Abstract
Background
Cassava brown streak disease (CBSD) has a viral aetiology and is caused by viruses belonging to the genus Ipomovirus (family Potyviridae), Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV). Molecular and serological methods are available for detection, discrimination and quantification of cassava brown streak viruses (CBSVs) in infected plants. However, precise determination of the viral RNA localization in infected host tissues is still not possible pending appropriate methods.
Results
We have developed an in situ hybridization (ISH) assay based on RNAscope® technology that allows the sensitive detection and localization of CBSV RNA in plant tissues. The method was initially developed in the experimental host Nicotiana rustica and was then further adapted to cassava. Highly sensitive and specific detection of CBSV RNA was achieved without background and hybridization signals in sections prepared from non-infected tissues. The tissue tropism of CBSV RNAs appeared different between N. rustica and cassava.
Conclusions
This study provides a robust method for CBSV detection in the experimental host and in cassava. The protocol will be used to study CBSV tropism in various cassava genotypes, as well as CBSVs/cassava interactions in single and mixed infections.
Keywords: Cassava brown streak virus, Virus localization, Ipomovirus, In situ hybridization, RNAscope®, Formalin-fixed paraffin-embedded tissue sections
Background
Cassava brown streak disease (CBSD) is caused by two distinct virus species, Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV), both members of the genus Ipomovirus in the family Potyviridae [1]. The viruses are the most devastating pathogens of cassava (Manihot esculenta) in Africa, threatening cassava cultivation particularly in East and Central Africa [2]. The viruses have single-stranded RNA genomes of about 9000 nt, and while genetically distinct, they cause similar symptoms in the leaves, stems and root tissues of cassava [1, 3, 4] including leaf chlorosis, brown streaks on stems and necrosis of root tubers [5]. The natural host range of CBSVs is restricted to M. esculenta and Manihot glaziovii, a perennial species related to cassava, but other natural hosts which could serve as viral inoculum sources may exist [2, 6].
Immunological and molecular techniques for the detection of CBSVs have been developed. Enzyme-linked immunosorbent assays (ELISA) based on monoclonal antibodies [7], reverse transcription-polymerase chain reaction (RT-PCR) [8–10] and quantitative RT-PCR [11–14] are routinely used in diagnosis of the viruses. While these methods are sensitive, reproducible and robust for virus detection in a given cassava sample, accurate quantification of CBSVs in cassava is hampered by the uneven distribution of the virus in the plant [15, 16], making comparative studies very difficult.
The disease caused by cassava brown streak viruses is the subject of intensive research in many institutes around the world, and research on the causative viruses is a key topic in the DSMZ Plant Virus Department. In particular, we are interested in following the movement of cassava brown streak viruses in cassava to study tissue invasion and the possible association of CBSV with specific plant tissues and organs. This approach aims to investigate a possible correlation between the virus loads in leaf, stem and tuberous root tissues and the extent of necrotic brown streak symptoms in root tissues. In situ hybridization of CBSV RNA in cassava tissue sections requires highly sensitive methods to detect even small amounts of RNA. To address this aim, we have developed an in situ hybridization (ISH) method which is based on RNAscope® technology, allowing detection and localization of RNA targets with high specificity and sensitivity [17, 18]. The RNAscope® technology developed by Advanced Cell Diagnostics (ACD; Hayward, CA, USA) is based on a unique probe design and signal amplification strategy that results in high specificity and sensitivity. RNAscope® has been mostly used in clinical studies with human and animal tissues [17, 19–22]. Recently, two studies have also used RNAscope® in plant tissues: for the sensitive localization of messenger RNAs (mRNAs) coding for C4 photosynthetic enzymes in maize leaves [23] and for the simultaneous visualization of two isolates of Citrus tristeza virus (CTV) in the petioles and root tissues of citrus [24].
In this manuscript, we present a protocol for preparation of tissue sections from CBSV-infected Nicotiana rustica and cassava plants. We describe the optimal conditions for the ISH assay and provide a robust method for CBSV detection in tissue sections of its experimental host and cassava. The method represents a significant technical advancement enabling studies of CBSV-infected cassava organs and tissues to further advance our understanding of the mechanisms of CBSV infection and disease development.
Methods
Plant material and virus inoculations
The experimental host N. rustica and the cassava cultivar Tropical Manihot esculenta 7 (TME7) were used for this study. The plants were grown in a glasshouse at 24 to 26 °C. Virus infections were established using the virus isolate CBSV-Mo83 (DSMZ PV-0949), which has been isolated from naturally infected cassava collected in Mozambique [1]. CBSV-Mo83 was transmitted to N. rustica by mechanical inoculation. N. rustica plants at the three-to-four leaf stage were inoculated with inoculum prepared by grinding CBSV-Mo83-infected N. benthamiana leaves in 0.05 M phosphate buffer (0.05 M Na3PO4, 1 mM EDTA, 5 mM DIECA, 5 mM thioglycolic acid, pH 7.0) in a ratio of 1:20 (w/v). Symptoms developed after 7 days, and virus infection was confirmed by RT-PCR [1]. In cassava, CBSV-Mo83 was routinely maintained in var. TMS 96/0304 and propagated by cuttings or, for new infections, by grafting. Virus infections in TME7 were established by grafting buds of CBSV-Mo83-infected cassava var. TMS 96/0304 onto cassava plants grown from tissue culture. Virus infections were confirmed by symptom development and RT-PCR approximately four weeks after inoculation.
Design and synthesis of CBSV-Mo83 probes
Probes against CBSV-Mo83 were custom-designed by ACD and are available in the ACD catalogue as V-CBSV-Mo83-P1 (Ref. 509,481). The probes were designed based on the CBSV-Mo83 genomic sequence (GenBank accession number FN434436) and were complementary to nucleotides at positions 127 to 1191 within the P1 sequence.
