Summary
Endothelial cells (ECs) are of great value for cell therapy, tissue engineering, and drug discovery. Obtaining high-quantity and -quality ECs remains very challenging. Here, we report a method for the scalable manufacturing of ECs from human pluripotent stem cells (hPSCs). hPSCs are expanded and differentiated into ECs in a 3D thermoreversible PNIPAAm-PEG hydrogel. The hydrogel protects cells from hydrodynamic stresses in the culture vessel and prevents cells from excessive agglomeration, leading to high-culture efficiency including high-viability (>90%), high-purity (>80%), and high-volumetric yield (2.0 × 107 cells/mL). These ECs (i.e., 3D-ECs) had similar properties as ECs made using 2D culture systems (i.e., 2D-ECs). Genome-wide gene expression analysis showed that 3D-ECs had higher expression of genes related to vasculature development, extracellular matrix, and glycolysis, while 2D-ECs had higher expression of genes related to cell proliferation.
Key words: human pluripotent stem cells, endothelial cells, 3D culture, thermoreversible hydrogel
Graphical Abstract
Highlights
-
•
hPSCs can be differentiated into endothelial cells in 3D thermoreversible hydrogels
-
•
The differentiation efficiency is similar to this in 2D cultures
-
•
The global gene expression and phenotypes are similar to ECs made in 2D cultures
Endothelial cells (ECs) differentiated from human pluripotent stem cells (hPSCs) are needed in large numbers for regenerative medicine. In this article, Lei and colleagues describe a simple and innovative method for the scalable production of ECs from hPSCs. The method has potential to make ECs broadly available and affordable for various applications.
Introduction
Endothelial cells (ECs) are major components of blood vessels (Carmeliet, 2001, Richards et al., 2010). They are of great value for disease modeling, drug screening, cell therapy, and tissue engineering (Heo et al., 2014, Huang et al., 2013, Kang et al., 2013, Leung et al., 2016, Medina et al., 2010, Moubarik et al., 2011, Patsch et al., 2015, Schwarz et al., 2012, Stroncek et al., 2012). However, obtaining large numbers of primary ECs for those applications, in particular for clinical applications (Arici et al., 2015, Chao et al., 2014, den Dekker et al., 2011, Granton et al., 2015, Matoba et al., 2008), is still challenging due to their limited proliferation capacity and phenotype changes during the in vitro culture (van Beijnum et al., 2008, de Carvalho et al., 2015, Gui et al., 2009, Gumbleton and Audus, 2001, Hayflick, 1965, Augustin-Voss et al., 1993). Human pluripotent stem cells (hPSCs) provide a potential solution to this challenge (Levenberg et al., 2007). hPSCs, including human embryonic stem cells (hESCs) (Thomson et al., 1998) and induced pluripotent stem cells (iPSCs) (Takahashi et al., 2007, Yu et al., 2007), have unlimited proliferation capacity and can be efficiently differentiated into ECs through 3D embryonic body (EB)-based (Condorelli et al., 2001, James et al., 2010, Levenberg et al., 2002, Levenberg et al., 2007, Li et al., 2009a, Li et al., 2009b, Nourse et al., 2010) or 2D monolayer culture-based protocols (Cao et al., 2013, Kane et al., 2010, Palpant et al., 2016, Patsch et al., 2015, Vodyanik et al., 2005). In addition, cells derived from patient-specific iPSCs have the patient's genetic information and can model many human diseases. Further, they induce minimal immune response in vivo (Lalit et al., 2014). These hPSC-derived ECs have the potential to provide unlimited cell sources for the applications.
While making small-scale hPSC-derived ECs in laboratories can be readily done (Giacomelli et al., 2017, Lian et al., 2014, Orlova et al., 2014, Palpant et al., 2016, Zhang et al., 2017a), generating or manufacturing large numbers of ECs from hPSCs has not been achieved. Current 2D culture methods, in which cells are cultured as adherent cells on 2D surfaces (e.g., cell culturing flasks), are labor, time, and cost intensive, and not suitable for culturing cells on a large scale (Jenkins and Farid, 2015, Kropp et al., 2017). 3D suspension culture methods (e.g., using stirred-tank bioreactors), in which cells are suspended in an agitated culture medium, have been considered a potential solution for scaling up the cell production (Jenkins and Farid, 2015, Kropp et al., 2017, Lei and Schaffer, 2013). However, recent research has shown that culturing cells on a large scale with 3D suspension cultures is also very challenging (Lei et al., 2014, Serra et al., 2012, Steiner et al., 2010, Wurm, 2004). hPSCs in 3D suspension cultures frequently aggregate to form large cell agglomerates (Kropp et al., 2017). The mass transport to cells located at the center of large agglomerates (e.g., >400 μm diameter) becomes difficult, leading to slow cell growth, cell death, and uncontrolled differentiation (Kropp et al., 2017). While agitating the culture can reduce cell agglomeration, it also generates hydrodynamic stresses, which are adverse to the cell's physiology (Fridley et al., 2012, Kinney et al., 2011, Kropp et al., 2017). As a result, 3D suspension culturing has significant cell death, low cell growth, and low volumetric yield (Lei and Schaffer, 2013). For instance, hPSCs typically expand 4-fold in 4 days to yield around 1.0 × 106 to 2.0 × 106 cells/mL, which occupy ∼0.4% of the bioreactor volume (Lei et al., 2014, Serra et al., 2012, Steiner et al., 2010, Wurm, 2004).
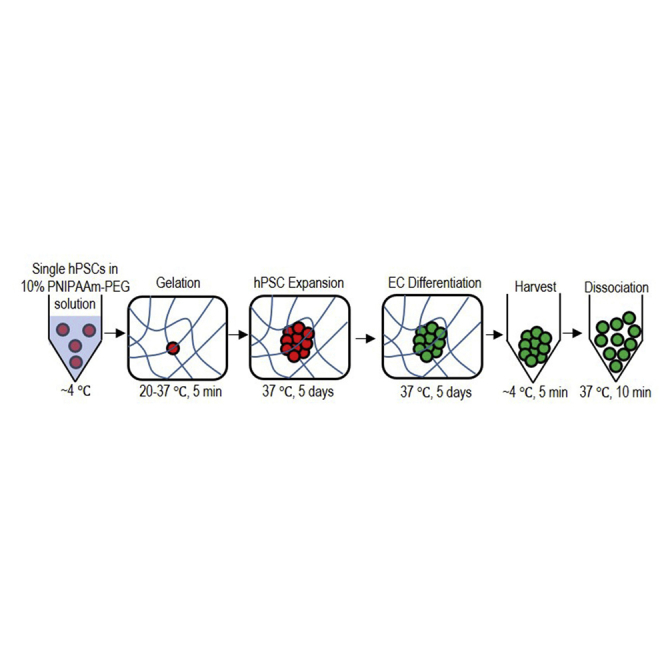
To address the challenge, we previously developed a scalable, efficient, and current Good Manufacturing Practice (cGMP)-compliant method for expanding hPSCs (Lei and Schaffer, 2013, Li et al., 2016, Lin et al., 2017). The method, which was successfully repeated in this study (Figures 1 and S2), uses a 3D thermoreversible hydrogel (Mebiol Gel) as the scaffold. Single hPSCs are first suspended in a liquid PNIPAAm-PEG polymer solution at low temperature (e.g., 4°C). Upon heating to 20°C–37°C, the polymer solution forms an elastic hydrogel matrix, resulting in single hPSCs encapsulated in the hydrogel matrix. After culturing for about 4–5 days, these single hPSCs clonally grow into spherical cell aggregates (spheroids) with very uniform size (Figures 1B, S2A, and S2D). The hydrogel can be quickly liquefied through cooling to ∼4°C to harvest the cells for the next passage (Figure 1A). The hydrogel scaffold protects cells from hydrodynamic stresses in the culture vessel and prevents cells from excessive agglomeration, leading to high culture efficiency. For instance, the hydrogel scaffold enables long-term, serial expansion of hPSCs with a high cell viability (e.g., >90%, Figures 1D, S2C, and S2F), growth rate (e.g., 20-fold/5days, Figure 1E), yield (e.g., 2.0 × 107 cells/mL, Figure 1F), and purity (>99%, Figure 1C, S2B, and S2E), all of which offer considerable improvements over 3D suspension cultures (Lei and Schaffer, 2013, Li et al., 2016, Lin et al., 2017). We hypothesize that hPSCs can also be differentiated into ECs in this culture system. In this paper, we successfully tested the hypothesis. Together, we developed a scalable bioprocess for making high-quality ECs with high volumetric yield, high viability, and high purity (>80%).
