Abstract
Background/Aim
GATA3, a transcription factor expressed in luminal breast epithelial cells, is required for mammary gland development. Heterozygous GATA3 mutations occur in up to 15% of estrogen receptor (ER)-positive breast tumors and have been proposed to be null alleles resulting in haploinsufficiency; however, the mutation spectrum of GATA3 in breast cancer is in sharp contrast to that found in HDR syndrome, a true GATA3 haploinsufficiency disease. Materials and
Methods
Transgenic mice, 3D cultures and xenografts were used to examine the effect of mutant GATA3 expression on mammary cell proliferation.
Results
Mutant GATA3 accelerated tumor growth of ZR751 cell xenografts and promoted precocious lobuloalveolar development in transgenic mouse mammary glands.
Conclusion
These data indicate that the GATA3 mutations, recently observed in breast cancer, encode active transcription factors, which elicit proliferative phenotypes in normal mammary epithelium and promote the growth of ER-positive breast cancer cell lines.
Keywords: GATA3, breast cancer, transcription factor, mammary gland biology
GATA3 is expressed in normal luminal mammary epithelial cells and is a key transcription factor in mammary gland development (1). Mice with mammary-specific deletion of GATA3 have significant defects in mammary terminal end bud formation, branching morphogenesis, and lactation (2, 3). Concerning human breast cancer, GATA3 is highly expressed in the majority of estrogen receptor-positive (ER+) tumors (4, 5), where GATA3 and ERα are involved in a positive cross-regulatory feedback loop (6). Somatic GATA3 mutations have recently been identified in breast cancer, primarily in Luminal A and Luminal B ER+ tumors (7–9). In the largest of these studies (8), GATA3 mutations were detected in 14% of Luminal A and 15% of Luminal B tumors, making it the second most frequently mutated gene in Luminal A tumors (after PIK3CA) and the third most frequently mutated gene in Luminal B tumors (after PIK3CA and TP53). The vast majority of mutations are frameshifts. Approximately 40% of the mutations identified in the TCGA study lack the second zinc finger, which is responsible for binding the canonical GATA motif (10, 11), while the remainder encode longer proteins containing both of the GATA3 zinc fingers.
Although the precise nature of the contribution of GATA3 mutation to tumorigenesis is unclear, its high mutation frequency indicates that this alteration plays an important role in breast tumor pathogenesis. Prior studies have found that GATA3 mutations in breast cancer are clustered in exons 4, 5 and 6, which encode the zinc finger domains necessary for DNA binding. In addition, the wild-type GATA3 allele tends to be retained and both alleles are expressed in these tumors (9, 12). The initial interpretation in the literature suggests that GATA3 is a haploinsufficient tumor suppressor gene in breast cancer and that the mutant alleles encode non-functional proteins (9); however, this model has not been rigorously tested. Previous studies (12–14) have noted that comparison of the mutation spectrum of GATA3 in breast cancer with that found in hypoparathyroidism, deafness and renal dysplasia (HDR) syndrome, a genuine human GATA3 haploinsufficiency disease, casts doubt on this interpretation. HDR syndrome is generally associated with GATA3 mutations throughout the open reading frame or whole gene deletions (15). However, the mutations found in breast cancer specimens cluster predominantly at the C-terminus of the protein. Data from HDR cases have clearly shown that GATA3 function can effectively be disrupted by mutations throughout the coding sequence, but these mutations are not selected for in breast cancer. Instead, there appears to be a selective pressure to retain and express a substantial proportion of the GATA3 open reading frame, which suggests that the encoded protein may have a residual or novel function rather than simply being a null allele. Consistent with this interpretation, a recent study has shown that CRISPR-mediated deletion of mutant GATA3 attenuates tumorigenesis in MCF7 cell line xenografts and that overexpression of mutant GATA3 increased tumor growth rate in CAMA-1 cell line xenografts (16).
Previous animal studies have relied on xenografts in immunocompromised mice to functionally characterize GATA3 mutant proteins. In this study, we developed a transgenic mouse with mammary-specific expression of human mutant GATA3 to more rigorously determine whether the truncating GATA3 alleles are truly null alleles or if the encoding proteins may retain residual or novel functions relevant to tumorigenesis. We showed that mutant GATA3 elicits hyperproliferative phenotypes during mammary gland development. Consistent with this phenotype, studies with two GATA3 hotspot mutations showed that ectopic expression of these proteins increased the growth rate of ZR751 cell line both in vitro and in xenografts. These findings suggest a model in which the studied recurrent breast cancer mutations do not result in haploinsufficiency or simple loss-of-function, but encode functional proteins, which actively contribute to tumor growth.
