activity in living cells and tissues (Gross and Rotwein, 2015; Regot et al., 2014; Vandame et al., 2014). Combined with time-lapse imaging and existing mouse mutants, they could illuminate unappreciated in vivo signaling domains, dynamics, and crosstalk. To date they have already been successfully utilized in cells, worms, flies, zebrafish, and mice (Aoki et al., 2017; Goto et al., 2013; Kamioka et al., 2012; Kumagai et al., 2015; la Cova et al., 2017; Yoo et al., 2010; Q. Zhang et al., 2018). They require a careful approach, as sensors first characterized in cells may not behave the same way in embryos, can be difficult to visualize and quantify. Moreover, depending on the sensor’s own dynamic range, sensitivity, timescale, and localization in the cell, they may report incompletely on a particular molecule’s relevant activity.
It is clear that RTKs can drive divergent cellular responses, depending on the cell type, the identity of the RTK itself, as well as ligand identity, concentration, and duration of exposure. A more complete understanding of how downstream signaling and transcriptional responses vary according to these factors will be key to untangling the complexities of growth factor signaling in NC development. The increasing practicality of mass spectrometry and RNA sequencing, including at the single-cell level, will help to address these questions. These approaches are already generating considerable insight into how NCCs are specified, migrate, and respond to signals (Lignell et al., 2017; J. A. Morrison et al., 2017; Rada-Iglesias et al., 2012).
In addition to techniques that enhance our ability to read out how NCCs are responding, there are several tools facilitating perturbation of signaling systems in more sophisticated ways. Genetic knock-in of point mutations in RTKs to disrupt particular downstream effector molecules has been an effective approach (Asai et al., 2006; Brewer et al., 2015; Hoch and Soriano, 2006; Jijiwa et al., 2004; Kimura et al., 2004; Klinghoffer et al., 2002), but one largely limited to the mouse and time consuming to undertake. CRISPR-Cas9 should extend this technique, and genetic disruption generally, to other systems (Gandhi et al., 2017; Lai et al., 2017; Z. Liu et al., 2016; Willems et al., 2015) and increase throughput where it already exists.
Last, optogenetic control of growth factor signaling is an exciting technique to probe signaling dynamics because of the exquisite spatial and temporal control it allows (Toettcher et al., 2013). It has already been used to great effect to study the ERK1/2 pathway in cells and flies (Johnson et al., 2017; Toettcher et al., 2013; Wilson et al., 2017), and a similar system for studying RAC signaling was recently described (Graziano et al., 2017). The system could be difficult to deploy in mice as samples must be cultured under microscopic observation and is mostly applicable to relatively transparent or superficial cells, but the approach is likely to be useful in culture or smaller embryos.
Out of 150 years of NC research, well over 100 have seen the use of RTK mutants (Dunn, 1937; Little, 1915) and this line of research shows no signs of slowing. Indeed, the past history heralds even more exciting things to come.
Highlights.
ERK1/2 and PI3K signaling play critical roles in neural crest development.
Recent publications have shed light on the in vivo roles of these pathways in multiple developmental models.
Acknowledgments
We thank Stuart Aaronson, Katherine Fantauzzo, Robert Krauss, Sergei Sokol, and our laboratory colleagues for discussions and critical comments on the manuscript. We apologize to authors whose work we have not cited due to space limitations. Work from the author’s laboratory is supported by NIH/NIDCR grants R01 DE022363 and R01 DE022778 to P.S. C.J.D. is supported by NIH/NIDCR fellowship F32 DE026678.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abu-Issa R, Smyth G, Smoak I, Yamamura KI, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 2.Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 3.Adams RH, Porras A, Alonso G, Jones M, Vintersten K, Panelli S, Valladares A, Perez L, Klein R, Nebreda AR. Essential Role of p38α MAP Kinase in Placental but Not Embryonic Cardiovascular Development. Mol Cell. 2000;6:109–116. doi: 10.1016/S1097-2765(05)00014-6. [DOI] [PubMed] [Google Scholar]
- 4.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 5.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 6.Allen M, Svensson L, Roach M, Hambor J, McNeish J, Gabel CA. Deficiency of the stress kinase p38alpha results in embryonic lethality: characterization of the kinase dependence of stress responses of enzyme-deficient embryonic stem cells. J Exp Med. 2000;191:859–870. doi: 10.1084/jem.191.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 8.Aoki K, Kondo Y, Naoki H, Hiratsuka T, Itoh RE, Matsuda M. Propagating Wave of ERK Activation Orients Collective Cell Migration. Dev Cell. 2017;43:305–317e5. doi: 10.1016/j.devcel.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Araki T, Nawa H, Neel BG. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. J Biol Chem. 2003;278:41677–41684. doi: 10.1074/jbc.M306461200. [DOI] [PubMed] [Google Scholar]
- 10.Asai N, Fukuda T, Wu Z, Enomoto A, Pachnis V, Takahashi M, Costantini F. Targeted mutation of serine 697 in the Ret tyrosine kinase causes migration defect of enteric neural crest cells. Development. 2006;133:4507–4516. doi: 10.1242/dev.02616. [DOI] [PubMed] [Google Scholar]
- 11.Aubin J, Davy A, Soriano P. In vivo convergence of BMP and MAPK signaling pathways: impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev. 2004;18:1482–1494. doi: 10.1101/gad.1202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagrodia S, Dérijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 13.Bahm I, Barriga EH, Frolov A, Theveneau E, Frankel P, Mayor R. PDGF controls contact inhibition of locomotion by regulating N-cadherin during neural crest migration. Development. 2017;144:2456–2468. doi: 10.1242/dev.147926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barriga EH, Trainor PA, Bronner M, Mayor R. Animal models for studying neural crest development: is the mouse different? Development. 2015;142:1555–1560. doi: 10.1242/dev.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 16.Belanger LF, Roy S, Tremblay M, Brott B, Steff AM, Mourad W, Hugo P, Erikson R, Charron J. Mek2 Is Dispensable for Mouse Growth and Development. Molecular and Cellular Biology. 2003;23:4778–4787. doi: 10.1128/MCB.23.14.4778-4787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci USA. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernex F, De Sepulveda P, Kress C, Elbaz C, Delouis C, Panthier JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122:3023–3033. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- 19.Bi L, Okabe I, Bernard DJ, Nussbaum RL. Early embryonic lethality in mice deficient in the p110β catalytic subunit of PI 3-kinase. Mammalian Genome. 2002;13:169–172. doi: 10.1007/s00335-001-2123-x. [DOI] [PubMed] [Google Scholar]
