Abstract
Key points
Slc4a4 (mouse) encodes at least five variants of the electrogenic sodium/bicarbonate transporter NBCe1. The initial 41 cytosolic amino acids of NBCe1‐A and ‐D are unique; NBCe1‐A has high activity. The initial 85 amino acids of NBCe1‐B, ‐C and ‐E are unique; NBCe1‐B and ‐C have low activity.
Previous work showed that deleting residues 1–85 or 40–62 of NBCe1‐B, or 1–87 of NBCe1‐C, eliminates autoinhibition. These regions also include binding determinants for IRBIT (inositol trisphosphate (IP3)‐receptor binding protein released with IP3), which relieves autoinhibition.
Here, systematically replacing/deleting residues 28–62, we find that only the nine amino acid cationic cluster (residues 40–48) of NBCe1‐B is essential for autoinhibition. IRBIT stimulates all but one low‐activity construct.
We suggest that electrostatic interactions – which IRBIT presumably interrupts – between the cationic cluster and the membrane or other domains of NBCe1 play a central role in tempering the activity of NBCe1‐B in the pancreas, brain and other organs.
Abstract
Variant B of the electrogenic Na+/HCO3 − cotransporter (NBCe1‐B) contributes to the vectorial transport of HCO3 − in epithelia (e.g. pancreatic ducts) and to the maintenance of intracellular pH in the central nervous systems (e.g. astrocytes). NBCe1‐B has very low basal activity due to an autoinhibitory domain (AID) located, at least in part, in the unique portion (residues 1–85) of the cytosolic NH2‐terminus. Previous work has shown that removing 23 amino acids (residues 40–62) stimulates NBCe1‐B. Here, we test the hypothesis that a cationic cluster of nine consecutive positively charged amino acids (residues 40−48) is a necessary part of the AID. Using two‐electrode voltage clamping of Xenopus oocytes, we assess the activity of human NBCe1‐B constructs in which we systematically replace or delete residues 28–62, which includes the cationic cluster. We find that replacing or deleting all residues within the cationic cluster markedly increases NBCe1‐B activity (i.e. eliminates autoinhibition). On the background of a cationic clusterless construct, systematically restoring Arg residues restores autoinhibition in two distinct quanta, with one to three Arg residues restoring ∼50%, and four or more Arg residues restoring virtually all autoinhibition. Systematically deleting residues before the cluster reduces autoinhibition by, at most, a small amount. Replacing or deleting residues after the cluster has no effect. For constructs with low NBCe1 activity (but good surface expression, as assessed by biotinylation), co‐expression with super‐IRBIT (lacking PP1‐binding site) restores full activity (i.e. relieves autoinhibition). In summary, the cationic cluster is a necessary component of the AID of NBCe1‐B.
Keywords: acid‐base transporters, SLC4A4, autoinhibition
Key points
Slc4a4 (mouse) encodes at least five variants of the electrogenic sodium/bicarbonate transporter NBCe1. The initial 41 cytosolic amino acids of NBCe1‐A and ‐D are unique; NBCe1‐A has high activity. The initial 85 amino acids of NBCe1‐B, ‐C and ‐E are unique; NBCe1‐B and ‐C have low activity.
Previous work showed that deleting residues 1–85 or 40–62 of NBCe1‐B, or 1–87 of NBCe1‐C, eliminates autoinhibition. These regions also include binding determinants for IRBIT (inositol trisphosphate (IP3)‐receptor binding protein released with IP3), which relieves autoinhibition.
Here, systematically replacing/deleting residues 28–62, we find that only the nine amino acid cationic cluster (residues 40–48) of NBCe1‐B is essential for autoinhibition. IRBIT stimulates all but one low‐activity construct.
We suggest that electrostatic interactions – which IRBIT presumably interrupts – between the cationic cluster and the membrane or other domains of NBCe1 play a central role in tempering the activity of NBCe1‐B in the pancreas, brain and other organs.
Introduction
The renal proximal tubule of the salamander was the site of the first detected activity of an electrogenic sodium/bicarbonate cotransporter (Boron & Boulpaep, 1983), and mRNA from the salamander kidney yielded the cDNA that encodes the cotransporter NBCe1‐A, a product of the slc4a4 gene (Romero et al. 1997). This latter discovery led to the cloning of the cDNAs encoding mammalian orthologues of NBCe1 (Burnham et al. 1997; Romero et al. 1998), as well as the other Na+‐coupled HCO3 − transporters (Parker & Boron, 2013; Romero et al. 2013). In mammals, Slc4a4 encodes at least five NBCe1 variants, A–E (Parker & Boron, 2013). NBCe1‐A is mainly expressed in kidney. NBCe1‐B is expressed in many tissues throughout the body, but is particularly abundant in pancreas. NBCe1‐C is mainly expressed in brain. NBCe1‐D and ‐E are comparatively minor variants, originally cloned from cDNAs from mouse reproductive tract tissues.
At the beginning of the cytosolic NH2‐terminus (Nt), NBCe1‐A/D share a unique 41‐amino acid (41aa) module that contains an autostimulatory domain (ASD). Indeed, truncating the 41aa module in NBCe1‐A reduces activity by half (McAlear et al. 2006). Replacing this 41aa module in NBCe1‐B/C/E is a unique 85aa module that includes at least part of an autoinhibitory domain (AID) that reduces baseline activity of NBCe1‐B to ∼10% of that of NBCe1‐A (Shirakabe et al. 2006). Indeed, truncating the 85aa module in NBCe1‐B (Lee et al. 2012), or truncating the initial 87aa in NBCe1‐C (McAlear et al. 2006), increases activity by many fold. In summary, NBCe1‐A has a high intrinsic (i.e. per molecule) activity due to the presence of an ASD in its early Nt, whereas NBCe1‐B/C have low intrinsic activities due to the presence of at least part of an AID in their early Nt.
Concurrent with the discovery that the initial 87aa of NBCe1‐C contains an AID, others reported that a similar region of NBCe1‐B includes binding determinants for IRBIT (inositol trisphosphate (IP3)‐receptor (IP3R) binding protein released with IP3) (Shirakabe et al. 2006). Although the initial ∼100aa of IRBIT are unique, the rest is homologous to S‐adenosylhomocysteine hydrolase, which catalyses the hydrolysis of S‐adenosylhomocysteine to adenosine and homocysteine (Devogelaere et al. 2008). IRBIT interacts with many transmembrane proteins, including the IP3R, cystic fibrosis transmembrane conductance regulator, Slc26a6 and sodium/hydrogen exchanger 3 (Ando et al. 2003; He et al. 2008; Yang et al. 2009; Hong et al. 2013; Park et al. 2013).
Because essential elements of the AID and the IRBIT‐binding domain encompass similar regions of NBCe1‐B/C, we might anticipate that IRBIT modulates autoinhibition. Indeed, several groups have found that IRBIT enhances the activities of NBCe1‐B and ‐C (Shirakabe et al. 2006; Yang et al. 2009; Thornell et al. 2010; Lee et al. 2012). That is, IRBIT appears to relieve autoinhibition. Preliminary work suggests that IRBIT also stimulates three electroneutral Na+‐coupled HCO3 − cotransporters NBCn1, NDCBE and NBCn2 (Parker et al. 2007b ). In secretory epithelia – such as those of the pancreatic and parotid‐salivary ducts, where NBCe1‐B is abundant – IRBIT is responsible for the coordinated stimulation of NBCe1‐B and other transporters that contribute to transepithelial ion and fluid secretion (Yang et al. 2009, 2011).
Clearly, understanding the action of IRBIT on NBCe1‐B/C/E will require elucidating the AID, at least part of which is somewhere in the first 85aa of the Nt. Our group found that deleting NBCe1‐B residues 2–4 (sequence: EDE) has no effect on autoinhibition, although it does markedly reduce the ability of IRBIT to stimulate NBCe1‐B. Thus, this short motif may be a key part of the IRBIT‐binding domain but not the AID. Further along the Nt, others have suggested that NBCe1‐B residues 32–36 (GVHVP) constitute a motif (Shcheynikov et al. 2015) that, in the presence of IRBIT, binds Cl− and inhibits NBCe1‐B. Slightly downstream, NBCe1‐B residues 40–48 (RRRRRHKRK) represent a cationic cluster that is followed by a KEKE motif (residues 52–62: KEKKEKERISE) of alternating positive (predominantly Lys) and negative (predominantly Glu) side chains. Others have suggested that KEKE motifs are important in protein–protein interactions (Realini et al. 1994; Kobayashi et al. 2000; Lee et al. 2004; Hamazaki et al. 2006; Motta et al. 2016). Previous work by Shcheynikov et al. showed that the deletion of residues 40–62 largely eliminates autoinhibition (Shcheynikov et al. 2015). Thus, the combination of the cationic cluster and the KEKE motif appear to be necessary for autoinhibition.
The goals of the present study were to determine which parts of the cationic cluster/KEKE motif (residues 40–62), as well as residues extending down to residue 28, are necessary parts of the AID. Our approach was to express NBCe1‐B substitution and deletion mutants in Xenopus oocytes, and assess cotransporter activity using a two‐electrode voltage clamp. We found that between residues 28 and 62, inclusive, the only essential part of the AID is the cationic cluster (residues 40–48). After deleting this 9aa cluster, we added back Arg residues one at a time, finding that half‐maximal autoinhibition occurs with only one positive charge.
