Abstract
Symbiotic ectomycorrhizal fungi have received increasing attention as regulators of below-ground organic matter storage. They are proposed to promote organic matter accumulation by suppressing saprotrophs, but have also been suggested to play an active role in decomposition themselves. Here we show that exclusion of tree roots and associated ectomycorrhizal fungi in a boreal forest increased decomposition of surface litter by 11% by alleviating nitrogen limitation of saprotrophs–a “Gadgil effect”. At the same time, root exclusion decreased Mn-peroxidase activity in the deeper mor layer by 91%. Our results show that ectomycorrhizal fungi may hamper short-term litter decomposition, but also support a crucial role of ectomycorrhizal fungi in driving long-term organic matter oxidation. These observations stress the importance of ectomycorrhizal fungi in regulation of below-ground organic matter accumulation. By different mechanisms they may either hamper or stimulate decomposition, depending upon stage of decomposition and location in the soil profile.
Introduction
Global meta-analyses [1, 2] have indicated that forests dominated by ectomycorrhizal associations tend to accumulate more organic matter below ground than other forests. A commonly proposed explanation is that ectomycorrhizal fungi retard decomposition by competing with free-living saprotrophs for nitrogen (N) [1, 3]. This idea has its origin in a well-known experiment, in which exclusion of living roots and associated mycorrhizal fungi by trenching was shown to elevate litter decomposition rates [4] – a phenomenon referred to as the ‘Gadgil effect’. Reduction in soil respiration in the presence of ectomycorrhizal fungi was recently demonstrated in temperate Tsuga forest [5]. In boreal Pinus/Picea forest, dominance of ectomycorrhizal fungi over saprotrophs was related to hampered organic matter decomposition and larger below-ground stocks [6]. However, stimulatory effects of ectomycorrhizal roots on soil respiration [7, 8] and enzyme activities [9] have also been observed, and ectomycorrhizal proliferation has been connected to lower below-ground carbon (C) storage [7, 10, 11]. A review of previous assessments [12] concluded that the ‘Gadgil effect’ is variable and inconsistent across systems.
Boreal forests represent a globally important sink for atmospheric C, and the bulk of the C sequestered in terrestrial ecosystems is stored below ground [13]. The magnitude of the below-ground C sink does not reflect ecosystem primary productivity but is largely regulated by the rate of organic matter decomposition in the rooting zone [14, 15]. Boreal forests are characterised by an often thick, purely organic layer overlying the mineral soil. The organic layer may be subdivided into an uppermost litter layer (L), with recognisable plant remains, a layer containing fragmented litter components (F), and a deeper humus layer (H), consisting of well-decomposed organic matter (Table 1). Plant roots inhabit the lower part (F + H = the mor layer) of the organic profile, and have, together with mycorrhizal fungal mycelium, been found to make a major contribution to long-term accumulation of organic matter [14].
Table 1.
Definitions of vertical layers in the organic horizon of a boreal forest (based on control samples)
| Horizon | Fraction of C in the organic horizon | Definition |
|---|---|---|
| Needle litter | 5% | Brown intact needles |
| Moss litter | 2% | Withered, brown mosses |
| Fragmented 1 | 19% | Top 2/3 of recognisable but partly degraded litter |
| Mor | ||
| Fragmented 2 | 17% | Bottom 1/3 of recognisable but partly degraded litter |
| Humus 1 | 34% | Top 2/3 of horizon with no recognisable litter parts |
| Humus 2 | 24% | Bottom 1/3 of horizon with no recognisable litter parts |
The mor layer of coniferous forests is relatively N poor and commonly colonised by diverse fungal communities. Fungal:bacterial ratios are usually higher than in other ecosystems [16]. Soil macrofauna, such as anecic earthworms, are largely absent in the acidic mor, and the low degree of physical disturbance enables accumulation of dense fungal communities, potentially leading to intense interference competition between fungal individuals, species and functional guilds. Agaricomycetes (i.e., “mushroom forming fungi”) often constitute a significant proportion of the total fungal communities in boreal forest soils [17], presumably favoured by their unique capacity to produce potent oxidative enzymes necessary to exploit the lignin-rich needle litter [18]. Many Agaricomycetes commonly build large and long-lived mycelia and compete intensely for space and resources, congruent with a categorisation as C-strategists [17, 19].
Agaricomycete communities in boreal forest soils contain both free-living, saprotrophic genera, such as Mycena, and ectomycorrhizal genera, notably Cortinarius, Piloderma and Russula. The two functional guilds usually occupy distinct vertical niches. Saprotrophs predominantly colonise recently fallen, cellulose-rich litter components closer to the surface, whereas mycorrhizal fungi obtain sugars directly from their symbiotic plant partner and proliferate in the deeper, more decomposed and cellulose-depleted mor layer, which has the highest density of roots [10, 17, 20]. A field experiment involving vertical transplantation of litter components showed that guild separation depended only partly on substrate preferences, and that antagonistic competition for soil space also played an important role [21], supporting previous observations of antagonism between saprotrophic and ectomycorrhizal Agaricomycetes in laboratory microcosms [22, 23]. Saprotrophic Agaricomycetes depend on upward reallocation of limiting N to efficiently decompose the N-poor needle litter [24], but antagonistic ectomycorrhizal fungi may prevent them accessing the deeper, more N-rich layers of the mor. As roots and mycorrhizal fungi continuously deposit new organic matter below ground, hampered decomposition due to exclusion of saprotrophs from the rooting zone may result in net accumulation of organic matter and a persistent ecosystem C sink in the mor [6, 14].
We tested the ‘Gadgil hypothesis’ in a trenching experiment in a mixed coniferous forest in central Sweden (Fig. 1), aiming to exclude symbiotic mycorrhizal fungi from 1 m2 forest plots, and studied short-term (1 year) and longer-term (4 years) effects on decomposition in the organic horizon. We followed vertical redistribution of N pools by stable isotope analysis, to test the hypothesis that removal of ectomycorrhizal competition for 15N-enriched pools deeper in the profile would lead to upward reallocation into the 15N-depleted litter layer [25] and accelerate litter decomposition. We conducted detailed vertical profiling of fungal communities, using DNA-based metabarcoding, to test the hypothesis that exclusion of living roots and ectomycorrhizal fungi would lead to a general stimulation and downward proliferation of saprotrophic Agaricomycetes [6, 26]. We also investigated long-term decomposition in the mor layer by measuring activities of enzymes involved in decomposition of plant- and fungal cell wall components. Since saprotrophic Agaricomycetes generally have a more developed genetic potential to produce enzymes involved in decomposition of plant cell wall components [27, 28], we expected that downward proliferation of saprotrophic Agaricomycetes after trenching would elevate enzyme activities deeper in the profile [6].
