Abstract
Background & Aims
The human gut microbiota is becoming increasingly recognized as a key factor in homeostasis and disease. The lack of physiologically relevant in vitro models to investigate host–microbe interactions is considered a substantial bottleneck for microbiota research. Organoids represent an attractive model system because they are derived from primary tissues and embody key properties of the native gut lumen; however, access to the organoid lumen for experimental perturbation is challenging. Here, we report the development and validation of a high-throughput organoid microinjection system for cargo delivery to the organoid lumen and high-content sampling.
Methods
A microinjection platform was engineered using off-the-shelf and 3-dimensional printed components. Microinjection needles were modified for vertical trajectories and reproducible injection volumes. Computer vision (CVis) and microfabricated CellRaft Arrays (Cell Microsystems, Research Triangle Park, NC) were used to increase throughput and enable high-content sampling of mock bacterial communities. Modeling preformed using the COMSOL Multiphysics platform predicted a hypoxic luminal environment that was functionally validated by transplantation of fecal-derived microbial communities and monocultures of a nonsporulating anaerobe.
Results
CVis identified and logged locations of organoids suitable for injection. Reproducible loads of 0.2 nL could be microinjected into the organoid lumen at approximately 90 organoids/h. CVis analyzed and confirmed retention of injected cargos in approximately 500 organoids over 18 hours and showed the requirement to normalize for organoid growth for accurate assessment of barrier function. CVis analyzed growth dynamics of a mock community of green fluorescent protein– or Discosoma sp. red fluorescent protein-expressing bacteria, which grew within the organoid lumen even in the presence of antibiotics to control media contamination. Complex microbiota communities from fecal samples survived and grew in the colonoid lumen without appreciable changes in complexity.
Conclusions
High-throughput microinjection into organoids represents a next-generation in vitro approach to investigate gastrointestinal luminal physiology and the gastrointestinal microbiota.
Keywords: Organoid, Microinjection, High-Throughput, Fecal Microbiota, Anaerobic, Barrier Function, High-Content Sampling
Abbreviations used in this article: CAG, chicken beta-actin promoter with CMV enhancer; CFU, colony-forming unit; CRA, CellRaft Array; CVis, computer vision; EGFP, enhanced green fluorescent protein; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; GI, gastrointestinal; HF, hydrogen fluoride; hiPS, Human Induced Pluripotent Stem Cell; OUT, operational taxonomic unit; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; QIIME, Quantitative Insights Into Microbial Ecology; rRNA, ribosomal RNA; 3D, 3-dimensional; 2D, 2-dimensional; WT, wild-type
Graphical abstract
See editorial on page 352.
Summary.
A high-throughput organoid microinjection platform was developed to study gastrointestinal physiology and the microbiome. Monitoring and quantification of injected microbes and other cargos was achieved by automated imaging. Human fecal microbiota including highly oxygen-sensitive anaerobic taxa were transplanted into the organoid lumen and maintained over time in stable monocultures or microbial communities.
The human gastrointestinal tract contains a remarkably dense and diverse microbial community.1, 2 The interactions between gut microbiota and host are becoming increasingly recognized as key factors in homeostasis and disease.3 Many studies have indicated that community imbalances, known as dysbioses, are associated with the onset and progression of diseases including diabetes,4 obesity,5, 6, 7 colorectal cancer,8, 9, 10 and inflammatory bowel disease.11 Despite tight statistical associations between dysbiosis and disease, the ability to formally test cause-and-effect relationships is severely limited by a lack of in vitro experimental models that enable controlled interrogation of host–microbe interactions.
Sequencing of the 16S ribosomal RNA (rRNA) gene is used routinely to characterize microbial communities and is a powerful tool to identify bacteria that may contribute to disease.12 Although 16S rRNA gene sequencing provides a signature of microbial composition within a community, alone it is insufficient to define specific microbial mechanisms that impact host biology. Germ-free (gnotobiotic) animal models commonly are used to investigate host–microbe interactions in a physiologically relevant system, but germ-free animal models often are impractical for researchers to use because of the scarcity of gnotobiotic facilities and the high cost of gnotobiotic experimentation.13 In addition, the inherent low-throughput nature of germ-free rodent studies limits the ability to decipher the individual role that each microbial species plays in health and disease.
A recent assessment of microbiota research in the United States identified the development of high-throughput tools as a key common unmet need for this field.14 With the recognition of this problem, concerted efforts now are being made to build a translational microbiome toolbox to create innovative and high-throughput approaches to test detailed mechanisms of host–microbe interactions.15 For instance, engineering Bacteriodes thetaiotaomicron, Bacteriodes fragilis, Bacteriodes vulgatus, Bacteriodes ovatus, Bacteriodes eggerthii, and Bacteriodes uniformis with 6 different fluorescent proteins enabled delineation of species within the gut of mice, and showed that the priority of gut colonization determines colonic crypt microbial occupancy.16 Similarly, engineering-inducible promoters in Bacteroides has enabled the study of host–microbe interactions through measurement of commensal sialidase activity and liberation of mucosal sialic acid, a nutrient for pathogens.17 Furthermore, new methods have been developed that permit genetic manipulation and analysis of genetically intractable bacteria from the intestine.18 As these types of tools are being increasingly developed to test mechanistic questions, and studies are expanded to interrogate the thousands of different microbes that inhabit the gut, high-throughput in vitro models will be essential for these next-generation microbiome studies.
Transplanting the complex microbial communities found in the gut lumen to a physiologically relevant system in vitro is particularly challenging because most microbes comprising the human gastrointestinal (GI) microbiota are highly sensitive to oxygen as shown by limited viability or proliferative capacity in the presence of oxygen.19 A recent study suggested that 50%–60% of the oxygen-sensitive bacterial genera in the GI microbiota can produce resilient spores and can be detected on specialized agar plates incubated in an anaerobic environment,20 however, nonsporulating anaerobes remain difficult or impossible to cultivate in vitro and require an environment sufficiently hypoxic for survival and growth.19 Minibioreactors without a host cellular component have been built to cultivate fecal samples in a hypoxic environment.20 This system increased throughput by allowing up to 48 communities to be cultivated per anaerobic chamber. Although this system proved sufficient to stably culture complex communities, only 15%–25% of the initial fecal operational taxonomic units (OTUs) were observed, suggesting that other system components are required to cultivate the full complexity of fecal microbiota.
Designing culture environments that possess a host cellular component in combination with the physiologically relevant luminal environment may enable more complex communities to be cultivated while facilitating the study of host–microbial interactions. Culturing techniques have been developed that permit growth of primary intestinal epithelium on 2-dimensional (2D) surfaces,21, 22, 23 however, generating a steep oxygen gradient over a single cell layer in monolayer cultures remains an engineering challenge owing to the requirement of a constantly intact monolayer, which is difficult to achieve in 2D cultures. Gut organoids, also known as enteroids or colonoids,24 represent an alternative in vitro system to culture fecal-derived microbiota by the virtue of their morphologic, cellular, and physiologic properties that are unavailable in 2D culture systems.
Organoids are microscale spherical structures composed of an epithelial monolayer that surrounds a hollow lumen containing mucous and cellular debris, and serves as a natural physical barrier to the ambient atmosphere.25 Organoids form in culture from isolated crypts or single intestinal stem cells,25, 26, 27, 28, 29 retain long-term self-renewal properties owing to stable intestinal stem cells compartments (eg, crypt buds) interspersed between zones of differentiated cells, and possess cell lineage ratios, polarization, and cell migration patterns that mimic those found in vivo.26 Clostridium difficile, a sporulating pathogenic anaerobe, has been microinjected and monocultured for 12 hours in the organoid lumen, suggesting that the organoid luminal environment can support oxygen-sensitive sporulating taxa.30, 31 Although organoids are a potentially suitable environment to model physiology of the gut lumen, microinjection currently represents the most direct method for delivering defined cargo to the lumen.32
In this study, we developed a high-throughput, organoid-specific platform to study luminal physiology and the gut microbiome. We leveraged advances in organoid culture, microfabricated culture devices,33, 34, 35 computer vision (CVis),33 and semiautomated microinjection technologies to enable high-content sampling of a number of microinjected cargos, including materials to study epithelial barrier integrity and fecal isolates from human donors. The platform can be extended to organoids derived from other tissues such as the lung and stomach, and injectable cargos can be tailored to study a broad range of topics related to gut epithelial biology including microbe–microbe and host–microbe interactions, nutrient transport, and barrier function.
Materials and Methods
Automated Imaging System
Microinjection hardware was fitted within a Precision Plastics (Beltsville, MD) enclosure mounted on an Olympus IX81 (Japan) inverted microscope outfitted with a ProScan (Prior, Cambridge, England) automated stage and controller. Images were acquired by an ORCA-03G (Hamamatsu, Hamamatsu City, Shizuoka, Japan) high-speed camera at parameters controlled through HF204 or HF202 (Prior) automated shutters and a HF108IX3 (Prior) filter wheel. The system was controlled using custom image acquisition software (MathWorks, Natick, MA) and MicroManager (Vale Lab, UCSF).36
Microinjection Hardware Custom Fittings
Custom adaptors were 3-dimensionally (3D) printed to house a commercially available MM-89 (Narashigi, Tokyo, Kantō, Japan) motor-drive manipulator within the confines of a microscope-mounted atmospheric regulating chamber. The physiologic chamber was necessary to preserve biological function of organoids during injection, high throughput, and time-lapse imaging. A spacer was 3D-printed to increase the clearance between the rear wall of the chamber and the NO-PIX-3 (Narashigi) manipulator mounting bar. Pilot studies showed that pneumatically driven microinjectors were best suited to remotely deliver nanoliter cargo loads within the chamber constraints. The Pico-spritzer III (Parker-Hannifan, Mayfield Heights, OH) was suitable for use within the IX81 chamber with a low rate of valve malfunction occurrences, which can lead to pressure build-up and needle rupturing. A custom control arm was 3D-printed to orient the injection needle holder perpendicular to the MM-89 shaft collar and parallel to the stage, facilitating vertical needle articulation. A stage insert with a compression fitting was 3D-printed to hold CellRaft Array (CRA) devices tightly and reduce stage position error when devices were moved in and out of the microinjection platform.36 All computer-aided design files for 3D-printed fittings are available in the Supplementary CAD files.
