Abstract
Disease-suppressive soils are ecosystems in which plants suffer less from root infections due to the activities of specific microbial consortia. The characteristics of soils suppressive to specific fungal root pathogens are comparable to those of adaptive immunity in animals, as reported by Raaijmakers and Mazzola (Science 352:1392–3, 2016), but the mechanisms and microbial species involved in the soil suppressiveness are largely unknown. Previous taxonomic and metatranscriptome analyses of a soil suppressive to the fungal root pathogen Rhizoctonia solani revealed that members of the Burkholderiaceae family were more abundant and more active in suppressive than in non-suppressive soils. Here, isolation, phylogeny, and soil bioassays revealed a significant disease-suppressive activity for representative isolates of Burkholderia pyrrocinia, Paraburkholderia caledonica, P. graminis, P. hospita, and P. terricola. In vitro antifungal activity was only observed for P. graminis. Comparative genomics and metabolite profiling further showed that the antifungal activity of P. graminis PHS1 was associated with the production of sulfurous volatile compounds encoded by genes not found in the other four genera. Site-directed mutagenesis of two of these genes, encoding a dimethyl sulfoxide reductase and a cysteine desulfurase, resulted in a loss of antifungal activity both in vitro and in situ. These results indicate that specific members of the Burkholderiaceae family contribute to soil suppressiveness via the production of sulfurous volatile compounds.
Introduction
The rhizosphere microbiome is our silent ally in various ecosystem services, including protection of plants against abiotic and biotic stresses. Exploration of this largely untapped microbial resource is of increasing importance for crop production to stimulate plant growth and health while reducing fertilizer and pesticide inputs. Composition and activity of the rhizosphere microbiome is influenced by the plant through the release of root exudates that recruit and/or activate commensal microorganisms for protection against invading pathogens, either directly via competition and antibiosis or indirectly via induction of the plant defense system [1–3].
The best examples of microbiome-mediated protection of plants against root pathogens are disease-suppressive soils. These are “soils in which a pathogen does not establish or persist, establishes but causes little or no disease, or establishes and causes disease at first but then the disease declines” [4]. Suppressive soils have been described for plant pathogenic fungi, oomycetes, bacteria, nematodes, and parasitic weeds. While soil physicochemical properties modulate the onset and intensity of the disease-suppressive state, functionality of the response is in most cases (micro)biological in nature [4, 5]. The specific suppression that operates in these soils can be eliminated by soil pasteurization or biocides and can be transferred to non-suppressive (conducive) soils via soil transplantations [6, 7]. The characteristics of soils that are suppressive to specific fungal root pathogens are comparable to the characteristics of innate and adaptive immunity in animals [5]. Similar to the requirement of specialized cell types in adaptive immunity in animals, induced suppressiveness of soils to fungal root pathogens requires the enrichment and activation of select microbial genera and antagonistic traits that interfere at some stages of the pathogen infection cycle [5]. For most disease-suppressive soils, however, the responsible microorganisms and underlying mechanisms involved in the soil suppressiveness are largely unknown.
Over the past 7 years, we have started to unravel the role and identity of microbial consortia in disease suppressiveness of soils to the fungus Rhizoctonia solani, a devastating root pathogen of multiple crops including sugar beet, wheat, potato, and rice. Initial PhyloChip-based taxonomic analyses of the rhizosphere of sugar beet plants grown in Rhizoctonia-suppressive soil pinpointed several bacterial genera that were consistently associated with the disease-suppressive state, in particular bacterial genera belonging to the Gammaproteobacteria, Actinobacteria, and Betaproteobacteria [6, 8]. Subsequent isolations and functional analyses showed that, for the Gammaproteobacteria, Pseudomonas species contribute to disease suppressiveness via the production of the NRPS-encoded lipopeptide thanamycin [9]. Work on the potential role of Lysobacter species (Gammaproteobacteria) demonstrated that, in spite of their extensive metabolic potential and antifungal activities in vitro [10], none of the species tested were able, either alone or in a consortium, to consistently suppress Rhizoctonia damping-off disease in situ [11, 12]. Among the Actinobacteria, the Streptomyces species showed significant inhibition of hyphal growth of R. solani via the production of volatile organic compounds (VOCs), in particular the VOCs methyl 2-methylpentanoate and 1,3,5-trichloro-2-methoxybenzene [13].
For the Betaproteobacteria, taxonomic analyses combined with metatranscriptomics of the rhizosphere microbiome showed that members of the Burkholderiaceae family were significantly more abundant and more active in Rhizoctonia-suppressive soil [14]. The Burkholderiaceae family is ubiquitous in diverse environments and associated with different plant species [15–17], as well as with fungi [18–22]. They are metabolically versatile and can inhibit fungal growth through the production of various secondary metabolites [23], including VOCs [24, 25]. To resolve the potential role of members of the Burkholderiaceae family in disease suppressiveness, we isolated, sequenced, and functionally characterized one Burkholderia and four Paraburkholderia species from the rhizosphere of sugar beet plants grown in Rhizoctonia-suppressive soil. Combining comparative genomics, VOC profiling and site-directed mutagenesis, we identified unique genes in Paraburkholderia graminis PHS1 associated with the production of sulfurous VOCs, with antifungal activity in vitro and disease suppression in situ.
Materials and methods
Semi-selective isolation of Burkholderiaceae family members
The suppressive soil used in this study was collected in 2003 and 2004 from an agricultural sugar beet field close to the town of Hoeven, the Netherlands (51°35′10″N 4°34″44′E). Briefly, sugar beet seeds (cultivar Alligator) were sown in square PVC pots containing 250 g of field soil with an initial moisture content of 10% (v/w). Plants were grown in a growth chamber (24 °C/24 °C day/night temperatures; 180 μmol light m−2 s−1 at plant level during 16 h/d; 70% relative humidity) and watered weekly with standard Hoagland solution (macronutrients only). After three weeks of plant growth, 1 g of sugar beet roots with tightly adhering soil was suspended in 5 mL of potassium-phosphate buffer (pH 7.0). Samples were vortexed and sonicated for 1 min and rhizospheric soil was stored in glycerol (40%, v/v) stocks at −80 °C [6]. Burkholderia and Paraburkholderia spp. were isolated by dilution plating these glycerol stocks on specific agar media. Selective media initially described as ‘Pseudomonas cepacia azelaic with and without acid tryptamine’ (PCAT and PCATm, respectively) was used to enrich for different species of Burkholderia and Paraburkholderia as previously described [26, 27]. Single colonies were picked based on morphology and purified on fresh agar plates. Isolates were stored in glycerol (40%, v/v) at −80 °C.
Identification and phylogeny of Burkholderia and Paraburkholderia spp.
Putative Burkholderia and Paraburkholderia isolates were initially selected and characterized based on colony morphology and based on 16S rRNA sequencing using a specific set of primers (Supplementary Table S1) for Burkholderia–Ralstonia [28, 29]. This set of primers allowed us to identify two Burkholderia and twenty-seven Paraburkholderia isolates out of 102 colonies. To further identify the species, we sequenced the housekeeping genes lepA (GTP binding protein) [30] and recA (Recombinase A) [31] (Supplementary Table S1) and performed phylogenetic analyses. For obtaining DNA, bacterial cells were disrupted by heating at 95 °C for 10 min. Suspensions were centrifuged at 13,000 rpm for 10 min. After centrifugation, 2 μl of the supernatants were used for the PCR reactions. PCR products were purified and sequenced at Macrogen Europe, Amsterdam. Isolates were characterized based on sequence identity with 16S rRNA gene sequences in the Greengenes database [32] (http://greengenes.lbl.gov/). Partial sequences of lepA (500 bp), recA (423 bp), and 16S rRNA (516 bp) genes of Burkholderia were concatenated to yield an alignment of 1439 sites. A concatenated phylogenetic tree supplemented with sequences of Burkholderia strains with a sequenced genome (NCBI database) was constructed using UPGMA with the Tamura-3 parameter calculation model with gamma distribution and 1000 bootstrap replicates.
