Abstract
Background
Bacterial endophytes are widespread inhabitants inside plant tissues that play crucial roles in plant growth and biotransformation. This study aimed to offer information for the exploitation of endophytes by analyzing the bacterial endophytes in different parts of Panax notoginseng.
Methods
We used high-throughput sequencing methods to analyze the diversity and composition of bacterial endophytes from different parts of P. notoginseng.
Results
A total of 174,761 classified sequences were obtained from the analysis of 16S ribosomal RNA in different parts of P. notoginseng. Its fibril displayed the highest diversity of bacterial endophytes. Principal coordinate analysis revealed that the compositions of the bacterial endophytes from aboveground parts (flower, leaf, and stem) differed from that of underground parts (root and fibril). The abundances of Conexibacter, Gemmatimonas, Holophaga, Luteolibacter, Methylophilus, Prosthecobacter, and Solirubrobacter were significantly higher in the aboveground parts than in the underground parts, whereas the abundances of Bradyrhizobium, Novosphingobium, Phenylobacterium, Sphingobium, and Steroidobacter were markedly lower in the aboveground parts.
Conclusions
Our results elucidated the comprehensive diversity and composition profiles of bacterial endophytes in different parts of 3-year-old P. notoginseng. Our data offered pivotal information to clarify the role of endophytes in the production of P. notoginseng and its important metabolites.
Electronic supplementary material
The online version of this article (10.1186/s13020-018-0198-5) contains supplementary material, which is available to authorized users.
Keywords: Panax notoginseng, Endophytes, Plant parts, High-throughput sequencing, 16S ribosomal RNA
Background
Panax notoginseng is renowned for its remarkable antihypertensive, antithrombotic, anti-atherosclerotic, and neuroprotective bioactivities, making it one of the most valuable ingredients in staple household medicines [1–3]. Protopanaxadiol and protopanaxatriol saponins are the main active compounds detected in the different parts of P. notoginseng [4, 5]. P. notoginseng is a perennial plant cultivated in fixed plots, and continuous cropping leads to decreased productivity, reduced tuber quality, and even seedling death [6, 7]. Approximately 8–10 years of crop rotation are necessary for improving the soil conditions for planted P. notoginseng [8]. P. notoginseng has a narrow ecological range, and its production mainly occurs in Wenshan, Yunnan Province, where the climatic and soil conditions are optimal for its cultivation. Nevertheless, arable soils available for P. notoginseng cultivation are becoming scarce.
Endophytic bacteria localized inside plant tissues have shown no negative effect on their host plants [9]. Bacteria inhabit different plant tissues, including rhizosphere, root, leaf, and stem [10]. Bacterial endophytes play key roles in improving plant growth, increasing tolerance against biotic factors, and producing secondary metabolites [11, 12]. Song et al. reported that endophytic Bacillus altitudinis isolated from Panax ginseng enhanced ginsenoside accumulation [13]. Gao et al. reported that Paenibacillus polymyxa isolated from P. ginseng leaves improved plant growth, increased ginsenoside concentration, and reduced morbidity [14]. Endophytes stimulated secondary metabolites and enhanced plant growth. Numerous works highlighted that the composition of the bacterial endophytes were influenced by plant species, parts, and growth stage [11, 15]. Analysis of the diversity and composition of endophytes in plant parts could provide valuable resources for plant growth promotion and biotransformation [16]. Although the diversity and composition of root endogenous bacteria in P. notoginseng have been described [17], limited information is available on the endophytic community in different parts of P. notoginseng. Thus, the diversity and composition of bacterial endophytes must be investigated to exploit the agronomical and metabolic potential of P. notoginseng.
Cultivation-independent methods can facilitate a rapid analysis of vast samples and provide reliable information on the diversity and composition of endophytic bacteria [18]. Many studies have explored rhizosphere and plant-associated bacterial communities by using high-throughput sequencing analysis [19–21]. Metagenetic methods for analyzing endophyte communities would provide deeper insight into the diversity and composition of bacterial endophytes, thereby leading to the potential discovery of new endophytes [22, 23]. Checcucci et al. reported the high taxonomic diversity of bacterial endophytes in the leaves of Thymus spp. by using 16S rRNA gene metagenomic sequencing [24]. A total of 29 culturable bacterial endophytes have been identified in the tissues of Aloe vera and characterized to 13 genera [25]. Nevertheless, culture-dependent biodiversity studies on endophytic bacterial communities remain scarce [19]. Pyrosequencing could detect low-abundance bacteria in leaf salad vegetables that could not be identified by culture-dependent methods [26]. Additional, high-throughput sequencing also used in the analysis of soil microbial communities, and this method effectively revealed the changes in diversity of soil microbial communities in soils during the cultivation of Panax plants [21, 27, 28]. In the present study, high-throughput sequencing analysis of 16S ribosomal RNA (rRNA) genes was conducted to describe the diversity and composition of associations among different parts of P. notoginseng. The results clarified the tissue-wise diversity of bacterial endophytes in the samples collected from P. notoginseng as well as expanded the knowledge on plant–microbe relationships and the potential properties for plant growth promotion and biotransformation.
