Abstract
The ribosome has recently transitioned from being viewed as a passive, indiscriminate machine to a more dynamic macromolecular complex with specialized roles in the cell. Here, we discuss the historical milestones from the discovery of the ribosome itself to how this ancient machinery has gained newfound appreciation as a more regulatory participant in the central dogma of gene expression. The first emerging examples of direct changes in ribosome composition at the RNA and protein level, coupled with an increased awareness of the role individual ribosomal components play in the translation of specific mRNAs, is opening a new field of study centered on ribosome-mediated control of gene regulation. In this Perspective, we discuss our current understanding of the known functions for ribosome heterogeneity including specialized translation of individual transcripts and its implications for the regulation and expression of important gene regulatory networks. In addition, we suggest what the crucial next steps are to ascertain the extent of ribosome heterogeneity and specialization and its importance for regulation of the proteome within subcellular space, across different cell types, and during multi-cellular organismal development.
Article Summary
The ribosome, while previously viewed as a homogenous and indiscriminate machine, has recently been recognized to play a central role in the regulation of gene expression. In this Perspective, we discuss ribosome heterogeneity, where ribosomes of distinct compositions may be present both within cells and across tissues, and the functions of specific classes of ribosomes in translational control.
A brief history of the ribosome and its heterogeneity or lack thereof
In the mid-1950s, future Nobel Prize winner George Palade became fascinated by “a small particulate component of the cytoplasm” (Palade, 1955) (Figure 1). While examining the structure of the cell by electron microscopy, he noticed small, roughly spherical granules both free-floating in the cytoplasm and associated with the endoplasmic reticulum (ER) membrane. These “microsomes,” as the ER-bound granules were originally termed, were determined to be composed of both protein and RNA and to be the sites of protein synthesis in the cell (reviewed in (Palade, 1958)). Because of confusion in the field over whether microsomes referred to just granules or also the ER lipid membrane, a new name was suggested at a 1958 symposium of the Biophysical Society: “The phrase ‘microsomal particles’ does not seem adequate, and ‘ribonucleoprotein particles of the microsome fraction’ is much too awkward. During the meeting the word ‘ribosome’ was suggested: this seems a very satisfactory name, and it has a pleasant sound” (Roberts, 1958).
Figure 1: Timeline of key discoveries on ribosome heterogeneity and specialization.
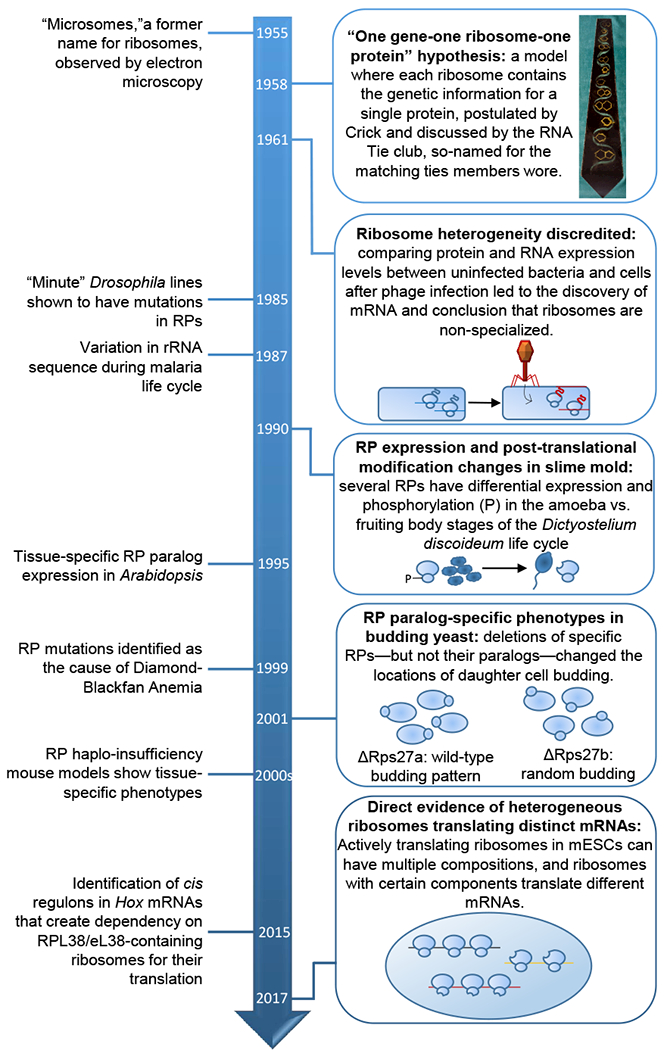
Image of the RNA Tie Club Tie accessed from the Wellcome Library, https://search.wellcomelibrary.org/iii/encore/record/C__Rb1998728.
Almost immediately subsequent to the discovery of the ribosome, several scientists postulated that there may be diversity in ribosome composition. Palade noted small differences in the size and shape of microsomes and suggested that there may be further heterogeneity that couldn’t be seen with the current resolution of electron microscopy (Palade, 1958). Perhaps the greatest proponent of ribosome heterogeneity in composition and activity at that time was Francis Crick, who after his discovery of the structure of DNA was eagerly discussing the genetic code with the RNA Tie Club, a private scientific social group with twenty primary members—one for each amino acid—and up to four honorary members—each designated by a nucleotide. In 1958 he sparked debate with his bold model for the flow of genetic information known as the “one gene-one ribosome-one protein hypothesis,” where each ribosome carries the genetic information required to encode a single protein (Crick, 1958). This model suggested that the cell contains thousands of distinct ribosomes, each tailored to the production of a single protein. However, this view rapidly fell out of favor when further biochemical characterization of the ribosome did not reveal pronounced differences in ribosomal rRNA (rRNA) size across individual ribosomes, so Crick altered his model “to the idea that only part of the ribosomal RNA acts as a template” (Crick and Brenner, 1959). However, work by Francois Jacob and Jacques Monod on the kinetics of β-galactosidase synthesis led Crick and his fellow RNA Tie club member Sydney Brenner to conclude that the template for protein synthesis is likely an unstable “genetic RNA” that allows the ribosome to make “one protein at one moment, and a quite different protein a few minutes later” (Brenner and Crick, 1960). In a series of elegant experiments, Brenner, Jacob, and Matthew Meselson showed that in E. coli after bacteriophage infection new RNA corresponding to the phage genome was synthesized, but no new ribosomes were made (Brenner et al., 1961). The phage RNA made post-infection associated with actively translating bacterial ribosomes that had been made prior to infection, revealing that bacterial ribosomes are capable of synthesizing phage proteins, leading to the conclusion that “ribosomes are non-specialized structures which synthesize, at a given time, the protein dictated by the messenger they happen to contain.” In three short years, the field had vacillated from the most extreme view of ribosome specialization—where each protein is translated by a different type of ribosome—to the most extreme view of ribosome homogeneity—where ribosomes are passive machines with no regulatory function.