- 20.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 21.Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rørth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- 22.Bolande RP. Neurocristopathy: its growth and development in 20 years. Pediatr Pathol Lab Med. 1997;17:1–25. doi: 10.1080/15513819709168343. [DOI] [PubMed] [Google Scholar]
- 23.Bolande RP. The Neurocristopathies. Human Pathology. 1974;5:409–429. [Google Scholar]
- 24.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC. Role of phosphoinositide 3-kinase regulatory isoforms in development and actin rearrangement. Molecular and Cellular Biology. 2005;25:2593–2606. doi: 10.1128/MCB.25.7.2593-2606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brewer JR, Mazot P, Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30:751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brewer JR, Molotkov A, Mazot P, Hoch RV, Soriano P. Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes Dev. 2015;29:1863–1874. doi: 10.1101/gad.264994.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britsch S, Li L, Kirchhoff S, Theuring F, Brinkmann V, Birchmeier C, Riethmacher D. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998;12:1825–1836. doi: 10.1101/gad.12.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush JO, Soriano P. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes Dev. 2010;24:2068–2080. doi: 10.1101/gad.1963210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campa CC, Ciraolo E, Ghigo A, Germena G, Hirsch E. Crossroads of PI3K and Rac pathways. Small GTPases. 2015;6:71–80. doi: 10.4161/21541248.2014.989789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cargnello M, Roux PP. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiology and Molecular Biology Reviews. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casar B, Crespo P. ERK Signals: Scaffolding Scaffolds? Front. Cell Dev Biol. 2016;4:87–11. doi: 10.3389/fcell.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;335:88–89. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 34.Chambard JC, Lefloch R, Pouysségur J, Lenormand P. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, Ishan M, Yang J, Kishigami S, Fukuda T, Scott G, Ray MK, Sun C, Chen S-Y, Komatsu Y, Mishina Y, Liu H-X. Specific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos. genesis. 2017;55 doi: 10.1002/dvg.23034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JY, Lin JR, Cimprich KA, Meyer T. A Two-Dimensional ERK-AKT Signaling Code for an NGF-Triggered Cell-Fate Decision. Mol Cell. 2012;45:196–209. doi: 10.1016/j.molcel.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christen B, Slack JM. Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development. 1999;126:119–125. doi: 10.1242/dev.126.1.119. [DOI] [PubMed] [Google Scholar]
- 39.Ciarlo C, Kaufman CK, Kinikoglu B, Michael J, Yang S, D Amato C, Blokzijl-Franke S, den Hertog J, Schlaeger TM, Zhou Y, Liao E, Zon LI. A chemical screen in zebrafish embryonic cells establishes that Akt activation is required for neural crest development. eLife. 2017;6:421. doi: 10.7554/eLife.29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, Azzolino O, Gonella C, Rubinetto C, Wu H, Dastrù W, Martin EL, Silengo L, Altruda F, Turco E, Lanzetti L, Musiani P, Rückle T, Rommel C, Backer JM, Forni G, Wymann MP, Hirsch E. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Science Signaling. 2008;1:ra3–ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copeland NG, Gilbert DJ, Cho BC, Donovan PJ, Jenkins NA, Cosman D, Anderson D, Lyman SD, Williams DE. Mast cell growth factor maps near the steel locus on mouse chromosome 10 and is deleted in a number of steel alleles. Cell. 1990;63:175–183. doi: 10.1016/0092-8674(90)90298-s. [DOI] [PubMed] [Google Scholar]
- 42.Corson LB, Yamanaka Y, Lai KMV, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 43.Cross MJ, Lu L, Magnusson P, Nyqvist D, Holmqvist K, Welsh M, Claesson-Welsh L. The Shb adaptor protein binds to tyrosine 766 in the FGFR-1 and regulates the Ras/MEK/MAPK pathway via FRS2 phosphorylation in endothelial cells. Mol Biol Cell. 2002;13:2881–2893. doi: 10.1091/mbc.E02-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuenda A, Rousseau S. p38 MAP-Kinases pathway regulation, function and role in human diseases. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Curran KL, Grainger RM. Expression of activated MAP kinase in Xenopus laevis embryos: evaluating the roles of FGF and other signaling pathways in early induction and patterning. Dev Biol. 2000;228:41–56. doi: 10.1006/dbio.2000.9917. [DOI] [PubMed] [Google Scholar]
- 46.del Barco Barrantes I, Coya JM, Maina F, Arthur JSC, Nebreda AR. Genetic analysis of specific and redundant roles for p38alpha and p38beta MAPKs during mouse development. Proc Natl Acad Sci USA. 2011;108:12764–12769. doi: 10.1073/pnas.1015013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 48.Di Yang, Tournier C, Wysk M, Lu H-T, Xu J, Davis RJ, Flavell RA. Targeted Disruption of the MKK4 Gene Causes Embryonic Death, Inhibition of c-Jun NH2-Terminal Kinase Activation, and Defects in AP-1 Transcriptional Activity. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.2307/41766?ref=search-gateway:325741a6806e4288fd4417e24ee663c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dummler B, Tschopp O, Hynx D, Yang ZZ, Dirnhofer S, Hemmings BA. Life with a single isoform of Akt: mice lacking Akt2 and Akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Molecular and Cellular Biology. 2006;26:8042–8051. doi: 10.1128/MCB.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunn LC. Studies on Spotting Patterns II. Genetic Analysis of Variegated Spotting in the House Mouse. Genetics. 1937;22:43–64. doi: 10.1093/genetics/22.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edouard T, Combier JP, Nédélec A, Bel-Vialar S, Métrich M, Conte-Auriol F, Lyonnet S, Parfait B, Tauber M, Salles JP, Lezoualc’h F, Yart A, Raynal P. Functional effects of PTPN11 (SHP2) mutations causing LEOPARD syndrome on epidermal growth factor-induced phosphoinositide 3-kinase/AKT/glycogen synthase kinase 3beta signaling. Molecular and Cellular Biology. 2010;30:2498–2507. doi: 10.1128/MCB.00646-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eferl R, Sibilia M, Hilberg F, Fuchsbichler A, Kufferath I, Guertl B, Zenz R, Wagner EF, Zatloukal K. Functions of c-Jun in liver and heart development. J Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eivers E, Demagny H, De Robertis EM. Integration of BMP and Wnt signaling via vertebrate Smad1/5/8 and Drosophila Mad. Cytokine & Growth Factor Reviews. 2009;20:357–365. doi: 10.1016/j.cytogfr.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Etchevers HC, Amiel J, Lyonnet S. Molecular bases of human neurocristopathies. Adv Exp Med Biol. 2006;589:213–234. doi: 10.1007/978-0-387-46954-6_14. [DOI] [PubMed] [Google Scholar]