Fig. 1.

A trenched experimental plot in a Swedish boreal forest. Trenching was performed by digging around a central plot (1 × 1 m) using an excavator, leaving the plot as undisturbed as possible. Steel barriers were thereafter pressed down round the plots to a depth of 0.7 m, and the trenches were refilled
Methods
Field experiment
The experiment was set up in a mixed boreal Pinus sylvestris L. (Scots pine) and Picea abies (L.) H. Karst. (Norway spruce) forest with an understory dominated by mosses (mostly Hylocomium splendens (Hedw.) Schimp.), ericaceous shrubs (mostly Vaccinium myrtillus L.) and herbs (mostly Oxalis acetosella L. and Convallaria majalis L.) near Sala in central Sweden (59°57'07.9“N 16°45'18.8“E; 77 m a.s.). The soil type was a podzol with a ca. 10 cm thick organic horizon overlying the mineral soil. In October 2009, the below-ground C flux into 1 × 1 m experimental plots was interrupted by severing root connections–a well-established method to reduce the impact of mycorrhizal associations on soil processes [29]. Eight trenched plots and eight control plots were distributed within an area of 1 ha. Trenching was performed by digging a ditch around the plot using an excavator. Plots were kept as undisturbed as possible and covered by a protective plastic sheet during trenching. Steel barriers were thereafter inserted around the plots to a depth of 0.7 m to isolate them from surrounding tree roots, and the trenches were refilled (Fig. 1). Sensors (Em50 data loggers, 5TM sensors, Decagon Devices, Pullman, USA) were inserted to a depth of 7 cm into the organic horizon of each plot, recording temperature and moisture once per hour from August to December, 2011.
Four soil cores (2.7 cm in diameter, to a depth of 10 cm) per plot were collected in October 2010 (1 year after trenching), and four additional soil cores were collected in October 2013 (4 years after trenching). Soil cores were kept intact in the plastic corers and stored at −20 °C. After thawing, each core was divided into six depth layers according to Table 1. Materials from each layer were pooled within each plot and year and mixed by hand, resulting in six composite samples per plot and year (2 times × 2 treatments × 6 layers × 8 replicates = 192 samples). All samples were weighed, and subsamples were kept at −20 °C for enzyme analyses (4 years samples only), determination of water content by oven-drying at 60 °C for 12 h and analysis of organic matter content by loss-on-ignition at 550 °C for 6 h. The remaining material was freeze-dried for DNA extraction as well as ergosterol and δ15N (1 year samples only) analyses. Prior to DNA analysis, roots larger than 2 mm in diameter were removed, and mor samples were milled to powder using a pestle and mortar, whereas litter samples were ground in a ball mill. For some samples, there was insufficient material available for all analyses. The number of replicates for each analysis-treatment-horizon combination is specified in Table S1.
Litter decomposition assay
Four mesh bags (10 × 2.5 cm) of 50 μm-pore size polyamide mesh (Sintab Produkt AB, Oxie, Sweden) were inserted horizontally in each plot below the mosses in June 2011. Each bag was filled with 2 g of pine litter (intact brown needles, picked from the ground and dried). In October 2011 (after 4 months incubation, two years after trenching), two mesh bags per plot were harvested, and in October 2012 (after 16 months incubation, 3 years after trenching) the remaining two bags were retrieved. Needle weight loss was recorded after freeze-drying, and the bag content was ground in a ball mill.
Sequencing of fungal communities
Fungal community composition was assessed by extraction of DNA and metabarcoding of ITS2 amplicons according to the protocol of ref. [30]. Approximately 50 mg of each organic layer from each plot (both harvests) was used for DNA extraction using the NucleoSpin Soil Genomic DNA extraction kit (Macherey-Nagel, Düren, Germany). Extracts were subjected to PCR-amplification of ITS2 markers with the primer combination ITS4a (consisting of 2/3 of the general eukaryote primer ITS4: CCTCCGCTTATTGATATGC [31], and 1/3 of primer CCTCGCCTTATTGATATGC, modified to cover Archaeorhizomycetes) and gITS7: GTGARTCATCGARTCTTTG [32]. Both primers were elongated by a unique sample-tag of nine and ten bases, respectively, designed using Barcrawl [33]. PCR cycle numbers were optimised for each sample and ranged between 27 and 33. Three PCR reactions from each sample were pooled and purified with the AMPure kit (Beckman Coulter, Brea, USA). Template-free negative controls were included in each PCR run. Concentrations of purified PCR products were established with a Qubit fluorometer (Thermo-Fisher, Waltham, USA), and equal amounts of DNA from each sample were pooled to provide a single pooled sample that was purified with Jetquick PCR Product Purification Spin Kit (Genomed, Löhne, Germany). Amplicon sequencing was performed on a PSII system (Pacific Biosciences, Menlo Park, USA) by SciLifeLab (Uppsala Genome Centre, Sweden) using 16 SMRT cells. Sequence data are archived at the Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) with accession number SRP132058.
Sequences were filtered and clustered using the SCATA pipeline (http://scata.mykopat.slu.se). Sequences were filtered for quality, removing data with an average quality score below 20 or with a score below 3 at any position, screened for primers (90% sequence similarity required) and sample identifying tags and complementary reversed when necessary. The sequences were then compared for similarity, using USEARCH [34]. Pairwise alignments were scored with equal penalty for mismatch and gaps. Homopolymers were collapsed to 3 bp before alignment. Sequences were clustered into species hypotheses (SHs) [35] by single linkage clustering, with a 1.5% maximum distance allowed for sequences to enter clusters.
To make careful manual annotation practically manageable, only SHs with more than 20 reads (491 out of a total of 1922) were assessed for identification. The most abundant sequence in each SH was used as a representative (after restoration of homopolymers). Representative sequences were compared against the UNITE and INSD databases [35] using the BLASTn massblaster in PlutoF [36] for taxonomic and functional annotation. Plant sequences were removed from the dataset. SHs were assigned to functional guilds according to Table 2, either based on taxonomic identity or depending on the origin (living roots or leaf litter) of closely related (>97% similarity) sequences in the reference databases. For each sample, the relative abundance of each fungal guild was calculated as the ratio between the added number of reads of all SHs in the guild and the total number of reads (with unidentified SHs included but plants and singletons excluded). A list of species hypotheses (Database S1) and a species hypothesis vs. sample matrix (Database S2) are supplemented, as well as species accumulation curves produced in R using the package Vegan (Fig. S1).