Methods
Processing of samples
Three-year-old P. notoginseng plants were collected from Wenshan, Yunnan Province, China, which is the main production area of P. notoginseng. These plant samples were used to analyze the bacterial endophytes in different parts of P. notoginseng. Six plants were randomly gathered from one plantation and served as one sample in our test sites of Wenshan Miaoxiang Notoginseng Technology, Co., Ltd. in August. There were three replicates from three plantations. The flowers (Fl), leaves (Le), stems (St), roots (Ro), and fibers (Fi) of all samples were separated, washed with running tap water, and rinsed thrice with distilled water. A single sample consisted of 1 g of each part from six plants as one sample. To sterilize the surface of the plant parts, the samples from each part were successively immersed in 70% ethanol for 5 min, 2.5% sodium hypochlorite for 1–2 min, and 70% ethanol for 1 min, and then rinsed five times with sterile Millipore water. The last portion of the washing water was inoculated in Luria–Bertani agar at 37 °C for 24 h to validate sterilization efficiency. A total of 15 samples were stored at − 80 °C until DNA extraction.
DNA extraction, polymerase chain reaction (PCR) amplification, and sequence processing
The total genomic DNA was extracted from all plant parts by using the MOBIO PowerSoil® Kit (MOBIO Laboratories, Inc., Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The DNA quality of each sample was confirmed by utilizing a NanoDrop spectrophotometer (Thermo Fisher Scientific, Model 2000, MA, USA) and stored at − 20 °C for further PCR amplification. Bacterial 16S rRNA V1 hypervariable region genes were amplified by using the universal primers 27F/338R [29]. The forward and reverse primers contained an 8 bp barcode (Additional file 1: Table S1). PCRs were performed as described by Dong et al. with slight modifications [21]. The reaction systems were denatured at 94 °C for 3 min and then amplified for 25 cycles at 94 °C for 45 s, 55 °C for 30 s, and 72 °C for 60 s. A final extension of 10 min was added at the end of the program. Negative controls (no templates) were included to check DNA contamination of the primer or the sample. PCR products from each sample were separated with 1% agarose gel, purified with a MinElute Gel Extraction Kit (Qiagen, Valencia, CA, USA), and quantified with a Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). The amplicons were pooled in equimolar ratios. The amplicon libraries were paired-end sequenced (2 × 250) by using an Illumina MiSeq platform in accordance with the manufacturer’s protocol.
Data analysis
The data were processed by utilizing the QIIME pipeline [30]. Bacterial sequences were trimmed and assigned to each sample based on their barcodes. Sequences were binned into operational taxonomic units (OTUs) at 97% similarity level by using UPARSE (version 7.1 http://drive5.com/uparse/). Chimeric sequences were identified and removed by using UCHIME. The phylogenetic affiliation of each 16S rRNA gene sequence was analyzed by using a RDP Classifier (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database at a confidence threshold of 70% [31]. Rarefaction analysis based on Mothur v.1.21.1 was conducted to reveal the diversity indices, including Chao 1 and Shannon [32]. Principal coordinate analysis (PCoA) was performed to examine dissimilarities in the community composition among samples on the basis of Bray–Curtis distance metrics [33]. Statistical analyses were performed by using the R package [34].
Statistical analyses
SPSS version 16.0 was used for the statistical analyses (SPSS Inc., Chicago, IL, USA). The data were presented as mean ± SD of n = 3. No adjustments were implemented for multiple comparisons. The parameters were obtained for all treatment replicates and subjected one-way ANOVA.
The Minimum Standards of Reporting Checklist contains details of the details of the experimental design, and statistics, and resources used in this study (Additional file 2).
Results
Alpha diversity of bacterial endophytes among P. notoginseng parts
A total of 174,761 reads, with an average of 11,650 sequences per sample, were obtained from 15 samples through high-throughput sequencing analyses of 16S rRNA gene sequences (Table 1). A total of 10,351 OTUs, which ranged from 254 to 964, were found in all sequences. Alpha diversity indices (Chao 1 and Shannon) presented differences among the plant parts of P. notoginseng (Fig. 1). Chao 1 indicated a high number of species in the fibril samples and a low number of species in the leaf and root samples (Fig. 1A). H’ revealed that the fibril samples had the highest diversity, whereas the root sample had the lowest diversity (Fig. 1B). The flower and stem displayed similar diversity levels.
Table 1.
Bacterial numbers of sequences and OUTs in each sample
| Samples | Sequences | OTUs |
|---|---|---|
| Flower-1 | 10,784 | 877 |
| Flower-2 | 6818 | 657 |
| Flower-3 | 23,711 | 908 |
| Leaf-1 | 666 | 254 |
| Leaf-2 | 7279 | 678 |
| Leaf-3 | 806 | 256 |
| Stem-1 | 22,538 | 849 |
| Stem-2 | 2683 | 533 |
| Stem-3 | 18,277 | 817 |
| Root-1 | 4402 | 388 |
| Root-2 | 12,424 | 726 |
| Root-3 | 5944 | 676 |
| Fibril-1 | 18,206 | 922 |
| Fibril-2 | 12,035 | 846 |
| Fibril-3 | 28,188 | 964 |
-1, -2, and -3 present three replicates of each P. notoginseng part
Fig. 1.