- 55.Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 56.Fantauzzo KA, Soriano P. PDGFRβ regulates craniofacial development through homodimers and functional heterodimers with PDGFRα. Genes Dev. 2016;30:2443–2458. doi: 10.1101/gad.288746.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fantauzzo KA, Soriano P. Receptor tyrosine kinase signaling: regulating neural crest development one phosphate at a time. Curr Top Dev Biol. 2015;111:135–182. doi: 10.1016/bs.ctdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fantauzzo KA, Soriano P. PI3K-mediated PDGFR signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev. 2014;28:1005–1017. doi: 10.1101/gad.238709.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng H, Hu B, Liu KW, Li Y, Lu X, Cheng T, Yiin JJ, Lu S, Keezer S, Fenton T, Furnari FB, Hamilton RL, Vuori K, Sarkaria JN, Nagane M, Nishikawa R, Cavenee WK, Cheng SY. Activation of Rac1 by Src-dependent phosphorylation of Dock180(Y1811) mediates PDGFRα-stimulated glioma tumorigenesis in mice and humans. J Clin Invest. 2011;121:4670–4684. doi: 10.1172/JCI58559. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Figueiredo AL, Maczkowiak F, Borday C, Pla P, Sittewelle M, Pegoraro C, Monsoro-Burq AH. PFKFB4 control of AKT signaling is essential for premigratory and migratory neural crest formation. Development. 2017;144:4183–4194. doi: 10.1242/dev.157644. [DOI] [PubMed] [Google Scholar]
- 61.Finegan KG, Wang X, Lee EJ, Robinson AC, Tournier C. Regulation of neuronal survival by the extracellular signal-regulated protein kinase. 2009;5(16):674–683. doi: 10.1038/cdd.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flanagan JG, Leder P. The kit ligand: a cell surface molecule altered in steel mutant fibroblasts. Cell. 1990;63:185–194. doi: 10.1016/0092-8674(90)90299-t. [DOI] [PubMed] [Google Scholar]
- 63.Focke PJ, Schiltz CA, Jones SE, Watters JJ, Epstein ML. Enteric neuroblasts require the phosphatidylinositol 3-kinase pathway for GDNF-stimulated proliferation. J Neurobiol. 2001;47:306–317. doi: 10.1002/neu.1037. [DOI] [PubMed] [Google Scholar]
- 64.Fort P, Theveneau E. PleiotRHOpic: Rho pathways are essential for all stages of Neural Crest development. Small GTPases. 2014;5:e27975. doi: 10.4161/sgtp.27975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Francavilla C, Rigbolt KTG, Emdal KB, Carraro G, Vernet E, Bekker-Jensen DB, Streicher W, Wikström M, Sundström M, Bellusci S, Cavallaro U, Blagoev B, Olsen JV. Functional proteomics defines the molecular switch underlying FGF receptor trafficking and cellular outputs. Mol Cell. 2013;51:707–722. doi: 10.1016/j.molcel.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Franco CA, Blanc J, Parlakian A, Blanco R, Aspalter IM, Kazakova N, Diguet N, Mylonas E, Gao-Li J, Vaahtokari A, Penard-Lacronique V, Fruttiger M, Rosewell I, Mericskay M, Gerhardt H, Li Z. SRF selectively controls tip cell invasive behavior in angiogenesis. Development. 2013;140:2321–2333. doi: 10.1242/dev.091074. [DOI] [PubMed] [Google Scholar]
- 67.Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, Lemmon MA. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell. 2017;171:683–695. e18. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frémin C, Saba-El-Leil MK, Lévesque K, Ang SL, Meloche S. Functional Redundancy of ERK1 and ERK2 MAP Kinases during Development. Cell Reports. 2015;12:913–921. doi: 10.1016/j.celrep.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuchs S, Herzog D, Sumara G, Büchmann-Møller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, Ricci R, Relvas JB, Brakebusch C, Sommer L. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell Stem Cell. 2009;4:236–247. doi: 10.1016/j.stem.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 73.Fukuhara S, Marinissen MJ, Chiariello M, Gutkind JS. Signaling from G protein-coupled receptors to ERK5/Big MAPK 1 involves Galpha q and Galpha 12/13 families of heterotrimeric G proteins. Evidence for the existence of a novel Ras AND Rho-independent pathway. J Biol Chem. 2000;275:21730–21736. doi: 10.1074/jbc.M002410200. [DOI] [PubMed] [Google Scholar]
- 74.Gandhi S, Piacentino ML, Vieceli FM, Bronner ME. Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo. Dev Biol. 2017;432:86–97. doi: 10.1016/j.ydbio.2017.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ganiatsas S, Kwee L, Fujiwara Y, Perkins A, Ikeda T, Labow MA, Zon LI. SEK1 Deficiency Reveals Mitogen-Activated Protein Kinase Cascade Crossregulation and Leads to Abnormal Hepatogenesis. Proc Natl Acad Sci USA. 1998;95:6881–6886. doi: 10.2307/45034?ref=search-gateway:cb6260adcbfaeeb40748d30e933677d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Z, Kim GH, Mackinnon AC, Flagg AE, Bassett B, Earley JU, Svensson EC. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137:1543–1551. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garg A, Bansal M, Gotoh N, Feng GS, Zhong J, Wang F, Kariminejad A, Brooks S, Zhang X. Alx4 relays sequential FGF signaling to induce lacrimal gland morphogenesis. PLoS Genet. 2017;13:e1007047–23. doi: 10.1371/journal.pgen.1007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Geary L, LaBonne C. FGF mediated MAPK and PI3K/Akt Signals make distinct contributions to pluripotency and the establishment of Neural Crest. eLife. 2018;7:139. doi: 10.7554/eLife.33845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988;55:185–192. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 80.Girardin SE, Yaniv M. A direct interaction between JNK1 and CrkII is critical for Rac1-induced JNK activation. EMBO J. 2001;20:3437–3446. doi: 10.1093/emboj/20.13.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giroux S, Tremblay M, Bernard D, Cardin-Girard JF, Aubry S, Larouche L, Rousseau S, Huot J, Landry J, Jeannotte L, Charron J. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9:369–372. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 82.Goicoechea SM, Awadia S, Garcia-Mata R. I’m coming to GEF you: Regulation of RhoGEFs during cell migration. Cell Adh Migr. 2014;8:535–549. doi: 10.4161/cam.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goto A, Sumiyama K, Kamioka Y, Nakasyo E, Ito K, Iwasaki M, Enomoto H, Matsuda M. GDNF and Endothelin 3 Regulate Migration of Enteric Neural Crest-Derived Cells via Protein Kinase A and Rac1. Journal of Neuroscience. 2013;33:4901–4912. doi: 10.1523/JNEUROSCI.4828-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, Smith AJH, Ridley AJ, Ruhrberg C, Gerhardt H, Vanhaesebroeck B. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 85.Graziano BR, Gong D, Anderson KE, Pipathsouk A, Goldberg AR, Weiner OD. A module for Rac temporal signal integration revealed with optogenetics. J Cell Biol. 2017;216:2515–2531. doi: 10.1083/jcb.201604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grego-Bessa J, Bloomekatz J, Castel P, Omelchenko T, Baselga J, Anderson KV. The tumor suppressor PTEN and the PDK1 kinase regulate formation of the columnar neural epithelium. eLife. 2016;5:e12034. doi: 10.7554/eLife.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Griffin JN, Compagnucci C, Hu D, Fish J, Klein O, Marcucio R, Depew MJ. Fgf8 dosage determines midfacial integration and polarity within the nasal and optic capsules. Dev Biol. 2013;374:185–197. doi: 10.1016/j.ydbio.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gross SM, Rotwein P. Akt signaling dynamics in individual cells. J Cell Sci. 2015;128:2509–2519. doi: 10.1242/jcs.168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grueneberg H, Truslove GM. Two closely linked genes in the mouse. Genetics Research. 1960;1:69–90. doi: 10.1017/S0016672300000094. [DOI] [Google Scholar]
