Abstract
Loss of brain glutathione has been associated with cognitive decline and neuronal death during aging and neurodegenerative diseases. However, whether decreased glutathione precedes or follows neuronal dysfunction has not been unambiguously elucidated. Previous attempts to address this issue were approached by fully eliminating glutathione, a strategy causing abrupt lethality or premature neuronal death that led to multiple interpretations. To overcome this drawback, here we aimed to moderately decrease glutathione content by genetically knocking down the rate-limiting enzyme of glutathione biosynthesis in mouse neurons in vivo. Biochemical and morphological analyses of the brain revealed a modest glutathione decrease and redox stress throughout the hippocampus, although neuronal dendrite disruption and glial activation was confined to the hippocampal CA1 layer. Furthermore, the behavioral characterization exhibited signs consistent with cognitive impairment. These results indicate that the hippocampal neurons require a large pool of glutathione to sustain dendrite integrity and cognitive function.
Abbreviations: CA1, Cornu Ammonis area 1; CA3, Cornu Ammonis area 3; CK2a, calmodulin kinase 2A; DG, dendate gyrus; DMEM, Dulbecco's modified Eagle's médium; DMSO, dimethylsulfoxide; DTNB, 5,5′-dithio-bis-(2-nitrobenzoic acid); FBS, fetal bovine serum; GCL, glutamate-cysteine ligase, catalytic subunit; GSH, glutathione, reduced form; GSSG, glutathione, oxidized form; MAP2, microtubule-associated protein-2; MEFs, mouse embryonic fibroblasts; shGCL, shRNA against GCL; shGCLSFL, switched flox shGCL; shGCLUFL, unswitched flox shGCL; shRNA, small hairpin RNA; SDS, sodium dodecyl sulfate; ROS, reactive oxygen species; PCR, polymerase chain reaction; TM, 4-hydroxy-tamoxifen; TUJ1, neuron-specific Class III β-tubulin
Keywords: Neurons, Glutamate-cysteine ligase, Glutathione, In vivo knockdown, Dendrite disruption, Memory impairment
Graphical abstract
Highlights
-
•
Whether glutathione fall precedes or follows neuronal dysfunction is unknown.
-
•
A genetic approach to downregulate glutathione in neurons in vivo was generated.
-
•
Systematic characterization reveals redox stress throughout the hippocampus.
-
•
Neuronal dendrite disruption was confined to neurons of the CA1 layer.
-
•
Behavioral characterization exhibits cognitive impairment.
1. Introduction
When compared with astrocytes, neurons are highly dependent on oxidative phosphorylation for correct function [2], [3], a situation linked with persistent production of reactive oxygen species (ROS) by the mitochondrial respiratory chain [4]. Being post-mitotic cells, neurons are exposed to ROS during longer time periods than dividing cells, which may contribute to aging and, if inappropriately controlled, to neurological diseases [5], [6], [7], [8], [9]. To modulate the possible deleterious actions of excess ROS, cells contain glutathione (γ-glutamyl-cysteinyl-glycine, GSH), an active and ubiquitous antioxidant cofactor responsible for ROS detoxification in neurons [10]. Accordingly, oxidative damage likely caused by progressive GSH deficiency is considered to be one of the earliest biochemical indicator of neuronal degeneration in aging, Parkinson's disease, and certain mental disorders [11], [12], [13]. In consistency with this notion, over-expression of glutamate-cysteine ligase (GCL), i.e. the first and rate-limiting step in GSH biosynthesis, in brain mitochondria protects neurons against neurological deficit in vivo [10].
Unfortunately, all clinical trials on antioxidants performed so far have not alleviated the neurological declines in a wide range of disorders [14], which might challenge the oxidative theory of aging and cognitive impairment [15], [16], [17]. Moreover, the lack of an experimental model that faithfully recreate, in neurons in vivo, the mild mode of oxidative damage occurring during neurological deterioration, has hampered understanding the actual role played by ROS in aging and neurological disorders. To address this issue, here we designed and generated a mouse model genetically engineered to knockdown GCL in neurons of the postnatal mouse brain in vivo. We found that GCL knockdown caused a moderate decrease in GSH in the neurons of the CA1 area of the hippocampus that led to dendrite disruption and cognitive impairment. These results reveal that mild oxidative damage in a discrete group of hippocampal neurons is sufficient to trigger the cellular and behavioral signs of neurological disorders.
2. Methods and materials
2.1. shGCLUFL DNA construct generation
A DNA construct was designed to harbor a LoxP-flanked small hairpin RNA (shRNA) precursor against the catalytic subunit of glutamate-cysteine ligase (GCL) (hereafter shGCL). Before recombination (unswitched flox or UFL), the shGCL sequence (5′-GAAGGAGGCTACTTCTATA-3′) [18] was separated from the Pol-III promoter (H1) by the LoxP and Lox2272 sequences in the appropriate positions and orientations. Importantly, the shGCL has opposite strand orientation to the H1 promoter. To clone the DNA fragment elements, we used pIRES2-EGFP (Clontech), pSuper-neo/gfp (Oligoengine), pCDNA3.1(+) (Invitrogen) and pEGFP-C1 (Addgene). The small sequence adaptors were designed and chemically synthesized (Thermo) to introduce the LoxP and Lox2272 sites in the correct orientation and the appropriate restriction enzymes sites. To construct shGCL [18], the oligonucleotides were annealed and phosphorylated before ligation with the corresponding linearized and dephosphorylated plasmids. Elements from plasmids were excised by restriction enzyme digestion and directly cloned into the linearized plasmids. The correct sequence and orientations of the intermediary vectors were checked by restriction enzyme assay and, for the final clone containing shGCL (shGSLUFL), by sequencing.
2.2. shGCLUFL/+ and CK2a-Cre/shGCLSLF mice generation
To generate mice harboring the shGCLUFL construct, the linearized DNA was microinjected in the pre-fertilized pro-nucleus of a one-cell state oocyte from the hybrid C57Bl/6J-CBA lineage at the Animal Service of the University of Salamanca following standard protocols. A founder, identified to harbor a unique integration site for shGCLUFL at the fourth (N4) generation, was successively backcrossed with C57Bl/6J until the congenic status (tenth generation) was achieved. Mice expressing Cre recombinase driven by the neuronal-specific promoter of mouse calcium-calmodulin kinase 2a (CK2a-Cre) were purchased from The Jackson Laboratories in a congenic C57BL/6 background. According to The Jackson Laboratories datasheet, Cre recombinase expression in this mouse is postnatal (from day 14–21 onwards) and confined mainly to the forebrain and hippocampus [19]. CK2a-Cre mice were backcrossed for two generations in a C57BL/6J background to obtain transgene homozygosis. To generate CK2a-Cre/shGCLSFL mice, heterozygous shGCLUFL/+ mice were bred with homozygous (CK2a-Cre/CK2a-Cre) mice. The efficacy of the Cre-mediated recombination in the CK2a-Cre/shGCLSFL mice, but not in the CK2a-Cre/+, was tested in the hippocampus by PCR.
2.3. Tissue extraction
Male mice (9 months-old) were euthanized with CO2 and tissues were extracted within the following 5 min. Mice were decapitated, and their brains were extracted and dissected. We collected the cerebellum, striatum, hippocampus and pre-frontal cortex. For DNA extraction, a small aliquot of tissue (tails or hippocampus) was placed in ice and frozen until further processing. DNA was extracted from tails, pelleted cells or tissue samples using the phenol-chloroform-isoamilic acid method followed by isopropanol precipitation. DNA concentrations were measured using NanoDrop 2000 (Thermo).
