Abstract
Statement of the Problem:
Periodontal diseases are complex oral diseases characterized by bacterial-induced inflammatory destruction of tooth-supporting tissues. Porphyromonas gingivalis (P. gingivalis) is a common gram-negative anaerobic oral bacteria strongly associated with periodontal disease.
Purpose:
The present study was conducted to estimate prevalence of P. gingivalis in patients with periodontal diseases by using meta-analysis method.
Martials and Method:
Different databases including PubMed, EmBase, Scopus, the Institute for Scientific Information (ISI) Web of Science, and the Cochrane Library were searched to identify original English-language studies addressing prevalence of P. gingivalis in periodontal diseases up to December 2014. The random effects model was applied in the meta-analysis and the heterogeneity between studies was assessed using a Cochran test and the I2 index. Funnel plots and Egger test were used to examine publication bias. Statistical analyses were performed using STATA version 12.
Results:
Forty-two eligible studies published during 1993- 2016 were selected for meta-analysis. Considering all the included studies, the total sample size was 5,884 individuals containing 2,576 healthy people with a mean age of 37.21±7.45 years and 3,308 periodontal patients with a mean age of 44.16±8.35 years. Overall, the prevalence of P. gingivalis was 78% [95% CI: 74-81] in periodontal diseases group and 34% [95% CI: 26-41] in healthy individuals. There was a significantly higher prevalence of P.gingivalis in individuals with periodontal diseases compared to healthy subjects [78% versus 34%, respectively].
Conclusion:
This study indicates that P. gingivalis is highly present in subjects with periodontal diseases and it also appears in periodontally healthy people, although to a lesser extent. Thus, the presence of P. gingivalis increases the chance of periodontal disease and it can be considered as a main potential risk factor.
Keywords: Porphyromonas gingivalis , Periodontal diseases , Chronic periodontitis , Aggressive periodontitis , Gingivitis
Introduction
Periodontal disease is an infectious clinical entity characterized by the destruction of supporting tissues of the teeth (periodontal ligament and alveolar bone) and can lead to gum recession, soft tissue damage, bone loss and tooth loss.[1-2] It is also an important risk factor for multiple systemic diseases, such as rheumatoid arthritis and cardiovascular disease, high blood pressure, diabetes, pulmonary disease, pregnancy, cancer and MS in later years of life. The scientific rationale for these problems would probably be related to chronic and long-term aspects of inflammation in periodontal disease.[3-11]
The severity and progression of periodontal disease is influenced by multiple risk factors, including genetic, environmental and host factors. Moreover, polymicrobial biofilms present in subgingival crevices are the most important etiological factor in the pathogenesis of periodontal disease.[12-13] Studies show that the oral cavity is a source of different microorganisms and more than 700 species of bacteria have been detected in subgingival biofilms.[2,12] Polymicrobial communities develop through interspecies interactions and adaptation within the surrounding microenvironments.[14] Some of the major periodontogenic pathogens are aggregatibacter actinomycetemcomitans, porphyromonas gingivalis, prevotella nigrescens, fusobacterium nucleatum, and treponema denticola.[15]
The most common bacteria associated with periodontal disease, porphyromonas gingivalis (P. gingivalis), is a gram-negative obligatory anaerobe, which resides in the mouth and is strongly associated with periodontal disease.[16-20] P. gingivalis is one of the species that constitute the red complex group and is the most important in the initiation or progression of periodontal disease.[21-22] The red complex group including P. gingivalis, T. denticola and T. forsythia from unattached subgingival plaque; occur in combination in periodontal pockets, appear in the developing biofilm and are considered the first pathogens involved in the clinical destruction of periodontal tissues in a cooperative manner.[16-22] Of the bacteria believed to be pathogenic in periodontal disease, P. gingivalis has been extensively studied due to its unique ability to evade the immune response;[23] as a result, it creates an environment that facilitates dysbiosis of subgingival microbiota, and the dysbiotic microbiota with increased pathogenicity overactivate inflammation in periodontal tissues.[24] gingivalis deregulates host immune systems by producing a number of virulence factors, such as lipopolysaccharide, fimbriae, and several proteases.[25] The various surface components of P. gingivalis enable the bacterium to formation of a biofilm that protects it against the host’s defense.[26-27]
Due to the high prevalence and complications of periodontal disease, planning to prevention and treatment of this disease seemed necessary. A major aspect of periodontal disease prevention is the identification of potential periodontal pathogens; in the other hand, determining the risk factors that affect the incidences of periodontal disease is crucial for preventive and management strategies.[28] Since P.gingivalis has been known as a major etiological agent in periodontal disease and a risk factor for periodontal disease, it is of particular importance to investigate the prevalence of this periodontal pathogen, which can be an important approach for prevention, and treatment of periodontal disease. In addition, study of prevalence of oral microbes in periodontal patients is an important effort to provide the basic data for further control of the oral complications in these patients.[29] P. gingivalis has been extensively studied for well over a century and extensive studies have been conducted to control this pathogen causing dental diseases; in order to authenticate conducted studies, performing a meta-analysis seems to be necessary. Since combination of different studies via meta-analysis leads to a suitable sample size and better resolution, it can provide an overall precise and valid understanding of a desired subject compared to the separated reported studies. Therefore, it seems that assessment of prevalence of P. gingivalis in patients with periodontal disease via meta-analysis can be a useful tool for an overall and clear understanding of this disease.[30] The aim of this study was providing an overall summary measure of the prevalence of P. gingivalis in patients with periodontal disease by synthesizing available studies.
Materials and Method
Search strategies
This article was written according to the PRISMA guidelines.[31] We performed a literature search of the Scopus, ISI web of Science, PubMed, EmBase, and the Cochrane Library databases for original articles that present the prevalence or incidence of P. gingivalis among patients with periodontal disease from 1993- 2016. The searches were applied by using the keywords porphyromonas gingivalis, chronic periodontitis, aggressive periodontitis, gingivitis, and related words; also, the study was limited to the English languages. We also used wildcard symbol ‘*’ and combined the search words or phrases using Boolean operators (‘AND’, ‘OR’, ‘NOT’) and also scanned bibliographies of retrieved articles to expand the search. In addition, relevant original articles noted in the reference lists of each selected article were also evaluated as a further search tool. Furthermore, review articles were manually searched for additional references.