- 90.Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJH, Okkenhaug K, Vanhaesebroeck B. The p110beta isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110gamma. Proc Natl Acad Sci USA. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hatano N, Mori Y, Oh-hora M, Kosugi A, Fujikawa T, Nakai N, Niwa H, Miyazaki JI, Hamaoka T, Ogata M. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells. 2003;8:847–856. doi: 10.1046/j.1365-2443.2003.00680.x. [DOI] [PubMed] [Google Scholar]
- 92.He F, Soriano P. Dysregulated PDGFRα signaling alters coronal suture morphogenesis and leads to craniosynostosis through endochondral ossification. Development. 2017;144:4026–4036. doi: 10.1242/dev.151068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He F, Soriano P. A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet. 2013;9:e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hedges JC, Dechert MA, Yamboliev IA, Martin JL, Hickey E, Weber LA, Gerthoffer WT. A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. Journal of Biological Chemistry. 1999;274:24211–24219. doi: 10.1074/jbc.274.34.24211. [DOI] [PubMed] [Google Scholar]
- 95.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 96.Hoch RV, Soriano P. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development. 2006;133:663–673. doi: 10.1242/dev.02242. [DOI] [PubMed] [Google Scholar]
- 97.Hong J, Zhou J, Fu J, He T, Qin J, Wang L, Liao L, Xu J. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Research. 2011;71:3980–3990. doi: 10.1158/0008-5472.CAN-10-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hosaka T, Biggs WH, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Houslay DM, Anderson KE, Chessa T, Kulkarni S, Fritsch R, Downward J, Backer JM, Stephens LR, Hawkins PT. Coincident signals from GPCRs and receptor tyrosine kinases are uniquely transduced by PI3Kβ in myeloid cells. Science Signaling. 2016;9:ra82–ra82. doi: 10.1126/scisignal.aae0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang C. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 101.Hui L, Bakiri L, Mairhorfer A, Schweifer N, Haslinger C, Kenner L, Komnenovic V, Scheuch H, Beug H, Wagner EF. p38α suppresses normal and cancer cell proliferation by antagonizing the JNK–c-Jun pathway. Nat Genet. 2007;39:741–749. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 102.Iwata JI, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-β signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest. 2012;122:873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jain S, Encinas M, Johnson EM, Milbrandt J. Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev. 2006;20:321–333. doi: 10.1101/gad.1387206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127:923–928. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jijiwa M, Fukuda T, Kawai K, Nakamura A, Kurokawa K, Murakumo Y, Ichihara M, Takahashi M. A Targeting Mutation of Tyrosine 1062 in Ret Causes a Marked Decrease of Enteric Neurons and Renal Hypoplasia. Molecular and Cellular Biology. 2004;24:8026–8036. doi: 10.1128/MCB.24.18.8026-8036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson HE, Goyal Y, Pannucci NL, Schüpbach T, Shvartsman SY, Toettcher JE. The Spatiotemporal Limits of Developmental Erk Signaling. Dev Cell. 2017;40:185–192. doi: 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan JD, Landau EM, Iyengar R. Signaling networks: the origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Carpino N, Wang D, Ihle JN. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Molecular and Cellular Biology. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kamakura S, Moriguchi T, Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J Biol Chem. 1999;274:26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- 110.Kamioka Y, Sumiyama K, Mizuno R, Sakai Y, Hirata E, Kiyokawa E, Matsuda M. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct Funct. 2012;37:65–73. doi: 10.1247/csf.11045. [DOI] [PubMed] [Google Scholar]
- 111.Kato Y, Tapping RI, Huang S, Watson MH, Ulevitch RJ, Lee JD. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature. 1998;395:713–716. doi: 10.1038/27234. [DOI] [PubMed] [Google Scholar]
- 112.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kazlauskas A, Feng GS, Pawson T, Valius M. The 64-kDa protein that associates with the platelet-derived growth factor receptor beta subunit via Tyr-1009 is the SH2-containing phosphotyrosine phosphatase Syp. Proc Natl Acad Sci USA. 1993;90:6939–6943. doi: 10.1073/pnas.90.15.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keshet Y, Seger R. The MAP Kinase Signaling Cascades: A System of Hundreds of Components Regulates a Diverse Array of Physiological Functions. In: Turksen K, editor. Planar Cell Polarity, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2010. pp. 3–38. [DOI] [PubMed] [Google Scholar]
- 115.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. BBA - Molecular Basis of Disease. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 117.Kimura Y, Jones N, Klüppel M, Hirashima M, Tachibana K, Cohn JB, Wrana JL, Pawson T, Bernstein A. Targeted mutations of the juxtamembrane tyrosines in the Kit receptor tyrosine kinase selectively affect multiple cell lineages. Proc Natl Acad Sci USA. 2004;101:6015–6020. doi: 10.1073/pnas.0305363101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- 119.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 120.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 121.Kuan CY, Yang DD, Roy DRS, Davis RJ, Rakic P, Flavell RA. The Jnk1 and Jnk2 Protein Kinases Are Required for Regional Specific Apoptosis during Early Brain Development. Neuron. 1999;22:667–676. doi: 10.1016/S0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 122.Kubota Y, Ito K. Chemotactic migration of mesencephalic neural crest cells in the mouse. Dev Dyn. 2000;217:170–179. doi: 10.1002/(SICI)1097-0177(200002)217:2<170::AID-DVDY4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 123.Kumagai Y, Naoki H, Nakasyo E, Kamioka Y, Kiyokawa E, Matsuda M. Heterogeneity in ERK activity as visualized by in vivo FRET imaging of mammary tumor cells developed in MMTV-Neu mice. Oncogene. 2015;34:1051–1057. doi: 10.1038/onc.2014.28. [DOI] [PubMed] [Google Scholar]