Fixation, embedding, and sectioning of N. rustica
For the ISH assays of N. rustica, stem sections were prepared from healthy and CBSV-infected plants at 14 dpi. Stem tissues (~ 5 mm in diameter and length) were cut using a sterile razor blade and placed into 10% neutral buffered formalin fixative solution (Sigma-Aldrich, St. Louis, MO, USA). Incubation was performed for 45 min at room temperature (RT) under vacuum conditions, followed by a fixative exchange and a 45 min incubation period, after which the fixative was exchanged and the samples were incubated for 16 h. The samples were subsequently washed two times in DEPC-treated phosphate-buffered saline (PBS, pH 7.4) for 15 min and dehydrated by incubation in increasing ethanol concentrations (30%, 50%, 70%, 95%, 100%) for 30 min at each concentration. After dehydration, the tissues were directly embedded into a low-melting agarose solution (5% w/v) (Serva Electrophoresis GmbH, Heidelberg, Germany). The agarose was melted in a microwave and cooled to approximately 40 °C, and tissue samples were placed into the gel in the desired orientation. Semi-thin (10 μm) cross-sections were cut using a Microm HM 650 V vibrating blade microtome (Thermo-Fisher Scientific, Pittsburgh, PA, USA) and applied to Superfrost Plus slides (Thermo-Fisher Scientific). Sections were allowed to dry on the slides overnight at RT and then baked in a hybridization oven for 1 h at 60 °C. Dried slides were stored in a covered box with silica gel, before proceeding with the ISH assays.
Fixation, embedding, and sectioning of cassava
Leaf, stem and petiole explants (~ 5 mm in length; stems diameter: ~ 4 mm; petioles diameter: ~ 1.5 mm) from healthy and CBSV-infected cassava were fixed following the same procedures as described for N. rustica. For infected plants, leaf and petiole samples were collected from symptomatic leaves showing chlorosis, and stem samples from stems showing brown streaks. Because of the nature of cassava stem tissues (hard cortex and soft central medulla), different embedding media were tested, including low-melting agarose (SERVA Electrophoresis GmbH, Heidelberg, Germany), low-melting polyester-wax (Plano GmbH, Wetzlar, Germany) and Paraffin Paraplast Plus (Sigma-Aldrich). Embedding of cassava samples in low-melting agarose and tissue sectioning was performed as described above for N. rustica. For embedding in low-melting wax, the tissues were infiltrated with wax using increasing concentrations of wax solubilized in ethanol. Tissues were incubated in ethanol/wax mixtures (2:1, 1:1, 1:2 (v/v), pure wax) at 40 °C for 1 h at each concentration and then transferred into pure low-melting polyester wax in Peel-A-Way molds (Sigma-Aldrich). Embedding of tissue samples in Paraplast Plus paraffin was performed using a sequential-steps protocol by infiltrating plant tissues at RT in ethanol/xylene mixtures at 2:1, 1:1, and 1:2 (v/v) and pure xylene for 45 min in each substitute mixture. The tissues were then infiltrated with xylene/paraffin substitutes at 2:1, 1:1, 1:2 (v/v), followed by pure paraffin, with 1 h at each step in an oven at 60 °C. The samples were then transferred and embedded into pure paraffin in Peel-A-Way molds. Samples in low-melting wax and paraffin molds were allowed to cool to RT and stored at 4 °C overnight prior to sectioning. The blocks were trimmed to a suitable size, and cross-sections of 10–15 μm were prepared using a Microm HM 355 rotary microtome (Thermo-Fisher Scientific). After sectioning, the obtained ribbons were placed in a water bath at 37 °C and then placed on Superfrost Plus slides. Sections were completely dried and baked for 1 h at 60 °C. After baking, sections were deparaffinized in xylene (two times, 5 min each wash), washed in absolute ethanol, and stored in a covered box with silica gel before proceeding with the ISH assays.
Optimization of RNAscope® ISH procedure
The ISH assay was performed using the ACD RNAscope® 2.5 HD Detection Reagent-RED kit (cat. no. 322360). A reference RNAscope® hybridization protocol provided by ACD (http://www.acdbio.com/technical-support/user-manuals) was essentially followed by modifying the pre-hybridization treatment conditions and the washing and signal amplification steps to achieve optimal results (Table 1). Prior to the ISH assay, slides were baked for 30 min at 60 °C. A hydrophobic barrier was created around the sections with an ImmEdge hydrophobic barrier pen (Biozol diagnostica Vertrieb GmbH, Eching, Germany). Sections were treated with hydrogen peroxide for 10 min at RT to inhibit endogenous peroxidases. A second pretreatment step was performed by incubation of the tissue sections in target retrieval buffer maintained at boiling temperature (100 °C to 102 °C) for 5 or 15 min, a step required for breaking crosslinks introduced upon fixation. The tissue sections were completely dried overnight (RT) and treated with a broad-spectrum Protease Plus solution at 40 °C to make the RNA accessible. The CBSV-Mo83 probes were hybridized at 40 °C in a HybEZ II oven for 2 h. As a control, the CBSV-Mo83 probe was hybridized with sections prepared from mock inoculated/grafted plants. Probe hybridization was followed by serial amplification steps (AMP1 to AMP6), as recommended by ACD, testing varying times (10, 15 or 30 min) for the AMP5 step. All washing steps following hybridization and during amplification consisted of two/three incubations in washing buffer (provided with the kit) for 5 min at each step. A final hybridization step using an alkaline phosphatase-labelled probe was followed by incubation with Fast-Red substrate that resulted in red precipitates. Slides were washed in water, counterstained with 50% hematoxylin (Sigma-Aldrich) for 2 min, then rinsed several times in distilled water. Sections were dried at 60 °C for 45 min, submerged in xylene, covered with EcoMount mounting media (Biocare Medical, Pacheco, CA, USA) and with 24x50mm microcover glass. Once mounted, the sections were air-dried for at least 10 min at RT prior to imaging.
Table 1.
RNAscope® in situ hybridization conditions tested for protocol definition
| Sample | Pretreatment conditions | Probe concentration | AMP5 incubation time | Washes after probe and AMPs incubation | Substrate incubation | Observations | ||
|---|---|---|---|---|---|---|---|---|
| Target Retrieval | Hydrogen Peroxide | Protease Plus | ||||||
| N. rustica (stem) | 15 min | 10 min | 30 min | not diluted | 30 min | 2 W, 2 min each | 10 min | Strong background |
| 5 min | 10 min | 10 min | not diluted | 15 min | 3 W, 5 min each | 2 min | Background reduced but low signal | |
| 15 min | 10 min | 15 min | not diluted | 15 min | 3 W, 5 min each | 2 min | Significant background | |
| 15 min | 10 min | 15 min | not diluted | 10 min | 3 W, 5 min each | 2 min | Significant background | |
| 15 min | 10 min | 15 min | 1:10 in diluent buffer | 10 min | 3 W, 5 min each | 2 min | Minimal background | |
| 15 min | 10 min | 15 min | 1:40 in diluent buffer | 10 min | 3 W, 5 min each | 2 min | Minimal background | |
| Cassava (stem) | 15 min | 10 min | 15 min | not diluted | 10 min | 3 W, 5 min each | 8 min | Absence of background |
| 15 min | 10 min | 15 min | not diluted | 15 min | 3 W, 5 min each | 8 min | Absence of background | |
| Cassava (leaf, petiole) | 15 min | 10 min | 15 min | not diluted | 10 min | 3 W, 5 min each | 8 min | Absence of background |
| 15 min | 10 min | 15 min | not diluted | 15 min | 3 W, 5 min each | 8 min | Absence of background | |
Imaging
Imaging of the tissue sections was performed using an Olympus SZX16 stereomicroscope (Olympus Deutschland GmbH, Hamburg, Germany) and a Zeiss Axioscope.A1 (Zeiss, Jena, Germany). To improve imaging, the “D” setting of the modulator disk of the Askioscope.A1 was also used for acquisitions in dark field. Fluorescence microscopy was performed using a Leica SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany) with a 20X/0.75 IMM objective using the following settings: 561 nm excitation and 631–651 nm emission window. Images from transmitted light were also collected during the acquisitions to allow overlay of different channels. Images were processed using the Huygens deconvolution software (Scientific Volume Imaging, Hilversum, The Netherlands).
