Abstract
The anaphase‐promoting complex/cyclosome (APC/C) is an E3 ubiquitin ligase and key regulator of cell cycle progression. Since APC/C promotes the degradation of mitotic cyclins, it controls cell cycle‐dependent oscillations in cyclin‐dependent kinase (CDK) activity. Both CDKs and APC/C control a large number of substrates and are regulated by analogous mechanisms, including cofactor‐dependent activation. However, whereas substrate dephosphorylation is known to counteract CDK, it remains largely unknown whether deubiquitinating enzymes (DUBs) antagonize APC/C substrate ubiquitination during mitosis. Here, we demonstrate that Cezanne/OTUD7B is a cell cycle‐regulated DUB that opposes the ubiquitination of APC/C targets. Cezanne is remarkably specific for K11‐linked ubiquitin chains, which are formed by APC/C in mitosis. Accordingly, Cezanne binds established APC/C substrates and reverses their APC/C‐mediated ubiquitination. Cezanne depletion accelerates APC/C substrate degradation and causes errors in mitotic progression and formation of micronuclei. These data highlight the importance of tempered APC/C substrate destruction in maintaining chromosome stability. Furthermore, Cezanne is recurrently amplified and overexpressed in numerous malignancies, suggesting a potential role in genome maintenance and cancer cell proliferation.
Keywords: APC/C, cell cycle, deubiquitinase, mitosis, ubiquitination
Subject Categories: Cell Cycle; Post-translational Modifications, Proteolysis & Proteomics
Introduction
During cell division, there are coordinated regulatory systems that together control hundreds of cell cycle genes and proteins at the transcriptional, translational, and post‐translational levels. This regulation is best exemplified by cyclins, which drive cell cycle progression by activating cyclin‐dependent kinases (CDKs). The activity of CDKs is temporally controlled by cyclin abundance, which is regulated through a balance between synthesis and degradation. Degradation of cyclins, and other important drivers of the cell cycle, is controlled by the ubiquitin‐proteasome system (UPS), in which the formation of ubiquitin chains on substrate proteins targets them for degradation by the proteasome.
The UPS utilizes a three‐step enzymatic cascade that involves E1, E2, and E3 enzymes. E3 ubiquitin ligases provide specificity in the UPS, designating substrates for ubiquitination. The polyubiquitination of a substrate occurs by directly conjugating ubiquitin onto a lysine residue in the target protein and then adding ubiquitin molecules to the first and subsequent ubiquitins to form a chain (Varshavsky, 2012). Since ubiquitin contains seven lysine residues, distinct chain topologies can be generated that adopt different conformations, and which produce varied cellular outcomes. This is collectively referred to as the ubiquitin code (Komander & Rape, 2012; Yau & Rape, 2016). The most well‐studied conformations are K48‐linked ubiquitin chains, which trigger substrate degradation, and K63‐linked ubiquitin chains, which serve as scaffolds for the assembly of protein complexes involved in various signaling cascades, including the DNA damage response (Komander & Rape, 2012).
K11‐linked ubiquitin chains promote degradation specifically during cell cycle progression (Matsumoto et al, 2010; Min et al, 2015). These atypical chains are the products of the anaphase‐promoting complex/cyclosome (APC/C; Jin et al, 2008; Williamson et al, 2009b; Wu et al, 2010). The APC/C is a 1.2 MDa multi‐subunit E3 ubiquitin ligase at the heart of the cell cycle. The APC/C becomes active in mid‐mitosis, leading to degradation of Cyclin B and Securin, resulting in anaphase onset [see reviews from (Pines, 2011; Sivakumar & Gorbsky, 2015)]. Then, in late mitosis and throughout G1, the APC/C targets a myriad of proteins, such as the Aurora A kinase and FoxM1 transcription factor, before being turned off at S‐phase entry (Sørensen et al, 2001; Floyd et al, 2008; Laoukili et al, 2008; Park et al, 2008; Cappell et al, 2016; Choudhury et al, 2016). During S, G2, and early mitosis, the APC/C is inactive, allowing the accumulation of cyclins and increasing CDK activity. During S‐phase and G2‐phase, APC/C is inactivated by several mechanisms, including phosphorylation and degradation of its co‐activators and E2s, as well as binding to Emi1 (Lukas et al, 1999; Reimann et al, 2001; Rape & Kirschner, 2004; Fukushima et al, 2013; Choudhury et al, 2016). During mitosis, APC/C is kept inactive by the spindle assembly checkpoint (SAC), which prevents anaphase onset until all chromosomes become properly attached to the mitotic spindle (Pines, 2011; Musacchio, 2015; Sivakumar & Gorbsky, 2015). The SAC promotes assembly of a mitotic checkpoint complex (MCC), a multi‐protein complex that binds APC/C and prevents its activation. Through feedback mechanisms, APC/C coordinates the state of the SAC by ubiquitinating MCC subunits (Reddy et al, 2007; Nilsson et al, 2008; Foster & Morgan, 2012; Uzunova et al, 2012).
Because of its crucial role in both completion of mitosis and maintenance of G1, numerous studies have dissected APC/C modes of action at the molecular level. Substrates are recognized through short, linear degron sequence motifs, termed D and KEN boxes, by APC/C co‐activator subunits Cdc20 and Cdh1/Fzr1 (He et al, 2013; Davey & Morgan, 2016). In humans, APC/C utilizes two E2‐conjugating enzymes, UBE2C/UbcH10 and UBE2S, to build polyubiquitin chains on substrates (Garnett et al, 2009; Williamson et al, 2009b; Wu et al, 2010). UBE2C serves as the priming factor and allows the APC/C to add single ubiquitin molecules onto lysine residues in a substrate. Then, UBE2S facilitates the formation of K11‐linked polyubiquitin chains on substrate proteins, leading to their degradation (Williamson et al, 2009b; Wu et al, 2010; Wickliffe et al, 2011; Brown et al, 2014, 2016; Kelly et al, 2014).
Like other post‐translational modifications, ubiquitination is reversible and removed from substrates by catalytically active proteases termed deubiquitinating enzymes (DUBs). In human cells, previous reports suggested that USP44 reverses ubiquitination of the MCC, thereby enforcing the SAC (Stegmeier et al, 2007). In addition, Cyclin A is stabilized in early S‐phase by USP37 (Huang et al, 2011). Nevertheless, human DUBs that antagonize APC/C substrate degradation during mitosis remain undescribed.
Screening of large collections of recombinant DUB enzymes against a panel of diubiquitin probes revealed that a subfamily containing the OTU (ovarian tumor) domain shows ubiquitin linkage specificity (Mevissen et al, 2013). However, conflicting reports on one of these enzymes, OTUD7B/Cezanne (hereafter referred to as Cezanne), indicated its ability to disassemble K11‐linked chains (Bremm et al, 2010; Mevissen et al, 2013, 2016), as well as K48‐linked and K63‐linked ubiquitin chains (Enesa et al, 2008; Wang et al, 2017). Cezanne has been implicated in the degradation of hypoxia‐inducible factor, independent of the proteasome, and in mTORC2 signaling through deubiquitinating K63‐linked chains in GβL (Moniz et al, 2015; Wang et al, 2017). Remarkably, despite biochemical and structural evidence for its role in disassembling K11‐linked ubiquitin chains, the significance of this specificity in cells has remained elusive.
Here, we show that Cezanne is a cell cycle‐regulated DUB. Our results demonstrate that Cezanne can bind, deubiquitinate, and stabilize APC/C substrates. By opposing APC/C activity and contributing to the kinetics of substrate degradation, Cezanne contributes to mitotic progression and proliferation. This represents the first demonstration of a DUB that removes K11‐linked ubiquitin chains from APC/C substrates at the exit of mitosis.
Results
Cezanne disassembles K11‐linked ubiquitin chains
Structural and biochemical studies suggested that Cezanne is a K11‐specific DUB (Bremm et al, 2010; Mevissen et al, 2013, 2016). However, others reported that Cezanne also disassembles both K48‐linked and K63‐linked ubiquitin chains (Enesa et al, 2008; Wang et al, 2017). To address this discrepancy, we used a bacterially purified, GST‐tagged version of Cezanne (amino acids 53–446; hereafter referred to as GST‐Cezanne) and analyzed its specificity in vitro toward K11‐linked, K48‐linked, and K63‐linked diubiquitin substrates. We observed a remarkable specificity for K11‐linked diubiquitin substrates in this assay (Fig 1A). We also monitored Cezanne activity toward longer, K11‐linked tetraubiquitin chains. Cezanne cleaves K11‐linked diubiquitin and tetraubiquitin probes with similar kinetics and efficiency (Fig 1B).
Figure 1. Cezanne is a cell cycle‐regulated, K11 linkage‐specific DUB .
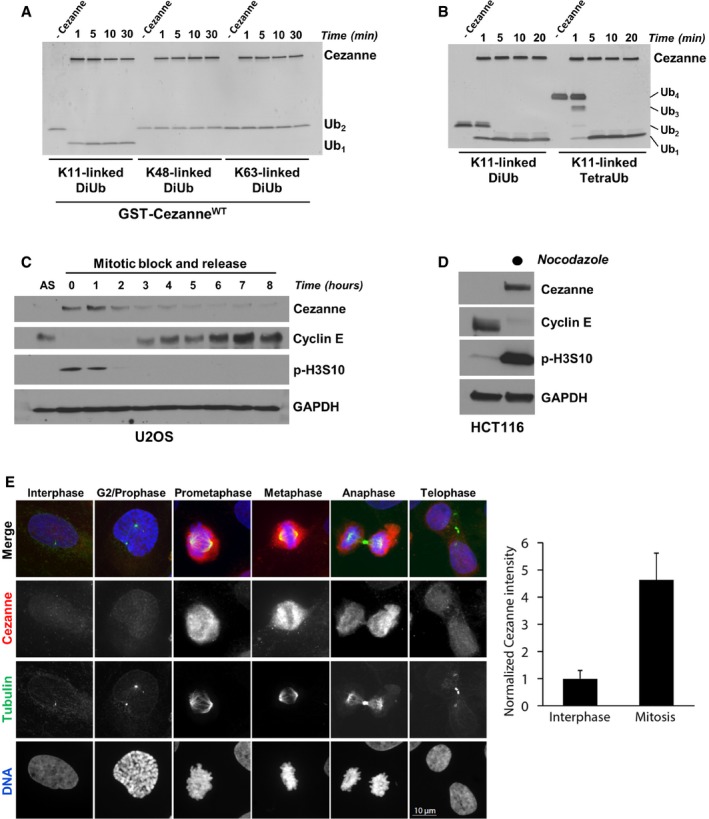
- Recombinant GST‐Cezanne (0.2 μM) was incubated with 1 μM of the indicated diubiquitin probes in DUB reaction buffer at room temperature. Aliquots were collected at the indicated time points and analyzed by silver stain.