Table 2.
Definitions of functional guilds among fungal taxa identified by high-throughput sequencing of amplified ITS markers in soil from trenched and control plots in a boreal forest
| Fungal guild | Definition of guild |
|---|---|
| Ectomycorrhizal fungi | Known ectomycorrhizal species |
| Other root basidiomycetes | Basidiomycetes associated with living plant roots, other than known ectomycorrhizal species |
| Root ascomycetes | Ascomycetes associated with living plant roots (e.g. ericoid mycorrhizal fungi), excluding known ectomycorrhizal species |
| Litter Agaricomycetes | Agaricomycetes previously detected in intact plant litter, mainly saprotrophic species |
| Litter ascomycetes | Ascomycetes previously detected in intact plant litter (saprotrophic fungi as well as endophytic species) |
| Yeasts and moulds | Species in the groups Penicillium, Trichoderma, Mortierella, Mucorales, Saccharomycetales, Tremellales, Sporidiales, and Microbotriomycetes |
| Unknown function | Fungi with unknown identity and/or uncertain ecological role |
Ergosterol analysis
Fungal biomass was assessed in each layer of each plot by extracting the fungal biomarker ergosterol from approximately 75 mg dried and milled material with 4 ml 10% KOH in methanol [37]. Extracted ergosterol was transferred to a cyclohexane phase, which was evaporated. Residues were dissolved in 500 µl methanol, filtered (0.5 ml, 0.45 μm PTFE syringe filter, Millex LCR-4; Millipore, Billerica, USA) and separated on a reversed-phase column (Nova-Pak C18, 3.9 × 150 mm, Waters Corporation, Milford, USA), preceded by a guard column (Nova-Pak C18, 3.9 × 22 mm) in a HPLC system (WatersTM 717 plus Auto sampler, WatersTM 600 Controller, WatersTM 996 Photodiode Array Detector) with a flow rate of 1 ml min−1. The ergosterol peak was detected at 282 nm.
Enzyme activity analyses
Potential activities of six extracellular enzymes involved in soil C and nutrient cycling [38] were assayed on thawed material collected in 2013 (4 years after trenching) from each of the six layers of each plot.
Activities of cellobiohydrolase, β-glucosidase, chitobiohydrolase and N-acetyl-glucosaminidase were assessed via the hydrolysis of 4-methylumbelliferyl β-D-cellobioside, 4-methylumbelliferyl β-D-glucopyranoside, 4-methylumbelliferyl β-D-N,N’-diacetylchitobioside and 4-methylumbelliferyl N-acetyl-β-D-glucosaminide, respectively. Organic materials were added to sodium acetate buffer (50 mM, pH 5) in the proportion 1 g of organic matter to 1 l of buffer, and the suspension was mixed with a mixer (Ultra-Turrax, IKA, Staufen, Germany) for 2 minutes. For each enzyme, a 200 μl sample aliquot (pipetted during continuous stirring) was added to 50 μl of fluorogenic substrate (dissolved in buffer). After incubation for 1 h, 10 μl 1 M NaOH was added to stop the reaction, and after an additional 10 minutes, fluorescence was measured (excitation wavelength 365 nm, excitation slit 2.5, emission wavelength 450 nm and emission slit 2.5) on a LS 50B Fluorescence Spectrometer (PerkinElmer, Waltham, USA) and related to 4-methylumbelliferone standards.
To analyze peroxidase and laccase activity, 0.5 g of fragmented/humus material or 0.05–0.15 g of litter was mixed with 50 ml sodium acetate buffer, finely fragmented with a mixer (Ultra-Turrax) for two minutes and centrifuged at 10,000×g for 10 min. Peroxidase activity was assessed via the oxidative coupling of 3-(dimethylamino)benzoic acid (DMAB) and 3-methyl-2-benzothiazolinone hydrazone (MBTH) in the presence of Mn2+ and H2O2. A 50 μl aliquot of extract was added to 150 μl of reaction solution (50 mM DMAB, 1 mM MBTH, 1 mM MnSO4, 100 mM sodium lactate, 100 mM sodium succinate, 67 µM H2O2, pH 4.5). Enzyme activities were estimated by regression of absorbance at 590 nm in a SpectraMax Plus 384 Microplate Reader (Molecular Devices, Sunnyvale, CA, USA) against time. Mn-peroxidase activity was estimated by subtracting the activity with MnSO4 substituted for 2 mM ethylenediaminetetraacetic acid (EDTA). Laccase activity was estimated by adding 100 μl of extract to 100 μl of reaction solution containing 50 mM 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)(ABTS) in sodium acetate buffer. Enzyme activities were estimated by regression of absorbance at 420 nm against time. Boiled aliquots served as negative controls for all enzyme measurements.
15N analysis
15N abundance in litter from the incubated mesh bags and in each of the six layers of soil cores collected in 2010 (1 year after trenching) was analysed using an elemental analyser (model EuroEA3024; Eurovector, Milan, Italy) connected to an Isoprime isotope-ratio mass spectrometer (GV-Instruments, Manchester, UK). The results are presented as δ15N values based on deviations in the ratios relative to atmospheric N2:
δ15N = [(Rsample − Ratmosphere) / Ratmosphere] × 1000‰,where R is the molar ratio between the heavy and light isotope. All isotope analyses were performed at the Isotope Laboratory at Örebro University, Sweden, and the results were corrected for instrument drift (the δ15N standard deviation among 13 replicated standards was 0.061‰).
Statistics
Differences in ergosterol concentration, relative abundance of fungal guilds, enzyme activities and 15N in organic soil samples were analysed by generalised linear mixed models. To test overall effects of trenching and layer, “treatment” (df = 1) and “layer” (df = 5) and their interaction (df = 5) were defined as fixed factors. To account for possible dependency between measures at different depths within individual plots, “layer” was treated as a repeated measure with “plot” as the subject and a first-order autocorrelation structure specified through random statements. The two sampling occasions were analysed separately. Satterthwaite-type degrees of freedom based on the Kenwood–Roger adjustment were used to calculate Wald F test statistics of the fixed factors, and results were evaluated using Tukey’s adjustment for multiple comparisons with α = 0.05. Relative abundance data were arcsine transformed and enzyme activities log10 transformed prior to analysis. Effects of trenching and incubation time on weight loss and δ15N of litter bag materials were analysed in a model specifying treatment (df = 1), time (df = 1), and their interaction (df = 1) as fixed factors. All statistical models were run using the GLIMMIX procedure in the SAS 9.3 package (Statistical Analysis System Institute, Cary, NC, USA). Effects of trenching on parameters across the entire mor layer were assessed based on mass-weighted averages of measurements in individual horizons.