Over the next few decades, the idea that all ribosomes are exactly the same largely prevailed, and is in fact a notion still prevalent in most modern biology textbooks. Any subsequent studies suggesting greater modularity in ribosome composition or activity, were met with skepticism as differences in laboratory procedures made published results from one group difficult to replicate by others, leading to confusion and a general disregard for proponents of ribosome heterogeneity (reviewed in (Dinman, 2016; Shi and Barna, 2015; Xue and Barna, 2012)). However, inklings of greater dynamics to ribosomes began to arise again in the 1980s and 1990s, as studies in multiple diverse invertebrate model organisms observed differential expression of individual ribosome components, albeit without a direct measure of whether such changes actually produced mature ribosomes that were of a distinct composition. For example, in the slime mold Dictyostelium discoideum, changes in the modification of several ribosomal proteins (RPs) occurred during the transition from unicellular growth to a multicellular body (Ramagopal, 1990). In Arabidopsis thaliana and Brassica napus, where RPs typically have multiple paralogs due to genome duplication events, RP paralogs are expressed in different regions of the plant (Weijers et al., 2001; Whittle and Krochko, 2009; Williams and Sussex, 1995). Different rRNA sequences were also identified in Plasmodium berghei, a parasite that causes malaria in rodents, at different stages of its life cycle (Gunderson et al., 1987). Mutations in ribosomal components also unexpectedly produced tissue-specific phenotypes in fruit flies (Kongsuwan et al., 1985; Marygold et al., 2005), zebrafish and mice (reviewed in (Shi and Barna, 2015)), and in human disease (Draptchinskaia et al., 1999). Additional studies suggested RP paralogue specific differences in control of cell function in S. cerevisiae, although additional moonlighting functions of RPs outside of the ribosome made such observations difficult to attribute to selective changes in the ribosome itself (Komili et al., 2007; Ni and Snyder, 2001; Segev and Gerst, 2018). While these correlative findings were suggestive of possibly greater regulation to the ribosome itself, it is only in the last few years where ribosome heterogeneity and specialization has undergone a renaissance, with modern techniques finally permitting the discovery of ribosomes with distinct compositions performing unique cellular functions.
Structural heterogeneity of the ribosome
Evidence of physical ribosome heterogeneity has been technologically challenging to obtain. Definitive determination of the stoichiometry of a macromolecular complex requires state-of-the-art quantitative mass spectrometry measurements, ideally of all protein components. Our lab recently measured the absolute abundance of 15 of the 80 core RPs in polysomes (actively translating ribosomes) from mouse embryonic stem cells (mESCs) using selected reaction monitoring (SRM) mass spectrometry, a technique where the abundance of a peptide from the protein of interest is measured relative to a heavy-labeled peptide spiked-in at a known concentration (Shi et al., 2017). Six of the 15 RPs measured were substoichiometric, with four of those present on only 60-70% of polysomal ribosomes, revealing for the first time that there are indeed actively translating ribosomes lacking at least one core RP. While this is direct evidence of ribosome heterogeneity, it may only be scratching the surface: the stoichiometry of the other 65 core RPs has yet to be quantified, and even small changes in abundance could have large effects on translation given that millions of ribosomes are present in a single mammalian cell. As this data is also from a single cell type, there may be additional changes in ribosome composition across cell types. Proteomics methods have yet to be brought to bear on this question, but RNA-seq of RP transcripts across mouse tissues (Kondrashov et al., 2011), human tissues (Guimaraes and Zavolan, 2016; Gupta and Warner, 2014), human hematopoietic cell types (Guimaraes and Zavolan, 2016), and human cancer cell lines (Guimaraes and Zavolan, 2016) suggests that RPs have many more cell type-specific expression patterns than would be expected for a machine deemed invariable across all cell types. However, more work at the protein level and in particular at the level of the ribosome itself is required to confirm whether these changes produce ribosomes which are heterogeneous, as RNA abundance may not correlate with protein amount or incorporation into functional ribosomes.
Tissue-specific expression patterns are also frequently observed for the paralogs of core RPs (Gupta and Warner, 2014; Wong et al., 2014). Unlike yeast or plants, where the majority of RPs have paralogs due to genome duplications, only a handful of RPs have paralogs in metazoans. As RP paralogs are highly homologous to each other, they are likely to compete for the same position on the ribosome, suggesting that different paralogs could be swapped out on the ribosome to varying degrees depending on the cell type. For some RPs, such as RPL22/eL22 and RPL22L/eL22L in Drosophila, one paralog appears to be ubiquitous while the other has tissue-specific expression (Kearse et al., 2017). In other cases, RP paralogs actually appear to anti-correlate in expression, suggesting coordinate regulation of their abundance on the ribosome. For instance, the paralog RPL3L/uL3L is most abundant at the RNA level in the human heart and skeletal muscle; in those same tissues, the canonical RP RPL3/uL3 mRNA is decreased in abundance relative to other regions of the body (Gupta and Warner, 2014). In fact, in response to a hypertrophic stimulus, RPL3L/uL3 mRNA abundance is decreased in skeletal muscle coordinately with an increase in RPL3/uL3 mRNA (Chaillou et al., 2014, 2016; Kirby et al., 2015). While the mechanism underlying this regulation of RPL3/RPL3L levels is unknown, RP paralogs have been known to regulate each other’s expression directly. For instance, RPL22/eL22 destabilizes the transcript of its paralog RPL22L1/eL22L1 via direct binding to a stem loop in its mRNA (O’Leary et al., 2013). Interestingly, GWAS studies have linked RPL3L/uL3 mutations to a remarkably specific disease state leading to increased risk of atrial fibrillation, but the consequence of these mutations on the relative activities of RPL3L/uL3L and RPL3/uL3 or on global translation are yet to be determined (Thorolfsdottir et al., 2018).
In addition to the core RPs, many other proteins associate with the ribosome and may be sources of heterogeneity. A recent study from our lab developed an affinity enrichment methodology for ribosome isolation from mESCs and used mass spectrometry to identify the “ribo-interactome,” which is comprised of hundreds of ribosomal associated proteins (RAPs) that fall into diverse functional categories such as mRNA binding proteins, mRNA/tRNA modifiers, RNA helicases, as well as regulators of metabolism and cell cycle control (Simsek et al., 2017). Intriguingly, one RAP, the glycolysis enzyme PKM2, was enriched on ribosomes at the ER compared to ribosomes in the cytosol, revealing the existence of subcellular ribosome heterogeneity within mESCs. Subcellular heterogeneity is an area requiring further investigation not only in mESCs, but particularly in polarized cells with localized translation like the intestinal epithelium (Moor et al., 2017) and neurons (reviewed in (Jung et al., 2014)). Ribosomes are in fact located in the dendrites as well as the soma of neurons, and another RAP, Fragile X mental retardation protein (FMRP), has been shown to localize to synaptosomal polysomes (Feng et al., 1997). What percentage of ribosomes contain FMRP, and whether FMRP is particularly enriched on ribosomes in certain cellular compartments is not yet established, but given FMRP’s known regulation of the translation of transcripts associated with synaptic signaling (Chen et al., 2014; Darnell et al., 2011), these findings suggest that FMRP could specialize ribosomes in key functional cellular regions for the translation of particular mRNAs.