- 124.de la Cova C, Townley R, Regot S, Greenwald I. A Real-Time Biosensor for ERK Activity Reveals Signaling Dynamics during C. elegans Cell Fate Specification. Dev Cell. 2017;42:542–553. e4. doi: 10.1016/j.devcel.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lai FPL, Lau ST, Wong JKL, Gui H, Wang RX, Zhou T, Lai WH, Tse HF, Tam PKH, Garcia-Barcelo MM, Ngan ESW. Correction of Hirschsprung-Associated Mutations in Human Induced Pluripotent Stem Cells Via Clustered Regularly Interspaced Short Palindromic Repeats/Cas9, Restores Neural Crest Cell Function. Gastroenterology. 2017;153:139–153. e8. doi: 10.1053/j.gastro.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 126.Larsson H, Klint P, Landgren E, Claesson-Welsh L. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain-containing adaptor protein Crk. J Biol Chem. 1999;274:25726–25734. doi: 10.1074/jbc.274.36.25726. [DOI] [PubMed] [Google Scholar]
- 127.Laugel-Haushalter V, Paschaki M, Marangoni P, Pilgram C, Langer A, Kuntz T, Demassue J, Morkmued S, Choquet P, Constantinesco A, Bornert F, Schmittbuhl M, Pannetier S, Viriot L, Hanauer A, Dollé P, Bloch-Zupan A. RSK2 is a modulator of craniofacial development. PLoS ONE. 2014;9:e84343. doi: 10.1371/journal.pone.0084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lavoie JN, L’Allemain G, Brunet A, Müller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 129.Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lechleider RJ, Sugimoto S, Bennett AM, Kashishian AS, Cooper JA, Shoelson SE, Walsh CT, Neel BG. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor. J Biol Chem. 1993;268:21478–21481. [PubMed] [Google Scholar]
- 131.Lelievre E, Bourbon PM, Duan LJ, Nussbaum RL, Fong GH. Deficiency in the p110alpha subunit of PI3K results in diminished Tie2 expression and Tie2(−/−)-like vascular defects in mice. Blood. 2005;105:3935–3938. doi: 10.1182/blood-2004-10-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lemmon MA, Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 1994;8:1875–1887. doi: 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 134.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 135.Li J, Molkentin JD, Colbert MC. Retinoic acid inhibits cardiac neural crest migration by blocking c-Jun N-terminal kinase activation. Dev Biol. 2001;232:351–361. doi: 10.1006/dbio.2001.0203. [DOI] [PubMed] [Google Scholar]
- 136.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci USA. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 138.Lignell A, Kerosuo L, Streichan SJ, Cai L, Bronner ME. Identification of a neural crest stem cell niche by Spatial Genomic Analysis. Nature Communications. 2017:1–11. doi: 10.1038/s41467-017-01561-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Little CC. The Inheritance of Black-Eyed White Spotting in Mice. The American Naturalist. 1915;59:727–740. [Google Scholar]
- 140.Liu Y, Jin Y, Li J, Seto E, Kuo E, Yu W, Schwartz RJ, Blazo M, Zhang SL, Peng X. Inactivation of Cdc42 in neural crest cells causes craniofacial and cardiovascular morphogenesis defects. Dev Biol. 2013;383:239–252. doi: 10.1016/j.ydbio.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 141.Liu Z, Cheng TTK, Shi Z, Liu Z, Lei Y, Wang C, Shi W, Chen X, Qi X, Cai D, Feng B, Deng Y, Chen Y, Zhao H. Efficient genome editing of genes involved in neural crest development using the CRISPR/Cas9 system in Xenopus embryos. Cell Biosci. 2016;6:22. doi: 10.1186/s13578-016-0088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life (International Union of Biochemistry and Molecular Biology: Life) 2006;58:621–631. doi: 10.1080/15216540600957438. [DOI] [PubMed] [Google Scholar]
- 143.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 144.Maina F, Panté G, Helmbacher F, Andres R, Porthin A, Davies AM, Ponzetto C, Klein R. Coupling Met to specific pathways results in distinct developmental outcomes. Mol Cell. 2001;7:1293–1306. doi: 10.1016/s1097-2765(01)00261-1. [DOI] [PubMed] [Google Scholar]
- 145.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 147.Mayor R, Theveneau E. The neural crest. Development. 2013;140:2247–2251. doi: 10.1242/dev.091751. [DOI] [PubMed] [Google Scholar]
- 148.Mazzucchelli C, Vantaggiato C, Ciamei A, Fasano S, Pakhotin P, Krezel W, Welzl H, Wolfer DP, Pagès G, Valverde O, Marowsky A, Porrazzo A, Orban PC, Maldonado R, Ehrengruber MU, Cestari V, Lipp HP, Chapman PF, Pouysségur J, Brambilla R. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron. 2002;34:807–820. doi: 10.1016/s0896-6273(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 149.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 150.McLennan R, Schumacher LJ, Morrison JA, Teddy JM, Ridenour DA, Box AC, Semerad CL, Li H, McDowell W, Kay D, Maini PK, Baker RE, Kulesa PM. Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development. 2015;142:2014–2025. doi: 10.1242/dev.117507. [DOI] [PubMed] [Google Scholar]
- 151.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Milet C, Monsoro-Burq AH. Neural crest induction at the neural plate border in vertebrates. Dev Biol. 2012;366:22–33. doi: 10.1016/j.ydbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 153.Miyamoto R, Jijiwa M, Asai M, Kawai K, Ishida-Takagishi M, Mii S, Asai N, Enomoto A, Murakumo Y, Yoshimura A, Takahashi M. Loss of Sprouty2 partially rescues renal hypoplasia and stomach hypoganglionosis but not intestinal aganglionosis in Ret Y1062F mutant mice. Dev Biol. 2011;349:160–168. doi: 10.1016/j.ydbio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 154.Monsoro-Burq AH, Fletcher RB, Harland RM. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- 155.Morrison JA, McLennan R, Wolfe LA, Gogol MM, Meier S, McKinney MC, Teddy JM, Holmes L, Semerad CL, Box AC, Li H, Hall KE, Perera AG, Kulesa PM. Single-cell transcriptome analysis of avian neural crest migration reveals signatures of invasion and molecular transitions. eLife. 2017;6:1021. doi: 10.7554/eLife.28415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mudgett JS, Ding J, Guh-Siesel L, Chartrain NA, Yang L, Gopal S, Shen MM. Essential Role for p38α Mitogen-Activated Protein Kinase in Placental Angiogenesis. Proc Natl Acad Sci USA. 2000;97:10454–10459. doi: 10.2307/123582?ref=search-gateway:5e951052ef230c57549c5a7f49e18f22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakamura T, Gulick J, Colbert MC, Robbins J. Protein tyrosine phosphatase activity in the neural crest is essential for normal heart and skull development. Proc Natl Acad Sci USA. 2009a;106:11270–11275. doi: 10.1073/pnas.0902230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nakamura T, Gulick J, Pratt R, Robbins J. Noonan syndrome is associated with enhanced pERK activity, the repression of which can prevent craniofacial malformations. Proc Natl Acad Sci USA. 2009b;106:15436–15441. doi: 10.1073/pnas.0903302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Napoli I, Noon LA, Ribeiro S, Kerai AP, Parrinello S, Rosenberg LH, Collins MJ, Harrisingh MC, White IJ, Woodhoo A, Lloyd AC. A Central Role for the ERK-Signaling Pathway in Controlling Schwann Cell Plasticity and Peripheral Nerve Regeneration In Vivo. Neuron. 2012;73:729–742. doi: 10.1016/j.neuron.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 160.Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- 161.Neben CL, Lo M, Jura N, Klein OD. Feedback regulation of RTK signaling in development. Dev Biol. 2017:1–19. doi: 10.1016/j.ydbio.