Optimized Microinjection Needle Processing
To generate microinjection needles tailored to organoid microinjection, 1-mm filament capillaries (World Precision Instruments, Sarasota, FL) were pulled to a fine point using a P-2000 (Sutter, Medical Technologies, Atlanta, GA) laser-based micropipette puller at parameters modified from the standard “bee-stinger” needle production protocol to reduce the diameter of the needle point.37 A 90° bend was formed in the needle body approximately 1.5 cm from the needle tip by heating the needle body with an open flame while horizontally suspended, allowing the bend to form by gravity. The needle pulling process produced microscopic defects that impaired efficient organoid microinjection. To remove the defects, the needle aperture was polished through 6 minutes of wet-etching in 5% hydrogen fluoride (HF). The needle tip subsequently was submerged in sodium bicarbonate to neutralize the HF coating the needle tip. To prevent HF and sodium bicarbonate from traveling up the internal filament into the needle body and contaminating cargos, compressed air was fed into the needle body as the tip was submerged in all solutions.
Colonoid Culture and Expansion
Crypts were isolated from the colons of adult (age, 10–16 wk) wild-type (WT) or chicken beta-actin promoter with CMV enhancer:DsRED (constitutively DsRED expressing) male mice reared on Bed-o’-cobs (Anderson’s Lab Bedding, Maumee, OH) and fed a soy-free, grain-based diet (Envigo, Huntingdon, Cambridgeshire, United Kingdom) as described.38 In brief, the whole colon was resected, flushed with cold phosphate-buffered saline (PBS), opened longitudinally, and then treated with isolation buffer.38 After 90 minutes, whole crypts were released from the tissue by gentle shaking and then collected by centrifugation at 1000 × g. Crypts were embedded in laminin-rich Cultrex hydrogel (Corning, USA) at approximately 4 crypts/μL. Colonoids were expanded in 50% L-WNR media and 50% LCM media produced according to established protocols with the minor modification of removing antibiotics during the conditioning phase to prevent carryover into organoid cultures.39 In these studies, we used media shown to produce colonoids containing all of the native differentiated lineages.39 For passaging, colonoids were dissociated into small cell aggregates by digesting in TrypLE (Gibco) supplemented with 10 μmol/L Y-27632 (Selleck) for 4–6 minutes. Cold serum (10% vol/vol) was added to the dissociated cells to quench enzymatic activity. For injection experiments, large aggregates were excluded before culture by passing dissociated colonoids through a 40-μm filter. Organoids were grown for 72 hours before injections were performed and sampled by 168 hours to prevent overgrowth that could compromise luminal integrity. Aggregate concentration was quantified visually (fragments/volume) by microscopy. All protocols for animal use were reviewed and approved by the University of North Carolina Institutional Animal Care and Use Committee.
Plating Colonoids on CRA Devices
Colonoid fragments were added to CRA reservoirs at 1.5 fragments per raft in L-WNR culture media and centrifuged at 51 × g for 5 minutes at 4°C to deposit fragments on the rafts. After centrifugation, the culture media was aspirated gently so as not to disrupt fragments, and 150 μL of Cultrex (Trevigen, Gaithersburg, MD) was added per cm2 of array surface. A second centrifugation for 5 minutes at 51 × g was used to ensure hydrogel-embedded fragments were deposited at the bottom of the raft. The hydrogel was allowed to polymerize at 37°C for 30 minutes before 200 μL of L-WNR culture media supplemented with 10 μmol/L Y-27632 was added per cm2 of array surface. Media was exchanged every 72 hours.39 Y-27632 was included only in the media at initial plating. Colonoids were grown until they occupied approximately 80% of each raft (typically 72 hours after plating) before injection assays.
Computer Vision Identification of Organoids
Organoids were cultured on CRAs (Cell Microsystems, Research Triangle Park, NC), which are microculture devices that contain approximately 15,000 wells in a 2.0-cm2 footprint.33, 40 Organoids were passaged and dissociated to small fragments, which were plated on CRAs such that there was a density of approximately 1 organoid/well. When organoids had grown to a point they showed a defined luminal space (∼3 days), the entire CRA was tile-imaged using a MATLAB (Mathworks, Natick, MA) automated imaging script, which operated the MMCore application programming interface component of the open-source MicroManager (Vale Lab, UCSF) automated microscope controller.36 Autofocus was used to correct for variations in the flatness of the CRAs. To determine whether an organoid suitable for injection was contained on a raft, individual images of each raft were evaluated by the novel CVis algorithm OrganoidMorph (see Supplementary Program Files). The OrganoidMorph image analysis pipeline was developed in the open-source computational image analysis platform CellProfiler (Broad Institute of MIT and Harvard).41 OrganoidMorph identifies the contents in each raft using a Gaussian mixture model that identifies positive from negative pixels.42 This creates a binary image mask of the fluorescent signal. The size and shape of the mask then is filtered using threshold cut-off values that are consistent with organoid morphology. The location of organoids that passed the OrganoidMorph filtering algorithm was logged automatically so that it could be returned to easily for microinjection. The luminal compartment was identified by inverting the signal within the colonoid area and segmenting by Otsu’s method thresholding (supplementary software: OrganoidMorph.cpproj).
Computer Vision Quantification of Injected Cargos
To identify the fluorescent cargos that were microinjected into the organoid lumen, images were analyzed by CVis (supplementary software: OrganoidCargo.cpproj). Specifically, the fluorescent signal area of each image was identified by Otsu thresholding, which is a computational method that decides positive and negative pixels.43 Masks were generated from the positive fluorescent signal and the area and mean pixel intensity within the mask was measured. These values (intensity × area) were normalized to t = initial values for each organoid, and integrated (mean intensity × area) to account for volumetric changes.
Assessing Efficiency of the Microinjection Device
Efficiency of the platform was assessed by targeting DsRED colonoids of various size and morphology for microinjection with 0.4 nL ± 0.8% of 70 kilodaltons fluorescein conjugated dextran (Thermo Fisher, Waltham, MA). Colonoids were grown for 72 hours on a CRA device and identified using the OrganoidMorph pipeline. Five hundred identified colonoids were randomly targeted and microinjection was attempted with the duration of each attempt recorded. Microinjection success was assessed by fluorescent microscopy and recorded immediately after each attempt. Microinjection success and duration was related to colonoid size and monolayer width post hoc based on the CRA address of colonoids targeted for microinjection.
Retrieving Bacteria From the Organoid Lumen
Live microbes were retrieved from the colonoid lumen by passing intact organoids through a 30-gauge needle (BD Biosciences, Franklin Lakes, NJ), disrupting the surrounding monolayer. Whole colonoids were retrieved from culture platforms by pipetting and suspended in sterile Dulbecco Phosphate Buffered Saline (Gibco, Waltham, MA) before loading into a standard syringe (BD Biosciences). Organoids transplanted with anaerobic species were suspended in anoxic DPBS intact and transferred into an anaerobic chamber (COY, Grass Lake, MI) before passage through a 30-gauge needle.
Bacterial Cultivation
Aerobic bacteria strains (Escherichia coli NC101 and Yersinia pseudotuberculosis YPIII) were cultivated in Luria broth shaken at 240 rpm or on broth agar plates. E coli NC101 transformed with pEGFP was grown at 37°C in medium containing 10 mg/mL carbenicillin. E coli NC101 transformed with pRZT3 was grown at 37°C in medium containing 10 mg/mL tetracycline. Y pseudotuberculosis YPIII containing the PfrdA::gfp vector was grown at 30°C on media supplemented with 20% tryptone and 10 mg/mL chloramphenicol. Bifidobacterium adolescentis was isolated from the stool of a healthy patient (National Center for Biotechnology Information BioProject ID PRJNA291486). The strain was maintained in anaerobic conditions until the time of injection.44 B adolescentis was grown from frozen glycerol stocks (-80°C) in oxygen-depleted De Man, Rogosa and Sharpe broth supplemented with 0.2% cysteine overnight in an anaerobic chamber (5% CO2, 10% H2, 85% N2) at 37°C without agitation. Fecal microbiota communities were processed or retrieved from the organoid lumen in an anaerobic chamber, plated on MRS medium and incubated in the anaerobic chamber (5% CO2, 10% H2, 85% N2) at 37°C, or were cultured aerobically at 37°C.
High-Resolution Imaging of Microinjected Colonoids
Green fluorescent protein (GFP)-expressing E coli NC101 was grown to stationary phase (overnight) before approximately 1500 colony-forming units (CFUs) were injected into 5 CAG:DsRED colonoids grown on 50-mm tissue culture dishes. Colonoids were fixed 2 hours after injections with 4.0% paraformaldehyde before injected E coli was observed within the colonoid lumen using an Olympus FV1000MPE upright confocal microscope. A 20×/1.00 W immersion objective was used to collect 1024 × 1024 pixel Z-stacks at 2.0-μm intervals through each organoid with sequential detection of E coli (GFP), colonoid epithelium (DsRED), and nonspecific debris (4′,6-diamidino-2-phenylindole) with Kalman averaging of 2–4× line scans. The detector and laser setting were adjusted to account for signal attenuation resulting from variable colonoid size and imaging depth. ImageJ (National Institutes of Health, Bethesda, MD) was used to adjust brightness and contrast for optimal visualization.45
Mock Community Response to Media Antimicrobials
WT colonoids were grown for 72 hours on a CRA device separated into 4 discrete media chambers. Chambers were treated with either chloramphenicol, tetracycline, both antibiotics, or no antibiotics. Lysogeny broth (LB) monocultures of E coli K12-expressing DsRED and Y pseudotuberculosis–expressing enhanced green fluorescent protein (EGFP) were grown to stationary phase (overnight for E.coli, 48 hours for Y pseudotuberculosis). Equal volumes of stationary phase cultures were mixed, diluted in LB to 20%, and washed repeatedly in LB by centrifugation at 18,000 × g for 15 minutes to remove residual antibiotics. A total of 20 μL of the mixed sample was loaded into injection needles and transferred to the injection apparatus. Injection droplets were collected in sterile media and plated on selective LB agar plates to measure microbial cargo load before targeting colonoids greater than 100 μm in diameter for microinjection. After injections, additional injection droplets were collected in sterile media and plated on LB agar plates to ensure similar microbial cargo loads were delivered during the injection process. Two hours after injection, successfully injected organoids were retrieved from each condition and luminal contents were cultured to assess transplant efficiency. The remaining colonoids were imaged fluorescently at 30-minute intervals for 24 hours before retrieval and luminal contents were cultured to assess microbial growth. As controls for antibiotic activity, 10-μL samples of the organoid culture media from each condition collected 2 hours, 24 hours, and 48 hours after injections were collected and used to seed conventional broth cultures treated with tetracycline or chloramphenicol.