Ribosome heterogeneity is not unique to the protein components of the ribosome: it can also occur at the level of rRNA. The mammalian ribosome contains four rRNAs, and each of these are encoded by hundreds of copies of ribosomal DNA (rDNA) on multiple chromosomes (Henderson et al., 1972; Little and Braaten, 1989; Stults et al., 2008). Because of the large copy number and repetitive nature of these genes, reliable sequencing of these loci has been technically challenging. Recent computational advances, however, have improved mapping of sequencing reads to rDNA loci and have suggested extensive human and mouse rRNA sequence variation both across and within individuals (Parks et al., 2018). Intriguingly, many rRNA alleles have tissue-specific expression in mice (Arnheim and Southern, 1977; Parks et al., 2018; Tseng et al., 2008), suggesting that rRNA variant usage may be developmentally regulated. In fact, all 4 rRNAs have also been shown to change from maternal to somatic ribosomes over the course of embryonic development in zebrafish (Locati et al., 2017a, 2017b). While the functional consequences of this rRNA variation is not yet established, in silico modeling has suggested that maternal and somatic rRNA variants may preferentially bind different mRNAs (Locati et al., 2017a), suggesting that rRNA may play a direct role in the regulation of translation, initially proposed as the ribosome filter hypothesis (Mauro and Edelman, 2002). In addition to mRNA selection, rRNA variation may also specialize ribosomes for programmed frameshifting, as was suggested for several yeast and Xenopus laevis rRNA alleles (Kiparisov et al., 2005). rRNA is furthermore extensively modified, adding an additional layer of heterogeneity, as described in other recent reviews on the topic (Mcmahon et al., 2015; Roundtree et al., 2017; Xue and Barna, 2012).
How heterogeneous are ribosomes?
While there is evidence of ribosomes with varying composition, what is the actual extent of ribosome heterogeneity? A candid answer to this question is that it really remains largely unknown. The stoichiometry of 15 RPs has been calculated in mESCs, and six RPs have been identified as substoichiometric, and these alone could produce dozens of distinct ribosome compositions lacking one or more of these RPs. It is not yet clear whether this handful of variations of the ribosome is all that can be tolerated, or if this is only the tip of iceberg. If one considers not only the other 65 core RPs but also the additional hundreds of RAPs and rRNA sequence variants—the complexity increases exponentially. While systematic assessment of the stoichiometry of each of these variants, using SRM or similar mass spectrometry methods, would be helpful for understanding ribosome heterogeneity, determining the actual number of ribosome types requires examination of their composition on a ribosome-by-ribosome basis. One potential strategy to accomplish this is to use native mass spectrometry, where intact macromolecular complexes as large as 9MDa are run on the spectrometer. This method has been successfully used to examine heterogeneity in bacterial ribosomes (van de Waterbeemd et al., 2017) as well as ribosomes from a human cancer cell line (van de Waterbeemd et al., 2018) and could be an effective means of determining not only which proteins are substoichiometric but also whether the incorporation of particular RPs is coordinately regulated. A potentially even more powerful but technically challenging method to address these questions would be cryo-electron microscopy (cryo-EM). Cryo-EM has been successfully employed to examine the structure of the mammalian ribosome (Anger et al., 2013) but has yet to be brought to bear on the question of heterogeneity. Heterogeneity may be difficult to visualize because cryo-EM data of many complexes are “class averaged” into an individual structure, so if there are many types of ribosomes each making up a smaller population of the total (as seems likely from the mass spectrometry data), it may be difficult to resolve them. One way to potentially reduce this complexity would be to first isolate ribosomes containing a specialized component of interest, such as by immunoprecipitation. Not only could this technique then be used to visualize ribosomes of diverse compositions, but it also would determine how the presence or absence of particular components affects conformational heterogeneity of the overall ribosome. For instance, specific core RPs could serve as docking points for multiple RAPs, or loss of certain RPs could cause rearrangements in rRNA or ribosome binding of neighboring proteins. Other proteins may even take the place of a missing RP, though this appears not to be the case for ribosomes in yeast lacking RPL38/eL38, which simply have a “hole” where the RP would usually be (Armache et al., 2010). While the most flexible regions of rRNA, known as expansion segments, that are likely to bind RAPs cannot currently be resolved, future advancements in cryo-EM technology will hopefully permit these questions to be addressed.
It is important to note that not only can ribosome composition be heterogeneous, but it also may be dynamic. Ribosome composition has to date been examined largely in mESCs, but it is interesting to speculate that different cell types may have distinct types of ribosomes (Figure 2). Even within a single cell type, ribosomes may change in composition in response to different stimuli, including stress, cell cycle progression, or activation of particular signaling pathways. Going even deeper, it is likely that certain types of ribosomes may reside in particular subcellular regions: for instance, ribosomes at the ER may have a specialized conformation to optimize their translation of luminal, membrane, or secreted proteins. It is even possible for specialized ribosomes and their mRNA targets to cluster together as their own cytoplasmic compartments in order to increase the efficiency of translation. An extreme example of this might be having specialized ribosomes present at synapses in neurons in order to rapidly translate co-localized synaptic mRNAs upon neuron firing, but such compartmentalization may also take place in less polarized cells.
Figure 2: Five outstanding questions on the nature of ribosome heterogeneity and its function.
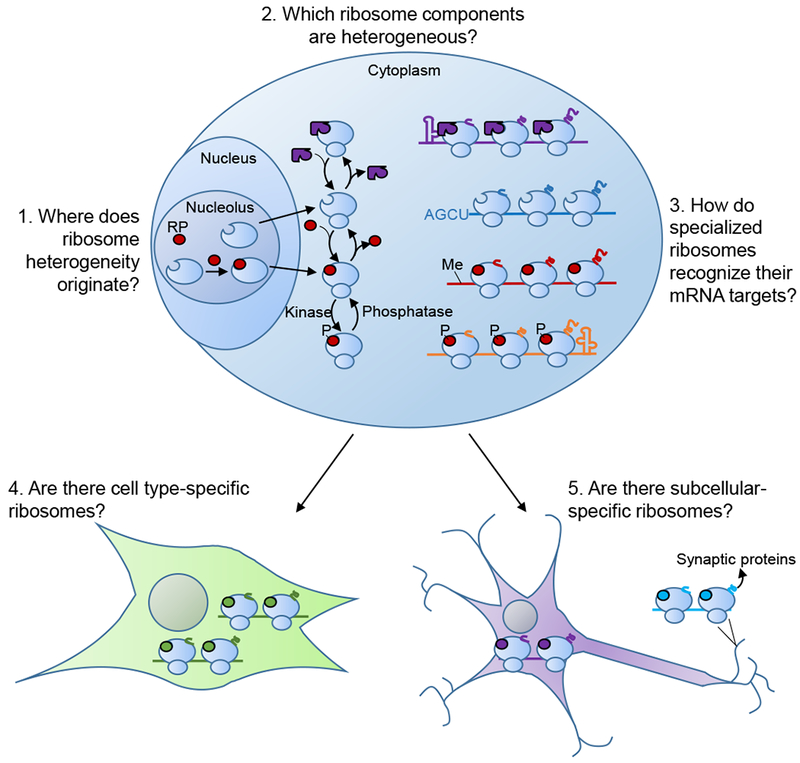
Ribosome heterogeneity can originate from many sources, from initial changes in incorporation of RPs (red) in the nucleus to cytoplasmic exchange of RPs and RAPs (purple) in the cytoplasm to post-translational modifications (such as phosphorylation, P). Each of these different ribosome compositions may be specialized for the translation of specific mRNAs demarcated by unique cis regulons. There additionally may be distinct compositions of ribosomes found in different cell types and in different subcellular locations to optimize the production of proteins necessary in those regions.
