SUMMARY
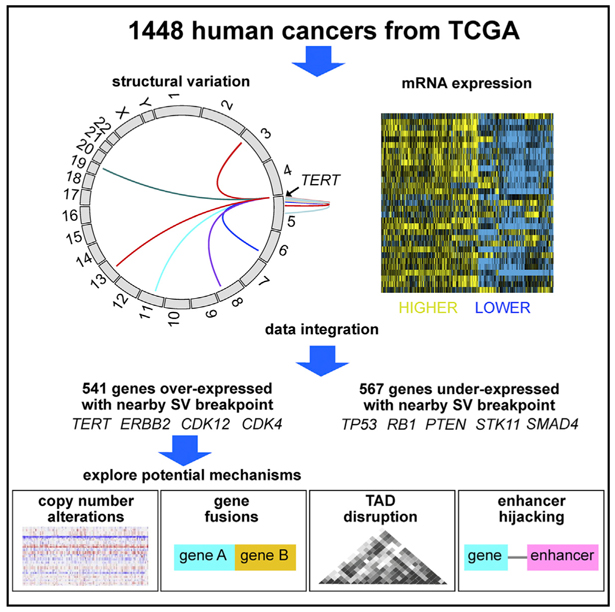
A systematic cataloging of genes affected by genomic rearrangement, using multiple patient co-horts and cancer types, can provide insight into cancer-relevant alterations outside of exomes. By integrative analysis of whole-genome sequencing (predominantly low pass) and gene expression data from 1,448 cancers involving 18 histopathological types in The Cancer Genome Atlas, we identified hundreds of genes for which the nearby presence (within 100 kb) of a somatic structural variant (SV) breakpoint is associated with altered expression. While genomic rearrangements are associated with widespread copy-number alteration (CNA) patterns, approximately 1,100 genes—including overexpressed cancer driver genes (e.g., TERT, ERBB2, CDK12, CDK4) and underexpressed tumor suppressors (e.g., TP53, RB1, PTEN, STK11)—show SV-associated deregulation independent of CNA. SVs associated with the disruption of topologically associated domains, enhancer hijacking, or fusion transcripts are implicated in gene upregulation. For cancer-relevant pathways, SVs considerably expand our understanding of how genes are affected beyond point mutation or CNA.
In Brief
Zhang et al. analyzed over 1,400 cancers by high- or low-pass whole-genome sequencing, focusing on patterns of structural variation. They saw a widespread impact of somatic structural variants on gene expression patterns, independent of copy-number alterations, involving key oncogenes and tumor suppressor genes.
Graphical Abstract
INTRODUCTION
Cancer genomes are characterized by widespread somatic genomic rearrangements, in addition to point mutations. Somatic structural variants (SVs) resulting from rearrangement—each SV involving two breakpoints representing different genomic coordinates from the unaltered genome being joined together—may represent important cancer driving events. Classes of SVs may include deletions, insertions, inversions, tandem duplications, translocations, and more complex rearrangements (Yang et al., 2013). SVs may exert a heavy influence on the expression of genes through various mechanisms, including CNAs, direct disruption of the gene by breakpoint falling within the coding region, formation of fusion transcripts involving two genes, disruption or repositioning of cis-regulatory elements near genes, formation of cryptic promoters, placement of genes into anomalous chromatin environments, and disruption of topologically associated domain (TAD) organization affecting long-range enhancer-promoter interactions (Dekker and Heard, 2015; Harewood and Fraser, 2014). Whole-genome sequencing (WGS) enables the accurate detection of somatic rearrangements in cancer; somatic SVs were characterized in several previous studies, for both individual cancer types (Bass et al., 2011; Berger et al., 2011; Campbell et al., 2010; Davis et al., 2014; Stephens et al., 2011) and pan-cancer studies (Alaei-Mahabadi et al., 2016; Drier et al., 2013; Hillmer et al., 2011; Yang et al., 2013). These studies typically involved a relatively modest number of cancer cases.
There is need for a systematic identification and cataloging of genes that are recurrently altered transcriptionally in cancer as a result of genomic rearrangement. To date, much effort has been made in better defining the set of recurrently and significantly mutated genes across human cancers (Chang et al., 2016; Gon-zalez-Perez et al., 2013; Kandoth et al., 2013; Lawrence et al., 2014; Martincorena et al., 2017). An analogous list of candidate driver genes resulting from rearrangement could provide further insight into cancer-related processes and pathways and could be relevant from the standpoint of personalized or precision medicine approaches, which typically focus primarily on point mutations within the coding region of genes. Previous studies have, for example, defined broad patterns of association involving genomic rearrangements and transcription collectively involving large groups of genes (Alaei-Mahabadi et al., 2016; Drier et al., 2013), defined the landscape of gene fusions in cancer (Hu et al., 2017; Stransky et al., 2014; Yoshihara et al., 2015), and documented cases of individual genes altered by the rearrangement of cis-regulatory elements within specific cancer types (Davis et al., 2014; Gröschel et al., 2014; Northcott et al., 2014; Peifer et al., 2015). However, a pan-cancer, gene-by-gene assessment of which ones appear recurrently deregulated by genomic rearrangement, using sizable sample numbers for greater power (Lawrence et al., 2014), remains to be carried out.
The Cancer Genome Atlas (TCGA) provides a common platform for the study of diverse cancer types (Cancer Genome Atlas Research Network et al., 2013b), with a sizable number of cases being sequenced at the whole-genome level. With TCGA gene expression data, large-scale integration between WGS and expression data is possible. TCGA WGS data were generated with either low-depth-of-coverage (low pass) or high-depth-of-coverage (high pass), with both types of WGS being used effectively to identify SVs in previous TCGA consortium-led studies focusing on a specific cancer type (Cancer Genome Atlas Network, 2012, 2015a, 2015b; Cancer Genome Atlas Research Network, 2013, 2014a, 2014b, 2015b, 2017a, 2017b; Davis et al., 2014; Robertson et al., 2017). While low-pass WGS may involve decreased sensitivity of detection, >1,200 cases in TCGA have low-pass data, representing a rich resource, with no pan-cancer study to date using these data. Previous studies have surveyed genomic rearrangements in TCGA using whole-exome or SNP array platforms (Weischenfeldt et al., 2017; Yang et al., 2016); the platforms are more limited in terms of sensitivity of detection as compared to WGS. The unified datasets and larger sample numbers offered by TCGA would allow us to identify robust associations between WGS-inferred SVs and expression that would cut across multiple cancer types of various lineages.
RESULTS
Somatic SVs across Cancer Types
We analyzed WGS data from 1,493 individuals across 18 cancer types represented in TCGA cohort (Tables 1 and S1), with cases including 114 bladder urothelial carcinomas (BLCAs), 89 breast-invasive carcinomas (BRCAs), 51 cervical squamous cell carcinomas and endocervical adenocarcinomas (CESCs), 120 colorectal adenocarcinomas (CRCs), 51 esophageal carcinomas (ESCAs), 108 head and neck squamous cell carcinomas (HNSCs), 50 kidney chromophobe renal cell carcinoma (KICHs), 41 kidney clear cell renal cell carcinomas (KIRCs), 38 kidney renal papillary cell carcinomas (KIRPs), 53 brain lower-grade gliomas (LGGs), 122 lung adenocarcinomas (LUADs), 50 ovarian serous cystadenocarcinomas (OVs), 116 prostate adenocarcinomas (PRADs), 118 skin cutaneous melanomas (SKCMs), 107 stomach adenocarcinomas (STADs), 100 thyroid carcinomas (THCAs), 114 uterine corpus endometrial carcinomas (UCECs), and 51 uveal melanomas (UVMs). Of the 1,493 cases, 1,448 had gene expression data by RNA-sequencing (RNA-seq) platform available. For 5 of the 18 cancer types studied (BRCA, OV, KICH, KIRC, KIRP, representing 268 cases), WGS was carried out with ~30–60× coverage, with the other cancer types sequenced at ~6–8× coverage.
Table 1.
