Abstract
Intervertebral disc (IVD) degeneration is the major cause of back pain. Notch signaling is activated in annulus fibrosus (AF) and nucleus pulposus (NP) tissues of degenerated IVDs, and induced by IL1-β and TNF-α in NP cells. However, the role of Notch activatin in the pathogenesis of IVD degeneration is largely unknown. In this study, we overexpressed the Notch1 intracellular domain (NICD1) in AF, NP, and chondrogenic ATDC5 cells via adenoviruses. Over-expression of NICD1 activated transcription of Notch signaling target genes in AF, NP, and ATDC5 cells, and caused cell type-specific effects on expression of matrix anabolic and catabolic genes. Activation of Notch signaling promoted expression of matrix catabolic genes and inhibited expression of matrix anabolic genes in both AF and ATDC5 cells, whereas its activation suppressed expression of matrix catabolic genes (including Mmp3, Mmp13, Adamts4, and Adamts5) and attenuated TNF-α and inflammatory macrophage-induced Mmp13 expression in NP cells. Consistently, sustained activation of Notch1 signaling in postnatal IVDs in mice severely disrupted growth plate and endplate cartilage tissues, but did not overly affect NP tissues. Together, these data indicated that activation of Notch signaling exerted differential and cell type-specific effects in intervertebral discs, and specific Notch signaling regulation may be considered during the treatment of IVD degeneration.
Keywords: annulus fibrosus, intervertebral disc degeneration, notch signaling, nucleus pulposus
1| INTRODUCTION
Back pain, a common musculoskeletal disease with a lifetime occurrence of about 80%, is the leading contributor of musculoskeletal disabilities (McCann, Tamplin, Rossant, & Seguin, 2012; Oh et al., 2016; Walker, 2000; Wang et al., 2013). Although the exact causes of back pain are still not completely understood, the intervertebral disc (IVD) degeneration is widely accepted as the major one (Risbud & Shapiro, 2014). Currently, no effective disease-modifying therapies are available for back pain, largely due to our limited understanding of the molecular mechanisms underlying disc homeostasis and degeneration. The IVD is a specialized fibrocartilaginous tissue that is composed of the nucleus pulposus (NP), the annulus fibrosus (AF), the cartilaginous endplate (CEP), and the growth plate cartilage (GP) (Oh et al., 2016). The centrally located NP is a hydrated gelatinous structure that is rich in proteoglycans and functions as a cushion against compressive load for the discs. While the fibrous AF surrounds NP laterally and provides tensile strength, GP and CEP cover NP superiorly and connect IVD to the two adjacent vertebral bodies. Degeneration of IVD is caused by an imbalance between anabolic and catabolic metabolism of IVD cells, which results in increased extracellular matrix degradation and/or decreased extracellular matrix synthesis (Kim et al., 2012, 2013; McCann et al., 2012). Therefore elucidating the mechanism controlling matrix catabolic and anabolic processes of the IVD cells is critical for developing potential therapies for IVD degeneration and back pain.
Notch signaling is an evolutionarily conserved pathway that plays important roles in development and homeostasis of multiple tissues (Hori, Sen, & Artavanis-Tsakonas, 2013), whereas its dysregulation is implicated in a wide variety of diseases. Notch signaling is primarily triggered by binding of a Notch receptor (Notch1–4 in mammals) by its ligand (including Dll1, Dll3, Dll4, Jag1, and Jag2 in mammals), both of which are expressed on the surfaces of two opposing cells. The Notch receptor–ligand engagement activates two consecutive enzymatic cleavages of Notch receptor, which release its intracellular domain (NICD) into the cytoplasm. The NICD subsequently translocates to the nucleus, where it associates with RBPjk and mastermind-like (MAML) to form a transcriptional complex to activate the transcription of the downstream targets, including genes in the Hes/Hey family. Prior studies have revealed that Notch signaling plays a dual role in joint maintenance and osteoarthritis in part through modulating anabolic and catabolic molecules in the cartilage matrix (Liu et al., 2015, 2016; Mead & Yutzey, 2009; Mirando et al., 2013). Physiological Notch signaling is required for the maintenance of articular cartilage, whereas sustained activation of Notch signaling leads to cartilage degeneration and osteoarthritis.
Similar to articular cartilage, expression of Notch signaling molecules and downstream target genes could be detected in both AF and NP cells of IVD tissues (Hiyama et al., 2011). Moreover, pharmacological inhibition of Notch signaling by γ-secretase inhibitor significantly decreased proliferation of both AF and NP cells in vitro (Hiyama et al., 2011). In line with the hypoxic environment where inner AF and NP cells reside in vivo, hypoxia activated the Notch signaling activity in these cells (Hiyama et al., 2011). These data suggest that Notch signaling may play an important role in IVD homeostasis under physiological conditions. It has been shown that the activity of Notch signaling was increased in AF and NP cells in degenerated disc tissues compared to those in normal discs in human (Hiyama et al., 2011; Wang et al., 2013). Moreover, the Notch signaling was induced by IL1-β and TNF-α, two key inflammatory cytokines involved in pathogenesis of IVD degeneration, in NP cells (Millward-Sadler, Costello, Freemont, & Hoyland, 2009; Wang et al., 2013). One potential explanation for these observations is that Notch signaling activation may be involved in IVD degeneration and inhibition of Notch signaling may decelerate IVD degeneration. Another possibility is that Notch activation could represent a compensatory response by resident cells attempting to repair the degenerated discs. To determine the role of Notch activation in IVD degeneration, we overexpressed the Notch1 intracellular domain (NICD1) in AF, NP, and ATDC5 cells via adenoviral infection, and examined the effects of Notch signaling activation on expression of matrix catabolic and anabolic genes in these cells. We also generated a mouse model with sustained activation of Notch signaling in postnatal IVD tissues. Using these in vitro and in vivo approaches, our studies revealed cell type-specific effects of Notch signaling activation on matrix anabolic and catabolic gene expression in the intervertebral discs, and suggested that modulation of Notch signaling may serve as a potential therapeutic approach for the treatment of IVD degeneration.