- Recombinant GST‐Cezanne (0.1 μM) was incubated with 1 μM of K11‐linked DiUb or TetraUb in DUB reaction buffer at room temperature. Aliquots were collected at the indicated time points and analyzed by silver stain.
- U2OS cells were synchronized in mitosis with nocodazole, isolated by “shake‐off”, and analyzed by immunoblot after release into the cell cycle.
- HCT116 cells grown asynchronously or synchronized in mitosis with nocodazole and isolated by “shake‐off” were analyzed by immunoblot with the indicated antibodies.
- Representative immunofluorescence images stained for Cezanne, Tubulin, and DNA during the cell cycle in U2OS. Quantification of Cezanne intensity between interphase and mitotic cells is shown on the right (error bars show standard deviation for n = 105 and 28 interphase and mitotic cells, respectively).
The recombinant Cezanne, used here and in the previous studies (Mevissen et al, 2013), lacks amino acids 1–52 and 447–843, as well as potential post‐translational modifications which could affect its activity or specificity (Zhao et al, 2018). We therefore characterized the specificity of a full‐length version of Cezanne expressed in mammalian cells. HA‐tagged Cezanne was expressed in HEK‐293T cells, immunoprecipitated, and incubated with K11‐linked, K48‐linked, and K63‐linked diubiquitin substrates. Consistently, the full‐length version of Cezanne, expressed in human cells, is remarkably specific toward K11‐linked diubiquitin substrates (Appendix Fig S1A). Deubiquitination by HA‐Cezanne is lost when using a catalytically dead mutant version (CezanneC194S), confirming that we are not co‐purifying and assaying a second DUB with K11 specificity (Appendix Fig S1B). The difference in electrophoretic mobility between CezanneWT and CezanneC194S is because the catalytically inactive mutant cannot deubiquitinate itself (Appendix Fig S1C).
Using a K11‐linked ubiquitin chain specific antibody (Appendix Fig S1D), we monitored the assembly of K11‐linked chains during the cell cycle. HeLaS3 was arrested at the G1/S boundary using a double‐thymidine synchronization and released into the cell cycle. K11‐linked chains appeared in mitosis, consistent with prior reports and the role of APC/C in forming these chains (Appendix Fig S1E; Matsumoto et al, 2010).
Cezanne is cell cycle‐regulated
Because APC/C activity and K11‐linked ubiquitin chain formation are temporally controlled during the cell cycle, we analyzed whether Cezanne levels fluctuate during cell cycle progression. We synchronized U2OS cells in pro‐metaphase by nocodazole treatment, then isolated cells by “shake‐off”, and analyzed Cezanne abundance by immunoblot after release into the cell cycle. Cezanne protein levels were greatly elevated in mitosis and then decreased as cells progress through G1 and into S (Fig 1C). Similarly, in non‐transformed RPE1 cells synchronized in G0 by serum withdrawal, and then re‐fed to initiate cell cycle progression, Cezanne protein levels peak in mitosis, coincident with the mitotic marker phosphorylated Serine 10 on Histone H3 (p‐H3S10; Fig EV1A). The elevated levels of Cezanne in mitotic cells were also observed in HCT116 cells (Fig 1D). U2OS cells were then arrested by nocodazole and released in medium containing the proteasome inhibitor MG132, which prevents mitotic exit, indicated by persistence of the mitotic marker p‐H3S10 (Fig EV1B). Cezanne expression remained stable compared to cells released into the cell cycle without MG132.
Figure EV1. Cezanne is cell cycle‐regulated.
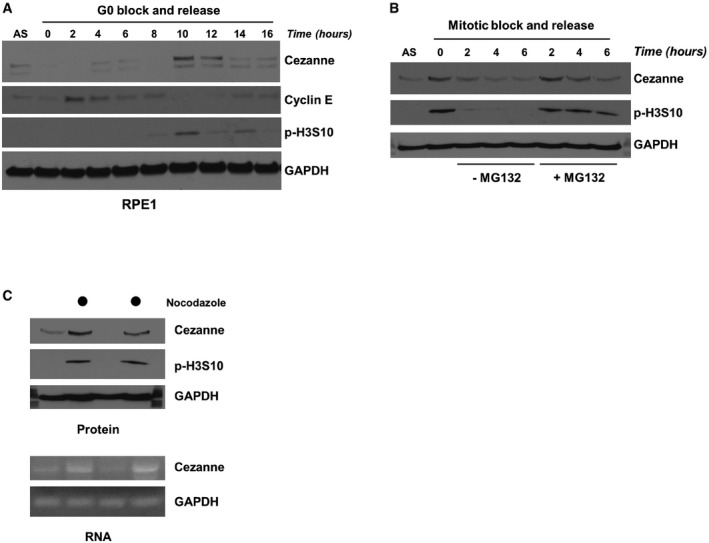
- RPE1 cells were synchronized by serum deprivation and then released into the cell cycle and analyzed by immunoblot.
- U2OS cells were synchronized in mitosis with nocodazole, isolated by “shake‐off”, and released into the cell cycle with or without the proteasomal inhibitor MG132. Samples were analyzed by immunoblot.
- U2OS cells grown asynchronously or synchronized in mitosis with nocodazole and isolated by “shake‐off” were analyzed by immunoblot and RT–PCR.
We next analyzed Cezanne abundance by immunofluorescence imaging. Asynchronous U2OS cells were fixed and stained for Cezanne and Tubulin and analyzed by confocal microscopy. Cezanne abundance was significantly elevated in mitotic cells relative to interphase cells (Fig 1E). Together, these data demonstrate that Cezanne protein levels are cell cycle‐regulated.
To determine how Cezanne levels are controlled, U2OS cells were treated with nocodazole or DMSO (control) and analyzed for both protein and RNA levels of Cezanne. RT–PCR results suggest that Cezanne upregulation occurs, at least in part, at the transcriptional level (Fig EV1C). We conclude that Cezanne is a cell cycle‐regulated DUB whose expression coincides with the timing of APC/C activation.
Cezanne interacts with several APC/C substrates
We next examined whether Cezanne can interact with established APC/C substrates. We performed co‐immunoprecipitation (co‐IP) experiments in HEK‐293T cells by ectopically expressing an HA‐tagged version of Cezanne, together with Myc‐tagged versions of either FoxM1 or Aurora A, or a Venus‐tagged version of Cyclin B. Following Cezanne purification on anti‐HA agarose, we observed interaction between Cezanne and FoxM1, Aurora A and Cyclin B (Fig 2A–C). Next, we incubated in vitro‐translated Cyclin B with recombinant GST‐Cezanne or GST alone. Pull‐down on glutathione beads showed that Cezanne interacts with Cyclin B (Fig 2D). Similarly, recombinant, full‐length hexa‐histidine (6HIS)‐tagged Aurora A interacts with GST‐Cezanne but not GST alone (Fig 2E). Finally, we performed a GST‐Cezanne pull‐down from extracts prepared from either asynchronous or nocodazole‐arrested mitotic U2OS cells. This showed an interaction between GST‐Cezanne and both endogenous Aurora A and Cyclin B (Fig 2F). This interaction appears cell cycle‐regulated, since Cyclin B and Aurora A were enriched in pull‐downs performed with mitotic extracts. Together, these observations show that APC/C substrates can interact with Cezanne.
Figure 2. Cezanne binds APC/C substrates in vivo and in vitro .
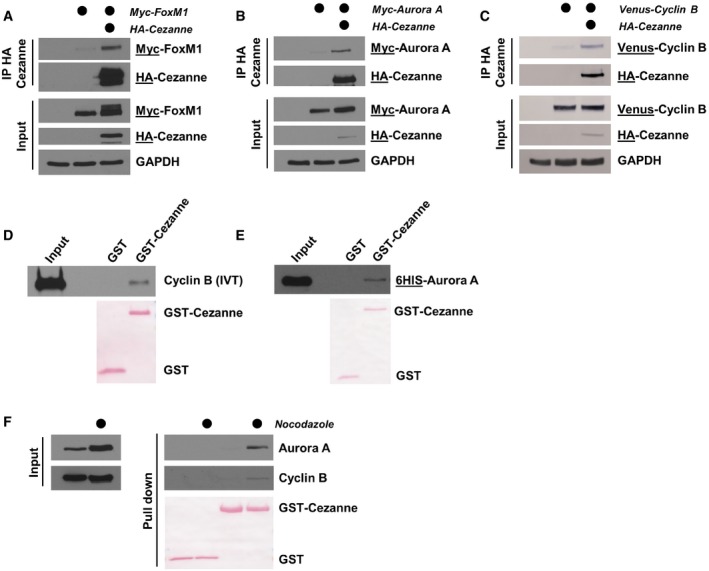
- HA‐Cezanne and Myc‐FoxM1 were ectopically expressed in HEK‐293T cells. After 24 h, cells were treated with 20 μM of MG132 for 4 h and Cezanne was immunoprecipitated on anti‐HA beads. Immunoblotted antigen is underlined to the right of blots.
- HA‐Cezanne and Myc‐Aurora A interaction was analyzed as in (A), except that no MG132 was added before the IP.
- HA‐Cezanne and Venus‐Cyclin B interaction was analyzed as in (B).
- GST‐Cezanne was incubated on beads with in vitro‐translated Cyclin B. In vitro binding was analyzed by immunoblot using anti‐Cyclin B antibodies. GST was used as a negative control.
- In vitro binding between Cezanne and Aurora A was analyzed as in (C), except that Aurora A was produced in bacteria and detected using anti‐6HIS antibodies.
- Lysates of U2OS cells grown asynchronously or synchronized in mitosis with nocodazole were incubated with GST‐Cezanne on beads. GST was used as a negative control and protein detected by immunoblot.
Interestingly, Cezanne also binds to the APC/C co‐activators Cdc20 and Cdh1. This could be observed by co‐IP after ectopically expressing HA‐Cezanne with either FLAG‐Cdc20 or FLAG‐Cdh1 (Fig EV2A and B). Similarly, GST‐Cezanne bound both FLAG‐Cdc20 and FLAG‐Cdh1 from lysates of transfected 293T cells (Fig EV2C and D).
Figure EV2. Cezanne binds APC/C co‐activators in vivo and in vitro.
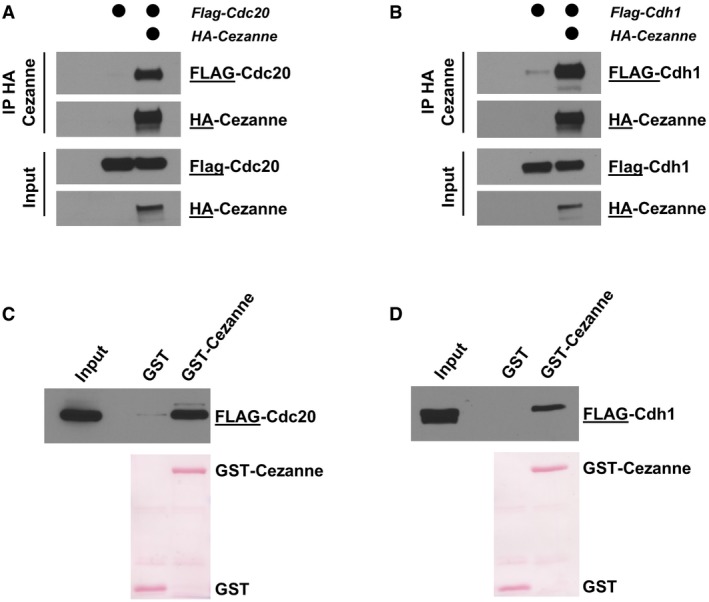
- HA‐Cezanne and FLAG‐Cdc20 were ectopically expressed in HEK‐293T cells, and Cezanne was immunoprecipitated on anti‐HA beads. Samples were analyzed by immunoblot with the indicated antibodies.