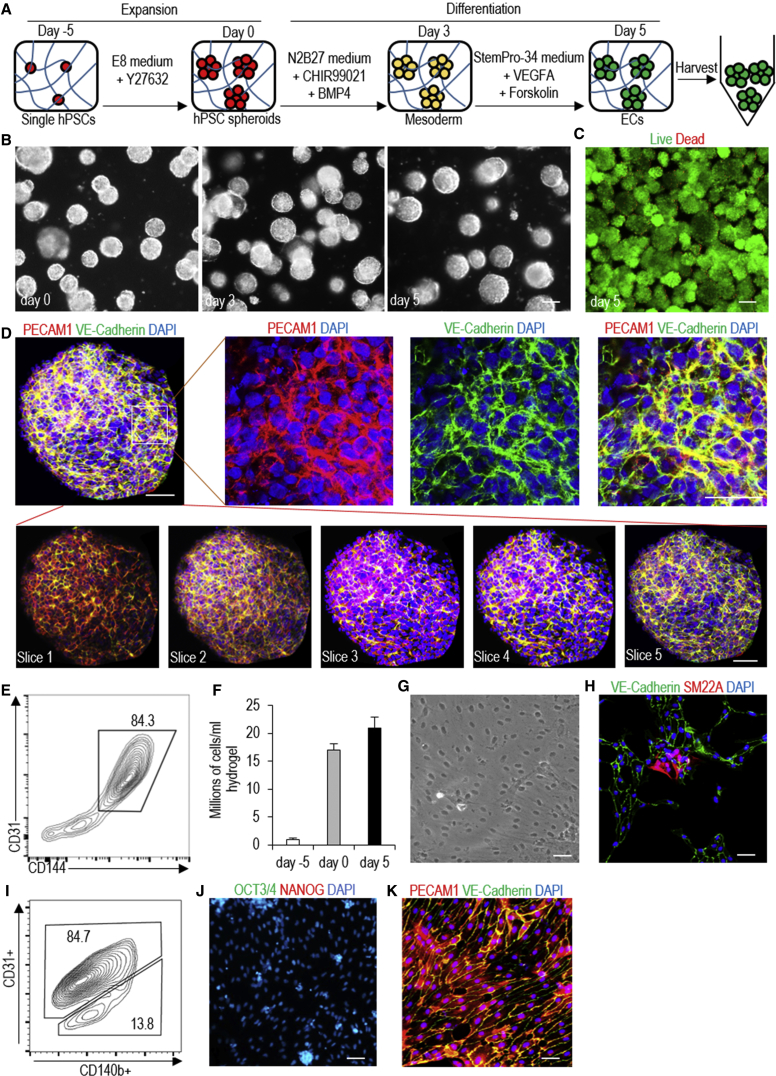
Figure 1.
Manufacture hPSC-Derived ECs in 3D Thermoreversible Hydrogels
(A) Illustration of the bioprocess. Single hPSCs are mixed with 10% PNIPAAm-PEG solution at low temperature (e.g., 4°C), which forms an elastic hydrogel at 37°C. Single hPSCs clonally expand into uniform spheroids in the hydrogel in 5 days. Upon cooling to 4°C, the hydrogel is liquefied, and spheroids are harvested and dissociated into single cells for the next expansion (i). Once the targeted cell number is reached, hPSCs are differentiated into ECs in the hydrogel (ii).
(B) Phase images of day 0, 1, 3, and 5 H9 hESCs. Scale bar, 200 μm.
(C) Immunostaining of day 5 H9 spheroids for pluripotency marker OCT3/4, NANOG, ALP, and SSEA4. Scale bar, 50 μm.
(D) Live/dead staining of harvested day 5 H9 spheroids. Scale bar, 200 μm.
(E and F) About 5-, 10-, and 20-fold expansion (E), yielding ∼5, 10, and 20 million cells per milliliter of hydrogel (F) on day 3, 4, and 5, is achieved for H9s, Fib-iPSCs, and MSC-iPSCs. Data are represented as mean ± SD of three independent replicates (n = 3).
Results
Differentiation of hPSCs into ECs in 2D Adherent Cultures
We used H9 hESCs, iPSCs reprogrammed from human dermal fibroblasts (i.e., Fib-iPSCs), and iPSCs reprogrammed from mesenchymal stem cells (i.e., MSC-iPSCs) (Park et al., 2008), for this study. All formed compact colonies when cultured on Matrigel-coated plates in the chemical-defined Essential 8 (E8) medium (Figures S1A, S1E, and S1I). They expressed pluripotency makers OCT3/4 and NANOG (Figures S1B, S1F, and S1J), and could be differentiated into all three germ layer cells (e.g., NESTIN+ ectodermal, α-SMA+ mesodermal, and HNF-3β+ endodermal cells) in EB assay (Figures S1C, S1G, and S1K). They also formed teratomas containing all three germ layer tissues in immunodeficient mice (Figures S1D, S1H, and S1L).
Patsch et al. (2015) recently reported a protocol that could efficiently generate ECs from hPSCs in 6 days in 2D cultures. This protocol is simple and quick, and thus it is very appealing for making high-quantity ECs. We successfully repeated this protocol with our H9s and iPSCs (Figure S3). The produced ECs had the typical EC cobblestone morphology (Figures S3B and S3H). Immunostaining showed that the majority of these cells expressed the EC markers PECAM1 (or CD31) and VE-Cadherin (or CD144) (Figures S3C and S3I). Flow cytometry analysis showed that about 80% cells were positive for the two markers (Figures S3D and S3J). A small fraction of produced cells was positive for SM22A and CD140b, markers for smooth muscle cells (Figures S3E, S3F, and S3K). We did not detect any undifferentiated OCT3/4+ and NANOG+ hPSCs (Figures S3G and S3L). H9s and iPSCs had similar outcomes (Figure S3). Our results were very similar to these reported by Patsch and Cowan, indicating the robustness of the differentiation protocol (Patsch et al., 2015). We termed ECs made in 2D culturing as 2D-ECs.
Differentiation of hPSCs into ECs in 3D Thermoreversible PNIPAAm-PEG Hydrogels
We then applied the protocol to differentiate hPSCs in the 3D thermoreversible hydrogels (Figure 2A). Single hPSCs were encapsulated into the gel and expanded for 5 days to generated hPSC spheroids with a diameter of around 150 μm. Differentiation was initiated on day 0 by switching the expansion medium to the differentiation medium (Figure 2B). Live/dead cell staining showed that the majority of cells on day 5 were live (Figure 2C). Immunostaining and confocal imaging showed that the majority of cells in the day 5 spheroids were positive for EC markers PECAM1 and VE-Cadherin (Figure 2D). ECs were uniformly distributed, and no cysts were found in the spheroids, indicating no or little cell death in the spheroids. Flow cytometry analysis found that about 84% of the cells were PECAM1+ and VE-Cadherin+ (Figure 2E). About 1.6 × 107 cells and 2.0 × 107 cells were produced in each milliliter of hydrogel on day 0 and 5, respectively (Figure 2F). Thus, about 20 cells were generated from one input hPSC on day 5. When the day 5 EC spheroids were dissociated into single cells and plated on Matrigel-coated plates at high density, they formed tight cell-cell interactions (Figure 2G). Immunostaining detected small numbers of SM22A+ cells (Figure 2H), and flow cytometry analysis found that about 13.8% of cells were CD140b+ (Figure 2I). No undifferentiated OCT3/4+ and NANOG+ hPSCs were detected (Figure 2J). The majority of cells were PECAM1+/VE-Cadherin+ (Figure 2K). Fib-iPSCs and MSC-iPSCs had similar outcomes (Figures S4 and S6). We found the differentiation efficiencies in the 3D hydrogel and the conventional 2D culture were very close (Figures 2 and S3). We termed ECs made in the hydrogel and that had not been cultured on any 2D surfaces as 3D-ECs.