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Newbern J, Zhong J, Wickramasinghe RS, Li X, Wu Y, Samuels I, Cherosky N, Karlo JC, O’Loughlin B, Wikenheiser J, Gargesha M, Doughman YQ, Charron J, Ginty DD, Watanabe M, Saitta SC, Snider WD, Landreth GE. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci USA. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Newbern JM, Li X, Shoemaker SE, Zhou J, Zhong J, Wu Y, Bonder D, Hollenback S, Coppola G, Geschwind DH, Landreth GE, Snider WD. Specific Functions for ERK/MAPK Signaling during PNS Development. Neuron. 2011;69:91–105. doi: 10.1016/j.neuron.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nishina H, Vaz C, Billia P, Nghiem M, Sasaki T, De la Pompa JL, Furlonger K, Paige C, Hui C, Fischer KD, Kishimoto H, Iwatsubo T, Katada T, Woodgett JR, Penninger JM. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development. 1999;126:505–516. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 167.O’Donnell A, Odrowaz Z, Sharrocks AD. Immediate-early gene activation by the MAPK pathways: what do and don’t we know? Biochem Soc Trans. 2012;40:58–66. doi: 10.1042/BST20110636. [DOI] [PubMed] [Google Scholar]
- 168.Obara Y, Nakahata N. The signaling pathway leading to extracellular signal-regulated kinase 5 (ERK5) activation via G-proteins and ERK5-dependent neurotrophic effects. Molecular Pharmacology. 2010;77:10–16. doi: 10.1124/mol.109.060236. [DOI] [PubMed] [Google Scholar]
- 169.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, Smith AJH, Vanhaesebroeck B. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 170.Ortuño MJ, Ruiz-Gaspà S, Rodríguez-Carballo E, Susperregui ARG, Bartrons R, Rosa JL, Ventura F. p38 regulates expression of osteoblast-specific genes by phosphorylation of osterix. J Biol Chem. 2010;285:31985–31994. doi: 10.1074/jbc.M110.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 172.Pages G, Guérin S, Grall D, Bonino F, Smith A, Anjuere F, Auberger P, Pouyssegur J. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 173.Pages G, Lenormand P, L’Allemain G, Chambard JC, Meloche S, Pouyssegur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Panayiotou R, Miralles F, Pawlowski R, Diring J, Flynn HR, Skehel M, Treisman R. Phosphorylation acts positively and negatively to regulate MRTF-A subcellular localisation and activity. eLife. 2016:5. doi: 10.7554/eLife.15460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Parada C, Han D, Grimaldi A, Sarrion P, Park SS, Pelikan R, Sanchez-Lara PA, Chai Y. Disruption of the ERK/MAPK pathway in neural crest cells as a potential cause of Pierre Robin sequence. Development. 2015;142:3734–3745. doi: 10.1242/dev.125328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pegoraro C, Monsoro-Burq AH. Signaling and transcriptional regulation in neural crest specification and migration: lessons from xenopus embryos. Wiley Interdiscip Rev Dev Biol. 2012;2:247–259. doi: 10.1002/wdev.76. [DOI] [PubMed] [Google Scholar]
- 178.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Pawson T, Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 179.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Phillips HM, Papoutsi T, Soenen H, Ybot-Gonzalez P, Henderson DJ, Chaudhry B. Neural crest cell survival is dependent on Rho kinase and is required for development of the mid face in mouse embryos. PLoS ONE. 2012;7:e37685. doi: 10.1371/journal.pone.0037685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta. 2011;1813:1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 183.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 184.Pumiglia KM, Decker SJ. Cell cycle arrest mediated by the MEK/mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 1997;94:448–452. doi: 10.1073/pnas.94.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Purvis JE, Lahav G. Encoding and Decoding Cellular Information through Signaling Dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM. TGF-β/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Res. 2015;3:15005. doi: 10.1038/boneres.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 189.Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L, Advani J, George B, Kini KR, Geetha NP, Prakash HS, Prasad TSK, Chang YJ, Chen L, Pandey A, Gowda H. A Network Map of FGF-1/FGFR Signaling System. J Signal Transduct. 2014;2014:962962. doi: 10.1155/2014/962962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Regan CP, Li W, Boucher DM, Spatz S, Su MS, Kuida K. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc Natl Acad Sci USA. 2002;99:9248–9253. doi: 10.1073/pnas.142293999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rice AB, Ingram JL, Bonner JC. p38 mitogen-activated protein kinase regulates growth factor-induced mitogenesis of rat pulmonary myofibroblasts. American Journal of Respiratory Cell and Molecular Biology. 2002;27:759–765. doi: 10.1165/rcmb.2002-0070OC. [DOI] [PubMed] [Google Scholar]
- 193.Saba-El-Leil MK, Vella FDJ, Vernay B, Voisin L, Chen L, Labrecque N, Ang S-L, Meloche S. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 2003;4:964–968. doi: 10.1038/sj.embor.embor939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M, Wagner EF. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech Dev. 1999;89:115–124. doi: 10.1016/S0925-4773(99)00213-0. [DOI] [PubMed] [Google Scholar]
- 195.Sabio G, Arthur JSC, Kuma Y, Peggie M, Carr J, Murray-Tait V, Centeno F, Goedert M, Morrice NA, Cuenda A. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 2005;24:1134–1145. doi: 10.1038/sj.emboj.7600578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Saldana-Caboverde A, Perera EM, Watkins-Chow DE, Hansen NF, Vemulapalli M, Mullikin JC, Pavan WJ, Kos L NISC Comparative Sequencing Program. The transcription factors Ets1 and Sox10 interact during murine melanocyte development. Dev Biol. 2015;407:300–312. doi: 10.1016/j.ydbio.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Samuels IS, Saitta SC, Landreth GE. MAP’ing CNS development and cognition: an ERKsome process. Neuron. 2009;61:160–167. doi: 10.1016/j.neuron.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 199.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 200.Sater AK, El-Hodiri HM, Goswami M, Alexander TB, Al-Sheikh O, Etkin LD, Akif Uzman J. Evidence for antagonism of BMP-4 signals by MAP kinase during Xenopus axis determination and neural specification. Differentiation. 2003;71:434–444. doi: 10.1046/j.1432-0436.2003.7107006.x. [DOI] [PubMed] [Google Scholar]
- 201.Sato A, Scholl AM, Kuhn EB, Stadt HA, Decker JR, Pegram K, Hutson MR, Kirby ML. FGF8 signaling is chemotactic for cardiac neural crest cells. Dev Biol. 2011;354:18–30. doi: 10.1016/j.ydbio.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 203.Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases--GEFs what’s the link. Cell Signal. 2006;18:1834–1843. doi: 10.1016/j.cellsig.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 204.Schramek D, Kotsinas A, Meixner A, Wada T, Elling U, Pospisilik JA, Neely GG, Zwick RH, Sigl V, Forni G, Serrano M, Gorgoulis VG, Penninger JM. The stress kinase MKK7 couples oncogenic stress to p53 stability and tumor suppression. Nat Genet. 2011;43:212–219. doi: 10.1038/ng.767. [DOI] [PubMed] [Google Scholar]
