Summary
Deciphering the mechanisms that regulate the quiescence of adult neural stem cells (NSCs) is crucial for the development of therapeutic strategies based on the stimulation of their endogenous regenerative potential in the damaged brain. We show that LeXbright cells sorted from the adult mouse subventricular zone exhibit all the characteristic features of quiescent NSCs. Indeed, they constitute a subpopulation of slowly dividing cells that is able to enter the cell cycle to regenerate the irradiated niche. Comparative transcriptomic analyses showed that they express hallmarks of NSCs but display a distinct molecular signature from activated NSCs (LeX+EGFR+ cells). Particularly, numerous membrane receptors are expressed on quiescent NSCs. We further revealed a different expression pattern of Syndecan-1 between quiescent and activated NSCs and demonstrated its role in the proliferation of activated NSCs. Our data highlight the central role of the stem cell microenvironment in the regulation of quiescence in adult neurogenic niches.
Keywords: neural stem cells, cell sorting, microarray, quiescence, syndecan-1, irradiation, adult neurogenic niches, neurogenesis
Highlights
-
•
Transcriptome analysis reveals molecular hallmarks of activated and quiescent NSCs
-
•
Data resource of putative markers and/or regulators of NSC quiescence
-
•
Quiescent NSCs integrate various signals from the microenvironment
-
•
Syndecan-1 is involved in proliferation of NSCs
In this article, Boussin, Mouthon, and colleagues provide transcriptomic profiles of quiescent and activated neural stem cells. They report on the use of Syndecan-1 as a marker to purify long-term proliferating stem cells.
Introduction
Adult stem cells reside within specialized microenvironments that integrate intricate signals critical for maintaining stem cell populations in an undifferentiated state, guiding cell fate decisions, and modulating the regenerative potential of the niche (Papanikolaou et al., 2008). In the adult mammalian brain, neural stem cells (NSCs) continuously generate neurons throughout life in two discrete regions: the subventricular zone (SVZ) along the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyrus. NSCs from the adult SVZ successively give rise to transit-amplifying cells and neuroblasts that differentiate into neurons once they have reached the olfactory bulbs (Lim and Alvarez-Buylla, 2014). A key feature of NSCs is their remarkable proliferative capacity that sustains regeneration of damaged tissue through the activation of quiescent stem cells (Codega et al., 2014, Daynac et al., 2013, Doetsch et al., 1999, Llorens-Bobadilla et al., 2015, Mich et al., 2014, Morshead et al., 1994).
In contrast to their progeny, most adult NSCs are quiescent and a tight regulation of the balance between their quiescent and proliferative states appears essential for their long-term maintenance in neurogenic niches (Fuentealba et al., 2015, Furutachi et al., 2015). Indeed, dysregulation and/or loss of quiescence often results in premature proliferation of NSCs ultimately leading to the depletion of neural stem and progenitor cells (Kippin et al., 2005, Mira et al., 2010, Molofsky et al., 2003, Ottone et al., 2014). Deciphering the functional properties of quiescent NSCs and the associated regulatory mechanisms is thus important to develop new approaches for NSC-based regenerative medicine.
The complexity of neurogenic niches, small number of resident NSCs, as well as the lack of specific cellular markers have long hampered the study of NSCs. The recent development of flow cytometry-based cell sorting strategies has enabled the identification and the isolation of quiescent and proliferating NSCs from their niche and opened new doors for the study of the regulation of stem cell quiescence (Codega et al., 2014, Daynac et al., 2013, Dulken et al., 2017, Llorens-Bobadilla et al., 2015, Mich et al., 2014). While all of the cell-sorting strategies rely on the use of the EGF receptor to discriminate the two subpopulations of NSCs, several combinations of markers have been used to identify the stem cell population, including CD133 (Beckervordersandforth et al., 2010, Codega et al., 2014), Glast (Llorens-Bobadilla et al., 2015, Mich et al., 2014), LeX (Capela and Temple, 2002, Daynac et al., 2013, Daynac et al., 2015), and GFAP (Codega et al., 2014).
With the advent of transcriptome analyses, it has also become possible to study molecular hallmarks and gene regulatory networks governing NSC behavior (Beckervordersandforth et al., 2010, Codega et al., 2014, Daynac et al., 2016a, Dulken et al., 2017, Llorens-Bobadilla et al., 2015). We have recently developed a cell-sorting strategy to prospectively isolate quiescent NSCs contained in the LeXbright population from the adult SVZ (Daynac et al., 2013, Daynac et al., 2015). Moreover, we have previously shown that the vast majority of LeXbright cells that survived to radiation exposure entered the cell cycle to regenerate the irradiated niche (Daynac et al., 2013). Herein, we show that LeXbright cells are slowly dividing cells in vivo. The comparative analysis of the transcriptomic profiles of LeXbright and LeX+EGFR+ cells further reveals that the quiescent state is tightly regulated by the microenvironment and provides a comprehensive data resource to investigate cellular quiescence in adult neurogenic niches. Finally, we unravel the role of Syndecan-1 in the proliferation of activated NSCs.
Results
LeXbright Cells Exhibit Properties of Slowly Dividing NSCs
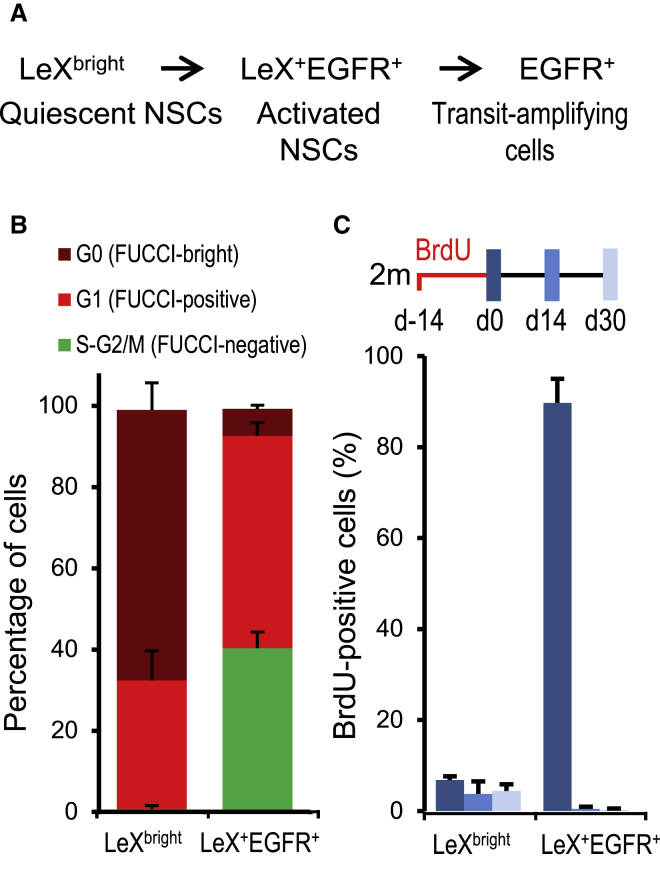
We recently developed a cell-sorting strategy based on the exclusion of CD24-positive cells and on the detection of the surface markers LeX and EGFR to simultaneously isolate quiescent NSCs (CD24−EGFR−LeXbright, hereafter LeXbright cells), activated NSCs (CD24−LeX+EGFR+, hereafter LeX+EGFR+ cells), and transit-amplifying cells (CD24−EGFR+, hereafter EGFR+ cells) from the adult SVZ (Figure 1A) (Daynac et al., 2013, Daynac et al., 2015). In contrast to the other neurogenic SVZ cell populations, we previously showed that the vast majority of LeXbright cells are not proliferating (Daynac et al., 2013). To explore their cell cycle in more detail, we used fluorescence ubiquitination cell cycle indicator (FUCCI)-Red mice (Sakaue-Sawano et al., 2008), which allow the visualization of cells in G1 with the presence of a G1 specific red-Cdt1 reporter (FUCCI-Redpositive cells), while it is absent in cells during the S-G2/M phases (FUCCI-Rednegative cells). In addition, FUCCI-Redbright cells have been shown to have exited the cell cycle (G0) (Daynac et al., 2014, Roccio et al., 2013). While most activated LeX+EGFR+ cells progressed through S-G2/M phases (40.3% ± 4.0% FUCCI-Rednegative), LeXbright cells were for the most part distributed in G0 (66.5% ± 6.5% FUCCI-Redbright) or in G1 (31.7% ± 7.3% FUCCI-Redpositive) in accordance with their quiescent state (Figure 1B).
Figure 1.