Figure 2.
Differentiate H9 hESCs into ECs in 3D Thermoreversible PNIPAAm-PEG Hydrogels
(A) Illustration of the differentiation protocol.
(B) Phase images of day 0, 3, and 5 cells. Scale bars, 100 μm.
(C) Live/Dead staining of harvested day 5 cells. Scale bars, 200 μm.
(D and E) Immunostaining (D) and flow cytometry analysis (E) of EC markers PECAM1 (or CD31) and VE-Cadherin (or CD 144) on day 5 cells. Immunostaining of five slices of one day 5 spheroid for EC markers PECAM1 and VE-Cadherin. Scale bar, 50 μm.
(F) ∼ 2 × 107 ECs are produced per milliliter of hydrogel on day 5. Data are represented as mean ± SD of three independent replicates (n = 3).
(G–K) The day 5 spheroids are dissociated into single cells and plated on 2D surface overnight. Phase image shows the cobblestone morphology (G). Immunostaining and flow cytometry shows a small fraction of SM22A+ cells (H) and CD140b+ cells (I), respectively, but no undifferentiated OCT3/4+/NANOG+ hPSCs (J). Majority cells are PECAM1+/VE-Cadherin+ (K). Scale bars, 50 μm.
Properties of hPSC-Derived ECs
Our culture system provides a 3D microenvironment for hPSC growth and differentiation. Recent studies found that the 3D microenvironment could alter the cell phenotype and functional properties compared with those cultured in 2D (Zhang et al., 2017b, Zujur et al., 2017). We thus asked if the 3D-ECs and 2D-ECs were similar in phenotype, function, and gene expression. Using fluorescently labeled acetylated LDL and microscope imaging, we found that they had a similar capacity to uptake lipids (Figure 3A). In the classical tube formation assay, they could form vascular network-like structures (Figure 3B). Through quantification with the Angiogenesis Analyzer of ImageJ (Fork et al., 2015), we found 3D-ECs had higher tube length and branching counts than 2D-ECs (Figure 3C). When ECs were co-cultured with vascular smooth muscle cells, they could arrange in highly organized structures (Figure 3D). The trans-endothelial electrical resistance (TEER) analysis (Srinivasan et al., 2015) revealed that both formed tight barriers as shown by the high TEER value. The barrier tightness was disrupted by tumor necrosis factor alpha, interleukin-1β (IL-1β), and vascular endothelial growth factor A (VEGFA), as shown by a sharp decrease of TEER values. Importantly, 3D-ECs and 2D-ECs performed very similar to primary human umbilical vein endothelial cells (HUVECs) (Figure 3E). To test the angiogenic potential of 3D-ECs and 2D-ECs in vivo, we injected them subcutaneously with a Matrigel matrix into immunodeficient mice. H&E staining and immunostaining revealed similar blood vessel density and structure in the matrix for 3D-ECs and 2D-ECs (Figures 3F and 3G). We also used qPCR to quantitatively analyze the expression of a few EC-specific genes including the surface markers (CD31, CD144, VWF, and CD34), growth factors (VEGFA, VEGFB, and VEGFC), and extracellular matrix (ECM) (FN and COL4A). The results showed that the 3D microenvironment enhanced the expression of these genes (Figures 3H and 3I). Similar results were found for iPSC-derived ECs (Figure S5).
Figure 3.
Properties of ECs Derived from H9 hESCs in 3D Hydrogel (3D-ECs) and 2D Culture (2D-ECs)
(A) Both ECs uptake fluorescence-labeled acetylated LDL (Ac-LDL). Scale bar, 25 μm.
(B–D) Both form tube network (B) when plated on Matrigel for 24 hr. The tube length (mm/field) and branches (C) are calculated using the Angiogenesis Analyzer of the ImageJ software. The co-plated vascular smooth muscle cells attach to the ECs (D). Data are represented as means ± SD of three independent replicates (n = 3). Scale bars, 50 μm. ∗p < 0.05, ∗∗p < 0.01.
(E) TEER properties of HUVECs, 3D-ECs and 2D-ECs, either untreated or treated with 100 ng/mL tumor necrosis factor alpha (TNF-α), 100 ng/mL IL-1β, or 100 ng/mL VEGFA, are similar. Data are represented as means ± SD of three independent replicates (n = 3). ∗∗∗p < 0.001.
(F and G) When transplanted subcutaneously with a Matrigel matrix, 2D-ECs (F) and 3D-ECs (G) form vascular structures. Scale bars, 100 and 25 μm.
(H and I) qRT-PCR shows that 3D-ECs have higher expression of some key genes related to ECs, including EC markers (H), growth factors and ECM genes (I). Data are represented as means ± SD of three independent replicates (n = 3). ∗∗∗p < 0.001.
Whole Transcriptome Analysis of 3D-ECs and 2D-ECs Derived from H9s
The above qPCR findings drove us to study the genome-wide gene expression difference using RNA sequencing (RNA-Seq). We sequenced the undifferentiated H9s, 3D-ECs, and 2D-ECs derived from H9s (three biological replicates for each). Hierarchical clustering analysis (Figure 4A) and principal component analysis (PCA) (Figure 4B) showed that 3D-ECs and 2D-ECs clustered closely and were very different from H9s. The genome-wide gene expression profile correlation coefficients between 3D-ECs and 2D-ECs were >0.83, indicating similar global gene expressions (Figures 4C and 4D). However, the separation of 2D-ECs and 3D-ECs in PC2 of the PCA indicated that these cells had some differences in gene expressions (Figure 4B), which drove us to perform detailed differential gene expression analysis.
Figure 4.
Whole Transcriptome Analysis of 3D-ECs and 2D-ECs Derived from H9s
(A and B) Global heatmap of expressed genes (A) and principal-component analysis (PCA) (B) of 3D-ECs and 2D-ECs.
(C and D) The global gene expression correlation coefficients (C) and scatterplot in log scale of gene expression (D) between 3D-ECs and 2D-ECs. Three biological replicates are used for each sample.
Differential gene expression analysis identified 919 genes upregulated in 3D-ECs, and 718 genes upregulated in 2D-ECs (Data S1). Gene ontology term analysis showed that genes enriched in 3D-ECs are related to vasculature development, ECM assembly and cell adhesion, glycolysis, and ephrin and Notch signaling. Genes enriched in 2D-ECs are related to mitotic cell-cycle process genes and ECM disassembly (Figure 5A). These results indicated that 2D-ECs adopted a proliferative phenotype, while the 3D environment promoted vascular morphogenesis.
Figure 5.
Differential Gene Expression Analysis between 3D-ECs and 2D-ECs Derived from H9s
(A) Gene Ontology terms that have significant differentially expressed genes.
(B–F) Log2 (expression level in 3D-ECs/expression level in 2D-ECs) of extracellular matrix genes including collagen (B), laminin (C), integrin (D), other ECM components (E) and proteases (F).
(G–L) Log2 (expression level in 3D-ECs/expression level in 2D-ECs) of genes related to EC secretome (G), growth factors (H), chemokines (I), cytokines (J), proliferation (K), and glycolysis (L). Three biological replicates are used for each sample.