- 205.Schuff M, Siegel D, Bardine N, Oswald F, Donow C, Knöchel W. FoxO genes are dispensable during gastrulation but required for late embryogenesis in Xenopus laevis. Dev Biol. 2010;337:259–273. doi: 10.1016/j.ydbio.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 206.Schwartz B, Marks M, Wittler L, Werber M, Währisch S, Nordheim A, Herrmann BG, Grote P. SRF is essential for mesodermal cell migration during elongation of the embryonic body axis. Mech Dev. 2014;133:23–35. doi: 10.1016/j.mod.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 207.Sheean ME, McShane E, Cheret C, Walcher J, Muller T, Wulf-Goldenberg A, Hoelper S, Garratt AN, Kruger M, Rajewsky K, Meijer D, Birchmeier W, Lewin GR, Selbach M, Birchmeier C. Activation of MAPK overrides the termination of myelin growth and replaces Nrg1/ErbB3 signals during Schwann cell development and myelination. Genes Dev. 2014;28:290–303. doi: 10.1101/gad.230045.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sheridan DL, Kong Y, Parker SA, Dalby KN, Turk BE. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J Biol Chem. 2008;283:19511–19520. doi: 10.1074/jbc.M801074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shinya M, Koshida S, Sawada A, Kuroiwa A, Takeda H. Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development. 2001;128:4153–4164. doi: 10.1242/dev.128.21.4153. [DOI] [PubMed] [Google Scholar]
- 210.Silver LM. Mouse Genetics. Oxford University Press; 1995. [Google Scholar]
- 211.Silvers WK. The Coat Colors of Mice. Springer Verlag; 1979. [Google Scholar]
- 212.Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Smith EA, Seldin MF, Martinez L, Watson ML, Choudhury GG, Lalley PA, Pierce J, Aaronson S, Barker J, Naylor SL. Mouse platelet-derived growth factor receptor alpha gene is deleted in W19H and patch mutations on chromosome 5. Proc Natl Acad Sci USA. 1991;88:4811–4815. doi: 10.1073/pnas.88.11.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Song Z, Liu C, Iwata J, Gu S, Suzuki A, Sun C, He W, Shu R, Li L, Chai Y, Chen Y. Mice with Tak1Deficiency in Neural Crest Lineage Exhibit Cleft Palate Associated with Abnormal Tongue Development. J Biol Chem. 2013;288:10440–10450. doi: 10.1074/jbc.M112.432286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 216.Soriano P. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 1994;8:1888–1896. doi: 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- 217.Srinivasan S, Anitha M, Mwangi S, Heuckeroth RO. Enteric neuroblasts require the phosphatidylinositol 3-kinase/Akt/Forkhead pathway for GDNF-stimulated survival. Molecular and Cellular Neuroscience. 2005;29:107–119. doi: 10.1016/j.mcn.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 218.Stephenson DA, Mercola M, Anderson E, Wang CY, Stiles CD, Bowen-Pope DF, Chapman VM. Platelet-derived growth factor receptor alpha-subunit gene (Pdgfra) is deleted in the mouse patch (Ph) mutation. Proc Natl Acad Sci USA. 1991;88:6–10. doi: 10.1073/pnas.88.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, Painter GF, Holmes AB, McCormick F, Hawkins PT. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 220.Storm EE, Garel S, Borello U, Hébert JM, Martínez S, McConnell SK, Martin GR, Rubenstein JLR. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 221.Stuhlmiller TJ, García-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012a;69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Stuhlmiller TJ, García-Castro MI. FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development. 2012b;139:289–300. doi: 10.1242/dev.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sun W, Wei X, Kesavan K, Garrington TP, Fan R, Mei J, Anderson SM, Gelfand EW, Johnson GL. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Molecular and Cellular Biology. 2003;23:2298–2308. doi: 10.1128/MCB.23.7.2298-2308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sørensen V, Zhen Y, Zakrzewska M, Haugsten EM, Wälchli S, Nilsen T, Olsnes S, Wiedlocha A. Phosphorylation of fibroblast growth factor (FGF) receptor 1 at Ser777 by p38 mitogen-activated protein kinase regulates translocation of exogenous FGF1 to the cytosol and nucleus. Molecular and Cellular Biology. 2008;28:4129–4141. doi: 10.1128/MCB.02117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tajan M, de Rocca Serra A, Valet P, Edouard T, Yart A. SHP2 sails from physiology to pathology. Eur J Med Genet. 2015;58:509–525. doi: 10.1016/j.ejmg.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 226.Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- 227.Tamura K, Sudo T, Senftleben U, Dadak AM, Johnson R, Karin M. Requirement for p38α in Erythropoietin Expression A Role for Stress Kinases in Erythropoiesis. Cell. 2000;102:221–231. doi: 10.1016/S0092-8674(00)00027-1. [DOI] [PubMed] [Google Scholar]
- 228.Tan PB, Kim SK. Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet. 1999;15:145–149. doi: 10.1016/s0168-9525(99)01694-7. [DOI] [PubMed] [Google Scholar]
- 229.Tanaka S, Ouchi T, Hanafusa H. Downstream of Crk adaptor signaling pathway: activation of Jun kinase by v-Crk through the guanine nucleotide exchange protein C3G. Proc Natl Acad Sci USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tangkijvanich P, Santiskulvong C, Melton AC, Rozengurt E, Yee HF. p38 MAP kinase mediates platelet-derived growth factor-stimulated migration of hepatic myofibroblasts. J Cell Physiol. 2002;191:351–361. doi: 10.1002/jcp.10112. [DOI] [PubMed] [Google Scholar]
- 231.Thomas PS, Kim J, Nunez S, Glogauer M, Kaartinen V. Neural crest cell-specific deletion of Rac1 results in defective cell-matrix interactions and severe craniofacial and cardiovascular malformations. Dev Biol. 2010;340:613–625. doi: 10.1016/j.ydbio.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ting MC, Wu NL, Roybal PG, Sun J, Liu L, Yen Y, Maxson RE. EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development. 2009;136:855–864. doi: 10.1242/dev.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tokuda M. AN EIGHTEENTH CENTURY JAPANESE GUIDE-BOOK ON MOUSE-BREEDING. Journal of Heredity. 1935;26:481–484. doi: 10.1093/oxfordjournals.jhered.a104011. [DOI] [Google Scholar]