Materials and Methods
Generation of MMTV-GATA3335fs mice
The GATA3335fs coding sequence was subcloned under the control of the steroid hormone-responsive mouse mammary tumor virus (MMTV) promoter into the MMTV-SV40-BSSK plasmid (kindly provided by Philip Leder, plasmid #1824, Addgene) (17). Constructs were linearized and injected into FVB pronuclei and implanted into surrogate FVB mice. Germline transmission was confirmed by PCR of tail DNA (5′-ACTCCAGCCACATGCTGAC and 5′-TCACACCACAGAAGTAAGGTTCC). Whole mount analysis of inguinal mammary glands from estrus-matched (18) 9-week old non-parous mice was performed as previously described (19). The extent of mammary gland development was quantified on a 0–3 scale as has previously been described (20).
Cell culture
ZR751 cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and identity was verified by short tandem repeat (STR) profiling at our institutional genomics core facility using the Genemarker 10 kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. ZR751 cells stably expressing FLAG-tagged GATA3308*, GATA3335fs, or the empty vector control were generated using pBM-IRES-Puro retroviral vector (21). 3D culture was performed as previously described (22).
Xenografts
Xenografts were performed in athymic mice (National Cancer Institute) and were approved by the Institutional Animal Use and Care Committee. Thirty nulliparous 5-week old female mice were implanted subcutaneously with 60-day slow-release pellets containing 0.72 mg of 17β-estradiol (Innovative Research, Sarasota, FL, USA). Two million ZR751 cells in 100 μl of a 1:1 mixture of RPMI 1640 (Thermo Fisher, Waltham, MA, USA) and Matrigel (BD Biosciences, San Jose, CA, USA) were injected orthotopically into the right and left 4th inguinal mammary fat pads using a 21½ gauge needle. Animals were checked daily for tumor formation by palpation. Tumor size was measured thrice per week using a caliper and was calculated using the formula ½ x length x width x height
Western Blot
Mammary glands from 9 week, 15.5 day pregnant, and lactating wildtype and transgenic mice were minced, resuspended in lysis buffer (25 mM Tris-HCl pH 7.6, 200 mM NaCl, 5 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS) and homogenized using a tissue grinder. Samples were resolved by SDS-PAGE on 4–20% gradient gels (Biorad Laboratories, Hercules, CA) and transferred to PVDF membranes. Antibodies used were against GATA3 (BD Biosciences, San Jose, CA, USA) and E-Cadherin (Cell Signaling Technology, Danvers, MA, USA) and were incubated overnight prior to washing and detection with HRP-conjugated secondary antibodies and the Super Signal Chemilumescent Substrate (Pierce, Waltham, MA, USA).
Statistical analysis
All statistical analyses were performed using GraphPad Prism (Graphpad Software, La Jolla, CA, USA). The frequencies of categories of mammary phenotypes in transgenic and matched control mice were compared using a Chi-squared test. Differences between median colony sizes in 3D culture were evaluated using the Mann-Whitney test. For xenograft studies, Kaplan-Meier analysis with log-rank test was used to compare time to tumor formation between groups, and differences between experimental and control xenograft volumes at each time point shown were evaluated using a t-test. Cell culture experiments were repeated twice, and animal experiments included the numbers of replicate animal indicated in each figure legend. P < 0.05 was the threshold for statistical significance in all cases.
Results
Two common GATA3 mutations were chosen for further analysis; a splice acceptor site deletion affecting codon 308, modeled as a stop codon (GATA3308*), and a frameshift at codon 335 (GATA3335fs), which, collectively, are reported in more than 100 breast cancer cases in cBioportal (7–9, 23, 24). Codon numbering was according to NCBI reference sequence NM_002051.2. The GATA3335fs mutant was evaluated in a transgenic model, and both mutants in cancer cell line xenografts.
GATA3335fs leads to precocious lobuloalveolar development in the mammary gland
To determine whether expression of the GATA3335fs protein might confer a competitive proliferative advantage on mammary epithelial cells in a physiologic setting, transgenic mice expressing this allele under the control of the MMTV promoter were generated (Figure 1A). This strategy also allowed the comparison of the present GATA3335fs-induced phenotype to prior reports of GATA3 manipulation in the mammary gland. Mammary-specific GATA3 deletion has been reported to cause a failure in mammary gland development (2, 3); a finding expected to be phenocopied if the mutant GATA3 allele encodes a dominant negative protein. In contrast, the overexpression of wild-type GATA3 in mice using the MMTV promoter resulted in precocious lobuloalveolar development reminiscent of early pregnancy in the mammary gland (25).