Inclusion and exclusion criteria
All papers with the selected keywords in their titles or abstracts were included in the initial list and other unrelated articles were eliminated. Accordingly, all original articles that reported the prevalence of P. gingivalis in periodontal disease were reviewed. The non-human studies were excluded. Studies that were conducted in patients with diseases other than periodontal diseases, non-epidemiologic studies, presented insufficient data, in languages other than English were excluded. In addition, review articles, congress abstracts, meta-analyses or systematic reviews and duplicate publication of the same study were omitted. In the necessary cases, authors were contacted for additional information. The STROBE (strengthening the reporting of observational studies in epidemiology) statement was used for quality control of the studies.32 Non-qualified studies were excluded.
Data Extraction
Abstracts and full articles were reviewed independently by two of the authors, and if results were discordant, papers were reviewed jointly until the differences were resolved.
The following items were extracted from the studies: first author, year of publication, study location, sample size, sample age, P. gingivalis screening method, sample specimens and percentage of P.gingivalis in patient and healthy individual (Table 1). Two of the authors independently reviewed the abstracts and full articles and extracted data according to a standard protocol. In which cases the results were discordant, papers were reviewed jointly until the differences were resolved. The data were entered into a standardized data extraction form and entered into Microsoft Excel.
Table 1.
Characteristics of the 48 trials included in the meta-analysis
| First author, [Reference] | Country [year of publication] | Case | Control | Mean age | Prevalence of P.g [%] | Type of disease | Sample specimens | Methods of Assay | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | |||||||
| [27] | Jamaica [2000] | 35 | 65 | 14-18 | 14-18 | 77 | 34 | Periodontitis | subgingival plaque | PCR method |
| [28] | Japan [2003] | 35 | 18 | 51.8 ±7.29 | 27.3±3.71 | 97.1 | 5.6 | chronic periodontitis | subgingival plaque | PCR method |
| [29] | Brazil [2011] | 33 | 32 | 35 | 0 | apical periodontitis | purulent exudate aspirates | PCR method | ||
| [1] | Japan [2013] | 85 | 20 | 57.4±13.1 | 45.9±17.0 | 65 | 40 | Chronic periodontitis | plaque samples | PCR method |
| [30] | Korea [2005] | 17 | 19 | 52±11.1 | 49±10.2 | 24.7 | 27.6 | Gingivitis | subgingival plaque | PCR method |
| [31] | USA [2009 | 39 | 40 | 52±11.1 | 49±10.2 | 77 | 40 | Periodontitis | subgingival plaque | stabilization P. gingivalis antibody seropositivity |
| [32] | Ohio State [1998] | 130 | 181 | 51.4±9.3 | 49.2± 9 | 79 | 25 | Periodontitis | subgingival plaque | PCR method |
| [33] | Brazil [2002] | 50 | 50 | 45.5±9.7 | 32.3±8.9 | 70 | 60 | Periodontitis | subgingival plaque | PCR method |
| [34] | Netherlands [2002] | 116 | 94 | 42.9±9.8 | 40.4±11.9 | 59.5 | 10.6 | Periodontitis | subgingival plaque | Anaerobic cultivation |
| [35] | Chile [2007] | 20 | 6 | 27±5.2 | 22.7±4.9 | 50 | 50 | Periodontitis/gingivitis | subgingival plaque | Bacterial culture |
| [36] | Japan [2013] | 139 | 380 | 87.1 | 36.8 | Periodontitis | Dental plaque | PCR method | ||
| [37] | Japan [2001] | 103 | 20 | 89.9 | 10 | Periodontitis | Saliva and subgingival plaque | PCR method | ||
| [38] | Brazil [2004] | 57 | 25 | 89.4 | 8 | periodontal attachment loss | subgingivalplaque | PCR method | ||
| [38] | Brazil [2004] | 20 | 25 | 30 | 8 | Gingivitis | subgingival plaque | PCR method | ||
| [39] | Chile [2007] | 115 | 136 | 81.7 | 22.1 | Chronic periodontitis | subgingival samples | PCR method | ||
| [40] | Taiwan [2004] | 407 | 91 | 85.7 | 23.1 | periodontal disease | subgingival plaque | indirect immunofluorescent assay | ||
| [41] | Korea [2000] | 29 | 20 | 96 | 18 | Periodontitis | subgingival plaque | PCR method | ||
| [42] | Japan [2000] | 29 | 15 | 61.8 | 0 | Periodontitis | subgingival plaque | culture | ||
| [43] | Italy [2013] | 66 | 46 | 48.9±18.2 | 31.6±18.6 | 52 | 83 | Periodontitis | periodontal pocket | PCR method |
| [44] | USA [1993] | 28 | 18 | 18-59 | 18-59 | 59 | 78 | Periodontitis | subgingival plaque | ELISA |
| [45] | Lebanon [2010] | 20 | 20 | 34.3±5.36 | 26.10 ±4.57 | 65 | 10 | Periodontitis | Oral plaque | PCR method |
| [46] | Germany [2009] | 46 | 21 | 55.2±11.2 | 66.6±1.5 | 76 | 62 | Chronic periodontitis | subgingival plaque | PCR method |
| [46] | Germany [2009] | 44 | 21 | 34.4 ± 6.5 | 66.6±1.5 | 65 | 62 | Aggressive periodontitis | subgingival plaque | PCR method |