- HA‐Cezanne and FLAG‐Cdh1 interaction was analyzed as in (A).
- 5 μg of GST‐Cezanne coated on GSH beads was incubated with lysate of HEK‐293T cells expressing a FLAG‐tagged version of Cdc20. In vitro binding was analyzed by immunoblot using the indicated antibodies. GST was used as a negative control.
- Interaction of GST‐Cezanne with Cdh1 was analyzed as in (C).
Cezanne deubiquitinates APC/C substrates
These observations prompted us to determine whether Cezanne can reverse APC/C‐dependent ubiquitination. We utilized a previously developed cell extract system that has several important advantages. This system fully recapitulates the degradation of APC/C substrates observed physiologically and is amenable to biochemical manipulations. Furthermore, this system alleviates concerns associated with examining APC/C substrate abundance and ubiquitination following experimental manipulations which could alter cell cycle progression (Williamson et al, 2009a). We prepared a G1 extract of HeLaS3 cells supplemented with UBE2C, UBE2S, and fluorescent ubiquitin. Incubation at 25°C leads to polyubiquitination of endogenous proteins, which is largely reversed by the addition of Emi1, indicating that polyubiquitin in this system is largely controlled by the APC/C (Fig 3A). The addition of recombinant Cezanne also significantly decreases polyubiquitination, suggesting that Cezanne antagonizes APC/C polyubiquitination activity. We next used this system to interrogate APC/C substrate ubiquitination. After 40 and 80 min, Aurora A ubiquitination is evident in control extracts. This is antagonized by the addition of CezanneWT, but not a catalytically dead mutant (CezanneC194S; Fig 3B). These results suggest that Cezanne can oppose APC/C activity by deubiquitinating substrates in a physiologically relevant, synchronized cell extract system.
Figure 3. Cezanne counteracts APC/C activity and deubiquitinates APC/C substrates.
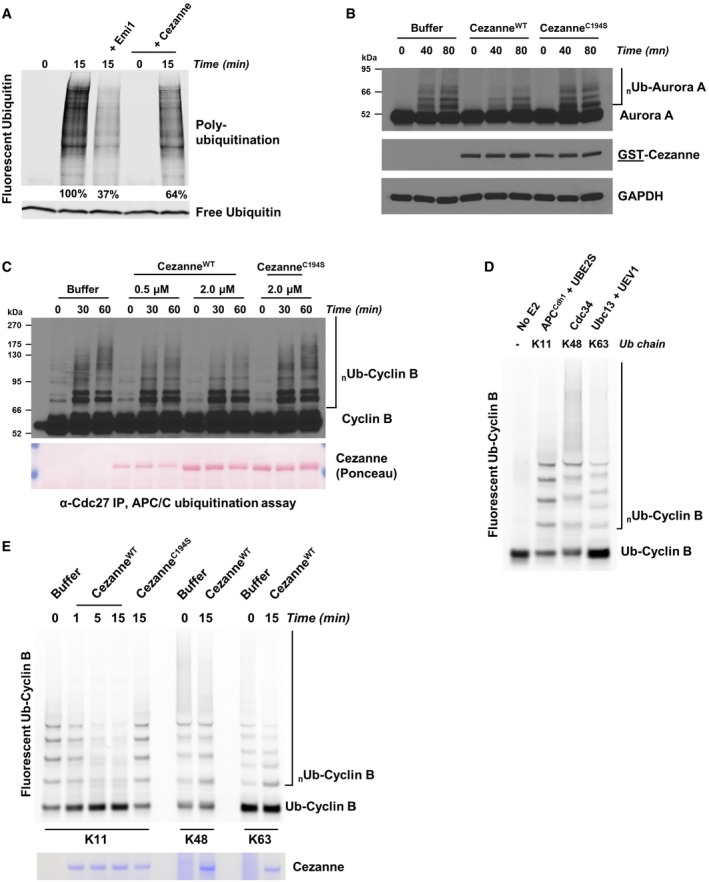
- G1 extract from HeLaS3 cells was prepared and mixed with ATP, UBE2C, UBE2S, and fluorescent ubiquitin. Where indicated, recombinant Emi1 or Cezanne was also added. Reactions were incubated at 25°C and analyzed by SDS–PAGE and fluorescence scanning.
- G1 extract from HeLaS3 cells was prepared and mixed with ATP, UBE2C, ubiquitin, and either 2 μM of recombinant CezanneWT or CezanneC194S. Reactions were incubated at room temperature and analyzed by SDS–PAGE and immunoblot.
- APC/C was immunopurified from mitotic HeLaS3 cell extracts using anti‐Cdc27 beads and then mixed with in vitro‐translated Cyclin B, E1, E2, ATP, ubiquitin, and indicated amounts of Cezanne. Aliquots were collected at the indicated time points and analyzed by immunoblot using Cyclin B antibodies.
- Ubiquitination reactions of a fluorescein‐labeled substrate N‐terminal fragment of Cyclin B fused to ubiquitin (Ub‐Cyclin B) were carried out in the presence of either recombinant APC/CCdh1, Cdc34, or Ubc13 and UEV1 to generate the indicated ubiquitin chain topologies. Reactions were quenched with EDTA and then analyzed by SDS–PAGE and fluorescence scanning.
- Fluorescein‐labeled Ub‐Cyclin conjugated to K11, K48, or K63 ubiquitin chains was incubated with recombinant Cezanne for the indicated times and then analyzed by SDS–PAGE, fluorescence scanning, and Coomassie staining.
To further characterize the ability of Cezanne to deubiquitinate APC/C substrates, we performed ubiquitination assays using APC/C from mitotic HeLaS3 cells that we immunopurified with anti‐Cdc27 antibodies. Ubiquitination was reconstituted in vitro by addition of E1, E2, ubiquitin, ATP, and in vitro‐translated Cyclin B as a substrate. The formation of polyubiquitin chains on Cyclin B can be observed over time as the accumulation of higher molecular weight species (Fig 3C, Appendix Fig S2A). Addition of Cezanne to these reactions significantly reduced Cyclin B ubiquitination (Fig 3C, Appendix Fig S2A). Importantly, the ability of Cezanne to inhibit Cyclin B ubiquitination in vitro was dependent on its catalytic activity (Fig 3C and Appendix Fig S2B).
Next, we reconstituted this reaction using a fully in vitro system, using APC/C complexes purified from insect cells and reconstituted in vitro (Brown et al, 2014, 2015, 2016). As a substrate, we used a fluorescein‐labeled, amino‐terminal fragment of Cyclin B fused to ubiquitin (Ub‐Cyclin B). By using this substrate, APC/C ubiquitination reactions can rely only on UBE2S which will build a chain on the proximal ubiquitin molecule fused to the N‐terminus of Cyclin B, and therefore, Cyclin B becomes exclusively polyubiquitinated with K11‐linked chains (Brown et al, 2014). In parallel, we generated K48‐linked and K63‐linked chains on Ub‐Cyclin B by using linkage‐specific E2 enzymes (Fig 3D). Significantly, CezanneWT but not CezanneC194S rapidly deubiquitinated K11‐polyubiquitinated Cyclin B (Fig 3E). However, Cezanne was unable to disassemble Cyclin B polyubiquitinated with K48‐linked and K63‐linked ubiquitin chains (Fig 3E). We observed a similar result using Ub‐Securin as a substrate (Fig EV3A and B). Importantly, since Cezanne was added after ubiquitination was complete, the loss of substrate ubiquitination cannot be accounted for by an effect of Cezanne on the activity of APC/C or its E2 enzymes. However, APC/CCdh1 was present in the reactions containing K11‐linked chains, but not in those where Cyclin B was polyubiquitinated with K48‐linked or K63‐linked chains. Nevertheless, the addition of APC/C to these reactions was insufficient to allow Cezanne to deubiquitinate K48‐linked or K63‐linked chains formed on Cyclin B (Fig EV3C). We therefore conclude that Cezanne specifically disassembles K11‐linked ubiquitin chains on established APC/C substrates.
Figure EV3. Cezanne regulates the ubiquitination of APC/C substrates.
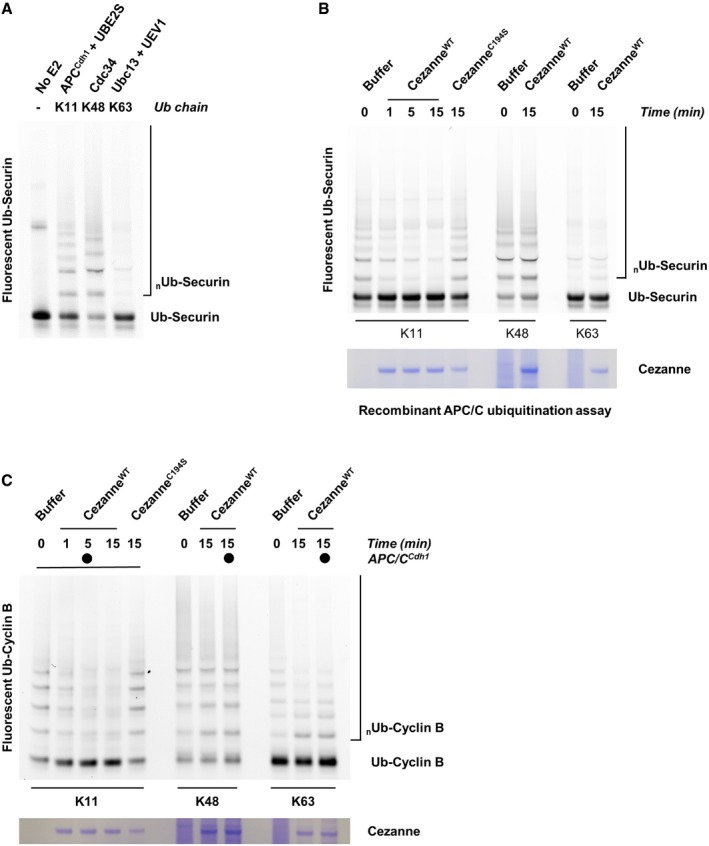
- Ubiquitination reactions of a fluorescein‐labeled Securin fused to ubiquitin (Ub‐Securin) were carried out in the presence of either recombinant APC/CCdh1, Cdc34, or Ubc13 and UEV1 to generate the indicated ubiquitin chain topologies. Reactions were quenched with EDTA and then analyzed by SDS–PAGE and fluorescence scanning.
