Abstract
Using the Hemagglutinin (HA) protein peptide array of H1N1 pdm09 and a panel of swine antisera against various swine influenza H1 and H3 clusters, we identified three immunoreactive epitopes with one (peptide 15) located in HA1 (amino acids 57-71) and two (peptides 121 and 139) in HA2 (amino acids 481-495 and 553-566). Further analysis showed that all swine antisera of H1 clusters efficiently recognized two HA2 epitopes; peptides 121 and 139, with only a subset of antisera reactive to HA1-derived peptide 15. Interestingly, none of these peptides were reactive to SIV H3 antisera. Finally, intranasal inoculation of peptides 15 and 121 into pigs revealed that peptide 121, not peptide 15, was able to generate antibody responses in some animals. The results of our experiments provide an important foundation for further analyzing the immune response against these peptides during natural viral infection and also provide peptide substrates for diagnostic assays.
Keywords: Swine influenza, H1 subtype, H3 subtype, hemagglutinin (HA) Protein, B cell epitope, peptide
1. Introduction
Swine influenza is caused by swine influenza A virus (SIV), which belongs to the Orthomyxoviridae family (Easterday, 1980). The pathogen is an important component of the porcine respiratory disease complex and by itself can have a significant economic impact in the swine industry (Vincent et al., 2008). Swine influenza is ranked among the top three major health challenges in the global swine industry (Vincent et al., 2017). An additional concern regarding SIV, is its zoonotic potential, as under certain circumstances, it can be transmitted to humans, which is best exemplified by 2009 H1N1 pandemic originated from swine source (Maines et al., 2009). Recently, transmissions of variant swine H3N2 virus (H3N2v) to humans have also been documented (Wong et al., 2013).
A typical outbreak of respiratory disease caused by SIV is characterized by sudden onset, and rapid spread within a herd. Clinical symptoms associated with swine influenza may include coughing, sneezing, nasal discharge, elevated rectal temperature, lethargy, breathing difficulty, and depressed appetite (Easterday, 1980; Vincent et al., 2008). While morbidity rates may reach 100% with SIV infections, mortality rates are generally low.
The genome of SIV is separated into 8 independently RNA segments at the negative-sense that allows for frequent reassortment when two different viruses infect and replicate within the same cell of a pig. Reassortment involving HA segment event often results in the production of new influenza virus (antigenic shift), which renders the current strain-specific vaccine strategy ineffective (Erbelding et al., 2018). In addition, influenza A virus has the unique capacity to undergo genetic variations (antigenic drift) by being able to mutate up to 50% of the amino acid sequence of its major surface protein, hemagglutinin (HA), without changing the function of the HA. Both antigenic drift and antigenic shift contribute to apparent failures of swine influenza vaccines when used in swine industry.
In the United States, SIV H1N1, H1N2, and H3N2 subtypes have emerged as the major causes of swine influenza, although two other subtypes of SIV (H3N1 and H2N3) have also been isolated from the diseased pigs on some occasions (Rajao et al., 2018). Each subtype consists of numerous linages or genetic clusters and these lineages of swine influenza virus circulate concurrently in pigs, which present a challenge to effective control and prevent of this important disease (Rajao et al., 2018).
The aim of this study was to determine the antigenic determinants of the hemagglutinin (HA) protein of H1N1 pandemic 2009 virus (H1N1 pdm09) by using the HA peptide array in Enzyme-linked immunosorbent assay coupled with immune sera from immunized pigs by SIVs. The peptide array contains 139 peptides spanning the HA protein of H1N1pdm09 and peptides are 14- or 15-mers with 11 amino acid overlaps. A panel of swine antisera against SIV H1 clusters α, β, γ, δ-1, δ-2, H1N1pdm09 (human A/CA/04/2009), and H3 cluster were used in this study (Hause BM et al., 2011; Hause et al., 2010). Our experiments identified two conserved peptide antigens (peptides 121 and 139) located in HA2 that were reactive to all tested SIV H1 antisera. In addition, peptide 15 in HA1 was recognized by a subset of H1 antisera. Interestingly, none of these identified peptides were recognized by SIV H3 reference antisera. The results of our experiments shall provide an important foundation for further analyzing the immune response against these peptides during natural SIV infections and also provide potential peptide substrates for design of diagnostic assays and vaccine strategies against influenza A virus infection of swine.
2. Materials and methods
2.1. Peptide array
Peptide array is derived from the hemagglutinin (HA) protein of influenza A virus H1N1 A/California/04/2009 (pdm09) with swine origin, which was provided by BEI Resources, NIAID, NIH (Cat# NR-15433). This is a 139-peptide array that spans the whole HA protein. Peptides are 14- to 15-mers in length, with 11 amino acid overlaps. Each peptide was dissolved with Dimethyl sulfoxide (DMSO) (Sigma-Aldrich) at a concentration of 1 mg/ml.