Detailed gene expression analysis (Data S2) showed the following differences: (1) 3D-ECs had higher expression of ECM genes including collagen (COL23A1, COL20A1, COL13A1, COL11A2, COL6A6, COL6A3, COL14A1, COL24A1, COL4A4, COL16A1, COL25A1, COL27A1, COL5A1, COL6A5, COL4A3, COL7A1, COL4A6, and COL6A2); laminin (LAMC3, LAMB3, and LAMB4); integrin (ITGA2, ITGB2, ITGB3, ITGA2B, ITGA6, ITGA10, ITGA11, ITGB8, ITGA9, ITGAM, and ITGA5); other ECM components (DCN, THBS2, HSPG2, FN1, and THSD1); and proteases (MMP9, MMP23B, MMP24, MMP17, and MMP16) (Figures 5B–5F); 2D-ECs had higher expression of ECM genes including collagen (COL12A1, COL2A1, COL8A2, COL21A1, and COL11A1); laminin (LAMC1); integrin (ITGAL, ITGB6, ITGA7, and ITGB4); other ECM components (EFEMP1, NTN1, and FBN1); and proteases (MMP11, MMP1, TIMP3, TIMP2, and MMP15) (Figures 5B–5F). (2) 3D-ECs had higher expression of genes for EC-secreted factors including MMP9, IGF1, TNFRSF10C, VEGFA, CCL5, IGFBP1, IGFBP2, IL6, CXCL16, ANGPT1, FGF10, ANGPT2, IL-1B, VEGFB, and ANG (Figures 5G–5J); 2D-ECs had higher expression of genes for EC-secreted factors including CXCL1, CCL7, CXCL10, TGFA, CTGF, CXCL2, CXCL6, HGF, CXCL11, CCL2, TIMP2, IL-8, and BMP6 (Figures 5G–5J). (3) 3D-ECs had higher expression of genes for glycolysis (Figure 5L), vasculature development (Figure S7A), angiogenesis (Figures S7B and S7C), hypoxia signaling (Figure S7D), ephrin (Figure S7E), NOTCH signaling (Figure S7F), and Wnt signaling (Figure S7G). (4) 2D-ECs had higher expression of genes for cell cycle and proliferation (Figure 5K).
We did further tests to study if the differences found in the RNA-seq data analysis could also be found in cell phenotype or function assays. Western blotting analysis showed that 3D-ECs had higher expressions for CD31, CD144, FN, NOTCH4, and ITGA2 than 2D-ECs (Figure 6A). Using Ki67 immunostaining and flow cytometry analysis, we found more cells were proliferating in 2D-ECs than in 3D-ECs (Figures 6B and 6C). Glycolysis analysis showed that 3D-ECs had higher glycolytic rate than 2D-ECs (Figure 6D). The in vitro tube formation assay already showed that 3D-ECs had longer tubes and more branches (Figure 3C). Similar results were found in ECs derived from iPSCs (Figures S5 and S6). These in vitro phenotypic analyses well supported the RNA-seq data (Figures 5 and S7). However, the in vivo Matrigel Plug Assay did not find significant differences in terms of the vascular density and branches for 3D-ECs and 2D-ECs (Figures 3F, 3G, and S5E). This indicates that the gene expression and in vitro phenotype differences are not sufficient to generate functional differences in vivo. Alternatively, the Matrigel Plug Assay is not sensitive enough to detect the functional differences in vivo. The functional differences of 3D-ECs and 2D-ECs, such as integration and potency, should be systematically studied with disease (e.g., limb ischemia model) or developmental models in the future (Cooke and Losordo, 2015, Tang et al., 2011).
Figure 6.
Functional Comparison of 2D-ECs and 3D-ECs
(A) Western blotting shows that 3D-ECs have higher expression of CD31, CD144, FN, NOTCH4, and ITGA2 than 2D-ECs.
(B and C) Immunostaining (B) and flow cytometry (C) of Ki67 shows more proliferating cells in 2D-ECs than in 3D-ECs. Scale bars, 50 μm.
(D) Glycolysis analysis shows that 3D-ECs produce more L-lactates than 2D-ECs. Data are represented as means ± SD of three independent replicates (n = 3). ∗∗∗p < 0.001.
Scalable Production of hPSC-ECs in Suspension with the Hydrogel Scaffold
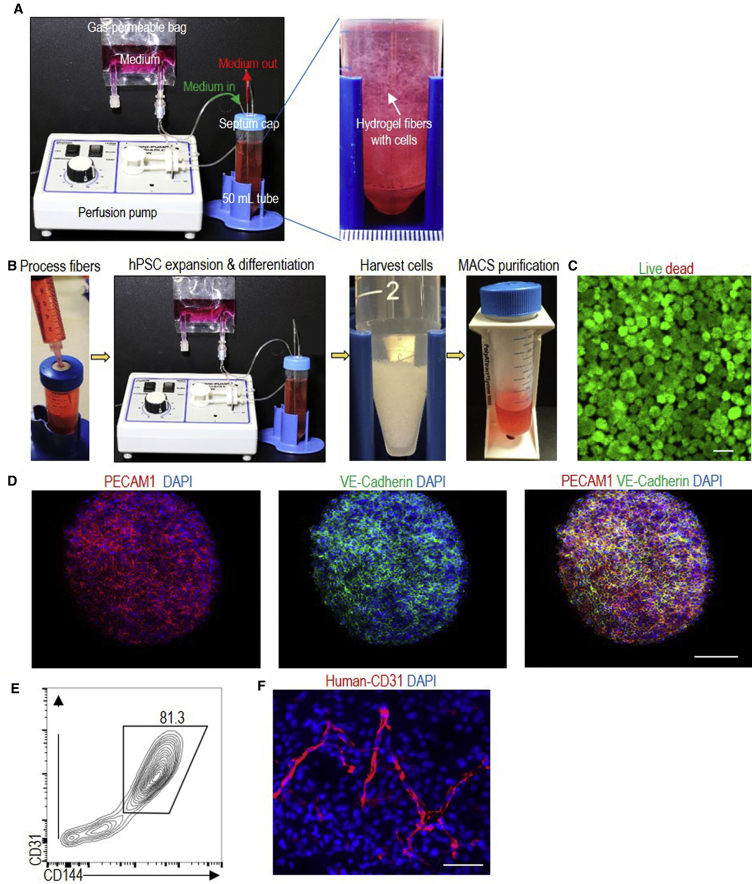
All the above studies were performed by casting the thermoreversible hydrogel (with cells encapsulated) as a thin layer (e.g., 500–1,000 μm thickness) on cell culture plates (e.g., 6-well plates). We also demonstrated that the hydrogel could be processed into small fibers (with cells encapsulated in the fibers) for suspension culturing (Figure 7A). It is generally accepted that 3D suspension culturing is more suitable for large-scale production of cells. On day 0, single hPSCs (∼4.0 × 106 cells) were mixed with 4 mL 10% PNIPAAm-PEG solution at 4°C and injected into room temperature E8 medium contained in a closed and sterile bioreactor (e.g., a 50-mL conical tube). Fibrous hydrogels were instantly formed with single hPSCs uniformly distributed in the hydrogels. Cells were cultured in a cell culture incubator at 37°C and 5% CO2. The medium was stocked in a gas-permeable bag and continuously perfused into the bioreactor. Perfusion instead of stirring was used to mix the medium since the hydrogel fibers were soft and stirring resulted in fiber breakage. Cells were expanded for 5 days and then differentiated for 5 days. On day 10, hydrogel scaffolds were liquefied, and cell spheroids were pelleted by spinning for 5 min at 100 × g. Spheroids were dissociated into single cells by treating with Accutase at 37°C for 10 min. About 8.0 × 107 ECs were produced. Magnetic beads coated with anti-CD144 antibodies were then added to the conical tube to pull down the ECs with a magnetic cell separator (Figure 7B). Live/dead cell staining showed that the majority of the cells were live (Figure 7C). Immunostaining and flow cytometry analysis showed that 81.3% of the day 10 cells were ECs (Figures 7D and 7E). When transplanted subcutaneously with a Matrigel matrix, ECs formed nice vascular structures (Figure 7F).