2.4. Genotyping
Genotyping was performed by PCR. To genotype the shGCLUFL transgene, we used the following forward and reverse oligonucleotides 5′-AAGTCGTGCTGCTTCATGTG-3′ and 5′-ACGTAAACGGCCACAAGTTC-3′, which generated a 200 bp band from the fragment placed within the LoxP and Lox2272 sites of shGCLUFL. An internal control was used to detect false negatives using the endogenous α-synuclein gene, the forward and reverse oligonucleotides of which were, respectively, 5′-ATCTGGTCCTTCTTGACAAAGC-3′ and 5′-AGAAGACCAAAGAGCAAGTGACA-3′, which generated a 150 bp band. To genotype the CK2a-Cre transgene, a 270 bp region of Cre recombinase was amplified by PCR. Forward and reverse oligonucleotides used were, respectively, 5′-GCATTTCTGGGGATTGCTTA-3′ and 5′-CCCGGCAAAACAGGTAGTTA-3′, using the same α-synuclein oligonucleotides as above as the internal control. PCR conditions were: 3 min at 94 °C, 35 cycles of 1) 1 min at 94 °C, 2) 1 min at 58 °C, 3) 1 min at 72 °C and a final step 10 min at 72 °C. To detect the non-recombined state of the construct, we used the following forward and reverse oligonucleotides, respectively, 5′-CATCGAGCTGAAGGGCATC-3′ and 5′-CGGTGGGAGGTCTATATAAGCA-3′, which amplified a 900 bp band. The recombined state was detected using the forward and reverse oligonucleotides, respectively, 5′-GGTCAGGGTGGTCACGAG-3′ and 5′-CGGTGGGAGGTCTATATAAGCA-3′, and which amplified a 300 bp band. PCR conditions for both PCRs were: 3 min at 94 °C, 35 cycles of 1) 1 min at 94 °C, 2) 1 min at 55 °C, 3) 1 min at 72 °C and a final step 10 min at 72 °C.
2.5. Southern blotting
Genomic DNA from the tail (5–10 µg) was digested with HindIII, the fragments were separated by agarose gel electrophoresis and transferred to a nylon membrane (Immobilon™ NY+, Millipore). After UV crosslinking (Hoefer), the membrane was pre-hybridized with denatured ssDNA and hybridized with the radiolabelled oligonucleotide probe within the LoxP and Lox2272 sites of shGCLUFL. The radioactive bands were visualized using a Phosphorimager screen (Fujifilm) and the images taken using a Bio-Rad Personal Molecular Imager.
2.6. Mouse embryonic fibroblasts (MEFs) immortalization
Fibroblasts were prepared from fetal shGCLUFL/+ mice (E13.5). Embryos were processed individually, and the liver and brain tissue were disregarded. The carcass was mechanically disaggregated in 0.25 g/ml of trypsin/1 mM EDTA and seeded in a 60 cm2 cell culture dish containing DMEM (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (Linus) and 25 mM D-glucose. Confluent cells were split 1:3 during 2 weeks, until the immortalization crisis occurred, which lasted for a further 2 weeks. During this period, fresh medium was replaced every 3 days. One week after the immortalization crisis, single cell colonies were spotted, picked and grew. MEFs were frozen in a mixture of FBS and DMSO (9:1). For experiments involving MEFs, dishes and plates were previously coated with poly-D-lysine (15 μg/ml). Cells were incubated at 37 °C in a humidified 5% CO2-containing atmosphere.
2.7. MEF transfections and treatments
Transfections of MEFs with plasmid vectors were performed using Lipofectamine2000™ (Invitrogen), following the manufacturer’ instructions. After 6 h, fresh medium was added containing either 1 µM 4-OH-Tamoxifen (TM) (Sigma) or vehicle (ethanol).
2.8. Western blotting
Cells were lysed in RIPA buffer (2% SDS, 2 mM EDTA, 2 mM EGTA and 50 mM Trizma base, pH 7.5), supplemented with phosphatase inhibitors (100 μM phenylmethylsulfonyl fluoride, 50 μg/ml antipapain, 50 μg/ml pepstatin, 50 μg/ml amastatin, 50 μg/ml leupeptin, 50 μg/ml bestatin, 1 mM o-vanadate, 50 mM NaF, and 50 μg/ml soybean trypsin inhibitor) and boiled for 5 min. Extracts were centrifuged at 13,000×g for 5 min at 4 °C, and aliquots of lysates (50 μg protein) were subjected to sodium docedyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% acrylamide gel (MiniProtean®, Bio-Rad) including Dual Colors™ (BioRad) as prestained protein ladder. The resolved proteins were transferred electrophoretically to nitrocellulose membranes (Hybond-ECL, Amersham Bioscience Europe GmbH). Membranes were blocked with 5% (w/v) low-fat milk in 20 mM Tris, 500 mM NaCl, and 0.1% (w/v) Tween 20, pH 7.5, for 1 h. After blocking, membranes were immunoblotted with 1:2000 rabbit anti-GCL (catalytic subunit) (Abcam), 1:500 anti-GAPDH (6C5; Ambion) and 1:1000 anti-Map2 (AP-20; Sigma-Aldrich) antibodies, overnight at 4 °C. After incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000 dilution, Pierce, Thermo Fischer Scientific) and/or goat anti-mouse IgG (1:10,000 dilution, BioRad), membranes were immediately incubated with the enhanced chemiluminescence kit SuperSignal West Dura (Pierce, Thermo Scientific, Illinois, USA) for 5 min, before exposure to Kodak XAR-5 film for 1–5 min, and the autoradiograms were scanned.
2.9. Glutathione concentration determination in MEFs
Oxidized (GSSG) and total glutathione (GSx; GSx=GSH+2GSSG) was measured in MEFs. For tissue samples, only total glutathione was assessed. For MEFs, 1 ml of 5% (w/v) 5-sulfosalicylic acid (SSA) per 106 cells was added. An equal volume of NaOH 0.1 M was added to the same amount of cells for protein quantification. In tissue samples, 20 µl of 5% (w/v) of SSA was added per mg of tissue, and was diluted 1:5 for subsequent glutathione measurement. In all cases, samples were pelleted and transferred to a fresh tube. For oxidized glutathione (GSSG) determination, 60 µl of sample or GSSG standard were incubated with 3.43 µl of 2-vinylpiridine and 2.75 µl of triethanolamine for 1 h at 4 °C. This derivatization reaction protects the sulfhydryl group forming a tioether, which does not react in the determination assay. Thus, only GSSG is measured in these samples. Reduced glutathione concentration (GSH) was calculated from the formula GSx=GSH+ 2GSSG, according to [20]. The assay uses NADPH(H+) and glutathione reductase (GSR) to reduce back the oxidized glutathione upon formation, a reaction that is coupled to the formation of TNB (λmax=405 nm) from GSH and DTNB. In view that this is, therefore, a cycling reaction, the slope is registered over a 10-min (20 iterations) period at 405 nm. The rates of TNB formation in the samples were extrapolated to those obtained with GSSG standards (Sigma). For GSx, GSSG standards were used in the range of 0–70 µM, whereas for GSSG, GSSG standards were used in the range 0–20 µM. The reaction composition consisted of 175 µl of 0.33 mM NADPH sodium salt prepared extemporarily in NaE buffer (143 mM sodium phosphate, 6.3 mM EDTA, pH 7.5), 35 µl of H2O, 25 µl of 6 nM DTNB (in NaE buffer), 10 µl of sample or standard and 50 µl of GSR (4 U/ml for GSx or 20 U/ml for GSSG determinations).
2.10. H2O2 determination
H2O2 assessment was performed in intact, seeded cells. 1.25 × 105 cells were incubated in 100 μM AmplexRed probe (Invitrogen) in Krebs-Ringer phosphate buffer (145 mM NaCl, 5.7 mM Na2PO4, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM MgSO4 and 5.5 mM glucose), containing horseradish peroxidase (0.5 U/ml). Luminescence was recorded for 2 h at 30 min intervals using a Varioskan Flash (Thermo Scientific, Rockford, IL, USA) fluorimeter (excitation: 538 nm, emission: 604 nm), and the slopes were compared with those obtained with H2O2 (0–10 µM) for the calculations.
2.11. Thiols concentration determination
Cells seeded in 96-well plates at 2 × 105 cells/cm2 were washed twice with PBS pH 7.4, incubated with 98 µl of SDS/EDTA buffer (SDS 2% (w/v) in 80 mM phosphate buffer pH 8, containing 1 mM EDTA), and the absorbance was read at 412 nm. Then, 2 µl of freshly prepared DTNB (10 mM in 0.1 mM phosphate buffer, pH 8) was added and absorbance was read again after 20 min. Glutathione standards were prepared in SDS/EDTA buffer. The absorbance at 0 min was subtracted from the absorbance at 20 min, and thiols concentrations were inferred from the glutathione standard curve.