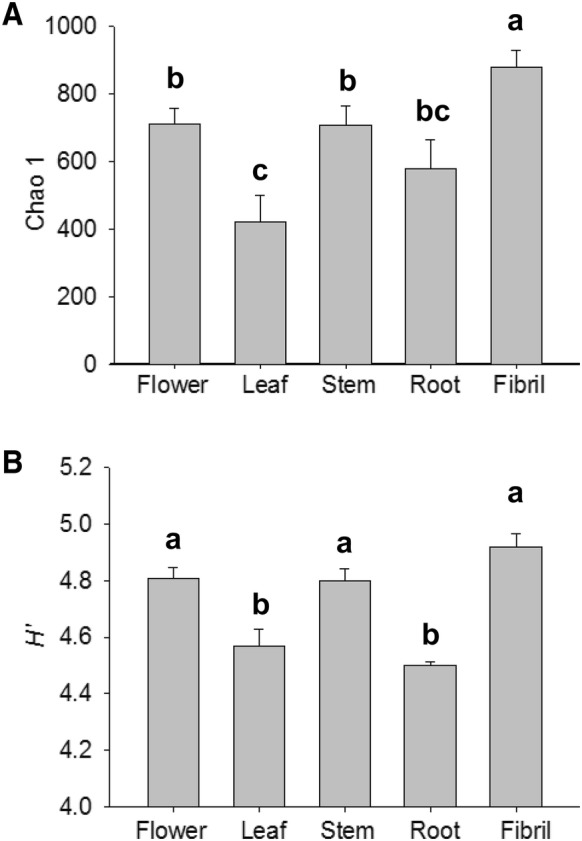
Changes of Chao 1 (A) and Shannon indices (B) for the 16S rRNA gene sequences from samples of P. notoginseng parts. The value of each bar represents the mean ± SD of n = 3. Different letters indicated significant difference at the 0.05 level
Beta diversity of bacterial endophytes among P. notoginseng parts
The Bray–Curtis dissimilarity matrix was calculated to differentiate the bacterial communities among P. notoginseng parts (Fig. 2). PCoA ordination showed a strong clustering of the bacterial communities of underground (root and fibril) and aboveground parts (flower, leaf, and stem). The first and second principal components explained 50.79 and 19.3% of the total variation. PCoA indicated that the samples collected from the aboveground parts had similar profiles, whereas the samples from the underground parts were clustered together at opposite sides of the plots of the aboveground parts. Bacterial diversity in the aboveground parts differed from that in the underground parts.
Fig. 2.
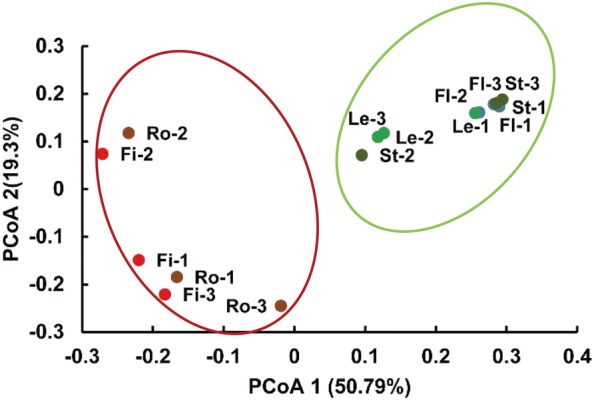
PCoA ordination plots of bacterial endophytes based on the Bray–Curtis distances of the classified 16S rRNA gene sequences. Fl, Le, St, Ro, and Fi denote the flower, leaf, stem, root, and fibril samples, respectively. -1, -2, and -3 indicate the first, second, and third replicates of each P. notoginseng part, respectively
Composition of bacterial endophytes among P. notoginseng parts
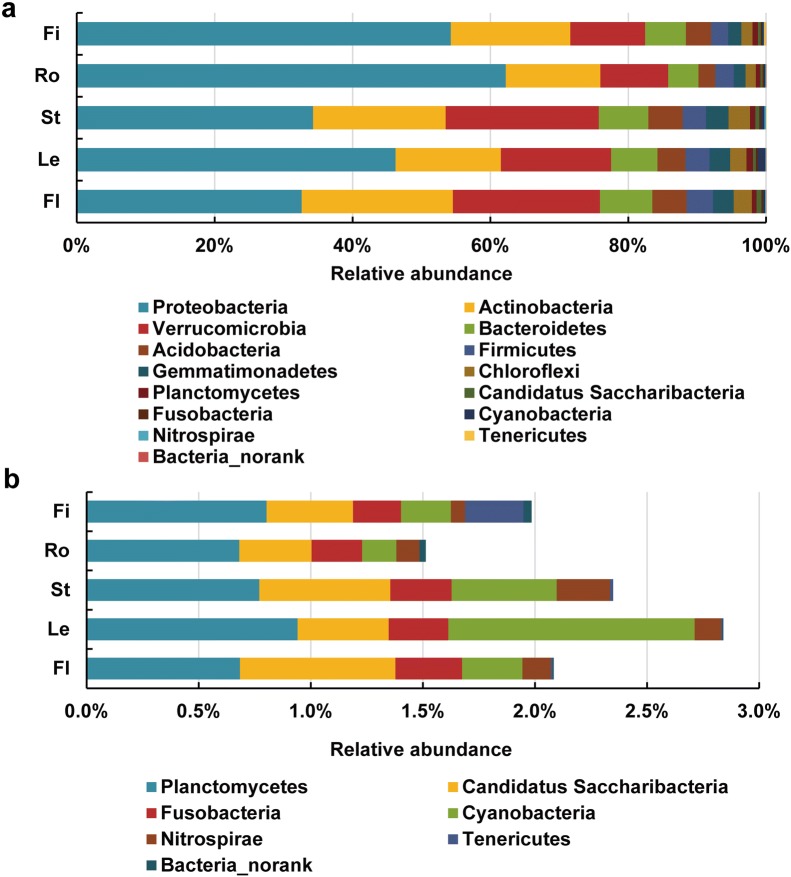
The reads from the 16S rRNA amplicon sequences that were generated from all samples mostly belonged to the 14 phyla (Fig. 3). The relative abundances of Proteobacteria, Actinobacteria, Verrucomicrobia, Bacteroidetes, Acidobacteria, Firmicutes, Gemmatimonadetes, and Chloroflexi reached 97.9, 97.2, 97.6, 98.2, and 97.7% in the samples from the flower, leaf, stem, root, and fibril, respectively (Fig. 3a). The relative abundance of Proteobacteria in the underground parts was significantly higher than in the aboveground parts. The abundance of Verrucomicrobia in the aboveground parts was markedly higher than in the underground parts. The relative abundances (< 1%) of Candidatus Saccharibacteria, Fusobacteria, Cyanobacteria, and Nitrospirae in the aboveground parts were higher than those in the underground parts (Fig. 3b). The relative abundance of bacterial endophytes varied and depended on the P. notoginseng parts at the phylum level.