Coupling Burkholderia and Paraburkholderia isolates to OTUs detected by PhyloChip
16S rRNA gene sequences of twenty-nine isolates were compared with the 16S rRNA gene sequences of Burkholderia OTUs previously identified by PhyloChip-based metagenomic analysis [6]. Phylogenetic analysis was performed with Muscle in MEGA6 [33] and iTOL [34] (http://itol.embl.de/). A neighbor-joining consensus tree [35] with 1000 bootstrap replicates [36] was constructed using the Tamura-Nei model [37] with gamma distribution.
In vitro antifungal activity of Burkholderiaceae family members
Antifungal activity against R. solani was determined by spotting 5 μl suspension of washed cells (1 × 108 CFU mL−1) of the Burkholderiaceae strains at the border of 90-mm-diameter Petri dishes containing R2A (Merck), Tryptic Soy Agar (TSA,Oxoid), potato dextrose agar (PDA; Difco), 1/5th strength PDA, or 1/10th strength TSA medium. Plates were incubated for three days at 25 °C. After that, a 5-mm-diameter mycelial plug of the fungus (grown on PDA medium for 2–3 days at 25 °C) was placed in the center of each plate. Hyphal growth inhibition was determined after 3–7 days of incubation at 25 °C. Plates containing only the fungal plug were used as control.
VOC-mediated antifungal activity
To test the effect of VOCs produced by B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 on hyphal growth of the fungus R. solani, the bottoms of two 90-mm-diameter Petri dishes were connected in a bottom-top-design to allow physical separation between the bacteria and the fungus, as previously described [13]. The bottom contained a bacterial isolate growing on PDA medium, previously pre-incubated at 25 °C for three days. The top plate contained a 5-mm-diameter plug of R. solani mycelium on PDA. Both Petri dishes were connected, sealed and incubated at 25 °C. As a control, the Petri dish containing R. solani was exposed to a Petri dish containing PDA medium only. Fungal growth inhibition was calculated by measuring the radial hyphal growth after one, two, and three days of incubation. Percentage of inhibition was calculated as ((diameter of fungus in control − diameter of fungus exposed to bacterial VOCs)*100/diameter of fungus in control) for each of the three replicates. Statistically significant differences were determined by one-way ANOVA and Tukey HSD analysis in R.
Collection and analysis of VOCs
Next to the antifungal assays, bacterial VOCs emitted by B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 were collected and analyzed. For trapping of VOCs emitted by the bacteria, a volume of 100 μl of bacterial cell suspension was spread on PDA (20 mL) in glass Petri dishes designed for headspace volatile trapping [38]. The Petri dishes were closed by a lid with an outlet connected to a steel trap containing 150 mg Tenax TA and 150 mg Carbopack B (Markes International, Ltd., Llantrisant, UK). All treatments were in triplicate. The VOCs were collected during four days of incubation and the Tenax steel traps were stored at 4 °C until GC-Q-TOF analysis. The trapped VOCs were desorbed from the traps using an automated thermodesorption unit (Unity TD-100, Markes International, Ltd., Llantrisant, UK) at 210 °C for 12 min (He flow 50 mL/min) and trapped on a cold trap at −10 °C. The trapped VOCs were introduced into the GC-QTOF (model Agilent 7890B GC and the Agilent 7200 A QTOF, Santa Clara, CA, USA) by heating the cold trap for 3 min to 280 °C. Split ratio was set to 1:10 and the column used was a 30 mm × 0.25 mm ID RXI-5MS, film thickness 0.25 μm (Restek 13424–6850, Bellefonte, PA, USA). Temperature program used was as follows: 39 °C for 2 min, from 39 to 95 °C at 3.5 °C min−1, then to 165 °C at 6 °C min−1, to 250 °C at 15 °C min−1, and finally to 300 °C at 40 °C min−1, hold 20 min. The VOCs were detected by the MS operating at 70 eV in EI mode. Mass spectra were acquired in full-scan-mode (30–400 AMU, 4 scans/s). Mass spectra were extracted with MassHunter Qualitative Analysis Software V B.06.00 Build 6.0.633.0 (Agilent Technologies, Santa Clara, CA, USA) using the GC-Q-TOF qualitative analysis module. GC-MS raw data were processed by an untargeted metabolomics approach. MetAlign software [39] was used to extract and align the mass features (s/n = 3). MSClust was used to remove signal redundancy per metabolite and to reconstruct compound mass spectra as previously described [40]. VOCs were tentatively annotated by comparing their mass spectra with those of reference databases (NIST14 and WILEY11th Edition). Linear retention indices (RI) of VOCs were calculated as previously described [41] and compared with those in the literature. Processed VOC data were log-transformed and pareto scaled [42]. The data were subjected to multivariate statistical analysis. Normality and homogeneity of variances were checked using Shapiro–Wilk test and Levene’s test. The transformed and scaled GC/MS data was converted to a BIOM file using QIIME and statistical analysis was performed, using the non-parametric Kruskal–Wallis test [43], to calculate statistically significant differences for any given peak between the medians of the different biological classes [44]. P-values were corrected by the Benjamini–Hochberg FDR procedure for multiple comparisons [45]. To consider VOCs significantly different between treatments, a FDR < 0.1 and the absolute ratio of the log2Ratio ≤ or ≥ 1.1 (i.e., at least 2.1 × higher than the control) were used as thresholds. To evaluate the chemical diversity of the VOCs produced by the five selected Burkholderiaceae family members, we employed diverse MS-based informatic approaches, including multivariate analyses, such as principal component analysis (PCA) and partial least squares discriminant analysis, (PLS-DA) [46, 47] using the R package MixOmics [48]. Figures were generated using several custom scripts and packages in Rstudio (R [49]). Data, scripts and code used for statistical and bioinformatic analyses are available at: https://zenodo.org/record/1163754#.WnJaFOdG1eW.
In vivo antifungal activity of Burkholderia and Paraburkholderia spp. and root colonization assay
Selected strains were grown in 10 mL of Luria-Bertani (LB, Difco) for two days at 25 °C on a rotary shaker at 200 rpm. Cultures were centrifuged, washed 3 times, and resuspended in 0.9% NaCl. Cell suspensions were mixed, with Rhizoctonia-conducive soil for the initial screening of the isolated Burkholderia and Paraburkholderia spp. or with gamma-irradiated conducive soil for comparison of P. graminis PHS1 and the cd and dsr mutants, at an initial density of 107 CFU g−1 soil and ~10% hydration (v/w). Plastic trays (19.5 × 6 × 3.5 cm) were filled with 250 g of the conducive soil (eight biological replicates per treatment) and 16 sugar beet seeds coated with thiram, hymexazol, and poncho-beta were sown in a row, 1 cm apart. Soil not inoculated with the Burkholderia or Paraburkholderia strains was used as a control. Trays were placed in boxes with transparent lids in a growth chamber at 24 °C with a 16 h photoperiod. After five days, seeds germinated and a single fresh plug (5-mm-diameter) of R. solani AG2-2 IIIB previously grown on 1/5th PDA agar was placed ~1 cm below the soil surface close to the roots of the first seedling, with the mycelial side toward the plant to allow infection of the first seedling. Spread of R. solani through the row of seedlings was scored at regular intervals during two weeks by scoring the number of diseased plants, as well as the distance between the inoculum and the most distal plant showing the typical damping-off symptoms caused by R. solani. For the root colonization assays with P. graminis and the cd and dsr mutants, rhizosphere samples were processed as described above. Serial dilutions were made and hundred microliters were plated on TSA agar dishes (without, for P. graminis PHS1 and with kanamycin at 25 µg mL−1, for the cd and dsr mutants) and incubated at 25 °C for one week and the amount of CFU per gram of root were determined by colony counting. Statistically significant differences between treatments were determined by One-way ANOVA and Tukey HSD in R.