2.12. Protein determination
Cells were lysed in 0.1 M NaOH and used for the determination of protein concentration by the BCA protein assay kit (Pierce), following the manufacturer's instructions. Bovine serum albumin was used as standard.
2.13. Protein carbonyl detection and proteomic analysis
These analyses were performed at the Proteomics Service of the University of Lleida. Protein carbonyl groups were detected in SDS-PAGE gels. Brain samples from 9 months-old mice were incubated in 20 mM Tris-HCl, pH 7.5, plus a cocktail of protease inhibitors (Roche) and disaggregated mechanically using an insulin syringe. For protein carbonylation analyses, a volume containing 50 µg of protein was treated with an equal volume of 15% SDS, 1% EDTA, 15% β-mercaptoethanol in 0125 M Tris-HCl pH 6.8. Samples were then derivatized with Bodipy-FL hydrazide (Invitrogen, D-2371) as previously reported [21]. Protein was then precipitated with 10% TCA for 30 min at 25 °C. Pellets were washed three times with 200 µl of ethanol: ethylacetate (1/1) solution and air-dried. Samples were resuspended in Laemmli buffer and analyzed in 10% PAGE-SDS electrophoresis gels. TCA-precipitated protein samples were also used for 2D-gel electrophoresis analyses, as described [22]. For this purpose, proteins were resuspended in 25 µl of 7 M urea, 2 M thiourea, 40 mM Tris, 1% C7BzO, pH = 10.4 Protein Extraction Reagent Type 4, (Sigma, C3056) at 25 °C under continuous shaking at 1400 rpm. Bodipy-derivatized proteins were separated in two-dimensional gels and visualized by epifluorescence using specific channel settings for Bodipy-FL. Total protein staining was performed with Flamingo Fluorescent gel stain (Bio-Rad). Images were captured with a VersaDoc MP4000 Imager and analyzed with PDQuest software (Bio-Rad). For each spot, a normalized value was obtained by establishing a ratio between Bodipy signal versus protein signal. Spots in 2D-gels samples displaying a ratio greater than 1.5 with respect to the matched control were selected for further identification by peptide mass fingerprinting using an Autoflex-Speed MALDI ToF/ToF mass spectrometer (Bruker Daltonics).
2.14. Mice perfusion and immunohistochemistry
Male mice (9 months-old) were anesthetized by intraperitoneal injection of a mixture of xylazine hydrochloride (Rompun; Bayer) and ketamine hydrochloride/chlorbutol (Imalgene; Merial) (1:4) at 1 ml per kg of body weight, and then perfused intraaortically with 0.9% NaCl followed by 5 ml/g body weight of paraformaldehyde (PFA, 4% (w/v)) in 0.1 M phosphate buffer (PB) at pH 7.4. After perfusion, brains were dissected out sagittally in two parts and post-fixed using 4% PFA overnight at 4 °C. Brain blocks were rinsed successively for 10 min, 30 min, and 2 h with 0.1 M PB solution and cryoprotected in 10%, 20% and 30% (w/v) sucrose in PB solution sequentially, until they sank. After cryoprotection, 30-μm-thick sagittal sections were obtained with a freezing-sliding cryostate (Leica; CM1950 AgProtect). The sections were collected serially in a 12 well plate in 0.1 M PB and rinsed 3 times for 10 min in 0.1 M PB and used for subsequent immunohistochemistry. The section-containing wells that were not used were kept in 0.05% sodium azide (w/v) in 0.1 M PB at 4 °C. For immunohistochemistry, sections were incubated sequentially in (i) 5 mg/ml sodium borohydride in PB for 30 min (to remove aldehyde autofluorescence); (ii) three PB washes 10 min each; (iii) 1:200 anti-CK2a (6G9; ENZO), 1:500 anti-CK2a (bs-1647R; Bioss Inc), 1:250 anti-GCL (sc22755; Santa Cruz), 1:500 anti-GFAP (G6171; Sigma-Aldrich), 1:200 anti-GSH (A001–50UG; Arbor Assays), 1:500 anti-IBA1 (019–19741; Wako), or 1:500 anti-MAP2 (AP-20; Sigma-Aldrich) in 0.02% Triton X-100 (Sigma-Aldrich) and 5% goat serum (Jackson Immuno-Research) in 0.1 M PB for 72 h at 4 °C; (iv) three PB washes 10 min each; (v) fluorophore conjugated secondary antibodies, 1:500 Cy2 goat anti-mouse, 1:500 Cy2 goat anti-rabbit, 1:500 Cy3 goat anti-mouse, or 1:500 Cy3 goat anti-rabbit (Jackson Immuno-Research) in PB for 2 h at room temperature; and (vi) 0.5 μg/ml DAPI in PB for 10 min at room temperature. After rinsing with PB, sections were mounted with Fluoromount (Sigma-Aldrich) aqueous mounting médium [23].
2.15. Nissl staining
Sections obtained as previously described were mounted in slides and left to dry overnight at room temperature. Slides were immersed in a Coplin jar in absolute ethanol and chlorophorm mix (1:1) overnight, and sequentially immersed for 5 min each in a battery of ethanol solutions at decreasing concentrations, starting from xylol down to distilled water for rehydration. Then, the slides were immersed again in Nissl dye, consisting of 0.1% (w/v) cresyl violet and 0.3% (v/v) glacial acetic in water, for 10 min at room temperature. Afterwards, sides were washed under tap water, distilled water and, finally, with 96% ethanol, and mounted in Entellan solution (Sigma).
2.16. Imaging and quantification
Sections were examined with epifluorescence and appropriate filter sets using an inverted microscope (Nikon; Eclipse Ti-E) equipped with a pre-centered fiber illuminator (Nikon; Intensilight C-HGFI) and B/W CCD digital camera (Hamamatsu; ORCA-ER). Confocal images were acquired using a spectral laser confocal microscope (TCS-SL; Leica Microsystems) and an inverted confocal microscope (TCS SP5; Leica Microsystems). Large fields of view were acquired with an HCX Plan Apo CS2 40× (N.A. 1.30) with a pixel size of ~300 nm at a scan speed of 400 Hz using three line averages. High-resolution images were acquired using an HCX Plan Apo CS2 63× oil objective (N.A. 1.40) with a pixel size of 100 nm and a z-step size of 290 nm at a scan speed of 400 Hz using three lines averages. Immunohistochemical digital images were used to analyze protein staining in the three most medial sagittal sections per animal from four different animals per condition (n = 4). Images were exported into the NIH image-processing package ImageJ (version 1.47) in tiff format for processing. Before image analysis, a maximum-intensity projection over z-series projections spanning 15–16 µm was performed. Images were converted to gray scale 8-bit images and brightness/contrast was adjusted using the ImageJ “auto” function. Staining was automatically delineated using the “auto setting threshold” (default method) and “dark background” functions of ImageJ. Thresholded images were subsequently quantified as percent area (area fraction) using the “analyze-measure” function, which represents the percentage of pixels in the image that have been highlighted (% area) [23]. We also quantified the number of CK2a-positive cells (neurons) that were GCL- or GSH-positive in images of the three most medial sagittal sections per animal from four different animals per condition (n = 4). In each studied area (CA1, CA3 and DG), we selected a specific area and were quantified in a blinded fashion using NIS Elements AR 3.22 (Nikon Inc.; Tokyo, Japan) the cells that were CK2a-positive and GCL- or GSH-positive (colocalization), and the data were expressed as number of GCL+/CK2a+ or GSH+/CK2a+ cells per µm2.
2.17. Behavioral tests
2.17.1. Motor balance and coordination
These were analyzed using the Rotarod test (Rotarod apparatus, Model 47600, Ugo Basile) every eight weeks in six male mice per condition. Male mice were previously trained during three consecutive days, two weeks before the first test. Each test was performed in three consecutive days at the same time of the day. The rotarod conditions were 300 s of continuous acceleration from 4 to 40 rpm with a net 4-rpm rise every 30 s, reaching the final speed at 270 s. Data from tests in which animals completed 3 turns without walking were disregarded.