Results and discussion
Localizing viruses in host tissues and organs can provide fundamental details on virus infection processes as well as on the host responses to virus invasion. Immunohistochemistry (IHC) and ISH are powerful techniques that allow the detection of target molecules in tissues and cells. ISH was originally developed to localize specific DNA sequences on chromosomes and later adapted using various probe modifications and labels to detect mRNAs and other RNA targets [25, 26]. ISH has been extensively used for in situ studies of plant DNA and RNA viruses in different hosts, and simple, rapid and inexpensive methods allow localization of viruses in plants and in insect vectors [27–39].
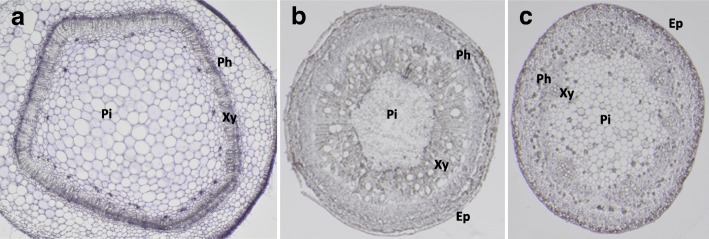
Because an in situ hybridization method for CBSV RNA has not been described, we developed an ISH protocol based on RNAscope®. Infection of CBSV-Mo83 in N. rustica resulted in stunting and leaf curling and chlorosis, while infected cassava plants showed typical brown streak symptoms on the stems and leaf chlorosis. In the two hosts, semi-thin cross sections of 10–15 μm were obtained using different embedding media. Stem sections of N. rustica were prepared from low-melting agarose-embedded tissues (Fig. 1), because this method was straightforward and did not require handling of hazardous chemicals. For cassava tissue samples, the low-melting agarose did not provide sufficient mechanical support to obtain sections of the desired thickness, as stem tissues have hard external cores and a soft internal medulla that present abrupt changes of resistance to the cutting knife, resulting in shattered cuts. The low-melting wax-based sectioning also did not result in satisfactory sections because of shredding and tearing of the wax ribbons, producing only partial, fragmentary sections. Finally, the paraffin-based method was successful for sectioning of cassava tissues, resulting in consistent and uniform sections for all tissue types (Fig. 1) and, therefore, was chosen for the ISH experiments.
Fig. 1.
Sections of Nicotiana rustica and cassava tissues. Cross-section of N. rustica stem tissues (a), cross-section of cassava stem tissues (b), cross-section of cassava petiole tissues (c). Tissue sections were stained by hematoxylin. Ep, Epidermis; Pi, pith; Xy, xylem; Ph, phloem. Sections diameter: stems, ~ 4 mm; petiole, ~ 1.5 mm
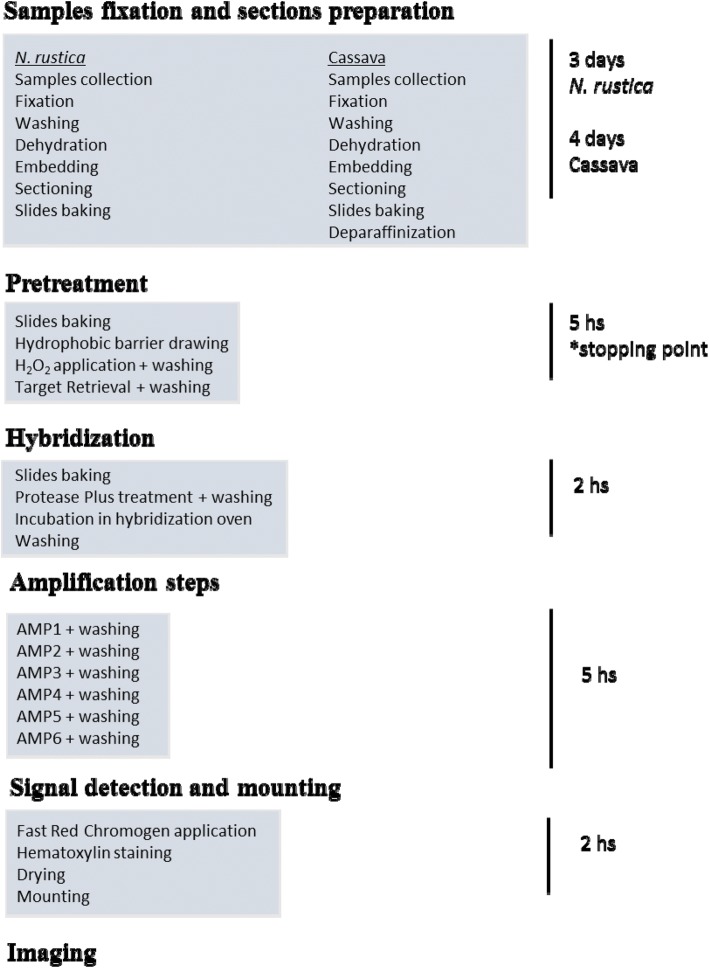
The ISH assay conditions were first optimized for CBSV detection in N. rustica and subsequently adapted for cassava experimentation. Table 1 summarizes the different ISH assay conditions tested. Key steps in the baseline ACD protocol were modified, including target retrieval, protease treatment, probe concentration, amplification steps, washing and substrate incubation. We found that reducing the protease incubation time and the AMP5 step and increasing the washing steps resulted in a significant reduction of the background. The concentration of the probes was also critical in N. rustica section hybridization, wherein 1:10 and 1:40 dilution of the stock provided within the kit resulted in minimal background in infected sections and no signal from healthy controls. In cassava, hybridizations were performed using undiluted probes, showing that the optimal probe concentration needs to be determined for each specific virus/host combination. The optimized incubation conditions for N. rustica, also applicable to ISH of cassava tissues, consisted of a 15 min target retrieval pretreatment, 10 min peroxidase treatment, 15 min protease incubation and 10 min AMP5 incubation. The optimal substrate reaction lasted 2 min for N. rustica and 8 min for cassava. The final protocol is summarized in Fig. 2 and allowed detection of CBSV RNA in the tissue sections of both hosts, without signal in healthy controls. The conditions determined for ISH are well-suited for sensitive and specific RNA detection, and the protocol provides a good reference for investigating other virus/host combinations.