Just how quickly ribosome composition can change is also an open question, as to date there is little understanding of how the cell creates and regulates ribosome heterogeneity (Figure 2). Regulation of ribosome composition likely occurs at least in part during ribosome biogenesis, a complex process involving hundreds of proteins and RNAs that begins in the nucleolus and progresses through the nucleus and into the cytoplasm. Changes in rRNA allele usage presumably would have to be specified at the earliest stage of ribosome biogenesis, the transcription of rRNA. Accordingly, switching between rRNA sequences is likely a slow process because it requires the entire process of ribosome biogenesis to occur before the new ribosomes are functional. While this may also be the case for RPs, most of which are assembled onto the pre-ribosomal subunits in the nucleolus, it also may be possible for them to exchange on and off the ribosomal subunits in the nucleus or cytoplasm. This is in fact the case for RPL13a/uL13, which upon interferon-γ signaling is phosphorylated and extracted from cytoplasmic ribosomes (Mazumder et al., 2003). Interestingly, while several of the known substoichiometric RPs in mESCsdo not seem to be required for ribosome biogenesis or cell viability, such as RPS25/eS25 (Hertz et al., 2013), others appear less dispensable, suggesting that posttranslational mechanisms—including possibly extraction from the ribosome in the cytoplasm—are required to explain their heterogeneous incorporation.
Phosphorylation and other post-translational modifications (PTMs) on RPs, as well as modifications on rRNA, are also potential dynamic sources of ribosome heterogeneity. In addition to phosphorylation of RPL13a/uL13, phosphorylation of RACK1 has also been reported in human cell lines infected with poxvirus infection (Jha et al., 2017). Regulatory ubiquitylation of several RPs occurs during the unfolded protein response (Higgins et al., 2015). More of these PTMs, as well as methylations, acetylations, and hydroxylations, have been identified on RPs in unbiased proteomics screens, but few have been further functionally validated (reviewed in (Simsek and Barna, 2017)). rRNA is also extensively modified by pseudouridylation, ribosylation, methylation, and acetylation, and several modifications have been previously shown to be substoichiometric or dynamically regulated (reviewed in (Sloan et al., 2017)). These modifications have yet to be explored systematically for their abundance and dynamics on the mammalian ribosome, but they may serve to modify ribosome function and connect the ribosome to other pathways in the cell. In fact, many RAPs function as protein or RNA modifiers, suggesting that there could be interplay between the presence of particular RAPs and the modification state of the core ribosome constituents.
Functional specialization or the enhanced regulatory potential of ribosomes
The most important question raised by the recent direct evidence of ribosome heterogeneity is what is the function of ribosomes that are distinct with respect to either protein or rRNA content? This is an outstanding question in the field and it is a logical hypothesis that ribosomes containing different sets of components could have distinct functional outcomes as wide-ranging as control of translation initiation, fidelity, mRNA translation selectivity, and translation speed. In all of these instances, ribosome diversity would likely culminate in some form of functional specialization or enhanced regulative capacity of ribosomes. We define “specialized ribosomes” as a preferential regulation of any aspect of translational control due to the enhanced regulative capacity of particular ribosomal components, even if the complex as a whole can bind and translate a larger portion of transcripts from across the genome (Box 1). Importantly, we believe that it is critical to consider dynamic changes to ribosomes throughout their lifetime and that further remodeling of ribosomes from their “birth” in the nucleolus to their maturation and their engagement with mRNAs in the cytoplasm as potential steps leading to the formation of specialized ribosomes. An important functional role of ribosome heterogeneity has been recently established as the ability of certain RPs to specialize the ribosome for the translation of particular mRNAs, although there are likely to be many more functions attributable to heterogeneity. Ribosomes containing RPS25/eS25 or RPL10a/uL1 (both of which are substoichiometric in mESCs), but not RPL22/eL22, were found to preferentially translate distinct sets of several hundred mRNAs (Shi et al., 2017). These transcripts encode proteins in related functional groups, indicating that ribosomes demarcated by substoichiometric RPs regulate the coordinated expression of distinct genetic networks including those important for metabolism, regulation of embryonic development, extracellular matrix organization, as well as many others. In fact, one unexpected and extreme version for this level of gene regulation is revealed by RPS25/eS25-containing ribosomes which preferentially translate the mRNAs encoding for the entirety of the vitamin B12 transport and uptake pathway, revealing that these specialized ribosomes simultaneously regulate the expression of all pathway components analogously to the bacterial operon system. RAPs additionally specialize their associated ribosomes for the translation of particular mRNAs, as is seen in the case of PKM2, which recruits mRNAs encoding ER and cell membrane constituents to the ER and promotes their translation (Simsek et al., 2017).
Box 1: What is a specialized ribosome?
As the perception of ribosomes as static, “housekeeping” molecular machinery has waned, a variety of definitions has been proposed to describe the regulative capacity of ribosomes (Dinman, 2016). We define “specialized ribosomes” as a ribosome with preferential regulation for any aspect of translational control. This may arise from the enhanced regulative capacity of particular ribosomal components, even if the complex as a whole binds and translates a larger portion of transcripts from across the genome. Ribosomes containing different sets of components could have distinct forms of specialization with functional outcomes as wide-ranging as control of translation initiation, fidelity, mRNA translation selectivity, and translation speed. To date, the most concrete example for ribosome specialization is with respect to heterogeneity in ribosomes that endows such translation machinery with a preference for the translation of particular mRNAs, and can be extended to encompass changes in ribosomal activity between cell types and cell states. In contrast to the classical biochemical “one enzyme-one substrate” view of specialization, one type of specialized ribosome does not necessarily exclusively translate only one set of genes. Rather, this definition of specialization is akin to the idea of specialized cells within an organism: specific cell populations are required, for instance, for the production of insulin, but this is not the only macromolecule that such cells synthesize. Analogously, specialized ribosomes may be required to synthesize certain proteins, but those ribosomes may also translate a larger portion of transcripts from across the genome.
If we accept this definition, then what components make a ribosome specialized? These could obviously include the core building blocks of the ribosome itself, namely the RPs and rRNAs. Importantly, we believe that it is critical to consider pathways to ribosome specialization that could include further remodeling of ribosomes after canonical ribosome biogenesis has been completed. For example, changes in ribosome activity could be caused by the association of RAPs or by post-translational modifications of core components at any step from when ribosome subunits are first made in the nucleolus to their export, maturation, and association with mRNAs in the cytoplasm—all of which constitute potential steps leading to the formation of specialized ribosomes.
Mechanistically, specialized ribosomes likely regulate the translation of particular transcripts via cis regulatory elements in their untranslated regions (UTRs). One cis regulon that appears to frequently rely on specialized ribosomes for its activity is the internal ribosome entry site (IRES) (Leppek et al., 2017). Canonical translation initiation in eukaryotes relies on the 5’cap on the mRNA to recruit translation complexes; IRESes, on the other hand, make more direct contacts with the ribosome itself and recruit the translation machinery to a site within the 5’UTR independently of the 5’ cap. While originally identified within viral RNAs, which lack 5’ caps and accordingly cannot employ the canonical cap-dependent mechanism, IRES elements are also present within eukaryotic mRNAs (Xue et al., 2015). Several transcripts that preferentially associate with RPL10a/uL1-containing ribosomes have RPL10a/uL1-dependent IRES activity (Shi et al., 2017). While mRNA elements have yet to be identified in transcripts enriched on RPS25/eS25-containing ribosomes, RPS25/eS25 has also been previously shown to regulate cellular IRES activity (Hertz et al., 2013). Another RP that is present on heterogeneous ribosomes, RPL38/eL38, additionally regulates the translation of several Hox genes, which are required for proper formation of the body plan, via IRES elements in their 5’UTRs. Haploinsufficiency of RPL38/eL38 in mouse models results in numerous homeotic transformations, the replacement of one skeletal element with another, and a selective decrease in the translation of several Hox genes that are required for axial skeletal patterning (Kondrashov et al., 2011). These Hox mRNAs were determined to contain IRESes that depend on RPL38/eL38 for their activity, and removal of these IRESes ablated Hox protein but not mRNA expression (Xue et al., 2015), revealing that these genes in fact rely on the IRES element for their translation. This is due to a second cis regulon present close to the 5’cap known as the translation inhibitory element (TIE), which inhibits cap-dependent translation initiation of these Hox mRNAs. The combination of these two elements (TIE+IRES) endows greater regulation by ribosomes on the expression of specific subsets of mRNA and represents a fascinating paradigm of translational control. Given the potential for hundreds of IRES elements being present in the human genome (Weingarten-Gabbay et al., 2016), such a mechanism may reflect a common regulatory strategy.