Relating Fluorescent Signal to Microbial Load
Colonoids were injected with increasing loads of fluorescent inert and microbial cargos and imaged before sampling the luminal contents. WT colonoids were grown in the presence of tetracycline for 72 hours in conventional culture devices allowing larger colonoids to be cultivated. E coli K12-expressing DsRED and E coli NC101-expressing EGFP were grown to stationary phase (overnight) in LB containing tetracycline. Equal volumes of stationary phase cultures were mixed, diluted in LB to 20%, and washed repeatedly in LB by centrifugation at 18,000× g for 15 minutes to remove residual antibiotics. The washed microbial sample was diluted to 50% in 70 kilodaltons of Alexa Fluor 647–conjugated dextran (Thermofisher). A total of 20 μL of the mixed sample was loaded into injection needles and transferred to the injection apparatus before 12 organoids were targeted for microinjection at 4 different durations (24 ms, 48 ms, 72 ms, and 96 ms). All injected organoids were imaged before 6 organoids from each microinjection duration were collected and luminal contents were cultured to assess the delivered microbial load. The remaining colonoids were fluorescently imaged at 30-minute intervals for 24 hours before retrieval and luminal contents were cultured to assess microbial growth.
Stool Sample Preparation
A stool sample (100 μL) from a healthy human host (University of North Carolina Institutional Review Board: 15-2133) was homogenized in sterile, anoxic PBS (1.0 mL) inside the confines of an anaerobic chamber. The homogenized stool sample then was subject to differential centrifugation (5 minutes at 1000 × g), filtration through 5.0-μm syringe filters (filter 1; Millipore, Darmstadt, Germany; filter 2; GE, Boston, MA), or left unprocessed. Samples (100 μL) were stored at -80°C until subjected to DNA isolation and 16S rRNA gene amplicon sequencing (see later).
Colonoid Fecal Microbiota Transplantation
A homogenized healthy donor stool sample was processed using a 5.0-μm Millipore syringe filter and loaded into injection needles in anaerobic conditions. Needles were transferred immediately to the injection apparatus and tested to verify reproducible cargo delivery before targeting a single colonoid in individual wells of a 96-well plate for injection. Colonoid injections that were off-target or showed observable leakage were used as controls, and wells with a single successful injection were sampled to assess microbial colonization by 16S rRNA gene amplicon sequencing and culturing. Colonoid injections were completed within an hour of removing needles from the anaerobic chamber. Retrieved fecal microbial communities were split by volume and cultured anaerobically and aerobically on MRS broth agar plates to determine the colonoid microbial load.
DNA Isolation
DNA from stool samples was isolated using a DNeasy kit (Qiagen, Venlo, Netherlands) as directed. DNA from microbe-associated colonoids was subject to the Qiagen DNeasy kit protocol with an additional 5-minute bead-beating step at room temperature to facilitate separation of bacteria from colonoids and improve cell lysis. Isolated DNA was subject to spectrophotometric quantification via NanoDrop (Thermo Fisher), assessing both quantity (ABS 260 nm) and purity (260/280 nm ratio).
16S rRNA Gene Amplicon Sequencing
Total DNA (12.5 ng per sample) was amplified using primers consisting of the locus-specific sequences targeting the V3–V4 region of the bacterial 16S rRNA gene.46 Primer sequences contained overhang adapters appended to the 5’ end of each primer for compatibility with the Illumina San Diego, CA sequencing platform. Complete primer sequences were as follows: forward: 5′-TCGTCGGCAG:CGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA-3′; reverse: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3′.
Polymerase chain reaction (PCR) mixes contained 12.5 ng of total DNA, 0.2 μmol/L of each primer, and 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Basel, Switzerland). The thermal profile for the amplification of each sample had an initial denaturing step at 95°C for 3 minutes, followed by 28 cycles of denaturing of 95°C for 30 seconds, annealing at 62°C for 30 seconds, and a 30-second extension at 72°C. After cycling, a 5-minute extension at 72°C was performed, and a final hold at 4°C. Each 16S rRNA gene amplicon was purified using the AMPure XP reagent (Beckman Coulter, USA). Illumina dual-index barcodes (index 1[i7] and index 2[i5]; Illumina) were added to each amplicon target using a limited-cycle PCR program. The thermal profile for the amplification of each sample had an initial denaturing step at 95°C for 3 minutes, followed by a denaturing cycle of 95°C for 30 seconds, annealing at 55°C for 30 seconds, and a 30-second extension at 72°C (8 cycles), a 5-minute extension at 72°C, and a final hold at 4°C. The final libraries were purified again using the AMPure XP reagent (Beckman Coulter Brea, CA), quantified via the Quant-IT PicoGreen dsDNA Assay Kit (Thermo Fisher), and normalized before pooling. The DNA library pool then was denatured with NaOH, diluted with hybridization buffer, and heat-denatured before loading on the MiSeq reagent cartridge (Illumina) and on the MiSeq instrument (Illumina). Automated cluster generation and paired-end sequencing with dual reads were performed according to the manufacturer’s instructions.
Sequencing Data Analysis
Multiplexed paired-end fastq files were produced from the sequencing results of the Illumina MiSeq using the Illumina software configureBclToFastq. The paired-end fastqs were joined into a single multiplexed, single-end fastq using the software tool fastq-join. Demultiplexing and quality filtering was performed on the joined results. Quality analysis reports were produced using the FastQC software. Bioinformatics analysis of bacterial 16S amplicon sequencing data was conducted using the Quantitative Insights Into Microbial Ecology (QIIME) software.46 OTU picking was performed on the quality-filtered results using pick_de_novo_otus.py. Chimeric sequences were detected and removed using ChimeraSlayer in the QIIME pipeline. α Diversity and β diversity analysis were performed on the data set using the QIIME routines: α_rarefaction.py and β_diversity_through_plots.py.47, 48 Summary reports of taxonomic assignment by sample and all categories were produced using QIIME summarize_taxa_through_plots.py and summarize_otu_by_cat.py.
Colonoid Anaerobe Monoculture Assay
B adolescentis were cultured until the cells reached late-log growth (OD, 600 nm; OD600nm = ∼0.6) before loading injection needles with 20 μL of culture inside an anaerobic chamber. Loaded needles were transferred immediately to the microinjection apparatus and tested to verify reproducible cargo delivery before targeting a single colonoid in individual wells of a 96-well plate for injection. Colonoid injections that were off target or showed observable leakage were used as controls while wells with a single successful injection were sampled over 60 hours and subjected to quantitative PCR targeting of the B adolescentis 16S rRNA gene and cultured in MRS broth access B adolescentis activity and validate activity organoids.
Statistics
Normalcy of colonoid morphology was assessed by D'Agostino–Pearson and Shapiro–Wilk normality tests. Monolayer width and the cross-sectional area of 1827 colonoids grown on 3 CRA devices and quantified by OrganoidMorph did not show normal distribution by either test at an α level of .05. Colony counts from agar plates were compared by the Tukey–Kramer method with an α level of .05. Droplet volumes were compared between needles by the Tukey–Kramer method at an α level of .05. No statistical method was used to predetermine sample size and experiments were nonrandomized. All analyses were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA).
Results
Development of an Organoid Injection System
The apical side of the gut epithelium is exposed to a complex mixture of luminal contents including nutrients, microbiota, metabolites, and indigestible material. Although there is merit to using the organoid lumen to model gut physiology, the apical surface of the organoid epithelium is inaccessible because the organoid epithelial monolayer creates a barrier to the luminal space (Figure 1A). Material can be introduced into the organoid lumen by incubating fragments of dissociated organoid with compounds that then passively are enveloped into the organoid as the lumen reforms.32 Although this technique can be effective, it lacks precision in that there is no control over the quantity of material that can be delivered to the organoid. Moreover, this strategy cannot be used to introduce materials into the lumen of mature organoids. Microinjection is an alternative approach that offers the ability to accurately, reproducibly, and precisely place a wide variety of cargos into the organoid lumen. However, microinjection is technically challenging and requires a high level of expertise that limits its broad use by investigators.
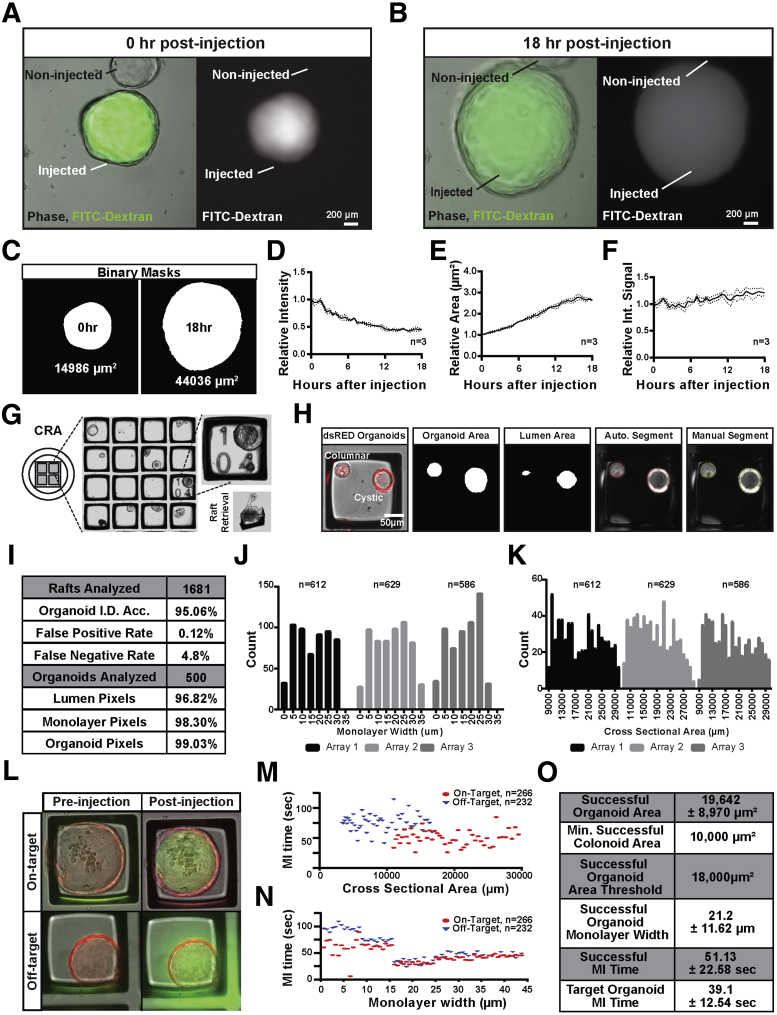
Figure 1.