Fig. 2.
Overview of the RNAscope® protocol for CBSV ISH in N. rustica and cassava tissues
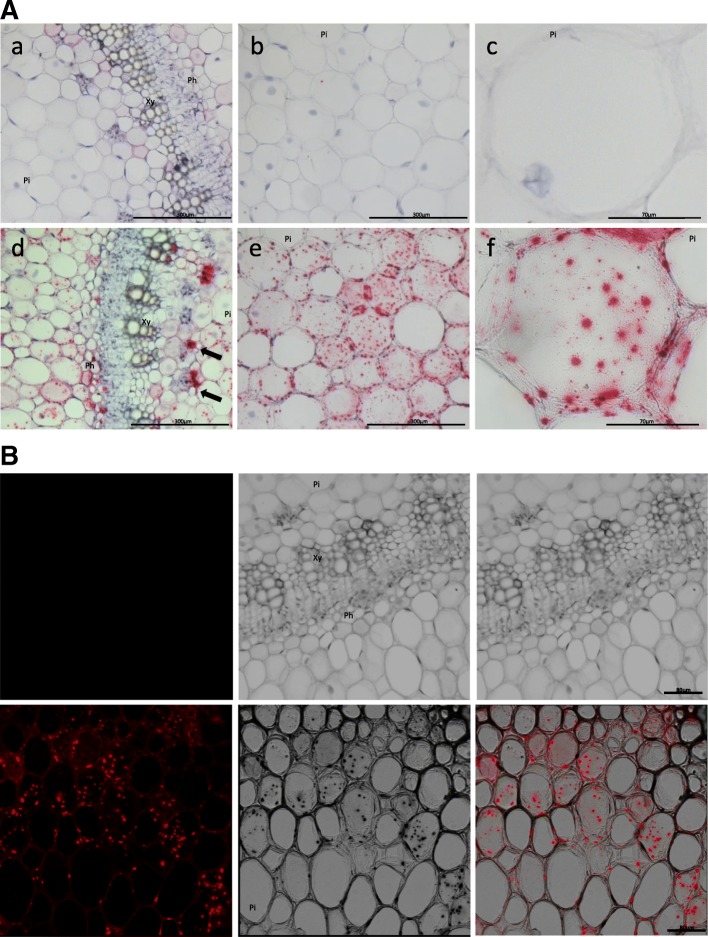
Imaging of at least 10 sections for each condition revealed that CBSV RNA was widely distributed throughout the stem tissues of infected N. rustica, as indicated by distinct red dots in different tissues, including the phloem, cortex, and pith cells, which occasionally formed clusters (Fig. 3A, panels d,e,f). The red signal was completely absent in healthy controls (Fig. 3A, panels a,b,c). Since the chromogenic red precipitate can be imaged by fluorescence microscopy, we examined the sections using confocal laser scanning microscopy, and CBSV RNA could be very clearly visualized (Fig. 3B). In cross-sections of infected cassava stem tissues, the red dot signal was less abundant and typically appeared as clusters of dots in phloematic tissue surrounding the xylem (Fig. 4c-h, arrows) and occasionally in cortical tissues (Fig. 4c). There was no signal detected in sections of the healthy controls, indicating absence of any background due to non-specific hybridization (Fig. 4a,b). In cross-sections of infected cassava leaf petioles, CBSV RNA was associated with phloematic tissues (Fig. 5c-f, open squares), and there was no signal detected in the healthy controls. In cross-sections of infected cassava leaves, viral RNA was detected in palisade, mesophyll and midrib tissues (Fig. 6c,d), while there was no signal in leaves of non-infected plants (Fig. 6a,b). To improve signal detection, we examined the sections using dark field acquisition settings, which significantly improved the signal detection and were particularly useful to detect signals in sections with a low number of chromogenic spots (Fig. 6g,h).
Fig. 3.
Localization of CBSV-Mo83 RNA in cross-sections of Nicotiana rustica stem tissues. A Acquisitions using light microscopy. Upper images: tissue from healthy control plants. Lower images: tissue from plants infected with CBSV-Mo83. Abundant presence of viral RNA in cells is visualized as single red dots or dots merged into signal clusters (arrows). B Acquisition by confocal laser scanning microscopy. Upper images: non-infected control. Lower images: CBSV-Mo83-infected section. The Fast-Red signal (left) laid over the image from transmitted light (centre) results in a merged picture (right) to demonstrate virus detection in infected cells. Pi, pith; Xy, xylem; Ph, phloem
Fig. 4.