Methods
Ethical approval and oocyte preparation
All procedures for the housing and handling of Xenopus laevis were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University. We obtained Xenopus oocytes as described previously (Parker et al. 2008; Musa‐Aziz et al. 2010). In brief, Xenopus were anaesthetized by immersion in 0.2% tricaine until the reflex withdrawal response was absent (∼20 min). Ovaries were surgically extracted and animals were exsanguinated by cardiac excision for termination. Ovaries were dissected to retrieve oocytes, which were subsequently defolliculated with 2 mg ml−1 collagenase Type 1A (Sigma‐Aldrich, St Louis, MO, USA). Following 24 h of incubation, oocytes were injected with H2O or cRNA (see below).
Creation of NBCe1‐B mutant constructs
Constructs were derived from the human NBCe1‐B‐EGFP.pGH19 construct used in our previous study (Lee et al. 2012). We refer to wild‐type NBCe1‐B, tagged at the cytosolic COOH‐terminus (Ct) with enhanced green fluorescent protein (EGFP), as BWT. In order to modify Nt of NBCe1‐B, we excised a part of NBCe1‐B‐EGFP.pGH19 with SmaI (occurring in pGH19, 5′ to the NBCe1 sequence) and StuI (naturally occurring in cDNA of NBCe1‐B). We PCR‐amplified two segments of NBCe1‐B‐EGFP.pGH19 with two primer sets. The first primer set included a forward primer that recognizes the region around the SmaI site, and a mutagenic reverse primer corresponding to residues 28–62 of NBCe1‐B. For the second primer set, we designed a forward primer that corresponds to mutated residues 28–62 in NBCe1‐B, and a reverse primer that recognizes the region around the StuI site. The resulting three DNA fragments were assembled by the In‐Fusion PCR cloning system (Takara Bio, Mountain View, CA, USA). Sequencing of NBCe1‐B‐EGFP.pGH19 mutants was performed by Eurofins MWG Operon (Louisville, KY, USA).
Expression in Xenopus oocytes
cDNA constructs in pGH19 were linearized with NotI (New England Biolabs, Ipswich, MA, USA) and then purified with the QIAquick PCR purification kit (Qiagen, Germantown, MD, USA). Capped mRNAs from linearized cDNA were transcribed as per manufacturer's instructions using the T7 Message Machine kit (Thermo Fisher Scientific, Waltham, MA, USA). Resulting cRNA was purified with the RNeasy MinElute RNA Cleanup kit (Qiagen). Using a micropipette (∼15 μm tip diameter) connected to a Nanoject II variable‐volume automatic injector (Drummond Scientific Company, Broomall, PA, USA), we injected purified cRNA (25 nl) into oocytes (prepared as described above) at a final concentration of 1 ng nl−1 NBCe1 cRNA (25 ng total) and/or 0.33 ng nl−1 cRNA (8 ng total) encoding super‐IRBIT (Lee et al. 2012), dissolved in sterile, RNase‐free H2O. The difference in cRNA concentrations represented an attempt to account for the threefold difference in open‐reading‐frame length. Injected oocytes were incubated in OR3 medium–L‐15 medium (Thermo Fisher Scientific) supplemented with 5 mm Hepes and penicillin–streptomycin (Thermo Fisher Scientific), diluted to ∼195 mosmol kg−1 (close to osmolality of amphibian plasma), and adjusted to pH 7.5 with NaOH.
Physiological solutions
For electrophysiological experiments, we used nominally CO2/HCO3 −‐free saline ‘ND96’ solution containing (in mm) 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, and 5 Hepes, pH 7.50. HCO3 −‐containing solutions included 33 mm NaHCO3 in place of 33 mm NaCl, and pH 7.50 was achieved by equilibration with 5% CO2–95% O2 for at least 30 min. The osmolality of all solutions was adjusted to ∼195 mosmol kg−1 H2O by adding H2O or mannitol as appropriate.
Electrophysiological measurements
A two‐electrode voltage clamp was used to measure whole‐cell ionic currents. Voltages and currents were recorded with a model OC‐725C oocyte clamp (Warner Instruments, Hamden, CT, USA). Electrodes were pulled from 2.0 mm o.d./1.56 mm i.d. thin‐walled borosilicate glass tubing (cat. no. BF200‐156‐10, Sutter Instrument, Navato, CA, USA) and had resistances of 0.5–2.0 MΩ when filled with 3 m KCl. In all experiments, the oocyte was placed in the recording chamber in ND96 solution and sequentially impaled, first with the voltage‐sensing and then with the current‐passing microelectrode. The cell was superfused with ND96 until membrane potential (V m) stabilized, indicating cell membrane sealing around sites of electrode placement. The voltage clamp was then turned on to hold V m at its stabilized, spontaneous value, and then the voltage‐clamp protocol was initiated. The voltage‐clamp protocol used to generate current−voltage (I−V) relationships stepped the V m from its spontaneous value to a holding potential of −160 mV for 100 ms and then back to the spontaneous V m for an additional 100 ms before the next step, which was 20 mV more positive than the last. This cycle was repeated until the final holding potential step shifted V m to +20 mV. After the first set of voltage‐clamp recordings, ND96 was switched to the CO2/HCO3 − solution, and the second set of voltage‐clamp recordings was obtained ∼30 s after the solution change. For each cDNA construct, data were collected from the oocytes of at least two different Xenopus, over at least 2 weeks, using a fresh batch of cRNA for each Xenopus. In order to avoid weekly bias of data, we did not collect data from more than about three oocytes in any 1 week for any one cDNA construct. However, when we tested the effect of super‐IRBIT on a NBCe1‐B mutant, we used one batch of cRNAs for a single Xenopus. All experiments were performed at room temperature (about 22°C).
Cell‐surface biotinylation
Biotinylation was performed using the Cell Surface Protein Isolation Kit (Thermo Fisher Scientific), using a protocol modified for oocytes (Lee et al. 2012, 2013). Briefly, groups of 10 oocytes were incubated for 1 h at 4°C in phosphate‐buffered saline (diluted to 200 mosmol kg−1 H2O) containing 0.24 mg ml−1 Sulfo‐NHS‐SS‐biotin (biotinylation reagent). Following incubation, unreacted biotinylation reagent was quenched, and cells were disrupted by trituration in Tris‐buffered saline with 1% Triton X‐100 and protease inhibitors (Roche Applied Biosciences, Indianapolis, IN, USA). Homogenates were incubated for 1 h in a NeutrAvidin agarose‐packed column (Thermo Fisher Scientific); unbound protein (i.e. non‐biotinylated protein) was washed from the column. Biotinylated protein was eluted from the column with SDS‐sample buffer that contained 50 mm dithiothreitol. Protein was resolved by SDS‐PAGE on TGX 4–20% Tris‐glycine gels (Bio‐Rad, Hercules, CA, USA), where each loading well had a sample volume (40 μl from entire 400 μl of eluate) that corresponded to one oocyte. Afterward, the protein was transferred onto polyvinylidene difluoride membranes using the tank blotting system (Bio‐Rad), and immunoblotted using an anti‐EGFP mouse‐monoclonal primary antibody (JL‐8, Takara Bio), followed by a horseradish peroxidase‐conjugated goat‐anti‐mouse polyclonal antibody (Thermo Fisher Scientific). Western blots were developed using ECL Plus reagents (Thermo Fisher Scientific), and imaging was performed using a ChemFluor E (Protein Simple, Santa Clara, CA, USA). Biotinylation experiments were repeated when there was a substantial change in the surface expression between mutants. BWT samples processed without biotinylation reagents were frequently included as negative controls.
Timing of oocyte experiments
To minimize week‐to‐week variability, we performed NBCe1 electrophysiological assays (terminal experiments, 1 oocyte each) 4 days after cRNA injection, when functional expression is optimal. In 1 day, we typically studied 20 oocytes over a 12 h period. We performed biotinylation assays (terminal experiments, 10 oocytes/condition), typically studying six conditions over a 5 h period. Because both the electrophysiological and biotinylation assays are labour intensive, it is impossible to do both on the same day. Therefore, we typically elected to perform the biotinylations on the day after the electrophysiology, that is, 5 days after cRNA injection.
Data analysis
Electrophysiology
Voltage‐clamp data were collected and analysed using pCLAMP and Clampfit software (version 10; Axon Instruments, Sunnyvale, CA, USA). Values are given as means ± SEM, and the n is defined as number of replicate experiments. Membrane conductance was calculated between −20 and +20 mV, where extracellular‐HCO3 −‐independent currents associated with NBCe1 expression are minimal (Lu & Boron, 2007). Statistical analyses (ANOVA with post hoc Tukey's comparison, and t tests) were performed using Minitab 18 (Minitab, State College, PA, USA) or Microsoft Excel 2010. The differences with a probability value of P < 0.05 were considered significant.
Alignment of deduced amino acid sequences
Entire coding sequences from the SLC4 family were aligned with NBCe1‐B amino acid sequences by using the web tool Clustal Omega (Sievers et al. 2011; https://www.ebi.ac.uk/Tools/msa/clustalo/), and then manually adjusted.