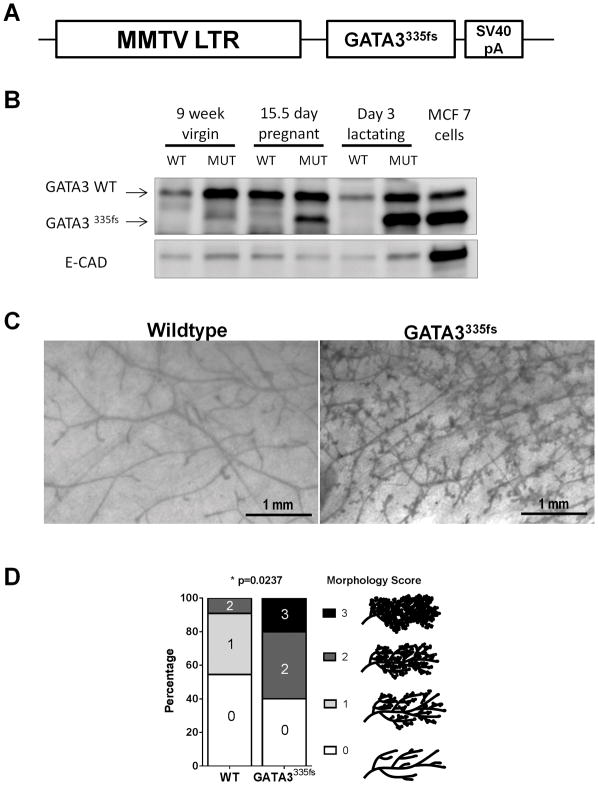
Figure 1.
Mammary specific expression of GATA3335fs in transgenic mice leads to precocious lobuloalveolar development. A. MMTV-GATA3335fs construct with GATA3335fs cloned downstream of the luminal cell-specific MMTV-LTR promoter. B. Expression of GATA3335fs transgenic protein in virgin, pregnant, and lactating mouse mammary glands analyzed by western blot. MCF7 cells, which express this mutant, were used as a positive control. E-cadherin, an epithelial cell marker, was used as a loading control. C. Representative carmine-stained mammary gland whole mounts in metestrus-matched 9 week MMTV-GATA3335fs and littermate wildtype controls. D. The extent of mammary gland development was quantified in 9 week old MMTV-GATA3335fs (n= 10) and wild-type (n=11) mice (p<0.0237, Fisher’s Exact Test).
Western blot analysis indicated that the mutant protein was expressed in the non-parous gland and that expression is increased during pregnancy and lactation (Figure 1B), consistent with control by the hormonally-regulated MMTV promoter (26). Lysate from MCF7 cells which endogenously express this mutant protein was used as a positive control. Transgenic female mice were indistinguishable from wild-type littermates in general health and body weight and were able to suckle pups normally. Whole mount analysis was performed on mammary glands from non-parous 9-week old mice, all of which were in metestrus. Representative examples showed significantly increased branching and precocious lobuloalveolar development in GATA3335fs glands compared to wild type glands (Figure 1C). Mammary glands were scored by the extent of their morphological development using a scale from 0 to 3 as described by Fata and colleagues (20). Glands were observed across the full range of developmental stages from Grade 0 (consisting mostly of primary ducts with a limited number of secondary ducts) to Grade 3 (fully branched with extensive alveolar development). Samples were scored blindly and independently by two individuals. A significant shift to a precocious lobuloalveolar phenotype was observed in the mammary glands expressing GATA3335fs. This precocious development of the mammary gland more closely resembled the phenotype observed in MMTV-GATA3WT mice (25) and was in stark contrast to the failure of mammary gland development in GATA3 conditional deletion mice (2, 3), suggesting that this mutant does not act in a dominant negative fashion, and is able to induce a proliferative phenotype.
Characterization of two GATA3 mutant proteins in human breast cancer cells
To further address the potential functions of these mutant proteins, stable cell lines were generated in the ZR751 cell line, an ER+ luminal breast cancer cell line that expresses endogenous wild-type GATA3. Specifically, ZR751 cells were transfected with either the empty vector, or with plasmids encoding FLAG-tagged GATA308* and GATA3335fs. This cell line was chosen since it more closely resembles the ER+ luminal tumors in which GATA3 mutations are observed in the clinic than the ER-negative cell lines typically used in prior studies. Accordingly, it may be a more physiologically relevant model for analyzing the role of GATA3 and its mutants in tumorigenesis.
