Abstract
Purpose
To develop a whole-body MRI technique at 3T with improved lesion conspicuity for metastatic cancer detection using fast, high-resolution and high signal-to-noise ratio T2-weighted (T2W) imaging with simultaneous fat and fluid suppression.
Theory and Methods
The proposed Dual-Echo T2-weighted acquisition for Enhanced Conspicuity of Tumors (DETECT) acquires four images, in-phase (IP) and out-of-phase (OP) at a short and a long echo time (TE) using single-shot turbo spin echo. The IP/OP images at the short and long TEs are reconstructed using the standard Dixon and shared-field-map Dixon reconstruction respectively, for robust fat/water separation. An adaptive complex subtraction between the two TE water-only images achieves fluid attenuation. DETECT imaging was optimized and evaluated in whole-body imaging of five healthy volunteers, and compared against diffusion-weighted imaging with background suppression (DWIBS) in five patients with known metastatic renal cell carcinoma.
Results
Robust fat/water separation and fluid attenuation were achieved using the shared-field-map Dixon reconstruction and adaptive complex subtraction, respectively. DETECT imaging technique generated co-registered T2W images with and without fat suppression, heavily T2W, and fat and fluid suppressed T2W whole-body images in less than 7 minutes. Compared to DWIBS acquired in 17 minutes, the DETECT imaging achieved better detection and localization of lesions in patients with metastatic cancer.
Conclusion
DETECT imaging technique generates T2W images with high resolution, high SNR, minimal geometric distortions, and provides good lesion conspicuity with robust fat and fluid suppression in less than 7 minutes for whole-body imaging, demonstrating efficient and reliable metastatic cancer detection at 3T.
Keywords: Whole-Body Imaging, Fat Suppression, Fluid Suppression, Metastatic Cancer
Introduction
Whole-body imaging using conventional techniques such as positron emission tomography combined with computed tomography (PET/CT) is routinely used clinically for whole-body cancer detection (1). A major concern with these techniques is the exposure to ionizing radiation (2-4), particularly in younger patients who need repeated exposures during long follow-up periods and staging in patients during post-treatment. Additionally, the spatial resolution of PET is limited and some tumors do not demonstrate uptake consistently with conventional radiotracers resulting in very low sensitivities reported for a variety of tumors, particularly when lesions are of smaller size (<1 cm) (5,6). Moreover, while these limitations may be partially compensated with contrast-enhanced computed tomography (CT) examinations, this leads to additional radiation exposure. Furthermore, repeated administrations of nephrotoxic iodinated contrast agents with CT is undesirable in patients with impaired renal function (7), a common occurrence in patients with metastatic disease.
In the past decade, whole-body magnetic resonance imaging (WB-MRI) has become a valuable alternative technique due to its excellent soft tissue contrast combined with high spatial resolution and the lack of ionizing radiation (8). WB-MRI, particularly using echo-planar based diffusion-weighted imaging (DW-EPI), and diffusion weighted imaging with background suppression (DWIBS) have shown improved sensitivity and specificity for metastatic cancer detection at 1.5 T (9). DWI offers increased conspicuity for lesions with restricted diffusion (e.g. high cellularity) by suppressing the confounding tissue signals such as fat and fluid (10,11). However, DWI techniques that rely on EPI sequences suffer from geometric distortions due to large B0 inhomogeneities, particularly using large field-of-views (FOV). Moreover, DWI is inherently signal-to-noise ratio (SNR) limited. Consequently, DWI acquisitions require reduced spatial resolution, multiple signal averages, or both, which results in an increase of the total scan time (10). While the inherent low SNR can be partly mitigated by performing WB-MRI at 3T, larger B0 inhomogeneities at 3T compared to 1.5T lead to worse geometric distortions (12). Alternatively, WB-MRI using short tau inversion recovery (STIR) has been shown to provide increased tumor conspicuity with limited image distortion (13,14). However, STIR also suffers from reduced SNR due to non-selective inversion and requires multiple signal averages resulting in increased total scan times (15).
Most metastatic lesions tend to have longer T2 relaxivity compared to their surrounding non-neoplastic tissues and therefore appear brighter on T2-weighted (T2W) images. However, fat has relatively long T2 relaxivity and fluid has very long T2 relaxivity and therefore, both also appear bright on most clinical T2W images and need to be suppressed to improve lesion conspicuity (16,17). T2W images with fat suppression, either using STIR or chemically selective suppression such as spectral pre-saturation using (adiabatic) inversion recovery (SPIR/SPAIR) (18), can generate fat-suppressed T2W images, but still carry fluid signal such as in cysts that often mimic lesions. Furthermore, STIR suffers from poor SNR, while SPIR/SPAIR suffers from inhomogeneous fat suppression particularly at 3T due to increased B0 inhomogeneities (12,15).
Thus, the purpose of this work was to develop a whole-body MRI technique at 3T with improved lesion conspicuity for metastatic cancer detection using fast, high-resolution and high SNR T2-weighted imaging with simultaneous fat and fluid suppression.
Theory
Imaging Sequence
The proposed imaging strategy is based on a single shot turbo spin echo (SShTSE), which is a routinely used T2W imaging sequence in the body due to its robustness, favorable SNR and minimal image distortion. SShTSE is often performed with fat suppression for improved lesion conspicuity, commonly using SPIR/SPAIR, due to its increased SNR compared to STIR. However, SPIR/SPAIR suffers from fat-suppression failures in areas with increased B0 inhomogeneities, particularly relevant at 3T. Moreover, when applied for whole-body imaging, the thoracic region is prone to fat-suppression failure due to increased B0 inhomogeneities. To overcome these challenges, we used a modified Dixon (mDixon) based SShTSE acquisition, which provided robust fat/water separation in the abdomen in a single acquisition (19). We combined this SShTSE-mDixon with a dual-echo acquisition to achieve fluid suppression. This sequence acquires two sets of images – one at a short echo time (TE) and the other at a long TE, following the same excitation. The non-neoplastic tissues with short T2 and the metastatic lesions with moderately prolonged T2 preferentially appear on the short TE image, while the fluids with very long T2 appear on both short and long TE images. Thus, subtraction of the long TE from the short TE preferentially suppresses fluid signal (20) and improves tumor conspicuity (21). We refer to this technique as, Dual Echo T2-weighted acquisition for Enhanced Conspicuity of Tumors (DETECT).
Figure 1 illustrates a schematic of the DETECT imaging sequence. Following a single 90° excitation pulse, short TE (TE1, ~60-80 ms) images and long TE (TE2, ~400 ms) images are acquired in the same repetition using variable refocusing flip angles (22) and partial phase-encoding acquisitions using a SShTSE. Between each pair of refocusing pulses, in-phase (IP) and out-of-phase (OP) echoes are acquired for both TEs, at all refocusing pulses, using a bipolar readout for mDixon reconstruction. Partial-echo readouts are implemented to balance the in-plane resolution and receiver bandwidth (RBW), while maintaining the optimal time interval (δt) of ~1.1 ms at 3T between IP and OP echoes for robust fat/water separation (19,23). An echo train length (ETL) of ~130 was used for both TEs, with 65 k-space lines for each TE. The RBW of the DETECT sequence was doubled to ~870 Hz/pixel, compared to ~440 Hz/pixel for the standard SShTSE. However, the reconstruction of water-only images using signal averaging of both IP/OP echoes generated comparable images to the standard SShTSE (19). Overall, four images are acquired with both partial phase-encoding and partial readout in a single repetition, including IP and OP images at both short and long TEs using DETECT.
Figure 1.
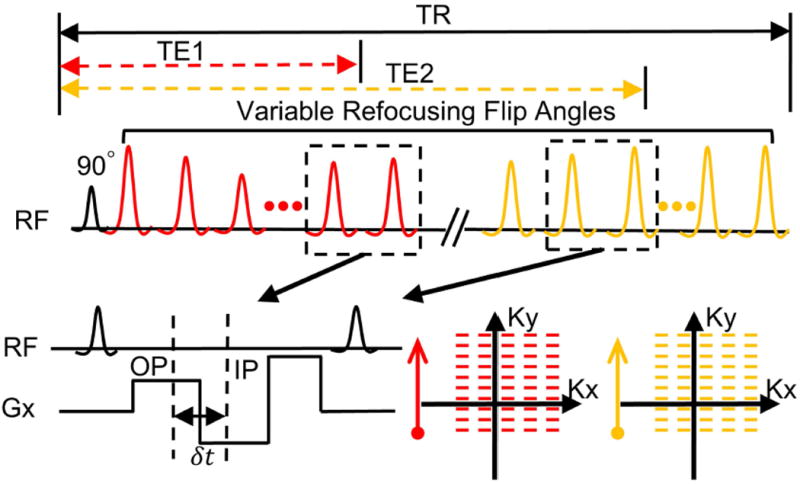
Schematic of the Dual Echo T2-weighted acquisition for Enhanced Conspicuity of Tumors (DETECT) using single-shot turbo spin echo. In each repetition, four images are acquired with variable refocusing flip angles, including out-of-phase (OP) and in-phase (IP) images at both short TE (TE1) and long TE (TE2). The IP and OP echoes are acquired using the bipolar readout gradients (Gx) with partial echo acquisitions between each pair of refocusing pulses, and at all refocusing pulses. After the readout gradients, a rewinder gradient with large gradient strength is used to minimize the echo spacing. δt is the time difference between the OP and IP acquisitions (e.g. 1.1 ms at 3T). For both TEs, a linear view-ordering with partial phase encoding is used to sample the k-space.