Fig. 3.
Relative abundances of the bacterial community at the phylum level. a All bacterial endophytes at the phylum level. b Bacterial endophytes with low relative abundance (< 1%) at the phylum level. Fl, Le, St, Ro, and Fi refer to the flower, leaf, stem, root, and fibril samples, respectively. The value of each bar represents the mean of n = 3
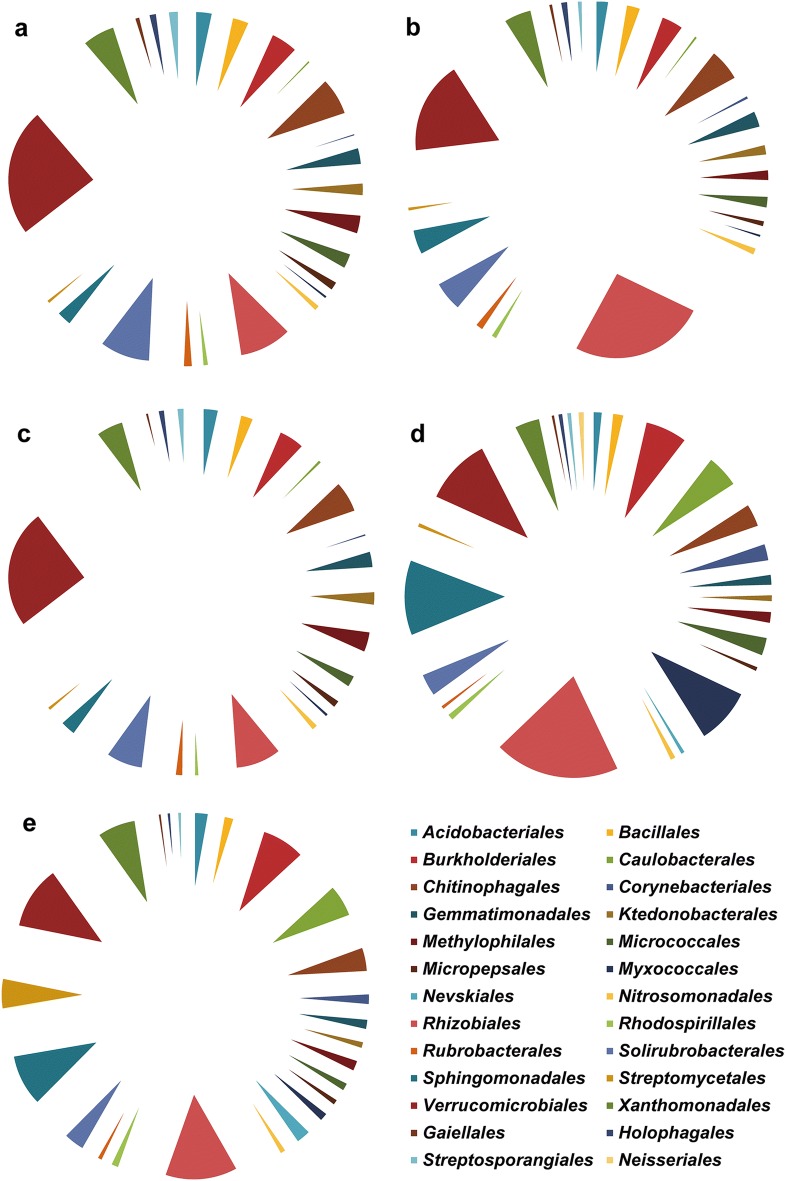
The relative abundances of the bacterial endophytes in the aboveground and underground parts showed considerable variation at the order level (Fig. 4 and Additional file 1: Table S2). The relative abundances of Bacillales, Chitinophagales, Gemmatimonadales, Solirubrobacterales, and Verrucomicrobiales in the aboveground parts were markedly higher than in the underground parts (P < 0.05). The abundances of Burkholderiales, Caulobacterales, Corynebacteriales, Myxococcales, and Sphingomonadales in the underground parts were significantly higher than those in the aboveground parts (P < 0.05). The bacterial endophytes in the samples from the aboveground parts with higher abundance at the order level were Chitinophagales, Rhizobiales, Solirubrobacterales, and Verrucomicrobiales. The bacterial endophytes in the samples from the underground parts with higher abundance at the order level were Burkholderiales, Rhizobiales, Sphingomonadales, and Verrucomicrobiales.
Fig. 4.
Relative abundances of bacterial community (> 1%) at the order level. a–e Abundances of the bacterial endophytes from Fl, Le, St, Ro, and Fi. The value of each bar represents the mean of n = 3
Heat map analysis of the relative abundances of bacterial endophytes at the genus level showed variations in the samples from the aboveground and underground parts (Fig. 5). The relative abundances of Conexibacter, Gemmatimonas, Holophaga, Luteolibacter, Methylophilus, Prosthecobacter, and Solirubrobacter in the aboveground parts were significantly higher than those in the underground