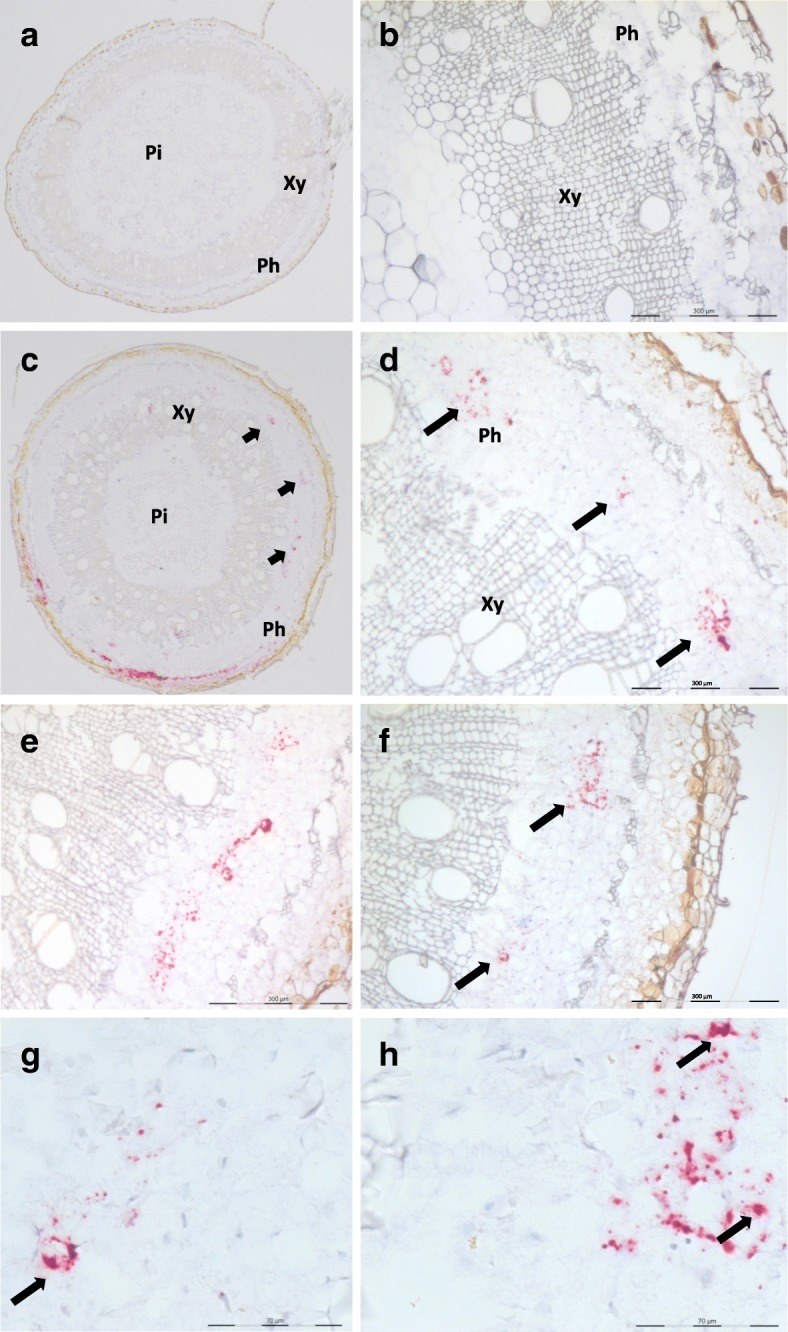
Localization of CBSV-Mo83 RNA in cross-sections of cassava stem tissues. Sections from a control healthy plant (a, b), stem sections from a CBSV-Mo83-infected cassava (c-h). Red dots indicate hybridization of the probes to viral RNA. Accumulation of dots mostly in phloematic cells (arrows in c, d ,f). Signal clusters were also detected (g, h). Pi, pith; Xy, xylem; Ph, phloem. Sections diameter: ~ 4 mm
Fig. 5.
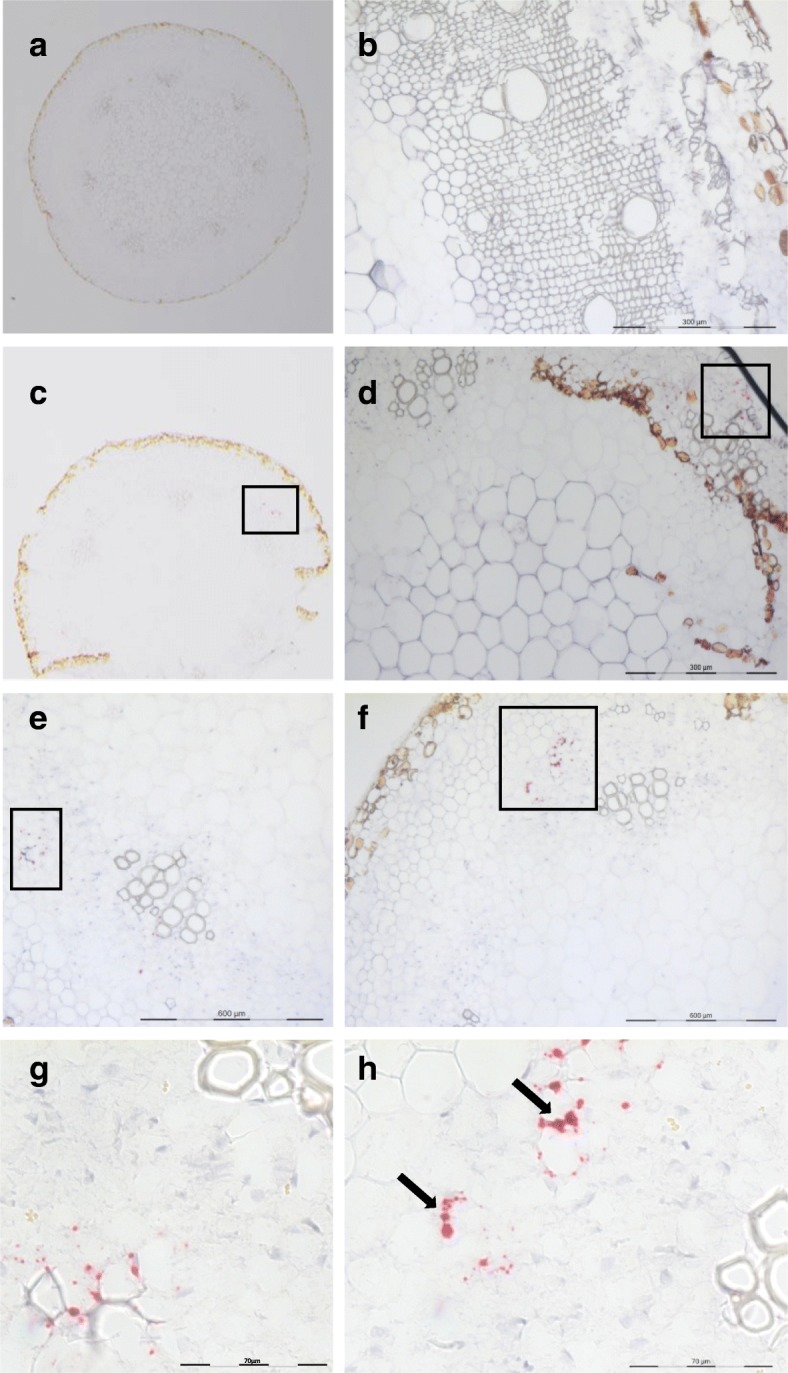
Localization of CBSV-Mo83 RNA in cross-sections of cassava petioles. Petiole sections from healthy control plants (a, b); sections from CBSV-Mo83-infected cassava (c-h). Red dots indicate hybridization of probes to viral RNA. Open squares indicate areas with hybridization signals (c-f), shown in lower panels (g, h) at a higher magnification. Accumulation of red dots is mostly evident in phloematic tissues. Signal clusters were also detected (h). Sections diameter: ~ 1.5 mm
Fig. 6.