Image Reconstruction
Fat suppression
A phase-preserved homodyne reconstruction was used to reconstruct the IP and OP images at both TEs, with zero-filling along the frequency-encoding direction and homodyne filtering along the phase-encoding direction (19,23). This facilitated the reconstruction of complex IP and OP images, which allowed standard mDixon reconstruction for fat/water separation (24). While this approach provided robust fat/water separated images at short TE, the fat/water separation failed at long TE due to the reduced SNR. To overcome this problem, a shared-field-map mDixon reconstruction was used, in which the B0 map estimated at the short TE was used for fat/water separation at the long TE. Considering that the B0 map changes slowly and all images are acquired within the same repetition, this shared-field-map mDixon reconstruction generates robust fat/water separation at the long TE (25). The standard mDixon reconstruction for the short TE images was performed on the scanner including the generation of the low-pass filtered B0 map. The shared-field-map mDixon reconstruction using this low-pass filtered B0 map for the long TE images was implemented in Matlab (Mathworks, Natick, MA).
Fluid Attenuation
The water-only images reconstructed at the short TE (WTE1) and the long TE (WTE2) represent T2-weighted and heavily T2-weighted images respectively, with uniform fat suppression. Given that the tissues with very long T2 (e.g. CSF and gall bladder) appear hyperintense on both short and long TE images, a subtraction between these two images was performed to achieve fluid attenuation. Specifically, a complex subtraction, enabled by the phase-preserved homodyne reconstruction, including a scaling factor (f) was used to perform fluid attenuation (Eq. 1).
| (1) |
where Ψ1is the phase of the WTE1 image and Wsub is the final subtracted water-only image with fat and fluid suppression. The scaling factor, f, was calculated using the following steps: First, the pixels that had signal intensities greater than 80% of the maximum signal intensity on the WTE2 image were selected. Next, the same pixels on the WTE1 image were identified. Finally, f was calculated as the mean value of the ratio of these pixels, i.e. f = mean(I1/I2), where I1 and I2 are the signal intensities of the reference pixels in WTE1 and WTE2 respectively. This scaling factor compensated the T2 decay of tissues with long T2; however, it overcompensated for tissues with very long T2 (e.g. when abs(f×WTE2) > abs(WTE1)). Thus, the demodulation of the phase, ψ1, and the final real operation in equation 1 preserved the sign after the complex subtraction and rectified this overcompensation by resetting those pixel values to zero. The complex subtraction including the scaling factor calculation and phase demodulation was implemented in Matlab. A flowchart showing the DETECT reconstruction including fat and fluid suppression is shown in Supporting Figure S1.
Simulations
The choice of the TEs determines the signal difference that can be achieved on the Wsub image to enhance the conspicuity of the tumors, while simultaneously suppressing the fluids. Several factors of a SShTSE acquisition determine the TE, including view-ordering, echo spacing, FOV, partial phase-encoding factor and refocusing flip angle scheme. In this study, we chose linear view-ordering and variable refocusing flip angle schemes (22) for both TEs, to match clinically used SShTSE acquisitions at short TE (i.e. TE1=60-80 ms) for T2-weighted imaging of the abdomen, while also reducing the total SAR. The variable refocusing flip angle scheme was defined by the minimum, and maximum refocusing flip angles (αmin, αmax), used to sample the beginning and end of the echo train respectively, along with the middle refocusing flip angle (αmid) used to sample the center of the k-space for short TE (26). The δt of about 1.1 ms, combined with receiver bandwidth and in-plane resolution forced the minimum echo spacing to be about 6.6 ms. Thus, the effective echo time (TEeff), defined as the TE when the center of k-space was sampled, and the equivalent echo time (TEequiv), defined as the TE that generates similar contrast as standard T2W image, were determined by the variable refocusing flip angle scheme in combination with the partial phase-encoding factor.
Bloch equation simulations were performed to investigate the influence of variable refocusing flip angle scheme and the partial phase-encoding factor on the signal difference to determine the optimal TEs. First, the partial phase-encoding factor was fixed at 0.6, similar to the standard clinical SShTSE acquisition, along with αmin at 90° and αmax at 180°, while the αmid was varied from 100° to 160° at 20° increments. This achieved the following TE values: TE1 = 60 ms and TE2 = 450 ms. Next, the partial phase-encoding factor was varied between 0.6 and 0.7 (which also varied the TE1 and TE2 times), with the αmin, αmid, and αmax fixed at 90°, 100°, and 120° respectively. These flip angles were chosen to achieve clinically equivalent TE, while reducing the total SAR (27) and sensitivity to motion (23). The other simulations parameters were: FOV = 520 mm (phase-encoding direction), voxel size = 1.8 mm, SENSE = 3, echo spacing = 6.6 ms. Signal evolution was calculated for a variety of tissues with the following T1 and T2 values at 3T (28-31): gray matter, T1/T2 = 1820/99 ms; white matter, T1/T2 = 1084/69 ms; liver, T1/T2 = 812/42 ms; kidney, T1/T2 = 1194/56 ms; fat, T1/T2 = 371/133 ms; synovial fluid, T1/T2 = 3620/767 ms; CSF, T1/T2 = 4500/2500 ms; and a generic tumor model with T1/T2 = 1000/150 ms. The signal differences between the two TEs were plotted against a range of T2 values.
Methods
Imaging Studies
All imaging was performed on a 3T MR scanner (Ingenia, Philips Healthcare, Best, The Netherlands). The DETECT was evaluated in 6 healthy volunteers, first in a dedicated abdominal imaging session of a healthy volunteer, followed by whole-body imaging protocol in 5 healthy volunteers. Subsequently, 5 patients with known metastatic renal cell carcinoma (mRCC) were enrolled for whole-body imaging evaluation of the DETECT sequence. The study protocol was approved by the institutional review board (IRB), Health Insurance Portability and Accountability Act (HIPAA)-compliant and all subjects provided written informed consent prior to their participation in the study.
Abdominal Imaging
To evaluate the shared-field-map mDixon reconstruction, one 40-year old healthy female volunteer was enrolled for abdominal imaging. The acquisition parameters for the DETECT sequence were: coronal orientation; FOV = 400 × 400 mm2; slice thickness/slice gap = 4 mm/0 mm; voxel size = 1.5 × 2 mm2; SENSE = 3; echo spacing = 6.2 ms; TE1eff/TE2eff = 69/340 ms; TEequiv1/TEequiv2 = 62/297 ms; TR = 1250 ms; δt = 1.1 ms; ETL = 130 for both TEs with 65 k-space lines for each TE; RBW = ~870 Hz/pixel; partial phase-encoding factor = 0.65 and partial readout factor = 0.7. A total of 42 slices were acquired with 14 slices each in a 16-second breathhold acquisition. A 16-channel phased-array anterior coil along with the 12-channel phased-array posterior coil, embedded in the table, were used for signal reception.
Whole-Body Imaging of Normal Volunteers
Five healthy volunteers (3 females, 2 males, age range: 24 – 61 years), including 2 volunteers for optimization and 3 volunteers for evaluation of whole-body DETECT imaging were enrolled. The three healthy volunteers were scanned in 5 stations (head, thorax, abdomen, pelvis, and thighs) to cover the whole body from the head to the knees. All images were acquired in the coronal plane at an acquisition time of about 1 minute per station for approximately 50 slices. The thoracic and abdominal regions were acquired in four, 15-second breathheld acquisitions each, which increased the scan time to about 2 minutes for each of these stations, including the breathhold instructions. A SENSE acceleration factor of 3, partial phase-encoding factor of 0.6, partial readout factor of 0.85, ETL of 130 for both TEs with 65 k-space lines for each TE, RBW of 870 Hz/pixel and δt of 1.1 ms were used. All images were acquired contiguous with no slice gap. The remaining acquisition parameters are listed in Table 1. The total scan time of whole-body DETECT imaging was approximately 7 minutes including the breathhold instructions.