Robotically articulated colonoid microinjection maintains atmospheric control facilitating long-term sampling of large batches of colonoids. (A) Organoids grown from adult stem cells in 3D culture form complex monolayers organized around a hollow, mucus-filled lumen cavity analogous to the colon lumen. (B) 3D-printed customized fittings were used to mount robotic microinjection hardware within an atmospheric imaging chamber of an automated imaging system. (C) A 90° bend in the injection needle allows for vertical articulation of the microinjection needle, minimizing hydrogel disruption and needle breaking during injections. (D) Wet etching mechanically pulled borosilicate capillaries produces clean, fine aperture needles capable of injecting large batches of organoids without disturbing monolayer integrity. (E) Computer vision made and measured the area of masks from images of the smallest droplets of fluorescent cargo delivered reproducibly by 5 replicate needles, facilitating volume estimation. (F) Optimized needles produce varying volumes at the same injection duration (black) but similar minimal volumes when the minimal duration reproducibly delivering cargo from each needle is used (blue). (G) Organoid-microbe injections can be performed visually by locating an organoid of interest (1), aligning the needle to the organoid lumen (2), articulating the needle against the organoid monolayer distorting its appearance (3), puncturing the monolayer and injecting cargo (4), and retracting the needle from the organoid lumen (5) to visualize specific transplantation of DsRED-expressing E coli within the lumen (needle tip is marked with an asterisk). EpCAM, Epithelial cell adhesion molecule; Muc2, Mucin 2; Ortho, Orthogonal view; RFP, Red Fluorescent Protein.
By using common hardware with some simple modifications, we developed a semi-automated, high-throughput organoid microinjection system that can be easily reproduced in many laboratories. To develop this system, conventional remote-controlled microinjection hardware and a fluorescent microscope fitted with an automated stage and imaging system were combined in a single platform (Figure 1B). The system is contained within a physiologic chamber to maintain appropriate temperature and CO2 levels to support long-term organoid viability for time-intensive experiments (Figure 1B). Three 3D-printed custom fittings were engineered to enable precise needle articulation over organoids cultured in conventional tissue culture plates or CRAs for high-throughput applications (Figure 1B) (CAD files are available in the Supplementary Materials).35, 49
Needle approach for pronuclear or zebrafish embryo microinjection typically occurs at an approximately 45° angle. These angles are incompatible with the use of multiwell plates because the walls of the wells prohibit needle articulation. In addition, diagonal needle approaches result in an unacceptable level of needle breakage when moving the needle through the hydrogel that surrounds the organoid. To circumvent these issues, microinjection needles were fashioned from glass capillaries, then heated and bent to a 90° angle, allowing for a vertical needle approach to organoid targets (Figure 1C). Positioning of the needle over the target organoid did not obstruct the microscope visual field owing to the inverted orientation of the objective lens and optical clarity of the needle.
Accurate and reproducible microinjection requires high-quality glass microneedles. We found that needle construction techniques for conventional zebrafish embryo microinjection produced needles that were not suitable for organoid microinjection and required substantial optimization.37 Specifically, needle taper and tip length were increased to promote smooth entry and exit from the organoid and rapid resealing. Needle bore diameter was optimized to 1 μm to reduce the size of epithelial puncture and enable reproducible nanoliter injections (Figure 1D). Preliminary attempts to inject organoids showed that microforged needles were highly susceptible to retention of cellular material, resulting in a high incidence of clogging and impaired insertion and removal of needles in subsequent injection attempts. At the microscopic level, the borosilicate needles are littered with microscopic defects,37 which we surmised caused retention of cellular material on the needles after injection. Wet etching using HF was adapted to remove these defects and produce a smooth external needle tip.37 A high internal pressure was maintained in the needle body during wet etching to reduce build-up of acid or salt residues that might cause clogging or contaminate cargos. These microneedle design features proved essential for efficient and reproducible high-throughput microinjection into organoids.
Validating the Organoid Injection System
Precise and reproducible microinjection volumes are critical for quantitative studies. The minimal microinjection volume was determined for the organoid-optimized needle design by injecting single droplets of aqueous fluorescein isothiocyanate (FITC)-conjugated dextran into oil (Figure 1E). The accuracy of 5 different microinjection needles was tested by imaging injection droplets, measuring the cross-sectional area of single-injection droplets using CVis, and calculating the spherical volumes of each droplet (Figure 1E). Optimized microinjection needles were capable of reproducibly delivering 0.2 nL ± 1.4% of cargo. Each 0.2-nL droplet is approximately half the theoretical lumen volume of a 100-μm–wide organoid. Injection duration is a property of pneumatic injection systems that changes injection volumes. Although the injection duration necessary to produce a 0.2-nL droplet varied from 24 to 51 ms between 5 different needles, there was little variability in droplet volume (<1%). This indicates that variation in needle tip geometry requires fine-tuning of injection durations to produce extremely small volumes. In contrast, longer injection durations of 200 ms produced consistent 2.0-nL droplets from 5 different needles with minimal variation (±2.1%) (Figure 1F). These data show that highly consistent volume droplets can be generated between 0.2 and 2.0 nL when modulating the injection duration.
When diagonal needle approaches are used for microinjection the needle tip can be observed in the focal plate of the target. When using a vertical needle approach the needle is not visible in the focal plane of the organoid until the puncture. The 90° articulation of the microinjection needle required a modified workflow to efficiently microinject organoids (Figure 1G). Visual cues caused by light refraction off of the needle tip enables alignment above the organoid (Figure 1G.1); the needle tip then is advanced until it touches the top surface of the organoid (Figure 1G.2). As the needle contacts the organoid exterior, deflection and bending of the epithelial monolayer are visualized as creasing and stretching of the organoid perimeter (Figure 1G.3). Once punctured, the organoid retracts to its original diameter and cargo is delivered by pneumatic pulse (Figure 1G.4). After needle retraction, creases in the organoid monolayer are no longer observed (Figure 1G.5). If cargo is fluorescent, imaging then can be performed (Figure 1G.6) (Supplementary Video 1).
Based on the diameter of the needle tip (1 μm), we estimate that an approximately 5- to 10-μm hole (about 1 cell thickness) is generated as a result of the puncture through the epithelium. To evaluate whether cargo remained in the organoid lumen after needle withdrawal, luminal retention of an inert 70-kilodalton fluorescein-dextran impermeable to the membranes and intercellular junctions was assessed.50 Immediately after injection, the fluorescent dextran quickly diffused and equilibrated throughout the luminal space (Figure 2A). Over time, signal intensity decreased as the organoid grew and expanded (Figure 2B; Supplementary Video 2). Although the decrease in fluorescent cargo might result from leakage, it was hypothesized that the reduction in fluorescent signal was owing to increased luminal volume, effectively diluting the fluorescent cargo with organoid growth. To test this, CVis measured the mean intensity and area of the florescent signal every 30 minutes for 18 hours (Figure 2C). Although the mean intensity decreased to approximately half of the original intensity during the 18-hour time course, the signal area doubled (Figure 2D and E). When the signal intensity was integrated over the observed area, the normalized signal was shown to remain constant over the 18 hours (Figure 2F). These data indicate that the injected cargo was effectively retained in the organoids and suggests there is unappreciable leakage from the needle puncture.
Figure 2.
Increasing microinjection throughput using computer vision to quantify cargo retention, organoid morphology, and injection success. (A) Inert high-molecular-weight fluorescent cargos fill the organoid lumen with no signal observed in the adjacent areas. (B) Inert cargos are retained long term as the organoid expands in size over 18 hours. (C) CellProfiler computational image analysis pipelines were used to create masks of the fluorescent signal of images collected during the 18-hour time course and quantified the area and mean intensity of the signal. (D) The mean signal intensity observed from fluorescent inert cargos deceased over time to approximately half of the original intensity. (E) The area of fluorescent signal doubled during the 18-hour time course as the organoid expanded in size. (F) Integrated fluorescent signal across the observed area shows relative stability, suggesting that the inert cargo was retained within the organoid lumen. (G) Culture protocols were optimized to grow organoids on 2.5-cm2 microfabricated culture array devices containing retrievable 200-μm2 rafts separated by 50-μm walls regularly addressed to facilitate downstream sampling. (H) Modified CellProfiler image cytometry pipelines can identify DsRED fluorescent organoids and automatically segment identified organoids of varying morphologies into lumen and epithelial monolayer compartments. (I) Automated organoid identification was >95% accurate (n = 1681 rafts), allowing for >96% accurate lumen identification. (J) Organoid cross-sectional area showed a bimodal distribution with an average of 18,511 ± 5916 μm2. (K) Organoid monolayer width also shows a bimodal distribution with an average of 16.4 ± 9.3 μm. (L) The area and morphology of 500 DsRED-expressing organoids was quantified before targeting each for microinjection with 0.36 nL of 7 kilodaltons of FITC-dextran solution, which could be observed specifically within the lumen of successfully injected organoids with fluorescence signal observed outside the organoid of unsuccessful injections. (M) Successfully injected organoids were larger (19,642 ± 8970 μm2) than unsuccessfully injected organoids (9142 ± 8970 μm2). (N) Organoids with wider monolayers were injected in shorter intervals (51.1 ± 35.6 s) than thinner organoids (74.1 ± 35.6 s). (O) Organoids larger than 18,000 μm2 with monolayers >15-μm wide were microinjected with the highest efficiency and accuracy with all targeted organoids of that range successfully microinjected in 39.1 ± 12.5 seconds. Auto, Automated; Int, Integrated; MI, Microinjection; Min, Minimum.