TCGA Samples Analyzed
| Cancer Type | TCGA Project | Cases with WGS | Cases with WGS + RNA-Seq | High Pass/Low Pass | Mean Detected SVs per Case |
|---|---|---|---|---|---|
| Bladder urothelial carcinoma | BLCA | 114 | 114 | low | 55.9 |
| Breast-invasive carcinoma | BRCA | 89 | 89 | high | 329.3 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 51 | 50 | low | 26.4 |
| Colorectal adenocarcinoma | CRC | 120 | 118 | low | 12.4 |
| Esophageal carcinoma | ESCA | 51 | 51 | low | 55.6 |
| Head and neck squamous cell carcinoma | HNSC | 108 | 108 | low | 28.7 |
| Kidney chromophobe renal cell carcinoma | KICH | 50 | 50 | high | 17.5 |
| Kidney clear cell renal cell carcinoma | KIRC | 41 | 41 | high | 46.0 |
| Kidney renal papillary cell carcinoma | KIRP | 38 | 38 | high | 20.1 |
| Brain lower-grade glioma | LGG | 53 | 53 | low | 19.8 |
| Lung adenocarcinoma | LUAD | 122 | 121 | low | 32.9 |
| Ovarian serous cystadenocarcinoma | OV | 50 | 23 | high | 225.3 |
| Prostate adenocarcinoma | PRAD | 116 | 116 | low | 25.4 |
| Skin cutaneous melanoma | SKCM | 118 | 118 | low | 58.6 |
| Stomach adenocarcinoma | STAD | 107 | 94 | low | 53.8 |
| Thyroid carcinoma | THCA | 100 | 100 | low | 5.9 |
| Uterine corpus endometrial carcinoma | UCEC | 114 | 113 | low | 22.5 |
| Uveal melanoma | UVM | 51 | 51 | low | 48.7 |
| Total | – | 1,493 | 1,448 | – | 57.3 |
With data from both tumor and germline samples for each patient to distinguish germline and somatic variants, a total of 85,560 high-confidence somatic SVs were detected, using the Meerkat algorithm (Yang et al., 2013, 2016) (Table S2). As would be expected, cases sequenced with higher coverage had more SVs detected. Cases with low-pass (~6–8×) WGS had 33.9 SVs detected on average, while cases with high-pass (~30–60×) WGS had 164.5 SVs detected on average. Although they were subjected to high-pass WGS, the three kidney cancer types showed relatively fewer detected SVs (average 27.3), which is consistent with previous findings (Chen et al., 2016; Yang et al., 2013, 2016). As compared to somatic SVs as detectable by whole-exome sequencing (Yang et al., 2016), low-pass WGS detected 10 times as many SVs on average. Based on comparisons between SV calls by either low-pass or high-pass WGS for a subset of cases (Table S2), ~20% of SVs identifiable by high-pass WGS were identified by low-pass WGS with the Meerkat algorithm, and ~75% of SVs identified by low-pass WGS were identifiable by high-pass WGS. Despite its decreased sensitivity, low-pass SV analysis would be likely to yield biologically meaningful associations through identification of recurrent patterns across multiple samples and through integration with other data platforms.
Widespread Impact of Somatic SVs on CNAs
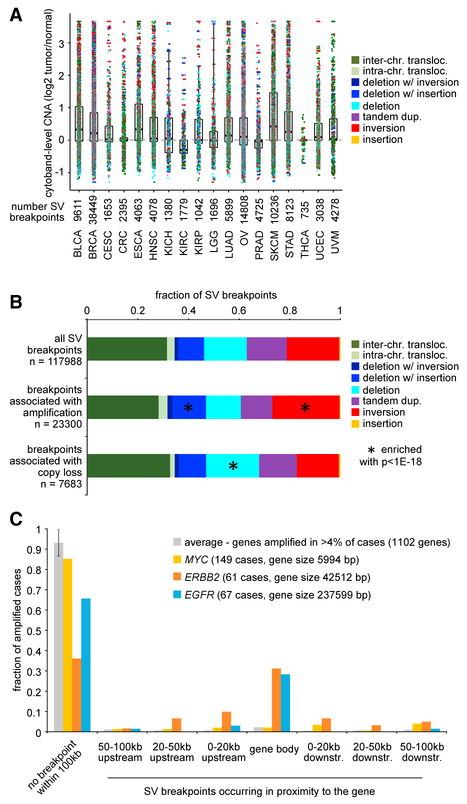
As may have been anticipated (Weischenfeldt et al., 2013; Yang et al., 2016), genomic rearrangements could be associated here with widespread patterns of CNAs. While SVs may be balanced or unbalanced in terms of CNAs within the immediate vicinity of the breakpoint (e.g., involving deletions, insertions, tandem duplications), here, we considered SV associations with CNAs at a broad level, by cytoband region. On the basis of an analysis of 1,465 cases with both WGS and SNP array data, tumor/ normal CNA log2 ratios were averaged by cytoband for each cancer case. For 117,988 SV breakpoints (counting SVs with both breakpoints occurring within the same cytoband only once), the corresponding cytoband-level CNAs were plotted by cancer type (Figure 1A; Table S3). For specific cancer types, including BLCA, BRCA, ESCA, LUAD, OV, SKCM, STAD, and UCEC, SV breakpoints on average tended to be associated with cytoband-level copy gain (while across all cases for the above types, the numbers of cytobands with gain versus loss tended to be approximate). In contrast to the other cancer types, KIRC had SV breakpoints on average associated with copy loss. As compared to all SV breakpoints, SV breakpoints associated with cytoband-level copy gain were significantly enriched for breakpoints involving inversion SVs or deletion with insertion SVs, while SV breakpoints associated with cytoband-level copy loss were significantly enriched for breakpoints involving deletion SVs (Figures 1A and1B).
Figure 1. CNAs Associated with Genomic Rearrangements in Human Cancers.
(A) By cancer type (denoted by TCGA project name), boxplot of cytoband-level CNA (log2 tumor/normal copy numbers) corresponding to structural variant (SV) breakpoints. SVs with both breakpoints occurring within the same cyto-band are represented only once. Cytobands in X and Y chromosomes are not represented. Foreach cancer type, the median log2 CNA across all cases and cytobands is approximately zero. Boxplots represent 5%, 25%, 50%, 75%, and 95%. Analysis involves 1,465 cases with both WGS and copy data. The maximum log2 tumor/normal CNA value is set to 3.6 by the SNP array analysis, approximating >24 copies.
(B) For SVs associated with cytoband-level gain (average log2 tumor/normal copy >1) or loss (average log2 tumor/normal copy <−0.5) breakdown by SV class. p values by chi-square test.
(C) Fraction of cancer cases with high-level amplifications for a given gene, according to SV breakpoint occurring within the gene body, upstream of the gene (0–20 kb, 20–50 kb, 50–100 kb), downstream of the gene (0–20 kb, 20–50 kb, 50–100 kb), or >100 kb from the gene. Results are shown for MYC, ERBB2, and EGFR, as well as the averages for 1,102 genes with amplification in >4% of the cases Where multiple breakpoints occur in proximity to a gene, the breakpoint closest to the gene is assigned to the given case. Error bars, SDs.
In addition to CNA at the cytoband level, we considered CNAs at the gene level to be associated with SV breakpoints. We separately considered SV breakpoints occurring 0–20 kb upstream of any gene, 20–50 kb upstream, 50–100 kb upstream, within a gene body, 0–20 kb downstream of a gene, 0–20 kb downstream, 20–50 kb downstream, and 50–100 kb downstream. According to specific SV classes—interchromosomal translocation, deletion with inversion, deletion with insertion, tandem duplication, and inversion classes in particular—SV breakpoints on average tended to be associated with gene-level copy gain (Figure S1). In considering 1,102 genes with high-level amplifications (approximating >5 copies) in >4% of cases, SV breakpoints tended to occur >100 kb away from the gene; however, for key genes such as EGFR and ERBB2, a substantial fraction of cases (~30%–40%) involved SV breakpoints occurring within the gene (Figure 1C); for ERBB2, 64% of amplified cases involved an SV breakpoint within 100 kb of the gene.