2.2. Pig sera
A previously described panel of porcine immune sera collected from experimentally inoculated pigs was used in this study (Hause BM et al., 2011; Hause et al., 2010). Three-week-old SIV-negative pigs were obtained from a commercial vendor. Groups of 3 pigs were used for antisera generation against a panel of swine influenza virus reference isolates, including H1 clusters (α, β, γ, δ-1, δ-2, and H1N1pdm09) and a H3 cluster. Note that human H1N1 A/CA/04/2009 was used to inoculate pigs to produce swine H1N1pdm09 antisera. Each of these viruses was propagated in swine testicles (ST) cells (reaching to 1280 to 2560 hemagglutination units per ml) followed by the inactivation with 5 mM binary ethyleneimine (BEI) (Sigma-Aldrich) for 24 hours at 37°C. After formulation with Trigen, a proprietary adjuvant (Newport Laboratories, Worthington, Minnesota), each of these viral antigens was inoculated into a group of three pigs (2 ml each pig) on days 0, 7, 14, and 21, intramuscularly. Sera were collected and pooled on days 0 and 35, respectively, prior to testing. Because sera collected from inoculated pigs had very high HI titers (homologous HI 1280 to > 20,480), sera were diluted four-fold to 16-fold in sera collected from cesarean-derived, colostrum-deprived pigs to achieve homologous HI titers of 1280. The diluted reference antisera stocks were aliquoted and frozen at −80°C. In addition, we generated negative control sera by injecting pigs with ST cell debris and adjuvant. Sera generated from the ST cell debris were not diluted. Non-immune pig sera obtained from 50 pigs known to be free from swine influenza exposure and infection (negative for IAV-specific antibodies in ELISA) were used as negative control sera to detect background immunoglobulin binding in peptide ELISAs.
2.3. Peptide ELISA
Flat bottom 96-well plates were coated with each of the synthetic HA peptides (100 μl/0.5μg each well) in coating buffer (10 mM NaHCO3 buffer, pH 9.6) for overnight incubation at 4°C. After washing six times (300 μl/well) with PBST wash buffer (PBS with 0.05% Tween 20), plates were blocked with blocking buffer (PBST containing 0.2% gelatin) for 2 hours at 37°C followed by additional washing. Then 100 μl of diluted serum samples (1:300 in PBST containing 0.01% gelatin) was added to each well and incubated at 37°C for 1 hour. Following six times washing, antibody binding was detected by incubation with 100 μL of goat anti-swine IgG-HRP conjugates (SeraCare) diluted 1:2000 in blocking buffer at 37°C for 30 min. The calorimetric reaction, indicative of the presence of bound antibody, was mediated by the addition of 100 μL freshly prepared tetramethylbenzidine (TMB) substrate solution (SeraCare). The reaction was stopped by addition of 50 μL of 4 M sulfuric acid and the optical density (OD) was measured at 450 nm in an ELISA microplate reader (BioTek). Considering that a relatively large pool of peptides was tested in our work, OD was reported instead of endpoint titer. We selected 1:300 dilution for ELISA throughout this work because our initial optimization work (i.e. test a range of serum dilutions) revealed that 1:300 dilution gave rise to a good and reproducible signal-to-noise signal. Under these conditions, ELISA can become saturated (end of the linear range) when OD reaches 1.5 or higher as determined in a plate reader.
The 50 negative control sera coupled with randomly selected 4 peptide pools (each peptide pool consisting of five sequentially overlapping peptides) were used to determine the cut-off value of the peptide ELISA, which was equal to the mean OD of the negative samples plus 3 standard deviations. The derived positive cut-off value of 0.18 or greater was used to determine whether a given SIV cluster-specific serum had positive serological reactivity to the peptides. To reduce the background (BG) noise reaction caused by the hydrophobic binding of immunoglobulin and immune-complexes in pig sera to plastic surfaces, all the OD values obtained in peptide(s)-coated wells were subtracted from the BG OD values in peptide(s)-uncoated wells.
2.4. Hemagglutination inhibition (HI) assay
HI assay was performed following the standard procedure (Hause et al., 2012). In brief, sera were treated with a receptor-destroying enzyme for 24 h at 37°C and then adsorbed with a 20% suspension of turkey erythrocytes in phosphate-buffered saline (PBS) for 30 min at room temperature. Influenza H1N1pdm09 virus suspensions containing 4 to 8 HA units of virus were incubated for 1 h with serial 2-fold dilutions of antiserum, and the HI titer was determined as the reciprocal of the highest dilution that showed complete inhibition of hemagglutination using 0.5% washed turkey erythrocytes.
2.5. Virus neutralization assay
Virus neutralization assays were performed as previously described (Ran et al., 2015), with slight modifications. Two-fold serial dilutions in PBS, from 1:4 to 1:32,768, were performed on the sera. Next, 100 μl of 40 to 200 median tissue culture infective doses (TCID50)/100 μl of influenza H1N1pdm09 virus was added to each serum dilution and incubated for 1 h at 37°C in 5% CO2. The serum-virus mixture was next transferred to a confluent monolayer of MDCK cells and incubated at 37°C in 5% CO2 for 4 days. Viral neutralization was assessed by a lack of cytopathic effects in each well by microscopy. Back titrations were performed on MDCK cells to confirm a virus concentration of 40 to 200 TCID50/100 μl.
2.6. Structural locations of peptides tested positive in ELISA
The Hemagglutinin structure (PDB 3LZG) from H1N1 pdm09 that derives the HA peptide array was chosen as the template to locate ELISA-positive peptides in the context of the HA structure.
2.7. Sequence logos
We used pLogo software (http://rth.dk/resources/plogo/) to determine the sequence conservation of the identified B-cell linear epitopes. For this purpose, we analyzed all available SIV cluster-specific full-length HA protein sequences of swine influenza A virus strains that circulated between 2006 to 2015 in pigs farms of the United States. Sequence logo provides better description of sequence conservation than consensus sequences and can reveal significantly structural and functional features of the identified epitopes. Each logo consists of stacks of letters (amino acids), one stack for each position in the sequence. The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding amino acid at that position. The residue position in the block is shown on the X-axis, and the information content is shown on the Y-axis. The default color scheme displaying different amino acids according to their different chemical properties is as follows: polar amino acids (G, S, T, Y, C, Q, N) colored with green, basic (K, R, H) with blue, acidic (D, E) with red, and hydrophobic (A, V, L, I, P, W, F, M) amino acids with black.