2.17.2. Cognitive performance
Animals were left to acclimatize in the room for not less than 15 min at the same time slot of the day (2–8 pm). Tracking was carried out once at a time and carefully cleaning the apparatus with 70% ethanol between trials to remove any odor cues. For the Open Field and Hole Board tests, an ANY-box® core was used, which contained a light gray base and an adjustable perpendicular stick holding a camera and an infrared photo-beam array to track the animal movement and to detect rearing behavior, respectively. Mouse movements were tracked with the ANY-maze® software and the AMi-maze® interface to register all parameters described subsequently. For the Open Field test, a 40 cm × 40 cm × 35 cm (w, d, h) black infrared transparent Perspex insert was used, and the arena was divided in three zones, namely border (8 cm wide), center (16% of total arena) and intermediate (the remaining area). The test lasted for 30 min, the distance travelled and the time spent in each zone were measured. The head dipping (nose pokes) on a Hole Board was used as an indicator of exploratory tendencies and locomotor activity-independent anxiety. To do this, a 40 cm × 40 cm (w, d) raised base with 16 holes (3 cm diameter each), arranged in a 4×4 grid raised 10 cm above the floor, fitted snugly inside, and a black Perspex insert infrared transparent (40 cm × 40 cm × 35 cm, w, d, h, respectively) was used. The test lasted for 5 min, and the total number of nose pokes was measured.
2.17.3. Spatial learning and memory
This was performed using the Radial Arm Maze test without any food restriction/food reward. A cued version was used instead taking advantage of the exploratory nature of mice that drives it to explore unvisited arms. The equipment consisted of eight equidistantly spaced arms radially placed from a small circular central platform (Stoelting). Each arm is 5 cm × 35 cm × 9 cm (w, l, h, respectively), geometric figures were placed in the arms as spatial cues, and the equipment was sub-divided in the following zones (in this order, anticlockwise), 1) empty arm, with no figures attached to the wall (start arm); 2) diamond arm, with 13 diamonds (6 per wall, plus one more at the end of the arm); 3) circle arm, with 11 circles (5 per wall, plus one more at the end of the arm); 4) square arm, with 11 squares (5 per wall, plus one more at the end of the arm); 5) cross arm, with 9 crosses (4 per wall, plus one more at the end of the arm); 6) triangle arm, with 13 triangles (6 per wall plus, plus one more at the end of the arm); 7) star arm, with 11 jagged circles (5 per wall, plus one more at the end of the arm); and 8) rectangle arm, with 11 rectangles (5 per wall, plus one more at the end of the arm). A “non-return” sequence was programmed in the software in order to count the number of non-return explorations (all arms visited without repeating a previously visited arm). The test duration was 15 min, and the number of “non-return” and broken sequences (errors) were measured.
2.17.4. Short-time memory
This was determined using the Novel Object Recognition test (Stoelting) in a 40 cm × 40 cm × 35 cm (w, d, h) core with black infrared transparent Perspex insert. Mice were accustomed to this environment for 10 min during two consecutive days, and the test was performed on the third day. Mice were left to explore two identical equidistant cubes for 3 min (the acquisition phase), and returned for 15 min into its cage. One cube was substituted for a similar size sphere and mice were left to explore the objects for other 3 min (the retention phase). The ability to recognize the sphere as a novel object was determined as the ratio time spent watching the sphere/time spent watching the cube.
2.18. Experimental design and statistical analysis
Immunocytochemical quantifications were performed in brain samples from four male mice (n = 4). The protein carbonyls assessment was performed in the brain samples from three male mice (n = 3). For the behavioral experiments, six male mice (n = 6) were used per condition. The values were expressed as mean ± standard error of the mean (SEM). The statistical comparisons between two groups of values were performed using the Student's t-test. In all cases, p < 0.05 values were considered significant. Statistics were performed using the IBM SPSS Statistics software.
2.19. Animal procedures
All procedures performed with mice were approved by the Bioethics Committee of the University of Salamanca in accordance with the Spanish guidelines and regulations (law 6/2013).
3. Results
3.1. Neuron-specific GCL knockdown causes mild GSH deficiency in mice in vivo
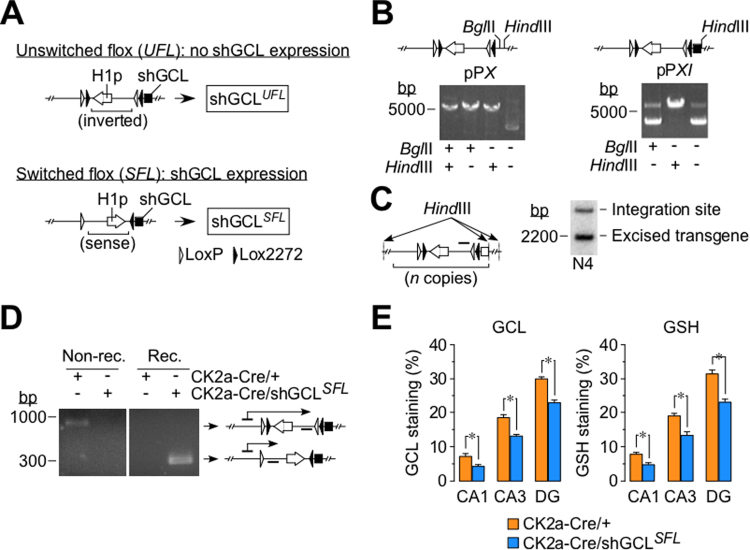
To induce cell-specific knockdown of GCL, we designed a DNA plasmid harboring a previously validated [18] small hairpin RNA (shRNA) sequence targeted against GCL (shGCL). The shGCL sequence was preceded by a LoxP/Lox2272-flanked RNA polymerase III (pol III) H1 promoter inserted in the inverse orientation (unswitched flox shGCL or shGCLUFL (Fig. 1A). This construct was designed to obtain switching of the LoxP/Lox2272-flanked region upon Cre recombinase-mediated activity to arrange the H1 promoter in the sense orientation (switched flox shGCL or shGCLSFL), thus amenable to trigger polymerase III-mediated shGCL transcription (Fig. 1A). During the construction of this DNA, an intermediary plasmid (pPX) harboring two unique restriction sites (BglII and HindIII) was generated, which is therefore ready for the insertion of any shRNA sequence (Fig. 1B; left panel). Insertion of the shGCL cDNA fragment into pPX yielded a plasmid (pPXI or shGCLUFL; Fig. 1B, right panel) that was used to generate transgenic mice under a C57Bl/6J-CBA hybrid lineage. A founder identified to harbor a unique integration site for shGCLUFL at the fourth (N4) generation (Fig. 1C), was successively backcrossed with C57Bl/6J until the congenic status (tenth generation) was achieved. To knockdown GCL in vivo, we next bred heterozygous shGCLUFL/+ mice with homozygous mice expressing Cre recombinase driven by the neuron-specific calcium-calmodulin kinase-2a (CK2a) promoter (CK2a-Cre/CK2a-Cre). Effective in vivo Cre-mediated recombination in the CK2a-Cre/shGCLSFL, but not in the CK2a-Cre/+ progeny was demonstrated by PCR in the hippocampus (Fig. 1D), i.e. the brain area where CK2a promoter is most active [19]. Furthermore, according to the immunofluorescence quantifications in the hippocampal CA1, CA3 and dentate gyrus (DG) regions, GCL and GSH abundances were moderately, albeit significantly reduced in the three analyzed regions, being the most prominent decrease observed in the CA1 layer (Fig. 1E; Supplementary Fig. S1A–E).
Fig. 1.