| [47] | Chine [2009] | 48 | 25 | 38.9 ± 9.9 | 23.6±1.8 | 93.8 | 32 | Periodontitis | subgingivalplaque | PCR method |
| [48] | Chine [2013] | 27 | 20 | 96.3 | 30 | Chronic periodontitis | subgingival | PCR and reverse hybridization assay | ||
| [49] | Chine [2013] | 80 | 56 | 93.8 | 4.7 | Aggressive periodontitis | gingival crevicular fluid | PCR method | ||
| [50] | Italy [1998] | 33 | 21 | 56.5 | 4.7 | severe periodontal disease | subgingival plaque | culture | ||
| [51] | Chine [2007] | 61 | 30 | 42.4±8.7 | 37.35±7.3 | 62.3 | 10 | chronic periodontitis | subgingival plaque | PCR method |
| [20] | Spain [2012] | 33 | 37 | 43.39±7.4 | 40.68±7.1 | 66.7 | 27 | chronic periodontitis | subgingival plaque | PCR method |
| [20] | Spain [2012] | 16 | 37 | 38.81±6.9 | 40.68±7.1 | 37.5 | 27 | Gingivitis | subgingival plaque | PCR method |
| [52] | China [2013] | 25 | 20 | 92 | 35 | Periodontitis | gingival crevicular fluid | PCR method | ||
| [53] | Colombia [2007] | 143 | 40 | 39.5±9.85 | 32.6±10.6 | 64.3 | 7.5 | Periodontitis | subgingival plaque | culture |
| [54] | Italy [2012] | 127 | 66 | 48.9±18.2 | 31.6 ±18.6 | 71.7 | 33.4 | Periodontitis | periodontal pocket microbiota | PCR method |
| [55] | China[2014] | 25 | 29 | 84 | 24.1 | Periodontitis | subgingival plaque | PCR method | ||
| [56] | Thailand[2009] | 20 | 20 | 95 | 45 | chronic periodontitis | subgingival plaque | PCR method | ||
| [57] | Iran [2007] | 61 | 40 | 43±11 | 41.35±9.8 | 83.6 | 4 | chronic periodontitis | subgingival plaque | PCR method |
| [58] | China [2006] | 55 | 17 | 81.8 | 17.6 | Aggressive periodontitis | subgingival plaque | PCR method | ||
| [59] | China [2005] | 152 | 30 | 91.5 | 3.3 | chronic periodontitis | Periodontal pocket and gingival sulcus | PCR method | ||
| [60] | Spain [2004] | 30 | 30 | 30 | 13.3 | Gingivitis | subgingival plaque | PCR method | ||
| [60] | Spain [2004] | 32 | 30 | 81.3 | 13.3 | Periodontitis | subgingival plaque | PCR method | ||
| [2] | China [2015] | 42 | 32 | 75 | 63 | chronic gingivitis | Gingival crevicular fluid | PCR method | ||
| [2] | China [2015] | 95 | 32 | 91 | 63 | chronic periodontitis | Gingival crevicular fluid | PCR method | ||
| [6] | Japan [2013] | 20 | 10 | 43.6±11.1 | 28.7 ± 3.2 | 75 | 0 | chronic periodontitis | subgingival plaque | PCR method |
| [10] | Korea [2013] | 284 | 128 | 48.3±9.5 | 42.3±13.5 | 97.5 | 57.5 | chronic periodontitis | subgingival plaque | PCR method |
| [61] | Germany [2012] | 33 | 20 | 33.39±10.47 | 37.65±10.88 | 51.6 | 10 | chronic periodontitis | subgingival plaque | PCR method |
| [62] | Colombia [2007] | 80 | 30 | 33.91±9.32 | 26.90± 7.17 | 79.8 | 10 | Periodontitis | subgingival plaque | PCR method |
| [63] | Switzerland[2004] | 17 | 33 | 53.1±8.53 | 26.8±5.3 | 71 | 90 | chronic periodontitis | subgingival plaque | PCR method |
Data Synthesis and Analysis
The main objective of the study was to evaluate the prevalence of P.gingivalis; therefore, the binomial distribution was used to calculate the variance of each study, since the prevalence of P.gingivalis and the sample number have been extracted in each study. To combine the prevalence of various studies, the average weight was used and each study was weighted in proportion to its variance. The heterogeneity between studies was assessed using a Cochran test and the I2 index. Considering the significant heterogeneity of the studies, the random effects model was applied in the meta-analysis and the findings are described in forest plots (the point estimations and their 95% CI). Sensitivity analyses were also performed. To examine publication bias, Funnel plots and Egger test were used. p Values <0.05 were considered significant in heterogeneity tests. Statistical analyses were performed using STATA version 12.
Results
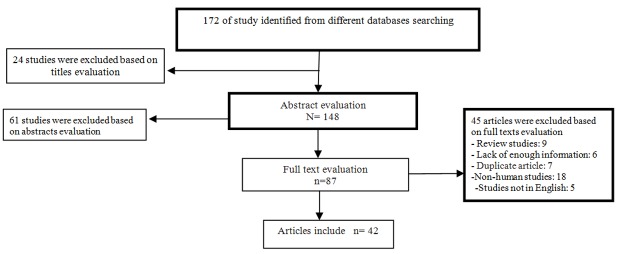
In the search process, 172 articles were identified through the literature search. The screening process of studies was completed based on titles, abstracts, and full texts evaluation in the first step and after the initial screening of abstracts and titles, 85 papers were excluded (of these, 24 were on non-periodontal diseases, 61 were unrelated) and 87 papers remained for full-text evaluation. In a secondary screening and after full text review, we excluded another 45 articles (Eighteen studies were non-human (animal) studies, six studies collected insufficient data, five studies was not published in English, seven were duplicated articles, nine studies were retrospective, review and meta-analyses studies. Finally, 42 case-control studies that were published between 1993 and 2015 selected for the final analysis. [1,2,12,16,26, 33-69] (Figure 1) The characteristics and extracted data from these studies are summarized in , including quality scores.
Figure1.