Widespread Impact of Somatic SVs on Gene Expression Patterns
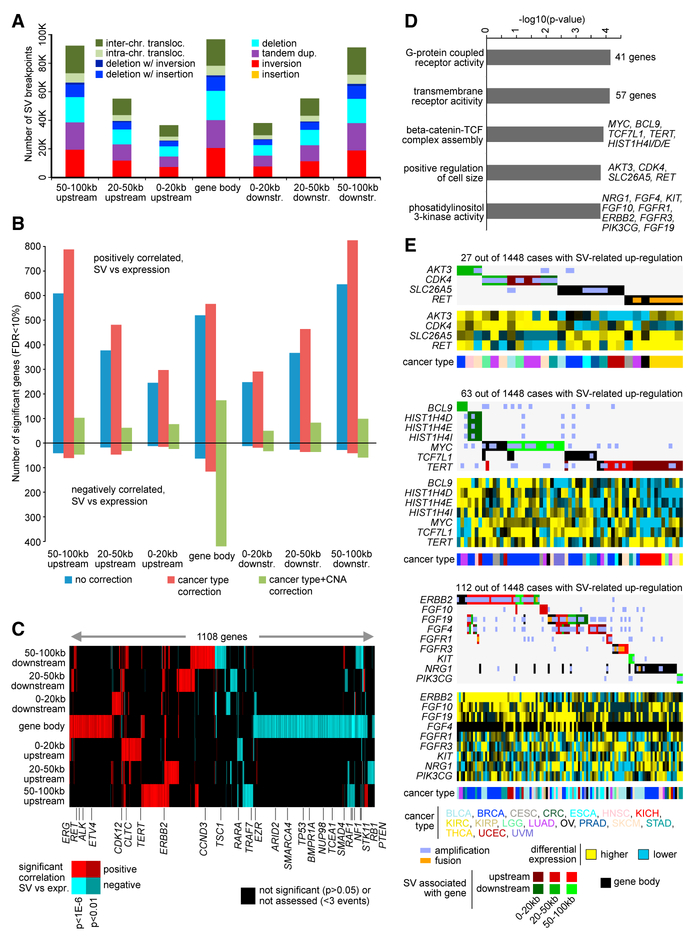
We carried out a systematic, pan-cancer analysis of all coding genes for patterns of expression affected by genomic rearrangements. We aimed to identify genes for which the nearby presence of an SV breakpoint could be significantly associated with changes in expression (based on an analysis of 1,448 cases with both WGS and RNA-seq data available). Because SV breakpoints in the region 0–20 kb upstream of TERT were previously associated with its upregulation in KICH (Davis et al., 2014), we considered fixed windows of genomic distance from each gene. Specifically, we considered SV breakpoints occurring 0–20 kb upstream of the gene, 20–50 kb upstream, 50–100 kb upstream, within the gene body, 0–20 kb downstream of a gene, 0–20 kb downstream, 20–50 kb downstream, and 50–100 kb downstream (Figure 2A). For each of the above regions, we assessed each gene for correlation between associated SV breakpoint occurrence and expression. Because each cancer type as a group would have a distinct molecular signature (Hoadley et al., 2014) and because genomic rearrangements may be involved in CNA (Figures 1A and S1), both were factored into our analysis using linear models, which also factored in differences in WGS coverage according to TCGA project.
Figure 2. Genes with Altered Expression Associated with Nearby SV Breakpoints.
(A) Numbers of SV breakpoints identified as occurring within a gene body, upstream of a gene (0–20 kb, 20–50 kb, 50–100 kb) or downstream of a gene (0–20 kb, 20–50 kb, 50–100 kb). For each SV set, the breakdown by alteration class is indicated. SVs located within a given gene are not included in the other upstream or downstream SV sets for that same gene.
(B) For each of the SV sets from (A), numbers of significant genes (FDR <0.1) showing correlation between expression and associated SV event. Numbers above and below the zero point of the y axis denote positively and negatively correlated genes, respectively. Linear regression models also evaluated significant associations when correcting for cancer type (red) and for both cancer type and gene-level CNA (green).
(C) Heatmap of significance patterns for genes from (B) (from the model correcting for both cancer type and CNA). Significant positive correlation (red), significant negative correlation (blue), not significant (p > 0.05) or not assessed (<3 SV events for given gene in the given genomic region) (black).
(D) Significantly enriched Gene Ontology (GO) terms for genes positively correlated (FDR <0.1, with corrections for cancer type and CNA) with occurrence of SV breakpoint in proximity to the gene (for any region considered). p values by one-sided Fisher’s exact test.
(E) Patterns of SV versus expression for selected gene sets from (D) (positive regulation of cell size [top], β-catenin-TCF complex assembly [middle], phosphatidylinositol 3-kinase activity [bottom]). Differential gene expression patterns relative to the median across sample profiles. Cases with genes associated with high-level gene amplification or with gene fusion event are respectively indicated.
For each of the genomic regions relative to genes that were considered (i.e., genes with at least three samples associated with an SV breakpoint within the given region), we found wide-spread associations between SV event and expression, after correcting for expression patterns associated with tumor type or CNA (Figures 2B and S2A; Table S4). For gene body, 0–20 kb upstream, 20–50 kb upstream, 50–100 kb upstream, 0–20 kb downstream, 20–50 kb downstream, and 50–100 kb downstream regions, the numbers of significant genes at false discovery rate (FDR) <0.1 (Storey and Tibshirani, 2003) (correcting for both cancer type and CNA) were 594, 101, 94, 150, 83, 119, and 158, respectively. For each of these gene sets, more genes were positively correlated with an SV event (i.e., expression was higher when SV breakpoint was present) than were negatively correlated, except for SV breakpoints occurring within the gene body, where many more genes were negatively versus positively correlated (420 versus 174 genes, respectively). Without correcting for copy number, even larger numbers of genes with SVs associated with increased expression were found (Figure 2B), reflecting many of these SV breakpoints as being strongly associated with copy gain (Figure 1A). Many of the genes found to be significant for one SV group were also significant for other SV groups (Figure 2C).
As an additional confirmation of the non-random associations observable between SV breakpoint events and gene expression, we carried out permutation testing. For the entire window of −100 to 100 kb in relation to genes, we constructed a somatic SV breakpoint matrix by annotating for every sample the presence or absence of at least one SV breakpoint within the given region. In each of 1,000 tests, we randomly shuffled the SV event profiles and computed correlations with expression. With the actual dataset, 599 genes were found to be significant (FDR <0.1) after correcting for cancer type and CNA. In contrast, the permutation results yielded an average of 25.4 “significant” genes with an SD of 7.5 (Figure S2B). These results indicate that despite the biological and technical noise involved in each of the two data platforms, the vast majority of the significant genes observed using the actual dataset would not be explainable by noise, chance, or multiple testing.
Key Driver Genes in Cancer Affected by SVs
Genes with altered expression associated with nearby SV breakpoints included many genes previously associated with cancer (Figure 2C). Genes with decreased expression associated with SV breakpoints located within the gene included PTEN (n = 39 cases with SV breakpoint of 1,448 cases with RNA-seq data available), STK11 (n = 7), TP53 (n = 12), RB1 (n = 33), and SMAD4 (n = 4), where genomic rearrangement would presumably have a role in disrupting important tumor suppressors; for other genes, SV breakpoints within the gene could affect intronic regulatory elements or they could represent potential fusion events. Examining the set of 541 genes positively correlated (FDR <0.1, with cancer type and CNA corrections) with occurrence of SV breakpoint upstream or downstream or within the gene (significant for any of the genomic regions in Figures 2A–2C), enriched gene categories by Gene Ontology (GO) analysis (Figure 2D) included G-protein-coupled receptor activity (41 genes), transmembrane receptor activity (57 genes), β-cate-nin-TCF complex assembly (MYC, BCL9, TCF7L1, TERT, HIST1H4I, HIST1H4D, and HIST1H4E), positive regulation of cell size (AKT3, CDK4, SLC26A5, and RET), and phosphatidyli-nositol 3-kinase activity (NRG1, FGF4, KIT, FGF10, FGFR1, ERBB2, FGFR3, PIK3CG, and FGF19). When taken together, SVs involving the above categories of genes could affect a substantial fraction of cancer cases, for example, on the order of 2%–8% of cases across various types (Figure 2E). “High-level” gene amplification events and gene fusion events could be observed for a number of genes and cases associated with SV breakpoints, but other cases showed elevated expression patterns without associated amplification or fusion (Figure 2E).