Isolation and sequencing of genomic DNA
A single colony of B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 was inoculated in LB broth medium and grown for two days at 25 °C. Bacterial cell suspensions were washed three times with 0.9 % NaCl and subsequently genomic DNA was extracted using the Master PureTM kit (Epicentre) according to manufacturer’s protocol, excluding the beat beating step. Genomic DNA of strains B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 was sequenced using the Pacific Biosciences (PacBio) RS sequencing platform. From the genomic DNA, 20 Kb insert size libraries were prepared and size selected using Blue Pippin™ and sequenced using C3 in combination with P5 polymerase chemistry for 2 SMRT cells per genome with 180 min or longer movie time and stage start. The 20-kb continuous long-read (CLR) data were de novo assembled using the PacBio hierarchical genome assembly process (HGAP3)/ Quiver software package version 2.2.0 [50]. The accession numbers for the genome sequences can be found the Supplementary Table S2.
Comparative genomics
Annotation was performed with a modified version of Prokka [51]. COG annotations for the predicted proteins were determined using KOGnitor [52]. Pan-core genome analyses were performed by clustering together the protein coding genes from the five Burkholderia and Paraburkholderia spp. using CD-HIT [53] with word length 3 (-n 3), global identity (-G 1), and a minimal alignment coverage of 60% for the shortest protein (-aS 0.6). CD-HIT clusters were parsed into a presence-absence matrix from which the pan, core, and variable genomes were inferred using custom scripts. Analyses of genes and/or gene clusters encoding the production of antibiotics and secondary metabolites were conducted using antiSMASH 3.0 [54]. 185 Burkholderia and Paraburkholderia genomes were downloaded from the NCBI database. A PHYML maximum likelihood phylogenetic tree of a concatenated alignment of 12 marker genes (dnaG, frr, nusA, pyrG, rplA, rplN, rpmA, rpsB, rpsE, rpsM, smpB, and tsf) was built using AMPHORA [55].
Construction of Paraburkholderia graminis PHS1 mutants
Single mutants in two selected genes putatively involved in sulfur metabolism (i.e., dimethyl sulfoxide reductase, dsr gene and the cysteine desulfurase, cd gene) were constructed in P. graminis PHS1 by allelic replacement of the targeted gene with a kanamycin resistant gene using the protocol described by Zumaquero et al. [56]. For the cd and dsr mutants, the mutation was confirmed by PCR amplification of the flanking regions using an external primer to the region involved in the double recombination and an internal primer from the kanamycin resistant gene introduced by double recombination into the target genes cd or dsr. The primers used for the mutagenesis and for confirming the integration are described in Supplementary Table S1.
Results and discussion
Diversity and abundance of Burkholderiaceae in Rhizoctonia-suppressive soil
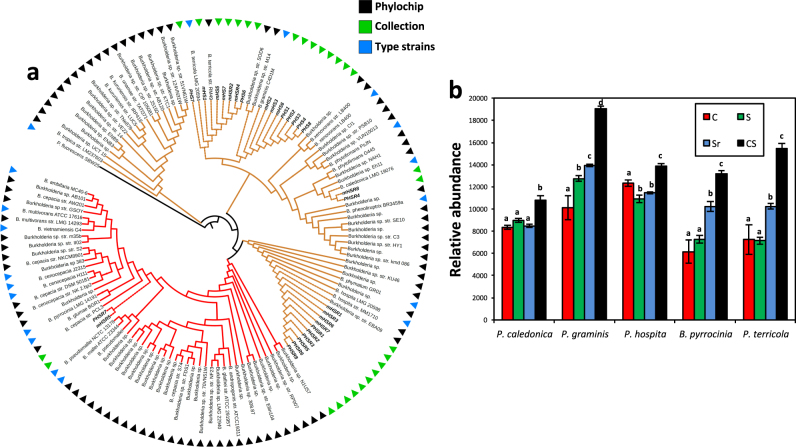
PhyloChip-based metagenomics detected more than 33,000 bacterial and archaeal OTUs in the rhizosphere of sugar beet seedlings grown in the Rhizoctonia-suppressive soil and revealed bacterial groups consistently associated with the disease-suppressive state [6]. Furthermore, metatranscriptomics [14] of the rhizosphere microbiome confirmed that members of the Burkholderiaceae family were significantly more abundant and more active on roots of sugar beet seedlings grown in Rhizoctonia-suppressive soil as compared to the conducive soil. Using the semi-selective media PCAT and PCATm [26, 27] followed by PCR-based analyses, twenty-nine Burkholderia and Paraburkholderia isolates were obtained from the rhizosphere of sugar beet plants grown in Rhizoctonia-suppressive soil. Based on sequence similarities (95–100%) to 16S rRNA gene sequences available in the Greengenes database (also used as reference in the original PhyloChip analysis by Mendes et al. [6], the isolates were classified into five species: B. pyrrocinia (n = 2), P. caledonica (n = 2), P. graminis (n = 8), P. hospita (n = 10) and P. terricola (n = 7) (Fig. 1a). To further support this phylogenetic delineation, we also sequenced the housekeeping genes lepA and recA. Concatenation of lepA, recA, and 16S rRNA sequences confirmed, at a higher resolution, the taxonomic classification of the isolates into five species (Supplementary Figure S1). Further phylogenetic analyses allowed crosslinking, based on sequence identity of at least 97%, these five Burkholderia and Paraburkholderia species with the Burkholderia OTUs (n = 83) originally detected in Rhizoctonia-suppressive soil by PhyloChip analysis (Fig. 1b; Supplementary Table S3). Using this crosslink threshold, we calculated the relative abundance of each of the five Burkholderia/Paraburkholderia species in the four different soil treatments used in the initial study by Mendes et al. [6]. The results showed that only P. graminis OTUs were significantly more abundant in Rhizoctonia-suppressive (S) than in conducive (C) soil (ANOVA, P < 0.005; Fig. 1b). The relative abundance of P. graminis further increased when the suppressive soil was challenged with the fungal pathogen R. solani (Sr); also the relative abundance of B. pyrrocinia and P. terricola increased significantly in Sr compared to their relative abundance in C and S soils (ANOVA, P < 0.005; Fig. 1b). In the transplantation soil (CS: conducive soil mixed with 10% (w/w) suppressive soil), the relative abundance of all five species was significantly higher than in the conducive (C) soil (Fig. 1b). Collectively, these results show that a higher relative abundance of several Burkholderia/Paraburkholderia species and in particular of P. graminis correlated with the disease-suppressive state of the soil. It should be noted, however, that the particular strains isolated in the present study may not constitute a full representation of each species in the soil. Additional phylogenetic markers will be needed in future experiments to determine the abundance of these species at strain level to better correlate their abundance with the disease-suppressive state of soils.
Fig. 1.
a Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences of the Burkholderia and Paraburkholderia collection obtained in this study (in green triangles), Burkholderia detected by Phylochip analysis (in black triangles) and Burkholderia and Paraburkholderia type strains (in blue triangles). Red lines represent isolates from the B. cepacia complex, brown: environmental isolates, black: the outgroup (P. fluorescens SBW25). b Relative abundance (based on phylochip signal intensity) of OTUs classified in Mendes et al. [6] as B. pyrrocinia, P. caledonica, P. graminis, P. hospita and P. terricola in conducive (C), suppressive (S), suppressive challenged with R. solani (Sr), conducive soil supplemented with 10% of suppressive soil (CS). Crosslinking of these five Burkholderia and Paraburkholderia species with the Burkholderia OTUs was based on a sequence identity of at least 97% (See Supplementary Table S3). Statistically significant differences between the relative abundance of each Burkholderia/Paraburkholderia species for the different soils as compared with the conducive soil were determined by one-way ANOVA (P < 0.05). Four replicates were used. Different letters indicate statistically significant differences (Fisher LSD test)
In vivo antifungal activity of Burkholderia and Paraburkholderia species
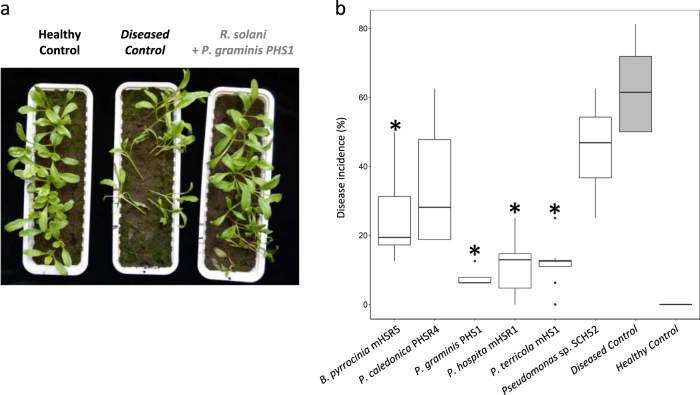
To resolve the potential role of each of the selected Burkholderia and Paraburkholderia species in Rhizoctonia-suppressive soil, three independent bioassays were performed to determine their efficacy to control Rhizoctonia damping-off disease of sugar beet seedlings. To this end, suspensions of selected strains of each of the five Burkholderia and Paraburkholderia species were introduced into the conducive soil to final densities of 1 × 107 CFU g−1 soil. The results showed that all tested strains significantly suppressed damping-off disease albeit at different levels (Fig. 2). P. graminis, P. terricola, and P. hospita provided the highest level of disease control compared to the B. pyrrocinia and P. caledonica strains, and compared to Pseudomonas spp. SH-C52, the strain previously shown to contribute to Rhizoctonia-suppressiveness via the production of the NRPS-encoded chlorinated lipopeptide thanamycin [6, 9, 57].