Results
Constructs with mutations targeting the cationic cluster (residues 40–48)
Design of mutants
Perhaps the most striking characteristic of the unique 85aa Nt of NBCe1‐B is the string of nine consecutive residues, the side chains of which have pK a (−log of the acid dissociation constant) values (Hass & Mulder, 2015) that, at a physiological intracellular pH (pHi) of ∼7.2, dictate a predominantly positive charge in the case of Arg (or R; pK a ≅ 13.5) and Lys (or K; pK a ≅ 10.4), or a possible positive charge in the case of His (or H; pK a ≅ 6.8). As shown in Fig. 1 A, the sequence of the cluster is RRRRRHKRK. In order to introduce a dramatic change in net charge, we first replaced the cluster with same number of Asp (D) residues (9D40–48), inasmuch as Asp has the lowest pK a (4.0) among the 20 naturally occurring amino acids found in proteins (Hass & Mulder, 2015). That is, Asp imparts the most negative charge to the protein. Subsequently, we designed and generated five additional NBCe1‐B mutants involving the cationic cluster. One has nine Ala (A) residues (9A40–48); the non‐polar methyl side chain in theory confers a high degree of conformational flexibility to the protein backbone (Morrison & Weiss, 2001). For a polar/uncharged side chain, we chose Asn (N) residues (9N40–48), which has one less –CH2– than Gln (Q), and avoids the potential complication of a phosphorylation (Pearlman et al. 2011) of Ser (S) or Thr (T). In a variation on the theme of the two previous substitutions, we replaced the cationic cluster with a stretch of Asn/Ala residues (9NA40–48), in which the side chains, overall, have an intermediate non‐polar/polar character. Finally, we deleted either just the cationic cluster (∆940–48), or the cluster and the following KEKE motif (∆2340–62).
Figure 1. Construct designs and current−voltage (I−V) relationships from experiments that explore the cationic cluster.
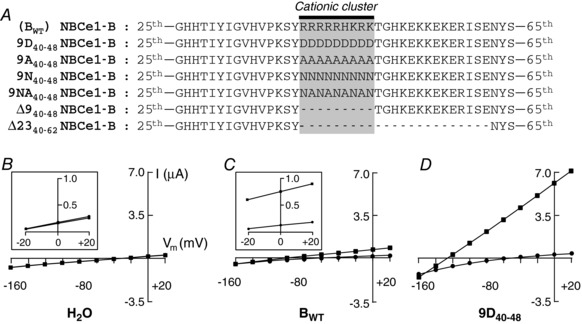
A, designs of constructs. NBCe1‐B was mutated into 6 different constructs: 9D40–48, 9A40–48, 9N40–48, 9NA40–48, ∆940–48 and ∆2340–62. B, representative I−V relationship from H2O‐injected Xenopus oocyte, exposed first to ND96 solution (filled circles), and then to CO2/HCO3 −‐containing solution (filled squares); lines are nearly super‐imposable. Inset shows magnification of region between −20 and +20 mV of the I−V relationship. C, representative I−V relationship from oocyte expressing BWT. D, representative I−V relationship from oocyte expressing 9D40–48.
Effect of replacing the cationic cluster with nine Asp residues
In our first study, we injected oocytes with H2O vs. cRNAs encoding BWT or 9D40–48, and assayed by two‐electrode voltage clamp. Fig. 1 B−D shows representative I−V relationships obtained from single oocytes, 4 days after injection. We obtained I−V relationships for each cell while exposed, sequentially, to the ND96 solution (filled circles) and, after ∼30 s, to the CO2/HCO3 −‐containing solution (filled squares). As described in Methods, we compute membrane conductance (G m) from the I−V relationship between −20 and +20 mV. In Fig. 1 B, the I−V relationships show that the G m of a H2O‐injected oocyte is almost independent of the presence of CO2/HCO3 − (see inset in Fig. 1 B for expanded scale). In Fig. 1 C, the I−V relationships for the oocyte expressing BWT show that G m is slightly higher in the presence than in the absence of CO2/HCO3 −. Fig. 1 D shows that when we express 9D40–48, G m is now substantially higher than for the previous two types of oocytes.
Effect of substituting or deleting the cationic cluster
Fig. 2 A summarizes the results for the seven constructs listed in Fig. 1 A, assessed as in Fig. 1 B−D on two separate experiment days. For each oocyte, we determined the HCO3 −‐dependent conductance (G HCO3) by subtracting G m in ND96 from G m in CO2/HCO3 −. Each experiment day we determined the average G HCO3 for the oocytes expressing Δ940–48, and normalized all other G HCO3 values to this average in order to compensate for week‐to‐week variations of Xenopus oocytes. As shown in Fig. 2 A, G HCO3 is virtually zero for H2O‐injected control oocytes, and ∼0.09 for BWT‐expressing oocytes, compared to the relative value of unity for oocytes expressing Δ940–48. All the other constructs – substitutions of the cationic cluster with D, A, N, NA or deletions of the entire cationic cluster ± the KEKE motif – had mean G HCO3 values statistically indistinguishable from unity. In other words, any of these approaches for disrupting the cationic cluster eliminates autoinhibition. Whether we replace the positively charged residues with negatively charged or neutral residues, or whether we delete the positive residues entirely, appears to be unimportant. Shcheynikov et al. (2015) also observed that Δ2340–62, as heterologously expressed in HeLa cells, eliminates autoinhibition.
Figure 2. Summary of data from experiments that explore the cationic cluster.
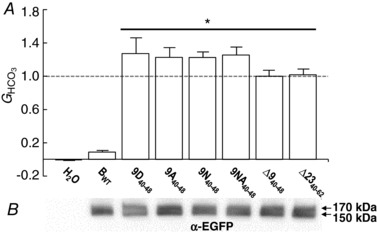
A, HCO3 −‐dependent conductances (G HCO3) computed between −20 and +20 mV of I−V relationships. G HCO3 values were normalized to the G HCO3 (dashed line) from an oocyte – studied the same day – expressing ∆940–48. The n for each group (i.e. construct) is 6. The asterisk over the straight bar indicates the six groups significantly different, by one‐way ANOVA followed by post hoc Tukey's comparison, from H2O or BWT oocytes. B, surface‐biotinylated proteins, analysed by western blotting. The 8 contiguous lanes (the H2O is virtually blank) of the blot are aligned with the labels in A.
Plasma membrane abundance
Fig. 2 B shows the results of a surface‐biotinylation experiment – performed 1 day after the electrophysiological studies – on oocytes expressing BWT and its mutants. Here we see doublets with molecular masses near 170 kDa (predicted mol. mass of EGFP‐tagged proteins: 146–149 kDa), consistent with the same degree of glycosylation of monomeric proteins as in a previous study from our group (Lee et al. 2012). Note that the surface‐biotinylation signal is similar for all seven constructs, indicating that the tenfold increase in G HCO3 between BWT and the six mutants is most likely due to an increase of intrinsic cotransporter activity, rather than an increase of surface protein expression.
Constructs with mutations upstream of the cationic cluster
Designs of mutants
In order to investigate the importance of residues upstream of the cationic cluster, we first attempted to replace three amino acids at a time (up to 12 amino acids total) with a stretch of Asn/Ala residues with BWT as the background construct. Unfortunately, we observed substantially decreased expression, as judged by surface biotinylation, for three of the four constructs. Therefore, we explored the region upstream of the cationic cluster by deleting three amino acids at a time up to a total of 12 amino acids (Fig. 3 A).
Figure 3. Construct designs and data summary from experiments that explore regions upstream of the cationic cluster.
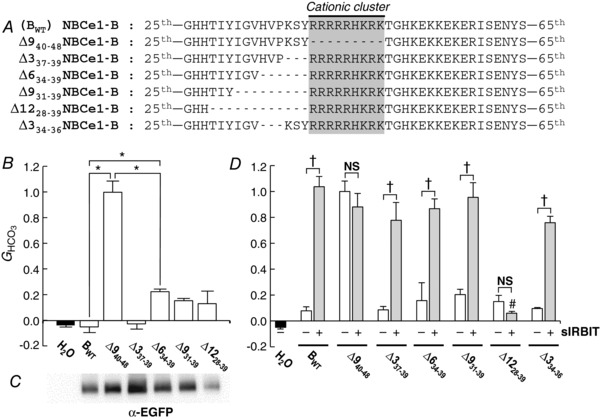
A, designs of constructs. In addition to Δ940–48 (see Fig. 1A), we created 5 additional deletion constructs, based on NBCe1‐B (BWT). B, HCO3 −‐dependent conductances (G HCO3) computed between −20 and +20 mV of I−V relationships. G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48. The n for each group (i.e. construct) is 6. Asterisks indicate significant differences, by one‐way ANOVA followed by post hoc Tukey's comparison. C, surface‐biotinylated proteins, analysed by western blotting. The 7 contiguous lanes (the H2O is virtually blank) of the blot are aligned with the labels in B. This blot is representative of two trials, which confirm that ∆1228–39 is present at relatively low abundance in the plasma membrane. D, HCO3 −‐dependent conductances (G HCO3) from oocytes expressing seven different constructs, without or with super‐IRBIT (sIRBIT). G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48 without sIRBIT. The n for each group is 3–6. †Pairs are significantly different in a one‐tailed, unpaired t test. NS, not significant. #Grey bar is significantly different from the other grey bars in an ANOVA with Tukey's post hoc comparison.
Effect of deleting residues upstream of the cationic cluster
We followed the same protocol as in Fig. 1 B−D. As shown in Fig. 3 B, in this data set, the G HCO3 of BWT was not different from zero. However, the G HCO3 of Δ940–48, which we used for normalization, was significantly greater than the G HCO3 of BWT. ∆337–39, the first deletion upstream from the cationic cluster, had a G HCO3 that, like BWT in this series of constructs, was not different from zero. On the other hand, the G HCO3 of ∆634–39 was significantly greater than that of BWT, but less than the G HCO3 of Δ940–48. We will see in Fig. 3 D that the deletion of only the second trio of amino acids (∆334–36) has no effect. Thus, although the deletion of all six residues produces a modest decrease in autoinhibition, the deletion of either set of three was ineffective. The upstream deletions of either 9 amino acids (∆931–39) or of 12 amino acids (∆1228–39) produced G HCO3 values that were not significantly different from that of BWT.