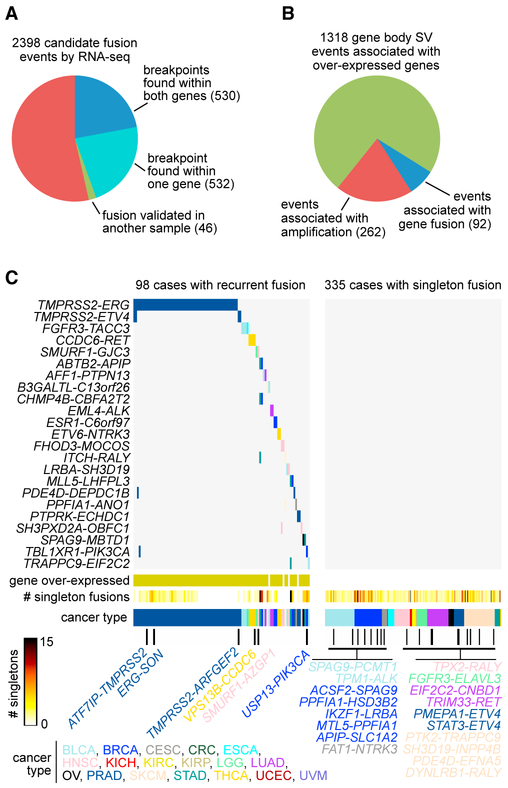
A substantial number of SV breakpoints identified within genes by WGS analysis represented gene fusions by RNA-seq analysis. From the TumorFusions database (Hu et al., 2017; Yoshi-hara et al., 2015), we obtained 20,731 high-confidence RNA fusion events identified in TCGA cases, of which 2,398 involved cases in our cohort of 1,448 with both WGS and RNA-seq data. Of these 2,398 fusion events by RNA-seq, 46% also had support from our WGS results (Figure 3A; Table S5). For the 174 genes with within-gene SV breakpoints associated with increased expression (FDR <0.1, correcting for cancer type and CNA), only a fraction — 7%—of the related 1,318 events involved fusion with the associated gene (Figure 3B). An additional 20% of the 1,318 events involved high-level gene amplification, which would plausibly contribute to overexpression, leaving a substantial number of events whereby other mechanisms of deregulation could conceivably be involved. The vast majority of gene fusions identified were not recurrent; in other words, the gene pairing represented by the fusion was unique to just one case in the analysis (Figure 3C). The most recurrent fusion identified (of 23 total recurrent fusions) was TMPRSS2-ERG fusion in PRAD (n = 58 cases), while other recurrent fusions involved two to four cases, including TMPRSS2-ETV4 (PRAD), FGFR3-TACC3 (BLCA and ESCA), CCDC6-RET (THCA), EML4-ALK (LUAD), ESR1-C6orf97 (BRCA), ETV6-NTRK3 (THCA), and TBL1XR1-PIK3CA (PRAD and BRCA). For a number of “singleton” fusions (i.e., fusions with gene pairing identified in only one case), one of the involved genes would be cancer associated or part of a recurrent fusion, including ALK, NTRK3, FGFR3, RET, ERG, ETV4, PTK2, and PIK3CA (Figure 3B).
Figure 3. Identification of Gene Fusions by Both RNA-Seq and WGS.
(A) Of 2,398 candidate fusion events identified by RNA-seq analysis (Yoshihara et al., 2015), numbers of events with support from WGS analysis are indicated (SV found within both genes, SV found within one gene, or fusion found to have both RNA-seq and WGS support in another sample).
(B) Of the 1,318 gene body SV events associated with overexpressed genes (from Figure 2C and Table S4, 174 genes with FDR <0.1 correcting for cancer type and CNA), the fractions of events associated with either gene fusion by RNA-seq analysis or high-level gene amplification are indicated.
(C) Across 433 cancer cases with at least one gene fusion identified (with both RNA-seq and WGS support), incidences for 20 recurrent fusions (fusions between two specific genes identified in more than one cancer case) are shown. Of the 433 cases, 98 harbored a recurrent fusion and the rest harbored at least one “singleton” fusion (i.e., a fusion between two specific genes being identified in a single case). Named singleton fusions involve at least one gene also involved in a recurrent fusion. For cases with recurrent fusion, the “gene overexpressed” track indicates whether at least one of the two involved genes also showed relatively higher mRNA levels (defined as >0.4 SDs from the median across all sample profiles). Cancer type (denoted by TCGA project) is indicated along the bottom and in the coloring of the recurrent fusion event, as well as in the coloring of the text in cases of highlighted singleton fusions.
See also Table S5.
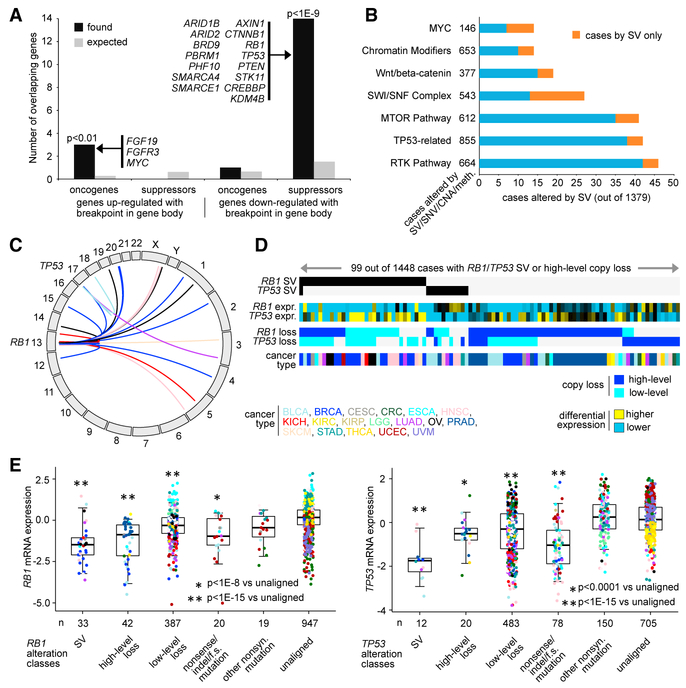
SV breakpoints within genes affected a number of tumor suppressors and associated pathways. Previously, pathway-level alterations—according to somatic mutation, CNA, or epigenetic silencing—were surveyed across TCGA, pathways including p53-related (e.g., TP53, RB1), the mammalian target of rapamy-cin (mTOR), receptor tyrosine kinase (RTK) signaling, chromatin modification, SWI/SNF complex, Wnt/β-catenin, and MYC (Chen et al., 2017, 2018; Zhang et al., 2017). When considering oncogene- or tumor suppressor-associated genes represented by the above pathways, a high overlap was observed between tumor suppressor genes and genes with decreased expression associated with SV breakpoints (Figure 4A, p < 1E–9, one-sided Fisher’s exact test). For each pathway, SV events alone could extend the number of affected cases beyond what would be observed by mutation or CNA or methylation data alone (Figure 4B; Table S6), with RTK, mTOR, and p53-related pathways showing the most SV-altered cases. SVs affecting gene suppressors of the mTOR pathway have been highlighted elsewhere (Zhang et al., 2017). Of 1,493 cases with WGS data, 48 cases (~3%) harbored an SV breakpoint within TP53 or RB1 tumor suppressor genes (39 and 12 cases, respectively; Figure 4C), the genes of which have been found to be altered by rearrangement in individual cancer types such as small-cell lung cancer (George et al., 2015). By SV or high-level copy loss (approximating near total loss) involving TP53 or RB1, 6.8% of cancers with both WGS and RNA-seq data were altered (Figure 4D); a number of cases with SV breakpoint showed only partial copy loss or no loss. In considering TP53 or RB1 expression according to alteration classes defined by SV breakpoint, copy loss, or mutation, cases with SV breakpoint showed the lowest expression for both genes (Figure 4E).