Neuron-specific GCL knock down causes mild GSH deficiency in mice in vivo. (A) Schematic representation of the DNA construct used to generate the unswitched (shGCLUFL) transgenic mouse (upper panel). The small hairpin RNA sequence against the catalytic subunit of glutamate cysteine ligase (shGCL) was preceded by a LoxP/Lox2272-flanked H1 promoter and inserted in the inverse orientation. Upon Cre recombinase-mediated activity, the H1 promoter is rearranged in the sense (switched) orientation (shGCLSFL) (bottom panel). (B) Restriction analysis of the pPX plasmid that harbors two unique restriction sites (BglII and HindIII) (left panel). Insertion of the shGCL cDNA fragment into pPX yielded the shGCLUFL plasmid (pPXI; right panel). (C) Southern blot of DNA isolated from the hippocampus of transgenic, shGCLUFL/+, adult mouse at the fourth generation (N4) showing the unique integration site and the excised transgene. (D) The N4 generation shGCLUFL/+ mice was backcrossed with C57Bl/6J until congenic status, and then bred with homozygous mice expressing Cre recombinase governed by the neuron-specific calcium-calmodulin kinase 2-a promoter (CK2a-Cre/ CK2a-Cre). The image shows the PCR analysis of the DNA isolated from the hippocampus of the adult CK2a-Cre/shGCLSFL and CK2a-Cre/+ mice. (E) Quantification of the GCL and GSH staining in the hippocampal CA1 and CA3 layers and the dentate gyrus (GD) of the 9 months-old CK2a-Cre/shGCLSFL male mice. Data are mean ± SEM values (n = 4). *p < 0.05 (Student's t-test).
3.2. GCL knockdown in neurons in vivo causes redox stress and oxidation of important proteins in the hippocampus
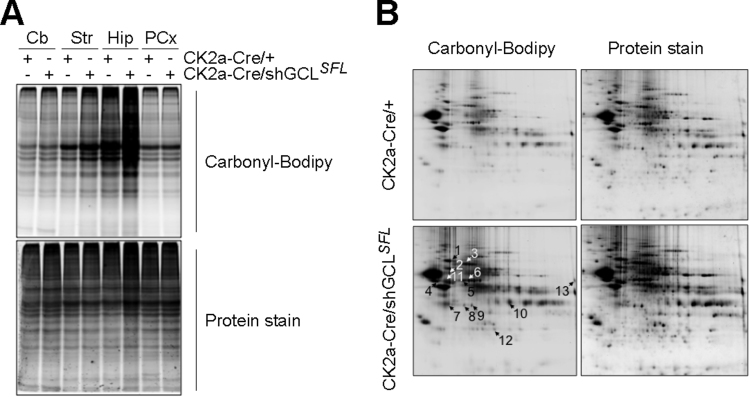
To test if GCL knockdown triggered redox stress, we first whether the decreased GSH and thiols were associated with increased ROS in shGCLSFL mouse embryonic fibroblasts (MEFs). Induction of Cre-mediated recombination in shGCLUFL MEFs efficiently decreased GCL protein abundance, as judged by western blotting, along with decreased total thiols and glutathione (total and reduced forms) (Supplementary Figs. S2A-C); in contrast, oxidized glutathione (GSSG) and ROS (H2O2) were increased (Supplementary Figs. S2C,D). These results, even not being a direct in vivo evidence, show that GCL knockdown causes redox stress in shGCLSFL cells. Next, we sought to investigate if GCL knockdown triggered redox stress in vivo. To do so, we assessed the abundance of protein carbonyls, i.e. a hallmark of ROS-mediated protein oxidation [24], in different brain areas. As shown in Fig. 2A and Supplementary Fig. S2E, protein oxidation was significantly increased in the hippocampus of CK2a-Cre/shGCLSFL mice when compared with controls (CK2a-Cre/+). However, this effect was not observed in the other brain regions analyzed, i.e. cerebellum, striatum and prefrontal cortex (Fig. 2A and Supplementary Fig. S2E). Given that the hippocampus was the brain region in which protein carbonylation was most affected, we next performed two-dimensional gel electrophoresis of these samples. As shown in Fig. 2B, we observed thirteen proteins that were oxidized by ~ 2–3-fold (see also Supplementary Fig. S2F). After purification and mass spectrometry analysis of these samples, we identified oxidized proteins involved in processes such as lysosomal trafficking (VATA, VATB), axonal growth and guidance (DPYL2), glycolysis (ENOG, ENOA, GAPDH and PGAM1), mitochondrial energy metabolism (PDHE1 and MDH1), energy homeostasis (KCRB) and protein translation (EF1A1) (Fig. 2B). Thus, knockdown of GCL leading to mild decrease in GSH in the hippocampus causes the oxidation of specific proteins important for neuronal functions.
Fig. 2.
GCL knock down in neurons in vivo causes oxidation of important proteins in the hippocampus. (A) Analysis of carbonylated proteins in different brain areas. Proteins obtained from different brain areas of the 9 months-old CK2a-Cre/+ and CK2a-Cre/shGCLSFL male mice were solubilized and carbonyl groups were derivatized with Bodipy-Hydrazide. Protein samples (10 μg) were separated in monodimensional polyacrylamide gel electrophoresis (PAGE). For total protein detection, gels were stained with Flamingo fluorescent stain. (B) Two dimensional PAGE (2D-PAGE) (pH range 3–10) of Bodipy-derivatized proteins and Flamingo staining. Arrows indicate spots corresponding to the most carbonylated proteins. Representative images are shown. Cb, cerebellum; Str, striatum, Hip, hippocampus; PCx, prefrontal cortex.
3.3. Neuron-specific GCL knockdown in vivo causes dendrite disruption in the CA1 region of the hippocampus
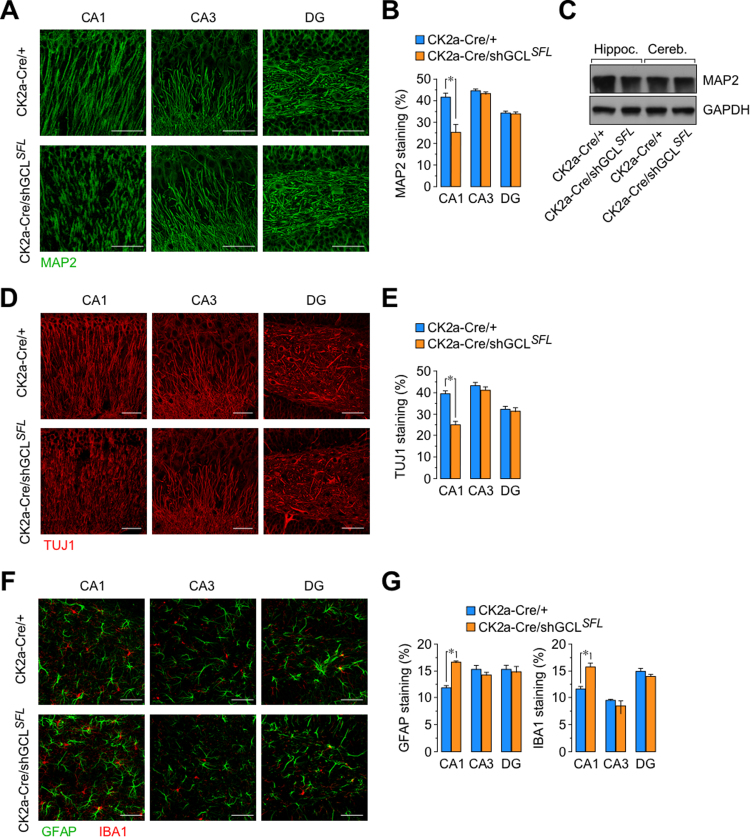
In view that the hippocampus of the CK2a-Cre/shGCLSFL mice was the brain area in which protein oxidation was strongest we further analyzed this area morphologically, immunohistochemically and biochemically. We found no evidence of the typical morphological changes associated with neuronal loss, including the DG, CA1 and CA3 width, the cortex layers length, or cell density in the striatum (Supplementary Figs. S3A and S3B). However, a highly significant dendrite disruption was observed in the CA1 region of the hippocampus. However, according to MAP2 staining, MAP2 protein abundance (Fig. 3A-C), and TUJ1 staining (Fig. 3D and E), our data are compatible with morphological changes in neurons compatible with dendrite disruption in the CA1 region of the hippocampus. Interestingly, dendrite organization in the CA3 or DG regions was unaffected (Fig. 3A, B, D and D). Since dendrite disruption might cause glial inflammation, we examined reactive astrocytes and microglia in these hippocampal regions. As shown in Fig. 3C and D, an increase in the GFAP and IBA1 staining -markers of astrocyte and microglial inflammation, respectively- was observed in the CA1, but not in the CA3 or DG regions of the hippocampus. Thus, mild GSH deficiency in the CK2a-Cre/shGCLSFL hippocampus causes neuronal dendrite disruption, specifically in the CA1 region, an effect that is anatomically linked with glial inflammation.