In sum, these findings reveal that ribosomes are not the homogeneous automatons, as they have historically been viewed, but in fact can vary in composition and function. This provides a novel layer of regulation to gene expression, enabling coordinate networks of genes to be simultaneously tuned at the level of translation. Given the prevalence of translational regulation across embryonic tissues in mammals (Fujii et al., 2017) and the importance of translation control for stem cell maintenance and differentiation (Sampath et al., 2008; Signer et al., 2014; Zhang et al., 2014), this process is likely to be fundamental for the proper functioning of cells and tissues. Many of these key findings, however, are the products of recent research, and many crucial questions remain unanswered.
Uncovering the full spectrum of mechanisms for translation by specialized ribosomes
In order for an mRNA to be translated by a particular specialized translation machinery, it is likely to be demarcated by some element that can be recognized by the appropriate ribosome. This cis-regulatory element could be a particular sequence, structure, base modification, or any combination of those items (Figure 2). Cis elements from each of those categories have been identified as regulators of translation, including IRES elements, the N6-methyladenosine modification (Meyer et al., 2015), and even the sequence context of the start codon: in a yeast strain with the RPS26B/eS26B paralog knocked out, RPS26A/eS26A endowed specificity for the canonical Kozak sequence flanking the start codon (Ferretti et al., 2017). In many cases, however, the exact connection between the cis element and the ribosome is not known. Only a minority of known targets of specialized ribosomes have even been assessed for the presence of RNA regulons, so there could be many novel motifs endowing translational regulation that have yet to be discovered. Even for cellular IRESes, one of the more common mechanisms of specialized ribosome recognition of their target mRNAs, there is little molecular detail on how they are able to recruit particular subsets of the translation machinery. Viral IRESes, which have been more thoroughly investigated, have been shown to have structural and functional conservation across closely related viruses even with divergent sequences (reviewed in (Fraser and Doudna, 2007)). This suggests that structure—not sequence—is the primary feature required for translation initiation at IRESes. Interestingly, this also appears to be the case for the RPL38/eL38-dependent cellular IRESes: the structures of both the Hoxa9 and Hoxa5 IRESes were solved by selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) and shown to have similar asymmetrical bulges (Xue et al., 2015). Point mutations in the Hoxa9 IRES that disrupted the secondary structure eliminated IRES activity, while the addition of compensatory mutations that restored base-pairing rescued IRES activity, revealing that structure is also the primary determinant of translation initiation on Hoxa9. Whether this will hold for the other Hox IRESes, let alone cellular IRESes that rely on different specialized ribosome components, has yet to be seen and would require more high-throughput approaches (reviewed in (Leppek et al., 2018)).
An alternative method that could provide RNA structure information in the context of protein binding is cryo-EM. Performing cryo-EM on ribosomes bound to specific transcripts, which could be either synthesized in vitro or isolated from cells, could pinpoint the exact sites of contact. While this is a challenging experiment, especially given the difficulties in accurately assigning structures to a complex made up of multiple components that are likely conformationally flexible, this would aid in characterization of known cis elements as well as potentially identifying novel regulons that promote specialized ribosome recruitment. In addition to identifying RNA structures, this would also provide important information on whether core RPs are direct binders of mRNAs or if additional trans-acting factors are involved. In the case of direct binders, it will be particularly interesting to determine which portions of RPs are required for mRNA recognition, and whether binding of the RP to the transcript alters its association with the ribosome or the structure of the ribosome as a whole. For instance, RPs could simply bind mRNAs to recruit them to the ribosome without any additional impact on translation, or mRNA binding could trigger additional remodeling that might further optimize the translation of the target mRNAs.
Organismal models and their interpretations
While specialized ribosomes have been shown to regulate translation, how important is this form of regulation for proper cellular function and organismal development? The multitude of animal models and human diseases with mutations in ribosomal machinery that cause tissue-specific phenotypes suggests that ribosome specialization plays a crucial role (Figure 3). However, the mechanisms underlying the tissue defects are often not yet elucidated; similarly, for many RPs with known functions in the translation of certain mRNA, the consequences of their loss in-vivo at the organismal level have yet to be explored. Several tissues appear particularly sensitive to RP mutations, as seen by mutations in multiple RPs in some instances causing overlapping phenotypes. For instance, mutations in RPS7/eS7, RPS19/eS19, RACK1, and RPL24/eL24 in mice produces a white belly spot, among other defects (reviewed in (Shi and Barna, 2015)). An even more striking example is the human disease Diamond Blackfan Anemia (DBA), a condition with bone marrow failure and tumor predisposition that has been linked to mutations in over 15 different RPs (reviewed in (Danilova and Gazda, 2015)). Recently, a theory-based perspective has suggested that the shared anemia phenotype observed in DBA could be caused by decreases in the total number of functional ribosomes upon mutation of these RPs, and thus lead to lower global rates of translation. Such an event might change the translation rate of some transcripts over others, thereby causing defects in particular cells, such as in hematopoietic cell types, but not across the entire body (Mills and Green, 2017). However, this hypothesis is based only on modeling and has yet to be validated experimentally. It also does not readily explain one of the most intriguing features of DBA: the distinct sets of congenital birth defects—including craniofacial defects, heart problems, and thumb malformations—that depend on which RP was mutated, which suggest that there are more specific requirements for certain RPs in selective tissues. DBA is also not the result of mutations in every RP. For example, mice lacking RPL29/eL29, which do have decreased protein synthesis, do not exhibit DBA symptoms and instead have a small size and bone fragility phenotype (Kirn-Safran et al., 2007; Oristian et al., 2009; Sloofman et al., 2010). In addition, a number of RP mutations result in unique tissue-specific phenotypes in humans, including asplenia upon loss of RPSA/uS2 (Bolze et al., 2013) and a hereditary form of hair loss upon mutation of RPL21/eL21 (Zhou et al., 2011), as well as in mice, such as the homeotic transformations upon RPL38/eL38 haploinsufficiency (Kondrashov et al., 2011) and male infertility in RPLP1 mutant mice (Perucho et al., 2014). Complications in our understanding of the significance of specialized translation in development and disease can arise from incomplete interpretation of phenotypes, such as the misconception that homeotic transformations of the axial skeleton, polydactyly, and bone marrow defects are equivalent because they all relate to bones, even though in actuality the relevant cell types derive from distinct embryonic tissues.