Figure 4. SVs Associated with Key Oncogenic or Tumor-Suppressive Pathways, Including p53 and Rb1.
(A) For the set of genes with differential expression patterns associated with SV breakpoints occurring within the gene (from Figure 2C and Table S4, FDR <0.1 with cancer type and CNA corrections, overexpressed and underexpressed gene sets considered separately), respective overlaps with predefined (Chen et al., 2018) sets of 31 oncogene-associated genes and 72 tumor suppressor-associated genes. p values by one-sided Fisher’s exact test.
(B) For selected predefined pathways, associated non-silent gene mutations (single nucleotide variant [SNV]), CNA events, and DNA methylation silencing were cataloged across 1,379 TCGA cancers as previously described (using cases with available exome sequencing, RNA-seq, and WGS data) (Chen et al., 2017). For each pathway, the number of cases also affected by SV is shown (for tumor suppressor genes, expression <−0.4 SDs from the median and within-gene SV breakpoints associated with underexpression per Figure 3C and Table S4; for oncogenes, SV breakpoint events and genes taken from Figure 2E, for which expression is >0.4 SDs from the median). Cases affected by SV but not by mutation or CNA are highlighted.
(C) Genomic rearrangements (represented in circos plot) involving TP53 or RB1, based on analysis of 1,493 cases with WGS data.
(D) Alterations involving TP53 or RB1 (somatic mutation, copy alteration, SV) found in the set of 1,448 cancer cases having both WGS and RNA-seq data available.
(E) Boxplots of expression for RB1 (left) and for TP53 (right) by their respective alteration classes. Boxplots represent 5%, 25%, 50%, 75%, and 95%. p values by t test on log-transformed values.
See also Table S6.
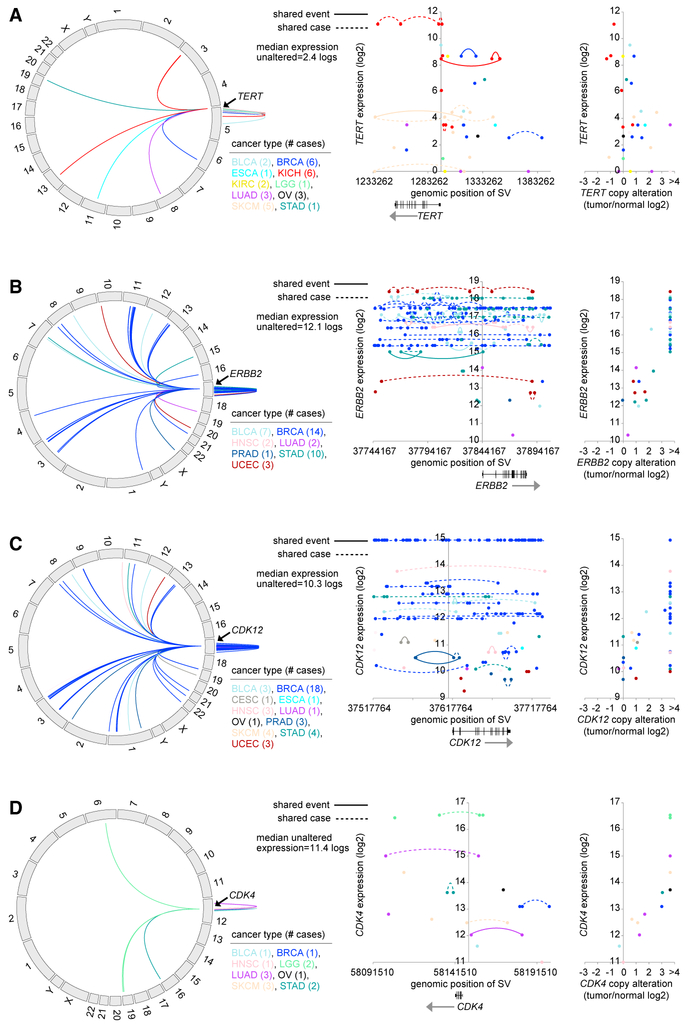
As another approach to identify cancer-relevant genes affected by SVs, we focused on genes in the Sanger Cancer Consensus Gene list (https://www.sanger.ac.uk/science/data/cancer-gene-census), for which SV breakpoints—either within the region 0–20 kb upstream, the region 20–50 kb upstream, or the region 50–100 kb upstream—were associated with increased expression after corrections for both cancer type and CNA (FDR <0.1). Eight genes (TERT, ERBB2, CDK12, CDK4, CLTC, SMARCE1, FGFR1, and TRIM33) met the above criteria, with the first four involving the most number of cases. Genomic rearrangements involving the region 0–100 kb up-stream of TERT (this gene was previously found to be affected by SVs in individual solid cancer types such as kidney [Davis et al., 2014] and neuroblastoma [Peifer et al., 2015]) included 47 SV breakpoints and 29 cancer cases (Figures 5A and S3A; Table S7), with cases showing elevated TERT expression (>0.4 SDs from the median, 17 cases) involving cancer types kidney (n = 7), breast (n = 3 cases), melanoma (n = 2), bladder (n = 2), esophageal or stomach (n = 2), and lung (n = 1). While some cases showed levels of copy number gain for TERT, CNA patterns overall did not account for the extent of deregulated expression observed. In contrast, most cases with overexpressed ERBB2 or CDK12 (36 and 27 cases, respectively, with breakpoint and associated expression >0.4 SDs from the me-dian)–both genes residing on cytoband 17q12–involved gene amplifications and complex genomic rearrangements (Figures 5B, 5C, S3B, and S3C). Although a plurality of cases with SV breakpoints and amplifications for these genes involved breast cancers, other involved cancer types included bladder, head and neck, stomach, and gastric. CDK12 is often encompassed by the ERBB2 amplicon in breast cancer, with phosphoproteo-mic profiling showing CDK12 and HER2 to be activated within the same tumors (Mertins et al., 2016). SV breakpoints associated with overexpression (>0.4 SDs from the median) of CDK4 involved 11 cases (Figures 5D and S3D), including melanoma (n = 3 cases), lung (n = 3), glioma (n = 2), breast (n = 1), stomach (n = 1), and ovarian (n = 1).
Figure 5. SVs Associated with CNA and Increased Expression of TERT, ERBB2, CDK12, and CDK4.
(A) Circos plot showing all intra- and inter-chromosomal rearrangements within TERT or 0–100 kb upstream (left). Gene expression levels of TERT corresponding to SVs located in the genomic region 20 kb downstream to 100 kb up-stream of the gene (47 SV breakpoints involving 29 cases) (middle); dotted lines denote breakpoints within the same sample and solid lines denote common SV event. Gene expression levels of TERT corresponding to CNA (log2 tumor/ normal ratio) (right). The maximum log2 tumor/ normal CNA value is set to 3.6 by the SNP array analysis, approximating >24 copies.
(B) Similar to (A), but for the ERBB2 gene (circos plot, rearrangements within ERBB2 or 0–100 kb upstream; scatterplot, genomic region 20 kb downstream to 100 kb upstream, 243 breakpoints involving 41 cases).
(C) Similar to (A), but for the CDK12 gene (circos plot, rearrangements within CDK12 or 0–100 kb upstream or 0–20 kb downstream; scatterplot, genomic region 20 kb downstream to 100 kb up-stream, 185 breakpoints involving 40 cases).
(D) Similar to (A), but for the CDK4 gene (circos plot, rearrangements within CDK4 or 0–20 kb upstream or 0–50 kb downstream; scatterplot, genomic region 0–50 kb downstream to 50 kb upstream, 22 breakpoints involving 13 cases).