parts (P < 0.05). The abundances of Bradyrhizobium, Novosphingobium, Phenylobacterium, Sphingobium, and Steroidobacter in the aboveground parts were markedly lower than those in the underground parts (P < 0.05). Among all samples, Prosthecobacter had the highest abundance at the genus level. The low abundance (< 1.0%) of bacterial endophytes was related to the samples from P. notoginseng parts at the genus level (Additional file 1: Table S3). The relative abundances of Agrobacterium, Sphingobium and Shinella in the root samples were significantly higher than in the fibril samples (P < 0.05). By contrast, the abundances of Burkholderia and Steroidobacter in the root samples were markedly lower than in the fibril samples (P < 0.05).
Fig. 5.
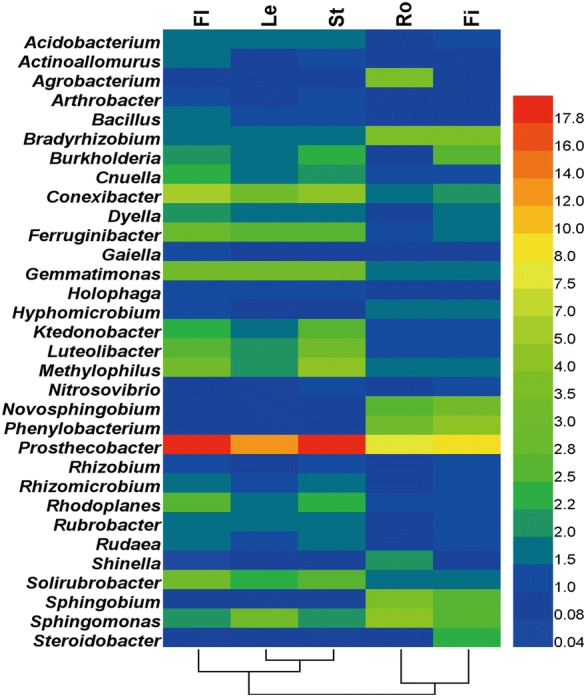
Relative abundances of the main bacterial community at the genus level. Bacterial endophytes with relative abundance (> 1%) in one treatment are shown. Fl, Le, St, Ro, and Fi refer to the flower, leaf, stem, root, and fibril samples, respectively. The value of each bar represents the mean of n = 3
Discussion
In this study, the diversity of bacterial endophytes was associated with the different parts of P. notoginseng. Chao 1 and H’ indices indicated that the fibril samples had the highest diversity among all of the samples from the different parts. Chao 1 revealed that Stellera chamaejasme L. displayed an increasing trend in species richness from the root samples to the stem and leaf samples [19]. The OTUs of the bacterial endophytes were randomly distributed among plant species and organs, and Chao 1 also revealed that the diversity of Santiria apiculate and Rothmannia macrophylla in the root samples was higher than that in the leaf samples [35]. PCoA showed that samples from the aboveground parts were distinguishable from those from the underground parts. Principal component analysis (PCA) revealed that the leaf and stem samples of S. chamaejasme L. were clustered together and were different from the plots for the root [19], and that the stem and leaf samples of poplar trees were distinguishable from the root samples [10]. The bacterial endophytes from the fibril had the highest diversity.