Fig. 3.
Neuron-specific GCL knock down in vivo causes dendrite disruption in the CA1 region of the hippocampus. (A) Immunostaining for the dendritic marker MAP2 (scale bars, 50 µm) showing dendrite disruption in the CA1 layer, but not in the CA3 layer and dendate gyrus (DG) of the hippocampus of the 9 months-old CK2a-Cre/shGCLSFL male mice when compared with the littermates controls (CK2a-Cre/+). Representative images are shown. (B) Quantification of the MAP2 staining images. Data are mean ± SEM values (n = 4). *p < 0.05 (Student's t-test). (C) Western blot against MAP2 in the hippocampus and cerebellum showing decreased MAP2 protein abundance in the hippocampus, but not in the cerebellum, of the 9 months-old CK2a-Cre/shGCLSFL male mice when compared with the littermates controls (CK2a-Cre/+). A representative western blot is shown. (D) Immunostaining for the dendritic marker TUJ1 (scale bars, 50 µm) showing dendrite disruption in the CA1 layer, but not in the CA3 layer and dendate gyrus (DG) of the hippocampus of the 9 months-old CK2a-Cre/shGCLSFL male mice when compared with the littermates controls (CK2a-Cre/+). Representative images are shown. (E) Quantification of the TUJ1 staining images. Data are mean ± SEM values (n = 4). *p < 0.05 (Student's t-test). (F) GFAP and IBA1 staining (scale bars, 50 µm), indicators of astrocyte and microgial inflammation, respectively, reveals inflammation in the CA1, but not in the CA3 or DG regions of the hippocampus of the CK2a-Cre/shGCLSFL mice when compared with the littermates controls (CK2a-Cre/+). Representative images are shown. (G) Quantification of the GFAP and IBA1 staining images. Data are mean ± SEM values (n = 4). *p < 0.05 (Student's t-test).
3.4. Knockdown of GCL in vivo triggers behavioral signs of cognitive decline
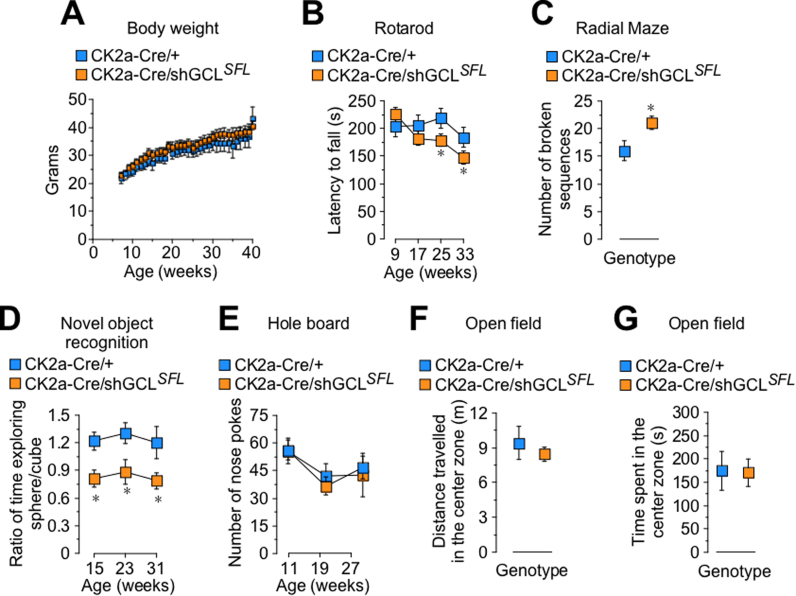
To assess whether oxidative stress in the hippocampus of the CK2a-Cre/shGCLSFL mice was sufficient to trigger signs of neurological damage, we performed a set of behavioral tests, blinded to the operator and digitally-registered. The body weight was unaltered at least up to 40 weeks of age in the CK2a-Cre/shGCLSFL mice when compared with their controls (CK2a-Cre/+) (Fig. 4A). However, we found an age-dependent decline in the performance of the rotarod test in the CK2a-Cre/shGCLSFL mice (Fig. 4B), reflecting a progressive deterioration of motor balance and coordination. Spatial memory, as revealed by the higher number of broken sequences in the radial arm maze test, was significantly impaired (Fig. 4C) as it was the performance at the novel object recognition test, indicating loss of short-term memory (Fig. 4D). Notably, these effects were absent in the shGCLUFL mice when compared with wild type mice (Supplementary Figs. S4A-G), thus ruling out any influence of the transgene insertion site on cognitive and motor functions. Fear/anxiety, which is reflected both by the nose pokes in the hole board test (Fig. 4E) and the distance travelled and time spent in the central zone of the open field test (Fig. 4F and G), were unaffected. Altogether, these findings indicate that a moderate decrease in GSH in the CK2a-Cre/shGCLSFL mice causes dendrite disruption in the CA1 layer of the hippocampus along with cognitive and motor dysfunctions.
Fig. 4.
Knock down of GCL in vivo triggers behavioral signs of cognitive decline. (A) Body weight was unaltered in CK2a-Cre/shGCLSFL male mice when compared with their controls (CK2a-Cre/+). (B) Age-dependent decline in the performance of the rotarod test in CK2a-Cre/shGCLSFL mice. (C) Higher number of broken sequences in the radial arm maze test in CK2a-Cre/shGCLSFL mice. (D) Lower performance at the novel object recognition test in CK2a-Cre/shGCLSFL mice. (E) Unaltered number of nose pokes in the Hole board test in CK2a-Cre/shGCLSFL mice. (F,G) Unaltered distance travelled and time spent in the central zone of the Open field test in CK2a-Cre/shGCLSFL mice. Data are mean ± SEM values (n = 6). *p < 0.05 (Student's t-test).
4. Discussion
Here we describe a transgenic mouse model that develops a moderate deficiency of GSH in the neurons of the adult brain in vivo. Our strategy to knockdown the catalytic subunit of the first rate-limiting enzyme of GSH biosynthesis, GCL, certainly differs with previous approaches aimed to extinguish GSH [25]. Thus, a GCL catalytic subunit ubiquitous knockout mouse model was first reported [26], [27], but it is not viable beyond embryonic day 8th [26]. Recently, a neuron-specific GCL knockout mouse model [28] was generated with the aim to eradicate, rather than attenuate, GSH abundance in neurons. Whereas the knockout strategy is appropriate to demonstrate fundamental roles of GSH in neurons [28], it does not seem to adequately resemble the moderate brain GSH deficiencies that take place during aging and neurodegenerative diseases [13]. Although this was also attempted in dopaminergic neurons by the full-length GCL cDNA antisense approach [29], it yielded a weak phenotype likely due to its promoter dependence to activation by doxycycline, a compound with poor penetrance across the blood brain barrier [30]. Accordingly, the mouse model of neuron-specific GSH attenuation that we herein report is the first to resemble the moderate brain GSH deficiency that is compatible with pathophysiological relevant situations.
The brain is particularly vulnerable to excess ROS [2], [3], [31], a fact that is likely due, amongst other possible factors, to the very low abundance of GCL and, accordingly, GSH [32]. In the brain, both GSH-synthesizing enzymes –GCL and GSH synthetase– are expressed in neurons and astrocytes, although more robustly in astrocytes [33], [34]. In fact, astrocytes actively synthesize GSH, which is cleaved to the extracellular space and shuttled in the form of GSH precursors into neurons [33], [35]. Thus, although astrocytes protect neurons from oxidative damage by supplying GSH precursors, neurons have to build up GSH by the de novo synthesis pathway [18], [35]. Hence, disruption of the GSH-synthesizing enzyme GCL in neurons is appropriate for affecting their GSH abundance even in the presence of neighboring astrocytes [18].