Fig. 2.
Suppression of Rhizoctonia damping-off disease by B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 in conducive field soil. a Representation of the in vivo bioassay to determine the ability of Burkholderia and Paraburkholderia strains to suppress damping-off disease of sugar beet seedlings caused by R. solani. At one side of the tray (indicated with the black dot), a mycelial plug of the fungal pathogen is point-inoculated at 1 cm underneath the soil surface at the edge of the tray. Within a time period of 2–3 weeks, R. solani progressively infects sugar beet seedlings positioned in a 20-cm row with a 1-cm distance between the seedlings. The level of disease suppression is quantified by measuring the distance of the disease spread as indicated by seedlings with damping-off symptoms. b Box plot showing the spread of damping-off disease of sugar beet seedlings in conducive soil that is untreated (Healthy Control), treated with the pathogen R. solani only (Diseased Control), treated with Pseudomonas sp. strain SHC52, or each of the five Burkholderia and Paraburkholderia strains inoculated in soil (107 cells g−1 soil) one day prior to pathogen inoculation (mean values ± SEM, n = 8). An asterisk indicates a statistically significant difference (Student’s t-Test, P < 0.05, n = 8) between the bacterial treatments and the treatment with the pathogen only (Diseased Control)
Functional characterization of Burkholderia and Paraburkholderia species
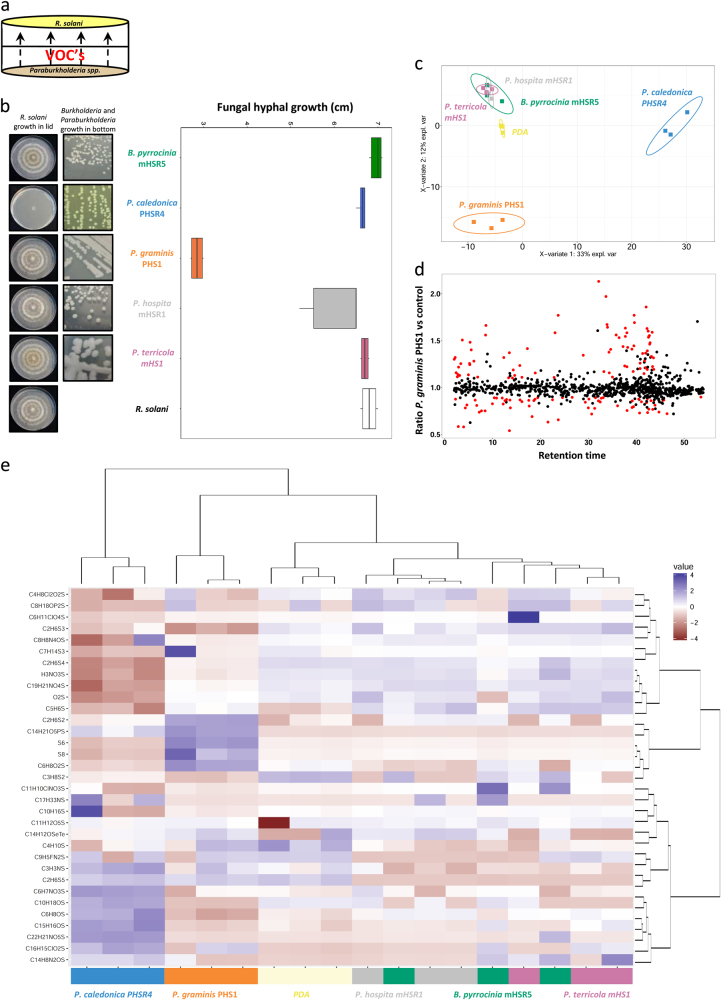
To elucidate the potential mechanism by which each of the five Burkholderia and Paraburkholderia species control R. solani, we first tested their extracellular enzymatic activities and their ability to inhibit hyphal growth of R. solani in vitro. None of the isolates showed extracellular chitinase activity, whereas all exhibited xylanase activity (Supplementary Table S4). The two B. pyrrocinia isolates were positive for four out of six extracellular enzymatic activities, whereas the other species were only positive for one or two enzymatic activities (Supplementary Table S4). The ability of the Burkholderia isolates to inhibit hyphal growth of R. solani in vitro was tested on various agar media in dual culture assay (Supplementary Table S4) and in an so-called sandwich plate assays to screen for antifungal activity via VOCs (Fig. 3a, b and Supplementary Table S4). Surprisingly, none of the isolates inhibited hyphal growth of R. solani in the dual culture assays via the production of diffusible antimicrobial compounds on the media tested (Supplementary Table S4). However, VOCs produced by P. graminis inhibited hyphal growth of R. solani by ~70%, as compared to the control, whereas none of the other species/strains tested exhibited antifungal activity via VOCs (Fig. 3a, b). Subsequent headspace analysis of cultures of representative strains of all five Burkholderia and Paraburkholderia species resulted in the detection of 1084 putatively annotated VOCs. Statistical analyses based on partial least squares discriminant analysis (PLS-DA) further revealed distinct differences between the VOCs emitted by P. graminis PHS1 and P. caledonica PHSR4 with the other species (Fig. 3c). Since P. graminis PHS1 was the only isolate that inhibited hyphal growth of R. solani via VOCs, we compared the VOC profile of this strain with the other strains. Out of the 1084 VOCs detected for all strains, 158 VOCs were significantly different for P. graminis PHS1 using a FDR < 0.1 and the absolute ratio of the log2Ratio ≤ or ≥ 1.1 (i.e., at least 2.1 × higher than the control) as thresholds (Fig. 3d). Among these 158 VOCs, the sulfurous VOCs were numerous and included, among others, dimethyl disulfide (C2H6S2), dimethyl trisulfide (C2H6S3), dimethyl tetrasulfide (C2H6S4), dimethyl pentasulfide (C2H6S5), hexasulfur (S6), and octasulfur (S8) (Fig. 3e). Sulfurous VOCs such as dimethyl di- and trisulfide have been previously shown to have an effect on growth of several plant pathogenic fungi [38, 58–60]. Dimethyl tetrasulfide and pentasulfide as well as octasulfur have been described to be produced by Streptomyces, Myxococcus, and Xanthomonas spp. [61, 62] based on a search in the database of microbial volatiles (mVOCs). However, dimethyl tetrasulfide (C2H6S4), dimethyl pentasulfide (C2H6S5), hexasulfur (S6), and octasulfur (S8) have not been previously shown to inhibit fungal growth. Moreover, there is no knowledge on the genes involved in the metabolism of these sulfurous VOCs, if these VOCs are produced and have a role in antifungal activity in vivo. Hence, we focused on the sulfurous VOCs produced by P. graminis PHS1 to elucidate the genetic basis of VOCs-mediated disease suppressiveness.