LeXbright Cells Are Slow Dividing Cells
(A) LeX/EGFR/CD24 triple staining strategy allowing the purification by flow cytometry of quiescent (LeXbright), activated NSCs (LeX+EGFR+), and transit-amplifying (EGFR+) cells (Daynac et al., 2013).
(B) Cell cycle analysis of NSCs from FUCCI-Red mice showed that almost all LeXbright cells were distributed in G1 (FUCCI-Redpositive) and G0 (FUCCI-Redbright).
(C) Quantification of cells positive for BrdU immediately after a 14-day exposure to BrdU at 2 months (dark blue) and after a chase of 2 weeks (medium blue) or 4 weeks (light blue) revealed that LeXbright cells are long-term BrdU label-retaining cells.
Data are represented as the mean ± SD and were obtained from independent experiments with several mice per group: (B), 9; (C), 3–4.
To further characterize the cell cycle dynamics of LeXbright cells, we administrated bromodeoxyuridine (BrdU) to mice for 2 weeks and assessed their ability to retain BrdU labeling for extended chase periods (Figure 1C). Immediately after BrdU treatment, the great majority of rapidly dividing LeX+EGFR+ cells had incorporated BrdU (89.7% ± 5.3%), while they had almost all lost the BrdU labeling after 2 and 4 weeks of chase (Figure 1C). By contrast, only 6.8% ± 0.7% of LeXbright cells had incorporated BrdU after 2 weeks, reflecting their much slower rate of division. Moreover, 64% ± 24% of LeXbright cells retained the BrdU labeling after 4 weeks of chase (Figure 1C). These data confirmed that LeXbright cells correspond to a subpopulation of slowly dividing NSCs in vivo.
LeXbright Cells Enter Oxidative Metabolism after Irradiation
We have previously shown that the vast majority of slowly dividing LeXbright cells that survived to radiation exposure entered the cell cycle to regenerate the irradiated niche (Daynac et al., 2013), recapitulating what is observed after antimitotic treatment with Ara-C (Doetsch et al., 1999).
Here, we performed a transcriptomic analysis of LeXbright cells sorted from 2-month-old control mice and 48 hr after mice were irradiated using whole-genome Affymetrix MOE430 2.0 arrays.
The obtained datasets are visualized as sets of coordinates using principal component analysis (PCA) in Figure S1A. PCA is an unsupervised pattern recognition and visualization tool used to reduce the dimensionality of datasets derived from transcriptomic arrays, making it possible to visually assess similarities and differences between cell populations (Ringner, 2008). This PCA illustrates the transcriptomic shift of quiescent NSCs induced by irradiation concomitantly to the entry in the cell cycle of a subset of these cells we reported before (Daynac et al., 2013) (Figure S1A). The comparative gene expression profile of LeXbright cells revealed an altered expression of 927 probes (Figure S1B). The resulting set of genes enriched in control LeXbright cells included 439 genes, whereas 409 genes were upregulated in irradiated LeXbright cells (Table S1). As expected, gene ontology (GO) term analysis revealed that genes upregulated after irradiation in LeXbright cells were mainly associated with the cell cycle and DNA/RNA processes (Figure S1C). Moreover, many of these genes were linked to translation and ribosomal activity (Figures S1C and S1D). Interestingly, gene set enrichment analysis (Subramanian et al., 2005) also showed enrichment in genes associated with the TCA (tricarboxylic acid) cycle and respiratory electron transport (Figure S1E). Therefore, the cell cycle entry of LeXbright cells after radiation was accompanied by a shift toward an oxidative metabolism that was consistent with that observed in stem cells during proliferation and differentiation (Huang et al., 2012).
Distinct Molecular Signatures of Quiescent and Activated NSCs
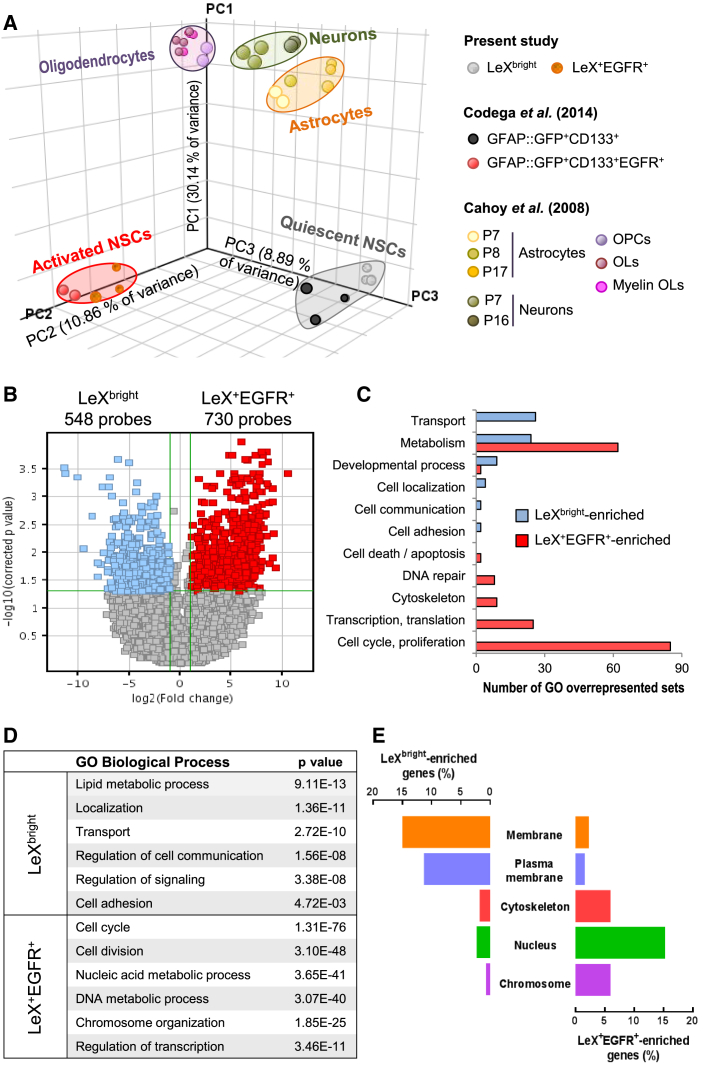
In order to gain insights into the mechanisms regulating stem cell quiescence, we performed a microarray analysis of LeXbright and LeX+EGFR+ cells (i.e., quiescent and activated NSCs) sorted from 2-month-old mouse SVZ. We compared their global mRNA expression patterns with those obtained from previous studies either characterizing NSCs (Codega et al., 2014) or differentiated cells (Cahoy et al., 2008) using PCA (Figure 2A). Direct comparison of LeXbright and LeX+EGFR+ transcriptome profiles revealed that they were clustered away from differentiated cells (astrocytes, oligodendrocytes, and neurons) (Figure 2A). Moreover, the clear separation of LeXbright cells from LeX+EGFR+ cells confirmed their distinct cellular identity (Figure 2A). Importantly, LeXbright and LeX+EGFR+ cells were closely clustered to GFAP:GFP+CD133+ and GFAP:GFP+CD133+EGFR+ cells, previously shown to correspond respectively to quiescent and proliferating NSCs in the adult SVZ (Codega et al., 2014), providing additional validation of our cell-sorting approach (Figure 2A). Besides, transcriptional hallmarks of NSCs such as Slc1a3/Glast, Prominin1/CD133, Nr2e1/Tlx, Hes5, and Sox2 were found substantially expressed in both LeXbright and LeX+EGFR+ cells (Table S2). It is noteworthy that our cell-sorting technique does not require transgene expression to identify the stem cell population and is thus easily transferable to any other mouse model.
Figure 2.
Comparative Transcriptome Analysis Reveals the Close Interactions between Quiescent NSCs and Their Microenvironment
(A) Principal component analysis (PCA) of gene expression datasets of freshly sorted LeXbright and LeX+EGFR+ cells compared with those obtained from studies either characterizing NSCs (Codega et al., 2014) or differentiated cells (Cahoy et al., 2008).
(B) Volcano plot of differentially expressed probes in LeXbright cells (blue) and LeX+EGFR+ cells (red).
(C) GO categories enriched in LeXbright and LeX+EGFR+ cells were identified using a statistical overrepresentation test and were hand curated into thematic categories.
(D) Selected sets of enriched GO categories in LeXbright and LeX+EGFR+ cells.
(E) Predicted cellular location of gene products differentially expressed in LeXbright and LeX+EGFR+ cells.