2.8. In vivo evaluation of immunogenicity of peptides 15 and 121
3-week-old commercial influenza antibody-negative pigs were obtained from a commercial vendor in Minnesota. A commercial ELISA kit (Swine Influenza NP Antibody Test Kit with Cat# 99-0000900, Idexx Laboratories Inc, Westbrook, Maine) was used to determine swine influenza antibody-negative status of these pigs. The pigs were acclimatized for 7 days before inoculation of peptide immunogens. Maintenance of pigs and all experimental procedures were conducted in accordance with the guidelines of the Institutional Laboratory Animal Care and Use Committee, South Dakota State University. Peptide antigens were conjugated with carrier protein KLH (500μg of each peptide dissolved in total volume of 1ml PBS). Each of six 4-week-old pigs was inoculated intranasally with one ml suspension of the KLH-conjugated peptides containing both peptides 15 and 121 along with heat labile enterotoxin (25 μg LT), while each of four pigs in control group received 1 ml PBS intranasally. Two weeks later, all piglets received a booster dosage. Blood samples were collected for the serum isolation via the jugular vein from piglets 2 weeks after the second round of immunization. Serum samples were also obtained by the same method prior to immunization. Peptide-specific antibodies were measured in the ELISA assay as described above.
3. Results
3.1. Identification of HA peptide pools reactive to SIV H1N1pdm09 reference serum
To localize linear B-cell epitopes of the HA protein of H1N1 pdm09 in swine, pig serum raised against H1N1 pdm09 virus was tested for its reactivity against pdm09 HA peptide array. We first screened antibody reactivity against peptide pools in ELISA. Each peptide contained five sequentially overlapping peptides that were arranged from the N-terminal end of the HA protein. Peptide pools were considered as positive for the SIV serum sample when the OD (optical density) value was higher than the cut-off value 0.18, which was determined at the mean + 3 standard deviations based on analysis of 50 SIV antibody-negative serum samples. Three out of the twenty-eight peptide pools were tested positively against the H1N1pdm09 antiserum (Table 1).
Table 1.
Summary of positive peptide pools identified in ELISA using swine antisera against homologous H1N1 pdm09 virus.
| Peptide | OD | Sequence and location |
|---|---|---|
| 1-5 | - | 1 mkailvvllytfatanadtlcigyhannstd 31 |
| 6-10 | - | 21 cigyhannstdtvdtvleknvtvthsvnlle 51 |
| 11-15 | 0.336 | 41 vtvthsvnlledkhngklcklrgvaplhlgk 71 |
| 16-20 | - | 61l rgvaplhlgkcniagwilgnpeceslstas 91 |
| 21-25 | - | 81 npeceslstasswsyivetpssdngtcypgd 111 |
| 26-30 | - | 101LSissdngtcypgdfidyeelreqlssvssferf 131 |
| 31-35 | - | 121 qlssvssferfeifpktsswpnhdsnkgvta 151 |
| 36-40 | - | 141 pnhdsnkgvtaacphagaksfyknliwlvkk 171 |
| 41-45 | - | 161 fyknliwlvkkgnsypklsksyindkgkevl 191 |
| 46-50 | - | 181 syindkgkevlvlwgihhpstsadqqslyqn 211 |
| 51-55 | - | 201 tsadqqslyqnadayvfvgssryskkfkpei 231 |
| 56-60 | - | 221 sryskkfkpeiairpkvrdqegrmnyywtlv 251 |
| 61-65 | - | 241 egrmnyywtlvepgdkitfeatgnlvvprya 271 |
| 66-70 | - | 261 atgnlvvpryafamernagsgiiisdtpvhd 291 |
| 71-75 | - | 281 giiisdtpvhdcnttcqtpkgaintslpfqn 311 |
| 76-80 | - | 301 qaintslpfqnihpitiqkcpkyvkstklrl 331 |
| 81-85 | - | 321 pkyvkstklrlatglrnipsiqsRGlfgaia 351 |
| 86-90 | - | 341 iqsRGlfqaiaqfieqqwtqmvdqwyqyhhq 371 |
| 91-95 | - | 361 mvdgqygyhhqneqgsgyaadlkstqnaide 391 |
| 96-100 | - | 381 dlkstqnaideitnkvnsviekmntqftavg 411 |
| 101-105 | - | 401 ekmntqftavgkefnhlekrienlnkkv 431 |
| 106-100 | - | 421 ienlnkkv fldiwtynaellvllenert 451 |
| 111-115 | - | 441 ellvllenertldyhdsnvknlyekvrsqlk 471 |
| 116-120 | - | 461 nlyekvrsqlknnakeigngcfefyhkcdnt 491 |
| 121-125 | 0.231 | 481 cfefyhkcdntcmesvkngtydypkyseeak 511 |
| 126-130 | - | 501 ydypkyseeaklnreeidgvklestriyqil 531 |
| 131-135 | - | 521 klestriyqilaiystvasslvlvvslgais 551 |
| 136-139 | 0.341 | 541 lvlvvslgaisfwmcsngslqcrici 566 |
Note: "-" sign indicates that peptide pools are not reactive to swine antisera raised against homologous H1N1 pdm09 virus (i.e., OD values less than the ELISA cut-off 0.18). Positive peptide pools are indicated with red font color. Sequences with yellow shades are located in HA1. Sequences with blue shades are located in HA2. Junction sequence between HA1 and HA2 are not shaded. Reported OD data represent average background-corrected OD values obtained from three independent experiments, each assayed in duplicate. See Materials and methods section for details.