In this study, Proteobacteria, Actinobacteria, Verrucomicrobia, Bacteroidetes, Acidobacteria, and Firmicutes were the main bacterial communities in P. notoginseng plants. A previous study detected Proteobacteria, Actinobacteria, Bacteroidetes, and Acidobacteria in the P. notoginseng root [17]. Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes were found in P. ginseng roots by using a culture-dependent method [16]. More than 300 endophytic actinobacteria and bacteria belonging to Rhodococcus, Brevibacterium, Nocardioides, Streptomyces, Microbacterium, Nocardiopsis, Brachybacterium, Tsukamurella, Arthrobacter, and Pseudonocardia were isolated from different tissues of Dracaena cochinchinensis L. [12]. The plant species influenced the selection of endophytes. The plant parts of P. notoginseng represented the ecological niches for bacterial endophytes.
The composition of bacterial endophytes from the aboveground parts varied from that of the underground parts. The composition of bacterial endophytes was associated with the plant compartments [10]. The relative abundances of the bacterial endophytes in all samples showed considerable variations at the phylum and genus levels [19]. The relative abundances of the bacterial endophytes, including Conexibacter, Gemmatimonas, Holophaga, Luteolibacter, Methylophilus, Prosthecobacter, Solirubrobacter, Bradyrhizobium, Novosphingobium, Phenylobacterium, Sphingobium, and Steroidobacter, in the aboveground and underground parts differed significantly. Evident strains are Gemmatimonas, Bradyrhizobium, Novosphingobium, and Sphingobium, which can solubilize insoluble elements, induce plant stress resistance or produce antifungal antibiotics [36–39]. Endophytic Bacillus altitudinis served as elicitors of biomass and ginsenoside production [13]. Bacterial endophytes from Zea displayed anti-fungal activity against two fungal pathogens [40]. Li et al. have reported that the domain genera included Rhizobium, Sulfurospirillum, Uliginosibacterium, Pseudomonas, Aeromonas and Bacteroides, all of which could fix nitrogen and improve plant growth [41]. The fungal endophytes communities in Monarda citriodora expressed anticancer and antimicrobial activities [42]. In view of the roles played by endophytes in plant growth and biotransformation, our findings contribute to the expansion of endophyte use in the production of P. notoginseng and its important metabolites. The information on the differences of endophytes in the aboveground and underground parts can serve as basis for the selection of functional bacteria. Importantly, higher saponins contents were detected in harvest 3-year-old P. notoginseng plants [43, 44]. Endophytes increased ginsenoside concentration and reduced morbidity [13, 14]. Thus, 3-year-old P. notoginseng plants served as the proper samples to analyze the endophytes. Additionally, the diversity of bacterial endophytes showed richness than fungal endophytes or exogenous bacteria in hour study (data not shown), and we focused on bacterial endophytes in different parts of P. notoginseng.
Conclusions
The diversity and composition of bacterial endophytes were associated with different plant parts of P. notoginseng, and bacterial endophytes from aboveground parts (flower, leaf, and stem) were distinguished from those from underground parts (root and fibril). Our results described the profiles of bacterial endophytes in P. notoginseng parts and provided insight into the exploitation of endophytes in the production of P. notoginseng and its important metabolites.
Additional files
Additional file 1: Table S1. Barcodes used to tag the PCR products. Table S2. Relative abundance (<1.0%) of the bacterial taxa at the order level. Table S3. Relative abundance (<1.0%) of the bacterial taxa at the genus level.
Additional file 2. Minimum Standards of Reporting Checklist.
Authors’ contributions
LD designed the work, analyzed the data, and wrote the manuscript. RC analyzed the data and collected samples. LX, GW and FW performed the experiment. JX and XG analyzed the data. YW and ZC performed the field experiment. SC designed the work and wrote this manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Jun Qian for analysis of 16S rRNA gene sequences.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The 16S rRNA sequences of endophytes used in this manuscript have been submitted to the NCBI and the Accession number is PRJNA416832. Most of the data generated of analyzed during the study are included in this article and its Additional file 1.
Consent to publish
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by the grants from the Fundamental Research Funds for the Central public welfare research institutes (No. ZXKT17049), Beijing Nova Program (No. Z181100006218020), the Major Science and Technology Programs in Yunnan Province (No. 2016ZF001-001), and the Science and Technology Project of Yantai (No. 2015ZH071).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Fl
flower
- Le
leaf
- St
stem
- Ro
root
- Fi
fibril
- rRNA
ribosomal RNA
- PCR
polymerase chain reaction
- PCoA
principal coordinates analysis
- RDP
ribosomal database project
- OTUs
operational taxonomic units
- H’
Shannon index
Contributor Information
Linlin Dong, Email: lldong@icmm.ac.cn.
Ruiyang Cheng, Email: rycheng@icmm.ac.cn.