Flow diagram of the studies identified in the systematic review and meta-analysis
Considering all the included studies, the total sample size was 5,884 individuals containing, 576 healthy people, ranging from 14 to 67 years of age with a mean age of 37.21±7.452 years and 3,308 periodontal patients, ranging from 14 to 59 years of age with a mean age of 44.16±8.35 years.
Among the 42 studies included in this meta-analysis, the prevalence of P. gingivalis in periodontal diseases group was 78% (95% CI: 74-81; Figure 2). Considering 45 included studies (three studies were excluded due to major difference in reported prevalence with the other studies), the prevalence of P. gingivalis in healthy individuals was 34% (95% CI: 26-41, Figure 3). The significant differences observed between the prevalence of P. gingivalis in periodontal diseases and healthy individuals (Figures 2 and 3). As seen in Figures 2 and 2. the prevalence of P.gingivalis in periodontal disease was significantly higher compared to healthy subjects (78% versus 34%, respectively).
Figure2.
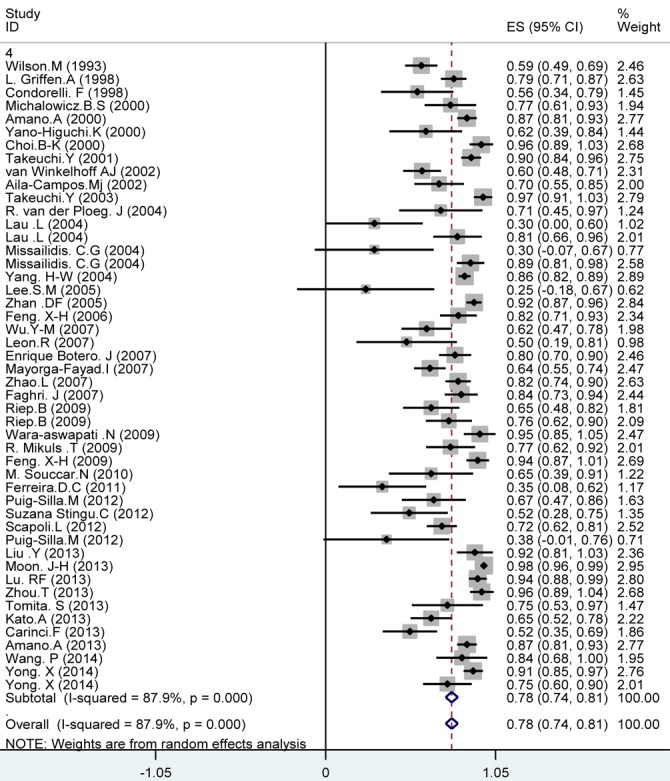
Meta-analysis of the prevalence of P. gingivalis in periodontal diseases: studies are sorted in order to their research author’s names and publication years
Figure3.
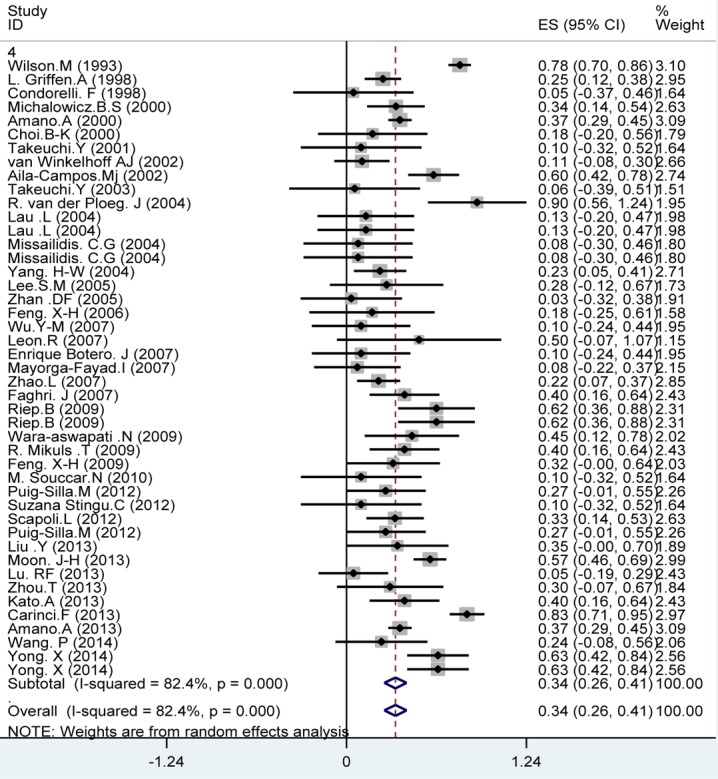
Meta-analysis of the prevalence of P. gingivalis in healthy individuals. Studies are sorted in order to their research author’s names and publication years
Figure 4 presents the Begg’s funnel plot of the included trials related to the prevalence of P. gingivalis in periodontal diseases. No sign of publication bias was observed, when the funnel plot was examined. In fact, most studies were located inside the Funnel Plot, and thus the results of most relevant studies were included into the analysis (p= 0.005). (Figure 4)
Figure4.
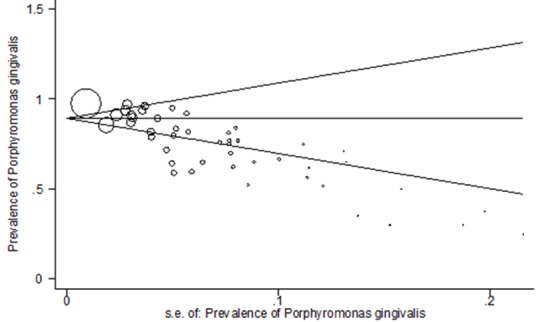
Begg’s funnel plot with pseudo 95% confidence limits
Discussion
The present study systematically reviewed the pub lished studies on P. gingivalis in periodontal diseases. Periodontal disease comprises a group of conditions that affect gingiva, periodontal ligament, cementum, alveolar bone, and tissue structures that support the teeth.[28] Initiation and progression of this disease are influenced by the interaction of a lot of genetic, environmental, host, and microbial factors.[2-3,10] Studies have revealed unexpectedly high diversity of microorganisms involved in periodontal disease; it has proven that the primary microbial factor contributing to this disease has been a shift in the content of the oral microflora.[49] The content of the microflora, associated with periodontal health and disease, has been intensely studied for well over a century and the available literature in this regard is rapidly growing in scope.[70-71] Since P. gingivalis has been known as a major etiological agent in periodontal diseases; we conducted a meta-analysis to comprehensively review previous studies and then quantitatively evaluate the prevalence of P. gingivalis in periodontal disease. This systematic review and meta-analysis included 5,884 individuals from 42 case-control studies (all studies received high quality ratings) demonstrated high prevalence of P. gingivalis in periodontal disease.