Figure 7.
A Prototype Bioreactor
(A) The bioreactor consists of a pump for medium perfusion, an oxygen-permeable plastic bag for stocking medium and a closed container (e.g., a 50-mL conical tube), where hydrogel fibers with cells are suspended.
(B) On day 0, single hPSCs are mixed with 10% PNIPAAm-PEG solution at 4°C and injected into the room-temperature E8 medium in the container. Fibrous hydrogels are instantly formed. Cells are cultured in E8 medium for 5 days, followed by an additional 5 days in EC differentiation medium. Medium is continuously perfused. On day 10, the hydrogel scaffold is liquefied, and spheroids are pelleted by centrifugation. Spheroids are dissociated into single cells by incubating in Accutase at 37°C for 10 min. Magnetic beads coated with anti-CD144 were added to pull down the CD144+ cells.
(C–E) Live/dead staining (C), immunostaining (D), and flow cytometry analysis (E) of day 10 cells. Scale bars, 200 and 100 μm.
(F) When transplanted subcutaneously with a Matrigel matrix, ECs form nice vascular structures. H9s are used in this figure. Scale bar, 50 μm.
Discussion
Currently, hPSC-derived ECs are made either in 2D culture or as EBs in 3D suspension culture. Both have difficulty to cost-effectively generate large numbers of high-quality ECs required for drug discovery, tissue engineering, and cell therapies. Both culture methods provide culturing conditions that are very different from the in vivo 3D microenvironments where cells reside (Chen et al., 2014a, Chen et al., 2014b, Kraehenbuehl et al., 2011, Thomson et al., 1998, Wong et al., 2010). 2D culturing is labor, space, and reagent consuming and considered only suitable for preparing low-scale cells (e.g., ≤109) (Kropp et al., 2017).
3D suspension culturing has been widely studied to scale up the production of hPSCs and their derivatives (Fridley et al., 2012, Kinney et al., 2011, Kropp et al., 2017). However, hPSCs in suspension culturing suffer from severe cellular agglomeration (Fridley et al., 2012, Kinney et al., 2011, Kropp et al., 2017). The strong cell-cell interactions make hPSCs form large agglomerates (i.e., agglomeration) that lead to culture inhomogeneity, impaired mass transport, and low culture efficiency (Fridley et al., 2012, Kinney et al., 2011, Kropp et al., 2017). Agitation (stirring or shaking) the culture can reduce agglomeration and enhance mass transport (Fridley et al., 2012, Kinney et al., 2011, Kropp et al., 2017). However, agitation generates complicate hydrodynamic conditions including medium flow direction, velocity, pressure, shear force, and chemical environment. These physical and chemical microenvironments vary spatially and temporally, generating critical stresses in some locations (e.g., near vessel wall and impeller tip), which induce cell death and phenotype changes (Fridley et al., 2012, Ismadi et al., 2014, Jenkins and Farid, 2015, Kinney et al., 2011, Kropp et al., 2017, Lei and Schaffer, 2013, Lei et al., 2014, Serra et al., 2012, Steiner et al., 2010).
These physical and chemical microenvironments are sensitive to many factors, such as the bioreactor design (e.g., impeller geometry, size and position, vessel geometry and size, positions of probes for pH, temperature, and oxygen), the medium viscosity and agitation rate (Ismadi et al., 2014, Kropp et al., 2017). They are currently not well understood and hard to control (Fridley et al., 2012, Ismadi et al., 2014, Kinney et al., 2011, Kropp et al., 2017). In addition, how these physical and chemical factors, individually or combined, influence different types of cells is not well understood and hard to study. These knowledge gaps lead to large culture variations between batches as well as difficulty in scaling up. The challenge of using 3D suspension culturing for large-scale cell production is well demonstrated by a few recent studies on producing hPSC-derived cardiomyocytes in stirred-tank bioreactors (Chen et al., 2015, Jara-avaca et al., 2014). For three independent batches (∼100 mL culture volume) with HES3 hESCs, both the yield and product purity varied in large ranges (e.g., from 40 million to 100 million cells for the yield, and from 54% to 84% for the cardiomyocyte purity). Using the same bioreactor and a different hPSC line, the final yield ranged from 89 million to 125 million cells, and final cardiomyocyte purity ranged from 28% to 88% (Chen et al., 2015, Jara-avaca et al., 2014). In addition, when culture volume was scaled from ∼100 to ∼1,000 mL, the yield and differentiation efficiency were significantly changed (Chen et al., 2015, Jara-avaca et al., 2014). This makes scaling up very challenging since optimizing multiple factors in large culture volumes is costly. To the best of our knowledge, the largest demonstrated culture volume for hPSCs and their derivative in literature is less than 10 L (Kempf et al., 2016, Kropp et al., 2017). In short, the uncontrolled cell aggregation and hydrodynamic stress/conditions make it difficult to culture hPSCs and their derivatives on a large scale with 3D suspension culturing.
The thermoreversible hydrogel scaffold used in this paper not only provides 3D space for cell growth, but also acts a physical barrier to prevent cell agglomeration and isolate the shear force (Figure 1). Eliminating these negative factors leads to significantly enhanced culture efficiency. We showed that ECs could be produced with high viability, high purity (>80%), and high yield (∼2.0 × 107 cells/mL hydrogel) within 10 days. For comparison, it usually yields 1.0 × 106 to 2.0 × 106 cells/mL in 3D suspension cultures (Lei et al., 2014, Serra et al., 2012, Steiner et al., 2010, Wurm, 2004). In addition, the synthetic hydrogel is chemically defined. Due to its thermoreversible nature, seeding and harvesting cells are simple and scalable. The system can be adapted to multiple scales—from the laboratory and toward the clinic—to support research in cell therapies, tissue engineering, and high-throughput drug discovery with hPSC-ECs. For instance, ∼50 mL hydrogel would be sufficient to produce 109 ECs for preclinical animal studies, and a bioreactor with ∼5 L of hydrogel could yield >1011 ECs for clinical studies. The ability to move a single-culture system through multiple scales may aid clinical development.
Conclusion
We developed a scalable and GMP-compliant method for manufacturing ECs from hPSCs with high viability, high purity (>80%), and high yield (∼2.0 × 107 cells/mL hydrogel). These ECs had similar properties as ECs made using 2D culture systems. The method will make ECs broadly available and affordable for various applications.
Experimental Procedures
hPSCs Culture
H9 hESCs (WiCell, no. WA09) were purchased from WiCell Research Institute. iPSCs were obtained from the Human Embryonic Stem Cell Core, Harvard Medical School. MSC-iPSCs and Fib-iPSCs were reprogrammed from mesenchymal stem cells and fibroblasts, respectively, by the George Q. Daley Lab (Children's Hospital Boston, MA), and have been well characterized and described in the literature (Park et al., 2008). hPSCs (H9s and iPSCs) were maintained in a six-well plate coated with Matrigel (BD Biosciences, no. 354277) in E8 medium (Invitrogen, no. A1517001). Cells were passaged every 4 days with 0.5 mM EDTA (Invitrogen, no. AM9260G). The medium was changed daily. Cells were routinely checked for the expression of pluripotency markers, OCT3/4 and NANOG, their capability to form teratomas in immunodeficient mice, their karyotypes, and bacterial contaminations. Human aortic vascular smooth muscle cells (no. CC-2571) and HUVECs (no. 00191027) were from Lonza.