Since the CK2a promoter is most active in the hippocampus [19], we focused in this brain area to analyze GCL and GSH abundances in the neurons. Consistently with this, we found that GCL and GSH levels moderately decreased throughout the hippocampus of the CK2a-Cre/shGCLSFL mice. Furthermore, protein carbonyls, which indicate protein oxidation during GSH deficiency [36], were specifically high in the hippocampus, without significant affection in the cortex, striatum or cerebellum, i.e. brain regions with lesser CK2a promoter activity [19]. Protein glutathionylation, which is another type of protein posttranslational modification that is consequence of GSH depletion, has been suggested to promote loss of cell survival [37]. However, the moderate GSH deficiency that we observed in the neurons of the hippocampus did not cause brain morphological alterations compatible with cell death. This contrasts with the abrupt neuronal death generated by GSH eradication in the GCL knockout model [28], further supporting the notion that mild GSH deficiency, such as that herein achieved, is a suitable approach to investigate functional roles of GSH in living neurons.
To further investigate the potential consequences of mild GSH deficiency for neuronal functions, we performed a mass spectrometry analysis of the oxidized proteins found in the hippocampus. Protein carbonylation is one of the markers of oxidative stress. Major pathways leading to protein carbonylation are the consequence of iron imbalance or lipid peroxidation. When available in the cell (mainly through the labile iron pool), iron would promote protein carbonylation through the well-known Fenton reaction. Since glutathione have been proposed as the main iron(II) chelator [1], [38], its deficiency could lead to increase free iron and catalyze Fenton reaction. In this paper, the decreased GSH synthesis caused by GCL knockdown, would favor protein oxidative modifications observed by decreasing GSH pool. In fact, we identified oxidized proteins known to regulate neuronal functions such as axonal growth and guidance, lysosomal trafficking and energy metabolism. Whilst future studies will be needed to define the repercussion that the oxidation of each of these proteins has for neuronal function, our data demonstrate that a moderate level of redox stress is sufficient to cause posttranslational protein modifications. The immunohistochemical characterization of the CK2a-Cre/shGCLSLF mice revealed neuronal dendrites disruption in a defined brain region, namely the CA1 layer of the hippocampus. Interestingly, GCL and GSH deficiencies were observed in the CA3 and DG regions, besides the CA1 layer, suggesting higher vulnerability of the CA1 neuronal dendrites. The occurrence glial inflammation limited to the CA1 layer strongly supports this notion. Moreover, it should be noted that the decrease in GCL and GSH staining was ~ 40% in neurons of the CA1 layer, in contrast with the ~ 30% loss observed in neurons of the CA3 and DG regions. Whether there is a threshold effect in dendrite disruption according to fine modulation of GSH levels is an interesting possibility worth pursuing. In sustain of this concept, in a previously reported threshold study, it was shown that the CA1 layer of the hippocampus is more vulnerable than the paramedian cortex against a metabolic stress [39].
According to the behavioral tests performed in the CK2a-Cre/shGCLSFL mice, both the spatial and short time memories were impaired. In principle, these findings are supported by previous studies performed in the ubiquitous knockout model for the modulatory GCL subunit [40], [41]. However, whether the massive liver failure occurring in that model explains the cognitive [40] and psychiatric [41] phenotypes due to hepatic encephalopathy is unknown. In a more recent study, neuron-specific loss of the catalytic GCL subunit was achieved causing memory impairment [28]. Whilst this is an interesting approach to investigate fundamental roles of GSH, it abolishes GSH thus triggering abrupt neurodegeneration [28], which likely accounted for the cognitive impairment. In contrast, our GCL knockdown strategy renders mice with spatial and short time memory loss despite GSH is still present -although at a slight low level. Furthermore, neuronal dendrite disruption was confined to the CA1 layer of the hippocampus, a region that is responsible for the control of the cognitive function [17], [42], [43]. These studies further reinforce the link between mild loss of hippocampal GSH and cognitive dysfunction in the CK2a-Cre/shGCLSFL mouse. The slight motor impairment that occurred in the CK2a-Cre/shGCLSFL mice reflects the possible affection of brain areas that control motor coordination. However, the striatum and cerebellum –brain regions controlling these tasks– showed no apparent signs of oxidative damage. Further studies would thus be required to address the specific link between motor unbalance and hippocampal redox stress in the CK2a-Cre/shGCLSFL mice.
In conclusion, here we show that moderate down modulation of neuronal glutathione leads to dendrite disruption in a concrete set of hippocampal neurons that cause behavioral symptoms compatible with cognitive impairment and loss of motor coordination in mice. These data suggest that finely tuning down glutathione is a suitable strategy to ascertaining novel functions of this cofactor for neuronal activity.
Acknowledgements
We are grateful to Ms. L. Martin-Martin and Ms. Estefania Prieto-Garcia (Fondo Social Europeo, Youth Employment Initiative, Junta de Castilla y León), from the Universidad de Salamanca, and Ms. Isabel Sanchez and Ms. Alba Sorolla, from the Proteomics facility at the University of Lleida, for technical support. We acknowledge Dr. R. Requejo-Aguilar for experimental support and Prof. D. Martin-Zanca and Dr. L. Parrilla for the help in the Southern blotting.
Acknowledgments
Conflict of interest
The authors declare that they have no competing financial interest.
Funding sources
This work was funded by the MINECO, Spain (SAF2016-78114-R to JPB; Red de Excelencia CONSOLREDOX SAF2015-71521-REDC to JPB and JR), H2020 European Commission, Belgium (BatCure Grant 666918 to JPB; PANA grant 686009 to AA), Instituto de Salud Carlos III, Spain (CIBERFES CB16/10/00282 to JPB; PI15/00473 and RD16/0019/0018 to AA), Junta de Castilla y León, Spain (IES007P17) and Fundación Ramón Areces, Spain (to AA). This work was funded by FEDER, European Commission, Belgium.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.08.003.