Plasma membrane abundance
Here we surface‐biotinylated proteins as in Fig. 2 B. As shown in Fig. 3 C, the surface‐expression levels of the constructs were similar to one another except for ∆1228–39, which had a much lower surface expression than the others. Taken together, the data of Fig. 3 B and C suggest that residues 34–39 contribute modestly to autoinhibition. The surface expression of ∆1228–39 was so low that it is difficult to draw conclusions about its effect on G HCO3.
Effect of super‐IRBIT on above constructs
We expressed the same six constructs as in Fig. 3 B – with the addition of ∆334–36 – without or with super‐IRBIT (sIRBIT). sIRBIT is an IRBIT mutant that lacks a consensus PP1‐binding site, and thus enhances NBCe1‐B activity more effectively than IRBIT (Lee et al. 2012). As shown in Fig. 3 D, the co‐expression of sIRBIT produced a marked increase in the G HCO3 of BWT, as observed previously (Lee et al. 2012), but had no effect on the G HCO3 of Δ940–48, which already had a near‐maximal value. Except for ∆1228–39 (which, as noted in Fig. 3 C, has a low surface expression), sIRBIT stimulated all low‐G HCO3 mutants. Except for ∆1228–39, all constructs co‐expressed with sIRBIT had statistically indistinguishable G HCO3 values. Because ∆1228–39 had a low surface expression and was not stimulated by sIRBIT, we cannot conclude whether 28TIY30 is a part of the AID.
Constructs with mutations at a proposed Cl−‐binding site
Designs of mutants
Shcheynikov et al., hypothesizing that the sequence GVHVP in NBCe1‐B (Fig. 4 A) is a Cl−‐binding site, generated three constructs with mutations within this motif, and expressed the constructs in HeLa cells (Shcheynikov et al. 2015). Each construct revealed markedly reduced autoinhibition, leading the authors to suggest that the mutations cause a conformational change in the AID that prevents autoinhibition. As summarized in Fig. 3, we already explored three deletion mutants that disrupt GVHVP. One (i.e. ∆634–39) modestly reduced autoinhibition, and the other two (∆931–39, ∆334–36) had no significant effect. In this part of our study, we tested two mutants of Shcheynikov et al. As summarized in Fig. 4 A, we converted GVHVP to GVAVP in one construct, and to AVHVA in another.
Figure 4. Construct designs and data summary from experiments that explore a proposed Cl−‐binding motif upstream of the cationic cluster.
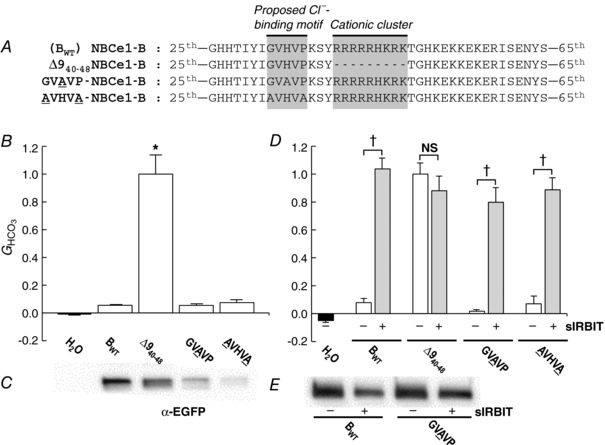
A, designs of constructs. In addition to Δ940–48 (see Fig. 1A), we created 2 additional constructs, based on NBCe1‐B (BWT). B, HCO3 −‐dependent conductances (G HCO3) computed between −20 and +20 mV of I−V relationships. G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48. The n for each group (i.e. construct) is 6. The asterisk indicates significant difference, by one‐way ANOVA followed by post hoc Tukey's comparison. C, surface‐biotinylation of proteins 5 days after cRNA injection (i.e. 1 day after electrophysiological experiments), and analysis by western blotting. The 5 lanes (the H2O is virtually blank) of the blot are aligned with the labels in B. This blot is representative of two trials, which confirm that GVAVP and AVHVA are present at relatively low abundance in the plasma membrane. Note that 3 lanes between ∆940–48 and GVAVP were deleted from the blot image in order to create the one‐to‐one correspondence between panels B and C. D, HCO3 −‐dependent conductances (G HCO3) from oocytes expressing four different constructs, without or with super‐IRBIT (sIRBIT). G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48 without sIRBIT. The n for each group is 3–6. †Pairs are significantly different in a one‐tailed, unpaired t test. NS, not significant. E, surface‐biotinylation of proteins 4 days after cRNA injection, and analysis by western blotting. BWT and GVAVP were expressed without and with sIRBIT.
Effect of disrupting the putative Cl−‐binding motif
Figure 4 B shows that, in contrast to previous observations (Shcheynikov et al. 2015), neither GVAVP nor AVHVA reduced autoinhibition (i.e. their G HCO3 values were indistinguishable from that of BWT).
Plasma membrane abundance in absence of sIRBIT (5 days after cRNA injection)
Here we processed biotinylated proteins as in Figs. 2 B and 3 C. As shown in Fig. 4 C, both GVAVP and AVHVA showed a major decrease in surface expression. Therefore, based only on the data of Fig. 4 B and C, the low activity of the two mutants could reflect decreased surface expression, decreased intrinsic activity, or both.
Effect of sIRBIT on above constructs
We expressed BWT, ∆940–48, GVAVP and AVHVA without or with sIRBIT. As shown in Fig. 4 D, sIRBIT had no effect on the G HCO3 of Δ940–48, which was already near‐maximal, but increased the G HCO3 of GVAVP and AVHVA to values indistinguishable from that of ∆940–48. We conclude that GVHVP, whether or not it interacts with cytosolic Cl−, is not necessary for autoinhibition. The biotinylation data suggest that, if anything, GVAVP and AVHVA have higher‐than‐normal intrinsic activities.
Plasma membrane abundance ± sIRBIT (4 days after cRNA injection)
Recall that the biotinylation signals for GVAVP and AVHVA in Fig. 4 C – obtained 5 days after cRNA injection (1 day after the electrophysiology experiments) – were very low. We hypothesized that either the cRNA or the protein for these constructs was especially labile. Therefore, we assessed biotinylation in a separate group of BWT and GVAVP oocytes, with or without sIRBIT, 4 days after cRNA injection. As shown in Fig. 4 E, both BWT and GVAVP showed robust surface expression in the absence of sIRBIT, and modestly decreased surface expression in the presence of sIRBIT. Thus, the sIRBIT‐dependent increase in G HCO3 for GVAVP in Fig. 4 D probably reflects a marked increase in intrinsic activity, and not an increase in surface expression.
Constructs with varying numbers of positive charges in the cationic cluster
Designs of mutants
In order to investigate the importance of the number of positively charged residues, we first attempted alanine‐scanning mutagenesis, employing a string of five Ala residues that we advanced one residue at a time across residues 40–48, with BWT as the background construct. Unfortunately, we observed inconsistent expression, as judged by surface biotinylation, among the five constructs. Therefore, as shown in Fig. 5 A, we generated a series of eight mutants, from Δ940–48 to 9R40–48.
Figure 5. Construct designs and data summary from experiments for varying numbers of positively charged residues in the cationic cluster.
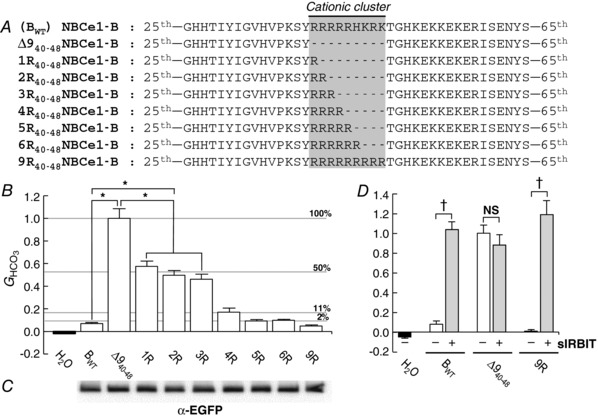
A, designs of constructs. In addition to Δ940–48 (see Fig. 1A), we created 7 additional constructs, based on NBCe1‐B (BWT). B, HCO3 −‐dependent conductances (G HCO3) computed between −20 and +20 mV of I−V relationships. G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48. The n for each group (i.e. construct) is 6. Asterisks indicate significant differences, by one‐way ANOVA followed by post hoc Tukey's comparison. Although not indicated by asterisks for the sake of clarity, 4R, 5R, 6R and 9R – although not different from one another – are all significantly different from Δ940–48, 1R, 2R and 3R. Labelled horizontal lines indicate percentage autoinhibition. C, surface‐biotinylated proteins, analysed by western blotting. The 9 contiguous lanes (the H2O sample was omitted due to the limited number of loading wells) of the blot are aligned with the labels in B. Biotinylated proteins were analysed by western blotting. D, HCO3 −‐dependent conductances (G HCO3) from oocytes expressing three different constructs, without or with super‐IRBIT (sIRBIT). G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48 without sIRBIT. The n for each group is 3–6. †Pairs are significantly different in a one‐tailed, unpaired t test. NS, not significant.