Table 1.
Parameters of the whole-body MRI sequences
| Parameter
|
||||||
|---|---|---|---|---|---|---|
| Sequences | FOV (cm2) Read-Phase | Voxel Size (mm3) Read-Phase-Slice | TR/TE (ms) | Flip Angle (degree) | Scan Time (min/station) | Total Scan Timea (min) |
| DETECT (Coronal) | 30×30 (head) | 1.2×1.2×5 (head) | 1250/70 (TE1), 450 (TE2) | 90 (min)- 100 (mid) - 120 (max) | 1:01 | 7:00 |
| 32×52 (body) | 1.3×1.8×5 (body) | |||||
|
| ||||||
| DWIBS (Coronal) | 30×30 (head) | 3.5×3.5×5 | 12000/70 | NA | 3:09(head) | 16:29 |
| 32×52 (body) | 3:20 (body) | |||||
|
| ||||||
| DWIBS (Axial) | 30×30 (head) | 3.5×3.5×5 | 12000/70 | NA | 3:09 (head) | 24:09 |
| 32×52 (body) | 5:15 (body) | |||||
Five stations and included breathhold instructions for thoracic and abdomen scans for DETECT.
For the most time-efficient imaging of larger FOV, coronal plane acquisitions are often preferred. However, DWIBS images are prone to increased image distortion due to gradient non-linearities in the coronal plane compared to the axial plane. Hence, DWIBS images are commonly acquired in the axial plane to minimize image distortions, but at the expense of increased acquisition times. To evaluate the image quality and acquisition efficiency, one healthy volunteer was scanned with whole-body DWIBS in both axial and coronal planes, compared to whole-body DETECT in the coronal plane. The scan parameters are listed in Table 1.
Whole-Body Imaging of Patients
Five patients (1 female, age: 58 years; and 4 males, age: 52 – 68 years) with known mRCC on prior clinical imaging, were scanned to evaluate the performance of DETECT for metastatic cancer detection. The whole-body MRI protocol included DETECT compared against DWIBS using 5 stations in the coronal plane. The acquisition parameters were similar to the above volunteer studies, except for the FOV along the anterior-posterior direction which varied among subjects between 300-400 mm for complete coverage of the body. To improve the visualization of the metastatic lesions and suppress the signals from complex fluids in the abdomen and bowel, an effective T2 map was generated using the two TE images (Eq. 2), with a threshold of 300 ms. This map effectively suppressed the signals from the complex fluid with moderate T2 values (~300 ms), which were longer than that of the metastatic lesions (~160 ms), but were not long enough to be visible on the long TE image (TE2equiv = ~400 ms) and therefore, were not suppressed on the DETECT image. Subsequently, a T2map-weighted subtracted image (Eq. 3) was generated to improve the conspicuity and localization of the lesions.
| (2) |
| (3) |
where TE1eff and TE2eff are the effective TEs.
Image Evaluation
In all 8 whole-body subjects, including 3 healthy volunteers and 5 mRCC patients, the signal reduction of long T2 tissues such as fluids on the DETECT sequence was measured compared to the short TE image, as described before (21). Additionally, the number of lesions identified on the proposed DETECT sequence compared to DWIBS was assessed in consensus by three board-certified radiologists with different levels of expertise in body MRI (A.P, fellow; N.M.R, 25 years and I.P, 16 years).
Results
Simulations
Figure 2 shows the simulated signal differences between the two TEs of the DETECT with varying refocusing flip angles (Fig. 2a) and partial phase-encoding factors (Fig. 2b). The signal difference for tissues of interest with T2 less than ~160 ms increases with larger αmid (Fig. 2a), but at the expense of increased SAR. The increasing partial phase-encoding factor also increases the relative signal difference for tissues with longer T2 (e.g. 200 ms vs. 150 ms) since it prolongs the TEeff for both TEs (Fig. 2b), but at the expense of reduced SNR. The majority of the tissues in the body (except for fat and fluid) have T2 less than 100 ms at 3T (28), while the tumors tend to have moderately prolonged T2 but still typically less than 200 ms (32). Thus, an αmid of 100° and partial phase-encoding factor of 0.6 were chosen to retain the T2 contrast of the normal tissues with T2 less than 100 ms on the subtracted DETECT, while maximizing the signal difference for tissues with targeted T2 values around 150 ms (Fig. 2c). This combination, along with αmin of 90° and αmax of 120°, provided a TEeff of 70 ms for the first TE, matching the clinical whole-body T2W imaging protocol and a TEeff of 450 ms for the second TE, with low SAR (~2.2 W/kg) and reduced sensitivity to motion (22). The simulated signal evolutions of the interested tissues are shown in Figure 2d. As expected, the fat and fluid appear bright on T2-weighed images and when suppressed using the DETECT, improve the visualization and conspicuity of the tumors.
Figure 2.
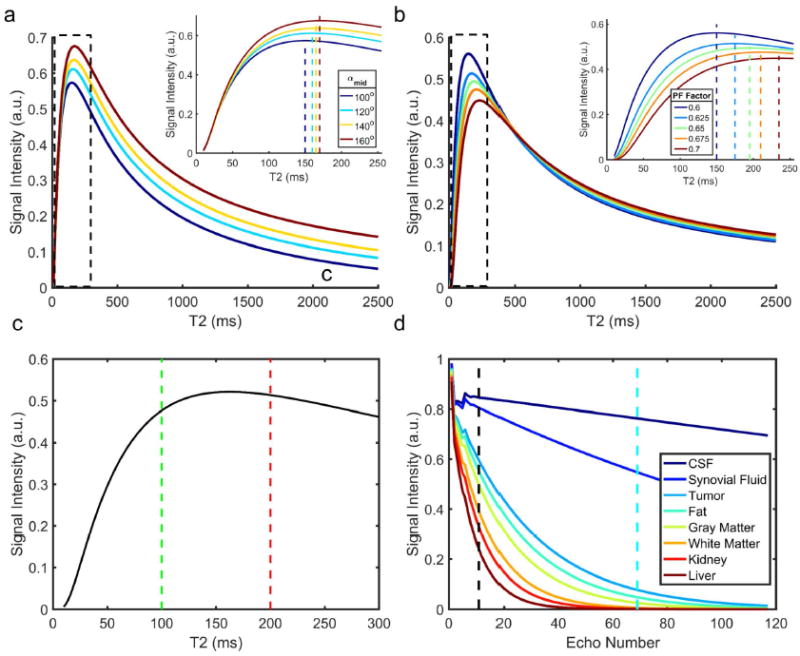
Simulated signal differences between the two different TEs against varying T2 for different refocusing flip angle schemes (a) and partial phase encoding factors (b). The simulation parameters are described in Theory. The maximum signal differences (dashed vertical color lines in insets) shift to longer T2 with the increase of either αmid (a) or partial phase encoding factor (b). A refocusing flip angle train of 90° (αmin) – 100° (αmid) – 120° (αmax) shows maximum signal difference for tissues with T2 values between 100 and 200 ms (c). The signal behavior with this scheme shows that the signals from tissues with very long T2 (e.g. CSF and synovial fluid) along with fat appear bright compared to tumor, and need to be suppressed to improve lesion conspicuity (d).
Shared Field-Map mDixon Reconstruction
Figure 3 shows the improved fat/water separation in the abdominal images of a 47-year old healthy female volunteer using the shared-field-map mDixon reconstruction. At the short TE (TE1), the standard mDixon reconstruction achieved robust fat/water separation throughout the imaging FOV, with minimum fat/water swaps at the edges of the large FOV (Figs. 3a, 3d). However, the fat/water separation failed significantly with the standard mDixon reconstruction at the long TE (TE2, Figs. 3b, 3e) due to the reduced SNR. The proposed shared-field-map mDixon reconstruction using the B0 map from the short TE achieved robust fat/water separation in the long TE images (Figs. 3c, 3f).
Figure 3.