Results
The total number of reads passing quality control was 177479 (59% of total sequencing output). Removal of non-fungal sequences left 144076 sequences (81% of high quality sequences), which clustered into 1922 local species hypotheses (SHs), of which 491 SHs (with >20 sequences) were assessed for taxonomic affiliation and 330 SHs (97853 sequences; 68% of fungal sequences) were assigned to known functional guilds (Table 2; Database S1). On average, fungal communities were represented by 833 reads per sample, ranging from 90 to 2737 (Database S2).
Different functional guilds of fungi occupied distinct vertical layers in the organic horizon (Fig. 2, Table 3), and this pattern was reflected in diverging activities of extracellular enzymes along depth profiles (Fig. 3, Table 3). Hydrolytic enzymes involved in decomposition of cellulose (Fig. 3a, b) and chitin (Fig. 3c, d), as well as phenol oxidising laccases (Fig. 3e), were most active in the surface litter, corresponding to the distribution of DNA-markers for saprotrophic Agaricomycetes and litter ascomycetes. In the deeper, more decomposed mor layer, where ectomycorrhizal fungi and other root-associated fungi dominated, activities of hydrolytic enzymes declined. In contrast, Mn-peroxidase activity remained high (in non-trenched controls; Fig. 3f), indicating a gradual transition with depth from hydrolysis to oxidation as the principal mechanism of decomposition.
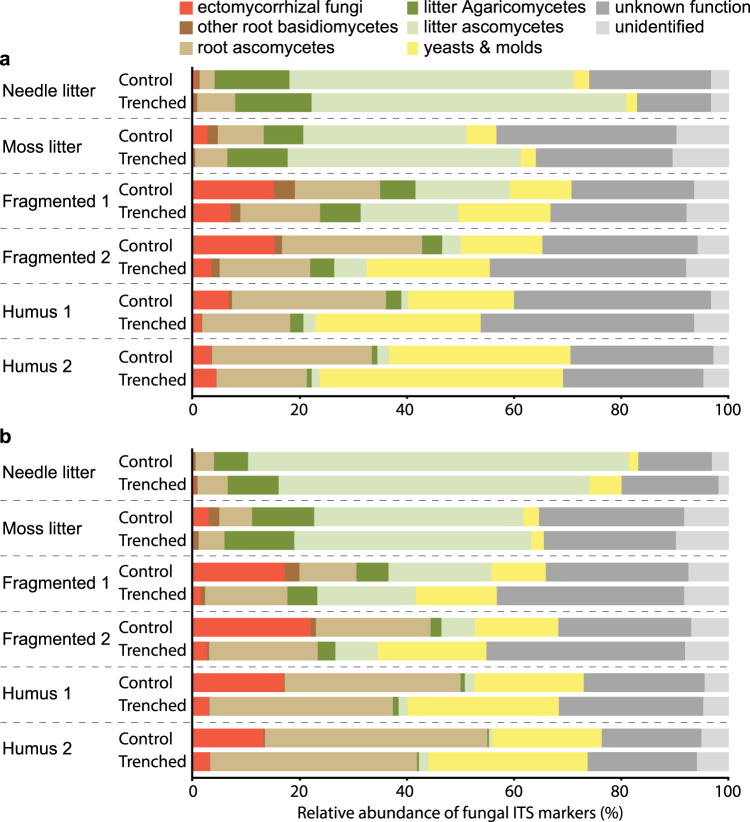
Fig. 2.
Depth profile of the relative abundance of ITS2 markers assigned to different functional guilds of fungi in the litter and mor layers of a boreal forest. Experimental plots where root connections were severed 1 year (a) or 4 years (b) prior to sampling (Trenched) are compared with non-trenched plots (Control). Data are means, n = 5–8
Table 3.
Results from mixed models evaluating the statistical significance of effects of trenching and layer on relative abundances of functional guilds of fungi, fungal biomass, 15N relative abundance and activities of selected extracellular enzymes in organic soil profiles of a boreal forest 1 year (fungal guilds, biomass and δ15N) and 4 years (all variables except δ15N) after disruption of root connections by trenching
| Variable | trenching | layer | trenching x layer | |||
|---|---|---|---|---|---|---|
| F df | P | F df | P | F df | P | |
| Fungal biomass (µg ergosterol gOM−1): | ||||||
| 1 year after trenching | 0.001,38 | 0.97 | 8.85,57 | <0.0001 | 1.85,57 | 0.14 |
| 4 years after trenching | 11.41,16 | 0.004 (−) | 4.85,54 | 0.001 | 3.05,54 | 0.02 |
| Fungal guilds (% of ITS markers), 1 year after trenching: | ||||||
| Ectomycorrhizal fungi | 14.31,22 | 0.001 (−) | 9.05,58 | <0.0001 | 1.45,58 | 0.23 |
| Root basidiomycetes | 1.81,23 | 0.19 | 7.45,57 | <0.0001 | 1.25,57 | 0.33 |
| Root ascomycetes | 3.71,24 | 0.07 | 18.95,59 | <0.0001 | 2.95,59 | 0.02 |
| Litter Agaricomycetes | 0.31,13 | 0.63 | 16.95,53 | <0.0001 | 0.25,53 | 0.98 |
| Litter ascomycetes | 7.21,27 | 0.01 (+) | 113.85,52 | <0.0001 | 0.65,52 | 0.74 |
| Yeasts and moulds | 5.31,27 | 0.03 (+) | 43.25,57 | <0.0001 | 1.55,57 | 0.20 |
| Fungal guilds (% of ITS markers), 4 years after trenching: | ||||||
| Ectomycorrhizal fungi | 147.81,31 | <0.0001 (−) | 29.95,60 | <0.0001 | 8.45,60 | <0.0001 |
| Root basidiomycetes | 3.81,24 | 0.06 | 6.55,55 | <0.0001 | 1.25,55 | 0.34 |
| Root ascomycetes | 0.01,23 | 0.95 | 61.15,59 | <0.0001 | 1.15,59 | 0.36 |
| Litter Agaricomycetes | 0.91,18 | 0.37 | 43.15,54 | <0.0001 | 0.75,54 | 0.66 |
| Litter ascomycetes | 0.31,33 | 0.60 | 115.65,48 | <0.0001 | 1.25,48 | 0.31 |
| Yeasts and moulds | 6.81,22 | 0.02 (+) | 38.25,58 | <0.0001 | 0.95,58 | 0.46 |
| Enzyme activities (nmol gOM−1 s−1), 4 years after trenching: | ||||||
| Total peroxidase | 59.31,30 | <0.0001 (− ) | 8.94,56 | <0.0001 | 6.34,56 | 0.0003 |
| Mn peroxidase | 20.91,31 | <0.0001 ( −) | 1.55,64 | 0.22 | 4.15,64 | 0.003 |
| Laccase | 0.11,27 | 0.82 | 47.35,65 | <0.0001 | 1.55,65 | 0.20 |
| BG | 3.31,24 | 0.08 | 8.35,65 | <0.0001 | 0.45,65 | 0.82 |
| CBH | 6.31,23 | 0.02 (−) | 13.15,63 | <0.0001 | 1.05,63 | 0.43 |
| NAG | 10.41,24 | 0.004 (−) | 10.75,67 | <0.0001 | 1.55,67 | 0.19 |
| CiBH | 6.81,25 | 0.02 (− ) | 4.45,66 | 0.002 | 0.75,56 | 0.64 |
| δ15N, 1 year after trenching: | ||||||
| 0.31,20 | 0.59 | 179.65,68 | <0.0001 | 1.85,68 | 0.13 | |
Significant values (P < 0.05) are presented in bold and the direction of response is indicated in parenthesis. Degrees of freedom (df) for F values are given as subscripts (effect, error) after each F value
Fig. 3.