SVs Associated with TAD Disruption and Enhancer Hijacking
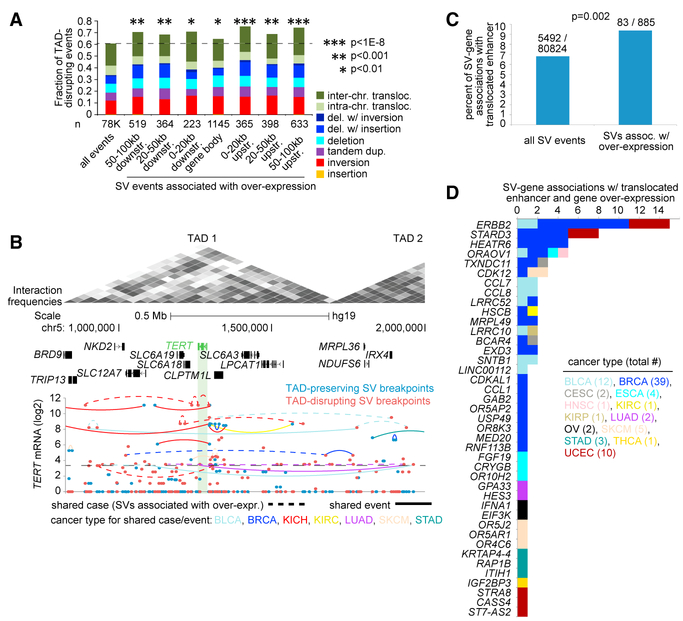
TADs can confine physical and regulatory interactions between enhancers and their target promoters, and disruption of TADs can result in ectopic expression of the associated genes (Dixon et al., 2012; Hnisz et al., 2016; Weischenfeldt et al., 2017). Using published data on TAD coordinates in human cells (Dixon et al., 2012), we categorized all SVs in our pan-cancer dataset by those that were TAD disrupting (i.e., the breakpoints span two different TADs) versus those that were non-disrupting (i.e., both breakpoints fell within the same TAD). Among all 78,496 SVs in the dataset (for cases with RNA-seq data), on the order of 61% were TAD disrupting (this percentage not being considered unusually high, see Experimental Procedures). For SVs with breakpoints located in proximity to a gene and associated with its overexpression (FDR <0.1 for the gene within the given region window, with corrections for cancer type and CNA, and expression >0.4 SDs from the median for the case harboring the breakpoint), an enrichment for TAD-disrupting SVs was observed (Figures 6A and S4; Table S8). In breaking down the SVs associated with overexpression, according to their breakpoint occurrence upstream of the gene (0–20, 20–50, and 50–100 kb) or down-stream of the gene (0–20, 20–50, and 50–100 kb), the percentages of TAD-disrupting SVs ranged from 68% to 75% (with corresponding enrichment p values ranging from 0.001 to <1E—12, one-sided Fisher’s exact test). SVs with breakpoints occurring within the gene body associated with overexpression also showed modest enrichment for TAD-disrupting SVs (65%, p = 0.003, one-sided Fisher’s exact test).
Figure 6. SVs Associated with Disruption of TADs and Translocated Enhancers.
(A) As compared to all SVs (based on cases with both WGS and RNA-seq data), a fraction of the SVs involving TAD disruption (i.e., SVs with breakpoints spanning TAD boundaries), for SVs with breakpoints located in proximity to a gene and associated with its overexpression (FDR <0.1 for the gene within the given region window, with corrections for cancer type and CNA, and expression >0.4 SDs from the median for the case harboring the breakpoint). SVs are broken down according to their breakpoint occurrence within the gene body, upstream of the gene (0–20 kb, 20–50 kb, 50–100 kb), and downstream of the gene (0–20 kb, 20–50 kb, 50–100 kb). p values by one-sided Fisher’s exact test.
(B) Depiction of the TERT locus and associated TADs and SVs. Top: TADs as Hi-C-based contact maps (Dixon et al., 2012), with gray shading indicating locus interactions (darker shading indicates stronger interactions as measured by Hi-C) (adapted from Weischenfeldt et al., 2017). Bottom: gene expression levels of TERT corresponding to SV breakpoints (involving 65 cases and 15 cancer types) located in the genomic region. SV breakpoints are annotated as TAD preserving (i.e., both breakpoints fall within the same TAD) or TAD disrupting; for SV breakpoints involving cases with high TERT expression (defined as expression >0.4 SDs from the median), dotted lines denote breakpoints within the same sample and solid lines denote common SV event. Of all of the genes listed, only TERT was associated with increased expression in proximity to SV breakpoints (Table S4).
(C) For the entire set of SV breakpoint associations occurring 0–100 kb upstream of a gene and with breakpoint mate on the distal side from the gene (for cases with WGS), as well as forthe subset of SV breakpoint associations involving gene overexpression (defined as expression >0.4 SDs from the median for the case harboring the breakpoint and FDR <0.1 for gene overexpression, with corrections for cancer type and CNA), the fraction of SV breakpoint associations involving the translocation of an active in vivo-transcribed enhancer (Andersson et al., 2014) within 0.5 Mb of the gene (where the unaltered gene had no enhancer within 1 Mb). p value by chi-square test.
(D) By gene and by cancer type, the number of SV breakpoint associations involving the translocation of an active in vivo-transcribed enhancer, which involved 41 genes and 83 SV events.
TAD-disrupting SVs include those associated with the TERT locus (Figure 6B), where, for example, for two of the six KICH cases, SVs previously associated with TERT overexpression — having breakpoints within 20 kb upstream of the gene (these SVs being validated by PCR [Davis et al., 2014])—were TAD disrupting. For two additional KICH cases, other TAD-disrupting SVs with breakpoints further upstream or downstream of TERT were observable here (Figure 6B). When evaluating SVs with breakpoints associated with gene underexpression, we observed a trend for SVs, with breakpoints located downstream of the gene showing modest enrichment for TAD-disrupting SVs and a significant enrichment for TAD-disrupting events within SVs having breakpoints associated within a gene body and underexpression (Figure S4A). While TAD disruption would represent one plausible mechanism for deregulated gene expression in many cases, a substantial fraction of SVs involved with deregulation are TAD preserving and could therefore involve other mechanisms.
We went on to examine potential enhancer hijacking events involving SVs, focusing here on a set of active, in vivo- transcribed enhancers as cataloged previously (Andersson et al., 2014). This provides enhancer-specificity information across a range of human cell types and tissues relevant to the respective tissues of origin represented by our pan-cancer cohort. For the entire set of 80,824 SV breakpoint associations occurring 0–100 kb up-stream of a gene and with breakpoint mate on the distal side from the gene, SV breakpoint associations involving the translocation of an active in vivo-transcribed enhancer within 0.5 Mb of the gene (assuming no other disruptions involving the region), where the unaltered gene had no enhancer within 1 Mb, were tabulated. For the subset of 885 SV breakpoint associations involving gene overexpression (FDR <0.1 for the gene, with corrections for cancer type and CNA, and expression >0.4 SDs from the median for the case harboring the breakpoint), there was a statistically significant percentage (9.4%) involving putative enhancer translocation events (Figure 6C; p = 0.002, chi-square test). A number of the genes involved with both enhancer trans-location and overexpression reside on either the 17q12 (e.g., ERBB2, CDK12, STARD3) or the 11q11-q13 (e.g., FGF19) cyto-band regions (Figure 6D).
DISCUSSION
Here, we have reported a comprehensive catalog of somatic rearrangements and their associated transcriptional patterns across >1,400 human cancers. DNA CNAs associated with SVs would show the most influence on gene expression. More than 400 genes, including many key tumor suppressor genes, were directly disrupted by SV breakpoints falling within the gene boundary. A small fraction of SVs associated with gene overexpression represented gene transcript fusions. For on the order of 500 genes—including important cancer driver genes—SV breakpoints in proximity to the gene or within non-coding elements of the gene were associated with overexpression independent of CNA; these events in most cases would likely represent disruption or repositioning of cis-regulatory elements. In considering SVs in addition to point mutations and CNA, our study results would considerably extend upon the types of alter-ations—potentially observable in cancer patients—leading to dysregulation of specific cancer genes and pathways.