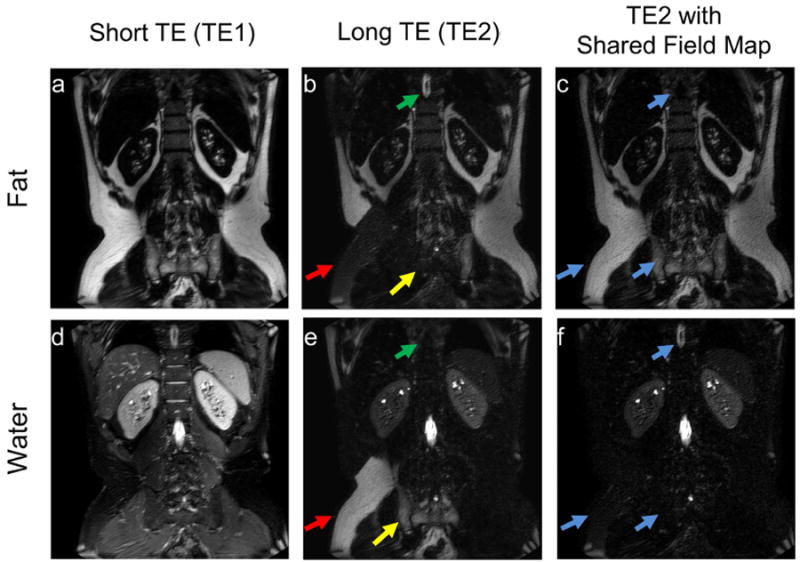
Coronal images of a 47-year-old healthy female volunteer’s abdomen showing robust fat/water separation using the standard mDixon reconstruction at the short TE (TE1, a, d), and failed fat/water separation at the long TE (TE2, b, e) involving multiple locations (e.g. the subcutaneous fat (red arrows), CSF (green arrow) and bone marrow (yellow arrow)). The shared-field-map mDixon reconstruction (c, f) using the B0 field map from TE1 achieved successful fat/water separation at TE2, even with reduced SNR (blue arrows). Some residual FID artifacts were observed on the fat images due to stimulated echoes, which were subsequently minimized in the whole-body images using stronger crusher gradients.
Complex Subtraction
The results of fluid attenuation, reconstructed with both the magnitude and complex subtraction are shown in Supporting Figure S2. The “dark-rim” artifacts, that are often observed around the edges of the tissues with relatively long T2 on magnitude subtraction, are the resultant of the modulation of the point spread function. Since the phase of the water signal is preserved with our phase-preserved homodyne and shared-field-map mDixon reconstruction, the complex subtraction eliminated these “dark-rim” artifacts and resulted in much smoother profiles.
Whole-Body Imaging of Normal Volunteers
The shared-field-map mDixon reconstruction and the complex subtraction achieved uniform fat and fluid suppression throughout the body (Fig. 4) and across all slices (Supporting Video S1). The whole-body images, acquired with DWIBS in both coronal and axial orientations and with DETECT in coronal orientation are shown in Figure 5. DWIBS images in the coronal orientation suffer from large geometric distortions (Fig. 5a). Although the coronal DWIBS images reformatted from the axial acquisitions (Fig. 5b) also suffer from geometric distortions, the originally acquired axial images show less in-plane distortions (not shown). However, the scan times for axial DWIBS acquisitions are generally longer compared to the coronal acquisitions. In our example, the total scan time for the axial DWIBS acquisitions was 19 minutes compared to the 13 minutes for coronal DWIBS acquisitions for 4-station WBMRI. Alternatively, the images acquired using DETECT exhibited minimal geometric distortions (Fig. 5c) in a 6:00 minute coronal plane acquisition for 4-station WBMRI, including breathhold instructions. DETECT images showed better quality with uniform fat and fluid suppression compared to DWIBS without geometric distortions and artifacts in the coronal plane over the entire volume (Supporting Video S2).
Figure 4.
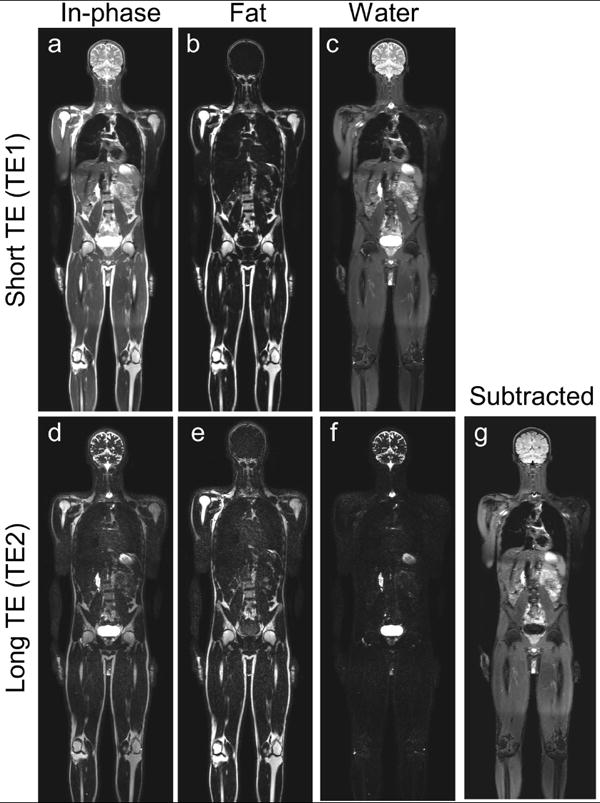
Whole-body MR DETECT images of a 28-year-old healthy male volunteer acquired in five stations in 7 minutes. The standard mDixon reconstruction demonstrates robust fat/water separation across the entire volume at the short TE (a-c), while the shared-field map mDixon reconstruction achieved uniform fat/water separation at long TE (d-f). The subtracted image (g) shows uniform fat and fluid suppression over the entire imaging volume and across all slices (Supporting Video S1).
Figure 5.
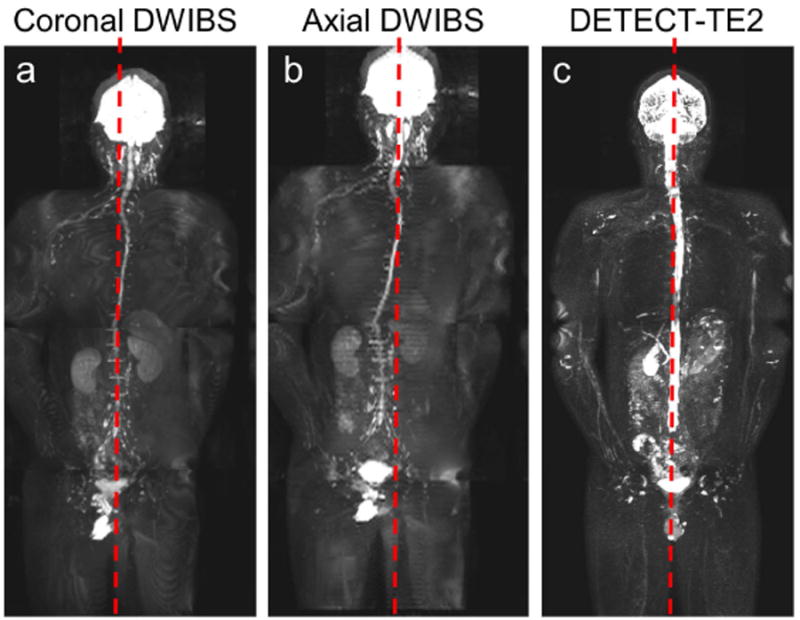
Whole-body 3D MIP reconstructions of a 34-year-old healthy male volunteer demonstrating increased robustness of DETECT to geometric distortions, compared to DWIBS. 3D MIP from coronal DWIBS at b = 800 s/mm2 (a) and the coronal reformat from the axial acquisition of DWIBS at b=800 s/mm2 (b) show distorted spinal cord from the midline of the image (red dashed line). 3D MIP of the long TE image from DETECT shows straight spinal canal compared to the midline (c). The DETECT and DWIBS images in the coronal plane across all slices are shown in Supporting Video S2.
Whole-Body Imaging of Patients
The total number of lesions identified, including lesions per station, on the WB-MRI with DETECT compared to WB-MRI with DWIBS is summarized in Table 2. Overall, the DETECT identified all metastatic lesions known on prior clinical imaging and several additional new lesions, that were not identified on DWIBS due to artifacts associated with severe geometric distortions. For example, both DETECT and DWIBS showed clear delineation of a RCC metastatic lesion in the left iliac bone (Fig. 6), for which the patient was receiving radiation treatment. However, the localization of the lesion with respect to the background anatomy was challenging on the DWIBS image due to the geometric distortions. The subtracted DETECT image, however, retained the lesion signal while suppressing the signals from fat and fluids with long T2 (Fig. 6b). The effective T2-map weighted image, generated with T2 values less than 300 ms, further increased the lesion conspicuity (Fig. 6c). In another mRCC patient with multiple metastatic lesions and right femoral metal implant, DETECT showed improved visualization of the lesions, while the visualization was significantly compromised on the DWIBS images (Fig. 7). Large metastases in the left lung are seen on both DWIBS and DETECT images (red, green arrows, Fig. 7), while DWIBS images suffer from geometric distortions and poor lesion localization. However, the metastatic lesion in the right femur is not visualized on the DWIBS image (yellow circle, Fig. 7a), while it is clearly identified on the DETECT images (yellow arrow, Figs. 7b, 7c). Similar behavior was observed throughout the entire volume in this patient (Supporting Video S3) as well as in other patients (Table 2). Furthermore, the availability of other image contrasts including fat-only image and in-phase image, all acquired in the same sequence and perfectly co-registered, improved the localization of the lesions on DETECT (Fig. 8).