Figure 3: Potential mechanisms underlying disease phenotypes caused by RP haploinsufficiency.
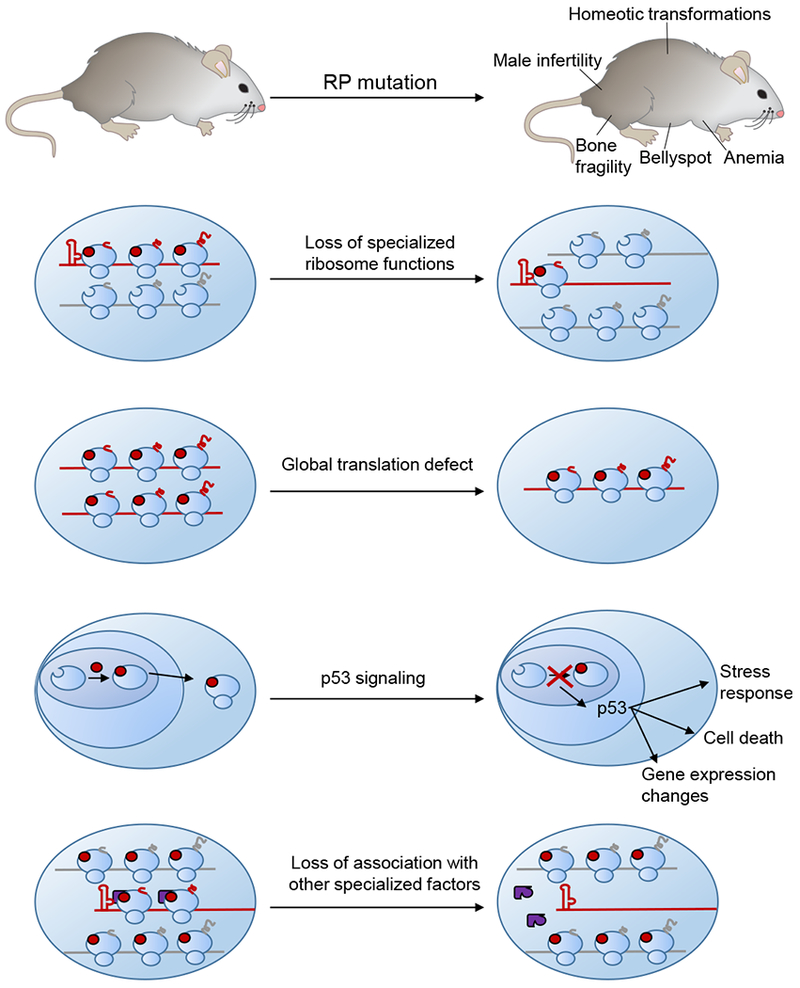
Depletion of an RP may cause tissue dysfunction via multiple direct and indirect effects. If the RP has a specialized function in recruiting particular mRNAs to the ribosome, the translation of those mRNAs may be decreased by the depletion of the RP. Certain RP mutations may also decrease the number of functional ribosomes in the cell, causing global decreases in protein synthesis. Perturbations in ribosome biogenesis can also trigger a stress response mediated by nucleolar stress and p53 activation, which could then have consequences on cellular function and gene expression. Finally, the RP may serve as a docking point for other specialized factors, such as RAPs, which upon RP depletion may no longer be able to associate with the ribosome.
It is also important to account for other potential contributors to the phenotype including the effect that mutation of one ribosomal component could have on the relative incorporations of other components and, in extreme cases where ribosome biogenesis is severely compromised, the role of nucleolar stress, the cellular response to defects in ribosome biogenesis (Zhang and Lu, 2009). Nucleolar stress involves activation of p53, and deletion of p53 in the background of several RP haploinsufficiency mutations can rescue some of the phenotypes. For instance, RPS6/eS6 haploinsufficient embryos die during gastrulation; embryos haploinsufficient for RPS6/eS6 and lacking p53 die at a later stage of embryonic development (Panić et al., 2006). This reveals that the inability to proceed through gastrulation observed upon depletion of RPS6/eS6 is due at least partially to more indirect effects from p53 signaling. However, the fact that RPS6/eS6 haploinsufficiency is still lethal even when p53 is removed reveals that there are additional p53-independent phenotypes. Parsing out the relative contributions of translation disruption and p53 signaling to the observed organismal phenotypes will lead to an improved understanding of the exact molecular consequences of ribosomal disruption.
Another important consideration when assessing an organismal phenotype is to consider whether the observed tissue-specific defects are the direct product of tissue-specific activities of specific ribosome components. Some RPs, for instance, may directly bind to certain mRNAs to regulate their translation, and thereby altering the RP’s abundance could reveal a form of specialization of an RP component, almost akin to a receptor-ligand interaction, even if that RP is a ubiquitous component of all ribosomes. Other RPs—including ones that might be stoichiometric or incapable of directly recruiting mRNA—could still exert tissue-specific effects due to their association with other factors. For example, an RP may serve as a docking point for a RAP with a restricted expression pattern. Mutating the RP might then eliminate RAP association with the ribosome, but this would only manifest in the tissues where the RAP is present. While this principle is currently speculative, it is quite likely that many of the ribosomal components are interconnected, with each one regulating the incorporation or activity of others.
Finally, it will be invaluable to expand these findings to other cell types and organisms. Over the years, many intriguing hypotheses have been proposed for specialized roles of ribosomes in different cell types, such as the idea of an immunoribosome (Yewdell and Nicchitta, 2006); now there are sufficient technological advances to put those models to the test. As studies in this field increase, it will be important to recognize that principles may arise that are generally applicable, but there also may be features unique to each cell type. In particular, much of the current literature has relied heavily on cancer cell lines, and it is not clear whether all findings from such studies will also apply to primary cell types within an organism. Cancer cells likely have their own distinct protein synthesis needs, and in fact multiple RP transcripts have been shown to be increased or decreased in particular cancer cell lines (Guimaraes and Zavolan, 2016), and multiple RP mutations are associated with cancers, including T-cell acute lymphoblastic leukemia (T-ALL) (De Keersmaecker et al., 2013). One such mutation in RPL10/uL16 that is found in T-ALL alters programmed ribosomal frameshifting on reporter constructs in yeast (Sulima et al., 2014) and on mouse and human genes in the JAK-STAT pathway, which promotes cell proliferation and is in fact hyper-activated in T-ALL (Girardi et al., 2017). These findings, as well as the association of other specialized translation machinery with the regulation of cancer progression (Truitt et al., 2015), suggests that cancer cells may have ribosomes that are tuned to infinite cellular growth and augmented protein synthesis capacity (Sulima et al., 2017).
Understanding how specialized ribosomes contribute to cancers as well as congenital disorders may lead to novel therapeutic strategies. Once the mechanisms underlying the construction of heterogeneous ribosomes are established, it may be possible to tweak ribosome composition. As most disease-causing mutations are haploinsufficient and therefore only affect one allele, there may be ways to increase the expression or incorporation of the other allele to ameliorate the phenotypes. These controlled changes in ribosome heterogeneity would then up- or down-regulate particular genetic networks. For instance, RPL10a/uL1 is required for translation of mRNAs that promote cell survival, while genes that contribute to cell death are depleted from RPL10a/uL1 containing ribosomes, suggesting that tuning the levels of RPL10a/uL1 could shift the balance between cell survival and death (Shi et al., 2017). As we improve our understanding of how ribosomes may be specialized for the translation of particular mRNAs, we also may be able to design ribosomes for the designated synthesis of proteins with therapeutic value (e.g. insulin or antibodies). These strategies would lead to not only improved synthetic biology tools and treatments but also would reveal the extent of our understanding of this novel layer of gene regulation.