Fig. 3.
a Schematic representation of the sandwich assay to screen for VOCs-mediated antifungal activity. Left pictures represent the fungal growth after four days of VOCs exposure and right pictures depict the colony morphology of the selected Burkholderia and Paraburkholderia strains. b Box plot showing the inhibition of fungal growth after four days of exposure to bacterial VOCs. Radial growth of the fungus exposed to the VOCs was expressed relative to the hyphal growth in the control. Boxes represent the mean with standard errors of six independent biological replicates. Asterisks indicate a statistical difference from the control (exposed to medium only) (Student’s t-Test, P < 0.05). c PLS-DA 2D-plot of GC-MS data of VOCs emitted by the different bacterial species after four days of incubation. d. Ratio between VOCs emitted by P. graminis PHS1 and the control (B. pyrrocinia mHSR5, P. caledonica PHRS4, hospita mHSR1, terricola mHS1, and PDA without bacteria (control)). Red dots indicate VOCs differentially produced (at least 2.1× higher than the control) by P. graminis PHS1 (Kruskal and Wallis, Benjamini–Hochberg P < 0.1). e Hierarchical cluster and heatmap analyses of sulfurous VOCs produced by the selected strains. Columns represent three replicate VOC measurements of each of the five species and the medium alone (PDA as control). Rows represent the different sulfurous VOCs (red: low abundance; blue: high abundance); several of which were tentatively annotated
Comparative genomics and identification of genes involved in sulfurous VOC production
To identify genes involved in the production of sulfurous VOCs, we sequenced the genomes of the five species by PacBio sequencing of size-selected 20-kb-insert libraries (Supplementary Table S5). Subsequent de novo assembly resulted in closed genome sequences for all five species. The results showed that the genome sizes ranged from 7.1–10.8 Mb with a GC content of 61.2–66.2 % (Supplementary Table S5). The number of replicons was also different between the species, with two chromosomes for P. terricola mHS1, two chromosomes and one plasmid for P. caledonica PHRS4 and P. graminis PHS1, two chromosomes and one megaplasmid for B. pyrrocinia mHSR5, and three chromosomes and one plasmid for P. hospita mHSR1. Phylogenetic analysis with 12 housekeeping genes of these 5 genomes, along with 197 genomes available in the NCBI database, showed five different clades: three clades containing the opportunistic human pathogenic Burkholderia species (B. cepacia, B. mallei, and B. thailandensis), one clade harboring plant pathogenic Burkholderia species (B. gladioli, B. plantarii, and B. glumae), and one clade with strains of environmental and plant-associated Burkholderia species, for which the new genus name Paraburkholderia has been proposed [63] (Supplementary Figure S2). Our B. pyrrocinia mHSR5 from the Rhizoctonia-suppressive soil clustered with other strains from the human pathogenic Burkholderia clade, whereas the strains representing the four other species (P. caledonica PHS1, P. graminis PHS1, P. hospita mHSR1, P. terricola mHS1) belong to the Paraburkholderia clade (Supplementary Figure S2).
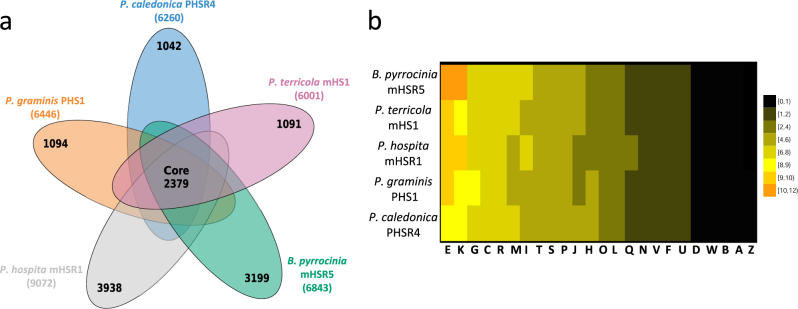
All-vs-all BLASTp analyses and subsequent orthology clustering of the predicted protein sequences of all five species/strains resulted in a pan-genome of 14,722 orthologous groups. The core genome consisted of 2379 orthologous groups (Fig. 4a). The heatmap based on COG annotation showed that the total number of genes belonging to each category was similar between Burkholderia spp., except for genes involved in amino acid transport and metabolism, transcription and carbohydrate transport, and metabolism (Fig. 4b). The number of Coding DNA Sequences (CDSs) ranged from 6001 in P. terricola mHS1 to 9072 in P. hospita mHSR1 (Supplementary Table S5). Secondary metabolites prediction using antiSMASH showed that all strains harbored biosynthetic gene clusters possibly involved in antifungal activity, such as terpenes, nonribosomal peptide synthetases (NRPS), polyketide synthases (PK), and bacteriocins (Supplementary Table S6). Three families of NRPS biosynthetic gene clusters were shared across all genomes (Supplementary Figure S3). The first two clusters appear to be two different families of siderophore biosynthetic gene clusters. The first cluster seems to encode the production of a lipopeptide, found in P. caledonica PHS1, P. graminis PHS1, and P. terricola mHS1 (Supplementary Figure S3a), which may function as a siderophore; it has distant homology to taiwachelin [64], lysobactin [65], pyoverdin [66], serobactins [67], and cupriachelin [68]. The second family detected in B. pyrrocinia mHSR5 and P. hospita mHSR1, contains homologs of the ornibactin NRPS (Supplementary Figure S3b), responsible for the production of a smaller peptide that acts as a siderophore [69]. The third cluster (Supplementary Figure S3c) and single-member found in P. hospita mHSR1, is a PKS-NRPS system that is co-conserved in a cluster with glycosyltransferases and acyltransferases, suggesting that it might encode the production of some kind of glycopeptidolipid. This gene cluster is homologous to a previously detected gene cluster in Burkholderia terrae strain BS001 [70]; however, no metabolic product has yet been described for this gene cluster.
Fig. 4.
Genomic diversity of B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1. a The number of unique coding sequences (CDSs) shared by the B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 representing the core genome is shown in the center. Numbers in non-overlapping portions of each oval show the number of CDSs unique to each strain. The total number of CDSs in the analysis was 32,019. b The heatmap depicts the average percentage of annotated proteins of each cluster belonging to each functional category for the B. pyrrocinia mHSR5, P. caledonica PHRS4, P. graminis PHS1, P. hospita mHSR1, and P. terricola mHS1 strains used in this study (y-axis) and the annotated proteins of each cluster belonging to each functional category (x-axis). Each COG category has been abbreviated D: cell cycle control, cell division, and chromosome partitioning, M: cell wall/membrane/envelope biogenesis, N: cell motility, O: post-translational modification, protein turnover, and chaperones, T: signal transduction mechanisms, U: intracellular trafficking, secretion, and vesicular transport, V: defense mechanisms, W: extracellular structures, Y: nuclear structure, Z: cytoskeleton, A: RNA processing and modification, B: chromatin structure and dynamics, J: translation, ribosomal structure, and biogenesis, K: Transcription, L: replication, recombination, and repair, C: energy production and conversion, E: amino acid transport and metabolism, F: nucleotide transport and metabolism, G: carbohydrate transport and metabolism, H: coenzyme transport and metabolism, I: lipid transport and metabolism, P: inorganic ion transport and metabolism, Q: secondary metabolites biosynthesis, transport, and catabolism, R: general function prediction only, and S: function unknown
For identification of genes involved in the production of the sulfurous VOCs, previous studies have described the dddD gene for its role in the production of dimethyl sulfide in different bacteria, including B. cepacia, from marine environments [71]. Bioinformatic analyses revealed that no homologs of the dddD gene or other genes of the biosynthetic pathway were present in the P. graminis PHS1 genome (data not shown), suggesting that other genes/pathways are involved in the biosynthesis of the sulfurous VOCs detected. To identify these genes, we selected the 1094 genes that were detected in P. graminis PHS1 only and were absent in the genomes of the other four Burkholderia and Paraburkholderia species. Based on annotation of these ‘unique’ P. graminis PHS1genes, 19 genes with a putative function in sulfur metabolism were identified (Table 1). Of these 19 genes, those that were predicted to have a role in transport (Table 1) were not considered for further functional analysis. Among the remaining eight genes with a putative role in sulfur metabolism, one was located on the first chromosome, six on the second chromosome, and one on the plasmid of P. graminis PHS1. Three of these eight genes were previously shown to encode enzymes with specific functions in sulfur metabolism: PHS1_B07225 is a flavin-dependent oxidoreductase involved in the production of methanesulfonic acid and formaldehyde [72]; PHS1_P00230 was shown to be involved in rearrangements of S-S bonds in proteins [73, 74], and PHS1_B04875 is required for the biosynthesis of the corrin macrocycle of coenzyme B12 in Pseudomonas denitrificans [75]. The gene predicted to encode a dimethyl sulfoxide reductase (PHS1_A1010) is presumably involved in the reduction of dimethyl sulfoxide to dimethyl sulfide [76], whereas the predicted cysteine desulfurase (PHS1_B13030) may be involved in transferring sulfur-containing groups derived from cysteine to various other acceptors, such as thio-nucleosides in tRNA, thiamine, biotin, lipoate, and pyranopterin [77]. Finally, three unique genes in strain PHS1 were predicted to encode alkyl sulfatases (genes PHS1_B05945, PHS1_B05965, and PHS1_B05970), all of which are located in the same region on chromosome 2.