To the best of our knowledge, there are no other meta-analyses similar to ours in terms of comparing the results; however, our findings were consistent with most previous studies that indicated high prevalence of P. gingivalis in patients with periodontal disease. The results of our quantitative meta-analysis showed the prevalence of P. gingivalis in periodontal disease was 78.7%. The high prevalence of P. gingivalis found here is similar to those in other studies examining periodontal disease; they report that P. gingivalis is very frequently present in periodontal patients, ranging from 50.3% to 89.4% of cases.[1,12,42,49,51, 61,67-68] Our findings and these results suggest that P. gingivalis is highly associated with incidence of periodontal disease; this bacterium increases the chance of periodontal disease and may be considered as a main potential risk factor. The authors of various studies on P. gingivalis reported that P. gingivalis was the marker of a destructive lesion; this pathogen is able to infect soft tissues along with virulence factors, such as lipopolysaccharide, fimbriae, and several proteases, and then flee the surgical debridement of periodontal lesions; this could account for some cases of resistant periodontitis or lesions. [72-73]
Our study also showed that the prevalence of P. gingivalis was 34% in periodontally healthy subjects. These results were in accordance with previous studies, which reported P. gingivalis appeared in periodontally healthy subjects, ranging from 22.1% to 36.8%.[18,48] The authors demonstrate that this bacterium does not appear exclusively in periodontal patients but is also present in periodontally healthy individual, although to a lesser extent.[58,60]
The results of our quantitative meta-analysis provided an overall estimate of the prevalence of P. gingivalis in periodontal patients and healthy individuals, and found that both these percentages were in the upper ranges. Whereas, the prevalence of P.gingivalis in periodontal disease was significantly higher than healthy subjects (78.7% versus 34%), in accordance with results in other studies examining the prevalence of P. gingivalis in both group and concluded that P. gingivalis was more prevalent among patients with periodontal disease than healthy people.[35,58, 74-75] Contrary to our results, some studies showed no differences in prevalence of P. gingivalis between the two groups.[52,76] This discrepancy may be associated with different conditions such as patients’ health status and types of periodontal disease. Furthermore, it should be noted the presence of periodontal pathogens in healthy people and patients might indicate that the presence of periodontal pathogens does not necessarily lead to periodontal disease.[2]
We observed that the prevalence of P. gingivalis was different in various included studies. Type of strain [ATCC 53978 and ATCC 33277], can be one of the causes of this difference. These two strains[77] are quite distinct as P. gingivalis ATCC 53978 has a capsule known as a major antigen associated with pathogenicity of the strain while P.gingivalis ATCC 33277 lacks this antigen and is minimally inflammatory. [78]
This meta-analysis had several limitations: First, in a meta-analysis of published studies, publication bias is an inevitable problem. Secondly, we were unable to evaluate the impact of some important factors such as age, gender, smoking and alcohol consumption because of insufficient data; these factors influence the prevalence of P. gingivalis and the incidence of periodontal disease because they may affect the ability of the bacteria to invade the gingival tissue and potentially impact the malignant process. Thirdly, the studies varied in types of periodontal diseases and demographic features of population (age, severity, complications) that could have influenced the results. Finally, some studies associated with the prevalence of P. gingivalis in periodontal disease were not accessible.
In summary, the results of the present study indicate that P. gingivalis is highly present in subjects with periodontal disease and it also appears in periodontally healthy individuals, although to a lesser extent. Thus, this bacterium increases the chance of periodontal disease and it can be considered as a main potential risk factor. This result suggests that further research is needed to investigate its pathogenicity.
Conclusion
This study indicates that P. gingivalis is highly present in subjects with periodontal diseases and it also appears in periodontally healthy people, although to a lesser extent. Thus, the presence of P. gingivalis increases the chance of periodontal disease and it can be considered as a main potential risk factor.
Acknowledgement
The authors extend their gratitude to the Student Research Committee, Ilam University of Medical Sciences for its support.