Depth profiles of activities of selected extracellular enzymes involved in organic matter decomposition (a–f) 4 years after root disruption by trenching (broken lines) and in non-trenched control plots (solid lines) and ergosterol concentration as a proxy for fungal biomass (g–h) one and 4 years after trenching in the litter layer and mor layers of a boreal forest. All data are means ± SE, n = 7-8 (in g, h, n = 3-8). 15N relative abundance (i) was measured 1 year after trenching without a significant treatment effect (treatments merged, n = 16)
Trenching increased mass loss of introduced needle litter significantly by 11% (Fig. 4a, Table 4). The δ15N values of introduced litter were significantly higher, on average by 1.2‰, in trenched plots but did not change significantly with increasing incubation time (Fig. 4b, Table 4).
Fig. 4.

Mass loss (a) and 15N relative abundance (b) in decomposing pine needles in litter bags incubated on the forest floor of a boreal forest in experimental plots with severed root connections (Trenched–open bars) or with intact root connections (Control–closed bars). Data are means ± SE, n = 8 (for control after 16 months n = 6)
Table 4.
Results from linear models evaluating effects of root trenching and litter incubation duration (4 months or 16 months) on mass loss (% of initial mass) and 15N relative abundance in litter incubated in the moss layer of a boreal forest
| Variable | Trenching | Incubation | Trenching × incubation | |||
|---|---|---|---|---|---|---|
| F df | P | F df | P | F df | P | |
| Mass loss (%) | 8.61,26 | 0.007 (+) | 661.81,26 | <0.0001 | 0.41,26 | 0.53 |
| δ15N | 79.11,26 | <0.0001 (+) | 2.51,26 | 0.13 | 0.11,26 | 0.75 |
Significant values (P < 0.05) are presented in bold and the direction of response is indicated in parenthesis. Degrees of freedom (df) for F values are given as subscripts (effect, error) after each F value
One year after trenching, there was no significant effect on fungal biomass (ergosterol) (Fig. 3g; Table 3). The relative abundance of ectomycorrhizal DNA-markers was significantly decreased, whereas relative abundances of yeasts and moulds, as well as litter ascomycetes, were elevated. The relative abundance and vertical distribution of saprotrophic Agaricomycetes was not significantly affected by trenching (Fig. 2a; Table 3).
Four years after trenching, total fungal biomass (ergosterol) in the mor layer had decreased by 62% (Fig. 3h, Table 3), and the relative abundance of DNA-markers attributed to ectomycorrhizal species had decreased by 84% (from 17% to 3%) (Fig. 2b, Table 3). The only other significant change in fungal functional guilds was a 35% increase in the relative abundance of moulds and yeasts in trenched plots, but this relative increase was counter-balanced by the overall decrease in fungal biomass. Notably, after 4 years, trenching led to an almost complete loss of Mn-peroxidase activity in the mor layer (Fig. 3f) and significantly decreased activities of enzymes involved in chitin degradation, as well as cellulose degrading cellobiohydrolases, but had no significant effect on β-glucosidases and laccases (Fig. 3, Table 3).
Four years after root exclusion the total organic stocks in the mor layer were on average lower in trenched plots (3.0 ± 0.3 kg/m2) than in control plots (3.8 ± 0.4 kg/m2), but the difference was not statistically significant (P = 0.1). We found no significant (P = 0.2) differences in soil moisture (13 ± 4% VWC in the trenched plots compared to 17 ± 7 % VWC in control plots) and similar soil temperatures (8.3 ± 0.4 °C in trenched plots compared to 8.7 ± 0.1 °C in control plots).
Discussion
Trenching was successful in specifically excluding the ectomycorrhizal component of the fungal community, as indicated by the decline in fungal biomass 4 years after trenching (Fig. 3h) combined with radically decreased abundance of ectomycorrhizal DNA-markers (Fig. 2b). However, 1 year after trenching fungal biomass was similar to controls, and opportunistic taxa and litter-associated ascomycetes had proliferated, presumably in response to the enrichment disturbance associated with death of roots and mycorrhizal mycelium (Fig. 2a). This finding is consistent with previously observed short-term (2 weeks) responses to root severing [39]. Presumably, opportunistic decomposition of 15N enriched mycorrhizal mycelium deeper in the profile (Fig. 2i) [40, 41] and concomitant mineralisation made N available for uptake and upward redistribution. Release and redistribution of N from dead mycorrhizal mycelium is in agreement with the elevated 15N abundance in surface litter incubated in trenched plots (Fig. 4b) and in line with previous observations of 15N enrichment in undergrowth vegetation after girdling of boreal forest trees [25].
Trenching stimulated decomposition in the uppermost litter layer (Fig. 4a), confirming the ‘Gadgil hypothesis’. Due to the low N content of freshly deposited needle litter, saprotrophs depend on upward reallocation of N to maintain high colonisation and decomposition rates [24], and faster litter decomposition after trenching supports the idea that, under normal circumstances, ectomycorrhizal fungi supress litter decomposition by competing with saprotrophs for N [1, 3–6, 12]. This result also agrees with the idea that ectomycorrhizal fungi may aggravate N-limitation of other organisms in boreal forest soils by retaining N in their extensive mycelia [6, 42].