While overall trends involving TAD disruption and enhancer hijacking involving genes deregulated by SVs have been identified here, multiple mechanisms of deregulation would likely be involved. For any given gene, there may be no single mechanism affecting all involved cases for us to confidently explain the observed deregulated expression patterns in every case; different cancer cases may have different types of alterations that achieve the same result. Unlike oncogenic point mutations, which typically need to affect specific domains or residues or regulatory motifs (Chang et al., 2016), SVs affecting non-coding regions near or within the gene can involve any one of a number of possible mechanisms, with evolutionary pressures not likely to favor one mechanism over another. In addition, there are limitations in using WGS data to infer mechanisms of SV-mediated deregulation, because SV breakpoint data primarily provides only genomic coordinates and whether the corresponding upstream or downstream sequence at each breakpoint would be involved at the breakpoint junction. Multiple breakpoints may be involved within a given genomic region, not all of which may be identifiable within a WGS profile, making it difficult to trace the precise upstream or downstream sequence near a gene resulting from a given SV breakpoint. Also, in many cases, the orientation of the SV breakpoint may not allow conjecture as to what new sequences are being positioned directly adjacent to the gene. Nevertheless, because our expression analysis approaches to identifying SV-deregulated genes do not assume a particular mechanism, we were able to identify recurrent patterns, regardless of the mechanisms involved.
Future work can further identify and refine the set of cancer-relevant SV-altered gene transcripts, which may involve larger sample numbers and deeper sequencing. As has been the case with pan-cancer somatic mutation analyses (Chang et al., 2016; Gonzalez-Perez et al., 2013; Kandoth et al., 2013; Lawrence et al., 2014; Martincorena et al., 2017), over time and as more data become available, additional WGS studies can build upon our own gene compendium involving SVs. While the low-pass WGS involving many of the cases in our cohort would entail lower sensitivity of SV detection, the larger sample numbers used also provided increased power, which is better able to tolerate false-negative events and other sources of noise in identifying recurrent patterns. To an extent, our analytical approach was also more tolerant of false negatives, whereby in our examining fixed genomic regions near a given gene, multiple SV breakpoints may exist, but only one would need to have been identified by WGS to contribute to associations found. Genes identified as significant tended to show patterns that cut across multiple cancer types, involving cases with low-pass as well as high-pass WGS. The integration involving WGS and RNA-seq in this study was a key step in identifying genes with significance levels (whether by statistical modeling or permutation testing) rising above noise. While we sought to take advantage of data resources offered by TCGA, future studies may use more cases with higher-coverage WGS in which we would expect the overall trends and phenomenon identified in this present study to be substantiated, with potentially even more genes being implicated.
Our study provides a valuable resource for future studies, including studies focusing on a specific gene or cancer type that seek to examine associated SV breakpoints and differential expression patterns. At the same time, limitations regarding low-pass WGS have been noted above, namely decreased sensitivity and specificity compared to deeply sequenced genomes resulting from missing coverage in the tumor and normal samples, respectively, with issues that may arise in conjunction with impurity and genetic heterogeneity in this context. In addition to the present study, the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium is in the process of comprehensively analyzing >2,800 cancer WGS profiles shared between TCGA and the International Cancer Genome Consortium (ICGC) (Campbell et al., 2017). At a future date, the PCAWG datasets are to be released to the research community at large. The number of cancer cases in the PCAWG cohort with both WGS and RNA-seq data is ~1,200, which would be on the order of the 1,448 cases analyzed in the present study. Of these 1,448 TCGA cases in our study, only 369 would be represented in the PCAWG cohort, leaving >1,000 cancer cases with SV data unique to our study. Differences involving the respective platforms and processing of the two studies may not facilitate a direct merging of our data and PCAWG data into a common dataset. However, as PCAWG data on SVs become available, over-all patterns and trends identified in our study can be re-evaluated in the PCAWG cohort and vice versa. Future analyses of SVs in cancer will continue to yield insights into the important role of genomic alterations occurring outside exome boundaries.
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Datasets
The results here are based upon data generated by TCGA Research Network. Whole-genome sequence analysis was carried out for 1,498 cases (with paired normal samples, high-pass coverage for BRCA, KICH, KIRC, KIRP, and OV cases, low-pass for BLCA, CESC, CRC, ESCA, HNSC, LGG, LUAD, PRAD, SKCM, STAD, THCA, UCEC, and UVM). All of the coordinates are based on the hg19 human reference genome.
WGS profiling and SV calling was carried for individual TCGA projects as previously described (Cancer Genome Atlas Network, 2012, 2015a, 2015b; Cancer Genome Atlas Research Network, 2014a, 2014b, 2015a, 2015b, 2017a, 2017b; Cancer Genome Atlas Research Network et al., 2013a; Chen et al., 2016; Davis et al., 2014; Robertson et al., 2017), as well as detailed in the Supplemental Experimental Procedures. Previous studies show that on the order of 96%–98% of high-confidence SVs from high-pass WGS data detected by the Meerkat algorithm are able to be validated by PCR (Davis et al., 2014; Yang et al., 2013). For a subset of 123 cases, both high-pass and low-pass data were available; in comparing Meerkat algorithm calls from both platforms (Table S2), 75% of SV calls by low-pass data were also identifiable by the high-pass data, and 20% of the SV calls by high-pass data were identified using the low-pass data.
Regarding CNA data, low-level gene gain, high-level gene amplification, low-level copy loss, or high-level copy loss were inferred using the “thresh-olded” calls as made by the Broad GDAC Firehose pipeline (http://gdac.broadinstitute.org/). High-level amplifications (approximating >5 copies) denote amplifications above the threshold and larger than the arm level amplifications observed for the given sample. Low-level copy deletions represent deletion above the threshold (approximating heterozygous deletions in the absence of whole-genome doubling); high-level copy deletions denote copy loss above the threshold and greater than the minimum arm-level deletion observed for the sample (approximating homozygous deletions in the absence of whole-genome doubling). Gene-level log base2 (tumor/normal) copy values were used to evaluate CNA as a continuous variable. The maximum log2 tumor/normal CNA value was set to 3.6 by the Firehose SNP array analysis, approximating >24 copies. For analysis of cytoband-level CNA, the gene-level log base 2 (tumor/normal) copy values were collapsed, or averaged, into cyto-band regions.
RNA-seq data were obtained from the Broad Institute Firehose pipeline. Gene fusion transcripts by RNA-seq analysis, as identified using the Pipeline for RNA Sequencing Data Analysis (PRADA) across 9,966 TCGA cancer samples, were obtained from http://tumorfusions.org/(Hu et al., 2017; Yoshihara et al., 2015). For fusion candidates by RNA-seq involving any of the 1,498 cases in our own cohort, SV breakpoints by WGS were examined for any that fell within at least one of the two genes.
Integrative Analyses between SVs and Gene Expression
For each of a number of specified genomic region windows in relation to genes, we constructed a somatic SV breakpoint matrix by annotating for every sample the presence or absence of at least one SV breakpoint within the given region. For the set of SVs associated with a given gene within a specified region in proximity to the gene (0–20 kb upstream, 20–50 kb upstream, 50–100 kb upstream, 0–20 kb downstream, 20–50 kb downstream, 50–100 kb downstream, or within the gene body), correlation between expression of the gene and the presence of an SV breakpoint was assessed using a linear regression model (with log-transformed expression values). (SV breakpoints located within a given gene were not included in the other upstream or downstream breakpoint sets for that same gene.) In addition to modeling expression as a function of SV event, models incorporating cancer type (one of the 18 major types listed in Table 1) as a factor in addition to SV and models incorporating both cancer type and CNA (using log2 tumor/normal values from Firehose) were considered. Using cancer type according to TCGA project as a covariate was carried out to factor in differences involving either WGS coverage or tissue-specific gene expression. For these linear regression models, genes with at least three samples associated with an SV within the given region were considered. Genes for which SVs were significant (FDR <0.1)after correct-ing for both cancer type and CNA were explored in downstream analyses.