Table 2.
Number of lesions identified on WB-MRI with DETECT compared to WB-MRI with DWIBS in patients
| Patient No. | WB-MRI with DETECT Total (per station) | WB-MRI with DWIBS Total (per station) |
|---|---|---|
| 1 | 1 (0/0/0/1/0) | 1 (0/0/0/1/0) |
| 2 | 8 (0/1/2/3/2) | 7 (0/1/2/3/1) |
| 3 | 3 (0/0/3/0/0) | 2 (0/0/2/0/0) |
| 4 | 40 (5/22/5/7/1) | 27 (2/17/3/5/0) |
| 5 | 3 (0/2/1/0/0) | - * |
Unable to complete WB-MRI DWIBS in patient 5, due to significantly long scan time (~50 minutes)
Figure 6.
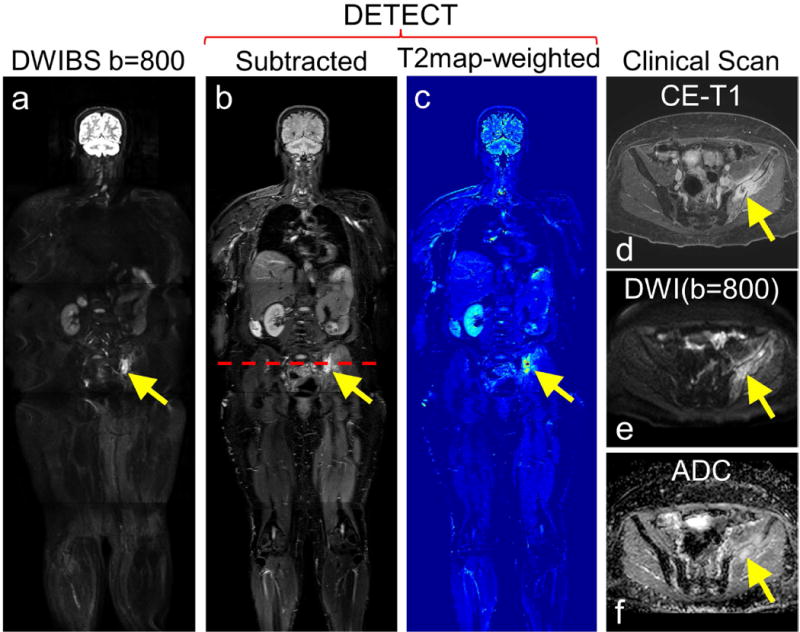
Whole-body MRI of a 58-year old female patient volunteer with advanced renal cell carcinoma and underwent radiation treatment to the left iliac bone metastatic lesion. DWIBS image at b=800 s/mm2 (a), subtracted DETECT image (b) and the effective T2map-weighted image (c) show conspicuous lesion. Clinical contrast-enhanced fat saturated T1-weighted image of the same patient reveals an enhancing left iliac bone lesion (d, yellow arrow), which also appeared hyperintense on clinical DWI image with b = 800 s/mm2 (e, yellow arrow), and ADC map (f) (calculated from 4 b-values; 0, 50, 400, 800 s/mm2), indicative of residual tumor with post-radiation effects.
Figure 7.
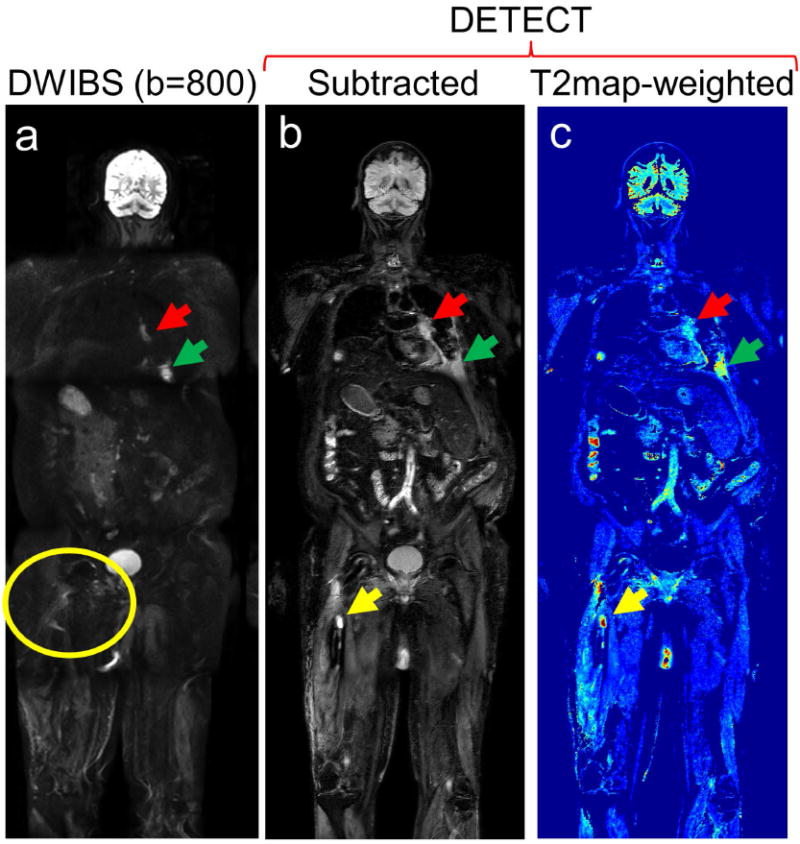
Whole-body MRI of a 68-year old male patient volunteer with advanced renal cell carcinoma with a history of prophylactic rod placement and radiation treatment for a right femur metastatic lesion: Coronal DWIBS image at b = 800 s/mm2 (a), subtracted DETECT image (b), and T2-map weighted image (c) demonstrate metastatic disease involving the left hilum (red arrows) and the left 8th rib (green arrows). While a right femur lesion (yellow arrow) is clearly identified on the DETECT images (b, c yellow arrows), it is not visualized on the DWIBS images due to image distortion from the metallic implant (a, yellow circle). All slices of this subject are shown in Supporting Video S3.
Figure 8.
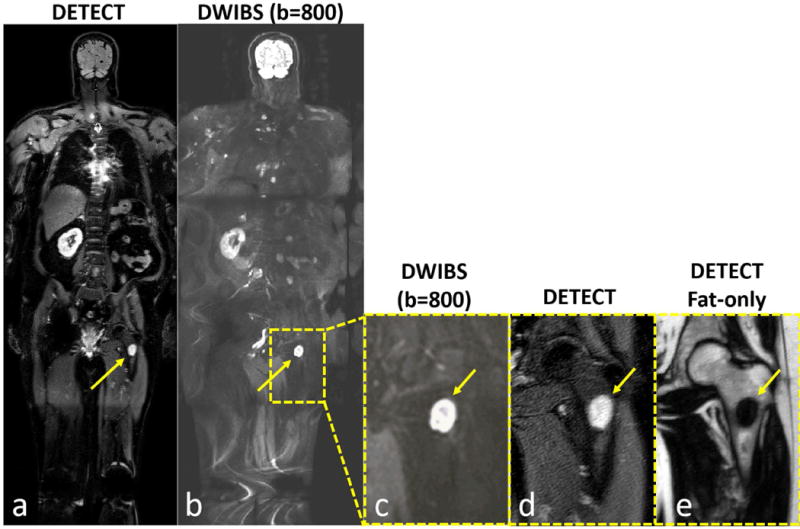
Whole-body MRI of a 64-year old male patient volunteer with advanced renal cell carcinoma showing improved lesion localization capability of DETECT. While both DETECT (a) and DWIBS (b) images show a left lower extremity lesion (arrows), the DWIBS image cannot localize the finding to bone, muscle or lymph node, even when zoomed in (c, arrow). However, DETECT clearly localizes the lesion within the left femur (a, d; arrows). (e) DETECT fat-only image, acquired in the same sequence, confirms the loss of normal marrow (arrow), increasing diagnostic confidence for both location and malignant nature of this lesion.
Additionally, across all 8 whole-body subjects, including 3 healthy volunteers and 5 mRCC patients, the signal of long T2 tissues including, CSF, bile and urine were suppressed by 98 ± 2 %, 89 ± 11 % and 86 ± 21 % respectively.