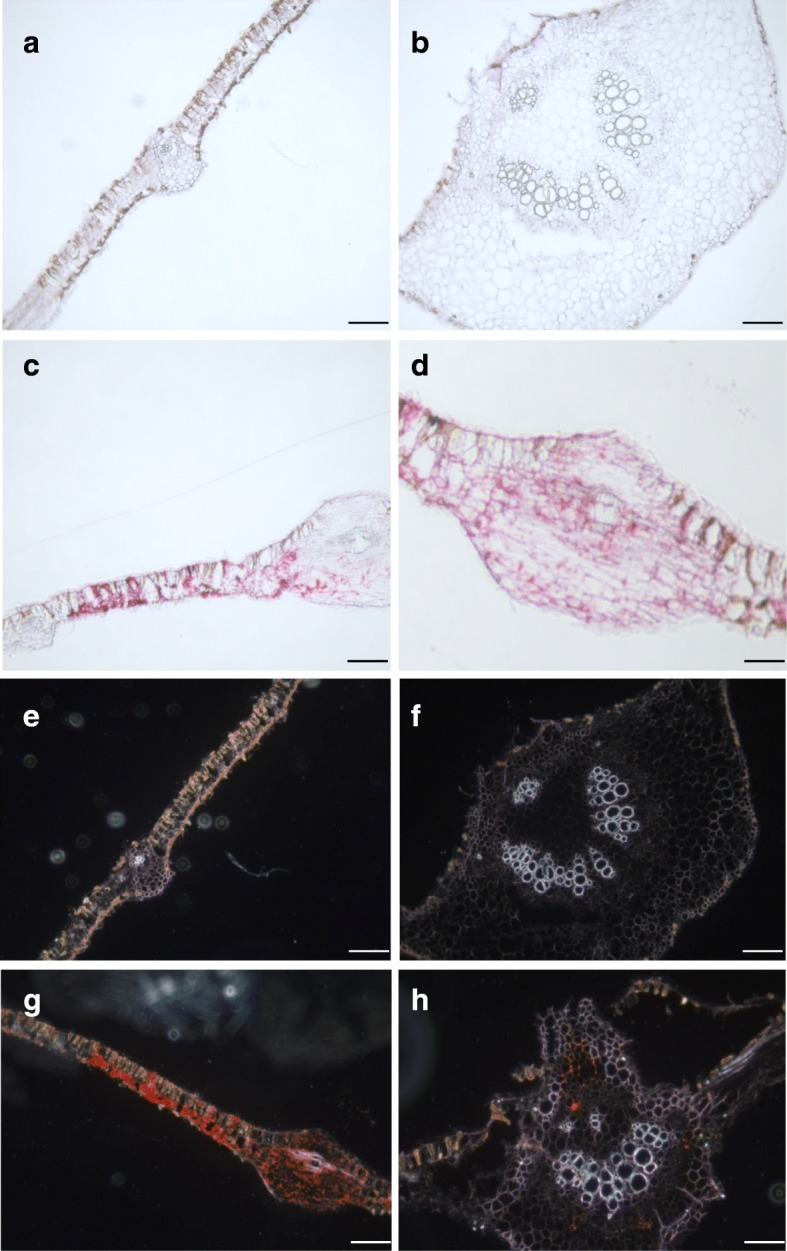
Localization of CBSV-Mo83 RNA in cross-sections of cassava leaves. Lamina and midrib tissues of healthy (a, b, e, f) and CBSV-infected leaves (c, d, g, h). Hybridization signals (evident as red dots or bright orange dots) show an abundant presence of viral RNA in infected cells, with an absence of signal in non-infected controls. Scale bars: 100 μm
Overall, a preliminary examination of CBSV-infected stem sections from N. rustica and cassava showed that viral RNA was highly abundant in the phloematic and non-phloematic tissues of N. rustica, while in cassava, CBSV RNA appeared more localized around phloematic tissues. It now remains to be investigated whether this difference was because of a considerably lower virus load in cassava compared to N. rustica or was a result of a different tissue tropism, as has also been shown for other viruses infecting cassava [39]. While further studies are pending, our results show that the ISH RNAscope® assay has the high resolution required to study virus invasion in cassava cultivars with differential responses to virus infection. The unique probe design and high resolution of RNAscope® also allows detection of multiple targets, as shown previously for two distinct citrus tristeza virus strains in double-infected plants [24]. We are now investigating mixed infections between cassava brown streak virus species and strains as well as mixed infections between CBSV and viruses causing cassava mosaic disease. It will be interesting to further combine RNAscope® ISH with IHC [40–42] to reach a more complete representation of the tissues and the interacting partner molecules.
Conclusions
We provide a protocol for the detection and localization of CBSV in tissue sections of N. rustica and cassava using a highly sensitive ISH technique based on RNAscope® technology. The assay allows in situ hybridization of CBSV RNA in different plant tissues and provides a platform to investigate CBSV tropism during plant infection and disease development.
Acknowledgements
We are grateful for all assistance in the experiments provided by the able staff of the DSMZ Plant Virus Department. We would like to thank Drs. Stephan Wagner and Monika Goetz (Julius-Kühn Institut, Braunschweig) for providing access to the rotary microtome and Drs. Morgane Reaoult and Jacqueline Ay (Advanced Cell Diagnostics, Hayward, CA, USA) for helpful discussion.
Funding
This work was supported by the Mikocheni Agricultural Research Institute (MARI) through the ‘Disease diagnostics for sustainable cassava productivity in Africa’ project of the Bill & Melinda Gates Foundation (Seattle, WA, USA). It was supported at DSMZ by the `New Sources for CBSD resistance` project of the Bill & Melinda Gates Foundation (Seattle, WA, USA).
Abbreviations
- CBSD
Cassava brown streak disease
- CBSV
Cassava brown streak virus
- DEPC
Diethylpyrocarbonate
- DIECA
Diethyldithiocarbamic acid
- DSMZ
Deutsche Sammlung von Mikroorganismen und Zellkulturen
- EDTA
Ethylenediaminetetraacetic acid
- ELISA
Enzyme-linked immunosorbent assay
- IHC
Immunohistochemistry
- ISH
In situ hybridization
- mRNA
Messenger RNA
- RT-PCR
Reverse-transcription polymerase chain reaction
- UCBSV
Ugandan cassava brown streak virus
Authors’ contributions
EM, PM and SW designed the study. EM, SS and PM performed the experiments and analyzed the data. EM, PM and SW wrote the manuscript and prepared all figures. All authors reviewed the manuscript, and read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Esperance Munganyinka, Email: esp.munganyinka@gmail.com.