To further define genes enriched in each cellular state, the transcriptomes of LeXbright and LeX+EGFR+ cells were compared. Probes were filtered by an average expression greater than 50 in at least one population, a differential expression of at least 2-fold, and a Student's t test corrected p value <0.05. As shown on the volcano plot, the comparative gene expression profile of LeXbright and LeX+EGFR+ cells revealed an altered expression of 1,278 probes (Figure 2B). The resulting set of LeXbright-enriched genes included 433 genes (548 probe sets, Table S2), whereas 563 genes were upregulated in LeX+EGFR+ cells (730 probe sets, Table S2) (Figure 2B).
GO term analysis was then performed using a statistical overrepresentation test to delineate the molecular features of quiescent and activated NSCs. In accordance with their proliferating state, the transcriptome of LeX+EGFR+ cells was enriched in genes linked to the cell cycle, DNA repair, DNA/RNA metabolism, transcription, and translation (Figures 2C and 2D, Tables S3 and S4). Strikingly, cellular component analysis also revealed a drastically different cellular location of the differentially expressed gene products. As expected due to their transcriptionally active state, 15.3% of the genes enriched in LeX+EGFR+ cells encoded proteins associated with the nucleus, as opposed to only 2.3% of those enriched in LeXbright cells (Figure 2E). In contrast, the vast majority of the genes enriched in LeXbright cells were related to GO categories linked to lipid metabolic process, transport, response to stimulus, cell localization, cell communication, and cell adhesion (Figures 2C and 2D, Tables S3 and S4). Importantly, most genes enriched in LeXbright cells encoded proteins associated with the membrane (Figure 2E), emphasizing the key role played by the microenvironment in the regulation of the quiescent state in the adult SVZ (Chaker et al., 2016).
Transcription Factors Enriched in Quiescent and Activated NSCs
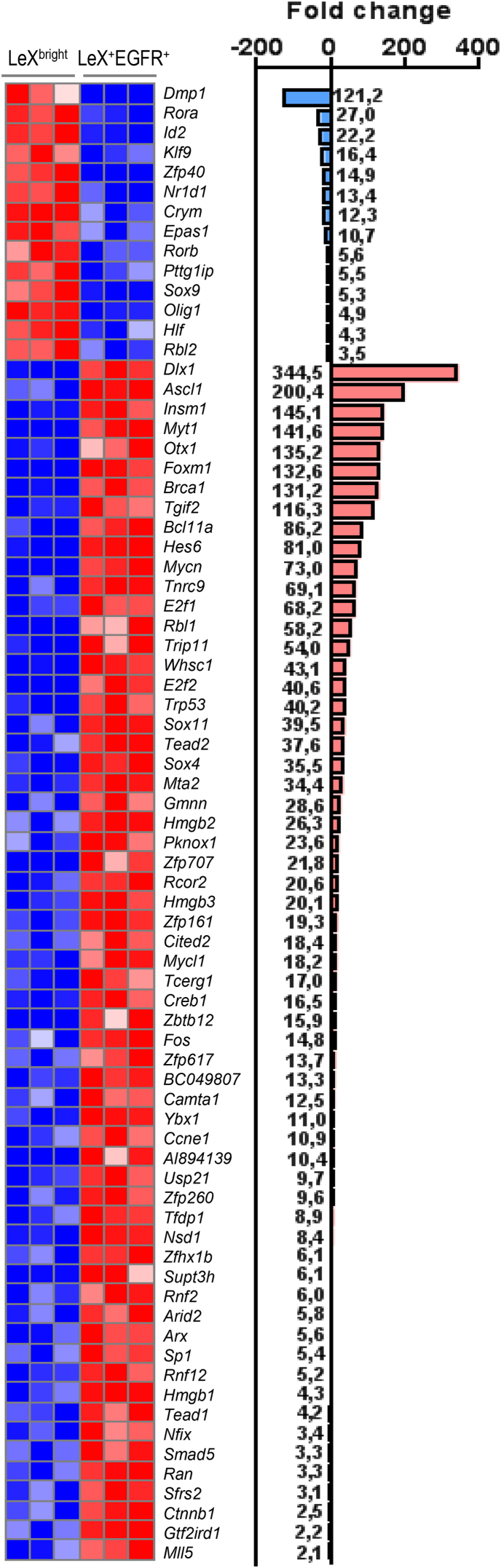
In order to identify putative transcriptional regulators of the quiescent and proliferative states of adult NSCs, we focused on transcription factors (TFs) and co-factors either enriched in LeXbright or LeX+EGFR+ cells. Analysis of our dataset using public databases (Zhang et al., 2012) revealed a total of 75 differentially expressed TFs, 14 of which were upregulated in LeXbright cells and the remaining 61 in LeX+EGFR+ cells (Figure 3).
Figure 3.
TFs and Co-factors Differentially Expressed in Quiescent and Activated NSCs
Heatmaps showing transcript expression levels for replicate samples of LeXbright and LeX+EGFR+ cells. Blue color indicates low expression and red high expression (log2 scale).
Among the TFs upregulated in LeXbright cells were Sox9 and Id2, which have been previously associated with quiescent NSCs (Llorens-Bobadilla et al., 2015) (Figure 3). Interestingly, Klf9, a member of the family of Kruppel-like TFs found upregulated in quiescent muscle satellite stem cells (Pallafacchina et al., 2010), was also enriched in LeXbright cells (Figure 3).
Among the TFs and co-factors that were the most enriched in LeX+EGFR+ cells, several were linked to the cell cycle (E2f1, E2f2, Rbl1, Ccne1, Trp53, and Tfdp1) (Figure 3). Of particular interest, LeX+EGFR+ cells expressed high levels of three members of the high-mobility group box (HMGB) protein family: Hmgb1/2/3 (Figure 3). Besides the broad role of HMGs in the control of transcription as well as replication, recent studies have linked HMGBs to the control of the proliferation and maintenance of embryonic and adult NSCs (Abraham et al., 2013). Additionally, transcripts for Ascl1 were 200 times higher in LeX+EGFR+ cells compared with LeXbright cells (Figure 3), in accordance with recent studies that have reported its key role in the proliferation of NSCs and in the exit of stem cells from quiescence in both the adult hippocampus and the SVZ (Urban et al., 2016). Finally, the proliferating state was also associated with the expression of the immediate-early gene Fos and SoxC factors (Sox4 and Sox11), which have been linked to stem cell activation (Adepoju et al., 2014, Foronda et al., 2014).
Quiescent LeXbright Cells Integrate Signals from the Microenvironment
We found that most genes enriched in LeXbright cells were linked to the cell membrane (Figure 2E).
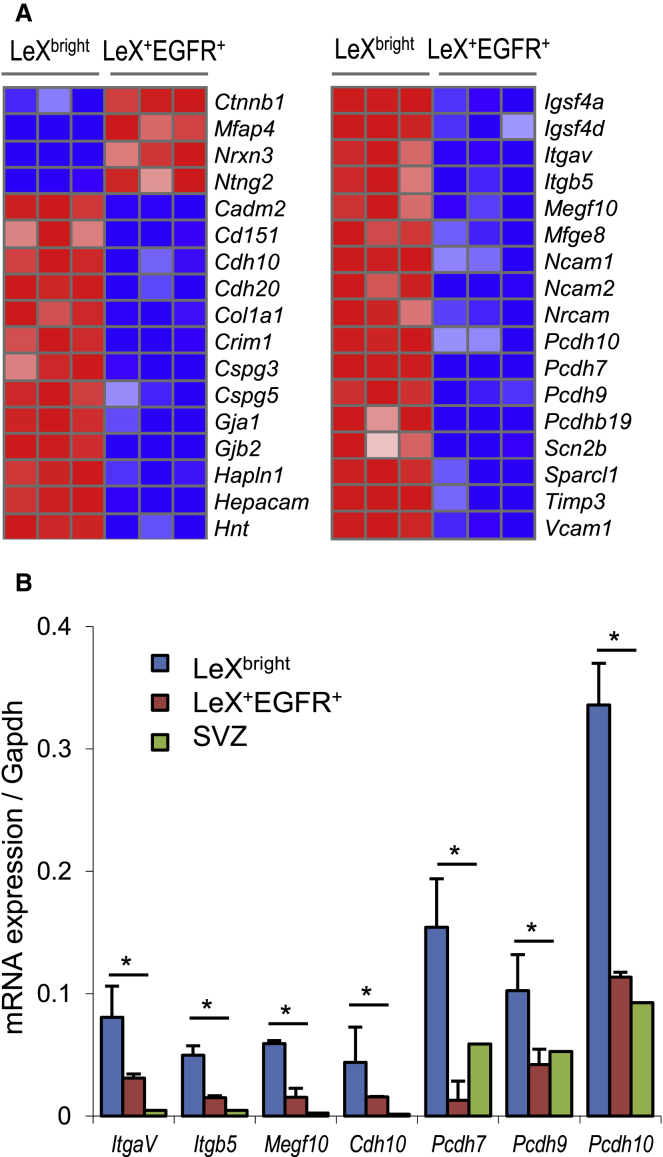
Various adhesion molecules, such as neural cell adhesion molecule 1 and 2 (Ncam1, Ncam2), and cadherins/protocadherins (Cdh10, Cdh20, Pcdh7, Pcdh9, Pcdh10, Pcdhb19) were found overexpressed in LeXbright cells (Figures 4A and 4B). Adhesion molecules have been shown to play a key role in the NSC niche by maintaining stem cell niche architecture and homeostasis (Marthiens et al., 2010). Indeed, disruption of Vcam1 was previously shown to lead to a massive activation of quiescent NSCs and consequent depletion of the NSC population (Kokovay et al., 2012). In addition, the proliferative status of NSC is dynamically modulated by the cleavage of Cdh2 (or N-cadherin) on NSCs in the adult SVZ (Porlan et al., 2014). Noticeably, both Vcam1 and Cdh2 were found upregulated in LeXbright cells (Figure 4A and Table 1).