Conflict of Interest:There is no benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1.Kato A, Imai K, Ochiai K, Ogata Y. Higher prevalence of Epstein-Barr virus DNA in deeper periodontal pockets of chronic periodontitis in Japanese patients. PLoS One. 2013; 8: e71990. doi: 10.1371/journal.pone.0071990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong X, Chen Y, Tao R, Zeng Q, Liu Z, Jiang L, et al. Periodontopathogens and human β-defensin-2 expression in gingival crevicular fluid from patients with periodontal disease in Guangxi, China. J Periodontal Res. 2015; 50: 403–410. doi: 10.1111/jre.12220. [DOI] [PubMed] [Google Scholar]
- 3.Linden GJ, Lyons A, Scannapieco FA. Periodontal systemic associations: review of the evidence. J Periodontol. 2013; 84(4 Suppl): S8–S19. doi: 10.1902/jop.2013.1340010. [DOI] [PubMed] [Google Scholar]
- 4.Michaud DS, Liu Y, Meyer M, Giovannucci E, Joshipura K. Periodontal disease, tooth loss, and cancer risk in male health professionals: a prospective cohort study. Lancet Oncol. 2008; 9: 550–558. doi: 10.1016/S1470-2045(08)70106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patra JK, Kim ES, Oh K, Kim HJ, Dhakal R, Kim Y, et al. Bactericidal effect of extracts and metabolites of Robinia pseudoacacia L. on Streptococcus mutans and Porphyromonas gingivalis causing dental plaque and periodontal inflammatory diseases. Molecules. 2015; 20: 6128–6139. doi: 10.3390/molecules20046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheikhi A, Jalali M, Gholamian M, Jafarzadeh A, Jannati S, Mousavifar N. Elimination of apoptotic spermatozoa by magnetic-activated cell sorting improves the fertilization rate of couples treated with ICSI procedure. Andrology. 2013; 1: 845–849. doi: 10.1111/j.2047-2927.2013.00140.x. [DOI] [PubMed] [Google Scholar]
- 7.Sheikhi A, Saadati K, Salmani R, Yahaghi N, Sheikhi A, Siemens DR. In vitro modulation of natural killer activity of human peripheral blood mononuclearcells against prostate tumor cell line. Immunopharmacol Immunotoxicol. 2011; 33: 700–708. doi: 10.3109/08923973.2011.561437. [DOI] [PubMed] [Google Scholar]
- 8.Sheikhi AK, Tayade C, Paffaro VA, Croy BA. Are natural killer cells distributed in relationship to nerve fibers in the pregnant mouse uterus? . Pak J Biol Sci. 2007;10:2885–2889. doi: 10.3923/pjbs.2007.2885.2889. [DOI] [PubMed] [Google Scholar]
- 9.Sharifi F, Sheikhi A, Behdad M, Mousavinasab N. Effect of garlic on serum adiponectin and interleukin levels in women with metabolic syndrome. Int J Endocrinol Metab. 2010; 8:68–73. [Google Scholar]
- 10.Jafarzadeh A, Minaee K, Farsinejad AR, Nemati M, Khosravimashizi A, Daneshvar H, et al. Evaluation of the circulating levels of IL-12 and IL-33 in patients with breast cancer: influences of the tumor stages and cytokine gene polymorphisms. Iran J Basic Med Sci. 2015; 18: 1189–1198. [PMC free article] [PubMed] [Google Scholar]
- 11.Amirghofran Z, Sheikhi AK, Kumar PV, Saberi Firouzi M. Soluble HLA class I molecules in malignant pleural and peritoneal effusions and its possible role on NK and LAK cytotoxicity. J Cancer Res Clin Oncol. 2002; 128: 443–448. doi: 10.1007/s00432-002-0371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomita S, Komiya-Ito A, Imamura K, Kita D, Ota K, Takayama S, et al. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb Pathog. 2013; 61-62: 11–15. doi: 10.1016/j.micpath.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010; 192: 5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheikhi A, Shakerian M, Giti H, Baghaeifar M, Jafarzadeh A, Ghaed V, et al. Probiotic Yogurt Culture Bifidobacterium Animalis Subsp. Lactis BB-12 and Lactobacillus Acidophilus LA-5 Modulate the Cytokine Secretion by Peripheral Blood Mononuclear Cells from Patients with Ulcerative Colitis. Drug Res (Stuttg) 2016; 66: 300–305. doi: 10.1055/s-0035-1569414. [DOI] [PubMed] [Google Scholar]
- 15.Borrell LN, Papapanou PN. Analytical epidemiology of periodontitis. J Clin Periodontol. 2005; 32 Suppl 6: 132–158. doi: 10.1111/j.1600-051X.2005.00799.x. [DOI] [PubMed] [Google Scholar]
- 16.Moon JH, Herr Y, Lee HW, Shin SI, Kim C, Amano A, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using newprimers. J Med Microbiol. 2013; 62(Pt 9): 1290–1294. doi: 10.1099/jmm.0.054247-0. [DOI] [PubMed] [Google Scholar]
- 17.Décaillet F, Giannopoulou C, Cionca N, Almaghlouth A, Mombelli A. Microbial profiles of patients seeking treatment for periodontitis. Influence of origin, smoking and age? Schweiz Monatsschr Zahnmed. 2012; 122: 198–204. [PubMed] [Google Scholar]
- 18.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012; 333: 1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi F, Okada M, Oda Y, Kojima T, Kozai K. Prevalence of Porphyromonas gingivalis fimA genotypes in Japanese children. J Oral Sci. 2012; 54: 77–83. doi: 10.2334/josnusd.54.77. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Chaparro PJ, Lafaurie GI, Gracieux P, Meuric V, Tamanai-Shacoori Z, Castellanos JE, et al. Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingivalplaque and blood sample during bacteremia. Biomedica. 2009; 29: 298–306. [PubMed] [Google Scholar]
- 21.Mineoka T, Awano S, Rikimaru T, Kurata H, Yoshida A, Ansai T, et al. Site-specific development of periodontal disease is associated with increased levelsof Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in subgingival plaque. J Periodontol. 2008; 79: 670–676. doi: 10.1902/jop.2008.070398. [DOI] [PubMed] [Google Scholar]