Paolo Margaria, Email: paolo.margaria@dsmz.de.
Samar Sheat, Email: samar.sheat@dsmz.de.
Elijah M. Ateka, Email: eateka@agr.jkuat.ac.ke
Fred Tairo, Email: fredtairo@gmail.com.
Joseph Ndunguru, Email: jndunguru2003@yahoo.co.uk.
Stephan Winter, Email: stephan.winter@dsmz.de.
References
- 1.Winter S, Koerbler M, Stein B, Pietruszka A, Paape M, Butgereitt A. Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J Gen Virol. 2010;91:1365–1372. doi: 10.1099/vir.0.014688-0. [DOI] [PubMed] [Google Scholar]
- 2.Mbanzibwa DR, Tian YP, Tugume AK, Mukasa SB, Tairo F, Kyamanywa S, Kullaya A, Valkonen JPT. Genetically distinct strains of cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch Virol. 2009;154:353–359. doi: 10.1007/s00705-008-0301-9. [DOI] [PubMed] [Google Scholar]
- 3.Hillocks R, Maruthi M, Kulembeka H, Jeremiah S, Alacho F, Masinde E, Ogendo J, Arama P, Mulwa R, Mkamilo G, Kimata B. Disparity between leaf and root symptoms and crop losses associated with cassava brown streak disease in four countries in eastern Africa. J Phytopathol. 2016;164:86–93. doi: 10.1111/jph.12430. [DOI] [Google Scholar]
- 4.Hillocks RJ, Jennings DL. Cassava brown streak disease: a review of present knowledge and research needs. Int J Pest Manag. 2003;49:225–234. doi: 10.1080/0967087031000101061. [DOI] [Google Scholar]
- 5.Patil BL, Legg JP, Kanju E, Fauquet CM. Cassava brown streak disease: a threat to food security in Africa. J Gen Virol. 2015;96:956–968. doi: 10.1099/vir.0.000014. [DOI] [PubMed] [Google Scholar]
- 6.Monger WA, Alicai T, Ndunguru J, Kinyua ZM, Potts M, Reeder RH, Miano DW, Adams IP, Boonham N, Glover RH, Smith J. The complete genome sequence of the Tanzanian strain of cassava brown streak virus and comparison with the Ugandan strain sequence. Arch Virol. 2010;155:429–433. doi: 10.1007/s00705-009-0581-8. [DOI] [PubMed] [Google Scholar]
- 7.Sheat S, Butgereitt A, Bonse S, Winter S. Virus Indexing of Cassava–Developing Standardised Serological Methods for Field Diagnosis. In: Tielkes E, editor. Management of land use systems for enhanced food security: conflicts, controversies and resolutions. Germany: Tropentag 2015; 2015. p. 184. [Google Scholar]
- 8.Abarshi MM, Mohammed IU, Wasswa P, Hillocks RJ, Holt J, Legg JP, Seal SE, Maruthi MN. Optimization of diagnostic RT-PCR protocols and sampling procedures for the reliable and cost-effective detection of cassava brown streak virus. J Virol Methods. 2010;163:353–359. doi: 10.1016/j.jviromet.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Mbanzibwa DR, Tian YP, Tugume AK, Mukasa SB, Tairo F, Kyamanywa S, Kullaya A, Valkonen JPT. Simultaneous virus-specific detection of the two cassava brown streak-associated viruses by RT-PCR reveals wide distribution in East Africa, mixed infections, and infections in Manihot glaziovii. J Virol Methods. 2011;171:394–400. doi: 10.1016/j.jviromet.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Monger WA, Seal S, Cotton S, Foster GD. Identification of different isolates of cassava brown streak virus and development of a diagnostic test. Plant Pathol. 2001;50:768–775. doi: 10.1046/j.1365-3059.2001.00647.x. [DOI] [Google Scholar]
- 11.Adams IP, Abidrabo P, Miano DW, Alicai T, Kinyua ZM, Clarke J, Macarthur R, Weekes R, Laurenson L, Hany U, Peters D, Potts M, Glover R, Boonham N, Smith J. High throughput real-time RT-PCR assays for specific detection of cassava brown streak disease causal viruses, and their application to testing of planting material. Plant Pathol. 2013;62:233–242. doi: 10.1111/j.1365-3059.2012.02622.x. [DOI] [Google Scholar]
- 12.Moreno I. GruissemW, Vanderschuren H. Reference genes for reliable potyvirus quantitation in cassava and analysis of cassava brown streak virus load in host varieties. J Virol Methods. 2011;177:49–54. doi: 10.1016/j.jviromet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Otti G, Bouvaine S, Kimata B, Mkamillo G, Kumar PL, Tomlins K, Maruthi MN. High-throughput multiplex real-time PCR assay for the simultaneous quantification of DNA and RNA viruses infecting cassava plants. J Appl Microbiol. 2016;120:1346–1356. doi: 10.1111/jam.13043. [DOI] [PubMed] [Google Scholar]
- 14.Shirima RR, Maeda DG, Kanju E, Ceasar G, Tibazarwa FI, Legg JP. Absolute quantification of cassava brown streak virus mRNA by real-time qPCR. J Virol Methods. 2017;245:5–13. doi: 10.1016/j.jviromet.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaweesi T, Kawuki R, Kyaligonza V, Baguma Y, Tusiime G, Ferguson ME. Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virol J. 2014;11:216. doi: 10.1186/s12985-014-0216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogwok E, Alicai T, Rey MEC, Beyene G, Taylor NJ. Distribution and accumulation of cassava brown streak viruses within infected cassava (Manihot esculenta) plants. Plant Pathol. 2014;64:1235–1246. doi: 10.1111/ppa.12343. [DOI] [Google Scholar]
- 17.Wang HW, Wang MXM, Su N, Wang LC, Wu X, Bui S, Nielsen A, Vo HT, Nguyen N, Luo Y, Ma XJ. RNAscope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. J Vis Exp. 2014;85:e51426. doi: 10.3791/51426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu XY, Vo HT, Ma XJ, Luo YL. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brostoff T, Dela Cruz FN, Church ME, Woolard KD, Pesavento PA. The raccoon polyomavirus genome and tumor antigen transcription are stable and abundant in neuroglial tumors. J Virol. 2014;88:12816–12824. doi: 10.1128/JVI.01912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter JM, Caron BL, Dogan A, Folpe AL. A novel chromogenic in situ hybridization assay for FGF23 mRNA in phosphaturic mesenchymal tumors. Am J Surg Pathol. 2015;39:75–83. doi: 10.1097/PAS.0000000000000290. [DOI] [PubMed] [Google Scholar]