Lina Xiao, Email: 425113948@qq.com.
Fugang Wei, Email: weifugang@live.com.
Guangfei Wei, Email: 874101744@qq.com.
Jiang Xu, Email: jxu@icmm.ac.cn.
Yong Wang, Email: ws-wangyong37@163.com.
Xiaotong Guo, Email: guoxtina@126.com.
Zhongjian Chen, Email: panaxnotoginseng@126.com.
Shilin Chen, Phone: 0086-10-57203877, Email: slchen@icmm.ac.cn.
References
- 1.Ng T. Pharmacological activity of sanchi ginseng (Panax notoginseng) J Pharm Pharmacol. 2006;58:1007–1019. doi: 10.1211/jpp.58.8.0001. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Cheng Y. Solid-phase extraction and liquid chromatography-electrospray mass spectrometric analysis of saponins in a Chinese patent medicine of formulated Salvia miltiorrhizae and Panax notoginseng. J Pharmaceut Biomed. 2006;40:429–432. doi: 10.1016/j.jpba.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Wang L, Chen X, Deng X, Cao Y, Wang Q. Simultaneous quantification of both triterpenoid and steroidal saponins in various Yunnan Baiyao preparations using HPLC-UV and HPLC-MS. J Sep Sci. 2008;31:3834–3846. doi: 10.1002/jssc.200800382. [DOI] [PubMed] [Google Scholar]
- 4.Du Q, Jerz G, Waibel R, Winterhalter P. Isolation of dammarane saponins from Panax notoginseng by high-speed countercurrent chromatography. J Chromatogr A. 2003;1008:173–180. doi: 10.1016/S0021-9673(03)00988-9. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Yang Z, Ye Y. Structure and biological activity of protopanaxatriol-type saponins from the roots of Panax notoginseng. Int Immunopharmac. 2006;6:14–25. doi: 10.1016/j.intimp.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Hu Z, You C, Zhang T. Discussing the obstacles caused by continuous notoginseng cropping. J Wenshan Univ. 2011;24:6–11. [Google Scholar]
- 7.Liu L, Liu D, Jing H, Feng G, Zhang J, Wei M, et al. Overview on the mechanisms and control methods of sequential cropping obstacle of Panax notoginseng F. H. Chen. J Mountain Agri Biol. 2011;30:70–75. [Google Scholar]
- 8.Ma C, Li S, Gu Z, Chen Y. Measures of integrated control of root rot complex of continuous cropping Panax notoginseng and their control efficacy. Acta Agri Shanghai. 2006;22:63–68. [Google Scholar]
- 9.Larousse M, Rancurel C, Syska C, Palero F, Etienne C, Industri B, et al. Tomato root microbiota and Phytophthora parasitica-associated disease. Microbiome. 2017;5:56. doi: 10.1186/s40168-017-0273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckers B, De Beek MO, Weyens N, Boerjan W, Vangronsveld J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome. 2017;5:25. doi: 10.1186/s40168-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan SED. Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J Adv Res. 2017;8:687–695. doi: 10.1016/j.jare.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh M, Kumar A, Singh R, Pandey K. Endophytic bacteria: a new source of bioactive compounds. Biotech. 2017;7:315. doi: 10.1007/s13205-017-0942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song X, Wu H, Yin Z, Lian M, Yin C. Endophytic bacteria isolated from Panax ginseng improved ginsenoside accumulation in adventitious ginseng root culture. Molecules. 2017;22:837. doi: 10.3390/molecules22060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Liu Q, Zang P, Li X, Ji Q, He Z, et al. An endophytic bacterium isolated from Panax ginseng C.A. Meyer enhances growth, reduces morbidity, and stimulates ginsenoside biosynthesis. Phytochem Lett. 2015;125:132–138. doi: 10.1016/j.phytol.2014.12.007. [DOI] [Google Scholar]
- 15.Van Overbeek L, van Elsas J. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.) FEMS Microbiol Ecol. 2008;64:283–296. doi: 10.1111/j.1574-6941.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury E, Jeon J, Rim S, Park Y, Lee S, Bae H. Composition, diversity and bioactivity of culturable bacterial endophytes in mountain-cultivated ginseng in Korea. Sci Rep. 2017;7:10098. doi: 10.1038/s41598-017-10280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, et al. Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous cropping practices. J Basic Microb. 2016;9999:1–8. doi: 10.1002/jobm.201600464. [DOI] [PubMed] [Google Scholar]
- 18.Bodenhausen N, Horton M, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H, Yang X, Yan Z, Liu Q, Li X, Chen J, et al. Characterization of rhizosphere and endophytic bacterial communities from leaves, stems and roots of medicinal Stellera chamaejasme L. Syst Appl Microbiol. 2014;37:376–385. doi: 10.1016/j.syapm.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Dong L, Xu J, Feng G, Li X, Chen S. Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Sci Rep. 2016;6:31802. doi: 10.1038/srep31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L, Xu J, Zhang L, Yang J, Liao B, Li X, et al. High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin Med. 2017;12:18. doi: 10.1186/s13020-017-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wani Z, Ashraf N, Mohiuddin T, Riyaz-Ul-Hassan S. Plant-endophyte symbiosis, an ecological perspective. Appl Microbiol Biotechnol. 2015;99:2955–2965. doi: 10.1007/s00253-015-6487-3. [DOI] [PubMed] [Google Scholar]