- 22.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the "red complex", a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005; 38: 72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 23.Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecularmechanisms and biological consequences. Future Microbiol. 2013; 8: 607–620. doi: 10.2217/fmb.13.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal diseasethrough the commensal microbiota and complement. Cell Host Microbe. 2011; 10: 497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zenobia C, Hajishengallis G. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence. 2015; 6: 236–243. doi: 10.1080/21505594.2014.999567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puig-Silla M, Dasí-Fernández F, Montiel-Company JM, Almerich-Silla JM. Prevalence of fimA genotypes of Porphyromonas gingivalis and other periodontalbacteria in a Spanish population with chronic periodontitis. Med Oral Patol Oral Cir Bucal. 2012; 17: e1047–e1053. doi: 10.4317/medoral.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis. J Periodontal Res. 2009; 44:1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 28.Sakai VT, Campos MR, Machado MA, Lauris JR, Greene AS, Santos CF. Prevalence of four putative periodontopathic bacteria in saliva of a group of Brazilian children with mixed dentition: 1-year longitudinal study. Int J Paediatr Dent. 2007; 17: 192–199. doi: 10.1111/j.1365-263X.2006.00813.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang MS, Oh JS, Kim HJ, Kim HN, Lee IK, Choi HR, et al. Prevalence of oral microbes in the saliva of oncological patients. Journal of Bacteriology and Virology. 2009; 39:277–285. [Google Scholar]
- 30.Steiner M. Postnatal depression: a few simple questions. Fam Pract. 2002; 19: 469–470. doi: 10.1093/fampra/19.5.469. [DOI] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009; 6: e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. Iniciativa STROBE. The Strengthening the Reporting of Observational Studies in Epidemiology [STROBE] statement: guidelines for reporting observational studies. Gac Sanit. 2008; 22: 144–150. doi: 10.1157/13119325. [DOI] [PubMed] [Google Scholar]
- 33.Michalowicz BS, Ronderos M, Camara-Silva R, Contreras A, Slots J. Human herpesviruses and Porphyromonas gingivalis are associated with juvenile periodontitis. J Periodontol. 2000; 71: 981–988. doi: 10.1902/jop.2000.71.6.981. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi Y, Umeda M, Ishizuka M, Huang Y, Ishikawa I. Prevalence of periodontopathic bacteria in aggressive periodontitis patients in a Japanese population. J Periodontol. 2003;74:1460–1469. doi: 10.1902/jop.2003.74.10.1460. [DOI] [PubMed] [Google Scholar]
- 35.Ferreira DC, Rôças IN, Paiva SS, Carmo FL, Cavalcante FS, Rosado AS, et al. Viral-bacterial associations in acute apical abscesses. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011; 112: 264–271. doi: 10.1016/j.tripleo.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Lee SM, Yoo SY, Kim HS, Kim KW, Yoon YJ, Lim SH, et al. Prevalence of putative periodontopathogens in subgingival dental plaques from gingivitis lesions in Korean orthodontic patients. J Microbiol. 2005; 43: 260–265. [PubMed] [Google Scholar]
- 37.Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int Immunopharmacol 2009; 9: 38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998; 36: 3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avila-Campos MJ, Velasquez-Melendez G. Prevalence of putative periodontopathogens from periodontal patients and healthy subjects in Sao Paulo, SP, Brazil. Rev Inst Med Trop Sao Paulo. 2002; 44: 1–5. doi: 10.1590/s0036-46652002000100001. [DOI] [PubMed] [Google Scholar]
- 40.van Winkelhoff AJ1, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontalpathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002; 29: 1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- 41.León R, Silva N, Ovalle A, Chaparro A, Ahumada A, Gajardo M, et al. Detection of Porphyromonas gingivalis in the amniotic fluid in pregnant women with a diagnosis of threatened premature labor. J Periodontol. 2007; 78: 1249–1255. doi: 10.1902/jop.2007.060368. [DOI] [PubMed] [Google Scholar]
- 42.Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontalhealth status. J Dent Res. 2000; 79: 1664–168. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol. 2001; 72: 1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 44.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004; 19: 224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontalhealth status in Chinese adults. J Periodontal Res. 2007; 42: 511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 46.Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004; 75: 1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- 47.Choi BK, Park SH, Yoo YJ, Choi SH, Chai JK, Cho KS, et al. Detection of major putative periodontopathogens in Korean advanced adultperiodontitis patients using a nucleic acid-based approach. J Periodontol. 2000; 71: 1387–1394. doi: 10.1902/jop.2000.71.9.1387. [DOI] [PubMed] [Google Scholar]
- 48.Yano-Higuchi K, Takamatsu N, He T, Umeda M, Ishikawa I. Prevalence of Bacteroides forsythus, Porphyromonas gingivalis and Actinobacillusactinomycetemcomitans in subgingival microflora of Japanese patients with adult and rapidly progressive periodontitis. J Clin Periodontol. 2000; 27: 597–602. doi: 10.1034/j.1600-051x.2000.027008597.x. [DOI] [PubMed] [Google Scholar]
- 49.Carinci F, Scapoli L, Girardi A, Cura F, Lauritano D, Nardi GM, et al. Oral microflora and periodontal disease: new technology for diagnosis in dentistry. Ann Stomatol (Roma) 2013; 4: 170–173. [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson M, Lopatin D, Osborne G, Kieser JB. Prevalence of Treponema denticola and Porphyromonas gingivalis in plaque from periodontally-healthy and periodontally-diseased sites. J Med Microbiol. 1993; 38: 406–410. doi: 10.1099/00222615-38-6-406. [DOI] [PubMed] [Google Scholar]
- 51.Souccar NM, Chakhtoura M, Ghafari JG, Abdelnoor AM. Porphyromonas gingivalis in dental plaque and serum C-reactive protein levels in pregnancy. J Infect Dev Ctries. 2010; 4: 362–366. [PubMed] [Google Scholar]
- 52.Riep B, Edesi-Neuss L, Claessen F, Skarabis H, Ehmke B, Flemmig TF, et al. Are putative periodontal pathogens reliable diagnostic markers? . J Clin Microbiol . 2009;47:1705–1711. doi: 10.1128/JCM.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng XH, Zhang L, Meng HX, Xu L, Chen ZB, Shi D, et al. Detection of 3 anaerobic microorganisms in saliva and subgingival plaque of patients with periodontitis. Beijing Da Xue Xue Bao Yi Xue Ban. 2009; 41: 44–48. [PubMed] [Google Scholar]