Figure 4.
Enrichment of Genes Implicated in Cell Adhesion in Quiescent NSCs
(A) Heatmaps showing transcript expression levels of genes coding for adhesion molecules and extracellular matrix components in LeXbright and LeX+EGFR+ cells. Blue color indicates low expression and red high expression (log2 scale). Replicate samples are shown for each group.
(B) Gene expression levels of indicated adhesion genes were confirmed by qRT-PCR. Data are represented as the mean ± SD and were obtained from three independent experiments. ∗p < 0.05.
Table 1.
Selected Membrane Receptors Upregulated on Quiescent NSCs
| Gene Symbol | Gene Title | Gene ID | Fold Change (qNSC/aNSC) | Signaling | Effect(s) on NSCs and Progenitors | Reference |
|---|---|---|---|---|---|---|
| Cdh2 | cadherin 2 | 12558 | 2.6 | Wnt | maintains NSC quiescence in the SVZ | Porlan et al., 2014 |
| Dner | delta/notch-like EGF-related receptor | 227325 | 7.3 | Notch | inhibits NPC proliferation and induces neuronal and glial differentiation | Hsieh et al., 2013 |
| Fgfr2 | fibroblast growth factor receptor 2 | 14183 | 4.8 | FGF | induces proliferation of adult NSCs | Zheng et al., 2004 |
| Fgfr3 | fibroblast growth factor receptor 3 | 14184 | 22.6 | FGF | expressed on non-proliferating SVZ cells | Frinchi et al., 2008 |
| Fzd6 | frizzled homolog 6 (Drosophila) | 14368 | 27.1 | Wnt | involves in proliferation of neuroblastoma cells | Cantilena et al., 2011 |
| Gabbr1 | gamma-aminobutyric acid (GABA-B) receptor, 1 | 54393 | 5.0 | GABA | GABA inhibits proliferation of quiescent NSCs | Daynac et al., 2013 |
| Gabrg1 | gamma-aminobutyric acid (GABA-A) receptor, subunit gamma 1 | 14405 | 39.7 | GABA | ||
| Lrig1 | leucine-rich repeats and immunoglobulin-like domains 1 | 16206 | 15.2 | EGFR | represses EGFR signaling | Gur et al., 2004 |
| Lrp4 | low-density lipoprotein receptor-related protein 4 | 228357 | 5.3 | Wnt | may be involved in the negative regulation of the Wnt signaling pathway | Styrkarsdottir et al., 2009 |
| Ptch1 | patched homolog 1 | 19206 | 2.4 | Hh pathway | orchestrates the balance between quiescent and activated NSCs | Daynac et al., 2016b |
| Ptprd | protein tyrosine phosphatase, receptor type, D | 19266 | 45.4 | CSPG | restricts the survival, migration, integration, and differentiation of NPCs following sciatic nerve injury | Dyck et al., 2015 |
| Sdc2 | Syndecan 2 | 15529 | 6.2 | upregulated during long-term expansion of human NSC | Oikari et al., 2016 | |
| Sdc4 | Syndecan 4 | 20971 | 9.9 | increases during neuronal differentiation | Oikari et al., 2016 | |
| VCAM-1 | vascular cell adhesion molecule 1 | 22329 | 8.7 | maintains NSC quiescence in the SVZ | Kokovay et al., 2012 |
See also Table S5. aNSC, activated NSC; qNSC, quiescent NSC.
We then looked for receptors that were differentially expressed between LeXbright and LeX+EGFR+ cells in an attempt to identify additional markers of quiescent and activated NSCs as well as signaling pathways involved in the regulation of NSC behavior (Tables 1 and S5). In the adult SVZ, gamma-aminobutyric acid (GABA) produced by neuroblasts decreased the proliferation of adult NSCs (Liu et al., 2005). Moreover, inhibition of GABA signaling through the GABAA receptors led to an entry in proliferation of LeXbright cells (Daynac et al., 2013). Interestingly, transcripts for several GABAA receptor chains (α4, β1, γ1, and γ3) were expressed in LeXbright cells, with the γ1 chain (Gabrg1) being almost 40 times higher in comparison with LeX+EGFR+ cells (Table 1). Moreover, Lrig1, a pan-ErbB inhibitor that has been used as a marker of quiescent stem cells in the epidermis as well as in the intestine and was shown to negatively regulate proliferation (Jensen and Watt, 2006, Powell et al., 2012), was also enriched in LeXbright cells (Table 1). Another receptor well described in neurogenesis is Ptch1, a member of the patched gene family and main receptor for sonic hedgehog (Shh), overexpressed in LeXbright cells (Ahn and Joyner, 2005, Balordi and Fishell, 2007, Ferent et al., 2014). We have recently shown that the activation of the SHH pathway through deletion of the Patched receptor in NSCs resulted in an increase of the pool of quiescent NSCs (Daynac et al., 2016b).
Receptors listed in Tables 1 and S5 could therefore be used as markers of quiescent NSCs and/or could act as putative regulators of the balance between quiescence and proliferation.
Syndecan-1 Is a Marker of Proliferating NSCs
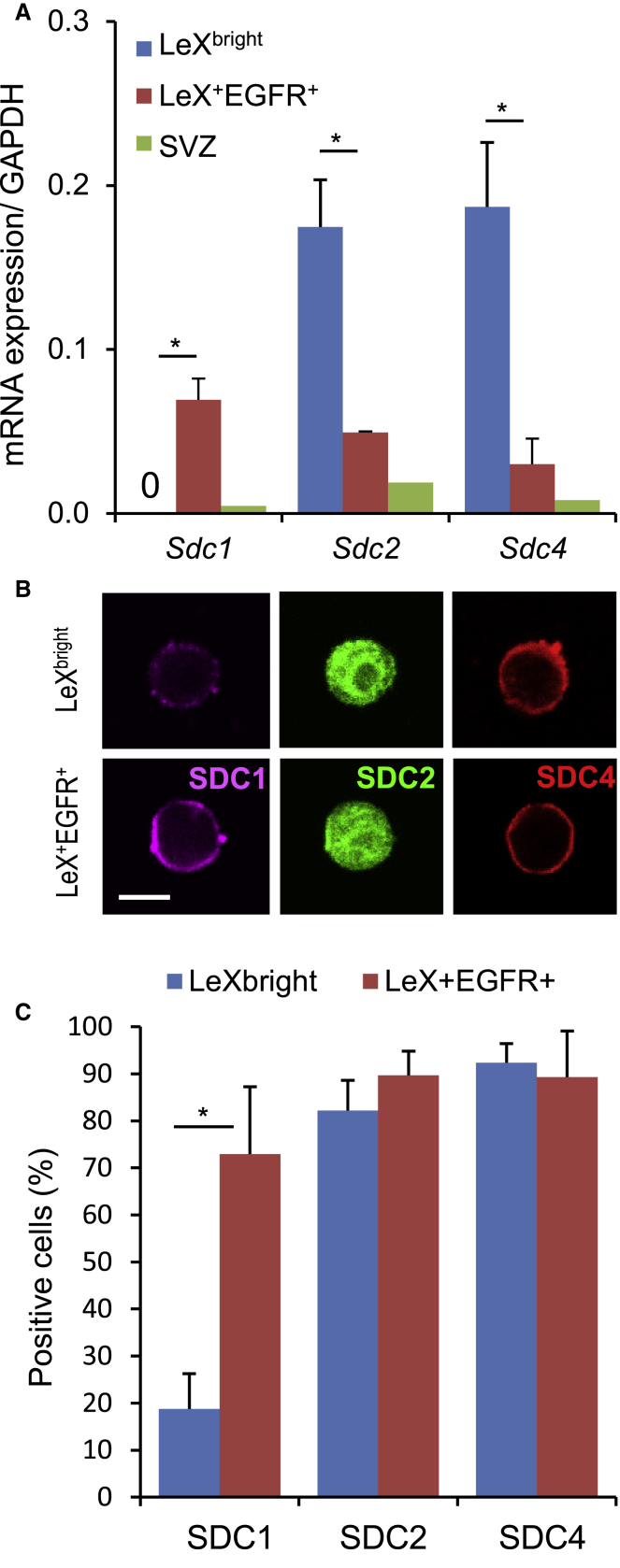
Syndecan family members were among the receptors that were found differentially expressed between LeXbright and LeX+EGFR+ cells (Table 1). The pattern of expression of three out of the four members of the syndecan family was particularly interesting as they were either enriched in LeXbright cells (Sdc2 and Sdc4) or strongly upregulated in LeX+EGFR+ cells (Sdc1), hinting at the possibility of their use as markers to discriminate quiescent NSCs from proliferating NSCs (Table 1, Figure 5A). However, SDC2 and SDC4 were found expressed on the vast majority of quiescent and activated NSCs at the protein level (Figures 5B and 5C), suggesting that they undergo complex post-transcriptional/post-translational regulation in these cells. By contrast, the specificity of SDC1 expression in actively dividing LeX+EGFR+ cells was confirmed at the protein level (Figures 5B and 5C).