3.2. Identification of individual peptides recognized by SIV H1N1pdm09 reference serum
To further determine individual peptide epitopes from the above positive peptide pools, each peptide was tested against swine H1N1pdm09 antiserum in ELISA under similar experimental conditions. The reactivity pattern was as follow (Table 2): peptide 15 from the positive peptide pool 11-15 reacted strongly with antiserum; similarly, peptide 121 from the positive peptide pool 121-125 was recognized by H1N1pdm 09 antiserum; interestingly, three overlapping peptides 137, 138, and 139 all reacted with antiserum. Significantly, the observation of an immunoreactive porcine B cell epitope represented by peptide 15 is in agreement with an early study demonstrating that this peptide was immunoreactive against convalescent sera from H1N1pdm09 patients (Zhao et al., 2011). Porcine B cell epitopes represented by peptides 121 and 137/138/139 have not been reported previously in the context of human infection by H1N1pdm09 virus. Three overlapping peptides (137/138/139) (Table 2) may represent a single epitope consisting of residues 553 to 559 (WMCSNGS). It is interesting to note that peptide 139, not other two peptides, contains a complete intracytoplasmic tail. Based on these reasons, we selected peptide 139 over other two overlapping peptides as a representative of this epitope for further analysis along with peptides 15 and 121.
Table 2.
Summary of Individual peptides reactive to swine antisera against H1N1 pdm09 virus.
| Peptide | OD | Sequence and position |
|---|---|---|
| 15 | 0.662 | 57 KLCKLRGVAPLHLGK 71 |
| 121 | 0.262 | 481 CFEFYHKCDNTCMES 495 |
| 137 | 1.101 | 545 VSLGAISFWMCSNGS 559 |
| 138 | 1.426 | 549 AISFWMCSNGSLQCR 563 |
| 139 | 0.636 | 553 WMCSNGSLQCRICI 566 |
Note: The underlined amino acid residues for peptides 137, 138, and 139 are those shared among three reactive peptides. Reported OD data represent average background-corrected OD values obtained from three independent experiments, each assayed in duplicate. See Materials and methods section for details.
3.3. Structural locations of positive peptides
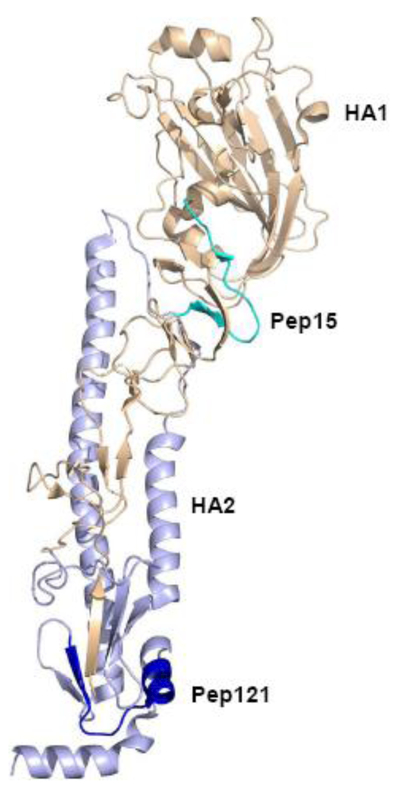
Peptide 15 (residues 57 to 71) is located in the HA1 subunit, while peptide 121 (residues 481 to 495) is located in the HA2 region. These two peptides are in the HA ectodomain. In contrast, peptide 39 (residues 553 to 566) is within the transmembrane (TM) domain and intracytoplasmic tail of HA. To visualize the location of the peptides on the HA protein, we mapped the peptides on the resolved H1N1 pdm09 HA’s ectodomain structure (Fig. 1). Peptide 15 comprises of a mixture and loop and β–sheet conformations, which links the stem and the globular domains and is largely surface exposed. Peptide 121 containing a mixture of α-helix, loop, and β–sheet is located in close proximity to the transmembrane region with the helical region fully exposed to the surface. The surface-exposed nature of peptides 15 and 121 may make their corresponding antibodies bind to the virions so antibody binding may have an effect on viral entry and replication. Visualization of peptide 139 could not been performed due to the unavailable structure of HA’s transmembrane region and intracytoplasm ic tail.
Figure 1. Locations of reactive peptides in the context of the hemagglutinin (HA) structure (PDB 3LZG) of H1N1 pdm09 virus.
The Hemagglutinin structure (PDB 3LZG) from H1N1 pdm09 that derives the HA peptide array was chosen as the template to locate ELISA-positive peptides in the context of the HA structure. Peptides 15 and 121 are colored with light green and dark blue, respectively.