- 23.Kaul S, Sharma T, Dhar M. “Omics” tools for better understanding the plant-endophyte interactions. Front Plant Sci. 2016;7:955. doi: 10.3389/fpls.2016.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Checcucci A, Maida I, Bacci G, Ninno C, Bilia A, Biffi S, et al. Is the plant-associated microbiota of Thymus spp. adapted to plant essential oil? Res Microbiol. 2016;3:276–287. doi: 10.1016/j.resmic.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Akinsanya M, Goh J, Lim S, Ting A. Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol Lett. 2015;362:184. doi: 10.1093/femsle/fnv184. [DOI] [PubMed] [Google Scholar]
- 26.Jackson C, Randolph K, Osborn S, Tyler H. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013;13:274. doi: 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong L, Xu J, Zhang L, Cheng R, Wei G, Su H, Yang J, Qian J, Xu R, Chen S. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm Sin B. 2018;8:272–282. doi: 10.1016/j.apsb.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong L, Xu J, Li Y, Fang H, Niu W, Li X, et al. Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol Biochem. 2018;125:64–74. doi: 10.1016/j.soilbio.2018.06.028. [DOI] [Google Scholar]
- 29.Fierer N, Jackson J, Vilgalys R, Jackson R. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso J, Bittinger K, Bushman F, DeSantis T, Andersen G, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Phylogenetics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katherine R, Carl J, Angela K, Nicoletta R, Franck C, Alejandro E, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister E, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Micro. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C, Lladser M, Dan K, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010;5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksanen J, Blanchet F, Kindt R, Simpson G. Vegan: community ecology package. Version 2.0-10. J Stat Softw. 2011;48:1–21. [Google Scholar]
- 35.Haruna E, Zin N, Kerfahi D, Adams J. Extensive overlap of tropical rainforest bacterial endophytes between soil, plant parts, and plant species. Microb Ecol. 2017 doi: 10.1007/s00248-017-1002-2. [DOI] [PubMed] [Google Scholar]
- 36.Mahanty T, Bhattacharjee S, Goswami M, Bhattacharyya P, Das B, Ghosh A, Tribedi P. Biofertilizers: a potential approach for sustainable agriculture development. Environ Sci Pollut Res Int. 2017;24:3315–3335. doi: 10.1007/s11356-016-8104-0. [DOI] [PubMed] [Google Scholar]
- 37.Berendsen RL, Pieterse CMJ, Bakker PHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Egamberdiyeva D. The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl Soil Ecol. 2007;36:184–189. doi: 10.1016/j.apsoil.2007.02.005. [DOI] [Google Scholar]
- 39.Verma JP, Yadav J, Tiwari KN, Lavakush SV. Impact of plant growth promoting rhizobacteria on crop production. Int J Agri Res. 2010;5:954–983. doi: 10.3923/ijar.2010.954.983. [DOI] [Google Scholar]
- 40.Shehata H, Lyons E, Jordan K, Raizada M. Relevance of in vitro agar based screens to characterize the anti-fungal activities of bacterial endophyte communities. BMC Microbiol. 2016;16:8. doi: 10.1186/s12866-016-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Liu Q, Liu Y, Zhu J, Zhang Q. Endophytic bacterial diversity in roots of Typha angustifolia L. in the constructed Beijing Cuihu Wetland (China) Res Microbiol. 2011;162:124–131. doi: 10.1016/j.resmic.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Katoch M, Phull S, Vaid S, Singh S. Diversity, Phylogeny, anticancer and antimicrobial potential of fungal endophytes associated with Monarda citriodora L. BMC Microbiol. 2017;17:44. doi: 10.1186/s12866-017-0961-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei G, Dong L, Yang J, Zhang L, Xu J, Yang F, et al. Integrated metabolomic and transcriptomic analyses revealed the distribution of saponins in Panax notoginseng. Acta Pharm Sin B. 2018;8:458–465. doi: 10.1016/j.apsb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Dong L, Wei G, Hu H, Zhu G, Zhang J, et al. Identification and quality of Panax notoginseng and Panax vietnamensis var. fuscidicus through integrated DNA barcoding and HPLC. Chin Herbal Med. 2018;10:177–183. doi: 10.1016/j.chmed.2018.03.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Barcodes used to tag the PCR products. Table S2. Relative abundance (<1.0%) of the bacterial taxa at the order level. Table S3. Relative abundance (<1.0%) of the bacterial taxa at the genus level.
Additional file 2. Minimum Standards of Reporting Checklist.
Data Availability Statement
The 16S rRNA sequences of endophytes used in this manuscript have been submitted to the NCBI and the Accession number is PRJNA416832. Most of the data generated of analyzed during the study are included in this article and its Additional file 1.