Table 1.
Unique genes detected in P. graminis PHS1 that are putatively involved in sulfur metabolism
| Gene ida | COG id | COG description | COG category | COG category description |
|---|---|---|---|---|
| PHS1_B03995 | COG1116 | ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component | P | Inorganic ion transport and metabolism |
| PHS1_A11055 | COG1116 | ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component | P | Inorganic ion transport and metabolism |
| PHS1_A11010 | COG1116 | ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component | P | Inorganic ion transport and metabolism |
| PHS1_B06885 | COG0715 | ABC-type nitrate/sulfonate/bicarbonate transport system, periplasmic component | P | Inorganic ion transport and metabolism |
| PHS1_B06855 | COG0715 | ABC-type nitrate/sulfonate/bicarbonate transport system, periplasmic component | P | Inorganic ion transport and metabolism |
| PHS1_B03985 | COG0715 | ABC-type nitrate/sulfonate/bicarbonate transport system, periplasmic component | P | Inorganic ion transport and metabolism |
| PHS1_A11020 | COG0715 | ABC-type nitrate/sulfonate/bicarbonate transport system, periplasmic component | P | Inorganic ion transport and metabolism |
| PHS1_A11045 | COG0715 | ABC-type nitrate/sulfonate/bicarbonate transport system, periplasmic component | P | Inorganic ion transport and metabolism |
| PHS1_A11005 | COG0600 | ABC-type nitrate/sulfonate/bicarbonate transport system, permease component | P | Inorganic ion transport and metabolism |
| PHS1_B04000 | COG0600 | ABC-type nitrate/sulfonate/bicarbonate transport system, permease component | P | Inorganic ion transport and metabolism |
| PHS1_A11050 | COG0600 | ABC-type nitrate/sulfonate/bicarbonate transport system, permease component | P | Inorganic ion transport and metabolism |
| PHS1_B05945 | COG2015 | Alkyl sulfatase BDS1 and related hydrolases, metallo-beta-lactamase superfamily | Q | Secondary metabolites biosynthesis, transport and catabolism |
| PHS1_B05970 | COG2015 | Alkyl sulfatase BDS1 and related hydrolases, metallo-beta-lactamase superfamily | Q | Secondary metabolites biosynthesis, transport and catabolism |
| PHS1_B05965 | COG2015 | Alkyl sulfatase BDS1 and related hydrolases, metallo-beta-lactamase superfamily | Q | Secondary metabolites biosynthesis, transport and catabolism |
| PHS1_A10100 | COG1018 | Ferredoxin-NADP reductase/putative dimethyl sulfoxide reductase chain YnfF | C | Energy production and conversion |
| PHS1_B07225 | COG2141 | Flavin-dependent oxidoreductase, luciferase family (includes alkanesulfonate monooxygenase SsuD and methylene tetrahydromethanopterin reductase) | H | Coenzyme transport and metabolism |
| PHS1_P00230 | COG2761 | Predicted dithiol-disulfide isomerase, DsbA family | O | Post-translational modification, protein turnover, chaperones |
| PHS1_B13030 | COG0520 | Selenocysteine lyase/Cysteine desulfurase | E | Amino acid transport and metabolism |
| PHS1_B04875 | COG0155 | Sulfite reductase, beta subunit (hemoprotein) | P | Inorganic ion transport and metabolism |
aPHS1: strain, A: Chromosome 1, B: Chromosome 2, and P: Plasmid
Site-directed mutagenesis and functional analysis of P. graminis PHS1 genes involved in the production of sulfurous VOCs
To resolve the role of the identified P. graminis PHS1 genes in sulfurous VOC production and antifungal activity in vitro and in situ, site-directed mutagenesis was successfully performed on the dimethyl sulfoxide reductase gene PHS1_A1010, referred to here as dsr, and the cysteine desulfurase gene PHS1_B13030, referred to as cd. For the cd and dsr mutants, the mutation was confirmed by PCR amplification of the flanking regions using an external primer to the region involved in the double recombination and an internal primer from the kanamycin resistant gene introduced by double recombination into the target genes cd or dsr (data not shown).
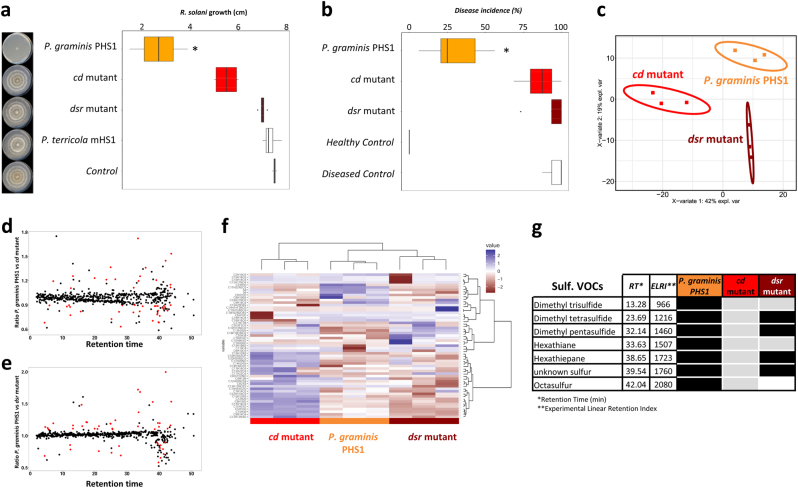
Subsequent in vitro antifungal activity via VOCs showed that the cd and dsr mutants grew equally well as wild-type P. graminis PHS1 but lost, to a large extent, their ability to inhibit hyphal growth of R. solani (Fig. 5a and Supplementary Table S7). When introduced into gamma-irradiated disease-conducive soil, the mutants established population densities in the rhizosphere of sugar beet seedlings of ~8 × 106 CFU g−1 root, very similar to the rhizosphere population densities of wild-type P. graminis PHS1 (1 × 107 CFU g−1, Supplementary Table S8). However, both mutants were not able to suppress Rhizoctonia damping-off disease of sugar beet, whereas the wild type significantly reduced disease incidence (Fig. 5b). These results indicate that the VOC-mediated antifungal activity of P. graminis PHS1 is largely determined by these two genes involved in sulfur metabolism in P. graminis PSH1. The gamma-irradiated soil that was used to test the effect of P. graminis PHS1 and its mutants was not checked for air-borne contamination with other bacteria during the experiment. Therefore, we cannot fully exclude that the suppressive effects are in part due to the activities of these other microorganisms. However, given that the suppressive effect of P. graminis PHS1 was consistently observed in three independent bioassays makes it unlikely that in every case a similar air-borne contamination contributed to this effect. Nevertheless, monitoring the composition of other microorganisms in these assays should shed light on their relative importance.
Fig. 5.