3.4. Cross reactivity of peptides to reference sera representing different H1 clusters/subtypes of swine influenza A virus
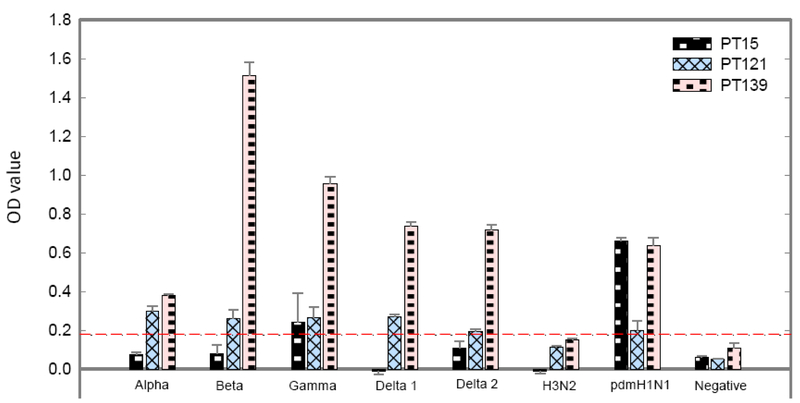
We next determined whether pig reference sera from different swine influenza H1 and H3 clusters recognized the identified peptides. For this experiment, we focused on peptides 15, 121, and 139. All reference sera used in this study were uniformly diluted into same HAI antibody titer 1:1280 against the viruses that were used to generate these sera. Fig. 2 summarizes the serological cross-reactivity of these peptides. Peptide 139 was highly reactive to all tested SIV H1 antisera, while peptide 121 was moderately reactive to all these sera. Strong inter-cluster reactivity demonstrated with peptide 139 and weak cross-reactivity (immunoreactive) with peptide 121 concurs with the results obtained with the referenc pdm09 serum on these peptides (Tables 1 and 2). Furthermore, in addition to H1N1 pdm09 reference serum, peptide 15 was weakly reactive to H1 γ cluster serum. Significantly, none of these three peptides were recognized by SIV H3 antiserum, despite that this antiserum has a HAI antibody titer similar to those of H1 clusters antisera. In summary, three peptides reactive to different H1N1 cluster sera displayed no antibody binding activity with the reference SIV H3 serum.
Figure 2. Recognition of three positive peptides by indicated swine antisera against different clusters of swine H1N1 and a H3N2 subtype.
Each of three peptides (15. 121, and 139) was used to coat 96-well plate. Swine antisera (1:300) from indicated H1 and H3 clusters of swine influenza were examined for the presence of antibodies to these three peptides using ELISA as detailed in materials and methods section. The background-corrected OD values of antisera for each peptide are calculated and plotted. The data presented in this figure are representative of three independent experiments, each assayed in duplicate.
3.5. Conservation analysis of the identified linear peptide epitopes in different H1 clusters and H3 subtype of swine influenza A virus
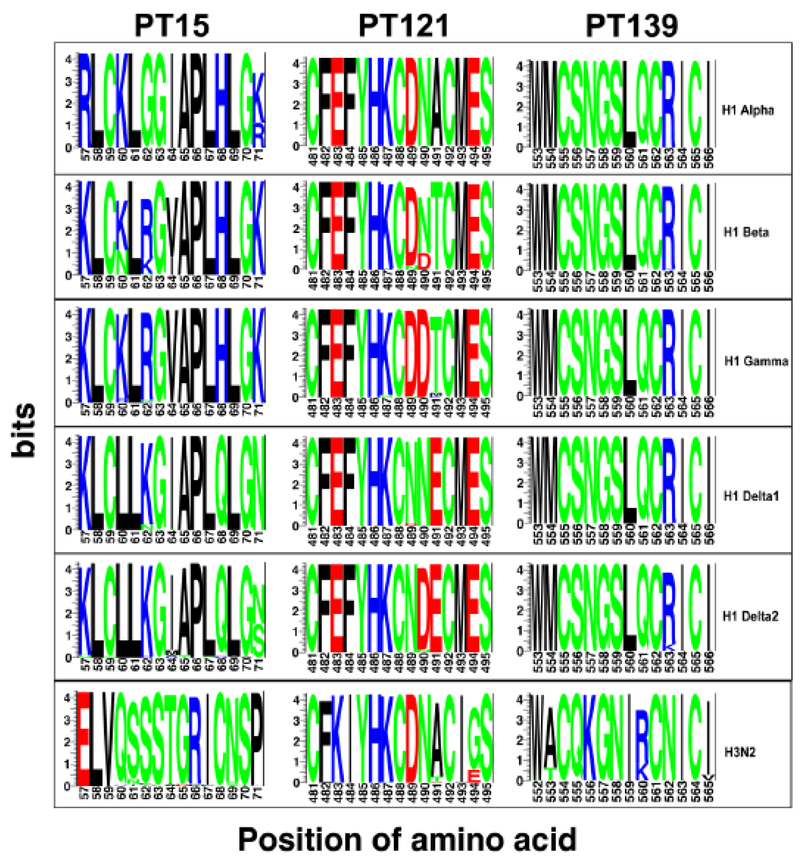
We used sequence logos (O’Shea et al., 2013) to visualize the conservation of individual amino acids within each identified epitope, with the height of each letter indicating the proportion of sequences that contain the residue at that site (Fig. 3). Sequence of peptide 139 (WMCSNGSLQCRICI) is completely identical among all H1 clusters except for δ-2 (Delta2) cluster. The vast majority (90%) of Delta2 strains have an arginine (R) at position 563 (R563), which is akin to SIV H1 strains in other clusters, while another 10% have a closely related lysine substitution (K563). This substitution seems not affecting the recognition of peptide 139 by Delta2 antisera used here. In contrast, 6 positions in the corresponding amino acid sequences of SIV H3 strains are occupied by different amino acids. Epitope conservation analysis of peptide 139 supports its universal recognition by all tested H1 cluster sera, not by a H3 subtype sera. Peptide 121 (CFEFYHKCDNTCMES) is highly conserved among all H1 clusters. Few amino acid variations in this epitope are centered on positions 489, 490, and 491. For example, all strains of swine influenza H1 alpha (α) cluster have an alanine (A) at position 491, while all strains of Beta (β) and 99% of Gamma (γ) strains have a closely related threonine substitution (T491). Interestingly, Glutamic acid (E) occupies this position at Delat1 and Delat2 strains. Changes at these three positions have no substantial effects on the universal recognition of peptide 121 by antisera of H1 clusters. Analysis also showed that SIV H3 strains differed from H1 strains by 8 positions in the corresponding region and this significant variation in the epitope may explain why peptide 121 is not recognized by SIV H3 antisera. Analysis of the sequence similarity in peptide 15 (KLCKLRGVAPLHLGK) displayed by various H1 clusters also supported a relation between epitope sequence conservation and antibody recognition. In addition to homologous sera (pdm09 H1N1), antiserum generated against Gamma (γ) cluster also recognized peptide 15. P15 peptide sequence is completely conserved among all analyzed strains of Gamma (γ) cluster. Interestingly, antisera generated against other clusters of SIV subtype H1 and a H3 subtype strain failed to be reactive to this epitope due to multiple changes in the region.