Effect of increasing the number of Arg residues
As demonstrated in previous figures in the present paper, Fig. 5 B shows that G HCO3 is substantially greater with Δ940–48 than with BWT. Figure 5 B also shows that the addition of only one Arg on a background of Δ940–48 produces ∼50% of maximal autoinhibition (i.e. half‐maximal G HCO3, considering BWT as representing 100% autoinhibition). The addition of a second and a third Arg has little effect. However, the addition of a fourth Arg produces ∼90% of full autoinhibition. The addition of the fifth Arg residue now increases autoinhibition to virtually 100%, and additional Arg residues have no further effect. We conclude that, from 0 to 1 and also from 3 to 4, the number of positive charges in the cationic cluster is a strong determinant of autoinhibition.
Plasma membrane abundance
Here we processed biotinylated proteins as in Figs. 2 B, 3 C and 4 C. As shown in Fig. 5 C, we did not see any substantial differences in surface expression among BWT, Δ940–48, or any of the “R” mutants. Thus, the differences in G HCO3 presumably reflect differences in intrinsic cotransporter activity.
Effect of sIRBIT on above constructs
We expressed BWT, ∆940–48 and 9R40–48 without or with sIRBIT. As shown in Fig. 5 D, sIRBIT markedly increased the G HCO3 of BWT and 9R40–48, but had no effect on the G HCO3 of Δ940–48, which was already near‐maximal. We conclude that the changes in the number of Arg residues reflect differences in intrinsic activity.
Constructs with mutations in the KEKE motif
Designs of mutants
We explored the region downstream of the cationic cluster by replacing residues 49–62 with a stretch of repeating Asn/Ala residues, or by deleting these 14 residues (Fig. 6 A).
Figure 6. Construct designs and data summary from experiments that explore the KEKE motif downstream of the cationic cluster.
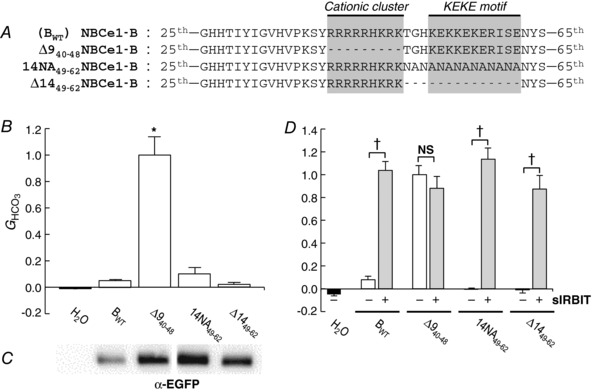
A, designs of constructs. In addition to Δ940–48 (see Fig. 1A), we created 2 additional constructs, based on NBCe1‐B (BWT). B, HCO3 −‐dependent conductances (G HCO3) computed between −20 and +20 mV of I−V relationships. G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48. The n for each group (i.e. construct) is 5–6. The asterisk indicates significant difference, by one‐way ANOVA followed by post hoc Tukey's comparison. C, surface‐biotinylated proteins, analysed by western blotting. The 5 lanes (the H2O is virtually blank) of the blot are aligned with the labels in B. Note that one lane between ∆940–48 and 14NA49–62 was deleted from the blot image in order to create the one‐to‐one correspondence between panels B and C. D, HCO3 −‐dependent conductances (G HCO3) from oocytes expressing four different constructs, without or with super‐IRBIT (sIRBIT). G HCO3 values were normalized to the G HCO3 from an oocyte – studied the same day – expressing ∆940–48 without sIRBIT. The n for each group is 3–6. †Pairs are significantly different in a one‐tailed, unpaired t test. NS, not significant.
Effect of replacing or deleting residues downstream of the cationic cluster
Fig. 6 B shows, as we have seen repeatedly, that Δ940–48 has a substantially greater G HCO3 than does BWT. Moreover, the G HCO3 of 14NA49–62 and Δ1449–62 was indistinguishable from that of BWT. We conclude that the replacement or deletion of the KEKE motif did not significantly reduce autoinhibition.
Plasma membrane abundance
We examined the above constructs for surface abundance as in previous figures. As shown in Fig. 6 C, the surface abundance of 14NA49–62 and Δ1449–62 were at least as great as for BWT. Therefore, the low G HCO3 of 14NA49–62 and Δ1449–62 presumably reflect low intrinsic cotransporter activity.
Effect of sIRBIT on above constructs
We expressed BWT, ∆940–48, 14NA49–62 and ∆1449–62 without or with sIRBIT. As shown in Fig. 6 D, sIRBIT had no effect on ∆940–48 (already maximally stimulated) but markedly increased the G HCO3 of BWT, 14NA49–62 and ∆1449–62. We conclude that the mutations in the KEKE motif do not affect autoinhibition.
Discussion
Three motifs in the amino acid range 28–62 of NBCe1‐B
Upstream domain that includes the Cl−‐binding motif (residues 28–39)
The only deletion in this range that had a statistically significant effect on autoinhibition was ∆634–39 (Fig. 3 B). However, neither of the two half mutants – ∆334–36 and ∆337–39 – by themselves affected autoinhibition. One explanation for these results is that aa34–36 and aa37–39 each contribute to autoinhibition, but that their individual contributions are too small to detect in isolation. Another explanation is that the important residues are partly in aa34–36 and partly in aa37–39. sIRBIT relieved the autoinhibition for all low‐G HCO3 constructs except ∆1228–39, which had an extremely low surface expression.
Putative Cl−‐binding motif (residues 32–36)
Previous work on CLC Cl− channels (Dutzler et al. 2002; Estévez & Jentsch, 2002; Faraldo‐Gómez & Roux, 2004) identified a three‐dimensional Cl−‐binding site consisting of three conserved GXXXP motifs, each near the N‐terminal end of an α‐helix, plus a tyrosine. Note that the three GXXXP/α‐helices and Tyr residue, which together form a single Cl−‐binding site, are distributed over nearly 350 residues of linear amino acid sequence. At a more granular scale, the coordination of a single Cl− requires: (1) the side‐chain O atom from the aforementioned Tyr residue, (2) the side‐chain O atom from a Ser residue in one XXX (i.e. the 3 interior residues of a GXXXP), and (3) and (4) two backbone N atoms contributed by another XXX. Meanwhile, helix–dipole interactions produce (5), (6) and (7) three partial positive charges near the three GXXXP motifs, and thus promote an electrostatic interaction with the Cl−. Thus, seven entities – two O atoms, two N atoms, and three partial positive charges near GXXXP/α‐helix structures – conspire to produce a complex, three‐dimensional site for the binding of one Cl− ion. Note that the partial positive charges on the three GXXXP motifs depend on the helix–dipole interactions of the associated α‐helices.
Shcheynikov et al. proposed that a single, isolated GXXXP motif in NBCe1‐B (32GVHVP36) – with no associated α‐helix – serves as a high‐affinity Cl−‐binding site that inhibits NBCe1‐B in the presence of IRBIT. They report that, even when [Cl−]i is presumably low and IRBIT is ostensibly absent (their Fig. 2), the mutants GVAVP and AVHVA each markedly stimulate an NBCe1‐B construct as expressed in HeLa cells. Moreover, the two mutant constructs are resistant to further stimulation by IRBIT. These data of Shcheynikov et al. are the opposite of those summarized in Fig. 4 B and D of the present paper, where we show that the GVAVP and AVHVA mutations have no effect on G HCO3 in the absence of added IRBIT, and leave the constructs fully responsive to the stimulatory effects of IRBIT. One possible explanation for the apparent discrepancy between the data of Shcheynikov et al. and our data is suggested by proteomics studies (Nagaraj et al. 2011; Grant et al. 2015) showing that the abundance of endogenous IRBIT is much higher in HeLa cells (1.5% of GAPDH) than in Xenopus oocytes (0.01% of GAPDH). If GVAVP and AVHVA were to have enhanced IRBIT affinities, then these constructs would each appear to relieve autoinhibition in the high‐IRBIT HeLa cells, but not in the low‐IRBIT oocytes.
Even if the above explanation were correct, it would not support the idea that GVHVP is a necessary part of the AID. In fact, six observations in the present study support the hypothesis that GVHVP is not part of the AID. As shown in Fig. 3, (1) Δ634–39, (2) Δ931–39 and (3) Δ334–36 – each of which disrupts GVHVP – all have low baseline activities that are markedly enhanced by sIRBIT. As shown in in Fig. 4, (4) GVAVP and (5) AVHVA also have low baseline activities that are markedly enhanced by sIRBIT. As shown in Fig. 4 E, (6) sIRBIT decreases the surface abundance of GVAVP (i.e. sIRBIT increases G HCO3 without increasing surface expression).
Finally, the structural data on CLC channels (see 7 entities in previous paragraph) suggest to us that it is most unlikely that a single, isolated GVHVP could be an effective Cl−‐binding site.
Cationic cluster (residues 40–48)
Statistical analyses show that positively charged residues of integral membrane proteins are more common on the cytosolic than on the extracellular side (Bogdanov et al. 2014). In addition to a potential role in determining membrane protein topology (Juretić et al. 2002), cytosolic cationic clusters can serve as docking sites for regulatory proteins (Obosi et al. 1997; Wang, 1997, 1999), and can anchor proteins to negatively charged head groups of phospholipids in the inner leaflet of lipid bilayer (van Klompenburg et al. 1997).