- Fluorescein‐labeled Ub‐Securin conjugated to K11, K48, or K63 ubiquitin chains was incubated with recombinant Cezanne for the indicated times and then analyzed by SDS–PAGE, fluorescence scanning, and Coomassie staining.
- Fluorescein‐labeled Ub‐Cyclin conjugated to K48 or K63 ubiquitin chains was incubated with recombinant Cezanne in the absence or presence of APCCdh1 for the indicated times and then analyzed by SDS–PAGE, fluorescence scanning, and Coomassie staining.
Cezanne controls APC/C substrate degradation
Given that Cezanne binds and deubiquitinates established APC/C substrates, we analyzed whether Cezanne could also alter their degradation kinetics. We performed degradation assays of APC/C substrates using G1‐phase cell extracts prepared from HeLaS3 cells as described above and previously (Williamson et al, 2009a). We monitored Aurora A, which is degraded over time once the assay has been initiated (Fig 4A). Adding GST‐CezanneWT prior to starting the reaction impaired Aurora A degradation, and this was dependent on its catalytic activity (Fig 4A). We also monitored degradation of Cyclin B under these conditions. Similarly, Cyclin B degradation was impaired by GST‐CezanneWT, but not GST‐CezanneC194S (Fig 4B).
Figure 4. Cezanne regulates APC/C substrates degradation.
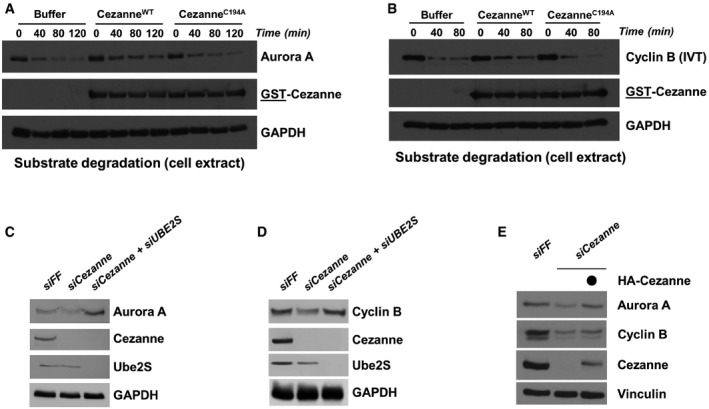
- Degradation assay of endogenous Aurora A using a G1 HeLaS3 cell extract supplemented with ATP, ubiquitin, UBE2C, and Cezanne. Aliquots were collected at the indicated time points and analyzed by immunoblot.
- Degradation assay was performed as in (A) except that in vitro‐translated Cyclin B was used as a substrate. Aliquots were collected at the indicated time points and analyzed by immunoblot.
- U2OS cells were transfected with the control firefly (FF), Cezanne, and Cezanne with UBE2S siRNAs for 48 h and analyzed by immunoblot with the indicated antibodies.
- U2OS cells were analyzed as in (C).
- U2OS cells were transfected with the indicated siRNAs, and after 48 h of knock down, an empty vector or an siRNA‐resistant Cezanne vector was introduced by transfection for an additional 24 h. Cells were analyzed by immunoblot.
Next, we monitored the degradation of endogenous proteins in cells. U2OS cells depleted for Cezanne using RNAi were synchronized in pro‐metaphase by nocodazole and then released into media containing cycloheximide to block translation. In Cezanne‐depleted cells, the abundance and stability of both Aurora A and Cyclin B were reduced, consistent with Cezanne preventing their degradation by promoting their deubiquitination (Appendix Fig S3A). Importantly, Cezanne‐depleted cells efficiently arrested in mitosis by nocodazole, and showed similar timing of mitotic exit, evidenced by the accumulation and disappearance of phospho Serine 10 on Histone 3. These experiments support a role for Cezanne in antagonizing the ubiquitination and degradation dynamics of APC/C substrates.
The effect of Cezanne depletion on endogenous Aurora A and Cyclin B was also evident in asynchronous cells, where depletion decreased the abundance of both proteins (Fig 4C and D). The decrease in Aurora A and Cyclin B was partially rescued by re‐expression of an RNAi resistant version of Cezanne, ruling out off‐target effects of RNAi, and consistent with the biochemical data above (Fig 4E). Significantly, co‐depletion of UBE2S, which adds K11‐linked ubiquitin chains via APC/C to these substrates, also reversed these effects (Fig 4C and D). Finally, co‐expression of Cezanne, but not the catalytically inactive mutant, led to an increase in Aurora A protein levels following transient transfection of 293T cells (Appendix Fig S3B). Altogether, these data strongly suggest that Cezanne regulates Aurora A and Cyclin B degradation.
Cezanne is required for proper mitotic progression
We next determined the consequences of Cezanne depletion on mitotic progression. U2OS cells were transfected with two independent siRNA oligonucleotides, either alone or in combination, fixed and stained for DNA and Tubulin and analyzed by confocal microscopy. Both siRNAs efficiently deplete Cezanne (Appendix Fig S5). Cezanne depletion significantly increased the percentage of cells with micronuclei (Fig 5A and B). Significantly, the increase in micronuclei observed in Cezanne‐depleted cells is almost entirely reversed by co‐depletion of UBE2S (Fig 5C).
Figure 5. Cezanne depletion impairs mitotic progression.
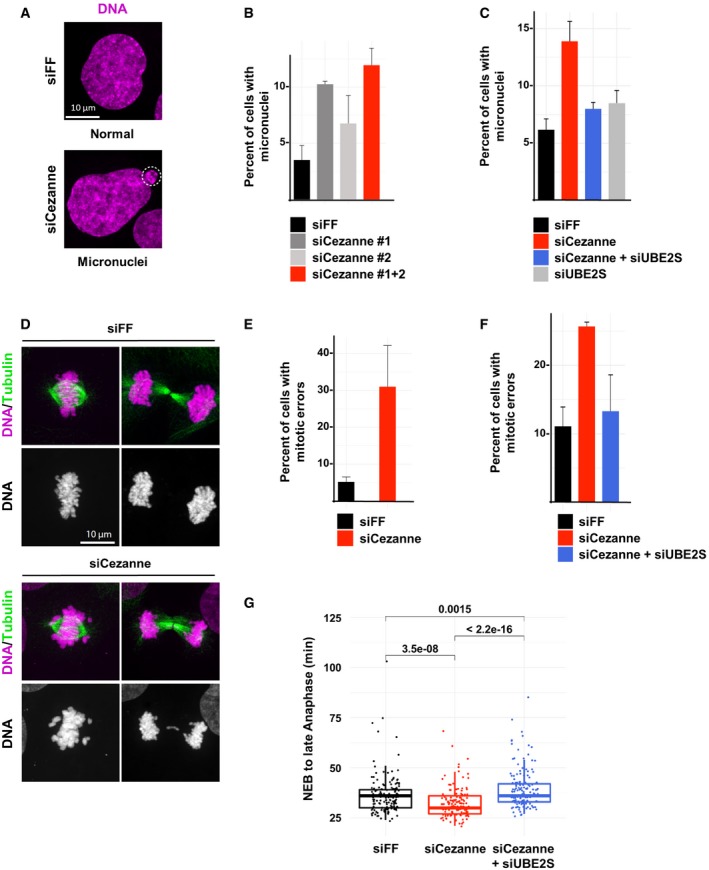
- Representative immunofluorescence images of micronuclei in control or Cezanne‐depleted U2OS cells (scale bar = 10 μm).
- Quantification of cells with micronuclei for each condition (error bars show standard deviation, duplicate experiments, n > 930 cells per condition).
- Quantification of U2OS cells with micronuclei from control cells (black), Cezanne only depleted (red), UBE2S only depleted (gray), or Cezanne and UBE2S co‐depleted cells (blue) (error bars show standard deviation, duplicate experiments, n > 980 cells per condition).
- Representative immunofluorescence images of metaphase and anaphase in control or Cezanne‐depleted U2OS cells (scale bar = 10 μm).
- Quantification of mitotic error frequency in control or Cezanne‐depleted cells (error bars show standard deviation, duplicate experiments, n > 62 cells per condition).
- Quantification of mitotic error frequency in control cells (black), Cezanne only depleted (red), or Cezanne and UBE2S co‐depleted cells (blue) (error bars show standard deviation, duplicate experiments, n > 140 cells per condition).
- GFP‐H2B U2OS cells transfected with control (black), Cezanne (red), or Cezanne and UBE2S siRNAs (blue) were subjected to live cell imaging. Cells were then manually tracked from the onset of visible chromosomal condensation in mitosis until late anaphase/early telophase. One hundred and fifty mitotic events were analyzed per siRNA condition, and P‐values between conditions were computed using the non‐parametric two‐sample Wilcoxon test (box and whisker plots represent the distribution of the values to allow visualization of the median, upper, and lower quartiles).
A likely cause of micronuclei formation is improper chromosome segregation. Notably, prior reports showed that restraining APC/C activity slows mitotic progression and reduces errors (Zeng et al, 2010; Sansregret et al, 2017). We analyzed lagging chromosomes, which cause aneuploidy and micronuclei, after depletion of Cezanne. Indeed, we observed a significant increase in the frequency of lagging chromosomes in Cezanne‐depleted cells (Fig 5D and E). In addition, in Cezanne‐depleted cells, chromosomes are less well aligned at the equator of the cell at metaphase. Importantly, co‐depletion of UBE2S completely reversed the increase in mitotic errors observed in Cezanne‐depleted cells (Fig 5F). Since APC/C promotes mitotic exit, we measured the time from nuclear envelope breakdown (NEB) until anaphase in U2OS cells expressing EGFP‐tagged Histone H2B. Cezanne‐depleted cells showed a small, but significant decrease in NEB to anaphase timing and this was reversed by depletion of UBE2S (Fig 5G). We also noted an increase in the number of apoptotic cells following Cezanne depletion (Appendix Fig S3C). Altogether, these observations point to a role for Cezanne in controlling mitotic progression and chromosome segregation fidelity by antagonizing APC/C activity.
Cezanne controls APC/C substrates in mitosis
To analyze the degradation kinetics of APC/C substrates in mitosis, we released U2OS cells from a mitotic block, using nocodazole, and monitored several substrates. FoxM1, Cyclin B, and Aurora A degradation rates were accelerated in Cezanne‐depleted cells (Fig 6A). However, the degradation of Aurora B and Geminin, other APC/C targets, was unaffected by Cezanne depletion (Appendix Fig S4). We further confirmed the role of Cezanne in controlling the degradation rate of Cyclin B, using live cell imaging of U2OS cells expressing Venus‐Cyclin B. Asynchronous cells were imaged, and the degradation of Cyclin B was analyzed during mitosis. We determined the time from mitotic entry until the intensity of Venus‐Cyclin B reached levels of background fluorescence. The timing of Cyclin B degradation is significantly accelerated in Cezanne‐depleted cells compared to control cells (Fig 6B and C), and this difference cannot be accounted for by changes in the duration of mitosis (Fig 5). Therefore, biochemical and cell biological evidence demonstrates that Cezanne antagonizes the destruction of APC/C substrates in mitosis.