Figure 3. Conservation of identified peptide epitopes among swine influenza A H1N1 and H3N2 viruses.
Logo analysis is shown for the frequency of each amino acids within each of three peptides in swine influenza strains from indicated H1 clusters and H3 that circulated between 2006 to 2015 in pigs farms of the United States. The height of the letter indicates the frequency of individual amino acid residue at that site. The residue position in the block is shown on the X-axis, and the information content is shown on the Y-axis. The default color scheme displaying different amino acids according to their different chemical properties is as follows: polar amino acids (G, S, T, Y, C, Q, N) colored with green, basic (K, R, H) with blue, acidic (D, E) with red, and hydrophobic (A, V, L, I, P, W, F, M) amino acids with black.
3.6. Evaluation of immunogenicity of peptides 15 and 121 in pigs
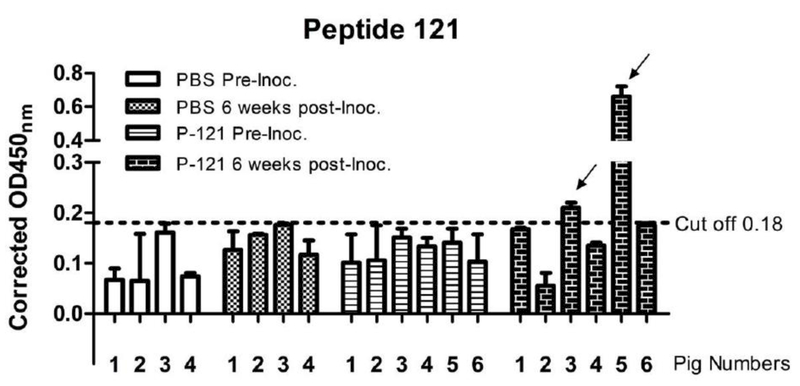
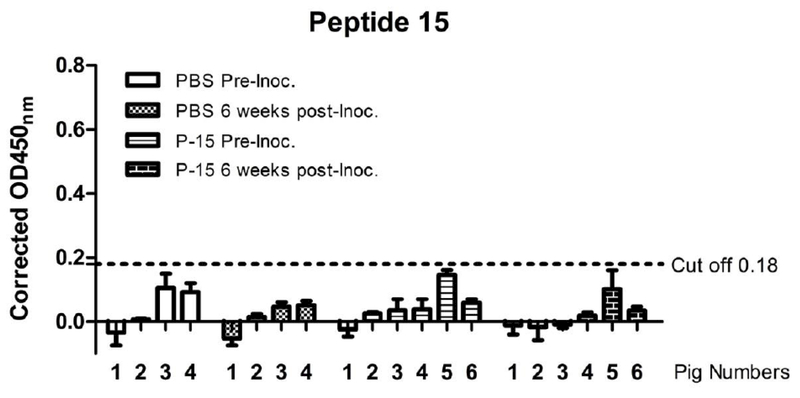
We finally determined the immunogenicity of the identified B cell epitopes displayed in peptides 15 and 121. Peptide 139 is not selected for this study because this peptide is not exposed in the HA protein surface and may not constitute a target for protective antibody response. Peptide antigens containing peptides 15 and 121 were conjugated with carrier protein KLH (500μg of each peptide dissolved in total volume of 1ml PBS). Each of six 4-week-old influenza antibody-negative pigs was inoculated intranasally with one ml suspension of the KLH-conjugated peptides along with heat labile enterotoxin (25 μg LT), while each of four control pigs received 1 ml PBS intranasally. Two weeks later, all piglets received a booster dosage. Blood samples were collected for the serum isolation via the jugular vein from piglets 2 weeks after the second round of immunization. Peptide-specific antibodies were measured in the ELISA assay as described above. As summarized in Figs. 4 and 5, 2 out of 6 inoculated pigs showed detectable antibodies against peptide 121 at six weeks following the first immunization (Fig. 4). All inoculated pigs, however, were found negative in antibody response to peptide 15 during the course of this study (Fig. 5). Taken together, peptide 121 appeared to be immunogenic to pigs; despite that the antibody response elicited by this peptide was not uniform among inoculated pigs.