Figure 5.
Expression of the Syndecan Family Members in Quiescent and Activated Adult NSCs
(A and B) (A) Variations in the expression of Sdc1, Sdc2, and Sdc4 in freshly sorted LeXbright, LeX+EGFR+, and total SVZ cells were confirmed by (A) qRT-PCR (0: undetectable) and (B) immunostaining (scale bar: 5 μm).
(C) Quantification of SDC1-, SDC2-, and SDC4-positive cells reveals higher expression of SDC1 by actively proliferating LeX+EGFR+ cells.
Data are represented as the mean ± SD from four independent experiments of three or four mice. ∗p < 0.05.
To further determine if Sdc1 was linked to the proliferative status of NSCs, we modeled NSC quiescence in culture with BMP4 as previously described (Martynoga et al., 2013, Mathieu et al., 2008) (Figures S2A and S2B). We confirmed that addition of BMP4 to proliferating SVZ cells drastically reduced the formation of neurospheres and that the total number of cells was decreased (Figures S2A and S2B). Interestingly, Sdc1 transcripts, highly enriched in proliferative cells, significantly decreased after BMP4 treatment (Figure S2C).
We then sought to evaluate whether SDC1 could be used as a prospective marker of proliferating NSCs using flow cytometry. Irrespective of the enzymatic cocktail used for cell dissociation, cell-membrane-bound SDC1 was shed, rendering its labeling impossible on adult NSCs (data not shown). As an enzymatic dissociation is required to obtain single-cell suspensions of NSCs from adult SVZ, we examined SDC1 expression on NSCs prepared by mechanical dissociation of postnatal day 10 (PN10) SVZ. Importantly, PN10 LeXbright, LeX+EGFR+ cells, as well as EGFR+, had similar fluorescence-activated cell sorting (FACS) profiles to those of adult SVZ, although some differences in their frequencies were observed (Figures S3A and S3B). Similarly to what was observed in adult NSCs, SDC4 was expressed on quiescent and proliferating NSCs at PN10, while SDC1 was present on most LeX+EGFR+ cells and absent from the vast majority of LeXbright cells (Figure S4).
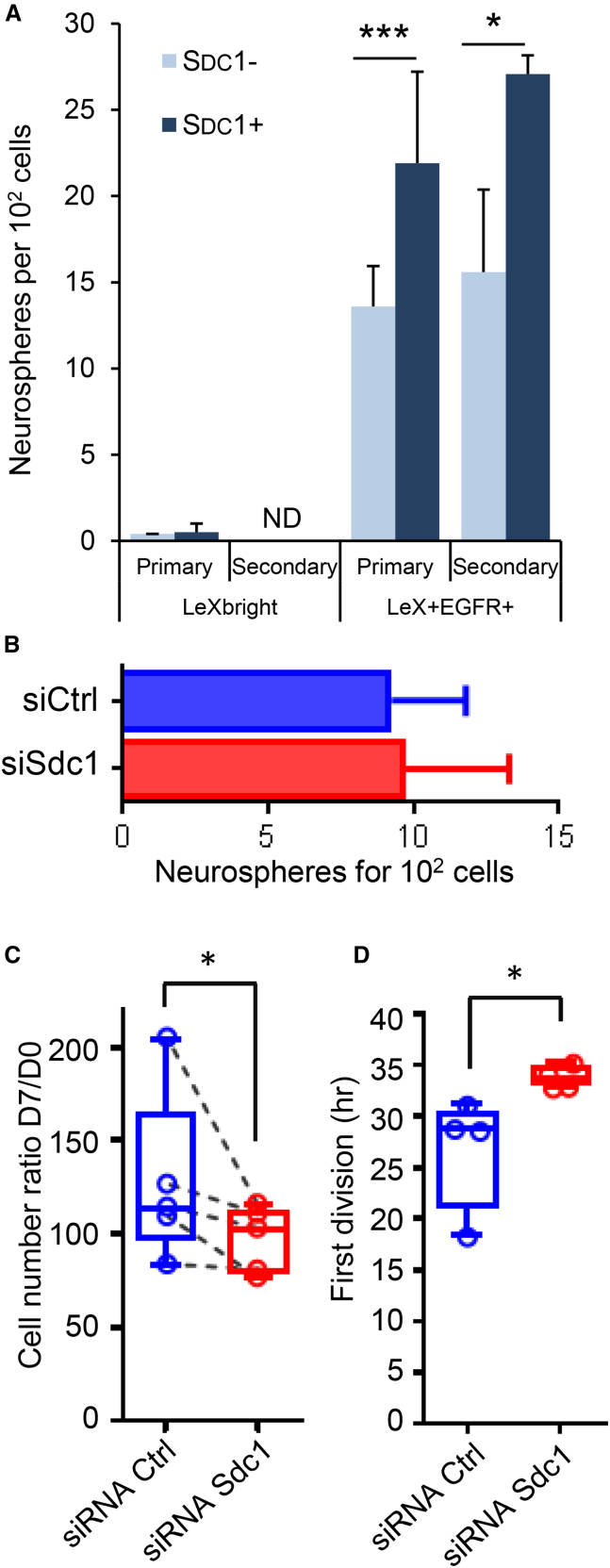
LeX+EGFR+ as well as LeXbright cells were then sorted according to SDC1 expression and a colony-forming neurosphere assay was performed to assess the clonogenic capacity of the different NSC subpopulations. In accordance with what was previously reported for LeXbright cells isolated from adult SVZ (Daynac et al., 2013), PN10 LeXbright cells very rarely formed neurospheres, regardless of the expression of SDC1 (Figure 6A). Importantly, a 1.6-fold increase in the number of primary neurospheres was observed for LeX+EGFR+SDC1+ cells as compared with their SDC1-negative counterparts (Figure 6A). This increase was also found when we performed secondary neurosphere formation, hinting at a higher long-term self-renewal potential of LeX+EGFR+SDC1+ cells (Figure 6A).
Figure 6.
Syndecan-1 Plays a Role in the Proliferation of Activated NSCs
(A) LeXbright and LeX+EGFR+ cells were purified by flow cytometry from PN10 SVZ according to SDC1 expression (SDC1 negative, light blue; SDC1 positive, dark blue) and their capacity to form primary then secondary neurospheres was assessed. The effect of Sdc1 silencing in LeX+EGFR+ cells sorted from adult SVZ was evaluated on neurosphere formation (B), cell proliferation (C), and the time required for the first cell division (D). Data are represented as the mean ± SD (A and B) and scattered dot plots with median ± minimum/maximum (C and D), and were obtained from independent experiments (N = 3–10 for primary, N = 3 for secondary, and N = 4 for siRNA experiments) with three or four pooled mice. Statistical analyses were performed using one-tailed Mann-Whitney (A, B, and D) and Wilcoxon matched-pairs signed rank test (C). ∗p < 0.05, ∗∗∗p < 0.0005. ND, not determined.