Figure 6. Cezanne depletion accelerates APC/C substrate destruction and reduces the percent of replicating cells.
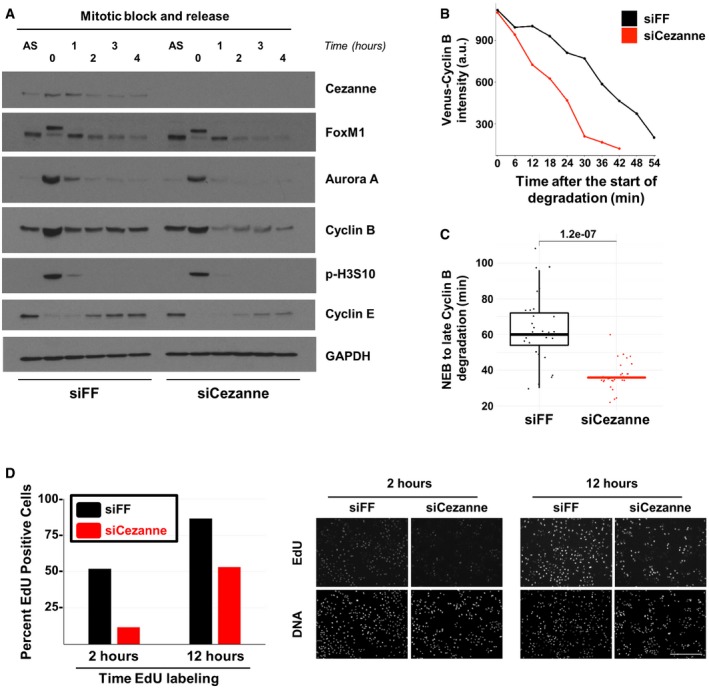
- U2OS cells transfected with firefly control or Cezanne siRNA (siFF and siCezanne, respectively) and synchronized in mitosis were released in fresh medium and analyzed at the indicated time points by immunoblot. Additional immunoblotting of these samples is shown in Appendix Fig S4, and the same Cezanne and GAPDH blots are shown in both figures.
- Representative in vivo degradation curves of Venus‐Cyclin B during mitosis from control (black) or Cezanne‐depleted cells (red).
- Quantification of Venus‐Cyclin B degradation curves from control U2OS cells (black) or Cezanne‐depleted cells (red). Thirty cells per condition were analyzed (box and whisker plots represent the distribution of the values to allow visualization of the median, upper, and lower quartiles—note that the box for the Cezanne plot is not immediately visible because the median and first and third quartiles are all equal to 36. This indicates the narrow distribution of points around the median).
- Cell cycle progression was analyzed using on plate EdU labeling in control and Cezanne‐depleted U2OS cells (scale bar = 400 μm).
In addition to its role in promoting mitotic exit, the APC/C plays an evolutionarily conserved role in restraining S‐phase entry. Since APC/C inactivation accelerates progression through G1‐phase, we hypothesized that depleting Cezanne, which antagonizes APC/C, would lead to a slower progression into S‐phase. Importantly, in cells proliferating after a mitotic block and release, the accumulation of Cyclin E, which indicates progression through G1, was significantly reduced in Cezanne‐depleted cells, indicating a slow progression toward S‐phase (Fig 6A). These results support the notion that Cezanne is involved in substrate degradation at mitotic exit and in progression through the cell cycle, by antagonizing APC/C. We depleted cells using control or Cezanne siRNA and monitored S‐phase using EdU incorporation in an asynchronous population. Cells were treated with EdU for different amounts of time prior to fixation to determine the percent of cells in S‐phase and as a measure of the accumulation of S‐phase cells over time. After 2 h of EdU labeling, 52% of control cells were EdU‐positive and this number increased to 87% after 12 h of EdU labeling (Fig 6D). In contrast, Cezanne depletion significantly decreased the number of EdU‐labeled cells, with only 12% of EdU‐positive cells after 2 h, and 53% after 12 h (Fig 6D). Accordingly, Cezanne depletion impaired the proliferation of U2OS, HCT116, and HeLaS3 cell lines, as analyzed by live cell imaging (Appendix Fig S5A–C). Altogether, these observations strongly suggest that Cezanne contributes to mitotic exit and that its function is crucial for cells to properly progress through the cell cycle and proliferate.
Discussion
The multi‐subunit E3 ubiquitin ligase APC/C plays a vital role in cell cycle progression. Since APC/C mediates the degradation of cyclins, oscillations in APC/C activity form the backbone of the eukaryotic cell cycle. The mechanisms controlling APC/C and CDK activity share many common features. Both are activated by obligate co‐activator subunits: cyclins in the case of CDK, and Cdc20 or Cdh1 for APC/C. Both enzymes are controlled by direct phosphorylation (Alfieri et al, 2017). In addition, similar mechanisms have evolved to restrain their activity. For CDK and APC/C, their activity is restrained by binding to protein inhibitors, and their co‐activators are tightly controlled by degradation (Kramer et al, 2000; Reimann et al, 2001; Rape & Kirschner, 2004; Williamson et al, 2009b; Fukushima et al, 2013; Choudhury et al, 2016).
Additionally, phosphatases indirectly control CDK phosphorylation networks, independent of direct regulation of CDK itself. Comparatively, much less is known about reversible ubiquitination on APC/C targets by DUBs. In yeast, UBP15 counteracts the degradation of the APC/C substrate Clb5 (Ostapenko et al, 2015). Similarly, Cyclin A is regulated by USP37, and USP37 is itself targeted for degradation by APC/C and other E3s (Huang et al, 2011; Burrows et al, 2012). In addition, USP44 deubiquitinates the MCC and promotes SAC signaling (Stegmeier et al, 2007).
The APC/C E2 enzyme UBE2S was identified as a vital E2 ubiquitin‐conjugating enzyme which is important for APC/C substrate degradation and mitotic exit (Garnett et al, 2009; Min et al, 2015). Interestingly, recent work found that, despite the physiological role of UBE2S in substrate ubiquitination, it is non‐essential for mitotic progression (Wild et al, 2016). Thus, like many other seemingly essential cell cycle regulators (e.g., cyclins and CDKs), there is surprising flexibility and redundancy in the signaling that drives mitotic progression. Nevertheless, while UBE2S forms K11‐linked ubiquitin chains on APC/C substrates to promote their degradation at mitotic exit (Williamson et al, 2009b; Wu et al, 2010), it was previously unknown whether a DUB counteracts the formation of APC/C‐driven, K11‐linked ubiquitin chains.
Komander et al reported that Cezanne has a preference for K11‐linked diubiquitin substrates using a recombinant fragment of Cezanne (Bremm et al, 2010; Mevissen et al, 2013, 2016). However, other studies suggested that Cezanne disassembles K63‐linked and K48‐linked ubiquitin chains. Our data, using the same bacterially purified Cezanne fragment as Komander et al, as well as full‐length Cezanne, support the conclusion that Cezanne has tremendous selectivity for K11‐linked chains. In addition, our in vitro data demonstrate, for the first time, the ability of Cezanne to directly deubiquitinate ubiquitin‐modified substrates (Cyclin B and Securin) harboring K11‐linked ubiquitin chains. Moreover, despite the ability of Cezanne to directly bind Cyclin B, it is unable to deubiquitinate Cyclin B when polyubiquitinated with either K48‐linked or K63‐linked ubiquitin chains.
Cezanne abundance is cell cycle‐regulated in multiple cell lines (U2OS, HCT116, and RPE1). Cezanne peaks in mitosis and is reduced in non‐mitotic cells. This raises the question of what drives changes in Cezanne abundance. Cezanne transcripts are elevated in synchronized mitotic cells, potentially accounting for its cell cycle dynamics. However, Cezanne was identified in only one of several studies that examined cell cycle transcriptional dynamics, indicating that it is likely not a core cell cycle gene, or alternatively, that the dynamics of its mRNA expression do not meet a threshold for considering it a cell cycle‐regulated transcript (Whitfield et al, 2002; Bar‐Joseph et al, 2008; Sadasivam et al, 2012; Grant et al, 2013; Peña‐Diaz et al, 2013). Nevertheless, several studies showed that the cell cycle transcription factors Myb and FoxM1 localize to the Cezanne gene promoter (Fischer et al, 2016). Cezanne abundance steadily decreases as cells exit mitosis. It remains unknown if Cezanne is subjected to cell cycle‐dependent degradation, and if so, what enzyme(s) coordinate its proteolysis.
APC/C and other E3 ligases recognize substrates through degron sequence motifs. However, for most DUBs, it is unknown how they engage substrates (Sahtoe & Sixma, 2015). Thus, an important question is how Cezanne specifically recognizes APC/C substrates to regulate their deubiquitination. Many other DUBs bind directly to E3 ubiquitin ligases (Sowa et al, 2009), and our data indicate that Cezanne can bind to the APC/C co‐activators. However, it is unknown whether Cezanne binds directly to the APC/C. Cezanne has not been identified in large‐scale protein interaction mass spectrometry studies, which surveyed a subset of APC/C subunits (Huttlin et al, 2015). Likewise, APC/C components were not identified in Cezanne pull‐down–mass spectrometry experiments (Sowa et al, 2009). It therefore seems unlikely that Cezanne is a core component of the APC/C complex. We showed that Cezanne binds directly to Cyclin B and Aurora A in vitro. In addition, co‐IP experiments from 293T cells showed that Cezanne could pull‐down unmodified Cyclin B, Aurora A, and FoxM1. Note that we did not detect interactions by endogenous IP, likely due to the quality of Cezanne antibodies and the transient nature of DUB‐substrate interactions. Nevertheless, these results strongly suggest that Cezanne engages targets without the need for K11‐linked chains or for an additional substrate receptor subunit. However, the underlying molecular features enabling substrate engagement are unknown. We predict that Cezanne could engage both a sequence motif and structure on substrates, while simultaneously forming a multivalent engagement with K11‐linked chains.
There is significant interest in the APC/C field in the concept of substrate ordering, that is, the observation that different substrates are degraded with different kinetics. This is likely due, in part, to complex effects associated with the affinity of APC/C for its substrates (Lu et al, 2014), as well as APC/C processivity (Rape et al, 2006). DUBs have been proposed to play a potential role in controlling substrate ordering. However, since there were previously no known DUBs that antagonize the degradation of APC/C substrates modified with K11‐linked ubiquitin chains, this contribution has been difficult to test.