Automated Computer Vision Facilitates High-Throughput Organoid Identification, Injection, and Quantification of Injected Cargos
Major limitations to efficient and high-throughput microinjection are manual identification of organoids suitable for injection, and the manual tracking and quantification of cargos after injection. To address these problems a CVis program was developed to automatically identify suitable organoids cultured on a CRA. CRAs are commercially available microculture well arrays that enable efficient imaging, identification, and position recall of thousands of organoids in a small footprint (Figure 2G).33, 35 There are approximately 15,000 microwells in a 2.0-cm2 area of the microfabricated device. Passaged organoids were randomly seeded using a Poisson distribution such that there was approximately 1 organoid per 200-μm2 microwell (Figure 2G). Organoids were cultured for 3 days to achieve a population of organoids that were of a practical size for injection. The CRA was imaged in brightfield and fluorescence channels using MATLAB scripts, which serially collected indexed images of individual rafts. Images of each raft then were processed through OrganoidMorph, a custom computational image analysis pipeline developed in the open-source platform CellProfiler with a simple user interface.33, 41 The total organoid area and luminal area were measured using CVis, and the monolayer width was determined by subtracting the overlapping area (Figure 2H; Supplementary Video 3). The organoid area and monolayer width metrics for each organoid were associated with a specific address on the CRA to enable automated recall for microinjection. To validate OrganoidMorph identification, CVis-identified organoids were compared with manually identified organoids (Figure 2I). Automated organoid identification in approximately 15,000 CRA wells took approximately 2 hours and was more than 95% accurate. Lumen segmentation was 96.8% accurate based on comparison with manual measurements (Figure 2H and I). These data show that a CVis pipeline can be used to accurately and efficiently identify the location of hundreds of organoids, and quantify their morphology to identify subsets of organoids suitable for injection.
Anecdotally, microinjection efficiency is highly variable between organoids of different sizes, shapes, luminal volumes, and monolayer widths. Average monolayer widths and cross-sectional areas from approximately 1800 organoids from CAG:DsRED mice were determined automatically by OrganoidMorph CVis analysis and the data were binned according to width and area (Figure 2J and K). To test whether distinct morphologies were more suitable for microinjection, approximately 500 organoids with different monolayer widths and cross-sectional areas were targeted for microinjection with 0.4 nL of membrane-impermeable 70-kilodalton fluorescein-dextran (Figure 2L). Microinjection was scored as successful if FITC signal was contained entirely within DsRED-positive cell boundaries immediately after injection, or as unsuccessful if FITC signal was detectable outside of organoid boundaries (Figure 2L). Organoid cross-sectional area, which is a proxy for luminal volume, was correlated positively with successful microinjection and injection efficiency (microinjection time vs area) (Figure 2M). The smallest organoid successfully injected measured 10,000 μm2. In general, organoids with cross-sectional areas of less than 10,000 μm2 were not suitable for microinjection. Approximately 50% of organoids with cross-sectional areas between 10,000 μm2 and approximately 18,000 μm2 were injected successfully; 100% of organoids with more than 18,000 μm2 cross-sectional areas were injected successfully. In contrast, there was no significant correlation between microinjection success and monolayer width (Figure 2N). However, there was a positive correlation between injection speed and monolayer width for organoids with monolayers wider than approximately 15 μm. When organoid monolayers were more than 15 μm, each injection took an average of 39.1 ± 12.5 seconds, whereas monolayers less than 15 μm2 took an average of 82.2 ± 14.4 seconds to inject (Figure 2O). These data provide important organoid morphology metrics and guidelines to increase the efficiency and success of organoid microinjection.
Growth Dynamics of Transplanted Microbial Communities and Colonoid Barrier Integrity Can Be Monitored by Computer Vision
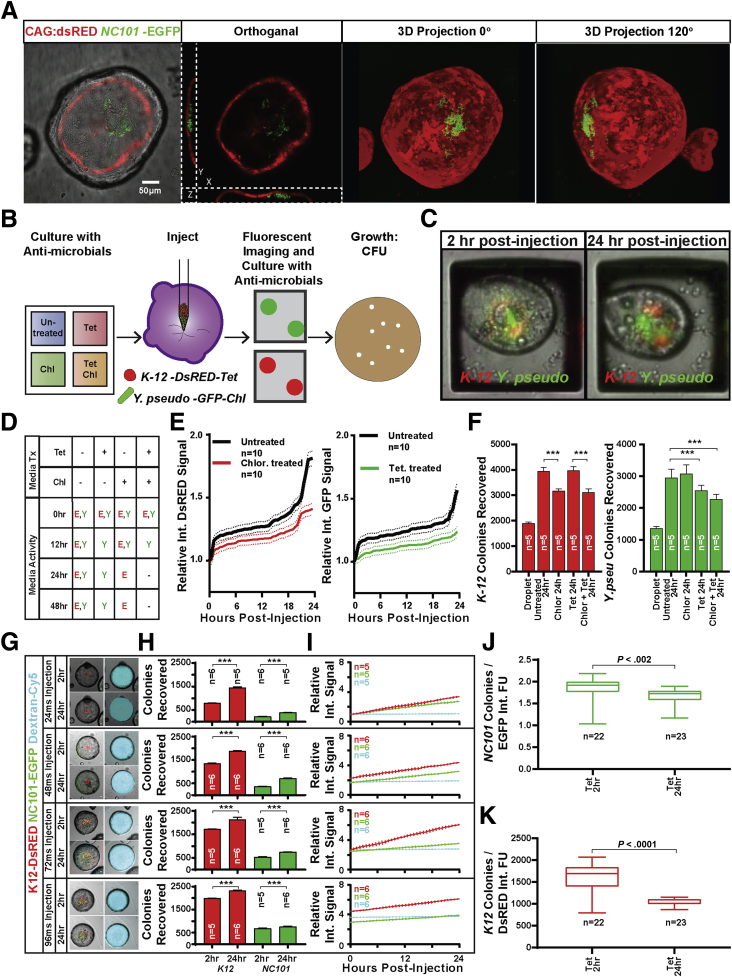
A number of studies have piloted monoculture of bacteria in the lumen of organoids.30, 50, 51, 52, 53, 54, 55, 56 We sought to explore whether our high-throughput microinjection system could be used to culture microbial communities in the lumen of colonoids, which are organoids derived from colonic epithelium, and whether CVis could be used to monitor growth dynamics of fluorescently labeled bacteria within a community. An inoculum of approximately 1500 nonpathogenic E coli NC101 constitutively expressing GFP was injected into 5 individual colonoids to assess whether the fluorescent signature could be detected by microscopy. Confocal microscopy showed that the GFP-expressing E coli NC101 could be detected easily (Figure 3A). Orthogonal views of image projections invariably showed a large E coli NC101-GFP signal in close proximity to the colonoid monolayer (Figure 3A; Supplementary Video 4). A mock community of 2 different traceable bacterial species, E coli–expressing DsRED and Y pseudotuberculosis–expressing GFP, was injected into colonoids, detected by fluorescence microscopy, and monitored over time to determine whether growth dynamics of each species could be quantified by CVis. Comparable with E coli NC101-GFP alone, E coli K12-DsRED and Y pseudotuberculosis–GFP could be readily imaged together in the same organoid by epifluorescence microscopy at 2, 12, and 24 hours after injection (Figure 3C). There were no appreciable changes observed in epithelial thickness, budding, or growth rate after organoid lumen colonization.
Figure 3.
The colonoid lumen forms a discrete compartment compatible with specific microbial growth. (A) GFP-expressing E coli can be visualized after microinjection into DsRED-expressing colonoids and appears to sit in the bottom of the lumen cavity in all colonoids observed. (B) The effects of antibiotics in the colonoid culture media on lumen microbe compatibility was investigated using a CRA device to culture colonoids in 4 discrete reservoirs treated with tetracycline and/or chloramphenicol. Colonoids from each well were targeted for microinjection with a mixed microbial culture of DsRED-expressing E coli resistant to tetracycline and GFP-expressing Y pseudotuberculosis resistant to chloramphenicol. Injected colonoids were monitored over time by live fluorescent imaging before lumen contents were harvested to assess microbial growth by colony formation on conventional agar plates. (C) Fluorescent signal from both microbes could be observed within the lumen of successfully injected colonoids during the entire time course. (D) Antibiotics were essential for preventing off-target growth by excess bacteria delivered to the media during microinjection with no active microbes discovered in culture media treated with chloramphenicol and tetracycline 24 hours after microinjection. (E) Computational analysis showed an increase in integrated DsRED and EGFP fluorescence signal of raft images containing successfully injected colonoids, suggesting an increase in DsRED- and EGFP-expressing microbes. (F) More E coli and Y pseudotuberculosis colonies were recovered from the lumen of colonoids from all media conditions compared with the input injection droplet, suggesting the colonoid lumen protected the injected microbes from chloramphenicol and tetracycline delivered in the culture media (n = 10 colonoids in each condition). Significantly more colonies were recovered from untreated colonoids, correlating with increased integrated fluorescence signal. (G) Fluorescent signal from both microbes as well as inert fluorescent cargo could be observed within the lumen of successfully injected colonoids during the entire time course. (H) More K12 and NC101 colonies were recovered from colonoids collected 24 hours after microinjection compared with those collected immediately after microinjection, suggesting that both microbes grew regardless of the delivered load (n = 5–6 colonoids from each injection duration). (I) Computational analysis showed an increase in integrated DsRED and EGFP fluorescence signal in injected colonoids, suggesting an increase in DsRED- and EGFP-expressing microbes. Computational analysis also showed stable integrated Alexa Fluor 647 signal, suggesting that delivered dextran was well retained. (J) The measured ratio of recovered NC101 colonies to integrated EGFP signal varied significantly between the 2-hour and 24-hour time points, suggesting that integrated EGFP signal cannot be used to directly measure E coli NC101-EGFP microbial load. (K) The measured ratio of recovered K12 colonies to integrated EGFP signal varied significantly between the 2-hour and 24-hour time points, suggesting that integrated EGFP signal cannot be used to directly measure E coli–DsRED microbial load. Chl, chloramphenicol; FU, follow-up evaluation; Int, integrated; Tet, tetracycline; Tx, treatment; Y pseudo, Y pseudotuberculosis.
Contamination of culture media owing to off-target injection, needle leakage, or minor leakage out of the colonoid lumen would compromise colonoid cultures and complicate analysis of injected microbial communities. To address this issue, 2 different antibiotics, tetracycline and chloramphenicol, were added to the colonoid media to inhibit growth of free bacteria outside of the organoid lumen. E coli K12-DsRED was resistant to tetracycline and Y pseudotuberculosis–GFP was resistant to chloramphenicol. To test the efficacy and specificity of antibiotics added to the media, an inoculum of approximately 1500 of each fluorescent strain was injected into 10 colonoids per antibiotic condition, 5 μL of media was collected over time, cultured for 24 hours, then cultures were assessed qualitatively for bacterial growth and fluorescent signature. Although there was no apparent bacterial contamination observed visually in the colonoid culture media at any time point, small amounts of active tetracycline-resistant and chloramphenicol-resistant microbes could be cultured from all conditions 2 hours after the injections (Figure 3D). Neither strain could be cultured from media collected at 24-hour or 48-hour time points when both tetracycline and chloramphenicol were used, indicating the antibiotics were effectively controlling contaminating bacteria outside of the colonoid lumen.