Concluding remarks
Since the first postulations of ribosome heterogeneity in the 1950s and its subsequent rapid demise a few years later the field has come far. Today, we now have direct evidence of at least a handful of ribosomes of distinct compositions translating specific subsets of mRNAs, revealing that ribosome heterogeneity plays an active role in the regulation of gene expression. Although Crick overshot ribosome heterogeneity with his original “one gene-one ribosome-one protein” model, the hypothesis of ribosome specialization has been resurrected. A more accurate though less pithy summary would be “one mRNA regulon-one ribosome-one protein network,” as each type of specialized ribosome appears to translate mRNAs, demarcated by particular cis regulatory elements, encoding proteins in a common pathway. Further work is needed for a detailed mechanistic understanding of the regulation and function of ribosome specialization within a cell and across an organism, but this novel layer of gene expression control clearly plays a crucial role in cellular function and organismal development. Having unique tissue-specific phenotypes upon mutating components of a complex vital in almost every cell may at first seem surprising, but this is actually also a feature of mutations in several other large macromolecular complexes such as the nuclear pore (Raices and D’Angelo, 2012), cilia (Waters and Beales, 2011), and subunits of large chromatin complexes such as SWI/SNF BAF (Ronan et al., 2013; Sokpor et al., 2017). For all of these complexes, selective components have evolved more specialized and distinct mechanisms of regulation within specific cell lineages during multicellular organismal development. For instance, several of the 30 or so nucleoporins that make up the nuclear pore—the passageway between the nucleus and the cytoplasm that is essential for all nucleated cells—show cell type-specific expression patterns, and mutations in these components affect only certain tissues. Mutations in Nupl55, for example, which is highly expressed in the human heart and several other tissues (Zhang et al., 1999), causes heart failure during childhood (Zhang et al., 2008). This suggests that several presumably ‘housekeeping’ molecular complexes may follow the same strategy of serving general cellular functions while also being specialized by the presence of heterogeneous components for the regulation of specific, developmentally important transitions. It will be exciting to see how this burgeoning field is shaped within the next decades.
Acknowledgements:
This work was supported by the New York Stem Cell Foundation NYSCF-R-I36 (M.B.), National Institutes of Health grant 1R01HD086634 (M.B.) and Pew Scholars Award (M.B.). N.R.G. is supported by National Science Foundation Graduate Research Fellowship DGE-114747. M.B. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anger AM, Armache J-P, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, pp.80–85. [DOI] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. (2010). Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci U S A 107, pp.19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, and Southern EM (1977). Heterogeneity of the Ribosomal Genes in Mice and Men. Cell 11, pp.363–370. [DOI] [PubMed] [Google Scholar]
- Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, et al. (2013). Haploinsufficiency in Humans with Isolated Congenital Asplenia. Science 340, pp.976–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, and Crick FHC (1960). What are the properties of genetic RNA? [Google Scholar]
- Brenner S, Jacob F, and Meselson M (1961). An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190, pp.576–581. [DOI] [PubMed] [Google Scholar]
- Chaillou T, Kirby TJ, and Mccarthy JJ (2014). Ribosome Biogenesis: Emerging Evidence for a Central Role in the Regulation of Skeletal Muscle Mass. J. Cell. Physiol 229, pp.1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T, Zhang X, and Mccarthy JJ (2016). Expression of Muscle-Specific Ribosomal Protein L3-Like Impairs Myotube Growth. J. Cell. Physiol 231, pp.1894–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Sharma MR, Shi X, Agrawal RK, and Joseph S (2014). Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, pp.407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC (1958). On Protein Synthesis. Symp. Soc. Exp. Biol 12, pp.138–163. [PubMed] [Google Scholar]
- Crick FHC, and Brenner S (1959). Some Footnotes on Protein Synthesis: a Note for the RNA TIE club. [Google Scholar]
- Danilova N, and Gazda HT (2015). Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model. Mech 8, pp.1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, pp.247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD (2016). Pathways to Specialized Ribosomes: The Brussels Lecture. J. Mol. Biol 428, pp.2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. (1999). The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet 21, pp. 169–175. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst C-A, Eberhart DE, Yi H, Warren ST, and Hersch SM (1997). Fragile X Mental Retardation Protein: Nucleocytoplasmic Shuttling and Association with Somatodendritic Ribosomes. J. Neurosci 17, pp.1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti MB, Ghalei H, Ward EA, Potts EL, and Karbstein K (2017). Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol 24, pp.700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, and Doudna JA (2007). Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol 5, pp.29–38. [DOI] [PubMed] [Google Scholar]
- Fujii K, Shi Z, Zhulyn O, Denans N, and Barna M (2017). Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun 8, pp.14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi T, Vereecke S, Sulima SO, Khan Y, Fancello L, Briggs JW, Schwab C, Beeck J. Op de, Verbeeck J, Royaert J, et al. (2017). The T-cell leukemia-associated ribosomal RPL10 R98S mutation enhances JAK-STAT signaling. Leukemia 32, pp.809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes JC, and Zavolan M (2016). Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol 17, pp.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP, and McCutchan TF (1987). Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238, pp.933–938. [DOI] [PubMed] [Google Scholar]
- Gupta V, and Warner JR (2014). Ribosome-omics of the human ribosome. RNA 20, pp.1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, and Atwood KC (1972). Location of Ribosomal DNA in the Human Chromosome Complement. Proc. Natl. Acad. Sci 69, pp.3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, and Thompson SR (2013). Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol 33, pp.1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. (2015). The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40S Ribosomal Proteins. Mol. Cell 59, pp.35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, and Walsh D (2017). Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546, pp.651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, and Holt CE (2014). Remote Control of Gene Function by Local Translation. Cell 157, pp.26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Chen AS, and Ware VC (2017). Expression of ribosomal protein L22e family members in Drosophila melanogaster : rpL22-like is differentially expressed and alternatively spliced 39, pp.2701–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. (2013). Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet 45, pp.186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiparisov S, Petrov A, Meskauskas A, Sergiev PV, Dontsova OA, and Dinman JD (2005). Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Mol. Genet. Genomics 274, pp.235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, Lee JD, England JH, Chaillou T, Esser K. a, and McCarthy JJ (2015).Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J. Appl. Physiol pp.jap.00296.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, and Carson DD (2007). Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn 236, pp.447–460. [DOI] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, and Silver PA (2007). Functional Specificity among Ribosomal Proteins Regulates Gene Expression. Cell 131, pp.557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, and Barna M (2011). Ribosome-Mediated Specificity in Hox mRNA Translation and Vertebrate Tissue Patterning. Cell 145, pp.383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuwan K, Yu Q, Vincent A, Frisardi MC, Rosbash M, Lengyel JA, and Merriam J (1985). A Drosophila Minute gene encodes a ribosomal protein. Nature 317, pp.555–558. [DOI] [PubMed] [Google Scholar]
- Leppek K, Das R, and Barna M (2017). 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, and Barna M (2018). Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol 19, pp.158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, and Braaten DC (1989). Genomic organization of human 5S rDNA and sequence of one tandem repeat. Genomics 4, pp.376–383. [DOI] [PubMed] [Google Scholar]