Suppression of Rhizoctonia damping-off disease by P. graminis PHS1 and the two mutants introduced into gamma-irradiated conducive soil. a Box plot showing VOCs-mediated inhibition of fungal growth after four days of exposure to P. graminis PHS1, the cd and dsr mutants. Boxes represent the mean with standard errors of 6 independent biological replicates. Asterisk indicates a statistically significant difference from wild-type P. graminis PHS1 (Student’s t-Test, p < 0.05, n = 6). Pictures depict the hyphal growth of the fungus exposed to VOCs from the wild type and the mutants or exposed to medium only (control). b Box plot showing the percent disease incidence of sugar beet seedlings in conducive soil untreated (Healthy Control), treated with R. solani (Diseased Control), and treated with R. solani and P. graminis PHS1 or its cd and dsr mutants. P. graminis PHS1 and the two mutants were inoculated in soil (107 cells g−1 soil) one day prior to pathogen inoculation (mean values ± SEM, n = 8). An asterisk indicates a statistically significant difference (Student’s t-Test, p < 0.05, n = 6) between the mutants and the wild-type strain P. graminis PHS1. c PLS-DA 2D-plot of GC-MS data of VOCs emitted by P. graminis PHS1 and its two mutants after four days of incubation. d, e ratio between volatiles emitted by P. graminis PHS1 and the mutants cd (d) and dsr genes (e), respectively. Red dots indicate VOCs differentially produced (at least 2.1× higher than the control) by P. graminis PHS1 (Kruskal and Wallis, Benjamini–Hochberg P < 0.1). f Hierarchical cluster and heatmap analyses of sulfurous VOCs of P. graminis PHS1 and its two mutants. Columns represent three replicate VOCs measurements per strain, mutant, or control (medium only). Rows represent the different VOCs (red, low abundance; blue, high abundance). g Table showing the sulfurous VOCs detected in the wild-type P. graminis PHS1 and its two mutants, black: same VOC level as the wild type, gray: lower VOC level than the wild type, white: VOC not detectable in the mutant
To identify the VOCs affected by the cd and dsr mutations, headspace analysis and subsequent PLS-DA analysis showed that the overall VOC profiles of the wild type and the mutants were very similar (Fig. 5c). When the absolute ratio of the log2Ratio ≤ or ≥ 1.1 (at least 2.1 × higher than the control) between the mutants and the wild type was compared, the abundances of 32 and 44 VOCs from cd and dsr mutants, respectively, were statistically significantly different (Fig. 5d–f). Among the differential VOCs that were not detected or detected at lower concentrations in the mutants (Fig. 5f, g and Supplementary Table S9), seven matched the sulfurous derivative compounds detected in our initial analysis (Fig. 3a–e and Fig. 5g). In the dsr mutant, the production of three sulfurous VOCs was significantly reduced as compared with the wild-type strain. Only octasulfur was not detected in the dsr mutant, suggesting that dsr, predicted to encode a dimethyl sulfoxide reductase, is involved in the production of this sulfurous VOC. For the cd mutant, none of the sulfurous VOCs were completely abolished but the production of seven VOCs was significantly reduced (Fig. 5g). The antifungal activity of VOCs from soils was first described in 1953 by Dobbs and Hinson [78] in the context of soil fungistasis, a widespread and more general characteristic of soils associated with the inhibition of spore germination. Also Hora and Baker [79] reported the involvement of a volatile factor in soil fungistasis, but the nature of the VOCs, as well as the identity of the producing microbes remained elusive. Our results show the involvement of sulfurous genes in the production of antifungal VOCs, as well as their involvement in the suppressiveness of soil to the fungal root pathogen R. solani.
Conclusions
Disease-suppressive soils provide intriguing examples of microbiome-mediated protection of plants against fungal infections [5]. Here we show that different Burkholderia and Paraburkholderia species contribute to this soil suppressiveness, with one species (P. graminis PHS1) operating via the production of sulfurous antifungal VOCs. Our study exemplifies that by integrating different “omics” technologies with classic microbiology and molecular biology, it is feasible to unravel highly complex ecosystem services provided by soil and plant-associated microorganisms. How the other four Burkholderia/Paraburkholderia species contribute to this soil suppressiveness remains to be investigated. Due to the lack of antifungal activity of these four species in our in vitro assays, other approaches need to be adopted to identify the underlying mechanisms and genes. These include screening of genome-wide random mutant libraries for reduced or loss of activity in situ, as recently demonstrated for identifying genes and mechanisms involved in plant growth promotion and induced systemic resistance by Pseudomonas [80]. Other approaches include targeted mutagenesis of a number of potential candidate gene clusters, in particular genes encoding PKS and nonribosomal peptide synthetases (NRPSs). Preliminary bioinformatic analyses of the genome sequences of these four other Burkholderia/Paraburkholderia species revealed various PKS and NRPS gene clusters of unknown function. Identifying the roles of these and other biosynthetic gene clusters, as well as their encoded metabolites in the specific soil suppressiveness will be subject of future studies.
Electronic supplementary material
Supplementary information is available at ISME’s website
Acknowledgements
J.R. and V.J.C. were supported by a grant from the Netherlands BE-Basic Foundation (project F07.003.01). VC was supported by the Dutch STW-program ‘Back to the Roots’. We thank Elizabeth Yanez Navarrete and Kazuki Fujiguara for their valuable help in the Burkholderia spp. phenotyping and Dr. Florian Rohart for his expert advice in using the R package mixOmics. This is publication number 6497 of the NIOO-KNAW.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0186-x) contains supplementary material, which is available to authorized users.
References
- 1.Jousset A, Rochat L, Lanoue A, Bonkowski M, Keel C, Scheu S. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol Plant Microbe Interact. 2010;24:352–8. doi: 10.1094/MPMI-09-10-0208. [DOI] [PubMed] [Google Scholar]
- 2.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–63. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 3.Rudrappa T, Czymmek KJ, Paré PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–56. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. Microbial populations responsible for specific soil suppressiveness. Annu Rev Phytopathol. 2002;40:309–48. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 5.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–3. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 6.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 7.Raaijmakers JM, Weller DM. Natural plant protection by 2, 4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant Microbe Interact. 1998;11:144–52. doi: 10.1094/MPMI.1998.11.2.144. [DOI] [Google Scholar]
- 8.Voort M, Kempenaar M, Driel M, Raaijmakers JM, Mendes R. Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol Lett. 2016;19:375–82. doi: 10.1111/ele.12567. [DOI] [PubMed] [Google Scholar]
- 9.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:E1743–52. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bruijn I, Cheng X, de Jager V, Expósito RG, Watrous J, Patel N, et al. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genom. 2015;16:1–16. doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez Expósito R, Postma J, Raaijmakers JM, de Bruijn I. Diversity and activity of Lysobacter species from disease suppressive soils. Frontiers in Microbiology. 2015;6:1243. doi: 10.3389/fmicb.2015.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez Expósito R, de Bruijn I, Postma J, Raaijmakers JM. Current Insights into the Role of Rhizosphere Bacteria in Disease Suppressive Soils. Frontiers in Microbiology. 2015;8:2529. doi: 10.3389/fmicb.2017.02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, Van Wezel GP, et al. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol. 2015;6:1081. doi: 10.3389/fmicb.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapelle E, Mendes R, PAHM Bakker, Raaijmakers JM. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10:265–8. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl L, Vandamme P. Members of the genus Burkholderia: Good and bad guys. F1000Res. 2016;5:F1000. doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott GN, Chou JH, Chen WM, Bloemberg GV, Bontemps C, Martínez‐Romero E, et al. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N‐limited conditions. Environ Microbiol. 2009;11:762–78. doi: 10.1111/j.1462-2920.2008.01799.x. [DOI] [PubMed] [Google Scholar]
- 17.Carlier AL, Eberl L. The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environ Microbiol. 2012;14:2757–69. doi: 10.1111/j.1462-2920.2012.02763.x. [DOI] [PubMed] [Google Scholar]