To confirm the role of SDC1 in the proliferation of activated NSCs, we performed silencing experiments using small interfering RNA (siRNA) directed against Sdc1. We verified the efficacy of Sdc1 silencing in neurosphere cultures both at the mRNA and protein levels 24 and 48 hr after electroporation (Figures S5A and S5B). Interestingly, a decrease in the diameter of neurospheres was observed after Sdc1 silencing in comparison with a scrambled control siRNA (Figure S5C). Subsequently, Sdc1 silencing was performed in LeX+EGFR+ cells freshly sorted from adult mice. While their clonogenic capacity was not altered, the total number of cells was reduced at day 7 (Figures 6B and 6C), suggesting a role of Sdc1 in the proliferation but not in the activation of activated NSCs. We further monitored the time required for the first cell division of LeX+EGFR+ cells after Sdc1 silencing by time-lapse videomicroscopy and found it significantly delayed by 7.1 hr (Figure 6D). Altogether, our data hint at a role of Sdc1 in the progression of activated NSCs through the cell cycle.
Discussion
Deciphering the molecular pathways involved in the regulation of quiescence and activation of NSCs is crucial to elucidate the mechanisms of maintenance of the neurogenic niches in the adult brain. Here, we used our previously described method to sort NSCs from the adult mouse SVZ (Daynac et al., 2013) to perform a comparative transcriptomic analysis of quiescent NSCs (LeXbright cells) and activated NSCs (LeX+EGFR+ cells). Strikingly, we found that most genes enriched in quiescent NSCs are linked to the cell membrane, emphasizing the importance of the microenvironment in the regulation of NSCs in the adult neurogenic niches.
Evidence from recent studies suggest that the balance of NSCs between quiescence and proliferation is tightly regulated by extrinsic signals from the stem cell niche through various signaling pathways. Interestingly, the two adhesion molecules Vcam1 and Cdh2 previously shown to play a role in quiescence of NSCs (Kokovay et al., 2012, Porlan et al., 2014) are found upregulated in quiescent NSCs. Furthermore, transcripts for the GABA receptor γ1 chain and Ptch1 are enriched in quiescent NSCs in accordance with their recognized role in quiescence (Daynac et al., 2013, Daynac et al., 2016b, Liu et al., 2005). Importantly, we provide a list of receptors enriched in quiescent NSCs (Tables 1 and S5) that could be used as additional markers of quiescent NSCs and/or could act as putative regulators of the balance between quiescence and proliferation of adult NSCs.
We further reveal the specific expression of Syndecan-1 on activated NSCs as compared with quiescent NSCs. Syndecan-1 belongs to a family of transmembrane heparan sulfate proteoglycans that have roles in cell-matrix interactions and recruit to the cell surface soluble growth factors known to promote the proliferation of neuronal precursors, such as FGF, EGF, VEGF, and HGF (Kwon et al., 2012). It has been shown that SDC1-positive cells sorted by flow cytometry from the embryonic telencephalon were enriched in neurosphere-forming cells (Nagato et al., 2005). Besides, sdc1 knockdown during cortical neurogenesis has been reported to reduce the maintenance and proliferation of NPCs (Wang et al., 2012). Here, we show that Syndecan-1 is a marker that allows the specific targeting of highly clonogenic/proliferating NSCs. Moreover, we demonstrate that Sdc1 plays a crucial role in the progression of activated NSCs through the cell cycle in the adult mouse SVZ. Expression of SDC1 was consistently found in malignant glioma cells but was undetectable in non-neoplastic brain tissues (Watanabe et al., 2006). Besides, higher SDC1 expression in malignant glioma has been associated with ascending tumor World Health Organization grades and poor diagnosis (Xu et al., 2012). Studying the regulation of adult NSC proliferation in relation to the expression of SDC1 could thus provide insight into adult NSC behavior and its modulation in health and disease.
Altogether, our transcriptomic study reveals specific and distinct interactions of quiescent and activated NSCs with their microenvironment. Our work provides a comprehensive data resource to investigate cellular quiescence and activation in adult neurogenic niches in the context of NSC-based brain tissue regeneration strategies.
Experimental Procedures
Animals and Treatments
Young adult C57Bl/6J mice (2–3 months) were maintained in standard cages with access to food and water ad libitum in a colony room kept at a constant temperature (19°C–22°C) and humidity (40%–50%) on a 12:12-hr light/dark cycle. Postnatal day 10 C57Bl/6J mice were produced in our animal facility by programmed breeding. For cell cycle analysis, we used FUCCI-Red transgenic mice (FUCCI for CDT1) (Sakaue-Sawano et al., 2008).
When indicated, mice received whole-brain irradiation (4 Gy) under anesthesia using a 60Co medical irradiator (Alcyon) as previously reported (Daynac et al., 2013).
Two-month-old mice were initially injected intraperitoneally with 100 mg BrdU/kg body mass then maintained with drinking water containing BrdU (1 mg/mL, 1% glucose) for 14 consecutive days followed by a 2- and 4-week chase period until sacrifice.
Animal experiments were performed in compliance with the European Communities Council Directive of 22th September 2010 (EC/2010/63) and were approved by our institutional committee on animal welfare (authorization #12–034; CEtEA-CEA DRF IdF).
SVZ Cell Preparation
Adult SVZs were dissected, dissociated, and labeled as previously described (Daynac et al., 2015). Briefly, dissected SVZs were digested with papain (1 mg/mL, Worthington) supplemented with 0.01 mg/mL DNase I (Sigma) for 10 min at 37°C. The minced tissue was then mechanically dissociated into a single-cell suspension using a P1000 micropipette in ovomucoid solution (0.7 mg/mL, Sigma). PN10 SVZs were prepared by mechanical dissociation. Papain solution was omitted for the preparation of PN10 SVZ. Aggregates were removed with 20 μm nylon filters (BD Biosciences) and cells were centrifuged at 250 × g for 20 min at 4°C without brake on a 22% Percoll gradient (GE Healthcare) to remove myelin. Finally, cells were incubated for 20 min with the following antibodies: CD24 phycoerythrin (PE)-conjugated (cat#561079; 1:50 BD Biosciences), CD15/LeX fluorescein isothiocyanate (FITC)-conjugated (clone MMA, mouse immunoglobulin M; 1:50 BD Biosciences), and Alexa 647-conjugated EGF ligand (1:250 Life Technologies). Rat anti-mouse Syndecan-1 PE-conjugated (cat#553714, BD Biosciences) and rat anti-mouse Syndecan-4 PE-conjugated (cat#550352, BD Biosciences) antibodies were used at 1:50 in combination with anti-CD24 PE-Cyanine7-conjugated antibody (cat#A14776, Molecular Probes). Immediately prior to FACS, Hoechst 33258 was added to a final concentration of 1 μg/mL to label dead cells. Adult SVZ cells were sorted on an INFLUX cell sorter equipped with an 86 μm nozzle at 40 psi and postnatal SVZ cells on an ARIA equipped with a 100 μm nozzle (BD Biosciences). Gates were set using fluorescence minus one controls on SVZ cells.
Immunofluorescence
Sorted cells were recovered in DMEM/F12 medium supplemented with 2% B27 then plated without mitogen on poly-D-lysine- and laminin-coated eight-well glass slides (Millicell) in an incubator at 37°C 5% CO2 for 2–4 hr and fixed in 2% paraformaldehyde. After 1 hr in blocking solution (PBS-0.1% Triton X-100–1% BSA) at room temperature (RT), cells were incubated overnight at 4°C with anti-mouse CD138/SDC1 (1:100, BD Pharmingen), anti-CD362/SDC2 (1:100, AF6585, R&D Systems), or anti-SDC4 (1:100, NB110-41551, NovusBio) primary antibodies. After three washes in PBS, cells were incubated with an Alexa Fluor donkey secondary antibody at 1:500 (Invitrogen). For BrdU detection, cells were permeabilized for 5 min at RT in 0.5% Triton X-100 in PBS. Incubation in blocking solution (PBS, 0.05% Tween 20, 4% BSA) for 1 hr was followed by a 30 min incubation at 37°C with the anti-BrdU antibody at 1/300 (GE Healthcare) in DNase incubation buffer (0.5× PBS, 30 mM Tris-HCl pH 8, 0.3 mM MgCl2, 0.5 mM 2-mercaptoethanol, 0.5% BSA, and 10 μg/mL DNase I). After several washes, cells were incubated with an Alexa Fluor 488-conjugated donkey secondary antibody at 1:500 (Invitrogen).