Discussion
Whole-body MRI has emerged as a promising clinical option for noninvasive detection of metastatic cancer. The major goals of WB-MRI for cancer detection include, fast imaging, high spatial resolution, and high SNR while simultaneously suppressing the signals from the background tissues to improve the conspicuity of the lesions. While the commonly used WB-MRI technique, DWIBS, provides improved conspicuity of the lesions, it often suffers from poor SNR, low spatial resolution and prolonged acquisition times (9,12,33). Additionally, DWIBS images suffer from geometric distortions, particularly at 3T, challenging the anatomical localization of the identified lesions. In this work, we have developed a dual-echo T2-weighted imaging technique for enhanced conspicuity of the tumors (DETECT), that generates fast, high-resolution, and high SNR images with simultaneous fat and fluid suppression, good tumor conspicuity and robustness of RF-refocused spin-echo acquisition in less than 7 minutes scan time for the whole-body imaging. Once the lesions are identified with the proposed DETECT technique, the lesions can be further characterized by dedicated functional MRI techniques such as diffusion, perfusion, hypoxia etc. Due to the use of mDixon acquisition, the DETECT technique also generates perfectly co-registered fat-suppressed T2-weighed images for improved anatomical localization of the lesions.
The multi-echo mDixon reconstruction combined with partial-echo acquisitions allowed high resolution T2W imaging with SShTSE in a truly single acquisition, without increasing the scan time compared to SShTSE acquisitions with and without fat suppression. Although, the slight increase in echo spacing (~2.2 ms) due to the acquisition of multi-echo mDixon increased T2 blurring, it was not substantial compared to the standard clinical SShTSE images due to the use of small partial phase encoding factor (0.6), and parallel imaging (x3). However, the fat/water separation often failed at long TE using the standard mDixon reconstruction. Although several algorithms have been developed to improve the fat/water separation, it is still challenging in images with low SNR (24,34,35). This is because the noise increases the uncertainty in solving the phasor ambiguity during the phase-map estimation, especially for dual-echo IP/OP mDixon reconstruction. The proposed shared-field-map mDixon reconstruction used the B0 map from the short TE to overcome this limitation, and achieved uniform fat/water separation on the long TE images. Additionally, the shared field map between the two TEs also reduces the reconstruction times since the phase map estimation is often time consuming and needs to be estimated only once with shared-field-map mDixon reconstruction. Nevertheless, occasional fat/water swaps were noticed on short TE images at the edges of the FOV along the right/left direction (~52 cm FOV). However, these fat/water swaps were minor and restricted to the subcutaneous fat at the far edges of the FOV without affecting the detection of metastatic lesions in our study. Future optimization with improved fat/water separation methods (36) may potentially overcome these fat/water swaps.
The complex signal modelled by our phase-preserved homodyne reconstruction and the shared-field-map mDixon reconstruction allowed complex subtraction between the two water-only images. This complex subtraction eliminated the “dark-rim” artifacts, observed in the magnitude-subtracted water-only images. Since the fluid-like tissues have longer T2s, the Gibbs ringing artifacts observed with Cartesian view ordering are larger in images acquired with partial phase-encoding and partial readout than those acquired with full k-space. This amplifies the side lobes in PSF on magnitude images, which are cancelled out by the complex subtraction and thus eliminating the “dark-rim” artifacts.
STIR is the most commonly used fat suppression method in WB-MRI due to its insensitivity to B0 inhomogeneities. However, STIR pulse imparts a mix of T1 and T2 contrast and thus, a T2W imaging sequence is typically included in the WB-MRI protocol. Compared to this, our DETECT imaging technique simultaneously provides standard T2W images with and without fat suppression in addition to the fat and fluid suppressed T2W images, all of them perfectly co-registered to each other, without increasing the total scan times as a consequence of the data for both TEs being acquired within the same excitation.
Whole-body DWIBS has been increasingly used for the detection of metastatic lesions, since the DWIBS images have increased lesion conspicuity (12,37). However, DWIBS images suffer from increased geometric distortions, combined with chemical shift artifacts and larger voxel size. Hence, the majority of DWIBS protocols are currently performed at 1.5T for whole-body imaging, necessitating longer scan times to compensate for the reduced SNR (12,38-40). Compared to DWIBS, our DETECT imaging technique provided images with superior SNR and higher spatial resolution in shorter scan times at 3T. Although the lesion-to-background conspicuity was not superior with DETECT compared to DWIBS, it generated images with fewer artifacts and minimal geometric distortions even in the presence of metal implants (e.g. Fig. 7), making it more reliable. Due to the shorter acquisition times of less than 7 minutes for whole-body imaging, the DETECT imaging technique can also be performed in multiple orientations, if needed, to improve the lesion localization. Furthermore, DETECT imaging technique can be an appealing alternative technique for whole-body imaging, with the increasing availability of 3T scanners.
Our study has several limitations. First, although not directly considered a limitation, the DETECT imaging technique is based on prolonged T2 of the metastatic lesions, instead of the restricted diffusion that DWIBS relies on. Nevertheless, most lesions tend to have longer T2 with restricted diffusion and appear bright on T2W images (e.g. Figs. 6-8). Some studies have shown better contrast with whole-body screening on lower b-value (50 s/mm2) than higher b-value (500 s/mm2) (11), where the contrast is primarily due to the longer T2 than the restricted diffusion. Next, the subtraction for fluid attenuation can reduce the contrast between the normal tissues and lesions, and may not be able to completely suppress the signals from complex fluids with moderate T2 values. However, the availability of water-only images at short TE and the use of effective T2 maps can offset these to improve the lesion conspicuity. Third, the DETECT acquisition of the thorax and abdomen required breath-holds. While this is an advantage compared to DWIBS to generate sharper images, it may be challenging in patients to perform multiple sequential breath-holds. In such scenario, DETECT images of the thorax and abdomen can be acquired using respiratory triggering, albeit at slightly prolonged acquisition times. Fourth, the use of variable refocusing flip angles reduced the signal intensities in the arteries, but veins appeared bright on the short TE image. The subtraction between the two TEs also did not reduce the vein vessel signal (reduced by ~5% only) since the T2 of the venous blood is relatively short (~70 ms) (41) and can obscure the lesion conspicuity. Future studies will consider implementation of motion-sensitized driven equilibrium to reduce the signal intensities in these blood vessels (21). Finally, the DETECT imaging technique was initially tested in only five mRCC patients and needs further evaluation in larger patient cohorts in multiple diseases.
Conclusion
In conclusion, we have developed a fast, high-resolution, and high SNR T2-weighted imaging with simultaneous fat and fluid suppression, called DETECT, for whole-body MRI at 3T. Compared to the commonly used DWIBS for whole-body MRI, DETECT can be performed in significantly shorter scan times (17 min. vs. 7 min) and generates images with good lesion conspicuity, and without the image distortion associated with EPI. This sequence can serve as an initial imaging technique for whole-body cancer detection, followed by characterization of selected tumors by dedicated functional MR imaging.
Supplementary Material
Whole-body MRI of the 28-year old healthy male volunteer shown in figure 4 using the DETECT sequence. Standard mDixon reconstruction provided robust fat/water separation at the short TE (TE1), while it failed at long TE (TE2) in multiple locations (red arrows), including right shoulder, hands, back and hip. The shared-field-map mDixon reconstruction corrected all these fat/water swaps and demonstrated the same degree of fat/water separation at the long TE as that at the short TE across the entire volume. Robust fat and fluid suppression across the entire volume was achieved in the subtracted images.
Whole-body DWIBS and DETECT images acquired in coronal plane of the 34-year-old healthy male volunteer shown in figure 5. DWIBS at b = 800 s/mm2 exhibited geometric distortions, low resolution and obvious artifacts, while the high resolution DETECT images demonstrated robust fat and fluid suppression with high SNR and without geometric distortions across the entire volume.
Whole-body DWIBS and DETECT images of the 68-year old male patient volunteer, shown in figure 7, with advanced renal cell carcinoma and a history of prophylactic rod placement and radiation treatment for a right femur metastatic lesion. DETECT images, including fat suppressed, fat-fluid suppressed and effective T2-map weighted images demonstrate metastatic disease involving the hilum, rib, pleura around the left lung (red arrows), a right retrocrural lymph node (brown arrows), the sacrum (white arrows), the right femur (yellow arrows) and the spleen (green arrows). While some of those lesions were visualized on DWIBS images, some right femur lesions (yellow arrows) and the sacrum lesion (white arrows) are not visualized due to image distortion. Due to the high resolution, high SNR and no geometric distortion, DETECT images provided better localization and characterization of these lesions. A splenic infarct on both DWIBS and DETECT images (orange arrows) were also found.
Supporting Figure S1. Flowchart of the image reconstruction for DETECT. The phase map, which is estimated using the standard mDixon with OP and IP images at short TE (TE1), is reused for fat/water separation at long TE (TE2) using the proposed shared-filed-map mDixon reconstruction. The water-only images at TE1 and TE2 are then subtracted to generate the final fat- and fluid-suppressed image.