- 236.Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ulsamer A, Ortuño MJ, Ruiz S, Susperregui ARG, Osses N, Rosa JL, Ventura F. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 239.Vacca B, Sanchez-Heras E, Steed E, Busson SL, Balda MS, Ohnuma S-I, Sasai N, Mayor R, Matter K. Control of neural crest induction by MarvelD3-mediated attenuation of JNK signalling. Sci Rep. 2018:1–15. doi: 10.1038/s41598-018-19579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Van Hoang T, Yan TJ, Cavanaugh JE, Flaherty PT, Beckman BS, Burow ME. Oncogenic signaling of MEK5-ERK5. Cancer Letters. 2017;392:51–59. doi: 10.1016/j.canlet.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Vandame P, Spriet C, Riquet F, Trinel D, Cailliau-Maggio K, Bodart JF. Optimization of ERK Activity Biosensors for both Ratiometric and Lifetime FRET Measurements. Sensors. 2014;14:1140–1154. doi: 10.3390/s140101140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 243.Vasudevan HN, Mazot P, He F, Soriano P. Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. eLife. 2015;4:503. doi: 10.7554/eLife.07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Vasudevan HN, Soriano P. A Thousand and One Receptor Tyrosine Kinases: Wherein the Specificity? Curr Top Dev Biol. 2016;117:393–404. doi: 10.1016/bs.ctdb.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Vasudevan HN, Soriano P. SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Dev Cell. 2014;31:332–344. doi: 10.1016/j.devcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB. An absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol. 2008;320:131–139. doi: 10.1016/j.ydbio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, Firulli AB. Twist1 Controls a Cell-Specification Switch Governing Cell Fate Decisions within the Cardiac Neural Crest. PLoS Genet. 2013;9:e1003405–14. doi: 10.1371/journal.pgen.1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Voisin L, Saba-El-Leil MK, Julien C, Fremin C, Meloche S. Genetic Demonstration of a Redundant Role of Extracellular Signal-Regulated Kinase 1 (ERK1) and ERK2 Mitogen-Activated Protein Kinases in Promoting Fibroblast Proliferation. Molecular and Cellular Biology. 2010;30:2918–2932. doi: 10.1128/MCB.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Volinsky N, Kholodenko BN. Complexity of Receptor Tyrosine Kinase Signal Processing. Cold Spring Harb Perspect Biol. 2013;5:a009043–a009043. doi: 10.1101/cshperspect.a009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang C, Chang JYF, Yang C, Huang Y, Liu J, You P, McKeehan WL, Wang F, Li X. Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. J Biol Chem. 2013;288:22174–22183. doi: 10.1074/jbc.M113.463620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 252.Wang X, Goh CH, Li B. p38 mitogen-activated protein kinase regulates osteoblast differentiation through osterix. Endocrinology. 2007;148:1629–1637. doi: 10.1210/en.2006-1000. [DOI] [PubMed] [Google Scholar]
- 253.Wang X, Merritt AJ, Seyfried J, Guo C, Papadakis ES, Finegan KG, Kayahara M, Dixon J, Boot-Handford RP, Cartwright EJ, Mayer U, Tournier C. Targeted Deletion of mek5 Causes Early Embryonic Death and Defects in the Extracellular Signal-Regulated Kinase 5/Myocyte Enhancer Factor 2 Cell Survival Pathway. Molecular and Cellular Biology. 2004;25:336–345. doi: 10.1128/MCB.25.1.336-345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 255.Watson FL, Heerssen HM, Bhattacharyya A, Klesse L, Lin MZ, Segal RA. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat Neurosci. 2001;4:981–988. doi: 10.1038/nn720. [DOI] [PubMed] [Google Scholar]
- 256.Watt KEN, Trainor PA. Neural Crest Cells. Elsevier Inc; 2013. Chapter 17. Neurocristopathies_ The Etiology and Pathogenesis of Disorders Arising from Defects in Neural Crest Cell Development. [DOI] [Google Scholar]
- 257.Wehrle-Haller B, Morrison-Graham K, Weston JA. Ectopic c-kit expression affects the fate of melanocyte precursors in Patch mutant embryos. Dev Biol. 1996;177:463–474. doi: 10.1006/dbio.1996.0178. [DOI] [PubMed] [Google Scholar]
- 258.Wickramasinghe SR, Alvania RS, Ramanan N, Wood JN, Mandai K, Ginty DD. Serum Response Factor Mediates NGF-Dependent Target Innervation by Embryonic DRG Sensory Neurons. Neuron. 2008;58:532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Willems B, Tao S, Yu T, Huysseune A, Witten PE, Winkler C. The Wnt Co-Receptor Lrp5 Is Required for Cranial Neural Crest Cell Migration in Zebrafish. PLoS ONE. 2015;10:e0131768. doi: 10.1371/journal.pone.0131768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE. Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell. 2017;67:757–769. e5. doi: 10.1016/j.molcel.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wong A, Bogni S, Kotka P, de Graaff E, D’Agati V, Costantini F, Pachnis V. Phosphotyrosine 1062 Is Critical for the In Vivo Activity of the Ret9 Receptor Tyrosine Kinase Isoform. Molecular and Cellular Biology. 2005;25:9661–9673. doi: 10.1128/MCB.25.21.9661-9673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wortzel I, Seger R. The ERK Cascade: Distinct Functions within Various Subcellular Organelles. Genes & Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xu J, Huang Z, Wang W, Tan X, Li H, Zhang Y, Tian W, Hu T, Chen YP. FGF8 Signaling Alters the Osteogenic Cell Fate in the Hard Palate. Journal of Dental Research. 2018:1–8. doi: 10.1177/0022034517750141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 265.Yan C, Liang Y, Nylander KD, Schor NF. TrkA as a life and death receptor: receptor dose as a mediator of function. Cancer Research. 2002;62:4867–4875. [PubMed] [Google Scholar]
- 266.Yang T, Moore M, He F. Pten regulates neural crest proliferation and differentiation during mouse craniofacial development. Dev Dyn. 2017:1–35. doi: 10.1002/dvdy.24605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 268.Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, Wahli W, Hemmings BA. Dosage-Dependent Effects of Akt1/Protein Kinase B (PKB) and Akt3/PKB on Thymus, Skin, and Cardiovascular and Nervous System Development in Mice. Molecular and Cellular Biology. 2005;25:10407–10418. doi: 10.1128/MCB.25.23.10407-10418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Yumoto K, Thomas PS, Lane J, Matsuzaki K, Inagaki M, Ninomiya-Tsuji J, Scott GJ, Ray MK, Ishii M, Maxson R, Mishina Y, Kaartinen V. TGF-β-activated Kinase 1 (Tak1) Mediates Agonist-induced Smad Activation and Linker Region Phosphorylation in Embryonic Craniofacial Neural Crest-derived Cells. J Biol Chem. 2013;288:13467–13480. doi: 10.1074/jbc.M112.431775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zaromytidou AI, Miralles F, Treisman R. MAL and Ternary Complex Factor Use Different Mechanisms To Contact a Common Surface on the Serum Response Factor DNA-Binding Domain. Molecular and Cellular Biology. 2006;26:4134–4148. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zeke A, Misheva M, Reményi A, Bogoyevitch MA. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Microbiology and Molecular Biology Reviews. 2016;80:793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zhang Q, Huang H, Lu Q, Wu R, Chung C-I, Zhang S-Q, Torra J, Schepis A, Coughlin SR, Kornberg TB, Shu X. Visualizing Dynamics of Cell Signaling In Vivo with a Phase Separation-Based Kinase Reporter. Mol Cell. 2018 doi: 10.1016/j.molcel.2017.12.008. [DOI] [Google Scholar]
- 274.Zhang S, Han J, Sells MA, Chernoff J, Knaus UG, Ulevitch RJ, Bokoch GM. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 275.Zsebo KM, Williams DA, Geissler EN, Broudy VC, Martin FH, Atkins HL, Hsu RY, Birkett NC, Okino KH, Murdock DC. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell. 1990;63:213–224. doi: 10.1016/0092-8674(90)90302-u. [DOI] [PubMed] [Google Scholar]