We disrupted the cationic cluster (residues 40–48) by replacing the positively charged residues with negatively charged or neutral residues, or by deleting the cationic cluster altogether (Fig. 1 B). As shown in Fig. 1 B–D, and summarized in Fig. 2 A, any of the above mutations completely eliminated autoinhibition. Particularly striking is our observation that replacing the positive charges with negative charges (i.e. 9D) leaves the construct fully active. Therefore, the positive charges are necessary for autoinhibition. Note that other parts of NBCe1‐B could also be necessary for autoinhibition.
On the background of Δ940–48, we systematically added up to nine Arg residues (Fig. 5). We were surprised to observe that the effect on G HCO3 appears to be quantal, with two distinct steps: (1) from 0 to 1 Arg, and (2) from 3 to 4 Arg residues. A question that arises is why NBCe1‐B has evolved to have nine positive charges in the cationic cluster, when only four are needed to produce near‐maximal autoinhibition. One possibility is that the additional positive charges are necessary for optimal interaction with a binding partner such as IRBIT.
KEKE motif (residues 51–62)
“KEKE” motifs are a kind of a spatial sign‐alternating charge cluster, in which the side chains of adjacent amino acids (primary structure) are in repetitive sequences that approximate Lys‐Glu‐Lys‐Glu‐… (Realini et al. 1994). Arg residues occasionally replace Lys, Asp residues occasionally replace Glu, and still other amino acids may occasionally interrupt the pattern. By this definition, residues 52–62 of NBCe1‐B are a KEKE motif. These motifs appear to play an important role in protein–protein interactions (Realini et al. 1994; Kobayashi et al. 2000; Lee et al. 2004; Hamazaki et al. 2006; Motta et al. 2016), and the same may be true for NBCe1‐B. However, in our assays we did not observe any functional defects in NBCe1‐B constructs in which we replaced or deleted the KEKE motif (Fig. 6 B–D). Our KEKE mutants traffic normally to the plasma membrane, exhibit normal autoinhibition, and reveal a normal responsiveness to sIRBIT.
Conservation of the three motifs among SLC4 family members
In the following analysis of the conservation of the three motifs – GXXXP, cationic cluster and KEKE – among human SLC4 family members, we adhere to the SLC4 nomenclature summarized in the review by Parker & Boron (2013). Our analysis of motif conservation may have implications for predicting the physiological regulation of SLC4 family members. Of the three motifs, the cationic cluster is the only one that appears to be of substantial importance for either autoinhibition or the relief of autoinhibition by IRBIT. Thus, the net number of positive charges in the cationic cluster may allow one to predict the strength of autoinhibition and, perhaps, even the likelihood that IRBIT may relieve this autoinhibition.
SLC4A1. Neither of the two variants of AE1 has any of the three motifs upstream of the SLC4 signature motif ETARWIKFEE (Romero et al. 1997), that is, corresponding to their positions in NBCe1‐B. However, downstream of the signature motif (which is EAARWVQLEE in AE1), AE1 does have four GXXXP motifs, three in the cytosolic Nt and one more in a cytosolic loop just before transmembrane segment 3. The absence of a cationic cluster is consistent with the idea that AE1 lacks an AID (i.e. has high baseline activity). Indeed, erythrocyte AE1 has a turnover rate of ∼105 anions s−1 (Jay & Cantley, 1986), which makes AE1 one of the fastest known transporters.
For the remainder of the analyses, all of the sequences are upstream of the SLC4 signature motif.
SLC4A2 (Fig. 7 A). The three known variants (AE2a, AE2b1, AE2b2) have a GXXXP motif, a weak cationic cluster (ERRR = net +2), but lack a KEKE motif. The weak cationic cluster is consistent with the idea that AE2 neither exhibits autoinhibition, nor is rescued by IRBIT. Indeed, IRBIT does not stimulate AE2 (Jeong & Hong, 2016).
SLC4A3 (Fig. 7 B). The two known variants (bAE3, cAE3) have a GXXXP motif, a strong cationic cluster (RRKKKKKK = net +8), and scattered residues reminiscent of a KEKE motif. The strong cationic cluster is consistent with the idea, thus far untested, that AE3 would exhibit substantial autoinhibition, but be rescued by IRBIT.
SLC4A5 (Fig. 7 C). The two known functional variants (NBCe2‐a, NBCe2‐c) have a GXXXP motif, followed by a weak cationic cluster (RKTDQK = net +2), but no KEKE motif. The weak cationic cluster suggests that this transporter should have high baseline activity, and be insensitive to IRBIT, as has been reported (Shcheynikov et al. 2015).
Figure 7. Alignments and analyses of three Nt motifs among SLC4 family members.
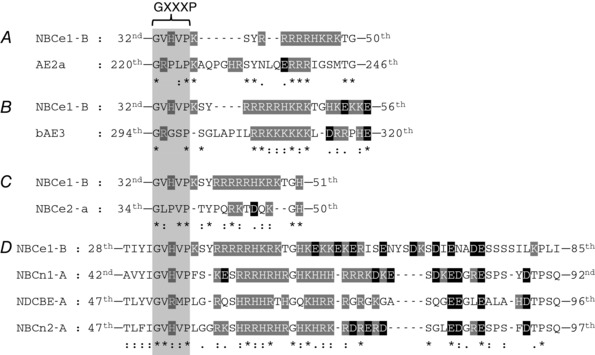
The GXXXP motif is highlighted with a light grey background. Positively charged residues are indicated by white characters on a dark grey background. Negatively charged residues are indicated by white characters on a black background. “*”, conserved; “:”, strongly similar; “.”, weakly similar.
Fig. 7 D shows the alignments of the three electroneutral Na+‐coupled HCO3 − transporters in the region of the three motifs.
SLC4A7. All known NBCn1 variants have all three motifs. The GXXXP motif is identical to the one in NBCe1‐B (GVHVP). NBCn1 also has a long cationic cluster and a weak KEKE motif.
SLC4A8. Three of five NDCBE variants (NDCBE‐A/B/E) have an intact GXXXP motif, whereas the other two (NDCBE‐C/D) lack the motif. All five variants have a long, interrupted cationic cluster. Although NDCBE does not have a clear KEKE motif, four scattered anionic residues are present.
SLC4A10. All known NBCn2 variants conserve the three motifs. The cationic cluster is long but interrupted, and the KEKE motif is unusual in being dominated by Arg and Glu residues (RDRERD).
The strong cationic clusters of NBCn1, NDCBE and NBCn2 suggest that these transporters exhibit autoinhibition that IRBIT rescues. Preliminary data from our group show that at least some variants of NBCn1, NDCBE and NBCn2 are stimulated by deletions in the Nt, or by co‐expression with wild‐type IRBIT (Parker et al. 2007a , b ).
The two remaining SLC4 family members are SLC4A9 and SLC4A11.
SLC4A9. The known AE4 variants have none of the three motifs. Despite the name “anion exchanger 4 (AE4)”, recent studies (Chambrey et al. 2013; Peña‐Münzenmayer et al. 2015) have shown that AE4 is Na+ dependent.
SLC4A11. One of the BTR1 variants has a GXXXP motif. None have cationic clusters or KEKE motifs.
Based on their lack of cationic clusters, we expect that neither AE4 nor BTR1 exhibit autoinhibition.
Single nucleotide polymorphisms in the motifs
We searched the NCBI database for single nucleotide polymorphisms (SNPs) of NBCe1‐B/C/E between residues 1 and 62, inclusive, finding a total of 23 polymorphisms. Table 1 summarizes the 14 non‐synonymous polymorphisms among these, 11 of which involve residues for which the data of the present paper yield definitive results (i.e. residues 31–62, inclusive).
Table 1.
Single nucleotide polymorphisms in residues 1–62
| Chromosome position | SNP ID | Major | Minor | Codon position | Amino acid change | MAF |
|---|---|---|---|---|---|---|
| 71236604 | rs765297615 | G | A | 1 | G10R | <0.0001 |
| 71236607 | rs753231947 | G | T | 1 | A11S | <0.0001 |
| 71255231 | rs749218443 | A | G | 1 | I29V | <0.0001 |
| 71255237 | rs1041064148 | A | G | 1 | I31V | ND |
| 71255240 | rs377031010 | G | A | 1 | G32R | 0.0002 |
| 71255262 | rs888297264 | A | G | 2 | Y39C | ND |
| 71255267 | rs747591863 | AGA | ∆AGA | NA | ∆R41 | <0.0001 |
| 71255268 | rs776904118 | G | A | 2 | R41K | <0.0001 |
| 71255276 | rs867931760 | C | A | 1 | R44S | ND |
| 71255295 | rs41265669 | G | C | 2 | G50A | 0.0014 |
| 71255316 | rs1037177466 | A | G | 2 | K57R | ND |
| 71255321 | rs560219064 | A | G | 1 | R59G | <0.0001 |
| 71255327 | rs763409420 | T | C | 1 | S61P | <0.0001 |
| 71255332 | rs764607132 | G | T | 3 | E62D | <0.0001 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; NA, not applicable; ND, no data.
Region near GXXXP motif (residues 28–39)
The polymorphism I29V is not in an area definitively addressed in the present study, inasmuch as the mutation Δ1228–39 yielded a very low surface expression (Fig. 3 C). Polymorphisms I31V (covered by Δ931–39 in Fig. 3 B–D), G32R (covered by Δ931–39 in Fig. 3 B–D and by AVHVA in Fig. 4 B–D), and Y39C (covered by Δ337–39 in Fig. 3 B–D) all involve residues that appear unimportant in our study. However, we note that G32R affects the highly conserved GXXXP motif.