Culturing hPSCs in 3D PNIPAAm-PEG Hydrogels
To transfer the culture from 2D to 3D PNIPAAm-PEG hydrogels (Cosmo Bio, no. MBG-PMW20-5005), hPSCs maintained in Matrigel-coated six-well plates were treated with Accutase (Life Technologies, no. A1110501) at 37°C for 5 min and dissociated into single cells (Lei and Schaffer, 2013, Lei et al., 2014). Dissociated cells were mixed with 10% PNIPAAm-PEG solution dissolved in E8 medium on ice and cast on tissue culture plates, then incubated at 37°C for 10 min to form hydrogels before adding warm E8 medium containing 10 μM ROCK inhibitor ([RI], Y-27632, LC Laboratories, no. Y5301). The medium was changed daily. Cells were passaged every 5 days. To passage cells, the medium was removed, and 2 mL ice-cold PBS was added to dissolve the hydrogel for 5 min. Cell spheroids were collected by spinning at 100 × g for 3 min. Cells were incubated in Accutase at 37°C for 10 min and dissociated into single cells.
EC Differentiation in 2D Cultures and in 3D Hydrogels
For 2D endothelial cell differentiation, hPSCs were dissociated with Accutase and plated on Matrigel at a density of 40,000 cells/cm2 in E8 medium with 10 μM RI. After 24 hr, the medium was replaced with differentiation medium, consisting of N2B27 medium (1:1 mixture of DMEM/F12 [HyClone, no. SH30004.04] with Glutamax-I [Life Technologies, no. 35050061] and Neurobasal medium [Life Technologies, no. 21103049] supplemented with N2 [Life Technologies, no. 17502048] and B27 minus vitamin A [Life Technologies, no. 12587010] with 8 μM CHIR99021 [LC Laboratories, no. C6556] and 25 ng/mL BMP4 [R&D Systems, no. 314BP010]). After 3 days, the differentiation medium was replaced by EC induction medium consisting of StemPro-34 SFM medium (Life Technologies, no. 10639011) supplemented with 200 ng/mL VEGFA (PeproTech, no. 100-20) and 2 μM Forskolin (Sigma, no. F3917). The induction medium was changed after 1 day. ECs were harvested for analysis on day 5.
For EC differentiation in 3D PEG hydrogel, single hPSCs were mixed with 10% PNIPAAm-PEG solution and cast on tissue culture plate, then incubated at 37°C for 10 min to form hydrogels before adding warm E8 medium containing 10 μM RI. Medium was changed daily. hPSCs were expanded for 5 days. E8 medium was removed and replaced with EC differentiation medium. After 3 days, the differentiation medium was replaced by EC induction medium. The induction medium was changed after 1 day. Cells were harvested for analysis on day 5.
Suspension Culturing in the Bioreactor
The bioreactor consists of a pump for medium perfusion, an oxygen-permeable plastic bag for stocking medium and a closed container (e.g., a 50-mL conical tube with a septa cap). On day 0, single iPSCs (∼4.0 × 106 cells) were mixed with 4 mL 10% PNIPAAm-PEG solution at 4°C and were injected into room-temperature E8 medium in the container. Fibrous hydrogels (with diameter <1.0 mm) were instantly formed. Cells were cultured in E8 medium for 5 days, followed by an additional 5 days of EC differentiation medium. Medium was continuously perfused. On day 10, hydrogel scaffolds were liquefied by perfusing ice-cold PBS. Spheroids were pelleted by centrifugation. Spheroids were dissociated into single cells by incubating in Accutase at 37°C for 10 min. Magnetic beads coated with anti-CD144 antibodies (Miltenyi Biotec, cat. no. 130-097-857) were added to pull down ECs with a magnetic cell separator. The bioprocess was repeated twice.
Immunocytochemistry and Flow Cytometry
For 2D immunostaining, cells were fixed with 4% paraformaldehyde (PFA) at room temperature for 20 min, permeabilized with 0.25% Triton X-100 for 30 min and blocked with 5% donkey serum for 1 hr before incubating with primary antibodies (Table S1) at 4°C overnight. After extensive washing, secondary antibodies (Table S1) and 10 μM, DAPI in 2% BSA was added and incubated at room temperature for 4 hr. Cells were washed with PBS three times before imaging.
For 3D immunostaining (spheroids), hPSCs were fixed with 4% PFA at room temperature for 30 min, and then incubated with PBS+ 0.25% Triton X-100+ 5% (vol/vol) goat serum+ primary antibodies at 4°C for 48 hr. After extensive washing, secondary antibodies in 2% BSA were added and incubated at 4°C for 24 hr. Cells were washed with PBS three times before imaging with a confocal microscope.
For flow cytometry analysis, the harvested cells were dissociated into single cells with Accutase. Single cells were fixed with 4% PFA and stained with primary antibodies (Table S1) at 4°C overnight. After washing (three times) with 1% BSA in PBS, secondary antibodies were added and incubated at room temperature for 2 hr. Cells were washed with 1% BSA in PBS and analyzed using Cytek flow cytometry. Single-color and isotype controls served as compensation and negative gating.
RNA Extraction, cDNA Synthesis, and qPCR
Total RNAs for qPCR and RNA-Seq were extracted from 2D-ECs and 3D-ECs on day 5 of the differentiation using TRIzol (Invitrogen, no. 15596018), according to the manufacturer's instructions. Reverse transcription is done with the Maxima First Strand cDNA Synthesis Kit (Life Technologies, no. K1642). Real-time qPCR was carried out in an Eppendorf MasterCycler RealPlex4 (Thermo Fisher Scientific) using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, no. 4367659), according to the manufacturer's instructions. The data were normalized to the endogenous GAPDH. Primer sequence was listed in Table S2.
EB Differentiation
hPSCs were suspended in DMEM+ 20% FBS+ 10 μM β-mercaptoethanol in a low adhesion plate for 6 days. The cell masses were then transferred into plates coated with 0.1% gelatin and cultured in the same medium for another 6 days, followed by fixation and staining as described above.
Teratoma Formation In Vivo
The animal experiments were carried out following the protocols approved by the University of Nebraska–Lincoln Animal Care and Use Committee. hPSCs (3.0 × 106) were suspended in 25 μL PBS+ 25 μL Matrigel and injected subcutaneously at the back of the neck of the non-obese diabetic/severe combined immunodeficiency mice (female, age 7 weeks, Charles River Laboratory). Teratoma was harvested when its size reached 2 cm. Teratoma was fixed with 4% PFA for 48 hr, dehydrated with 70%, 95%, and 100% ethanol sequentially, and de-fated with xylene for 2 hr before embedded in paraffin. A 10-μm-thick section was cut and stained with H&E.
Statistical Analysis
Statistical analyses were done using the statistical package Instat (GraphPad Software, La Jolla, CA). For multiple comparisons, the means of triplicate samples were compared using the Tukey multiple comparisons analysis with the alpha level indicated in the figure legend.
Author Contributions
Y.L. and H.L. conceived the idea, designed the experiments, and wrote the manuscript. H.L., Q.L., and O.W. performed the experiments. H.L., Q.D., K.L., and C.Z. analyzed the data. C.E. contributed to confocal images. N.S. and H.L. did the western blotting. Z.W., S.C., and B.D. revised the manuscript.
Acknowledgments
This work was partially funded by the UNL-UNMC Science Engineering and Medicine (SEM) initiative grant. Confocal microscope imaging was done in the Morrison Microscopy Core Research Facility at University of Nebraska, Lincoln. Dr. You Zhou assisted the confocal imaging. Flow cytometry was done in the Morrison center, the Flow Cytometry core, University of Nebraska, Lincoln with the assistance of Dirk Anderson. RNA sequencing was done at the UNMC deep sequencing core.
Published: August 2, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, two tables, and two data files and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.07.001.
Accession Numbers
The accession numbers for the data reported in this paper are GEO: GSE99776 and GSE109683. All other data supporting the findings of this study are available within the paper and its Supplemental Information.