In 3D culture, the size of colonies formed by the ZR751 cell line was estrogen-dependent, with the control cells having an 88% higher median colony size when cultured in the presence of 100 nM estradiol. Interestingly, ectopic expression of both mutant GATA3 proteins in the ZR751 cell line led to a larger colony size in the absence of estradiol (gray boxes, Figure 2). The magnitude of the proliferative response to estrogen was not as large in the GATA3308* and GATA3335fs expressing cells; 29.5 % larger (p<0.0001) and 22 % larger (p<0.0001), respectively. These data demonstrated that the expression of the single zinc finger mutants in ZR751 cells caused increased colony size in the 3D culture in the absence of estrogen. This outcome approached the effect size observed when the control cells were treated with estradiol.
Figure 2.
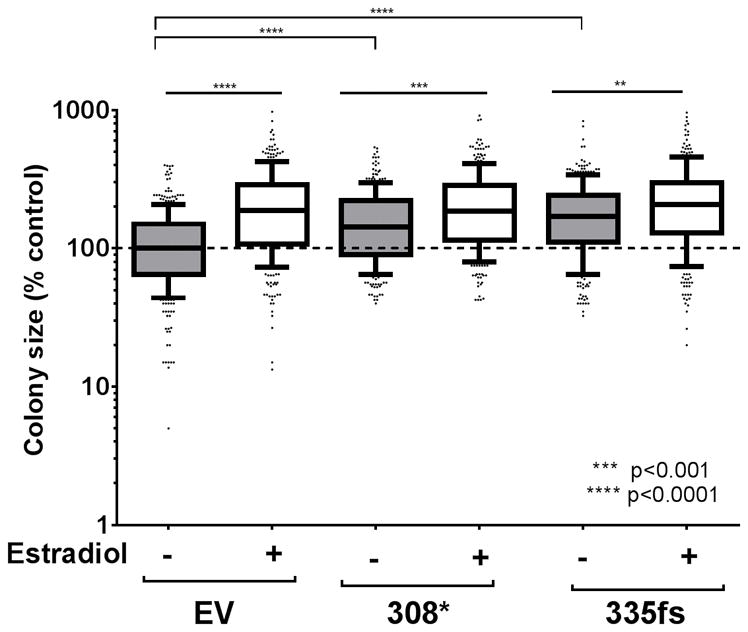
In vitro growth of ZR751 cells expressing ectopic mutant GATA3.
3D culture colony growth of the ZR751 stable cell lines in the absence or presence of 100 nM estradiol. The graph represents quantification of two independent experiments, each normalized to the median colony size in the control cultures at the time of quantification (**p<0.01, ***p<0.001 and ****p<0.0001, Mann-Whitney test). EV, ZR751 cells transfected with empty vector control; 308*, ZR751 cells expressing FLAG-tagged GATA3308*; 335fs, ZR751 cells expressing FLAG-tagged GATA3335fs
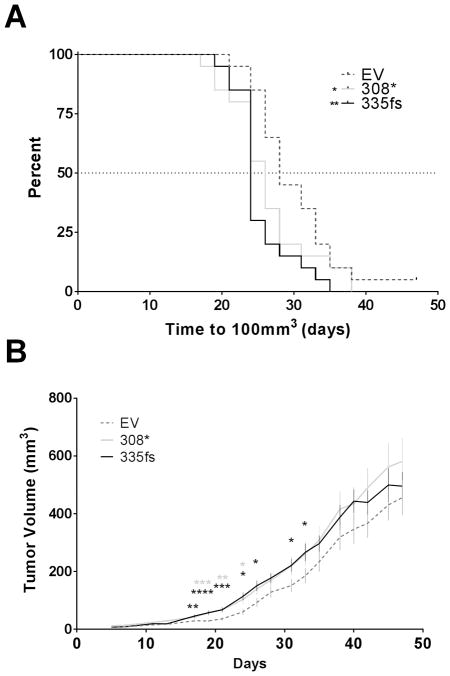
To determine whether the GATA3 mutants conferred a similar growth advantage in vivo, these cell lines were xenografted into the fat pads of athymic nude mice implanted with slow-release estrogen pellets (n=20 xenografts per cell line). Mice were followed up until the larger tumors reached a volume of 600 mm3 (48 days). The time required to reach 100 mm3 volume was measured and it was found that the GATA3308* and GATA3335fs expressing tumors reached the target volume significantly more quickly than the controls (Figure 3A). The GATA3308* and GATA3335fs expressing tumors were all significantly larger compared to the control tumors at the indicated time-points (Figure 3B).