Region of cationic cluster (residues 40–48)
Based on the results summarized in Fig. 5 B–D, ∆R41 (shortens cationic cluster by 1), R41K (no net change in charge) and R44S (reduces net + charge by 1), we expect that these polymorphisms would have a minimal effect on autoinhibition. It is possible that the Ser of R44S could be the subject of phosphorylation, which could disrupt certain protein–protein interactions.
Region near KEKE motif (residues 51–62)
Polymorphism G50A, plus four polymorphisms in the KEKE motif – K57R, R59G, S61P and E62D – all affect residues that appear unimportant in the present study (Fig. 6 B–D). Moreover, K57R and E62D conserve charge. However, we note that R59G and S61P change the charge status or physical structure of the KEKE motif.
Proposed models of the autoinhibition of NBCe1‐B
Recently, Huynh et al. (2018) determined the structure at 3.9 Å of the membrane‐domain dimer of human NBCe1 by cryo‐electron microscopy (cryo‐EM). However, they could not model the cytosolic Nt, part of extracellular loop 3 (EL3), EL4, intracellular loop 5 (IL5), and cytosolic Ct due to the dynamic nature of these regions, which nevertheless could be very important for protein regulation. Therefore, we fed the amino acid sequence of the residues 444–1079 of NBCe1‐B – the transmembrane domain (TMD) plus the Ct – into I‐TASSER (Iterative Threading ASSEmbly Refinement; Roy et al. 2010) in order to model an NBCe1 monomer, including the parts missing in the cryo‐EM structure. Based on the published cryo‐EM structure of the NBCe1 dimer and the I‐TASSER output, we used PyMOL to generate a Protein Data Bank (PDB) file of the full NBCe1 dimer from residues 444 to 1003 (left panel of Fig. 8 A), ignoring residues 1004–1079 due to poor model predictions. Subsequently, we calculated an electrostatic potential map from the PDB file, using the PBEQ solver from CHARMM‐GUI (Jo et al. 2008). The right panel of Fig. 8 A shows the electrostatic potential of the cytosolic surface. By comparison with the extracellular surface (not shown), the cytosolic surface of the TMD is dominantly positively charged, due to the transmembrane helices and ILs.
Figure 8. Proposed models.
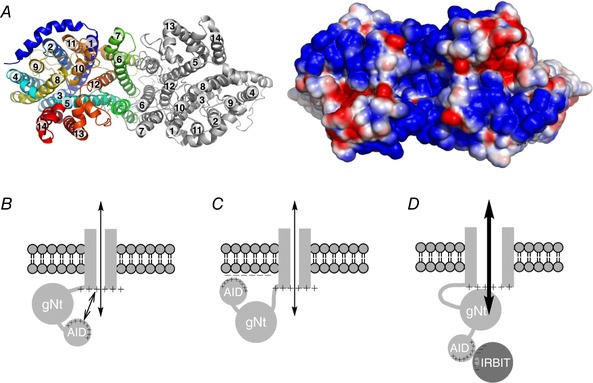
A, model of NBCe1 dimeric transmembrane domain, viewed from the cytosolic side. Left panel shows the modelled structure created by I‐TASSER. Each transmembrane helix is identified by a number. Right panel shows an electrostatic potential map, corresponding to the structure on the left. This map is limited to the solvent‐accessible surface. The thresholds for red (−) or blue (+) were set at ±0.6 kcal/(mole × e 0), where e 0 is the elementary charge. B, one possible mechanism of autoinhibition. Here, repulsion between (a) positive charges in the cationic cluster of the AID and (b) dominant positive charges from the cytosolic surface prevents the interaction between (c) the globular/structured portion of the Nt (gNt) and (d) the TMD, and thereby produces autoinhibition. C, another possible mechanism of autoinhibition. Here, attraction between (a) positive charges in the cationic cluster and (b) negative charges on the leaflet of the plasma membrane prevents the interaction between the gNt and the TMD. D, possible mechanism of relief of autoinhibition by IRBIT. Here, negative charges on IRBIT mask the positive charges in the cationic cluster, and thus allow the gNt to interact with – and thereby stimulate – the TMD.
Preliminary work (Parker et al. 2012) suggests that the Nt must interact with the TMD to produce the electrogenic activity of NBCe1‐A. Considering this observation, the data of the present paper, and the electrostatic potential map in Fig. 8 A, we propose one possible mechanism of autoinhibition (Fig. 8 B), in which the mutual electrostatic repulsion between the cationic cluster (residues 40–48) of the Nt and the positive cytosolic surface of the TMD impedes the interaction between the large structured portion of the Nt (Gill & Boron, 2006a , b ) and the TMD. In another possible mechanism of autoinhibition (Fig. 8 C), the cationic cluster anchors to the negatively charged inner leaflet of lipid bilayer, as suggested for other proteins (van Klompenburg et al. 1997). Finally, we propose that IRBIT relieves autoinhibition as its negatively charged domain masks the cationic cluster, and thereby allows the large structured portion of the Nt to interact with the TMD.
Additional information
Competing interests
None declared.
Author contributions
S.‐K.L. contributed to the conception and design of the research, performed the experiments, analysed data, interpreted results, wrote the first draft of manuscript, prepared the figures and edited the manuscript. W.F.B. contributed to the conception and design of the research, interpreted results and edited the manuscript. Both authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by NIH grant NS18400 (to W.F.B.), and Office of Naval Research Grant N00014‐11‐1‐0889 and N00014‐15‐1‐2060 (to W.F.B.).
Acknowledgements
We thank Dale E. Huffman for computer support. We acknowledge the assistance of Gerald T. Babcock in his role as laboratory manager. We thank Jessica M. Berthiaume and Mark D. Parker for their helpful discussions. We specially thank Vivien Yee for her help in generating Fig. 8 A. W.F.B. gratefully acknowledges the support of the Myers/Scarpa endowed chair.
Biography
Seong‐Ki Lee received his BS degrees in Biological Sciences and Physics from Seoul National University, Korea in 2005. He came to the USA in order to combine the two different disciplines in 2007. After studying potential substrates of the sodium/bicarbonate cotransporter NBCe1 as well as the effect of IRBIT on NBCe1‐B, he received his PhD degree in Physiology & Biophysics from Case Western Reserve University, USA in 2013. As a postdoctoral scholar, he has been studying the functional regulation of NBCe1, using the tools of electrophysiology and structural biology.

Edited by: Peying Fong & Dennis Brown
References
- Ando H, Mizutani A, Matsu‐ura T & Mikoshiba K (2003). IRBIT, a novel inositol 1,4,5‐trisphosphate (IP3) receptor‐binding protein, is released from the IP3 receptor upon IP3 binding to the receptor. J Biol Chem 278, 10602–10612. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W & Vitrac H (2014). Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta 1843, 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron WF & Boulpaep EL (1983). Intracellular pH regulation in the renal proximal tubule of the salamander. Basolateral HCO3 − transport. J Gen Physiol 81, 53–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham CE, Amlal H, Wang Z, Shull GE & Soleimani M (1997). Cloning and functional expression of a human kidney Na+:HCO3 − cotransporter. J Biol Chem 272, 19111–19114. [DOI] [PubMed] [Google Scholar]
- Chambrey R, Kurth I, Peti‐Peterdi J, Houillier P, Purkerson JM, Leviel F, Hentschke M, Zdebik AA, Schwartz GJ, Hübner CA & Eladari D (2013). Renal intercalated cells are rather energized by a proton than a sodium pump. Proc Natl Acad Sci U S A 110, 7928–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaere B, Sammels E & De Smedt H (2008). The IRBIT domain adds new functions to the AHCY family. BioEssays 30, 642–652. [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT & MacKinnon R (2002). X‐ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294. [DOI] [PubMed] [Google Scholar]
- Estévez R & Jentsch TJ (2002). CLC chloride channels: correlating structure with function. Curr Opin Struct Biol 12, 531–539. [DOI] [PubMed] [Google Scholar]
- Faraldo‐Gómez JD & Roux B (2004). Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli . J Mol Biol 339, 981–1000. [DOI] [PubMed] [Google Scholar]
- Gill HS & Boron WF (2006a). Expression and purification of the cytoplasmic N‐terminal domain of the Na/HCO3 cotransporter NBCe1‐A: structural insights from a generalized approach. Protein Expr Purif 49, 228–234. [DOI] [PubMed] [Google Scholar]
- Gill HS & Boron WF (2006b). Preliminary X‐ray diffraction analysis of the cytoplasmic N‐terminal domain of the Na/HCO3 cotransporter NBCe1‐A. Acta Crystallograph Sect F Struct Biol Cryst Commun 62, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant IM, Balcha D, Hao T, Shen Y, Trivedi P, Patrushev I, Fortriede JD, Karpinka JB, Liu L, Zorn AM, Stukenberg PT, Hill DE & Gilchrist MJ (2015). The Xenopus ORFeome: A resource that enables functional genomics. Dev Biol 408, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S‐I, Natsume T, Yashiroda H, Tanaka K & Murata S (2006). A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25, 4524–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass MAS & Mulder FAA (2015). Contemporary NMR studies of protein electrostatics. Annu Rev Biophys 44, 53–75. [DOI] [PubMed] [Google Scholar]