Supplemental Information
References
- Arici V., Perotti C., Fabrizio C., Del Fante C., Ragni F., Alessandrino F., Viarengo G., Pagani M., Moia A., Tinelli C. Autologous immuno magnetically selected CD133+ stem cells in the treatment of no-option critical limb ischemia: clinical and contrast enhanced ultrasound assessed results in eight patients. J. Transl. Med. 2015;13:1–10. doi: 10.1186/s12967-015-0697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin-Voss H.G., Voss A.K., Pauli B.U. Senescence of aortic endothelial cells in culture: effects of basic fibroblast growth factor expression on cell phenotype, migration, and proliferation. J. Cell. Physiol. 1993;157:279–288. doi: 10.1002/jcp.1041570210. [DOI] [PubMed] [Google Scholar]
- Cao N., Liang H., Huang J., Wang J., Chen Y., Chen Z., Yang H. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119–1132. doi: 10.1038/cr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Cardiovascular biology: creating unique blood vessels. Nature. 2001;412:868–869. doi: 10.1038/35091178. [DOI] [PubMed] [Google Scholar]
- Chao T.H., Tseng S.Y., Chen I.C., Tsai Y.S., Huang Y.Y., Liu P.Y., Ou H.Y., Li Y.H., Wu H.L., Cho C.L. Cilostazol enhances mobilization and proliferation of endothelial progenitor cells and collateral formation by modifying vasculo-angiogenic biomarkers in peripheral arterial disease. Int. J. Cardiol. 2014;172:e371–e374. doi: 10.1016/j.ijcard.2013.12.295. [DOI] [PubMed] [Google Scholar]
- Chen K.G., Mallon B.S., Johnson K.R., Hamilton R.S., Mckay R.D.G., Robey P.G. Developmental insights from early mammalian embryos and core signaling pathways that influence human pluripotent cell growth and differentiation. Stem Cell Res. 2014;12:610–621. doi: 10.1016/j.scr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.G., Mallon B.S., McKay R.D.G., Robey P.G. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14:13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.C., Ye J., Shukla P., Hua G., Chen D., Lin Z., Liu J., Chai J., Gold J., Wu J. Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Res. 2015;15:365–375. doi: 10.1016/j.scr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G., Borello U., De Angelis L., Latronico M., Sirabella D., Coletta M., Galli R., Balconi G., Follenzi A., Dejana E. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: implications for myocardium regeneration. Proc. Natl. Acad. Sci. USA. 2001;98:10733–10738. doi: 10.1073/pnas.191217898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J.P., Losordo D.W. Modulating the vascular response to limb ischemia angiogenic and cell therapies. Circ. Res. 2015;116:1561–1578. doi: 10.1161/CIRCRESAHA.115.303565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho J.L., Zonari A., de Paula A.C., Martins T., Gomes D.A., Goes A.M. Production of human endothelial cells free from soluble xenogeneic antigens for bioartificial small diameter vascular graft endothelization. Biomed. Res. Int. 2015;2015:652474. doi: 10.1155/2015/652474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dekker W.K., Houtgraaf J.H., Onuma Y., Benit E., de Winter R.J., Wijns W., Grisold M., Verheye S., Silber S., Teiger E. Final results of the HEALING IIB trial to evaluate a bio-engineered CD34 antibody coated stent (Genous™Stent) designed to promote vascular healing by capture of circulating endothelial progenitor cells in CAD patients. Atherosclerosis. 2011;219:245–252. doi: 10.1016/j.atherosclerosis.2011.06.032. [DOI] [PubMed] [Google Scholar]
- Fork C., Gu L., Hitzel J., Josipovic I., Hu J., Wong M.S., Ponomareva Y., Albert M., Schmitz S.U., Uchida S. Epigenetic regulation of angiogenesis by JARID1B-induced repression of HOXA5. Arterioscler. Thromb. Vasc. Biol. 2015;35:1645–1652. doi: 10.1161/ATVBAHA.115.305561. [DOI] [PubMed] [Google Scholar]
- Fridley K.M., Kinney M.A., Mcdevitt T.C. Hydrodynamic modulation of pluripotent stem cells. Stem Cell Res. Ther. 2012;3:45. doi: 10.1186/scrt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Bellin M., Sala L., van Meer B.J., Tertoolen L.G.J., Orlova V.V., Mummery C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008–1017. doi: 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granton J., Langleben D., Kutryk M.B., Camack N., Galipeau J., Courtman D.W., Stewart D.J. Endothelial NO-synthase gene-enhanced progenitor cell therapy for pulmonary arterial hypertension: the PHACeT trial. Circ. Res. 2015;117:645–654. doi: 10.1161/CIRCRESAHA.114.305951. [DOI] [PubMed] [Google Scholar]
- Gui L., Ph D., Muto A., Ph D., Chan S.A., Breuer C.K., Niklason L.E., Ph D. Development of decellularized human umbilical arteries as small-diameter vascular grafts. Tissue Eng. Part A. 2009;15:2665–2676. doi: 10.1089/ten.tea.2008.0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbleton M., Audus K.L. Progress and limitations in the use of in vitro cell cultures to serve as a permeability screen for the blood-brain barrier. J. Pharm. Sci. 2001;90:1681–1698. doi: 10.1002/jps.1119. [DOI] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Heo S.C., Kwon Y.W., Jang I.H., Jeong G.O., Yoon J.W., Kim C.D., Kwon S.M., Bae Y.-S., Kim J.H. WKYMVm-induced activation of formyl peptide receptor 2 stimulates ischemic neovasculogenesis by promoting homing of endothelial colony-forming cells. Stem Cells. 2014;32:779–790. doi: 10.1002/stem.1578. [DOI] [PubMed] [Google Scholar]
- Huang X.-T., Zhang Y.-Q., Li S.-J., Li S.-H., Tang Q., Wang Z.-T., Dong J.-F., Zhang J.-N. Intracerebroventricular transplantation of ex vivo blood-brain barrier integrity and promotes angiogenesis of mice with traumatic brain injury. J. Neurotrauma. 2013;30:2080–2088. doi: 10.1089/neu.2013.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismadi M., Gupta P., Fouras A., Verma P., Jadhav S. Flow characterization of a spinner flask for induced pluripotent stem cell culture application. PLoS One. 2014;9:e106493. doi: 10.1371/journal.pone.0106493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D., Nam H., Seandel M., Nolan D., Janovitz T., Tomishima M., Studer L., Lee G., Lyden D., Benezra R. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFβ inhibition is Id1 dependent. Nat. Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara-avaca M., Kempf H., Olmer R., Kropp C., Ru M., Robles-diaz D., Franke A., Elliott D.A., Wojciechowski D., Fischer M. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Reports. 2014;3:1132–1146. doi: 10.1016/j.stemcr.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins M.J., Farid S.S. Human pluripotent stem cell-derived products: advances towards robust, scalable and cost-effective manufacturing strategies. Biotechnol. J. 2015;10:83–95. doi: 10.1002/biot.201400348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N.M., Meloni M., Spencer H.L., Craig M.A., Strehl R., Milligan G., Houslay M.D., Mountford J.C., Emanueli C., Baker A.H. Derivation of endothelial cells from human embryonic stem cells by directed differentiation analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 2010;30:1389–1397. doi: 10.1161/ATVBAHA.110.204800. [DOI] [PubMed] [Google Scholar]
- Kang K.T., Coggins M., Xiao C., Rosenzweig A., Bischoff J. Human vasculogenic cells form functional blood vessels and mitigate adverse remodeling after ischemia reperfusion injury in rats. Angiogenesis. 2013;16:773–784. doi: 10.1007/s10456-013-9354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H., Andree B., Zweigerdt R. Large-scale production of human pluripotent stem cell derived cardiomyocytes. Adv. Drug Deliv. Rev. 2016;96:18–30. doi: 10.1016/j.addr.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Kinney M.A., Sargent C.Y., Mcdevitt T.C. The multiparametric effects of hydrodynamic environments on stem cell culture. Tissue Eng. Part B Rev. 2011;17:249–262. doi: 10.1089/ten.teb.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraehenbuehl T.P., Langer R., Ferreira L.S. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat. Methods. 2011;8:731–736. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- Kropp C., Massai D., Zweigerdt R. Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochem. 2017;59:244–254. [Google Scholar]