- 18.Lewenza S, Sokol PA. Regulation of ornibactin niosynthesis and N-Acyl-l-Homoserine lactone production by cepR in Burkholderia cepacia. J Bacteriol. 2001;183:2212–8. doi: 10.1128/JB.183.7.2212-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–8. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 20.Partida‐Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, et al. Production of the antifungal compound pyrrolnitrin is quorum sensing‐regulated in members of the Burkholderia cepacia complex. Environ Microbiol. 2009;11:1422–37. doi: 10.1111/j.1462-2920.2009.01870.x. [DOI] [PubMed] [Google Scholar]
- 22.Stopnisek N, Zuhlke D, Carlier A, Barberan A, Fierer N, Becher D, et al. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2016;10:253–64. doi: 10.1038/ismej.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol. 2016;100:5215–29. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 24.Elshafie H, Camele I, Racioppi R, Scrano L, Iacobellis N, Bufo S. In vitro antifungal activity of Burkholderia gladioli pv. agaricicola against some phytopathogenic fungi. Int J Mol Sci. 2012;13:16291. doi: 10.3390/ijms131216291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groenhagen U, Baumgartner R, Bailly A, Gardiner A, Eberl L, Schulz S, et al. Production of bioactive volatiles by different Burkholderia ambifaria strains. J Chem Ecol. 2013;39:892–906. doi: 10.1007/s10886-013-0315-y. [DOI] [PubMed] [Google Scholar]
- 26.Estrada-De Los Santos P, Bustillos-Cristales RO, Caballero-Mellado J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl Environ Microbiol. 2001;67:2790–8. doi: 10.1128/AEM.67.6.2790-2798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burbage D, Sasser M. A medium selective for Pseudomonas-cepacia. Phytopathology. 1982;72:706. [Google Scholar]
- 28.Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martínez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl Environ Microbiol. 2007;73:5308–19. doi: 10.1128/AEM.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perin L, Martínez-Aguilar L, Castro-González R, Estrada-de los Santos P, Cabellos-Avelar T, Guedes HV, et al. Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl Environ Microbiol. 2006;72:3103–10. doi: 10.1128/AEM.72.5.3103-3110.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estrada-de los Santos P, Vinuesa P, Martínez-Aguilar L, Hirsch AM, Caballero-Mellado J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol. 2013;67:51–60. doi: 10.1007/s00284-013-0330-9. [DOI] [PubMed] [Google Scholar]
- 31.Payne GW, Ramette A, Rose HL, Weightman AJ, Jones TH, Tiedje JM, et al. Application of a recA gene-based identification approach to the maize rhizosphere reveals novel diversity in Burkholderia species. FEMS Microbiol Lett. 2006;259:126–32. doi: 10.1111/j.1574-6968.2006.00257.x. [DOI] [PubMed] [Google Scholar]
- 32.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Bork P. Interactive Tree Of Lifev2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 38.Garbeva P, Hordijk C, Gerards S, De Boer W. Volatiles produced by the mycophagous soil bacterium Collimonas. FEMS Microbiol Ecol. 2014;87:639–49. doi: 10.1111/1574-6941.12252. [DOI] [PubMed] [Google Scholar]
- 39.Lommen A, Kools HJ. MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics. 2011;8:719–26. doi: 10.1007/s11306-011-0369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tikunov YM, Laptenok S, Hall RD, Bovy A, Vos RCH. MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics. 2011;8:714–8. doi: 10.1007/s11306-011-0368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. Retention index thresholds for compound matching in GC–MS metabolite profiling. J Chromatogr B. 2008;871:182–90. doi: 10.1016/j.jchromb.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 42.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 46.Worley B, Halouska S, Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem. 2013;433:102–4. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metab. 2013;1:92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed]
- 49.Core Team R. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 50.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 51.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 52.Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA. 2002;99:5890–5. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zumaquero A, Macho AP, Rufián JS, Beuzón CR. Analysis of the role of the type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448a in interaction with the plant. J Bacteriol. 2010;192:4474–88. doi: 10.1128/JB.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Voort M, Meijer H, Schmidt Y, Watrous J, Dekkers E, Mendes R, et al. Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front Microbiol. 2015;6:693. doi: 10.3389/fmicb.2015.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, et al. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012;193:997–1008. doi: 10.1111/j.1469-8137.2011.04001.x. [DOI] [PubMed] [Google Scholar]
- 59.Kai M, Haustein M, Molina F, Petri A, Scholz B, Piechulla B. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–12. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- 60.Wang C, Wang Z, Qiao X, Li Z, Li F, Chen M, et al. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD−1. FEMS Microbiol Lett. 2013;341:45–51. doi: 10.1111/1574-6968.12088. [DOI] [PubMed] [Google Scholar]
- 61.Hunziker L, Bönisch D, Groenhagen U, Bailly A, Schulz S, Weisskopf L. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl Environ Microbiol. 2015;81:821–30. doi: 10.1128/AEM.02999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–42. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 63.Sawana A, Adeolu M, Gupta RS. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 2014;5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kreutzer MF, Nett M. Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis. Org Biomol Chem. 2012;10:9338–43. doi: 10.1039/c2ob26296g. [DOI] [PubMed] [Google Scholar]
- 65.Hou J, Robbel L, Marahiel Mohamed A. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem Biol. 2011;18:655–64. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–31. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosconi F, Davyt D, Martínez V, Martínez M, Abin-Carriquiry JA, Zane H, et al. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ Microbiol. 2013;15:916–27. doi: 10.1111/1462-2920.12075. [DOI] [PubMed] [Google Scholar]
- 68.Kreutzer MF, Kage H, Nett M. Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. J Am Chem Soc. 2012;134:5415–22. doi: 10.1021/ja300620z. [DOI] [PubMed] [Google Scholar]
- 69.Agnoli K, Lowe CA, Farmer KL, Husnain SI, Thomas MS. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the fur regulon. J Bacteriol. 2006;188:3631–44. doi: 10.1128/JB.188.10.3631-3644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haq IU, Graupner K, Nazir R, van Elsas JD. The genome of the rungal-interactive soil bacterium Burkholderia terrae BS001—a plethora of outstanding interactive capabilities unveiled. Genome Biol Evol. 2014;6:1652–68. doi: 10.1093/gbe/evu126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science. 2007;315:666–9. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- 72.Wicht DK. The reduced flavin-dependent monooxygenase SfnG converts dimethylsulfone to methanesulfinate. Arch Biochem Biophys. 2016;604:159–66. doi: 10.1016/j.abb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 73.De Lorenzo F, Goldberger RF, Steers E, Givol D, Anfinsen CB. Purification and properties of an enzyme from beef liver which catalyzes sulfhydryl-disulfide interchange in proteins. J Biol Chem. 1966;241:1562–7. [PubMed] [Google Scholar]
- 74.Fuchs S, De Lorenzo F, Anfinsen CB. Studies on the mechanism of the enzymic catalysis of disulfide interchange in proteins. J Biol Chem. 1967;242:398–402. [PubMed] [Google Scholar]
- 75.Debussche L, Thibaut D, Cameron B, Crouzet J, Blanche F. Biosynthesis of the corrin macrocycle of coenzyme B12 in Pseudomonas denitrificans. J Bacteriol. 1993;175:7430–40. doi: 10.1128/jb.175.22.7430-7440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNicholas PM, Chiang RC, Gunsalus RP. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol Microbiol. 1998;27:197–208. doi: 10.1046/j.1365-2958.1998.00675.x. [DOI] [PubMed] [Google Scholar]
- 77.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 78.Dobbs CG, Hinson WH. A widespread fungistasis in soils. Nature. 1953;172:197–9. doi: 10.1038/172197a0. [DOI] [PubMed] [Google Scholar]
- 79.Hora TS, Baker R. Volatile factor in soil fungistasis. Nature. 1970;225:1071–2. doi: 10.1038/2251071a0. [DOI] [PubMed] [Google Scholar]
- 80.Cheng X, Etalo DW, Mortel JEvd, Dekkers E, Nguyen L, Medema MH, Raaijmakers JM. Genome‐wide analysis of bacterial determinants of plant growth promotion and induced systemic resistance by Pseudomonas fluorescens. Environmental Microbiology. 2017;19:4638–4656. doi: 10.1111/1462-2920.13927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is available at ISME’s website