- He P, Zhang H & Yun CC (2008). IRBIT, inositol 1,4,5‐triphosphate (IP3) receptor‐binding protein released with IP3, binds Na+/H+ exchanger NHE3 and activates NHE3 activity in response to calcium. J Biol Chem 283, 33544–33553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Yang D, Shcheynikov N, Ohana E, Shin DM & Muallem S (2013). Convergence of IRBIT, phosphatidylinositol (4,5) bisphosphate, and WNK/SPAK kinases in regulation of the Na+‐HCO3 − cotransporters family. Proc Natl Acad Sci U S A 110, 4105–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KW, Jiang J, Abuladze N, Tsirulnikov K, Kao L, Shao X, Newman D, Azimov R, Pushkin A, Zhou ZH & Kurtz I (2018). CryoEM structure of the human SLC4A4 sodium‐coupled acid‐base transporter NBCe1. Nat Commun 9, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay D & Cantley L (1986). Structural aspects of the red cell anion exchange protein. Annu Rev Biochem 55, 511–538. [DOI] [PubMed] [Google Scholar]
- Jeong YS & Hong JH (2016). Governing effect of regulatory proteins for Cl−/HCO3 − exchanger 2 activity. Channels (Austin) 10, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Vargyas M, Vasko‐Szedlar J, Roux B & Im W (2008). PBEQ‐Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res 36, W270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretić D, Zoranić L & Zucić D (2002). Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci 42, 620–632. [DOI] [PubMed] [Google Scholar]
- Kobayashi YM, Alseikhan BA & Jones LR (2000). Localization and characterization of the calsequestrin‐binding domain of triadin 1. Evidence for a charged beta‐strand in mediating the protein‐protein interaction. J Biol Chem 275, 17639–17646. [DOI] [PubMed] [Google Scholar]
- Lee JM, Rho S‐H, Shin DW, Cho C, Park WJ, Eom SH, Ma J & Kim DH (2004). Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J Biol Chem 279, 6994–7000. [DOI] [PubMed] [Google Scholar]
- Lee S‐K, Boron WF & Parker MD (2013). Substrate specificity of the electrogenic sodium/bicarbonate cotransporter NBCe1‐A (SLC4A4, variant A) from humans and rabbits. Am J Physiol Renal Physiol 304, F883–F899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S‐K, Boron WF & Parker MD (2012). Relief of autoinhibition of the electrogenic Na‐HCO3 cotransporter NBCe1‐B: role of IRBIT vs.amino‐terminal truncation. Am J Physiol Cell Physiol 302, C518–C526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J & Boron WF (2007). Reversible and irreversible interactions of DIDS with the human electrogenic Na/HCO3 cotransporter NBCe1‐A: role of lysines in the KKMIK motif of TM5. Am J Physiol Cell Physiol 292, C1787–C1798. [DOI] [PubMed] [Google Scholar]
- McAlear SD, Liu X, Williams JB, McNicholas‐Bevensee CM & Bevensee MO (2006). Electrogenic Na/HCO3 cotransporter (NBCe1) variants expressed in Xenopus oocytes: functional comparison and roles of the amino and carboxy termini. J Gen Physiol 127, 639–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison KL & Weiss GA (2001). Combinatorial alanine‐scanning. Curr Opin Chem Biol 5, 302–307. [DOI] [PubMed] [Google Scholar]
- Motta M, Chillemi G, Fodale V, Cecchetti S, Coppola S, Stipo S, Cordeddu V, Macioce P, Gelb BD & Tartaglia M (2016). SHOC2 subcellular shuttling requires the KEKE motif‐rich region and N‐terminal leucine‐rich repeat domain and impacts on ERK signalling. Hum Mol Genet 25, 3824–3835. [DOI] [PubMed] [Google Scholar]
- Musa‐Aziz R, Boron WF & Parker MD (2010). Using fluorometry and ion‐sensitive microelectrodes to study the functional expression of heterologously‐expressed ion channels and transporters in Xenopus oocytes. Methods 51, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S & Mann M (2011). Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obosi LA, Hen R, Beadle DJ, Bermudez I & King LA (1997). Mutational analysis of the mouse 5‐HT7 receptor: importance of the third intracellular loop for receptor‐G‐protein interaction. FEBS Lett 412, 321–324. [DOI] [PubMed] [Google Scholar]
- Park S, Shcheynikov N, Hong JH, Zheng C, Suh SH, Kawaai K, Ando H, Mizutani A, Abe T, Kiyonari H, Seki G, Yule D, Mikoshiba K & Muallem S (2013). Irbit mediates synergy between Ca2+ and cAMP signaling pathways during epithelial transport in mice. Gastroenterology 145, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD & Boron WF (2013). The divergence, actions, roles, and relatives of sodium‐coupled bicarbonate transporters. Physiol Rev 93, 803–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Daly C, Skelton L‐A & Boron W (2007a). IRBIT functionally enhances the electroneutral Na+‐coupled bicarbonate transporter NCBE by sequestering an N‐terminal autoinhibitory domain. FASEB J 21, 10.1096/fasebj.21.6.A1285-b. [DOI] [Google Scholar]
- Parker MD, Musa‐Aziz R, Rojas JD, Choi I, Daly CM & Boron WF (2008). Characterization of human SLC4A10 as an electroneutral Na/HCO3 cotransporter (NBCn2) with Cl− self‐exchange activity. J Biol Chem 283, 12777–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Skelton L‐A, Daly CM & Boron W (2007b). IRBIT binds to and functionally enhances the electroneutral Na+‐coupled bicarbonate transporters NBCn1, NDCBE and NCBE. FASEB J 21, 10.1096/fasebj.21.6.A1285-a. [DOI] [Google Scholar]
- Parker MD, Wass AB, Lee S‐K, Rahman F, Grant C & Boron WF (2012). Functional reassembly of NBCe1‐A from co‐expressed cytosolic and transmembrane domains. FASEB J 26, 10.1096/fasebj.26.1_supplement.882.2. [DOI] [Google Scholar]
- Pearlman SM, Serber Z & Ferrell JE (2011). A mechanism for the evolution of phosphorylation sites. Cell 147, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña‐Münzenmayer G, Catalán MA, Kondo Y, Jaramillo Y, Liu F, Shull GE & Melvin JE (2015). Ae4 (Slc4a9) anion exchanger drives Cl− uptake‐dependent fluid secretion by mouse submandibular gland acinar cells. J Biol Chem 290, 10677–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini C, Rogers SW & Rechsteiner M (1994). KEKE motifs. Proposed roles in protein‐protein association and presentation of peptides by MHC class I receptors. FEBS Lett 348, 109–113. [DOI] [PubMed] [Google Scholar]
- Romero MF, Chen A‐P, Parker MD & Boron WF (2013). The SLC4 family of bicarbonate (HCO3 −) transporters. Mol Aspects Med 34, 159–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero MF, Fong P, Berger UV, Hediger MA & Boron WF (1998). Cloning and functional expression of rNBC, an electrogenic Na+‐HCO3 − cotransporter from rat kidney. Am J Physiol 274, F425–F432. [DOI] [PubMed] [Google Scholar]
- Romero MF, Hediger MA, Boulpaep EL & Boron WF (1997). Expression cloning and characterization of a renal electrogenic Na+/HCO3 − cotransporter. Nature 387, 409–413. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A & Zhang Y (2010). I‐TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheynikov N, Son A, Hong JH, Yamazaki O, Ohana E, Kurtz I, Shin DM & Muallem S (2015). Intracellular Cl− as a signaling ion that potently regulates Na+/HCO3 − transporters. Proc Natl Acad Sci U S A 112, E329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K, Priori G, Yamada H, Ando H, Horita S, Fujita T, Fujimoto I, Mizutani A, Seki G & Mikoshiba K (2006). IRBIT, an inositol 1,4,5‐trisphosphate receptor‐binding protein, specifically binds to and activates pancreas‐type Na+/HCO3 − cotransporter 1 (pNBC1). Proc Natl Acad Sci U S A 103, 9542–9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD & Higgins DG (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell IM, Wu J & Bevensee MO (2010). The IP3 receptor‐binding protein IRBIT reduces phosphatidylinositol 4,5‐bisphosphate (PIP2) stimulation of Na/bicarbonate cotransporter NBCe1 variants expressed in Xenopus laevis oocytes. FASEB J 24. [Google Scholar]
- van Klompenburg W, Nilsson I, von Heijne G & de Kruijff B (1997). Anionic phospholipids are determinants of membrane protein topology. EMBO J 16, 4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL (1997). Basic amino acids at the C‐terminus of the third intracellular loop are required for the activation of phospholipase C by cholecystokinin‐B receptors. J Neurochem 68, 1728–1735. [DOI] [PubMed] [Google Scholar]
- Wang HL (1999). A conserved arginine in the distal third intracellular loop of the μ‐opioid receptor is required for G protein activation. J Neurochem 72, 1307–1314. [DOI] [PubMed] [Google Scholar]
- Yang D, Li Q, So I, Huang C‐L, Ando H, Mizutani A, Seki G, Mikoshiba K, Thomas PJ & Muallem S (2011). IRBIT governs epithelial secretion in mice by antagonizing the WNK/SPAK kinase pathway. J Clin Invest 121, 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Shcheynikov N, Zeng W, Ohana E, So I, Ando H, Mizutani A, Mikoshiba K & Muallem S (2009). IRBIT coordinates epithelial fluid and HCO3 − secretion by stimulating the transporters pNBC1 and CFTR in the murine pancreatic duct. J Clin Invest 119, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]