- Lalit P.A., Hei D.J., Raval A.N., Kamp T.J. Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circ. Res. 2014;114:1328–1345. doi: 10.1161/CIRCRESAHA.114.300556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Schaffer D.V. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc. Natl. Acad. Sci. USA. 2013;110:E5039–E5048. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Jeong D., Xiao J., Schaffer D.V. Developing defined and scalable 3D culture systems for culturing human pluripotent stem cells at high densities. Cell. Mol. Bioeng. 2014;7:172–183. doi: 10.1007/s12195-014-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung O.M., Zhou B., Lui K.O. Vascular development and regeneration in the mammalian heart. J. Cardiovasc. Dev. Dis. 2016;3:23. doi: 10.3390/jcdd3020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S., Golub J.S., Amit M., Itskovitz-Eldor J., Langer R. Endothelial cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2002;99:4391–4396. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S., Zoldan J., Basevitch Y., Langer R. Endothelial potential of human embryonic stem cells. Blood. 2007;110:806–814. doi: 10.1182/blood-2006-08-019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wilson K.D., Smith B., Kraft D.L., Jia F., Huang M., Robbins R.C., Gambhir S.S., Weissman I.L., Wu J.C. Functional and transcriptional characterization of human embryonic stem cell-derived endothelial cells for treatment of myocardial infarction. PLoS One. 2009;4:e8443. doi: 10.1371/journal.pone.0008443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Han Z., Wu J.C. Transplantation of human embryonic stem cell-derived endothelial cells for vascular diseases. J. Cell. Biochem. 2009;106:194–199. doi: 10.1002/jcb.22003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lin H., Wang O., Qiu X., Kidambi S., Deleyrolle L.P., Reynolds B.A., Lei Y. Scalable production of glioblastoma tumor-initiating cells in 3 dimension thermoreversible hydrogels. Sci. Rep. 2016;6:31915. doi: 10.1038/srep31915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Bao X., Al-Ahmad A., Liu J., Wu Y., Dong W., Dunn K.K., Shusta E.V., Palecek S.P. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Reports. 2014;3:804–816. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Li Q., Lei Y. An integrated miniature bioprocessing for personalized human induced pluripotent stem cell expansion and differentiation into neural stem cells. Sci. Rep. 2017;7:40191. doi: 10.1038/srep40191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Tatsumi T., Murohara T., Imaizumi T., Katsuda Y., Ito M., Saito Y., Uemura S., Suzuki H., Fukumoto S. Long-term clinical outcome after intramuscular implantation of bone marrow mononuclear cells (Therapeutic Angiogenesis by Cell Transplantation [TACT] trial) in patients with chronic limb ischemia. Am. Heart J. 2008;156:1010–1018. doi: 10.1016/j.ahj.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Medina R.J., Neill C.L.O., Humphreys M.W., Gardiner T.A., Stitt A.W. Outgrowth endothelial cells: characterization and their potential for reversing ischemic retinopathy. Retina. 2010;51:5906–5913. doi: 10.1167/iovs.09-4951. [DOI] [PubMed] [Google Scholar]
- Moubarik C., Guillet B., Youssef B., Codaccioni J., Piercecchi M., Sabatier F., Lionel P., Dou L., Foucault-bertaud A., Velly L. Transplanted late outgrowth endothelial progenitor cells as cell therapy product for stroke. Stem Cell Rev. 2011;7:208–220. doi: 10.1007/s12015-010-9157-y. [DOI] [PubMed] [Google Scholar]
- Nourse M.B., Halpin D.E., Scatena M., Mortisen D.J., Tulloch N.L., Hauch K.D., Torok-storb B., Ratner B.D., Pabon L., Murry C.E. VEGF induces differentiation of functional endothelium from human embryonic stem cells: implications for tissue engineering. Arterioscler. Thromb. Vasc. Biol. 2010;30:80. doi: 10.1161/ATVBAHA.109.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova V.V., van den Hil F.E., Petrus-Reurer S., Drabsch Y., Ten Dijke P., Mummery C.L. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat. Protoc. 2014;9:1514–1531. doi: 10.1038/nprot.2014.102. [DOI] [PubMed] [Google Scholar]
- Palpant N.J., Pabon L., Friedman C.E., Roberts M., Hadland B., Zaunbrecher R.J., Bernstein I., Zheng Y., Murry C.E. Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nat. Protoc. 2016;12:15–31. doi: 10.1038/nprot.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Patsch C., Challet-Meylan L., Thoma E.C., Urich E., Heckel T., O’Sullivan J.F., Grainger S.J., Kapp F.G., Sun L., Christensen K. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O.C., Raines S.M., Attie A.D. The role of blood vessels, endothelial cells, and vascular pericytes in insulin secretion and peripheral insulin action. Endocr. Rev. 2010;31:343–363. doi: 10.1210/er.2009-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz T.M., Leicht S.F., Radic T., Rodriguez-arab I., Hermann P.C., Berger F., Saif J., Bo W., Ellwart J.W., Aicher A. Vascular incorporation of endothelial colony-forming cells is essential for functional recovery of murine ischemic tissue following cell therapy. Arterioscler. Thromb. Vasc. Biol. 2012;32:e13–e21. doi: 10.1161/ATVBAHA.111.239822. [DOI] [PubMed] [Google Scholar]
- Serra M., Brito C., Correia C., Alves P.M. Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol. 2012;30:350–358. doi: 10.1016/j.tibtech.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Srinivasan B., Kolli A.R., Esch M.B., Abaci H.E., Shuler M.L., Hickman J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner D., Khaner H., Cohen M., Even-Ram S., Gil Y., Itsykson P., Turetsky T., Idelson M., Aizenman E., Ram R. Derivation, propagation and controlled differentiation of human embryonic stem cells in suspension. Nat. Biotechnol. 2010;28:361–364. doi: 10.1038/nbt.1616. [DOI] [PubMed] [Google Scholar]
- Stroncek J., Ren L., Klitzman B., Reichert W. Patient-derived endothelial progenitor cells improve vascular graft patency in a rodent model. Acta Biomater. 2012;8:201–208. doi: 10.1016/j.actbio.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tang Z.C.W., Liao W.Y., Tang A.C.L., Tsai S.J., Hsieh P.C.H. The enhancement of endothelial cell therapy for angiogenesis in hindlimb ischemia using hyaluronan. Biomaterials. 2011;32:75–86. doi: 10.1016/j.biomaterials.2010.08.085. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van Beijnum J.R., Rousch M., Castermans K., van der Linden E., Griffioen A.W. Isolation of endothelial cells from fresh tissues. Nat. Protoc. 2008;3:1085–1091. doi: 10.1038/nprot.2008.71. [DOI] [PubMed] [Google Scholar]
- Vodyanik M.A., Bork J.A., Thomson J.A., Slukvin I.I. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–627. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., Reijo Pera R.A. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-otto K., Antosiewicz-bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chu L., Hou Z., Schwartz M.P., Hacker T., Vickerman V. Functional characterization of human pluripotent stem cell-derived arterial endothelial cells. Proc. Natl. Acad. Sci. USA. 2017;114:E6072–E6078. doi: 10.1073/pnas.1702295114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schwartz M.P., Hou Z., Bai Y., Ardalani H., Swanson S., Steill J., Ruotti V., Elwell A., Nguyen B.K. A genome-wide analysis of human pluripotent stem cell-derived endothelial cells in 2D or 3D culture. Stem Cell Reports. 2017;8:907–918. doi: 10.1016/j.stemcr.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zujur D., Kanke K., Lichtler A.C., Hojo H. Three-dimensional system enabling the maintenance and directed differentiation of pluripotent stem cells under defined conditions. Sci. Adv. 2017;3:e1602875. doi: 10.1126/sciadv.1602875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.