- 54.Zhou T, Xie H, Yue Z. Relationships of five periodontal pathogens causing subgingival plaque in patientswith chronic periodontitis under different periodontal conditions. Hua Xi Kou Qiang Yi Xue Za Zhi. 2013; 31: 518–521. [PubMed] [Google Scholar]
- 55.Lu RF, Feng L, Gao XJ, Meng HX, Feng XH. Relationship between volatile fatty acids and Porphyromonas gingivalis and Treponema denticola in gingival crevicular fluids of patients with aggressive periodontitis. Beijing Da Xue Xue Bao Yi Xue Ban. 2013; 45: 12–16. [PubMed] [Google Scholar]
- 56.Condorelli F, Scalia G, Calì G, Rossetti B, Nicoletti G, Lo Bue AM. Solation of Porphyromonas gingivalis and detection of immunoglobulin A specific to fimbrial antigen in gingival crevicular fluid. J Clin Microbiol. 1998; 36: 2322–2325. doi: 10.1128/jcm.36.8.2322-2325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu YM, Yan J, Chen LL, Gu ZY. Association between infection of different strains of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in subgingival plaque and clinical parametersin chronic periodontitis. J Zhejiang Univ Sci B. 2007; 8: 121–131. doi: 10.1631/jzus.2007.B0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Zhang Y, Wang L, Guo Y, Xiao S. Prevalence of Porphyromonas gingivalis four rag locus genotypes in patients of orthodontic gingivitis and periodontitis. PLoS One. 2013; 8: e61028. doi: 10.1371/journal.pone.0061028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayorga-Fayad I, Lafaurie GI, Contreras A, Castillo DM, Barón A, Aya Mdel R. Subgingival microbiota in chronic and aggressive periodontitis in Bogotá, Colombia: an epidemiological approach. Biomedica. 2007; 27: 21–33. [PubMed] [Google Scholar]
- 60.Scapoli L, Girardi A, Palmieri A, Testori T, Zuffetti F, Monguzzi R, et al. Microflora and periodontal disease. Dent Res J (Isfahan) 2012; 9 Suppl 2: S202–S206. doi: 10.4103/1735-3327.109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang P, Duan D, Zhou X, Li X, Yang J, Deng M, et al. Relationship between expression of human gingival beta-defensins and levels of periodontopathogens in subgingival plaque. J Periodontal Res. 2015; 50: 113–122. doi: 10.1111/jre.12187. [DOI] [PubMed] [Google Scholar]
- 62.Wara-aswapati N, Pitiphat W, Chanchaimongkon L, Taweechaisupapong S, Boch JA, Ishikawa I. Red bacterial complex is associated with the severity of chronic periodontitis in a Thaipopulation. Oral Dis. 2009; 15: 354–359. doi: 10.1111/j.1601-0825.2009.01562.x. [DOI] [PubMed] [Google Scholar]
- 63.Faghri J, Moghim Sh, Abed AM, Rezaei F, Chalabi M. Prevalence of Porphyromonas gingivalis and Bacteroides forsythus in chronic periodontitis by multiplex PCR. Pak J Biol Sci. 2007; 10: 4123–4127. doi: 10.3923/pjbs.2007.4123.4127. [DOI] [PubMed] [Google Scholar]
- 64.Feng XH, Zhang L, Meng HX, Xu L, Chen ZB, Shi D. Prevalence of putative periodontal microorganisms in Chinese patients with aggressive periodontitis. Zhonghua Kou Qiang Yi Xue Za Zhi. 2006; 41: 344–347. [PubMed] [Google Scholar]
- 65.Zhan DF, Liu ZW, Xia XP, Hu JC, Chen LL, Yan J. Study on the detection of P. gingivalis, A. actinomycetemcomitans and T denticola and the correlation between coinfections of the microbes and levels of chronic periodontitis lesion] Zhonghua Liu Xing Bing Xue Za Zhi 2005; 26: 120–123. [PubMed] [Google Scholar]
- 66.Lau L, Sanz M, Herrera D, Morillo JM, Martín C, Silva A. Quantitative real-time polymerase chain reaction versus culture: a comparison between two methods for the detection and quantification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in subgingival plaque samples. J Clin Periodontol. 2004; 31: 1061–1069. doi: 10.1111/j.1600-051X.2004.00616.x. [DOI] [PubMed] [Google Scholar]
- 67.Stingu CS, Jentsch H, Eick S, Schaumann R, Knöfler G, Rodloff A. Microbial profile of patients with periodontitis compared with healthy subjects. Quintessence Int. 2012; 43: e23–e31. [PubMed] [Google Scholar]
- 68.Botero JE, Contreras A, Lafaurie G, Jaramillo A, Betancourt M, Arce RM. Occurrence of periodontopathic and superinfecting bacteria in chronic and aggressive periodontitis subjects in a Colombian population. J Periodontol. 2007; 78: 696–704. doi: 10.1902/jop.2007.060129. [DOI] [PubMed] [Google Scholar]
- 69.van der Ploeg JR, Giertsen E, Lüdin B, Mörgeli C, Zinkernagel AS, Gmür R. Quantitative detection of Porphyromonas gingivalis fimA genotypes in dental plaque. FEMS Microbiol Lett. 2004; 232: 31–37. doi: 10.1016/S0378-1097(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 70.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006; 42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 71.Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006; 42: 180–218. doi: 10.1111/j.1600-0757.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- 72.Meyer DH, Lippmann JE, Fives-Taylor PM. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect Immun. 1996; 64: 2988–2997. doi: 10.1128/iai.64.8.2988-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haffajee AD, Cugini MA, Dibart S, Smith C, Kent RL Jr, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997; 24: 324–334. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 74.Liu H, Sun J, Dong Y, Lu H, Zhou H, Hansen BF, et al. Periodontal health and relative quantity of subgingival Porphyromonas gingivalisduring orthodontic treatment. Angle Orthod. 2011; 81: 609–615. doi: 10.2319/082310-352.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periodontal.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82: 338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 76.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shiftsas assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44: 3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, et al. The capsule of Porphyromonas gingivalis leads to a reduction in the hostinflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun. 2011;79: 4533–4542. doi: 10.1128/IAI.05016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large Europeanprospective cohort study. Gut. : 1764–1770. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]