Figure 3.
In vivo growth of ZR751 cells expressing ectopic mutant GATA3. A. Kaplan-Meier curve showing the time to 100 mm3 xenograft tumor volume (*p<0.05, **p<0.01, Log-rank test). B. Comparison of growth rates of ZR751 xenograft tumors. Graph shows tumor volumes and standard error of the mean (n=20 per group, (*p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001, Student’s t-test). EV, ZR751 cells transfected with empty vector control; 308*, ZR751 cells expressing FLAG-tagged GATA3308*; 335fs, ZR751 cells expressing FLAG-tagged GATA3335fs
Discussion
In this study, we challenged the existing paradigm that haploinsufficiency is the main outcome for breast cancer-specific GATA3 mutations. We show that expression of mutant GATA3 leads to precocious lobuloalveolar development in non-parous mice in the presence of two wild-type GATA3 alleles, and increased xenograft growth of a luminal breast cancer cell line in vivo.
We considered the possibility that the mutant protein might somehow antagonize the transcriptional activity of the wild-type protein in a dominant-negative fashion; however, we did not see any dominant negative effects of the mutants on wildtype GATA3 in luciferase assays (data not shown). Other studies that have recently tested three different GATA3 mutants in a similar reporter assay also did not show any dominant-negative effect with truncated mutants (G314fs and R330fs) that lack the second zinc finger, but detected a modest dominant-negative effect in two other mutants (R330del and P408fs) that retained both zinc fingers, albeit only in one of the two cell lines they studied (12). A more rigorous test of dominant-negative activity than these artificial luciferase reporter constructs is to test whether expression of the mutant protein in the mouse mammary gland can phenocopy the defects in mammary outgrowth described in the mammary-specific GATA3 knockout models (2, 3). Unlike the substantial reduction in mammary epithelial tree growth that was reported in those mice (2, 3), mice expressing the GATA3335fs mutant transgene developed full mammary glands with a precocious lobuloalveolar development phenotype. These data indicate that – even in the presence of the two endogenous wild-type GATA3 alleles – expression of this truncated mutant is sufficient to confer a proliferative phenotype. This mammary phenotype strongly resembles that of MMTV-GATA3WT mice (25). In our cell line model, the expression of mutant GATA3 was sufficient to accelerate tumor growth. These findings argue against both the simple haploinsufficiency model and the dominant-negative model for the role of mutant GATA3 in breast cancer and are more consistent with the mutant protein having a residual or novel function.
Genes mutated in cancer can be considered to fall into one of four categories – passenger mutations, classical two-hit tumor suppressor genes, haploinsufficient tumor suppressor genes or proto-oncogenes. The prevalence of GATA3 mutations in ER+ breast cancer and their restriction to this breast cancer subtype argues strongly against a passenger mutation hypothesis. It is also clear from earlier work (9) that wild-type GATA3 and mutant GATA3 are co-expressed, indicating that GATA3 does not follow the classical two-hit tumor suppressor gene model. The haploinsufficiency model has received most attention (7, 9) so far, and while our data do not fully exclude a potential contribution for haploinsufficiency, one might expect a far greater prevalence of single allele GATA3 deletion and/or mutations throughout the open reading frame if this was the primary mechanism, and we have demonstrated in this study that the mutants are not null alleles.
Instead, given the facts that (i) the mutant proteins are expressed in GATA3mut tumors, (ii) the selective pressure in tumor evolution appears to favor truncating mutations rather than gene deletions, and (iii) the mutant proteins exert proliferative phenotypes on both normal and malignant mammary epithelial cells, we believe that the data are most consistent with GATA3 being a mammary proto-oncogene. This contention would be strengthened by demonstrating mammary tumorigenesis in the MMTV-GATA3335fs mice. We observed no tumors in a cohort of 13 non-parous females that we followed (median age 553 days, range 371–685 days), although in several other MMTV-driven mammary tumor models tumor incidence increases with parity, an interaction we have not addressed in our study. Tests to determine whether mammary tumorigenesis is accelerated in other models by crossing with MMTV-GATA3335fs mice would also provide a rigorous test for the proto-oncogene hypothesis.