- Locati MD, Pagano JFB, Girard G, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Spaink HP, Rauwerda H, Jonker MJ, et al. (2017a). Expression of Distinct Maternal and Somatic 5.8S, 18S, and 28S rRNA Types during Zebrafish Development. RNA 23, pp.1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati MD, Pagano JFB, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Zhu K, Spaink HP, Girard G, Rauwerda H, et al. (2017b). Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA 23, pp.446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Coelho CMA, and Leevers SJ (2005). Genetic Analysis of RpL38 and RpL5, Two Minute Genes Located in the Centric Heterochromatin of Chromosome 2 of Drosophila melanogaster. Genetics 169, pp.683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro VP, and Edelman GM (2002). The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U. S. A 99, pp. 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, and Fox PL (2003). Regulated Release of L13a from the 60S Ribosomal Subunit as A Mechanism of Transcript-Specific Translational Control. Cell 115, pp.187–198. [DOI] [PubMed] [Google Scholar]
- Mcmahon M, Contreras A, and Ruggero D (2015). Small RNAs with big implications: New insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip. Rev. RNA 6, pp.173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, and Jaffrey SR (2015). 5’UTR m6A Promotes Cap-Independent Translation. Cell 163, pp.999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, and Green R (2017). Ribosomopathies: There’s strength in numbers. Science 358, pp.eaan2755. [DOI] [PubMed] [Google Scholar]
- Moor AE, Golan M, Massasa EE, Lemze D, Weizman T, Shenhav R, Baydatch S, Mizrahi O, Winkler R, Golani O, et al. (2017). Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science 2399, pp.1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, and Snyder M (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, pp.2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MN, Schreiber KH, Zhang Y, Duc ACE, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, et al. (2013). The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1. PLoS Genet 9, pp.e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oristian DS, Sloofman LG, Zhou X, Wang L, Farach-carson MC, and Kirn-safran CB (2009). Ribosomal Protein L29 / HIP Deficiency Delays Osteogenesis and Increases Fragility of Adult Bone in Mice. J. Orthop. Res 27, pp.28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE (1955). A small particulate component of the cytoplasm. J. Cell Biol 1, pp.59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE (1958). Microsomes and ribonucleoprotein particles. In Microsomal Particles and Protein Synthesis, pp. 36–61. [Google Scholar]
- Panić L, Tamarut S, Sticker-Jantscheff M, Barkić M, Solter D, Uzelac M, Grabusić K, and Volarević S (2006). Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol 26, pp.8880–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, and Blanchard SC (2018). Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv 4, pp.eaao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho L, Artero-Castro A, Guerrero S, Ramón Y Cajal S, LLeonart ME, and Wang ZQ (2014). RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS One 9, pp.e99956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, and D’Angelo MA (2012). Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol 13, pp.687–699. [DOI] [PubMed] [Google Scholar]
- Ramagopal S (1990). Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem. Cell Biol 68, pp.1281–1287. [DOI] [PubMed] [Google Scholar]
- Roberts RB (1958). Introduction. In Microsomal Particles and Protein Synthesis, pp. vii–viii. [Google Scholar]
- Ronan JL, Wu W, and Crabtree GR (2013). From neural development to cognition: Unexpected roles for chromatin. Nat. Rev. Genet 14, pp.347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, pp.1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, and Murry CE (2008). A Hierarchical Network Controls Protein Translation during Murine Embryonic Stem Cell Self-Renewal and Differentiation. Cell Stem Cell 2, pp. 448–460. [DOI] [PubMed] [Google Scholar]
- Segev N, and Gerst JE (2018). Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J. Cell Biol 217, pp. 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, and Barna M (2015). Translating the Genome in Time and Space: Specialized Ribosomes, RNA Regulons, and RNA-Binding Proteins. Annu. Rev. Cell Dev. Biol 31, pp.annurev-cellbio-100814-125346. [DOI] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, and Barna M (2017). Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 67, pp.71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, pp.49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, and Barna M (2017). An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol 45, pp.92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Xu AF, Chang HY, Barna M, Simsek D, Tiu GC, Flynn RA, Byeon GW, et al. (2017). The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 169, pp.1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, and Bohnsack MT (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14, pp. 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloofman LG, Verdelis K, Spevak L, Zayzafoon M, Yamauchi M, Opdenaker LM, Farach-Carson MC, Boskey AL, and Kirn-Safran CB (2010). Effect of HIP/ribosomal protein L29 deficiency on mineral properties of murine bones and teeth. Bone 47, pp.93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G, Xie Y, Rosenbusch J, and Tuoc T (2017). Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci 10, pp.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, and Pierce AJ (2008). Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18, pp.13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, and Dinman JD (2014). Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci 111, pp.5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima SO, Hofman IJF, De Keersmaecker K, and Dinman JD (2017). How ribosomes translate cancer. Cancer Discov 7, pp.1069–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Nielsen J, Jonsson S, Halldorsson GH, Melsted P, Ivarsdottir EV, Davidsson OB, Kristjansson RP, et al. (2018). Coding variants in RPL3L and MYZAP increase risk of atrial fi brillation. Commun. Biol 1, pp.DOI: 10.1038/s42003-018-0068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, and Ruggero D (2015). Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 162, pp.59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Chou W, Wang J, Zhang X, Zhang S, and Schultz RM (2008). Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS One 3, pp.e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, and Heck AJR (2017). High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, pp.283–286. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd M, Tamara S, Fort KL, Damoc E, Franc V, Bieri P, Itten M, Makarov A, Ban N, and Heck AJR (2018). Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nat. Commun 9, pp.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AM, and Beales PL (2011). Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol 26, pp.1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint a, Hooykaas P, and Offringa R (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, pp.4289–4299. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, and Segal E (2016). Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, pp.aad4939. [DOI] [PubMed] [Google Scholar]
- Whittle CA, and Krochko JE (2009). Transcript Profiling Provides Evidence of Functional Divergence and Expression Networks among Ribosomal Protein Gene Paralogs in Brassica napus. Plant Cell 21, pp.2203–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, and Sussex IM (1995). Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J 8, pp.65–76. [DOI] [PubMed] [Google Scholar]
- Wong QW-L, Li J, Ng SR, Lim SG, Yang H, and Vardy LA (2014). RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol 11, pp.33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, and Barna M (2012). Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol 13, pp.355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, and Barna M (2015). RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature 517, pp.33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW, and Nicchitta CV (2006). The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol 27, pp.368–373. [DOI] [PubMed] [Google Scholar]
- Zhang Y, and Lu H (2009). Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, pp.369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shalaby N. a, and Buszczak M (2014). Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 343, pp.298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang H, Corydon MJ, Zhang X, Pedersen S, Korenberg JR, Chen XN, Laporte J, Gregersen N, Niebuhr E, et al. (1999). Localization of a human nucleoporin 155 gene (NUP155) to the 5p13 region and cloning of its cDNA. Genomics 57, pp.144–151. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. (2008). Mutation in Nuclear Pore Component NUP155 Leads to Atrial Fibrillation and Early Sudden Cardiac Death. Cell 135, pp.1017–1027. [DOI] [PubMed] [Google Scholar]
- Zhou C, Zang D, Jin Y, Wu H, Liu Z, Du J, and Zhang J (2011). Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum. Mutat 32, pp.710–714. [DOI] [PubMed] [Google Scholar]