Ectomycorrhizal fungi have been hypothesised to constrain downward proliferation of saprotrophic fungi by antagonistic interactions [6, 20, 21, 26, 43]. In the present study, contrary to our expectations, saprotrophic Agaricomycetes were consistently limited to the upper layers, and their vertical distribution was unaltered by impeded ectomycorrhizal competition (Fig. 2), indicating that ectomycorrhizal fungi did not impose any major constraint on the realised niche of the saprotrophs. This result contradicts previous observations of increased abundance of saprotrophic Agaricomycetes after clear-cutting of boreal pine forests [26] or under more nutrient rich conditions [6]. Here, the shallow distribution of litter saprotrophs rather seemed to reflect a limited fundamental niche. Saprotrophic colonisation of more decomposed organic matter may be restricted by a low energy return of decomposition, as indicated by the decreasing ratio between energy providing cellulose decomposition (Fig. 3a, b) and energy demanding oxidation (Fig. 3f) at greater soil depth. Laboratory studies have shown that saprotrophs may leave potentially usable, well-decomposed organic matter in the presence of freshly deposited litter [24]. Another possibility is that deeper proliferation of litter saprotrophs after trenching was hindered by other functional guilds, e.g., root ascomycetes associated with field layer vegetation.
We analysed enzyme activities to target the long-term turnover of organic matter at later stages of decomposition, and the most obvious effect of removing roots and associated ectomycorrhizal fungi was a drastic decline in Mn-peroxidase activity in the mor layer (Fig. 3f). Mn-peroxidase activity in the mor layer was previously identified as a principal predictor of below-ground organic matter accumulation in a boreal forest [6]. Further, in an inventory of >2000 forest plots across Sweden, extractable manganese was singled out as the best (negative) predictor of organic stocks in the mor layer, overshadowing other chemical, climatic and biological parameters [44]. These studies highlight the importance of Mn-dependent processes for overall organic matter dynamics in boreal forest soils. The capacity of ectomycorrhizal fungi to exploit organic matter has been questioned due to lack of solid empirical evidence [45], and the general trend is that ectomycorrhizal fungi have lost their genetic capacity to decompose plant litter during their evolution from saprotrophic ancestors [27, 28]. Here we provide evidence from a manipulative field experiment in boreal forest, suggesting that Mn-dependent oxidation of the large pool of organic matter in the mor layer was driven primarily by root-derived energy inputs to ectomycorrhizal fungi. The reduced activities of chitinolytic enzymes after trenching (Fig. 3c,d) could further indicate an active role of ectomycorrhizal fungi in decomposition of fungal necromass (c.f. [9]), but could also be an effect of the over-all decline in fungal biomass.
The idea that ectomycorrhizal fungi may play a central role as facilitators of decomposition was put forward more than a century ago during the initial description of the symbiosis [46]. Recently, spectroscopic analyses have supported biochemical alteration of soil-derived organic compounds exposed to pure cultures of ectomycorrhizal fungi [47]. Field studies have previously suggested a central role of ectomycorrhizal fungi in organic matter dynamics, as DNA-based analyses indicate strong dominance of root-associated fungi over saprotrophs in the mor layers of boreal forests [14, 20]. Mn-peroxidases are extracellular enzymes that have evolved uniquely within the Agaricomycetes [18], including some ectomycorrhizal taxa [48], and play a pivotal role in decomposition of lignin and other phenol-rich macromolecules [49]. On the basis of genetic evidence, gene expression in field samples and spatial correlation between enzyme activity and DNA-markers, it was earlier proposed that production of Mn-peroxidases by certain ectomycorrhizal fungi would enable them to participate actively in mor decomposition in boreal forests [26, 50].
Ectomycorrhizal fungi have been proposed to exploit organic matter in a manner fundamentally different to free-living saprotrophs, primarily as a source of N rather than metabolic C [51]. Here, high activities of cellulolytic enzymes were only observed in fresh litter components close to the surface (Fig. 3a, b), where saprotrophs are known to depend on enzymatic release of glucose and other compounds to support their metabolic demand [52]. In the mor layer, where root-associated fungi dominated (Fig. 2), activities of hydrolytic enzymes were low, regardless of trenching, supporting the conventionally accepted view that mycorrhizal fungi have little saprotrophic capacity (i.e. metabolism based on dead organic matter; [27, 28]). However, we here show that production of Mn-peroxidases may, nevertheless, enable some ectomycorrhizal fungi to play an active role in oxidation of organic matter, while remaining dependent on their biotrophic association with plants for their C supply. Oxidation of complex aromatic macromolecules has been described as a co-metabolic process, involving a net energetic cost for the fungi [53]. With access to host-derived sugars, mycorrhizal fungi would be well suited to perform co-metabolic oxidation of complex, humified organic matter, potentially enabling mobilisation of N sequestered in non-hydrolysable organic complexes [50, 54]. It is, however, important to acknowledge that not all ectomycorrhizal taxa are capable of organic matter oxidation [45]. The genetic capacity to produce Mn-peroxidases appears to be limited to certain evolutionary clades of ectomycorrhizal fungi [27], in particular, the genus Cortinarius [50]. Cortinarius is a highly diverse and ubiquitous mycorrhizal fungal genus in most boreal forests [17], and high abundance of these fungi has been found to correlate negatively with soil C stocks [10]. In the present study, the genus Cortinarius accounted for 27% of the ectomycorrhizal species and 9% of the ectomycorrhizal DNA pool in the mor layer of the control plots. Four years of trenching led to a 76% decrease in the relative abundance of Cortinarius amplicons in the mor layer.
In conclusion, we found that roots and associated mycorrhizal fungi hampered litter decomposition by constraining availability of N to saprotrophs. The results clearly support the ‘Gadgil effect’ (i.e., enhanced decomposition after removal of competing mycorrhizal fungi) in a boreal forest and its underlying mechanism. However, saprotrophs did not exploit the empty niche in the deeper organic layers left by mycorrhizal fungi after trenching, confining the ‘Gadgil effect’ to the surface litter. We propose that saprotrophs were unable to exploit these well decomposed substrates due to a low energy return of decomposition of material with low proportion of hydrolysable compounds, leading to accumulation of organic matter. Decomposition of deeper material is more important than decomposition of surface litter in regulating below-ground storage of organic matter [6, 14]. Since the ‘Gadgil effect’ here was limited to the uppermost litter layer, its importance for the over-all below-ground C sink may be questioned for this system.