To determine if the growth of the mock community could be monitored quantitatively over time, 40 colonoids were injected with approximately 1500 E coli K12-DsRED and approximately 1500 Y pseudotuberculosis–GFP bacteria, and fluorescence signal was quantified by time-lapse CVis every 30 minutes (Figure 3B). The integrated DsRED and GFP intensity within the colonoid lumen steadily increased over the 24-hour time course (Figure 3E), suggesting both strains of bacteria in the community were growing. Although colonoids were exposed to antibiotics in the media, the data show only a modest decrease in fluorescence. A powerful feature of the CRA platform is that the contents of each raft can be retrieved individually for downstream analyses.35 To determine whether bacteria were active and growing within the colonoid lumen, each colonoid injected with the mock community was retrieved from the CRA, and the number of live bacteria was quantified by colony formation assays (Figure 3F). No significant difference was observed in the number of bacteria cultured from the luminal contents of colonoids retrieved 2 hours after injection, suggesting that the majority of the microbial load was retained in the colonoid lumen (data not shown). Twenty-four hours after injection of the mock community, significantly more bacterium were retrieved from all colonoids, indicating growth within the colonoid lumen (Figure 3F). These findings show that growth dynamics of different bacterial species within a community can be monitored in the colonoid lumen by fluorescent signature and colony forming assays.
We next wanted to determine whether the signal level of fluorescent proteins expressed by bacteria could be correlated directly to the microbial load. Forty-eight colonoids were transplanted with 24-, 48-, 72-, or 96-ms injections consisting of approximately 800, approximately 1350, approximately 1700, or approximately 2000 K12-DsRED bacteria, approximately 200, approximately 300, approximately 500, or approximately 700 NC101-EGFP bacteria, as measured respectively, and 70 kilodalton dextran as an internal reference to monitor barrier function. Forty-five of the 48 attempted injections were successful (93.75%). Signal from both E coli strains as well as the inert dextran could be observed immediately after microinjection and throughout the 24-hour incubation time. There was no apparent loss of barrier function as determined by a consistent signal from fluorescent dextran (Figure 3G). The transplanted bacteria showed increased growth as measured by CFUs over the 24-hour period, and the fluorescent signal from each strain also showed a steady increase in the integrated DsRED and GFP intensity regardless of starting load (Figure 3H). As predicted from the visual observations, the integrated signal produced by the inert dextran remained steady during the 24-hour time course, indicating that epithelial barrier function was preserved (Figure 3I). The ratio of CFU to intensity showed a nonlinear relationship, indicating that integrated fluorescence signals cannot be used to directly measure luminal microbial loads, but can serve as a proxy for growth (Figure 3J and K).
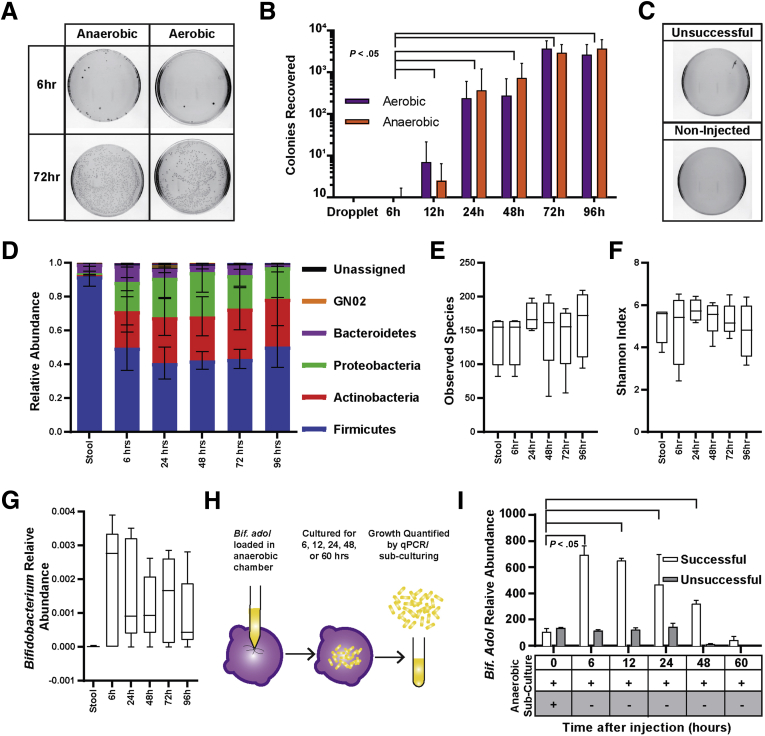
The Colonoid Lumen Supports the Growth of Aerobic and Obligately Anaerobic Human Microbiota Taxa
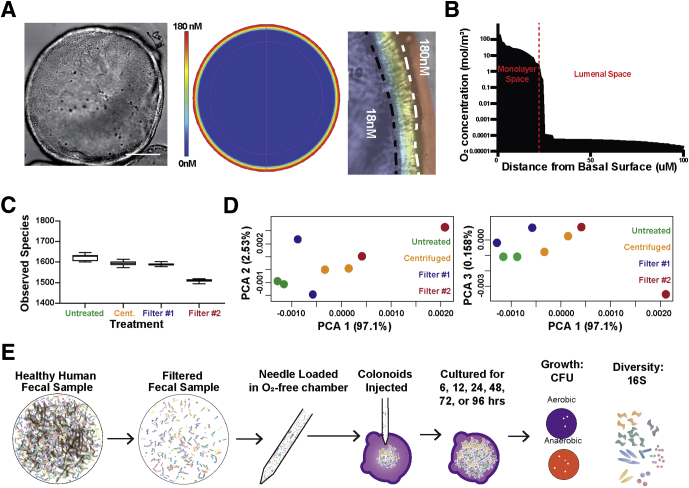
Investigating interactions of native microbial communities with the host epithelium in culture is technically challenging and many times impossible because native enteric microbial communities are dominated by obligate anaerobic species, which are difficult or impossible to cultivate in vitro.19 Because of the relatively small colonoid lumen volume, which is surrounded by oxygen-metabolizing epithelium, it was hypothesized that the colonoid lumen might be sufficiently hypoxic to cultivate the complex anaerobic microbial communities found in the human gut. Direct measurement of luminal oxygen in very large human organoids derived from induced pluripotent stem cells has been performed using an oxygen microprobe,55 however, these microprobe tips are far too large to directly measure oxygen in most organoids derived from primary tissues. Therefore, a computational model was developed to estimate the oxygen concentration using dimensions of a representative colonoid (Figure 4A). Monolayer thickness and luminal volume was modeled based on this colonoid, and empirically determined oxygen diffusion and oxygen consumption rates were applied to the model57 (Figure 4A). COMSOL multiphysics modeling suggested that the lumen of colonoids has an estimated 10% atmospheric O2 (∼180 nmol/L) reaching the apical surface of the organoid owing to respiration by the enclosing epithelial monolayer (Figure 4A and B). The model is remarkably consistent with oxygen levels measured in Human Induced Pluripotent Stem Cell-derived organoids where luminal oxygen was measured at approximately 8% and media levels at 18%.55
Figure 4.
Monolayer respiration makes the colonoid lumen a hypoxic environment capable of supporting the growth of anaerobic enteric microbes. (A) COMSOL modeling suggests the lumen of the average colonoid (left panel) is maintained in a state of hypoxia (right panel) resulting from respiration by the colonoid monolayer. (B) Modeling suggests that O2 levels decrease rapidly from the basal to the apical colonoid surface to approximately 10% of atmospheric O2 levels (180 mmol/L). (C) Stool filtered using a 5-μm polyethylene glycol membrane (filter 1) was compatible with microinjection needles and contained the majority of species present in the unfiltered or conventionally processed (centrifuged) stool. (D) Stool filtered using a 5-μm polyethylene glycol membrane (filter 1) showed greater similarity to the unfiltered and conventionally processed stool than stool filtered using a 5-μm polypropylene membrane. (E) Colonoid compatibility with human microbial populations was investigated by loading a filtered healthy human fecal sample into a microinjection needle under anaerobic conditions before 30 colonoids were microinjected with approximately 0.2 nL of filtered stool. Injected colonoids were harvested across a 96-hour time course and assessed for growth by colony formation in anaerobic and aerobic conditions. Cent, centrifuged; PCA, principle component analysis.
Although complex communities of human GI microbes can be easily acquired by stool sampling from donors, unprocessed stool samples contain large particles that can clog microinjection needles. We developed and validated a method for processing stool samples such that they could be loaded into microinjection needles without clogging the small needle aperture or causing appreciable loss of community diversity (Figure 4C). Although centrifugation of stool diluted in saline is a conventional method to prepare fecal samples for transplantation into gnotobiotic animal models, it was not sufficient to remove enough of the large material that causes needle clogging. Two different commercially available filters were used to filter stool samples diluted in PBS, and the flow-through was compared with untreated samples and centrifuged samples by 16S amplicon sequencing. The results show little difference in observed species between untreated, centrifuged, and filter 1, whereas there was an appreciable decrease in observable species when filter 2 was used. Similar trends were seen by principal component analysis of the OTUs in which filter 1 samples closely clustered with OTUs in untreated samples (Figure 4D). No needle clogging was observed when using stool flow-through from filter 1, which was used to process all stool samples before microinjection.
To determine whether aerobic and anaerobic communities could be transferred and cultivated within the colonoid lumen, a simple workflow was developed that enabled anaerobically collected stool samples to be efficiency injected into colonoids similar to the high-throughput applications described earlier. Essentially, stool samples were filtered and loaded into the microinjection needles in an anaerobic chamber, followed by microinjection of a 0.2-nL inoculum into the lumen of colonoids (Figure 4E). This workflow was used to evaluate the compatibility of the colonoid lumen with fecal-derived microbial communities (Figure 5).
Figure 5.