Since CDK activity plays a critical role in proliferation, CDK regulators and CDK substrates are often dysregulated in cancer. For example, both the CDK4/6 inhibitor p16INK4A, and the CDK4/6 substrate RB (retinoblastoma protein), are recurrently inactivated in cancer. Likewise, Cyclin E is frequently amplified in malignancy. In addition, many UPS enzymes are aberrantly regulated in cancer. This includes FbxW7, a component of the E3 ligase that controls Cyclin E, and Mdm2, the cognate E3 ligase for the tumor suppressor p53. Together, this points to a vital role for UPS enzymes in controlling normal and malignant cell proliferation. Whereas few UPS enzymes are mutated in cancer, the expression of many is altered through changes in gene expression and genome amplification.
Previous studies have examined transcriptional and DNA copy‐number changes in large cohorts of patient tumors. Among these studies, breast cancer remains one of the most well‐studied. Among the ~90 DUBs encoded by the human genome, the three that are most significantly amplified/overexpressed in breast cancer are USP21, and two ovarian tumor domain family enzymes, OTUD6B and OTUD7B/Cezanne (Cancer Genome Atlas Network, 2012; Ciriello et al, 2015). Remarkably, Cezanne is amplified and/or overexpressed in more than 30% of breast tumors and is localized in a amplicon on chromosome 1q that lacks a known oncogene (Silva et al, 2015).
Much of the focus on APC/C regulation centers on its role in mitosis, where it drives anaphase progression and mitotic exit. However, APC/C is active throughout G1‐phase, where it restricts G1‐S progression (Sørensen et al, 2001; Cappell et al, 2016; Choudhury et al, 2016). Consistently, Cdh1 collaborates with the retinoblastoma tumor suppressor protein to restrict S‐phase entry (Fay et al, 2002; Binné et al, 2007; Buttitta et al, 2010; The et al, 2015), and single allelic loss of the Cdh1 gene promotes tumorigenesis in mice (García‐Higuera et al, 2008). We recently showed that Cdh1 degradation could be driven by upstream signaling through the oncogenic AKT kinase (Choudhury et al, 2017), which is activated in numerous cancers (Manning & Toker, 2017). Since, Cezanne is recurrently amplified/overexpressed in cancers, it could promote proliferation and perhaps impact chromosome stability, by antagonizing APC/C.
Materials and Methods
Mammalian cell culture, transfection, antibody reagents, and synchronization
HEK‐293T, HCT‐116, RPE1, HeLaS3, and U2OS cells were obtained from ATCC and grown in DMEM complete medium (Gibco) supplemented with 10% fetal bovine serum (Atlanta Biologicals). All DNA transfection experiments were performed in HEK‐293T using Lipofectamine 2000 (Life Technologies) and cultured for 24 h prior to analysis. Samples for protein analysis by immunoblot were lysed in NETN buffer [20 mM Tris‐Cl (pH 8.0), 100 mM NaCl, 0.5 mM EDTA, and 0.5% (v/v) Nonidet P‐40 (NP‐40)] supplemented with 2 μg/ml pepstatin, 2 μg/ml apoprotinin, 10 μg/ml leupeptin, 1 mM AEBSF [4‐(2 Aminoethyl) benzenesulfonyl fluoride], and 1 mM Na3VO4. Lysis was performed on ice for ~10 min, and then, protein concentration was determined using Bradford (Bio‐Rad). Standard immunoblotting procedures were followed. Membranes were blocked in 5% non‐fat dried milk [diluted in Tris‐buffered saline 0.05% Tween‐20 (TBST)].
All siRNA transfection experiments were performed using Lipofectamine RNAiMAX reagent (Life Technologies) following the manufacturer's instructions.
The following antibodies, and the indicated concentrations, were used in this study. Anti‐FLAG‐HRP was purchased from Sigma (A8592; 1:5,000). The following antibodies were purchased from Santa Cruz Biotechnology: c‐Myc (sc‐40; 1:2,500), GAPDH (sc‐25778; 1:5,000), Tubulin (sc‐32293; 1:5,000), His‐probe (sc‐803; 1:1,000), Vinculin (sc‐25336; 1:2,000), FoxM1 (sc‐502; 1:2,000), Geminin (sc‐13015; 1:1,000), and Cdc27 (sc‐9972). The following antibodies were purchased from Cell Signaling Technologies: Cezanne/OTUD7B (Cat#14817; 1:2,000), phospho‐Histone H3 (Cat#3377; 1:2,500), Cyclin E (Cat#4129S; 1:2,500), Aurora A (Cat#14475; 1:2,000), HA‐tag (Cat#3724; 1:2,000), ubiquitin (Cat#3933; 1:2,500), and UBE2S (Cat#9630; 1:2,000). Cyclin B1 (ab32053; 1:10,000), Aurora B (ab2254; 1:2,000), and GFP (ab6556; 1:10,000) were from Abcam. GST (A190‐122A; 1:10,000) was from Bethyl, and ubiquitin K11 linkage (MABS107‐I; 1:1,000) was from Millipore. All antibodies were diluted in 5% non‐fat dried milk [prepared in tris‐buffered saline, 0.05% Tween‐20 (TBST)] and incubated overnight at 4°C and detected using HRP‐conjugated secondary antibodies (Jackson Immuno Research Laboratories Inc; 1:5,000 dilutions) following standard procedures.
Non‐transformed RPE1 cells were serum‐starved for 24 h to synchronize them in G0/G1. Serum was added back to monitor their progression through the cell cycle. HeLaS3 cells were treated with 2 mM thymidine for 24 h, washed, and then released for 4 h before being treated with 100 ng/ml of nocodazole in DMEM for 11 h to obtain a mitotic population. In order to get a G1 population, cells were washed with warm PBS once, then twice with DMEM and released for 2 h before harvest. U2OS cells were synchronized in mitosis using either 200 ng of nocodazole per ml of DMEM overnight or by using a thymidine/nocodazole treatment similar to the one described for HeLaS3, except that nocodazole treatment was 150 ng/ml of DMEM for 16 h. HTC116 cells were synchronized in mitosis using 200 ng of Nocodazole per ml of DMEM overnight.
Molecular biology
pOPINK‐Cezanne (OTU, aa 53–446) was a gift from David Komander (Addgene plasmid # 61581; Mevissen et al, 2013). Flag‐HA‐OTUD7B was a gift from Wade Harper (Addgene plasmid # 22550; Sowa et al, 2009). pDONR223‐AURKA was a gift from William Hahn & David Root (Addgene plasmid # 23532; Johannessen et al, 2010) and was subcloned using gateway recombination cloning into a pDEST N‐Myc vector. The C194S Cezanne mutant was generated by using site‐directed mutagenesis and verified by sequencing.
In vitro Cezanne DUB assay
GST‐Cezanne was diluted to a final concentration of 0.2 or 0.1 μM in DUB reaction buffer (50 mM Tris pH 7.5, 50 mM NaCl, and 5 mM DTT) and incubated for 5–10 min at room temperature. The reaction was started by adding di‐tetraubiquitin or tetraubiquitin at a final concentration of 1 μM. Aliquots were collected at indicated time points, stopped in sample buffer (60 mM Tris pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, and 0.1% Bromophenol Blue), and boiled. Ubiquitin cleavage was detected by silver staining.
In vivo ubiquitylation of Cezanne
The in vivo ubiquitylation assay was performed as described previously (Bonacci et al, 2014). Briefly, HEK‐293T cells were transfected in 10‐cm dishes using Lipofectamine2000 (Thermo Fisher Scientific) and harvested the next day in PBS. Eighty percent of cell suspension was lysed in 6 M guanidine‐HCl‐containing buffer to pull‐down His6‐Ubiquitinated proteins on Ni2+‐NTA beads, while the remaining 20% was used to prepare inputs. Ni2+ pull‐down eluates and inputs were separated through SDS–PAGE and analyzed by immunoblot.
Immunoprecipitation
Standard procedures of immunoprecipitation (IP) were used. HA‐tagged Cezanne was co‐expressed with Myc‐tagged FoxM1 or Aurora A in HEK 293T cells for 24 h. Cells were washed and scrapped in PBS (DIFCO) and then lysed in NETN for 10 min at 4°C with occasional vortexing. Cell debris was removed by centrifugation at 20,000 g for 10 min at 4°C. Anti‐HA beads (15 μl per IP, Sigma, Cat No E6779) were mixed with the clarified lysate and incubated on a rotary shaker for 2 h at 4°C. The beads were washed with lysis buffer five times and then incubated with sample buffer (60 mM Tris pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, and 0.1% Bromophenol Blue) at 95°C for 5 min.
GST‐Cezanne pull‐down assays
Pull‐down experiments were performed by coating 15 μl of GSH beads with 5 μg of GST‐Cezanne in GST pull‐down buffer (20 mM Tris pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% NP40) for 1 h at 4°C. Beads were incubated with IVT Cyclin B, recombinant Aurora A, or lysates from asynchronous or mitotic U2OS for 2 h at 4°C. The beads were washed five times in GST pull‐down buffer and then incubated with sample buffer at 95°C for 5 min.
In vitro APC/C ubiquitination assay using Cdc27 immunopurification
Cyclin B was translated in vitro in rabbit reticulocyte lysate (Promega). Mitotic HeLaS3 extract was prepared as described previously (Williamson et al, 2009b). APC/C was captured on Protein A/G beads coupled with an anti‐Cdc27 antibody (Santa Cruz Biotech), by rotating the bead slurry with a mitotic extract on a rotary mixer for ~4 h. In vitro ubiquitin assays were performed by incubating 1.5 μl of in vitro‐translated substrate in 45 μl reactions containing UBAB buffer (2.5 mM Tris–HCL pH 7.5, 5 mM NaCl, and 1 mM MgCl2), energy mix (15 mM creatine phosphate, 2 mM ATP, and 2 mM MgCl2 pH 8.0), 4 mM DTT, 1.5 μg purified ubiquitin, 100 nM E1 (UBE1), 100 nM of E2 (UbcH10), 5 μl of the APC/C‐bead slurry, and the indicated amount of GST‐Cezanne. Ubiquitin and E1 and E2 enzymes were purchased from Life Sensors. Aliquots of 12 μl were collected at desired time points, mixed in sample buffer, and boiled.
Generation of K11, K48, and K63 ubiquitin chains on Ub‐Cyclin B and Ub‐Securin
The Ub‐CyclinBNTD (residues 1‐95) and Ub‐Securin (CC197‐8AA) constructs were expressed as N‐terminal GST fusions with a C‐terminal Cys‐His6 tag and purified by nickel affinity chromatography, treated with TEV to remove tags and then polished with size exclusion chromatography (Brown et al, 2014).