- 21.Carossino M, Loynachan AT, MacLachlan NJ, Drew C, Shuck KM, Timoney PJ, Del Piero F, Balasuriya UB. Detection of equine arteritis virus by two chromogenic RNA in situ hybridization assays (conventional and RNAscope(a (R))) and assessment of their performance in tissues from aborted equine fetuses. Arch Virol. 2016;161:3125–3136. doi: 10.1007/s00705-016-3014-5. [DOI] [PubMed] [Google Scholar]
- 22.Patel K, Liu TC, Vaccharajani N, Chapman WC, Brunt EM. Characterization of inflammatory (lymphoepithelioma-like) hepatocellular carcinoma: a study of 8 cases. Arch Pathol Lab Med. 2014;138:1193–1202. doi: 10.5858/arpa.2013-0371-OA. [DOI] [PubMed] [Google Scholar]
- 23.Bowling AJ, Pence HE, Church JB. Application of a novel and automated branched DNA in situ hybridization method for the rapid and sensitive localization of mRNA molecules in plant tissues. Appl Plant Sci. 2014;2:1400011. doi: 10.3732/apps.1400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergua M, Phelan DM, Bak A, Bloom DC, Folimonova SY. Simultaneous visualization of two Citrus tristeza virus genotypes provides new insights into the structure of multi-component virus populations in a host. Virology. 2016;491:10–19. doi: 10.1016/j.virol.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Bishop R. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Biosci Horizons. 2010;3:85–95. doi: 10.1093/biohorizons/hzq009. [DOI] [Google Scholar]
- 26.Hsi BL, Xiao S, Fletcher JA. Chromogenic in situ hybridization and FISH in pathology. In: Walker JM, editor. Molecular Cytogenetics, Protocols and Applications, Methods in Molecular Biology, vol. 204. Humana Press; 2002. p. 343–351. [DOI] [PubMed]
- 27.Kliot A, Ghanim M. Fluorescent in situ hybridization for the localization of viruses, bacteria and other microorganisms in insect and plant tissues. Methods. 2016;98:74–81. doi: 10.1016/j.ymeth.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Ghanim M, Brumin M, Popovski S. A simple, rapid and inexpensive method for localization of tomato yellow leaf curl virus and potato leafroll virus in plant and insect vectors. J Virol Methods. 2009;159:311–314. doi: 10.1016/j.jviromet.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Cillo F, Roberts IM, Palukaitis P. In situ localization and tissue distribution of the replication-associated proteins of cucumber mosaic virus in tobacco and cucumber. J Virol. 2002;76:10654–10664. doi: 10.1128/JVI.76.21.10654-10664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao RM, Liu P, Wong SM. Identification of a plant viral RNA genome in the nucleus. PLoS One. 2012;7:e48736. doi: 10.1371/journal.pone.0048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gambino G, Vallania R, Gribaudo I. In situ localization of grapevine fanleaf virus and phloem-restricted viruses in embryogenic callus of Vitis vinifera. European J Plant Pathol. 2010;127:557–570. doi: 10.1007/s10658-010-9619-8. [DOI] [Google Scholar]
- 32.Horns T, Jeske H. Localization of Abutilon mosaic-virus (Abmv) DNA within leaf tissue by in situ hybridization. Virology. 1991;181:580–588. doi: 10.1016/0042-6822(91)90891-E. [DOI] [PubMed] [Google Scholar]
- 33.Kong LJ, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, Hanley-Bowdoin L. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 2000;19:3485–3495. doi: 10.1093/emboj/19.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latham JR, Saunders K, Pinner M, Stanley J. Induction of plant cell division by beet curly top virus gene C4. Plant J. 1997;11:1273–1283. doi: 10.1046/j.1365-313X.1997.11061273.x. [DOI] [Google Scholar]
- 35.Lucy AP, Boulton MI, Davies JW, Maule AJ. Tissue specificity of Zea mays infection by maize streak virus. Mol Plant-Microbe Interact. 1996;9:22–31. doi: 10.1094/MPMI-9-0022. [DOI] [Google Scholar]
- 36.Mochizuki T, Ohki ST. Detection of plant virus in meristem by immunohistochemistry and in situ hybridization. In: Uyeda I, Masuta C, editors. Plant virology protocols, methods in molecular biology. New York, NY: Humana Press; 2015. pp. 275–287. [DOI] [PubMed] [Google Scholar]
- 37.Shargil D, Zemach H, Belausov E, Lachman O, Kamenetsky R. Development of a fluorescent in situ hybridization (FISH) technique for visualizing CGMMV in plant tissues. J Virol Methods. 2015;223:55–60. doi: 10.1016/j.jviromet.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Wege C, Saunders K, Stanley J, Jeske H. Comparative analysis of tissue tropism of bipartite geminiviruses. J Phytopathol. 2001;149:359–368. doi: 10.1046/j.1439-0434.2001.00640.x. [DOI] [Google Scholar]
- 39.Rothenstein D, Krenz B, Selchow O, Jeske H. Tissue and cell tropism of Indian cassava mosaic virus (ICMV) and its AV2 (precoat) gene product. Virology. 2007;359:137–145. doi: 10.1016/j.virol.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Maidji E, Somsouk M, Rivera JM, Hunt PW, Stoddart CA. Replication of CMV in the gut of HIV-infected individuals and epithelial barrier dysfunction. PLoS Pathog. 2017;13:e1006202. doi: 10.1371/journal.ppat.1006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turkekul M, Barlas A, Yarilin D, Fujisawa S, Fan N, Brendel M, Manova-Todorova K. Automated double in situ detection of mouse Lgr5 mRNA and lysozyme protein in examining the neighboring cell types of the mouse intestinal crypt. Methods Mol Biol. 2017;1554:263–272. doi: 10.1007/978-1-4939-6759-9_19. [DOI] [PubMed] [Google Scholar]
- 42.Grabinski TM, Kneynsberg A, Manfredsson FP, Kanaan NM. A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS One. 2015;10:e0120120. doi: 10.1371/journal.pone.0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]