The colonoid lumen is compatible with patient-derived microbial communities and nonsporulating anaerobes. (A) Few colonies were recovered from colonoids retrieved 6 hours after microinjection grown under anaerobic or aerobic conditions, with increasing microbial loads recovered over time. (B) Significantly more anaerobic and aerobic colonies were recovered from injected colonoids 12 hours after microinjection, with microbial loads peaking 72 hours after microinjection. (C) Samples (10 μL) of the filtered stool collected and cultured under anaerobic conditions grew robustly as expected (top panel), whereas unsuccessfully and noninjected samples produced no colonies in either atmospheric condition (lower panels). (D) The composition of the microbial communities changed quickly after injection but remained stable throughout the 96-hour time course with no significant shifts in 5 dominant phylum. (E) No significant changes were observed in the number of species present in the microbial community after injection or incubation within the colonoid lumen. (F) The even composition of the microbial community remained after injection and did not change significantly during the 96-hour time course. (G) Bifidobacteria, a genus containing nonsporulating anaerobes and aerobes, was a minor member of the healthy stool sample and was detected at increased levels in the communities retrieved from colonoids. (H) Colonoid microinjection compatibility with anaerobic nonsporulating microbes was verified by monoculture microinjection of B adolescentis quantified by 16s quantitative PCR (qPCR) and culturing. (I) An increased abundance of B adolescentis was detected in successfully injected colonoids by targeted qPCR 6 hours after microinjection and was maintained for 48 hours with no increase observed in unsuccessfully injected colonoids. Active B adolescentis was recovered specifically from successfully injected colonoids by anaerobic culturing throughout the 60-hour time course, suggesting that increases in 16S abundance resulted from growth within the colonoid lumen. Bif. adol, B adolescentis.
Fecal transplants were performed by injecting approximately 0.2 nL of the filtered stool sample into 30 murine colonoids. At each specified time point, 5 of the colonoids were collected and mechanically disrupted under anaerobic conditions to release bacteria into a solution, which was applied to broth plates for CFU assays (Figure 5A). There was no apparent effect of colonization on colonoid viability throughout the time course. After fecal transplantation, increasing numbers of colonies were observed over time on plates incubated in both aerobic and anaerobic conditions (Figure 5B). Few or no colonies were recovered from off-target injections, indicating that growth was specific to the lumen environment (Figure 5C). To assess whether the colonoid lumen could support stable fecal communities over time, the remaining 5 colonoids from each time point were subjected to 16S amplicon sequencing to monitor taxonomic changes (Figure 5D). Although the Firmicutes represented the most abundant phylum in the stool samples and colonoids at each time point, there was a notable expansion of the relative abundance in Actinobacteria and Proteobacteria phyla in the colonoids. Compared with stool, microbial communities injected into colonoids and cultured over time showed no significant difference in the number of observed species or Shannon index, which is a quantitative metric to describe the numbers and dominance of a species within a community (Figure 5E and F). Together, these data show that complex bacterial communities isolated from donor fecal samples can be microinjected into the colonoid lumen efficiently and cultivated over a 4-day period with little change in the relative composition of the communities.
Obligate anaerobes can be subclassified as sporulating anaerobes, which show some oxygen tolerance, or nonsporulating anaerobes, which are highly intolerant to oxygen.57 The diarrheal pathogen C difficile produces highly resistant spores that can be transmitted through an oxygen-rich environment to colonize a new host.19 C difficile has been injected into organoids derived from hiPSc to study the impact of C difficile toxins on host epithelium.30, 58 We wanted to test whether nonsporulating and highly oxygen-intolerant species could survive and grow inside the colonoid lumen. Bifidobacterium is a gram-positive genus often associated with anaerobic bacteria and was represented in the healthy fecal microbiota sample. When the fecal microbial community was microinjected into colonoids this genus increased in relative abundance, suggesting it was growing in the colonoid lumen (Figure 5G).
Although 16S rRNA gene amplicon sequencing indicates the presence and relative abundance of bacterial taxa, it does not access their viability. To validate whether nonsporulating obligately anaerobe species of the Bifidobacterium genera could survive and grow within the colonoid lumen, a monoculture of B adolescentis, a highly oxygen-intolerant species that is an early colonizer of the infant gut,59 was microinjected into colonoids (Figure 5H). Survival and growth of B adolescentis in the colonoid lumen was evaluated at 6, 12, 24, 48, or 60 hours after microinjection. Because B adolescentis was not capable of growing on plates60 activity was evaluated by culture capacity in anaerobic broth and growth was evaluated by copy number quantitative PCR for the B adolescentis 16S rRNA gene, which can be used as a proxy for microbial load. Although control injections exposed to oxygen were unable to grow in culture at the earliest time point tested (6 hours), B adolescentis could be cultured in broth through 3 days of cultivation in the colonoid lumen (Figure 5I). There was an approximately 4.5-fold increase in the relative abundance of B adolescentis within the colonoid lumen between injection (t = 0) and 6 hours of cultivation (Figure 5I). Although B adolescentis could survive through 3 days after injection, there was a significant loss of 16S rRNA gene abundance at 2 days after injection, suggesting factors required for growth had diminished. Together, these results indicate that the colonoid lumen is sufficiently hypoxic to support survival and growth of nonsporulating anaerobes found within the larger community of fecal microbiota.
Conclusions
In response to the call for high-throughput platforms and physiologically relevant in vitro culture models to study ‘unculturable’ taxa61, 62 and their effect on GI physiology, we developed and validated a semi-automated high-throughput microinjection device that enables highly efficient and reproducible injection of cargos into the lumen of gut organoids. The microinjection device was married to CVis algorithms and microculture arrays (CRAs) for high-content sampling of inert and biological cargos injected into the organoid lumen. The microinjection platform was used to monitor and quantify the growth dynamics of mock microbial communities harboring fluorescent reporter genes. With the knowledge that the organoid lumen was sufficiently hypoxic to support cultivation of more oxygen-tolerant sporulating anaerobes such as C difficile,30, 50 we predicted that complex microbial communities containing highly oxygen-sensitive nonsporulating anaerobes might be able to be cultivated in the colonoid lumen. Indeed, computational models suggested a 10-fold decrease in oxygen concentration at the apical membrane of the organoid owing to the oxygen-consumption rate of colon epithelial cells and known oxygen diffusion properties. New methods to prepare human fecal samples for microinjection were developed and validated to functionally test whether complex microbial communities would survive and persist in the colonoid lumen. Microinjection of fecal samples into the colonoid lumen showed efficient fecal microbial transplantation based on similar relative abundance of taxa compared with the transplanted fecal sample. In addition, the microbial communities persisted over a 4-day period with relatively little change in relative composition. As a benchmark for functional anoxia of the colonoid lumen we monotransplanted B adolescentis, a highly oxygen-sensitive anaerobe, into the colonoid lumen and showed growth and survival over a 4-day period. Together, these findings strongly support the concept that the environment of the organoid lumen can serve as a suitable model to investigate many aspects of human fecal microbial communities.
Aside from the utility of this platform for the study of the microbiome, our findings indicate that robust retention of cargos injected into the organoid lumen will be useful for screening compounds and toxins that impair epithelial barrier function. Our high-content CVis applications show the ability to monitor retention of membrane-impermeable compounds in hundreds or thousands of organoids using time-lapse imaging. A pioneering study using hiPSC-derived organoids to study barrier function showed that C difficile toxins disrupted the epithelial barrier resulting in a loss of fluorescent membrane-impermeable probe.30 Although these studies and our findings indicate that retention of membrane-impermeable probes are useful for measuring epithelial membrane integrity, our results indicate that the fluorescent signal should be normalized to the organoid growth volume to account for loss of signal that could be misinterpreted as a leaky epithelial barrier.
Organoids are not all created equal. Studies at the single-organoid level have shown large variations in cell lineage composition, gene expression, and function of transporters.33, 59 In a previous study we isolated 96 organoids at different stages of ontogeny and showed that although there was a stereotypical pattern of lineage gene expression at early organoid development, mature organoids showed highly variable gene expression consistent with heterogeneous ratios of cell lineages within the organoid.33 Functional heterogeneity in organoids also has been shown in high-throughput analysis of single organoids using a swelling assay, which commonly is used to assess the function of the cystic fibrosis transmembrane conductance receptor.59, 60 Interestingly, at the single-organoid level, 2 categories of organoids were observed: responders to forskolin-induced swelling and nonresponders. Even within each of these populations substantial variation in swelling dynamics was documented. These findings highlight the importance of designing organoid experiments with sufficient biological and technical replicates to provide adequate statistical power. Our microinjection and CVis platform currently enables microinjection of approximately 90 organoids per hour with subsequent automated time-lapse monitoring of injected cargos. Because the organoids are injected in a physiologic chamber, data can be collected continuously over several days, or the CRAs can be returned to the conventional incubator and data can be sampled by returning the CRA to the automated stage for data sampling at specified time points. Although this throughput likely is adequate for most experiments, the data indicate that optimum throughput can be achieved by culturing organoids to a point that the cross-sectional area is more than 18,000 μm. Further optimization of automated needle positioning in the center of the organoid, and z-plane articulation during the puncture sequence, are predicted to increase microinjection throughput substantially.
The ability to efficiently and reproducibly introduce a variety of cargos into the organoid lumen is a significant methodologic advance that will be useful to investigate a broad range of topics related to luminal–epithelial physiology, such as barrier function, nutrient absorption, drug transport and metabolism, and the impact of dietary compounds or pharmaceuticals on stem cell maintenance and differentiation. Although this study focused on the analysis of bacterial communities, our system could be easily adapted to studies focused on the fungi, parasites, and viruses that impact human GI health.
Acknowledgments
The authors thank the University of North Carolina Microbiome Core for help with anaerobic microbial culture and sample processing, Jeff Roach of the University of North Carolina Research Computing for assistance in processing sequencing data, and Jennifer Marks for input on designing the device.
Footnotes
Author contributions Ian A. Williamson, Jason W. Arnold, Scott T. Magness, and M. Andrea Azcarate-Peril were responsible for conceptualization; Ian A. Williamson, Jason W. Arnold, Liam Gaynor, and Jordan L. Cocchiaro were responsible for the methodology; Ian A. Williamson, Jason W. Arnold, and Liam Gaynor performed the investigation; Ian A. Williamson and Matthew DiSalvo were responsible for the software; Ian A. Williamson and Leigh Ann Samsa performed the formal analysis; M. Andrea Azcarate-Peril, Ian Carroll, John F. Rawls, and Nancy L. Allbritton were responsible for resources; M. Andrea Azcarate-Peril was responsible for data curation; Ian A. Williamson, Leigh Ann Samsa, and Scott T. Magness wrote the original draft; Ian A. Williamson and Leigh Ann Samsa were responsible for visualization; M. Andrea Azcarate-Peril, John F. Rawls, M. Andrea Azcarate-Peril, Nancy L. Allbritton, and Scott T. Magness supervised the study; and Ian A. Williamson was responsible for project administration.