Integrative Analyses Using TAD and Enhancer Genomic Coordinates
To identify breakpoints associated with TAD disruption, we used recently published TAD data from the IMR90 cell line (Dixon et al., 2012), where TADs have been found to be largely invariant across cell types (Weischenfeldt et al., 2017). TAD-disrupting SVs were defined as those SVs for which the two breakpoints did not fall within the same TAD. Of all the SVs in the entire dataset, on the order of 61% were found to be TAD disrupting (taking the number of SVs with breakpoints in the same TAD and subtracting this from the total number of SVs, based on cases with gene expression data). The above percentage (61%) was not considered unusually high, given that all interchromosomal SVs are by definition TAD disrupting. In addition, for just the intrachromosomal SVs on chromosome 1,100 simulations were performed, with the SVs coordinates randomly shuffled; on averagethe simulations had a number of TAD-dis-rupting SVs that slightly exceeded the number found using the actual data (average of 2,994 compared to the actual 2,728).
For each SV breakpoint association 0–100 kb upstream of a gene (each association involving unique breakpoint and gene pairing), the potential for the translocation of an active in vivo-transcribed enhancer near the gene that would be represented by the rearrangement was determined (based on the orientation of the SV breakpoint mate). We used the enhancer annotations as provided by Andersson et al. (2014). Their study categorized a set of ~40,000 enhancers according to tissue- or cell-specific expression, with a small subset of enhancers categorized as “ubiquitous” or associated with expression in the majority of tissue and cell types examined. The ubiquitous enhancers were therefore applied to all of the cases in our TCGA cohort. In addition, for each one of the 18 TCGA cancer types in our study, any applicable Andersson group tissue- or cell-specific enhancer subsets for that particular cancer type were also applied (e.g., mammary epithelial cell-specific enhancers for TCGA-BRCA, epithelial cell of prostate and prostate gland for TCGA-PRAD; see Table S8). Only enhancers that were either ubiquitous or with tissue or cell specificity relevant to a given cancer type were applied to the SVs found for cases of that cancer type.
SV breakpoint-to-gene associations involving the translocation of an active in vivo-transcribed enhancer within 0.5 Mb of the gene (assuming no other dis-ruptions involving the region), where the unaltered gene had no enhancer within 1 Mb, were tabulated. Only SVs with breakpoints on the distal side from the gene were considered in this analysis; in other words, for genes on the negative strand, the upstream sequence of the breakpoint (denoted as positive orientation) should be fused relative to the breakpoint coordinates, and for genes on the positive strand, the downstream sequence of the break-point (denoted as negative orientation) should be fused relative to the break-point coordinates.
Statistical Analysis
All p values were two-sided unless otherwise specified. Linear regression models were used to associate the expression of genes with nearby SV breakpoints, as described above. One-sided Fisher’s exact tests or chi-square tests were used to determine the significance of the overlap between two given feature lists. The method of Storey and Tibshirani (2003) was used to estimate FDR for significant genes.
Highlights
Whole-genome analysis of >1,400 cancer cases by high- or low-pass sequencing
Hundreds of genes with overexpression associated with somatic structural variants
Structural variant breakpoints within specific tumor suppressors disrupt expression
No single mechanism involved with structural variant-mediated gene deregulation
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant P30CA125123 (to C.J.C.) and the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP120713 C2 (to C.J.C.).
Footnotes
DATA AND SOFTWARE AVAILABILITY
All molecular data are available through the Genome Data Commons: https://gdc.cancer.gov/.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and eight tables and can be found with this article online at https://doi.org/10.1016Zj.celrep.2018.06.025.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alaei-Mahabadi B, Bhadury J, Karlsson JW, Nilsson JA, and Larsson E (2016). Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc. Natl. Acad. Sci. USA 113, 13768–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. (2011). Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. (2011). The genomic complexity of primary human prostate cancer. Nature 470,214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. (2010). The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Getz G, Stuart J, Korbel J, and Stein L (2017). Pan-cancer analysis of whole genomes. bioRxiv. 10.1101/162784. [DOI]
- Cancer Genome Atlas Network (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015a). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015b). Genomic classification of cutaneous melanoma. Cell 161, 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014a). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014b). Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2015a). Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med 372, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2015b). The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2017a).). Integrated genomic and molecular characterization of cervical cancer. Nature 543, 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2017b). Integrated genomic charac-terization of oesophageal carcinoma. Nature 541, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network; Kandoth C, Schultz N, Cherniack A, Akbani R, Liu Y, Shen H, Robertson A, Pashtan I, Shen R, et al. (2013a). Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network; Weinstein J, Collisson E, Mills G, Shaw K, Ozenberger B, Ellrott K, Shmulevich I, Sander C, and Stuart J (2013b). The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genet. 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, et al. (2016). Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Şenbabaoğlu Y, Ciriello G, Yang L, Reznik E, Shuch B, Micevic G, De Velasco G, Shinbrot E, et al. (2016). Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 14, 2476–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Bossé D, Lalani AA, Hakimi AA, Hsieh JJ, Choueiri TK, Gibbons DL, Ittmann M, and Creighton CJ (2017). Pan-urologic cancer genomic subtypes that transcend tissue of origin. Nat. Commun. 8, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Gibbons DL, Deneen B, Kwiatkowski DJ, Ittmann M, and Creighton CJ (2018). Pan-cancer molecular classes transcending tu-mor lineage across 32 cancer types, multiple data platforms, and over 10,000 cases. Clin. Cancer Res. 24, 2182–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Bu-hay C, Kang H, Kim SC, Fahey CC, et al. ; The Cancer Genome Atlas Research Network (2014). The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, and Heard E (2015). Structural and functional diversity of topolog-ically associating domains. FEBS Lett. 589, 2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Meyerson M, Beroukhim R, and Getz G (2013). Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 23, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al. (2015). Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A, and Lopez-Bigas N (2013). IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BAM, Erpelinck C, van der Velden VHJ, Havermans M, Avellino R, van Lom K, et al. (2014). A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157, 369–381. [DOI] [PubMed] [Google Scholar]
- Harewood L, and Fraser P (2014). The impact of chromosomal rearrangements on regulation of gene expression. Hum. Mol. Genet. 23, R76–R82. [DOI] [PubMed] [Google Scholar]
- Hillmer AM, Yao F, Inaki K, Lee WH, Ariyaratne PN, Teo AS, Woo XY, Zhang Z, Zhao H, Ukil L, et al. (2011). Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 21, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Gold-mann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. ; Cancer Genome Atlas Research Network (2014). Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wang Q, Barthel F, Tang M, Amin S, Yoshihara K, Lang F, Lee S, Zheng S, and Verhaak R (2017). TumorFusions: an integrative resource for reporting cancer-associated transcript fusions in 33 tumor types. bioRxiv. 10.1101/162180. [DOI] [PMC free article] [PubMed]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, and Getz G (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine K, Gerstung M, Dawson K, Haase K, Van Loo P, Davies H, Stratton M, and Campbell P (2017). Universal patterns of selec-tion in cancer and somatic tissues. Cell 171, 1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. ; NCI CPTAC (2016). Proteoge-nomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. (2014). Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krámer A, Roncaioli JL, Sand F, Heuckmann JM, et al. (2015). Telome-rase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. ; TCGA Research Network (2017). Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, and Tibshirani R (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Cerami E, Schalm S, Kim JL, and Lengauer C (2014). The landscape of kinase fusions in cancer. Nat. Commun. 5, 4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, and Korbel JO (2013). Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14, 125–138. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, Waszak SM, Bosco G, Halvorsen AR, Raeder B, et al. (2017). Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, Zhang C, Ren X, Protopopov A, Chin L, et al. (2013). Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lee MS, Lu H, Oh DY, Kim YJ, Park D, Park G, Ren X, Bristow CA, Haseley PS, et al. (2016). Analyzing somatic genome rearrangements in human cancers by using whole-exome sequencing. Am. J. Hum. Genet. 98, 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, and Verhaak R (2015). The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34, 4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang Y, de Velasco G, Jeong K, Akbani R, Hadjipanayis A, et al. (2017). A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.