Cell Culture
Sorted NSCs or total SVZ cell suspensions were grown at 37°C in 5% CO2 in neurosphere medium composed of DMEM/F12 (Life Technologies) supplemented with 0.6% glucose (Sigma), 2 μg/mL heparin (STEMCELL Technologies), 1× insulin-selenium-transferrin (Life Technologies), N-2 supplement (Life Technologies), and B-27 without vitamin A supplement (Life Technologies), and in the presence of 20 ng/mL EGF (Millipore) and 10 ng/mL FGF2 (Millipore).
After 7 days, neurospheres were counted under an inverted microscope. Neurospheres were centrifuged and incubated for 5 min in the presence of Accutase (Sigma) then were mechanically dissociated. Dissociated cells were plated in neurosphere medium at a density not exceeding 1.4 cells/μL in 12- or 24-well plates.
Quiescence was induced in vitro by removing growth factors and by adding 25 ng/mL hBMP4 (R&D Systems).
For RNA measurement, cells were counted on day 3 and lysed in RLT buffer (Qiagen) for RNA isolation and qRT-PCR experiments.
SiRNA Silencing
Immediately after sorting or 1 week after initiation of neurosphere cultures, LeX+EGFR+ SVZ cells were electroporated using the Neon kit according to the manufacturer's instructions (Thermo Fisher). Briefly, dissociated cells (1.2 × 103 to 12 × 103) were suspended in 20 μL of R resuspension buffer (Thermo Fisher) and split into two vials containing 1 μL of siRNA at 20 μM. Cells were electroporated at 1,300 V for three pulses for 10 ms then transferred immediately into neurosphere medium. SiRNA was purchased from Qiagen: control nontargeting/scrambled control (cat#1027280) and a mix of four siRNAs against Sdc1 (cat#1027416). Different concentrations of siRNA (10, 20, and 100 nM) were tested in a first set of experiments on neurosphere cultures initiated with total adult SVZ cells.
Live Cell Imaging
Freshly sorted LeX+EGFR+ cells (2 × 103) from adult FUCCI-Red mice were electroporated with siRNA (control or Sdc1) at a final concentration of 50 nM. Brightfield and fluorescent images for Cdt1-red were captured through a Plan Apo VC 320 differential interference contrast objective (numerical aperture, 0.75) on a Nikon A1R confocal laser scanning microscope system attached to an inverted ECLIPSE Ti (Nikon, Tokyo, Japan) thermostated at 37°C under 5% CO2/20% O2 atmosphere as previously reported (Daynac et al., 2014). Recording was started 4 hr after electroporation. Proliferating cells (n = 20–30 cells) were individually followed and the time of the first cell division was determined in four independent experiments.
RNA Isolation, Microarrays, and qRT-PCR
NSCs were sorted into tubes containing RLT lysis buffer and total RNAs were isolated with the RNeasy Micro Kit with DNase treatment (Qiagen). For microarray experiments, RNA transcripts were converted into cDNAs and amplified using the Ovation Pico WTA System (NuGEN). cDNAs were fragmented and biotinylated; then, labeled cRNAs were hybridized to Affymetrix MOE430 2.0 arrays according to the manufacturer's protocol at PartnerShip (Evry, France). The data were normalized with the MAS5 algorithm and quality controlled with the Expression Console software (Affymetrix).
For qRT-PCR experiments, total RNAs were reverse transcribed into cDNA using the Reverse Transcription High Capacity Master Mix (Applied Biosystems) with specific primers listed in Table S6 (Sigma-Aldrich). q-PCR was performed on an ABI PRISM 7900 Sequence Detector System using SYBR Green for RT-PCR. Expression levels were normalized to GAPDH (glyceraldehyde 3-phosphate dehydrogenase).
Microarray Analysis
Data were normalized with GC-robust multi-array analysis (GC-RMA) using log2 transformed expression levels in Genespring GX12 (Agilent Technologies). For comparative analysis of datasets, differentially expressed probes were filtered by an average expression greater than 50 in at least one population, at least 2-fold change, and a Student's t test p value <0.05 (control LeXbright versus irradiated LeXbright) or corrected p value <0.05 (LeXbright versus LeX+EGFR+). An overrepresentation analysis of GO biological processes (p < 0.05) with Bonferroni correction and cellular component ontology analysis were carried out with PANTHER software (http://www.pantherdb.org). Statistically enriched GO terms were then hand curated into thematic categories. Heatmaps were generated with Gene-E (https://software.broadinstitute.org/GENE-E/).
Statistical Analyses
The data are expressed as the mean ± SD. Non-parametric Mann-Whitney test was conducted to compare qRT-PCR data using GraphPad PRISM software (GraphPad, San Diego, CA). Significance was set at p < 0.05.
Author Contributions
L.M., conception and design, collection and/or assembly of data, bioinformatic analysis, data analysis and interpretation, and manuscript writing. A.C., L.R.G., and M.D., collection and/or assembly of data. M.A.M., conception and design, collection and/or assembly of data, data analysis interpretation, and manuscript writing. F.D.B., conception and design, financial support, data analysis and interpretation, and manuscript writing.
Acknowledgments
We are indebted to S. Vincent-Naulleau, V. Neuville, V. Barroca, and the staff of the animal facilities; to T. Kortulewski for videomicroscopy; to J. Baijer and N. Dechamps for cell sorting; and to A. Gouret and A. Leliard for their administrative assistance. Flow cytometry and cell sorting were performed at the iRCM Flow Cytometry Shared Resource, established by equipment grants from DIM-Stem-Pôle, INSERM, Foundation ARC, and CEA. This work was supported by grants from Electricité de France (EDF) and CEA (Segment Radiobiologie). L.M. has a fellowship from Région Ile-de-France (DIM Biothérapies) and M.D. from La Ligue Contre le Cancer.
Published: July 5, 2018
Footnotes
Supplemental Information includes five figures and six tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.06.005.
Contributor Information
François D. Boussin, Email: boussin@cea.fr.
Marc-André Mouthon, Email: marc-andre.mouthon@cea.fr.
Accession Numbers
All data are deposited in NCBI GEO under accession number GEO: GSE99777.