Acknowledgments
This study was supported by a Peter T. Rowley Breast Cancer Research Grant from the New York State Department of Health (C028250) to PAK and by the Gundersen Medical Foundation. EAP was supported by an NIH NIGMS IRACDA K12 (1K12GM102779-01). Institutional core facilities were supported by the Albert Einstein Cancer Center (NIH P30 CA013330). PK holds the Dr. Jon & Betty Kabara Endowed Chair in Precision Oncology.
References
- 1.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 3.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- 5.Voduc D, Cheang M, Nielsen T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev. 2008;17:365–373. doi: 10.1158/1055-9965.EPI-06-1090. [DOI] [PubMed] [Google Scholar]
- 6.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 7.Arnold JM, Choong DY, Thompson ER, Waddell N, Lindeman GJ, Visvader JE, Campbell IG, Chenevix-Trench G. Frequent somatic mutations of GATA3 in non-BRCA1/BRCA2 familial breast tumors, but not in BRCA1-, BRCA2- or sporadic breast tumors. Breast Cancer Res Treat. 2010;119:491–496. doi: 10.1007/s10549-008-0269-x. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, Lonning PE, Strausberg RL, Chanock S, Borresen-Dale AL, Perou CM. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 10.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaynor KU, Grigorieva IV, Allen MD, Esapa CT, Head RA, Gopinath P, Christie PT, Nesbit MA, Jones JL, Thakker RV. GATA3 Mutations Found in Breast Cancers May Be Associated with Aberrant Nuclear Localization, Reduced Transactivation and Cell Invasiveness. Horm Cancer. 2013;4:123–139. doi: 10.1007/s12672-013-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adomas AB, Grimm SA, Malone C, Takaku M, Sims JK, Wade PA. Breast tumor specific mutation in GATA3 affects physiological mechanisms regulating transcription factor turnover. BMC Cancer. 2014;14:278. doi: 10.1186/1471-2407-14-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenny PA, Chandiramani N. Transgenic expression of a breast-cancer specific GATA3 mutant leads to mammary hyperplasia. Cancer Res. 2012;72(24 Suppl) Abstract nr P2-04-06. [Google Scholar]
- 15.Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, Beetz R, Bilous RW, Holdaway I, Shaw NJ, Fryns JP, Van de Ven W, Thakker RV, Devriendt K. GATA3 haplo-insufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- 16.Gustin JP, Miller J, Farag M, Rosen DM, Thomas M, Scharpf RB, Lauring J. GATA3 frameshift mutation promotes tumor growth in human luminal breast cancer cells and induces transcriptional changes seen in primary GATA3 mutant breast cancers. Oncotarget. 2017;8:103415–103427. doi: 10.18632/oncotarget.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci U S A. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caligioni CS. Assessing reproductive status/stages in mice. Curr Protoc Neurosci. 2009 doi: 10.1002/0471142301.nsa04is48. Appendix 4: Appendix 4I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen SB, Young LJ, Smith GH. Preparing mammary gland whole mounts from mice. In: MM, Asch BB, editors. Methods in Mammary Gland Biology and Breast Cancer Research. New York: Kluwer; 2000. pp. 75–85. [Google Scholar]
- 20.Fata JE, Chaudhary V, Khokha R. Cellular turnover in the mammary gland is correlated with systemic levels of progesterone and not 17beta-estradiol during the estrous cycle. Biol Reprod. 2001;65:680–688. doi: 10.1095/biolreprod65.3.680. [DOI] [PubMed] [Google Scholar]
- 21.Garton KJ, Ferri N, Raines EW. Efficient expression of exogenous genes in primary vascular cells using IRES-based retroviral vectors. Biotechniques. 2002;32:830, 832, 834. doi: 10.2144/02324rr01. passim. [DOI] [PubMed] [Google Scholar]
- 22.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer C, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asselin-Labat ML, Sutherland KD, Vaillant F, Gyorki DE, Wu D, Holroyd S, Breslin K, Ward T, Shi W, Bath ML, Deb S, Fox SB, Smyth GK, Lindeman GJ, Visvader JE. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol. 2011;31:4609–4622. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otten AD, Sanders MM, McKnight GS. The MMTV LTR promoter is induced by progesterone and dihydrotestosterone but not by estrogen. Mol Endocrinol. 1988;2:143–147. doi: 10.1210/mend-2-2-143. [DOI] [PubMed] [Google Scholar]