In addition, we found experimental evidence for a driving role of ectomycorrhizal fungi in enzymatic oxidation of organic matter at later stages of decomposition. We propose that this process releases N for the benefit of the fungi and their host plants, but the extent to which enzymatic oxidation leads to N release or C solubilisation remains to be investigated. From this study, we cannot draw any conclusions about the overall effect of trenching on decomposition in the mor layer, since the disturbance not only affected C-losses but also disrupted below-ground C input via root litter. Four years of root exclusion had no significant net effect on total C stocks in the mor layer. We postulate that ectomycorrhizal fungi may either hamper or stimulate decomposition by different mechanisms, depending upon stage of decomposition and location in the soil profile.
Electronic supplementary material
Acknowledgements
Bengt Söderström and Gösta Hedberg are acknowledged for providing the study site, Olga Vinnere-Pettersson at SciLifeLab for advice on sequencing, Mikael Brandström-Durling for assistance with sequencing analysis and David Wardle, for commenting on the manuscript.
Compliance with ethical standards
Conflict of interest
The work behind this manuscript was supported by The Swedish Research Council FORMAS, grants 2007-1365 and 2011-1747 to BDL. The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0181-2) contains supplementary material, which is available to authorised users.
References
- 1.Averill C, Turner BL, Finzi AC. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature. 2014;505:543–5. doi: 10.1038/nature12901. [DOI] [PubMed] [Google Scholar]
- 2.Lin G, McCormack ML, Ma C, Guo D. Similar below-ground carbon cycling dynamics but contrasting modes of nitrogen cycling between arbuscular mycorrhizal and ectomycorrhizal forests. New Phytol. 2016;213:1440–51. doi: 10.1111/nph.14206. [DOI] [PubMed] [Google Scholar]
- 3.Orwin KH, St Kirschbaum MUF, John MG, Dickie IA. Organic nutrient uptake by mycorrhizal fungi enhances ecosystem carbon storage: a model‐based assessment. Ecol Lett. 2011;14:493–502. doi: 10.1111/j.1461-0248.2011.01611.x. [DOI] [PubMed] [Google Scholar]
- 4.Gadgil RL, Gadgil PD. Mycorrhiza and litter decomposition. Nature. 1971;233:133. doi: 10.1038/233133a0. [DOI] [PubMed] [Google Scholar]
- 5.Averill C, Hawkes CV. Ectomycorrhizal fungi slow soil carbon cycling. Ecol Lett. 2016;19:937–47. doi: 10.1111/ele.12631. [DOI] [PubMed] [Google Scholar]
- 6.Kyaschenko J, Clemmensen KE, Karltun E, Lindahl BD. Guild interaction within fungal communities regulates below-ground organic matter accumulation along a boreal forest fertility gradient. Ecol Lett. 2017 doi: 10.1111/ele.12862. [DOI] [PubMed] [Google Scholar]
- 7.Hartley IP, Garnett MH, Sommerkorn M, Hopkins DW, Fletcher BJ, Sloan VL, et al. A potential loss of carbon associated with greater plant growth in the European Arctic. Nature. Clim Change. 2012;2:875–9. doi: 10.1038/nclimate1575. [DOI] [Google Scholar]
- 8.Subke JA, Voke NR, Leronni V, Garnett MH, Ineson P. Dynamics and pathways of autotrophic and heterotrophic soil CO2 efflux revealed by forest girdling. J Ecol. 2011;99:186–93. doi: 10.1111/j.1365-2745.2010.01740.x. [DOI] [Google Scholar]
- 9.Brzostek ER, Dragoni D, Brown ZA, Phillips RP. Mycorrhizal type determines the magnitude and direction of root-induced changes in decomposition in a temperate forest. New Phytol. 2015;206:1274–82. doi: 10.1111/nph.13303. [DOI] [PubMed] [Google Scholar]
- 10.Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD. Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol. 2015;205:1525–36. doi: 10.1111/nph.13208. [DOI] [PubMed] [Google Scholar]
- 11.Parker T, Subke JA, Wookey PA. Rapid carbon turnover beneath shrub and tree vegetation is associated with low soil carbon stocks at a subarctic treeline. Glob Change Biol. 2015;21:2070–81. doi: 10.1111/gcb.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez CW, Kennedy PG. Revisiting the ‘Gadgil effect’: do interguild fungal interactions control carbon cycling in forest soils? New Phytol. 2016;209:1382–94. doi: 10.1111/nph.13648. [DOI] [PubMed] [Google Scholar]
- 13.Pan Y, Birdsey RA, Fang J, Houghton R, Kauppi PE, Kurz WA, et al. A large and persistent carbon sink in the World’s forests. Science. 2011;333:988–93. doi: 10.1126/science.1201609. [DOI] [PubMed] [Google Scholar]
- 14.Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, et al. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science. 2013;339:1615–8. doi: 10.1126/science.1231923. [DOI] [PubMed] [Google Scholar]
- 15.Wardle DA, Hörnberg G, Zackrisson O, Kalela-Brundin M, Coomes DA. Long-term effects of wildfire on ecosystem properties across an island area gradient. Science. 2003;300:972–5. doi: 10.1126/science.1082709. [DOI] [PubMed] [Google Scholar]
- 16.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecol Lett. 2009;12:1238–49. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 17.Sterkenburg E, Bahr A, Brandström–Durling M, Clemmensen KE, Lindahl BD. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015;207:1145–58. doi: 10.1111/nph.13426. [DOI] [PubMed] [Google Scholar]
- 18.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, et al. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–9. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 19.Cooke RC, Rayner ADM. Ecology of saprotrophic fungi. London: Longman; 1984. [Google Scholar]
- 20.Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Högberg P, Finlay RD. Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 2007;173:611–20. doi: 10.1111/j.1469-8137.2006.01936.x. [DOI] [PubMed] [Google Scholar]
- 21.Bödeker ITM, Lindahl BD, Olson Aring, Clemmensen KE. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct Ecol. 2016;30:1967–78. doi: 10.1111/1365-2435.12677. [DOI] [Google Scholar]
- 22.Leake JR, Donnelly DP, Saunders EM, Boddy L, Read DJ. Rates and quantities of carbon flux to ectomycorrhizal mycelium following 14C pulse labeling of Pinus sylvestris seedlings: effects of litter patches and interaction with a wood-decomposer fungus. Tree Physiol. 2001;21:71–82. doi: 10.1093/treephys/21.2-3.71. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl B, Stenlid J, Finlay R. Effects of resource availability on mycelial interactions and 32P-transfer between a saprotrophic and an ectomycorrhizal fungus in soil microcosms. FEMS Microbiol Ecol. 2001;38:43–52. doi: 10.1111/j.1574-6941.2001.tb00880.x. [DOI] [Google Scholar]