K11 ubiquitin chains on Ub‐Cyclin B and Ub‐Securin were generated in a 200 μl reaction containing 20 nM of recombinant APC/C, 1 μM Cdh1, 0.1 μM E1 (UBE1), 1 μM E2 (UBE2S), 125 μM ubiquitin, 5 mM MgCl2, 0.82 μM of either Ub‐Cyclin B or Ub‐Securin, 0.25 mg/ml BSA, 20 mM HEPES, and 200 mM NaCl. Ubiquitination reactions were conducted at room temperature for 30 min and then quenched with 50 μM EDTA. K48 ubiquitin chains were generated in a 200 μl reaction containing 0.82 μM of either Ub‐Cyclin B or Ub‐Securin, 1 μM E1 (UBE1), 10 μM E2 (Cdc34), 100 μM ubiquitin, 5 mM MgCl2, 0.25 mg/ml BSA, 20 mM HEPES, and 200 mM NaCl. Ubiquitination reactions were conducted at 37°C for 16 h and then quenched with 50 μM EDTA. K63 ubiquitin chains were generated exactly as described for K48 chains, except that 40 μM of E2s was used (20 μM of Ubc13 and 20 μM of UEV1).
EdU incorporation analysis
S‐phase entry was monitored by EdU (5‐Ethynyl‐2′‐deoxyuridine; Sigma # T511285) incorporation. U2OS cells transfected with siRNA were plated in duplicate in 12‐well plates at 30,000 cells/well 24 h post‐transfection and allowed to attach overnight. Cells were labeled in medium containing 10% fetal bovine serum in the presence of 10 μM EdU or vehicle (DMSO) for the indicated times. Cells were fixed on plates in 3.7% formaldehyde and washed in PBS prior to EdU labeling by azide‐alkyne cycloaddition click chemistry. Click reactions are a total of 500 μl in PBS and include Alexa Fluor 488 Azide (final concentration of 1 μM), CuSO4 (final concentration of 1 mM), and ascorbic acid (final concentration of 100 mM). The labeled cells were counterstained for DNA with 7‐AAD or Hoechst. Images were captured on an EVOS™ FL Auto Imaging System (Life Technologies) and processed in ImageJ to obtain the percentage of EdU‐positive S‐phase cells within the total number of cells in the field.
Silver staining
Gel was rinsed twice in water for 10 min and then fixed for 20 min in 150 ml of fixation buffer (50% methanol + 5% acetic acid in water). The gel was first washed in 150 ml of water containing 50% methanol for 10 min and then in water for 10 min. Sensitization was performed by incubating the gel in 150 ml of water containing 0.02% sodium thiosulfate for 1 min followed by two washes in water for 1 min each. Silver staining was performed by submerging the gel in 150 ml of water containing 0.1% silver nitrate and 0.08% formalin for 20 min, and then, gel was rinsed twice in water for 1 min. Development was done by incubation in 150 ml of water containing 2% sodium carbonate and 0.04% formalin until desired intensity of staining occurred. Reaction was stopped by washing the gel in 150 ml of water containing 5% acetic acid for 10 min before scanning the gel.
Sample preparation for immunofluorescence
U2OS cells were grown on acid washed #1.5 coverslips (Corning). Cells were fixed by pre‐warmed 3% PFA (freshly made from paraformaldehyde powder (Sigma)) at 37°C in PHEM buffer (120 mM Pipes, 50 mM HEPES, 20 mM EGTA, 4 mM magnesium acetate, pH 7.0) for 15 min. Fixed samples were permeabilized by 0.5% NP40 (Roche) in PHEM buffer and incubated in BGS (Boiled Goat Serum) for 30 min at room temperature (Suzuki et al, 2015). Coverslips were incubated for 1.5 h at 37°C with anti‐Tubulin antibodies (Cytoskeleton, Inc.). After primary antibody incubation, samples were incubated for additional 1.5 h at 37°C with secondary antibodies with SiR‐DNA (Cytoskeleton, Inc.). Secondary antibodies were conjugated with Alexa488 (Jackson ImmunoResearch). After DNA staining by DAPI, samples were mounted using Prolong Gold Antifade (Molecular Probe).
Imaging
For image acquisition, 3D stacks of 70–90 frame of green and far‐red fluorescent images were obtained sequentially at 200‐nm steps along the z‐axis through the cell using MetaMorph 7.8 software (Molecular Devices) and a high‐resolution Nikon Ti inverted microscope equipped with an Orca AG cooled CCD camera with gain set to zero (Hamamatsu) and an 100X/1.4NA (PlanApo) DIC oil immersion objective (Nikon). Stage movement was controlled by MS2000‐500 (ASI) with piezo stage for z‐axis stepping (Suzuki et al, 2015). Solid‐state laser (Andor) illumination at 488 and 647 nm was projected through Borealis (Andor) for uniform illumination of a spinning disk confocal head (Yokogawa CSU‐10; Perkin Elmer).
Live cell imaging
GFP–Histone expressing U2OS cells were plated on glass bottom plates (Cellvis) #1.5 in Fluorobrite DMEM (Invitrogen) supplemented with FBS and l‐glutamine the day before imaging. After ~16 h of growth, cells were treated with siLuciferase, siCezanne, or siCezanne and siUBE2S, or transfected with the indicated expression plasmids and siRNAs, and then allowed to grow for another 8 h before being imaged on a Nikon Ti Eclipse inverted microscope using a Plan Apochromat 40× dry objective lens (NA 0.95) and the Nikon Perfect Focus System. Images were collected every 3 min with an Andor Zyla 4.2 sCMOS detector with 12‐bit resolution. Cells were imaged in a 37°C humidified chamber (Okolabs) with 5% CO2 for 48 h using NIS‐Elements software. The filter set was from Chroma, FITC 480/30 nm; 505 nm; 535/40 nm (excitation; beam splitter; emission filter) CFP ‐ 436/20 nm; 455 nm; 480/40 nm, and YFP ‐ 500/20 nm; 515 nm; 535/30 nm. No photobleaching or phototoxicity was observed under these imaging conditions. Cells were manually tracked from the onset of visible chromosomal condensation in mitosis until late anaphase/early telophase. One hundred and fifty mitotic events were analyzed per siRNA treatment. Nuclei were tracked using mTurquoise2‐H2B expression pattern during the cell cycle. mVenus‐Cyclin B levels were quantified from the regions of interest defined by the mTurquoise2‐H2B signal. Thirty cells were tracked for each condition, and P‐values between conditions were computed using the non‐parametric two‐sample Wilcoxon test (Hollander & Wolfe, 1973). This test was implemented in R using the wilcox.test() method. Distributions are visualized with the open source ggplot2 package.
GST‐Cezanne production and purification
pOPINK‐Cezanne WT and C194S were transformed in the Escherichia coli strain Rosetta2 (DE3) pLacl cells. Cezanne expression was induced at OD600 of 0.6 with 500 μM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG), and cells were collected after an overnight incubation at 18°C. Cells were resuspended in GST lysis buffer (20 mM Tris pH 7.5, 10 mM EDTA, 5 mM EGTA, 150 mM NaCl, supplemented with 1 mg/ml of lysozyme, 1 mM PMSF, and 10 mM β‐mercaptoethanol) and incubated on ice for 20–30 min. Lysis was performed by sonicating cells on ice at 50% power three times for 30 s with a 1‐min break between pulses. Lysate was supplemented with 0.5% Triton X100, kept on ice for 15 min and then centrifuged at 30,000 g for 45 min. Lysate was subsequently filtered on a 0.45‐μm syringe filter and then incubated with 500 μl of GSH beads pre‐washed with lysis buffer for 2 h at 4°C. Beads were washed for 5 min three times with GST washing buffer (20 mM Tris pH 7.5, 10 mM EDTA, 150 mM NaCl, and 0.5% Triton X100, supplemented with 1 mM PMSF and 10 mM β‐mercaptoethanol). Beads were then transferred to an Eppendorf tube and equilibrated for 10 min in 50 mM Tris pH 8.0. Cezanne was eluted in two sequential steps of 30 min each by incubating beads in 50 mM Tris pH 8.0 supplemented with 5 mM of reduced L‐Glutathione. Free l‐Glutathione from eluates was removed during an overnight dialysis at 4°C in 50 mM Tris pH 8.0. Aliquots were flash frozen and kept at −80°C.
Proliferation assays using the IncuCyte system
U2OS or HeLaS3 cells were transfected with the indicated siRNAs in 60‐mm plates. The day after, cells were trypsinized and seeded in 12‐well plates at a confluence of 30,000 cells per well in triplicates. The remaining cells were used for immunoblot analysis to confirm efficient knock down of Cezanne. Cells in 12‐well plates were used to follow proliferation using the IncuCyte system. For each condition, nine pictures per well were taken every 2 h and used to quantify proliferation for a time period of 3–4 days. Representative results of three independent experiments are shown.
Author contributions
TB and MJE conceptualized the project, conceived of experiments, and analyzed data. TB carried out all of the biochemical experiments. TB initially assembled figures and wrote the first draft of the manuscript. TB and MJE edited the manuscript and figures. NGB assisted with experiments using recombinant and purified APC/C complexes. AS performed cell biological analysis of mitotic cells and all experiments using confocal microscopy. JGC and GDG performed live cell imaging experiments, and GDG performing the quantification of mitosis in GFP‐H2B U2OS cells. NS generated all the plots and performed all statistical analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
Special thanks to the Emanuele laboratory for feedback throughout this study. We thank Brenda Schulman (St. Jude's/HHMI/Max Planck Society) for providing reagents and expertise, Catherine Lindon (University of Cambridge, UK) for providing reagents, and Ted Salmon (UNC‐Chapel Hill) for significant expertise, input, and support. This work was supported by start‐up funds from the University Cancer Research Fund to both MJE and NGB, grants to MJE from the Susan G. Komen Foundation (CCR14298820), Jimmy‐V Foundation, and National Institute of Health (R01GM120309), funding from the NIH to JGC (GM083024), and to GDG (T32CA009156) plus additional funding provided by the W.M. Keck Foundation to JGC and a K22 award to NGB (CA 216327‐01).