Appendix A. Supplementary material
Supplementary material
References
- 1.Hider R.C., Kong X. Iron speciation in the cytosol: an overview. Dalton Trans. 2013;42:3220–3229. doi: 10.1039/c2dt32149a. [DOI] [PubMed] [Google Scholar]
- 2.Bolaños J.P. Bioenergetics and redox adaptations of astrocytes to neuronal activity. J. Neurochem. 2016;139:115–125. doi: 10.1111/jnc.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Fernandez S., Almeida A., Bolaños J.P. Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem. J. 2012;443:3–11. doi: 10.1042/BJ20111943. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Fabuel I., Le Douce J., Logan A., James A.M., Bonvento G. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA. 2016;113:13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Temple M.D., Perrone G.G., Dawes I.W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005;15:319–326. doi: 10.1016/j.tcb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Levy O.A., Malagelada C., Greene L.A. Cell death pathways in Parkinson's disease: proximal triggers, distal effectors, and final steps. Apoptosis: Int. J. Program. Cell Death. 2009;14:478–500. doi: 10.1007/s10495-008-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolaños J.P., Moro M.A., Lizasoain I., Almeida A. Mitochondria and reactive oxygen and nitrogen species in neurological disorders and stroke: therapeutic implications. Adv. Drug Deliv. Rev. 2009;61:1299–1315. doi: 10.1016/j.addr.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Halliwell B. Free radicals and antioxidants - quo vadis? Trends Pharmacol. Sci. 2011;32:125–130. doi: 10.1016/j.tips.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Quintana-Cabrera R., Fernandez-Fernandez S., Bobo-Jimenez V., Escobar J., Sastre J. g-Glutamylcysteine detoxifies reactive oxygen species by acting as glutathione peroxidase-1 cofactor. Nat. Commun. 2012;3:718. doi: 10.1038/ncomms1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenner P., Schapira A.H., Marsden C.D. New insights into the cause of Parkinson's disease. Neurology. 1992;42:2241–2250. doi: 10.1212/wnl.42.12.2241. [DOI] [PubMed] [Google Scholar]
- 12.Jenner P., Olanow C.W. Oxidative stress and the pathogenesis of Parkinson's disease. Neurology. 1996;47:S161–S170. doi: 10.1212/wnl.47.6_suppl_3.161s. [DOI] [PubMed] [Google Scholar]
- 13.Liu H., Wang H., Shenvi S., Hagen T.M., Liu R.M. Glutathione metabolism during aging and in Alzheimer disease. Ann. N.Y. Acad. Sci. 2004;1019:346–349. doi: 10.1196/annals.1297.059. [DOI] [PubMed] [Google Scholar]
- 14.Kamat C.D., Gadal S., Mhatre M., Williamson K.S., Pye Q.N., Hensley K. Antioxidants in central nervous system diseases: preclinical promise and translational challenges. J. Alzheimers Dis. 2008;15:473–493. doi: 10.3233/jad-2008-15314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahn S., Cohen G. The oxidant stress hypothesis in Parkinson's disease: evidence supporting it. Ann. Neurol. 1992;32:804–812. doi: 10.1002/ana.410320616. [DOI] [PubMed] [Google Scholar]
- 16.Vina J., Borras C., Abdelaziz K.M., Garcia-Valles R., Gomez-Cabrera M.C. The free radical theory of aging revisited: the cell signaling disruption theory of aging. Antioxid. Redox Signal. 2013;19:779–787. doi: 10.1089/ars.2012.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettio L.E.B., Rajendran L., Gil-Mohapel J. The effects of aging in the hippocampus and cognitive decline. Neurosci. Biobehav. Rev. 2017;79:66–86. doi: 10.1016/j.neubiorev.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 18.Diaz-Hernandez J.I., Almeida A., Delgado-Esteban M., Fernandez E., Bolaños J.P. Knockdown of glutamate-cysteine ligase by small hairpin RNA reveals that both catalytic and modulatory subunits are essential for the survival of primary neurons. J. Biol. Chem. 2005;280:38992–39001. doi: 10.1074/jbc.M507065200. [DOI] [PubMed] [Google Scholar]
- 19.Minichiello L., Korte M., Wolfer D., Kuhn R., Unsicker K. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 20.Tietze F. Enzyme method for quantitative determination of nanogram amounts of total and oxidized glutathione: application to mammalian blood and other tissues. Anal. Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 21.Tamarit J., de Hoogh A., Obis E., Alsina D., Cabiscol E., Ros J. Analysis of oxidative stress-induced protein carbonylation using fluorescent hydrazides. J. Proteom. 2012;75:3778–3788. doi: 10.1016/j.jprot.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 22.Ros-Simo C., Moscoso-Castro M., Ruiz-Medina J., Ros J., Valverde O. Memory impairment and hippocampus specific protein oxidation induced by ethanol intake and 3, 4-methylenedioxymethamphetamine (MDMA) in mice. J. Neurochem. 2013;125:736–746. doi: 10.1111/jnc.12247. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez C., Sobrino T., Agulla J., Bobo-Jimenez V., Ramos-Araque M.E. Neovascularization and functional recovery after intracerebral hemorrhage is conditioned by the Tp53 Arg72Pro single-nucleotide polymorphism. Cell Death Differ. 2017;24:144–154. doi: 10.1038/cdd.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irazusta V., Moreno-Cermeno A., Cabiscol E., Tamarit J., Ros J. Proteomic strategies for the analysis of carbonyl groups on proteins. Curr. Protein Pept. Sci. 2010;11:652–658. doi: 10.2174/138920310794557664. [DOI] [PubMed] [Google Scholar]
- 25.Dalton T.P., Chen Y., Schneider S.N., Nebert D.W., Shertzer H.G. Genetically altered mice to evaluate glutathione homeostasis in health and disease. Free Radic. Biol. Med. 2004;37:1511–1526. doi: 10.1016/j.freeradbiomed.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 26.Dalton T.P., Dieter M.Z., Yang Y., Shertzer H.G., Nebert D.W. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem. Biophys. Res. Commun. 2000;279:324–329. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 27.Shi Z.Z., Osei-Frimpong J., Kala G., Kala S.V., Barrios R.J. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc. Natl. Acad. Sci. USA. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng W., Rosca M., Fan Y., Hu Y., Feng P. Gclc deficiency in mouse CNS causes mitochondrial damage and neurodegeneration. Hum. Mol. Genet. 2017;26:1376–1390. doi: 10.1093/hmg/ddx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chinta S.J., Kumar M.J., Hsu M., Rajagopalan S., Kaur D. Inducible alterations of glutathione levels in adult dopaminergic midbrain neurons result in nigrostriatal degeneration. J. Neurosci. 2007;27:13997–14006. doi: 10.1523/JNEUROSCI.3885-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 31.Bolaños J.P., Almeida A., Stewart V., Peuchen S., Land J.M. Nitric oxide-mediated mitochondrial damage in the brain: mechanisms and implications for neurodegenerative diseases. J. Neurochem. 1997;68:2227–2240. doi: 10.1046/j.1471-4159.1997.68062227.x. [DOI] [PubMed] [Google Scholar]
- 32.Makar T.K., Nedergaard M., Preuss A., Gelbard A.S., Perumal A.S., Cooper A.J.L. Vitamin E, ascorbate, glutathione, glutathione disulfide, and enzymes of glutathione metabolism in cultures of chick astrocytes and neurones: evidence that astrocytes play an important role in antioxidative processes in the brain. J. Neurochem. 1994;62:45–53. doi: 10.1046/j.1471-4159.1994.62010045.x. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez-Blasco D., Santofimia-Castano P., Gonzalez A., Almeida A., Bolaños J.P. Astrocyte NMDA receptors' activity sustains neuronal survival through a Cdk5-Nrf2 pathway. Cell Death Differ. 2015;22:1877–1889. doi: 10.1038/cdd.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dringen R., Brandmann M., Hohnholt M.C., Blumrich E.M. Glutathione-dependent detoxification processes in astrocytes. Neurochem. Res. 2015;40:2570–2582. doi: 10.1007/s11064-014-1481-1. [DOI] [PubMed] [Google Scholar]
- 35.Dringen R., Pfeiffer B., Hamprecht B. Synthesis of the antioxidant glutathione in neurons: supply by astrocytes of CysGly as precursor for neuronal glutathione. J. Neurosci. 1999;19:562–569. doi: 10.1523/JNEUROSCI.19-02-00562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butterfield D.A., Gu L., Di Domenico F., Robinson R.A. Mass spectrometry and redox proteomics: applications in disease. Mass Spectrom. Rev. 2014;33:277–301. doi: 10.1002/mas.21374. [DOI] [PubMed] [Google Scholar]
- 37.Franco R., Cidlowski J.A. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 38.Hider R.C., Kong X.L. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals. 2011;24:1179–1187. doi: 10.1007/s10534-011-9476-8. [DOI] [PubMed] [Google Scholar]
- 39.Davey G.P., Canevari L., Clark J.B. Threshold effects in synaptosomal and nonsynaptic mitochondria from hippocampal CA1 and paramedian neocortex brain regions. J. Neurochem. 1997;69:2564–2570. doi: 10.1046/j.1471-4159.1997.69062564.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen B., Bai Y., Sun M., Ni X., Yang Y. Glutathione S-transferases T1 null genotype is associated with susceptibility to aristolochic acid nephropathy. Int. Urol. Nephrol. 2012;44:301–307. doi: 10.1007/s11255-011-0082-z. [DOI] [PubMed] [Google Scholar]
- 41.Kulak A., Cuenod M., Do K.Q. Behavioral phenotyping of glutathione-deficient mice: relevance to schizophrenia and bipolar disorder. Behav. Brain Res. 2012;226:563–570. doi: 10.1016/j.bbr.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 42.Wu W.W., Oh M.M., Disterhoft J.F. Age-related biophysical alterations of hippocampal pyramidal neurons: implications for learning and memory. Ageing Res. Rev. 2002;1:181–207. doi: 10.1016/s1568-1637(01)00009-5. [DOI] [PubMed] [Google Scholar]
- 43.Okuyama T. Social memory engram in the hippocampus. Neurosci. Res. 2017;129:17–23. doi: 10.1016/j.neures.2017.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material