Figure 4. Antibody responses against peptide 121 in pigs.
Pig sera collected at the date of inoculation and 6 weeks post-inoculation of KLH-conjugated peptides 15 and 121 or PBS were examined by ELISA for their ability to bind to peptide 121 antigen as described in details in materials and methods section. The horizontal broken line represents the OD cut-off value. Background-corrected OD values above this line are seropositive for peptide antigen. The data presented in this figure are representative of three independent experiments, each assayed in duplicate.
Figure 5. Antibody responses against peptide 15 in pigs.
Pig sera collected at the date of inoculation and 6 weeks post-inoculation of KLH-conjugated peptides 15 and 121 or PBS were examined by ELISA for their ability to bind to peptide 15 antigen as described in details in materials and methods section. The horizontal broken line represents the OD cut-off value. Background-corrected OD values above this line are seropositive for peptide antigen. The data presented in this figure are representative of three independent experiments, each assayed in duplicate.
4. Discussion
A limitation of the peptide array-based approach is that it cannot identify discontinuous conformation-dependent epitopes that may in fact represent important antigenic determinants of the HA protein of swine influenza A viruses. This approach, however, has been extensively used to determine B-cell linear epitopes in viral glycoproteins of several enveloped viruses including human immunodeficiency virus (HIV), equine infectious anemia virus (a lentivirus), and influenza A virus (Ball et al., 1992; Forsstrom et al., 2015; Neurath et al., 1990; Prabakaran et al., 2009; Zhao et al., 2011; Zhou et al., 2014). The effectiveness of this approach in the identification of specific B-cell linear epitopes that are immunogenic in the native protein antigen may be influenced by three major factors. First, the structural and biophysical properties of a short protein fragment displayed between an isolated peptide and a native protein may be variable, which may lead to differential antibody binding to the target (Fieser et al., 1987). Second, the major histocompatibility complex polymorphisms present in an outbred population may influence the recognition of a particular peptide antigen by antibodies (Wieczorek et al., 2017). Third, host genetic variability may play a role in generation of differential activities of specie-specific antibodies to same B-cell epitopes (Rahman et al., 2015).
In this study, we used a synthetic peptide array covering the complete sequence of the HA protein of swine-originated influenza H1N1pdm09 virus to map distinct B-cell epitopes reactive with experimentally infected porcine immune sera. The most significant results of this study are the identification of two HA2-derived peptides (peptide 121 and 139) universally reactive to all tested SIV H1 cluster sera. Of two peptides, peptide 139, located in the HA’s intracytoplasmic tail, is highly reactive B cell linear determinant. In addition to peptide 121, another surface-exposed epitope represented by peptide 15 also emerged in our study but this B cell epitope differs from peptide 121 in that it is only recognized by a limited number of H1 cluster sera. It is intriguing to note that all the identified epitopes fail to be recognized by a H3 subtype serum, indicating a subtype-specific nature of the identified antigenic determinants. Furthermore, peptide 121-specific antibodies generated from pigs receiving a mixture of peptides 15 and 121 are not effective in neutralizing homologous influenza H1N1pdm09 virus in both hemagglutination inhibition (HI) and virus neutralization assays (data not shown).
The results of our study reveal several interesting observations. First, peptide antigen, represented by peptide 139 derived from the intracytoplasmic tail of the HA protein, is highly immunereactive in swine. This finding is similar to what have been reported previously in retroviral studies showing that the intracytoplasmic tail and transmembrane region of retrovirus’s envelope protein is strongly antigenic determinant (Ball et al., 1992). Second, a B cell epitope resided in the peptide 15 has been demonstrated in patients recovered from influenza H1N1pdm09 infection (Zhao et al., 2011). Despite the similarity, this early study revealed that this B cell epitope is capable of inducing high titer non-neutralizing antibodies in mice, while our study failed to detect measurable antibodies against peptide 15 in inoculated pigs. Different adjuvant formulation used between these two studies and host factor may play a significant role in causing this contrasting outcome, which warrant further investigation. Third, it is unfortunate to observe that antibodies generated from peptide 121 in some animals possessed non-neutralizing capability. It will be interesting to determine whether peptide 121-specific antibody can protect swine from influenza A virus infection through other antibody-mediated protective mechanism such as antibody-dependent cell-mediated cytotoxicity, which recently emerge as one of universal influenza vaccine targets (Erbelding et al., 2018). Fourth, Fourth, we noted that of five reactive peptides to swine H1 subtype sera, only peptide 15 is found to be immunoreactive in H1N1 infected patients or mice. The other four peptides (#121, #137, #138, and #139) seem to be swine-dependent as their antigenicity is not detected in other species such as humans and poultry (http://www.iedb.org). Finally, neither peptides derived the HA2’s helix A nor peptides from the HA2’s long CD helical domain were reactive to pig antisera against various H1 clusters in our ELISA assay. It has been reported that low antibody titers against the CD region of the HA2 protein were found in some influenza A viruses-infected patients (Wang et al., 2010). These data suggest that using a peptide-based approach to generate antibodies against two conserved targets (A and CD helices) may run into a challenge in swine. Considering that antibodies to these regions have a universal protection of experimental animals against various influenza subtypes, delivery of these targets into swine for effective induction of such cross-reactive antibodies may seek a different approach such as expression of these antigens through a nanoparticle platform or using a effective adjuvant.