Conflicts of interest These authors disclose the following: Scott T. Magness and Nancy L. Allbritton have a financial interest in Cell Microsystems. The remaining authors disclose no conflicts.
Funding Supported by the Duke Translational Medicine Institute/North Carolina Translational and Clinical Sciences Institute (S.T.M. and J.F.R.), National Institutes of Health R01 DK081426 (J.F.R.), R01 DK109559 (N.L.A.), R01 DK091427 02S1 (S.T.M.), R01 DK091427 (S.T.M.), and UL1TR001111 (North Carolina Translational and Clinical Sciences Institute), UL1TR001117 (Duke Clinical and Translational Science Institute), and P30 DK034987 (Center for Gastrointestinal Biology and Disease). I.A.W. received partial salary support from the National Institutes of Health through R01 DK091427 04S1.
Supplementary Material
References
- 1.Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S., Gordon J.I., Relman D.A., Fraser-Liggett C.M., Nelson K.E. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li K., Bihan M., Yooseph S., Methé B.A. Analyses of the microbial diversity across the human microbiome. PLoS One. 2012;7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones R.M. The influence of the gut microbiota on host physiology: in pursuit of mechanisms. Yale J Biol Med. 2016;89:285–297. [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X., Ma C., Han L., Nawaz M., Gao F., Zhang X., Yu P., Zhao C., Li L., Zhou A., Wang J., Moore J.E., Cherie Millar B., Xu J. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol. 2010;61:69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- 5.Lau E., Carvalho D., Pina-Vaz C., Barbosa J.-A., Freitas P. Beyond gut microbiota: understanding obesity and type 2 diabetes. Hormones. 2015;14:358–369. doi: 10.14310/horm.2002.1571. [DOI] [PubMed] [Google Scholar]
- 6.Petersen C., Round J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barlow G.M., Yu A., Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30:787–797. doi: 10.1177/0884533615609896. [DOI] [PubMed] [Google Scholar]
- 8.Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P., Gérard C., van Nhieu J.T., Furet J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchesi J.R., Dutilh B.E., Hall N., Peters W.H.M., Roelofs R., Boleij A., Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Smits A.M.M., Bos J.L. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 11.Kabeerdoss J., Jayakanthan P., Pugazhendhi S., Ramakrishna B.S. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res. 2015;142:23–32. doi: 10.4103/0971-5916.162091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartor R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 13.Arrieta M.C., Walter J., Finlay B.B. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Integrative HMP (iHMP) Research Network Consortium, The Integrative HMP (iHMP) Research Network The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joglekar P., Segre J.A. Building a translational microbiome toolbox. Cell. 2017;169:378–380. doi: 10.1016/j.cell.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Whitaker W.R., Shepherd E.S., Sonnenburg J.L. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell. 2017;169:538–546.e12. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim B., Zimmermann M., Barry N.A., Goodman A.L. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell. 2017;169:547–558.e15. doi: 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae S., Mueller O., Wong S., Rawls J.F., Valdivia R.H. Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc Natl Acad Sci U S A. 2016;113:14127–14132. doi: 10.1073/pnas.1612753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D., Goulding D., Lawley T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auchtung J.M., Robinson C.D., Britton R.A. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs) Microbiome. 2015;3:1–15. doi: 10.1186/s40168-015-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jabaji Z., Brinkley G.J., Khalil H.A., Sears C.M., Lei N.Y., Lewis M., Stelzner M., Martín M.G., Dunn J.C.Y. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.In J., Foulke-Abel J., Zachos N.C., Hansen A.-M., Kaper J.B., Bernstein H.D., Halushka M., Blutt S., Estes M.K., Donowitz M., Kovbasnjuk O. Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol. 2016;2:48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon C., VanDussen K.L., Miyoshi H., Stappenbeck T.S. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2014;7:818–828. doi: 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzner M., Helmrath M., Dunn J.C.Y., Henning S.J., Houchen C.W., Kuo C., Lynch J., Li L., Magness S.T., Martin M.G., Wong M.H., Yu J. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T., Vries R.G., Snippert H.J., Van De Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 26.Sato T., Clevers H., van der Flier L.G., Clevers H., Cheng H., Leblond C.P., Barker N., Potten C.S., Sangiorgi E., Capecchi M.R., Takeda N., Montgomery R.K., Powell A.E., Buczacki S.J., Snippert H.J., Lopez-Garcia C., Klein A.M., Ledford H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:241–260. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 27.Gracz A.D., Ramalingam S., Magness S.T. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramalingam S., Daughtridge G.W., Johnston M.J., Gracz A.D., Magness S.T. Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol. 2012;302:G10–G20. doi: 10.1152/ajpgi.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gracz A.D., Fuller M.K., Wang F., Li L., Stelzner M., Dunn J.C.Y., Martin M.G., Magness S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31:2024–2030. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leslie J.L., Huang S., Opp J.S., Nagy M.S., Young V.B., Spence J.R. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill D.R., Spence J.R. Gastrointestinal organoids : understanding the molecular basis of the host – microbe interface. Cell Mol Gastroenterol Hepatol. 2017;3:138–149. doi: 10.1016/j.jcmgh.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Gracz A.D., Williamson I.A., Roche K.C., Johnston M.J., Wang F., Wang Y., Attayek P.J., Balowski J., Liu X.F., Laurenza R.J., Gaynor L.T., Sims C.E., Galanko J.A., Li L., Allbritton N.L., Magness S.T. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W., Sims C.E., Allbritton N.L. Microcup arrays for the efficient isolation and cloning of cells. Anal Chem. 2010;82:3161–3167. doi: 10.1021/ac100434v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Phillips C., Xu W., Pai J.-H., Dhopeshwarkar R., Sims C.E., Allbritton N. Micromolded arrays for separation of adherent cells. Lab Chip. 2010;10:2917. doi: 10.1039/c0lc00186d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advance methods of microscope control using μManager software. J Biol Methods. 2015;1:1–18. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oesterle A. Pipette cookbook. 2015;2015:100. [Google Scholar]
- 38.Ahmad A.A., Wang Y., Gracz A.D., Sims C.E., Magness S.T., Allbritton N.L. Optimization of 3-D organotypic primary colonic cultures for organ-on-chip applications. J Biol Eng. 2014;8:8–11. doi: 10.1186/1754-1611-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells as organoids. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attayek P.J., Waugh J.P., Hunsucker S.A., Grayeski P.J., Sims C.E., Armistead P.M., Allbritton N.L. Automated microraft platform to identify and collect non-adherent cells successfully gene-edited with CRISPR-Cas9. Biosens Bioelectron. 2017;91:175–182. doi: 10.1016/j.bios.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delley R. Series for the exponentially modified Gaussian peak shape. Anal Chem. 1985;57:388. [Google Scholar]
- 43.Otsu N.A. Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 44.Monteagudo-Mera A.A., Arthur J.C., Jobin C., Keku T., Bruno-Barcena J.M., Azcarate-Peril M.A. High purity galacto-oligosaccharides enhance specific Bifidobacterium species and their metabolic activity in the mouse gut microbiome. Benef Microbes. 2016;7:247–264. doi: 10.3920/BM2015.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eliceiri K., Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lozupone C., Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lozupone C., Hamady M., Knight R. UniFrac - an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:1–14. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gach P.C., Wang Y., Phillips C., Sims C.E., Allbritton N.L. Isolation and manipulation of living adherent cells by micromolded magnetic rafts. Biomicrofluidics. 2011;5:1–12. doi: 10.1063/1.3608133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engevik M.A., Engevik K.A., Yacyshyn M.B., Wang J., Hassett D.J., Darien B., Yacyshyn B.R., Worrell R.T. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol. 2015;308:G497–G509. doi: 10.1152/ajpgi.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson S.S., Tocchi A., Holly M.K., Parks W.C., Smith J.G. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8:352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forbester J.L., Goulding D., Vallier L., Hannan N., Hale C., Pickard D., Mukhopadhyay S., Dougan G. Interaction of salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlaermann P., Toelle B., Berger H., Schmidt S.C., Glanemann M., Ordemann J., Bartfeld S., Mollenkopf H.J., Meyer T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wroblewski L.E., Piazuelo M.B., Chaturvedi R., Schumacher M., Aihara E., Feng R., Noto J.M., Delgado A., Israel D.A., Zavros Y., Montrose M.H., Shroyer N., Correa P., Wilson K.T., Peek R.M. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut. 2015;64:720–730. doi: 10.1136/gutjnl-2014-307650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill D.R., Huang S., Nagy M.S., Yadagiri V.K., Fields C., Mukherjee D., Bons B., Dedhia P.H., Chin A.M., Tsai Y.H., Thodla S., Schmidt T.M., Walk S., Young V.B., Spence J.R. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. Elife. 2017;6:e29132. doi: 10.7554/eLife.29132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paredes-Sabja D., Shen A., Sorg J.A. Clostridium difficile spore biology: sporulation, germination, and spore structural. Trends Microbiol. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engevik M.A., Yacyshyn M.B., Engevik K.A., Wang J., Darien B., Hassett D.J., Yacyshyn B.R., Worrell R.T. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol. 2015;308:G510–G524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunasekara D.B., Disalvo M., Wang Y., Nguyen D.L., Reed M.I., Speer J., Sims C.E., Magness S.T., Allbritton N.L. Development of arrayed colonic organoids for screening of secretagogues associated with enterotoxins. Anal Chem. 2018;90:1941–1950. doi: 10.1021/acs.analchem.7b04032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dekkers J.F., Wiegerinck C.L., De Jonge H.R., Bronsveld I., Janssens H.M., De Winter-De Groot K.M., Brandsma A.M., De Jong N.W.M., Bijvelds M.J.C., Scholte B.J., Nieuwenhuis E.E.S., Van Den Brink S., Clevers H., Van Der Ent C.K., Middendorp S., Beekman J.M. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 61.Hiergeist A., Reischl U. Multicenter quality assessment of 16S ribosomal DNA-sequencing for microbiome analyses reveals high inter-center variability. Int J Med Microbiol. 2016;306:334–342. doi: 10.1016/j.ijmm.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 62.Stulberg E., Fravel D., Proctor L.M., Murray D.M., LoTempio J., Chrisey L., Garland J., Goodwin K., Graber J., Harris M.C., Jackson S., Mishkind M., Porterfield D.M., Records A. An assessment of US microbiome research. Nat Microbiol. 2016;1:1–7. doi: 10.1038/nmicrobiol.2015.15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.