Supplemental Information
References
- Abraham A.B., Bronstein R., Reddy A.S., Maletic-Savatic M., Aguirre A., Tsirka S.E. Aberrant neural stem cell proliferation and increased adult neurogenesis in mice lacking chromatin protein HMGB2. PLoS One. 2013;8:e84838. doi: 10.1371/journal.pone.0084838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adepoju A., Micali N., Ogawa K., Hoeppner D.J., McKay R.D. FGF2 and insulin signaling converge to regulate cyclin D expression in multipotent neural stem cells. Stem Cells. 2014;32:770–778. doi: 10.1002/stem.1575. [DOI] [PubMed] [Google Scholar]
- Ahn S., Joyner A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Balordi F., Fishell G. Hedgehog signaling in the subventricular zone is required for both the maintenance of stem cells and the migration of newborn neurons. J. Neurosci. 2007;27:5936–5947. doi: 10.1523/JNEUROSCI.1040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantilena S., Pastorino F., Pezzolo A., Chayka O., Pistoia V., Ponzoni M., Sala A. Frizzled receptor 6 marks rare, highly tumourigenic stem-like cells in mouse and human neuroblastomas. Oncotarget. 2011;2:976–983. doi: 10.18632/oncotarget.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capela A., Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Chaker Z., Codega P., Doetsch F. A mosaic world: puzzles revealed by adult neural stem cell heterogeneity. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:640–658. doi: 10.1002/wdev.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., Deleo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynac M., Chicheportiche A., Pineda J.R., Gauthier L.R., Boussin F.D., Mouthon M.A. Quiescent neural stem cells exit dormancy upon alteration of GABAAR signaling following radiation damage. Stem Cell Res. 2013;11:516–528. doi: 10.1016/j.scr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Daynac M., Morizur L., Chicheportiche A., Mouthon M.A., Boussin F.D. Age-related neurogenesis decline in the subventricular zone is associated with specific cell cycle regulation changes in activated neural stem cells. Sci. Rep. 2016;6:21505. doi: 10.1038/srep21505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynac M., Morizur L., Kortulewski T., Gauthier L.R., Ruat M., Mouthon M.A., Boussin F.D. Cell sorting of neural stem and progenitor cells from the adult mouse subventricular zone and live-imaging of their cell cycle dynamics. J. Vis. Exp. 2015 doi: 10.3791/53247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynac M., Pineda J.R., Chicheportiche A., Gauthier L.R., Morizur L., Boussin F.D., Mouthon M.A. TGFbeta lengthens the G1 phase of stem cells in aged mouse brain. Stem Cells. 2014;32:3257–3265. doi: 10.1002/stem.1815. [DOI] [PubMed] [Google Scholar]
- Daynac M., Tirou L., Faure H., Mouthon M.A., Gauthier L.R., Hahn H., Boussin F.D., Ruat M. Hedgehog controls quiescence and activation of neural stem cells in the adult ventricular-subventricular zone. Stem Cell Rep. 2016;7:735–748. doi: 10.1016/j.stemcr.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Garcia-Verdugo J.M., Alvarez-Buylla A. Regeneration of a germinal layer in the adult mammalian brain. Proc. Natl. Acad. Sci. USA. 1999;96:11619–11624. doi: 10.1073/pnas.96.20.11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken B.W., Leeman D.S., Boutet S.C., Hebestreit K., Brunet A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18:777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck S.M., Alizadeh A., Santhosh K.T., Proulx E.H., Wu C.L., Karimi-Abdolrezaee S. Chondroitin sulfate proteoglycans negatively modulate spinal cord neural precursor cells by signaling through LAR and RPTPsigma and modulation of the Rho/ROCK pathway. Stem Cells. 2015;33:2550–2563. doi: 10.1002/stem.1979. [DOI] [PubMed] [Google Scholar]
- Ferent J., Cochard L., Faure H., Taddei M., Hahn H., Ruat M., Traiffort E. Genetic activation of hedgehog signaling unbalances the rate of neural stem cell renewal by increasing symmetric divisions. Stem Cell Rep. 2014;3:312–323. doi: 10.1016/j.stemcr.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda M., Martinez P., Schoeftner S., Gomez-Lopez G., Schneider R., Flores J.M., Pisano D.G., Blasco M.A. Sox4 links tumor suppression to accelerated aging in mice by modulating stem cell activation. Cell Rep. 2014;8:487–500. doi: 10.1016/j.celrep.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frinchi M., Bonomo A., Trovato-Salinaro A., Condorelli D.F., Fuxe K., Spampinato M.G., Mudo G. Fibroblast growth factor-2 and its receptor expression in proliferating precursor cells of the subventricular zone in the adult rat brain. Neurosci. Lett. 2008;447:20–25. doi: 10.1016/j.neulet.2008.09.059. [DOI] [PubMed] [Google Scholar]
- Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 2015;18:657–665. doi: 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- Gur G., Rubin C., Katz M., Amit I., Citri A., Nilsson J., Amariglio N., Henriksson R., Rechavi G., Hedman H. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh F.Y., Ma T.L., Shih H.Y., Lin S.J., Huang C.W., Wang H.Y., Cheng Y.C. Dner inhibits neural progenitor proliferation and induces neuronal and glial differentiation in zebrafish. Dev. Biol. 2013;375:1–12. doi: 10.1016/j.ydbio.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Huang T.T., Zou Y., Corniola R. Oxidative stress and adult neurogenesis–effects of radiation and superoxide dismutase deficiency. Semi. Cell Dev. Biol. 2012;23:738–744. doi: 10.1016/j.semcdb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.B., Watt F.M. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. USA. 2006;103:11958–11963. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin T.E., Martens D.J., van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E., Wang Y., Kusek G., Wurster R., Lederman P., Lowry N., Shen Q., Temple S. VCAM1 is essential to maintain the structure of the SVZ niche and acts as an environmental sensor to regulate SVZ lineage progression. Cell Stem Cell. 2012;11:220–230. doi: 10.1016/j.stem.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Kwon M.J., Jang B., Yi J.Y., Han I.O., Oh E.S. Syndecans play dual roles as cell adhesion receptors and docking receptors. FEBS Lett. 2012;586:2207–2211. doi: 10.1016/j.febslet.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Lim D.A., Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci. 2014;37:563–571. doi: 10.1016/j.tins.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang Q., Haydar T.F., Bordey A. Nonsynaptic GABA signaling in postnatal subventricular zone controls proliferation of GFAP-expressing progenitors. Nat. Neurosci. 2005;8:1179–1187. doi: 10.1038/nn1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Marthiens V., Kazanis I., Moss L., Long K., Ffrench-Constant C. Adhesion molecules in the stem cell niche–more than just staying in shape? J. Cell Sci. 2010;123:1613–1622. doi: 10.1242/jcs.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B., Mateo J.L., Zhou B., Andersen J., Achimastou A., Urban N., van den Berg D., Georgopoulou D., Hadjur S., Wittbrodt J. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu C., Sii-Felice K., Fouchet P., Etienne O., Haton C., Mabondzo A., Boussin F.D., Mouthon M.A. Endothelial cell-derived bone morphogenetic proteins control proliferation of neural stem/progenitor cells. Mol. Cell. Neurosci. 2008;38:569–577. doi: 10.1016/j.mcn.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Mich J.K., Signer R.A., Nakada D., Pineda A., Burgess R.J., Vue T.Y., Johnson J.E., Morrison S.J. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H., Andreu Z., Suh H., Lie D.C., Jessberger S., Consiglio A., San Emeterio J., Hortiguela R., Marques-Torrejon M.A., Nakashima K. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead C.M., Reynolds B.A., Craig C.G., McBurney M.W., Staines W.A., Morassutti D., Weiss S., van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Nagato M., Heike T., Kato T., Yamanaka Y., Yoshimoto M., Shimazaki T., Okano H., Nakahata T. Prospective characterization of neural stem cells by flow cytometry analysis using a combination of surface markers. J. Neurosci. Res. 2005;80:456–466. doi: 10.1002/jnr.20442. [DOI] [PubMed] [Google Scholar]
- Oikari L.E., Okolicsanyi R.K., Qin A., Yu C., Griffiths L.R., Haupt L.M. Cell surface heparan sulfate proteoglycans as novel markers of human neural stem cell fate determination. Stem Cell Res. 2016;16:92–104. doi: 10.1016/j.scr.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Ottone C., Krusche B., Whitby A., Clements M., Quadrato G., Pitulescu M.E., Adams R.H., Parrinello S. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat. Cell Biol. 2014;16:1045–1056. doi: 10.1038/ncb3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G., Francois S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D., Buckingham M. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Papanikolaou T., Lennington J.B., Betz A., Figueiredo C., Salamone J.D., Conover J.C. In vitro generation of dopaminergic neurons from adult subventricular zone neural progenitor cells. Stem Cells Dev. 2008;17:157–172. doi: 10.1089/scd.2007.0090. [DOI] [PubMed] [Google Scholar]
- Porlan E., Marti-Prado B., Morante-Redolat J.M., Consiglio A., Delgado A.C., Kypta R., Lopez-Otin C., Kirstein M., Farinas I. MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 2014;16:629–638. doi: 10.1038/ncb2993. [DOI] [PubMed] [Google Scholar]
- Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M. What is principal component analysis? Nat. Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Roccio M., Schmitter D., Knobloch M., Okawa Y., Sage D., Lutolf M.P. Predicting stem cell fate changes by differential cell cycle progression patterns. Development. 2013;140:459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Snorradottir S., Center J.R. New sequence variants associated with bone mineral density. Nat. Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban N., van den Berg D.L., Forget A., Andersen J., Demmers J.A., Hunt C., Ayrault O., Guillemot F. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353:292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yang L., Alexander C., Temple S. The niche factor syndecan-1 regulates the maintenance and proliferation of neural progenitor cells during mammalian cortical development. PLoS One. 2012;7:e42883. doi: 10.1371/journal.pone.0042883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Mabuchi T., Satoh E., Furuya K., Zhang L., Maeda S., Naganuma H. Expression of syndecans, a heparan sulfate proteoglycan, in malignant gliomas: participation of nuclear factor-kappaB in upregulation of syndecan-1 expression. J. Neurooncol. 2006;77:25–32. doi: 10.1007/s11060-005-9010-3. [DOI] [PubMed] [Google Scholar]
- Xu Y., Yuan J., Zhang Z., Lin L., Xu S. Syndecan-1 expression in human glioma is correlated with advanced tumor progression and poor prognosis. Mol. Biol. Rep. 2012;39:8979–8985. doi: 10.1007/s11033-012-1767-9. [DOI] [PubMed] [Google Scholar]
- Zhang H.M., Chen H., Liu W., Liu H., Gong J., Wang H., Guo A.Y. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Nowakowski R.S., Vaccarino F.M. Fibroblast growth factor 2 is required for maintaining the neural stem cell pool in the mouse brain subventricular zone. Dev. Neurosci. 2004;26:181–196. doi: 10.1159/000082136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.