In summary, we have identified two highly conserved epitopes contained in peptide peptides 121 and 139 located in HA2 that reacted to all tested SIV H1 antisera. This study also found another epitope in HA1 peptide 15 that was recognized by a subset of H1 antisera. None of these HA1/HA2 peptides of H1N1pdm09 were recognized by SIV H3 reference antisera. Results of our study shall provide an important foundation for further analyzing the immune response against these peptides during natural SIV infection and also provide potential peptide substrates for diagnostic assays and for vaccine strategies.
Acknowledgments
We thank BEI Resources, NIAID, NIH for providing the Peptide Array, NR-15433, which was used in this study.
Funding information
This work was partially supported by SDSU AES 3AH-477, by National Science Foundation/EPSCoR (http://www.nsf.gov/od/iia/programs/epscor/index.jsp) award IIA-1335423, and by the state of South Dakota Governor’s Office of Economic Development as a South Dakota Research Innovation Center. Radhey S. Kaushik was funded and supported by USDA NIFA SDSU Agricultural Experiment Station Hatch grant # SD00H547-15.
Footnotes
Declaration of interest:
The authors have read the journal’s policy and have the following conflicts: BMH is employed by Cambridge Technologies, a company that produces swine influenza virus vaccines. This does not alter the authors’ adherence to all the Journal policies on sharing data and materials.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ball JM, Rushlow KE, Issel CJ, Montelaro RC, 1992. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J Virol 66, 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterday BC, 1980. The epidemiology and ecology of swine influenza as a zoonotic disease. Comp Immunol Microbiol Infect Dis 3, 105–109. [DOI] [PubMed] [Google Scholar]
- Erbelding EJ, Post D, Stemmy E, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, Fauci AS, 2018. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieser TM, Tainer JA, Geysen HM, Houghten RA, Lerner RA, 1987. Influence of protein flexibility and peptide conformation on reactivity of monoclonal antipeptide antibodies with a protein alpha-helix. Proc Natl Acad Sci U S A 84, 8568–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsstrom B, Axnas BB, Rockberg J, Danielsson H, Bohlin A, Uhlen M, 2015. Dissecting antibodies with regards to linear and conformational epitopes. PLoS One 10, e0121673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Oleson TA, Stine DL, Bey RF, RR S, 2011. Genetic and antigenic characterization of recent human-like H1 (δ-cluster) swine influenza virus isolates. J. Swine Health Prod. 19:268–276. [Google Scholar]
- Hause BM, Oleson TA, Bey RF, Stine DL, Simonson RR, 2010. Antigenic categorization of contemporary H3N2 Swine influenza virus isolates using a high-throughput serum neutralization assay. J Vet Diagn Invest 22, 352–359. [DOI] [PubMed] [Google Scholar]
- Hause BM, Stine DL, Sheng Z, Wang Z, Chakravarty S, Simonson RR, Li F, 2012. Migration of the swine influenza virus delta-cluster hemagglutinin N-linked glycosylation site from N142 to N144 results in loss of antibody cross-reactivity. Clin Vaccine Immunol 19, 1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM, 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325, 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath AR, Strick N, Lee ES, 1990. B cell epitope mapping of human immunodeficiency virus envelope glycoproteins with long (19- to 36-residue) synthetic peptides. J Gen Virol 71 (Pt 1), 85–95. [DOI] [PubMed] [Google Scholar]
- O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D, 2013. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods 10, 1211–1212. [DOI] [PubMed] [Google Scholar]
- Prabakaran M, Ho HT, Prabhu N, Velumani S, Szyporta M, He F, Chan KP, Chen LM, Matsuoka Y, Donis RO, Kwang J, 2009. Development of epitope-blocking ELISA for universal detection of antibodies to human H5N1 influenza viruses. PLoS One 4, e4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman KS, Chowdhury EU, Poudel A, Ruettger A, Sachse K, Kaltenboeck B, 2015. Defining species-specific immunodominant B cell epitopes for molecular serology of Chlamydia species. Clin Vaccine Immunol 22, 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL, 2018. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, Ma J, Bawa B, Liu Q, Liu H, Quast M, Sexton G, Krammer F, Hause BM, Christopher-Hennings J, Nelson EA, Richt J, Li F, Ma W, 2015. Domestic pigs are susceptible to infection with influenza B viruses. J Virol 89, 4818–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Richt JA, 2008. Swine influenza viruses a North American perspective. Adv Virus Res 72, 127–154. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR, Lewis NS, 2017. Influenza A virus vaccines for swine. Vet Microbiol 206, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, Garcia-Sastre A, Moran TM, Palese P, 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A 107, 18979–18984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, Noe F, Freund C, 2017. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol 8, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Gambhir M, Finelli L, Swerdlow DL, Ostroff S, Reed C, 2013. Transmissibility of variant influenza from Swine to humans: a modeling approach. Clin Infect Dis 57 Suppl 1, S16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Cui S, Guo L, Wu C, Gonzalez R, Paranhos-Baccala G, Vernet G, Wang J, Hung T, 2011. Identification of a highly conserved H1 subtype-specific epitope with diagnostic potential in the hemagglutinin protein of influenza A virus. PLoS One 6, e23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Gao X, Wang Y, Zhou H, Wu C, Paranhos-Baccala G, Vernet G, Guo L Wang J, 2014. Conserved B-cell epitopes among human bocavirus species indicate potential diagnostic targets. PLoS One 9, e86960. [DOI] [PMC free article] [PubMed] [Google Scholar]