Supporting Figure S2. Subtracted brain images of a 62-year-old healthy male volunteer showing “dark-rim” artifacts with magnitude subtraction (a, white arrows), which are eliminated with complex subtraction (b). In long TE images, the first side lobes of the point spread function of hyper-intense tissues are often negative due to the heavy signal decay of the surrounding tissues. The magnitude operation converts these negative local minima into positive local maxima (red arrows, c), which manifest as local minima on magnitude-subtracted images (black arrow, c) and create “dark-rim” artifacts (white arrows, a). The signal profiles (c, d) corresponding to the dashed red lines in (a, b) show the local maxima of the magnitude profile on TE2 image (red arrow, c), that create the local minima on the magnitude subtracted profile (black arrow, c), and are rectified in complex subtraction (d).
Acknowledgments
The authors thank Kelli Key and Trevor Wigal for their help in human imaging. This work was partly supported by the NIH/NCI grants: P50CA196516, R01CA154475, and U01CA207091.
References
- 1.Antoch G, Saoudi N, Kuehl H, Dahmen G, Mueller SP, Beyer T, Bockisch A, Debatin F, Jr, Freudenberg LS. Accuracy of whole-body dual-modality fluorine-18-2-fluoro-2-deoxy-D-glucose positron emission tomography and computed tomography (FDG-PET/CT) for tumor staging in solid tumors: comparison with CT and PET. J Clin Oncol. 2004;22(21):4357–4368. doi: 10.1200/JCO.2004.08.120. [DOI] [PubMed] [Google Scholar]
- 2.Schöder H, Gönen M. Screening for cancer with PET and PET/CT: potential and limitations. J Nucl Med. 2007;48(1 suppl):4S–18S. [PubMed] [Google Scholar]
- 3.Hall E, Brenner D. Cancer risks from diagnostic radiology. Br J Radiol. 2008;81(965):362–378. doi: 10.1259/bjr/01948454. [DOI] [PubMed] [Google Scholar]
- 4.Klenk C, Gawande R, Uslu L, Khurana A, Qiu D, Quon A, Donig J, Rosenberg J, Luna-Fineman S, Moseley M. Ionising radiation-free whole-body MRI versus 18 F-fluorodeoxyglucose PET/CT scans for children and young adults with cancer: a prospective, non-randomised, single-centre study. Lancet Oncol. 2014;15(3):275–285. doi: 10.1016/S1470-2045(14)70021-X. [DOI] [PubMed] [Google Scholar]
- 5.Park J-W, Kim JH, Kim SK, Kang KW, Park KW, Choi J-I, Lee WJ, Kim C-M, Nam BH. A prospective evaluation of 18F-FDG and 11C-acetate PET/CT for detection of primary and metastatic hepatocellular carcinoma. J Nucl Med. 2008;49(12):1912–1921. doi: 10.2967/jnumed.108.055087. [DOI] [PubMed] [Google Scholar]
- 6.Nomori H, Watanabe K, Ohtsuka T, Naruke T, Suemasu K, Uno K. Evaluation of F-18 fluorodeoxyglucose (FDG) PET scanning for pulmonary nodules less than 3 cm in diameter, with special reference to the CT images. Lung Cancer. 2004;45(1):19–27. doi: 10.1016/j.lungcan.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Kim SM, Cha R-h, Lee JP, Kim DK, Oh K-H, Joo KW, Lim CS, Kim S, Kim YS. Incidence and outcomes of contrast-induced nephropathy after computed tomography in patients with CKD: a quality improvement report. Am J Kidney Dis. 2010;55(6):1018–1025. doi: 10.1053/j.ajkd.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt GP, Reiser MF, Baur-Melnyk A. Whole-body MRI for the staging and follow-up of patients with metastasis. Eur J Radiol. 2009;70(3):393–400. doi: 10.1016/j.ejrad.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 9.Takenaka D, Ohno Y, Matsumoto K, Aoyama N, Onishi Y, Koyama H, Nogami M, Yoshikawa T, Matsumoto S, Sugimura K. Detection of bone metastases in non-small cell lung cancer patients: Comparison of whole-body diffusion-weighted imaging (DWI), whole-body MR imaging without and with DWI, whole-body FDG-PET/CT, and bone scintigraphy. J Magn Reson Imaging. 2009;30(2):298–308. doi: 10.1002/jmri.21858. [DOI] [PubMed] [Google Scholar]
- 10.Kwee TC, Takahara T, Ochiai R, Nievelstein RA, Luijten PR. Diffusion-weighted whole-body imaging with background body signal suppression (DWIBS): features and potential applications in oncology. Eur Radiol. 2008;18(9):1937–1952. doi: 10.1007/s00330-008-0968-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eiber M, Holzapfel K, Ganter C, Epple K, Metz S, Geinitz H, Kubler H, Gaa J, Rummeny EJ, Beer AJ. Whole-body MRI including diffusion-weighted imaging (DWI) for patients with recurring prostate cancer: Technical feasibility and assessment of lesion conspicuity in DWI. J Magn Reson Imaging. 2011;33(5):1160–1170. doi: 10.1002/jmri.22542. [DOI] [PubMed] [Google Scholar]
- 12.Koh D-M, Blackledge M, Padhani AR, Takahara T, Kwee TC, Leach MO, Collins DJ. Whole-body diffusion-weighted MRI: tips, tricks, and pitfalls. Am J Roentgenol. 2012;199(2):252–262. doi: 10.2214/AJR.11.7866. [DOI] [PubMed] [Google Scholar]
- 13.Walker R, Kessar P, Blanchard R, Dimasi M, Harper K, DeCarvalho V, Yucel E, Patriquin L, Eustace S. Turbo STIR magnetic resonance imaging as a whole-body screening tool for metastases in patients with breast carcinoma: Preliminary clinical experience. J Magn Reson Imaging. 2000;11(4):343–350. doi: 10.1002/(sici)1522-2586(200004)11:4<343::aid-jmri1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Punwani S, Taylor SA, Bainbridge A, Prakash V, Bandula S, De Vita E, Olsen OE, Hain SF, Stevens N, Daw S. Pediatric and Adolescent Lymphoma: Comparison of Whole-Body STIR Half-Fourier RARE MR Imaging with an Enhanced PET/CT Reference for Initial Staging 1. Radiology. 2010;255(1):182–190. doi: 10.1148/radiol.09091105. [DOI] [PubMed] [Google Scholar]
- 15.Del Grande F, Santini F, Herzka DA, Aro MR, Dean CW, Gold GE, Carrino JA. Fat-suppression techniques for 3-T MR imaging of the musculoskeletal system. Radiographics. 2014;34(1):217–233. doi: 10.1148/rg.341135130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz L, Seltzer S, Tempany C, Silverman S, Piwnica-Worms D, Adams D, Herman L, Herman L, Hooshmand R. Prospective comparison of T2-weighted fast spin-echo, with and without fat suppression, and conventional spin-echo pulse sequences in the upper abdomen. Radiology. 1993;189(2):411–416. doi: 10.1148/radiology.189.2.8210368. [DOI] [PubMed] [Google Scholar]
- 17.Jackson A, Sheppard S, Johnson AC, Annesley D, Laitt RD, Kassner A. Combined fat-and water-suppressed MR imaging of orbital tumors. Am J Neuroradiol. 1999;20(10):1963–1969. [PMC free article] [PubMed] [Google Scholar]
- 18.Lauenstein TC, Sharma P, Hughes T, Heberlein K, Tudorascu D, Martin DR. Evaluation of optimized inversion-recovery fat-suppression techniques for T2-weighted abdominal MR imaging. J Magn Reson Imaging. 2008;27(6):1448–1454. doi: 10.1002/jmri.21350. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Greer JS, Pedrosa I, Rofsky NM, Madhuranthakam AJ. Robust abdominal imaging with uniform fat suppression using Dixon based single shot turbo spin echo. Proceedings of the 24th Annual Meeting of ISMRM Singapore. 2016:573. [Google Scholar]
- 20.Essig M, Deimling M, Hawighorst H, Debus J, van Kaick G. Assessment of cerebral gliomas by a new dark fluid sequence, high intensity REduction (HIRE): a preliminary study. J Magn Reson Imaging. 2000;11(5):506–517. doi: 10.1002/(sici)1522-2586(200005)11:5<506::aid-jmri6>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.Madhuranthakam AJ, Lee KS, Yassin A, Brittain JH, Pedrosa I, Rofsky NM, Alsop DC. Improved short tau inversion recovery (iSTIR) for increased tumor conspicuity in the abdomen. Magn Reson Mater Phy. 2014;27(3):245–255. doi: 10.1007/s10334-013-0410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loening AM, Saranathan M, Ruangwattanapaisarn N, Litwiller DV, Shimakawa A, Vasanawala SS. Increased speed and image quality in single-shot fast spin echo imaging via variable refocusing flip angles. J Magn Reson Imaging. 2015;42(6):1747–1758. doi: 10.1002/jmri.24941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Harrison C, Mariappan YK, Gopalakrishnan K, Chhabra A, Lenkinski RE, Madhuranthakam AJ. MR Neurography of Brachial Plexus at 3.0 T with Robust Fat and Blood Suppression. Radiology. 2017;283(2):538–546. doi: 10.1148/radiol.2016152842. [DOI] [PubMed] [Google Scholar]