The EMBO Journal (2018) 37: e98701
References
- Alfieri C, Zhang S, Barford D (2017) Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol 7: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar‐Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I (2008) Genome‐wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci USA 105: 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binné UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Näär AM, Dyson NJ (2007) Retinoblastoma protein and anaphase‐promoting complex physically interact and functionally cooperate during cell‐cycle exit. Nat Cell Biol 9: 225–232 [DOI] [PubMed] [Google Scholar]
- Bonacci T, Audebert S, Camoin L, Baudelet E, Bidaut G, Garcia M, Witzel II, Perkins ND, Borg JP, Iovanna JL, Soubeyran P (2014) Identification of new mechanisms of cellular response to chemotherapy by tracking changes in post‐translational modifications by ubiquitin and ubiquitin‐like proteins. J Proteome Res 13: 2478–2494 [DOI] [PubMed] [Google Scholar]
- Bremm A, Freund SMV, Komander D (2010) Lys11‐linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 17: 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, Watson ER, Weissmann F, Jarvis MA, VanderLinden R, Grace CRR, Frye JJ, Qiao R, Dube P, Petzold G, Cho SE, Alsharif O, Bao J, Davidson IF, Zheng JJ, Nourse A, Kurinov I, Peters J‐M, Stark H, Schulman BA (2014) Mechanism of polyubiquitination by human anaphase‐promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell 56: 246–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, VanderLinden R, Watson ER, Qiao R, Grace CRR, Yamaguchi M, Weissmann F, Frye JJ, Dube P, Ei Cho S, Actis ML, Rodrigues P, Fujii N, Peters J‐M, Stark H, Schulman BA (2015) RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase‐promoting complex. Proc Natl Acad Sci USA 112: 5272–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu KP, Zhang W, Yu S, Mercredi PY, Harrison JS, Davidson IF, Qiao R, Lu Y, Dube P, Brunner MR, Grace CRR, Miller DJ, Haselbach D, Jarvis MA, Yamaguchi M et al (2016) Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165: 1440–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AC, Prokop J, Summers MK (2012) Skp1‐Cul1‐F‐box ubiquitin ligase (SCF(βTrCP))‐mediated destruction of the ubiquitin‐specific protease USP37 during G2‐phase promotes mitotic entry. J Biol Chem 287: 39021–39029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta LA, Katzaroff AJ, Edgar BA (2010) A robust cell cycle control mechanism limits E2F‐induced proliferation of terminally differentiated cells in vivo . J Cell Biol 189: 981–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T (2016) Irreversible APCCdh1 inactivation underlies the point of no return for cell‐cycle entry. Cell 166: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Bonacci T, Arceci A, Lahiri D, Mills CA, Kernan JL, Branigan TB, DeCaprio JA, Burke DJ, Emanuele MJ (2016) APC/C and SCF(cyclin F) constitute a reciprocal feedback circuit controlling S‐phase entry. Cell Rep 16: 3359–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Bonacci T, Wang X, Truong A, Arceci A, Zhang Y, Mills CA, Kernan JL, Liu P, Emanuele MJ (2017) The E3 ubiquitin ligase SCF(Cyclin F) transmits AKT signaling to the cell‐cycle machinery. Cell Rep 20: 3212–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GMK, Factor RE, Collins LC, Allison KH, Chen YY et al (2015) Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163: 506–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Morgan DO (2016) Building a regulatory network with short linear sequence motifs: lessons from the degrons of the anaphase‐promoting complex. Mol Cell 64: 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enesa K, Zakkar M, Chaudhury H, Luong LA, Rawlinson L, Mason JC, Haskard DO, Dean JLE, Evans PC (2008) NF‐κB suppression by the deubiquitinating enzyme cezanne: a novel negative feedback loop in pro‐inflammatory signaling. J Biol Chem 283: 7036–7045 [DOI] [PubMed] [Google Scholar]
- Fay DS, Keenan S, Han M (2002) fzr‐1 and lin‐35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev 16: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Grossmann P, Padi M, DeCaprio JA (2016) Integration of TP53, DREAM, MMB‐FOXM1 and RB‐E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 44: 6070–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S, Pines J, Lindon C (2008) APC/CCdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol 18: 1649–1658 [DOI] [PubMed] [Google Scholar]
- Foster SA, Morgan DO (2012) The APC/C subunit Mnd2/Apc15 Promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol Cell 47: 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Ogura K, Wan L, Lu Y, Li V, Gao D, Liu P, Lau AW, Wu T, Kirschner MW, Inuzuka H, Wei W (2013) SCF‐mediated Cdh1 degradation defines a negative feedback system that coordinates cell‐cycle progression. Cell Rep 4: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Higuera I, Manchado E, Dubus P, Cañamero M, Méndez J, Moreno S, Malumbres M (2008) Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 10: 802–811 [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR (2009) UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol 11: 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Brooks L, Zhang X, Mahoney JM, Martyanov V, Wood TA, Sherlock G, Cheng C, Whitfield ML (2013) Identification of cell cycle‐regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell 24: 3634–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Chao WCH, Zhang Z, Yang J, Cronin N, Barford D (2013) Insights into degron recognition by APC/C coactivators from the structure of an Acm1‐Cdh1 complex. Mol Cell 50: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M, Wolfe D (1973) Nonparametric statistical methods, pp 200–208. New York, NY: John Wiley & Sons; [Google Scholar]
- Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM (2011) Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell 42: 511–523 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D et al (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162: 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin‐chain formation by the human anaphase‐promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi‐Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S et al (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M (2014) Ubiquitin chain elongation requires E3‐dependent tracking of the emerging conjugate. Mol Cell 56: 232–245 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters J (2000) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11: 1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez‐Fernandez M, Stahl M, Medema RH (2008) FoxM1 is degraded at mitotic exit in a Cdh1‐dependent manner. Cell Cycle 7: 2720–2726 [DOI] [PubMed] [Google Scholar]
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO (2014) Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J Cell Biol 207: 23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Sørensen CS, Kramer E, Santoni‐Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J (1999) Accumulation of cyclin B1 requires E2F and cyclin‐A‐dependent rearrangement of the anaphase‐promoting complex. Nature 401: 815–818 [DOI] [PubMed] [Google Scholar]
- Manning BD, Toker A (2017) AKT/PKB signaling: navigating the network. Cell 169: 381–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM (2010) K11‐linked polyubiquitination in cell cycle control revealed by a K11 linkage‐specific antibody. Mol Cell 39: 477–484 [DOI] [PubMed] [Google Scholar]
- Mevissen TET, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SMV, Ovaa H, Komander D (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154: 169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TET, Kulathu Y, Mulder MPC, Geurink PP, Maslen SL, Gersch M, Elliott PR, Burke JE, van Tol BDM, Akutsu M, El Oualid F, Kawasaki M, Freund SMV, Ovaa H, Komander D (2016) Molecular basis of Lys11‐polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538: 402–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min M, Mevissen TET, De Luca M, Komander D, Lindon C (2015) Efficient APC/C substrate degradation in cells undergoing mitotic exit depends on K11 ubiquitin linkages. Mol Biol Cell 26: 4325–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz S, Bandarra D, Biddlestone J, Campbell KJ, Komander D, Bremm A, Rocha S (2015) Cezanne regulates E2F1‐dependent HIF2α expression. J Cell Sci 128: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A (2015) The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25: R1002–R1018 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J (2008) The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol 10: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapenko D, Burton JL, Solomon MJ (2015) The Ubp15 deubiquitinase promotes timely entry into S phase in Saccharomyces cerevisiae . Mol Biol Cell 26: 2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P (2008) Anaphase‐promoting complex/cyclosome‐CDH1‐mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol 28: 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña‐Diaz J, Hegre SA, Anderssen E, Aas PA, Mjelle R, Gilfillan GD, Lyle R, Drabløs F, Krokan HE, Sætrom P (2013) Transcription profiling during the cell cycle shows that a subset of Polycomb‐targeted genes is upregulated during DNA replication. Nucleic Acids Res 41: 2846–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J (2011) Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 12: 427–438 [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW (2004) Autonomous regulation of the anaphase‐promoting complex couples mitosis to S‐phase entry. Nature 432: 588–595 [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW (2006) The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell 124: 89–103 [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW (2007) Ubiquitination by the anaphase‐promoting complex drives spindle checkpoint inactivation. Nature 446: 921–925 [DOI] [PubMed] [Google Scholar]
- Reimann JDR, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK (2001) Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105: 645–655 [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA (2012) The MuvB complex sequentially recruits B‐Myb and FoxM1 to promote mitotic gene expression. Genes Dev 26: 474–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, Sixma TK (2015) Layers of DUB regulation. Trends Biochem Sci 40: 456–467 [DOI] [PubMed] [Google Scholar]
- Sansregret L, Patterson JO, Dewhurst S, López‐García C, Koch A, McGranahan N, Chao WCH, Barry DJ, Rowan A, Instrell R, Horswell S, Way M, Howell M, Singleton MR, Medema RH, Nurse P, Petronczki M, Swanton C (2017) APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov 7: 218–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GO, He X, Parker JS, Gatza ML, Carey LA, Hou JP, Moulder SL, Marcom PK, Ma J, Rosen JM, Perou CM (2015) Cross‐species DNA copy number analyses identifies multiple 1q21‐q23 subtype‐specific driver genes for breast cancer. Breast Cancer Res Treat 152: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ (2015) Spatiotemporal regulation of the anaphase‐promoting complex in mitosis. Nat Rev Mol Cell Biol 16: 82–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J (2001) A conserved cyclin‐binding domain determines functional interplay between anaphase‐promoting complex‐Cdh1 and cyclin A‐Cdk2 during cell cycle progression. Mol Cell Biol 21: 3692–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ (2007) Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446: 876–881 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Salmon ED (2015) A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun 6: 8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The I, Ruijtenberg S, Bouchet BP, Cristobal A, Prinsen MBW, van Mourik T, Koreth J, Xu H, Heck AJR, Akhmanova A, Cuppen E, Boxem M, Muñoz J, van den Heuvel S (2015) Rb and FZR1/Cdh1 determine CDK4/6‐cyclin D requirement in C. elegans and human cancer cells. Nat Commun 6: 5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, Mechtler K, Hyman AA, Stark H, Schulman BA, Peters J‐M (2012) APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol 19: 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2012) The ubiquitin system, an immense realm. Annu Rev Biochem 81: 167–176 [DOI] [PubMed] [Google Scholar]
- Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, Guo J, Zhang J, North BJ, Dai X, Cheng X, Bian X, Zhang L, Harper JW, Sun S‐C, Wei W (2017) TRAF2 and OTUD7B govern a ubiquitin‐dependent switch that regulates mTORC2 signalling. Nature 545: 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D, Carolina N (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M (2011) The mechanism of linkage‐specific ubiquitin chain elongation by a single‐subunit E2. Cell 144: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Larsen MSY, Narita T, Schou J, Nilsson J, Choudhary C (2016) The spindle assembly checkpoint is not essential for viability of human cells with genetically lowered APC/C activity. Cell Rep 14: 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Jin L, Rape M (2009a) Preparation of synchronized human cell extracts to study ubiquitination and degradation. Methods Mol Biol 545: 301–312 [DOI] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M (2009b) Identification of a physiological E2 module for the human anaphase‐promoting complex. Proc Natl Acad Sci USA 106: 18213–18218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW (2010) UBE2S drives elongation of K11‐linked ubiquitin chains by the anaphase‐promoting complex. Proc Natl Acad Sci USA 107: 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R, Rape M (2016) The increasing complexity of the ubiquitin code. Nat Cell Biol 18: 579–586 [DOI] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh D‐C, Hathaway N, Dimova N, Cuny GD, King RW (2010) Pharmacologic inhibition of the anaphase‐promoting complex induces a spindle checkpoint‐dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mudge MC, Soll JM, Rodrigues RB, Byrum AK, Schwarzkopf EA, Bradstreet TR, Gygi SP, Edelson BT, Mosammaparast N (2018) OTUD4 is a phospho‐activated K63 deubiquitinase that regulates MyD88‐dependent signaling. Mol Cell 69: 505–515.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