- 24.Boberg JB, Finlay RD, Stenlid J, Ekblad A, Lindahl BD. Nitrogen and carbon reallocation in fungal mycelia during decomposition of boreal forest litter. PLoS One. 2014;9:e92897. doi: 10.1371/journal.pone.0092897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhupinderpal-Singh NordgrenA, Ottosson Löfvenius M, Högberg MM, Mellander PE, Högberg P. Tree root and soil heterotrophic respiration as revealed by girdling of boreal Scots pine forest: extending observations beyond the first year. Plant Cell Environ. 2003;26:1287–96. doi: 10.1046/j.1365-3040.2003.01053.x. [DOI] [Google Scholar]
- 26.Kyaschenko J, Clemmensen KE, Hagenbo A, Karltun E, Lindahl BD. Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME J. 2017;11:863–74. doi: 10.1038/ismej.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–5. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 28.Martin F, Kohler S, Murat C, Veneault-Fourrey C, Hibbett DS. Unearthing the roots of ectomycorrhizal symbioses. Nat Rev Microbiol. 2016;14:760–73. doi: 10.1038/nrmicro.2016.149. [DOI] [PubMed] [Google Scholar]
- 29.Romell LG. A trenching experiment in a spruce forest and its bearing on problems of mycotrophy. Sven Bot Tidskr. 1938;32:89–99. [Google Scholar]
- 30.Clemmensen KE, Ihrmark K, Brandström-Durling M, Lindahl BD. Sample preparation for fungal community analysis by high-throughput sequencing of barcode amplicons. In: Martin F, Uroz S, editors. Microbial Environmental Genomics (MEG) New York: Springer; 2016. pp. 61–88. [Google Scholar]
- 31.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. A Guide to Methods and Applications. Cambridge: Elsevier; 1990. pp. 315–22. [Google Scholar]
- 32.Ihrmark K, Bödeker IT, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, et al. New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 33.Frank DN. BARCRAWL and BARTAB: software for design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinforma. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 36.Abarenkov K, Tedersoo L, Nilsson RH, Vellak K, Saar I, Veldre V, et al. PlutoF-a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol Bioinform. 2010;6:189–96. doi: 10.4137/EBO.S6271. [DOI] [Google Scholar]
- 37.Nylund JE, Wallander H. Ergosterol analysis as a means of quantifying mycorrhizal biomass. Method Microbiol. 1992;24:77–88. doi: 10.1016/S0580-9517(08)70088-6. [DOI] [Google Scholar]
- 38.Saiya-Cork KR, Sinsabaugh RL, Zak DR. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem. 2002;34:1309–15. doi: 10.1016/S0038-0717(02)00074-3. [DOI] [Google Scholar]
- 39.Lindahl BD, de Boer W, Finlay RD. Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 2010;4:872–81. doi: 10.1038/ismej.2010.19. [DOI] [PubMed] [Google Scholar]
- 40.Högberg P, Högbom L, Schinkel H, Högberg M, Johannisson C, Wallmark H. 15N abundance of surface soils, roots and mycorrhizas in profiles of European forest soils. Oecologia. 1996;108:207–14. doi: 10.1007/BF00334643. [DOI] [PubMed] [Google Scholar]
- 41.Hobbie EA, Colpaert JV. Nitrogen availability and colonization by mycorrhizal fungi correlate with nitrogen isotope patterns in plants. New Phytol. 2003;157:115–26. doi: 10.1046/j.1469-8137.2003.00657.x. [DOI] [PubMed] [Google Scholar]
- 42.Näsholm T, Högberg P, Franklin O, Metcalfe D, Keel SG, Campbell C, et al. Are ectomycorrhizal fungi alleviating or aggravating nitrogen limitation of tree growth in boreal forests? New Phytol. 2013;198:214–21. doi: 10.1111/nph.12139. [DOI] [PubMed] [Google Scholar]
- 43.Baldrian P, Kolařík M, Stursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–58. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stendahl J, Berg B, Lindahl BD. Manganese availability is negatively associated with carbon storage in northern coniferous forest humus layers. Sci Rep. 2017;7:15487. doi: 10.1038/s41598-017-15801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellitier PT, Zak DR. Ectomycorrhizal fungi and the enzymatic liberation of nitrogen from soil organic matter: why evolutionary history matters. New Phytol. 2017; 10.1111/nph.14598. [DOI] [PubMed]
- 46.Frank B. Die Bedeutung der Mykorhiza Pilze für die gemeine Kiefer. Forstwiss Cent. 1894;16:185–90. doi: 10.1007/BF01842241. [DOI] [Google Scholar]
- 47.Shah F, Nicolás C, Bentzer J, Ellström M, Smits M, Rineau F, et al. Ectomycorrhizal fungi decompose soil organic matter using oxidative mechanisms adapted from saprotrophic ancestors. New Phytol. 2016;209:1705–19. doi: 10.1111/nph.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bödeker ITM, Nygren CMR, Taylor AFS, Olson Aring, Lindahl BD. ClassII peroxidase encoding genes are present in a wide phylogenetic range of ectomycorrhizal fungi. ISME J. 2009;3:1387–95. doi: 10.1038/ismej.2009.77. [DOI] [PubMed] [Google Scholar]
- 49.Sinsabaugh RL. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol Biochem. 2010;42:391–404. doi: 10.1016/j.soilbio.2009.10.014. [DOI] [Google Scholar]
- 50.Bödeker ITM, Clemmensen KE, de Boer W, Martin F, Olson Aring, Lindahl BD. Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytol. 2014;203:245–56. doi: 10.1111/nph.12791. [DOI] [PubMed] [Google Scholar]
- 51.Lindahl BD, Tunlid A. Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015;205:1443–7. doi: 10.1111/nph.13201. [DOI] [PubMed] [Google Scholar]
- 52.Šnajdr J, Valášková V, Merhautová V, Herinková J, Cajthaml T, Baldrian P. Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil. Soil Biol Biochem. 2008;40:2068–75. doi: 10.1016/j.soilbio.2008.01.015. [DOI] [Google Scholar]
- 53.Kirk TK, Farrell RL. Enzymatic combustion: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 54.Rineau F, Shah F, Smits MM, Persson P, Johansson T, Carleer R, et al. Carbon availability triggers the decomposition of plant litter and assimilation of nitrogen by an ectomycorrhizal fungus. ISME J. 2013;7:2010–22. doi: 10.1038/ismej.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.