- 24.Eggers H, Brendel B, Duijndam A, Herigault G. Dual-echo Dixon imaging with flexible choice of echo times. Magn Reson Med. 2011;65(1):96–107. doi: 10.1002/mrm.22578. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Greer JS, Pinho MC, Lenkinski RE, Madhuranthakam AJ. Volumetric T2-weighted and FLAIR Imaging of Spine with Uniform Fat Suppression in a Single Acquisition. Proceedings of the 25th Annual Meeting of ISMRM; Honolulu, Hawaii, USA. 2017. p. 191. [Google Scholar]
- 26.Busse RF, Brau A, Vu A, Michelich CR, Bayram E, Kijowski R, Reeder SB, Rowley HA. Effects of refocusing flip angle modulation and view ordering in 3D fast spin echo. Magn Reson Med. 2008;60(3):640–649. doi: 10.1002/mrm.21680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar SN, Alsop DC, Madhuranthakam AJ, Busse RF, Robson PM, Rofsky NM, Hackney DB. Brain MR imaging at ultra-low radiofrequency power. Radiology. 2011;259(2):550–557. doi: 10.1148/radiol.11092445. [DOI] [PubMed] [Google Scholar]
- 28.Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, Henkelman RM. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn Reson Med. 2005;54(3):507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- 29.Gold GE, Han E, Stainsby J, Wright G, Brittain J, Beaulieu C. Musculoskeletal MRI at 3.0 T: relaxation times and image contrast. Am J Roentgenol. 2004;183(2):343–351. doi: 10.2214/ajr.183.2.1830343. [DOI] [PubMed] [Google Scholar]
- 30.Smith SA, Edden RA, Farrell JA, Barker PB, Van Zijl P. Measurement of T1 and T2 in the cervical spinal cord at 3 tesla. Magn Reson Med. 2008;60(1):213–219. doi: 10.1002/mrm.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230(3):652–659. doi: 10.1148/radiol.2303021331. [DOI] [PubMed] [Google Scholar]
- 32.Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M. MR Fingerprinting of Adult Brain Tumors: Initial Experience. Am J Neuroradiol. 2017;38(3):492–499. doi: 10.3174/ajnr.A5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inada Y, Matsuki M, Nakai G, Tatsugami F, Tanikake M, Narabayashi I, Yamada T, Tsuji M. Body diffusion-weighted MR imaging of uterine endometrial cancer: is it helpful in the detection of cancer in nonenhanced MR imaging? Eur J Radiol. 2009;70(1):122–127. doi: 10.1016/j.ejrad.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 34.Ma J, Son JB, Hazle JD. An improved region growing algorithm for phase correction in MRI. Magn Reson Med. 2016;76(2):519–529. doi: 10.1002/mrm.25892. [DOI] [PubMed] [Google Scholar]
- 35.Berglund J, Ahlström H, Johansson L, Kullberg J. Two-point dixon method with flexible echo times. Magn Reson Med. 2011;65(4):994–1004. doi: 10.1002/mrm.22679. [DOI] [PubMed] [Google Scholar]
- 36.Stinson EG, Trzasko JD, Fletcher JG, Riederer SJ. Dual echo Dixon imaging with a constrained phase signal model and graph cuts reconstruction. Magn Reson Med. 2017;78(6):2203–2215. doi: 10.1002/mrm.26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padhani AR, Koh D-M, Collins DJ. Whole-body diffusion-weighted MR imaging in cancer: current status and research directions. Radiology. 2011;261(3):700–718. doi: 10.1148/radiol.11110474. [DOI] [PubMed] [Google Scholar]
- 38.Lavdas I, Rockall AG, Castelli F, Sandhu RS, Papadaki A, Honeyfield L, Waldman AD, Aboagye EO. Apparent diffusion coefficient of normal abdominal organs and bone marrow from whole-Body DWI at 1.5 T: the effect of sex and age. Am J Roentgenol. 2015;205(2):242–250. doi: 10.2214/AJR.14.13964. [DOI] [PubMed] [Google Scholar]
- 39.Jambor I, Kuisma A, Ramadan S, Huovinen R, Sandell M, Kajander S, Kemppainen J, Kauppila E, Auren J, Merisaari H. Prospective evaluation of planar bone scintigraphy, SPECT, SPECT/CT, 18F-NaF PET/CT and whole body 1.5 T MRI, including DWI, for the detection of bone metastases in high risk breast and prostate cancer patients: SKELETA clinical trial. Acta Oncol. 2016;55(1):59–67. doi: 10.3109/0284186X.2015.1027411. [DOI] [PubMed] [Google Scholar]
- 40.Kwee TC, Takahara T, Ochiai R, Katahira K, Van Cauteren M, Imai Y, Nievelstein RA, Luijten PR. Whole-body diffusion-weighted magnetic resonance imaging. Eur J Radiol. 2009;70(3):409–417. doi: 10.1016/j.ejrad.2009.03.054. [DOI] [PubMed] [Google Scholar]
- 41.Lu H, Ge Y. Quantitative evaluation of oxygenation in venous vessels using T2-Relaxation-Under-Spin-Tagging MRI. Magn Reson Med. 2008;60(2):357–363. doi: 10.1002/mrm.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole-body MRI of the 28-year old healthy male volunteer shown in figure 4 using the DETECT sequence. Standard mDixon reconstruction provided robust fat/water separation at the short TE (TE1), while it failed at long TE (TE2) in multiple locations (red arrows), including right shoulder, hands, back and hip. The shared-field-map mDixon reconstruction corrected all these fat/water swaps and demonstrated the same degree of fat/water separation at the long TE as that at the short TE across the entire volume. Robust fat and fluid suppression across the entire volume was achieved in the subtracted images.
Whole-body DWIBS and DETECT images acquired in coronal plane of the 34-year-old healthy male volunteer shown in figure 5. DWIBS at b = 800 s/mm2 exhibited geometric distortions, low resolution and obvious artifacts, while the high resolution DETECT images demonstrated robust fat and fluid suppression with high SNR and without geometric distortions across the entire volume.
Whole-body DWIBS and DETECT images of the 68-year old male patient volunteer, shown in figure 7, with advanced renal cell carcinoma and a history of prophylactic rod placement and radiation treatment for a right femur metastatic lesion. DETECT images, including fat suppressed, fat-fluid suppressed and effective T2-map weighted images demonstrate metastatic disease involving the hilum, rib, pleura around the left lung (red arrows), a right retrocrural lymph node (brown arrows), the sacrum (white arrows), the right femur (yellow arrows) and the spleen (green arrows). While some of those lesions were visualized on DWIBS images, some right femur lesions (yellow arrows) and the sacrum lesion (white arrows) are not visualized due to image distortion. Due to the high resolution, high SNR and no geometric distortion, DETECT images provided better localization and characterization of these lesions. A splenic infarct on both DWIBS and DETECT images (orange arrows) were also found.
Supporting Figure S1. Flowchart of the image reconstruction for DETECT. The phase map, which is estimated using the standard mDixon with OP and IP images at short TE (TE1), is reused for fat/water separation at long TE (TE2) using the proposed shared-filed-map mDixon reconstruction. The water-only images at TE1 and TE2 are then subtracted to generate the final fat- and fluid-suppressed image.
Supporting Figure S2. Subtracted brain images of a 62-year-old healthy male volunteer showing “dark-rim” artifacts with magnitude subtraction (a, white arrows), which are eliminated with complex subtraction (b). In long TE images, the first side lobes of the point spread function of hyper-intense tissues are often negative due to the heavy signal decay of the surrounding tissues. The magnitude operation converts these negative local minima into positive local maxima (red arrows, c), which manifest as local minima on magnitude-subtracted images (black arrow, c) and create “dark-rim” artifacts (white arrows, a). The signal profiles (c, d) corresponding to the dashed red lines in (a, b) show the local maxima of the magnitude profile on TE2 image (red arrow, c), that create the local minima on the magnitude subtracted profile (black arrow, c), and are rectified in complex subtraction (d).