2 | MATERIALS AND METHODS
2.1 | Cell culture, adenoviral infection, and treatments
Rat nucleus pulposus (rNP) and annulus fibrosus (rAF) cell lines were generated by Prof. Di Chen (Rush University) (Oh et al., 2016), and maintained in DMEM/F-12(1:1) containing 10% FBS and 1% penicillin/ streptomycin. Murine ATDC5 cells were grown in DMEM/F-12(1:1) supplemented with 5% FBS and 1% penicillin/streptomycin. Murine RAW264.7 cells (a macrophage cell line) were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37°C in a humidified 5% CO2 incubator, and the culture medium was refreshed every 2–3 days.
To prepare conditioned medium of inflammatory macrophages, RAW264.7 cells were seeded in 10 cm culture dishes at a density of 4× 106/dish. After overnight culture, the cells were cultured in 10 ml fresh growth medium supplemented with 100 ng/ml LPS. A 1 day after LPS treatment, LPS-containing medium was removed, and cultured in normal growth medium for 24 hr, culture medium was then collected and centrifuged briefly to remove cell debris. The remaining supernatant was designated as 100% conditioned medium of inflammatory macrophages (M3CM).
For adenoviral infections, cells were seeded in 6-well plates at a density of 2 × 105 cells per well. After overnight culture, the cells were infected with adenoviruses expressing GFP (Ad-GFP) or Notch1 intracellular domain (NICD1) (Ad-NICD1) with a multiplicity of infection (MOI) of 100 in the presence of 8 μg/ml polybrene. At 72 hr after adenoviral infection, cells were harvested for qPCR or Western blot experiments.
For TNF-α or M3CM treatments, rNP cells were first infected with Ad-GFP or Ad-NICD1 for 24 hr, and then switched to culture medium with or without either 20 ng/ml TNF-α or 30% M3CM. At 48 hr after treatments, cells were harvested for RNA or protein analyses
2.2 | Quantitative RT-PCR (qPCR)
qPCR analyses were performed as we previously reported (Gu et al., 2018). Briefly, total RNA was recovered from cells by isopropanol precipitation following extraction by the Trizol reagent (Invitrogen, Carlsbad, CA). Subsequently, cDNA was reverse transcribed from 1 μg total RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), and then subjected to qPCR amplification using universal SYBR green supermix (Bio-Rad). Relative gene expression was calculated by 2−ΔΔCt method after normalizing first to house-keeping genes (Gapdh, Actb, or 18S rRNA as indicated), and then to control groups. The DNA sequences for primers used in this study will be provided upon request.
2.3 | Western blot
Western blot analyses were performed as we previously described (Gu et al., 2018). Briefly, total protein extracts were prepared from cells using 1 × RIPA buffer supplemented with protease and phosphatase inhibitors (Roche, Mannheim, Germany). Protein concentration was determined with the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL) following its instruction. Equal amounts of protein samples were resolved on 10% SDS-polyacrylamide gels and blotted onto PVDF membranes (Perkin Elmer). After being blocked with 5% Blotting-Grade Blocker Nonfat Dry Milk (Bio-Rad) in TBST, the blots were incubated overnight with rabbit primary antibodies against Mmp13 (Abcam, Cambridge, MA, 1:3000) or β-actin (Cell Signaling, Danvers, MA, 1:1000) at 4°C, followed by 1 hr incubation with HRP-conjugated goat anti-rabbit secondary antibody (Cell Signaling, 1:2500). Subsequently, the blots were developed using the Clarity Western ECL Substrate (Bio-Rad), and imaged with the ChemiDoc Touch Imaging System (Bio-Rad).
2.4 | Mouse strains
Mouse strains used in this study, including Col2a1Cre, tetO-NICD1, Rosa-rtTAf/+, and R26-mT/mG mice, have been previously described (Belteki et al., 2005; Long, Zhang, Karp, Yang, & McMahon, 2001; Murtaugh, Stanger, Kwan, & Melton, 2003; Muzumdar, Tasic, Miyamichi, Li, & Luo, 2007; Stanger, Datar, Murtaugh, & Melton, 2005). For induction of NICD1 expression in postnatal intervertebral discs, 100 μg/g doxycycline(Dox) was injected intraperitoneally to 1 month-old Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+ mice and their littermate controls three times per week for four consecutive weeks as previously described (Liu et al., 2015), mice were sacrificed for analyses after 4 weeks of Dox administration. Animal studies were approved by the Animal Studies Committee at Soochow University.
2.5 | H&E staining and Alcian blue staining
For histology-based analyses, IVD samples were isolated, fixed in 10% formalin, and then decalcified in Formic acid Bone Decalcifier (Immunocal, Decal Chemical Corp., Tallman, NY) for 10 days. The decalcified IVD samples were processed for paraffin embedding before being sectioned at 6 μm thicknesses. One set of paraffin sections were then stained with hematoxylin and eosin (H&E) to assess the general morphology of IVDs as described previously (Jiang, Fu, Yang, Long, & Chen, 2017). In parallel experiments, the adjacent sections were incubated with Alcian blue solution (10% Alcian blue 8GX in 3% acetic acid, pH 2.5) to evaluate the proteoglycan level in IVDs.
2.6 | Statistical analysis
All quantitative results were expressed as mean ± standard deviation (S.D.) calculated from triplicates. Statistical difference between two groups was determined by Student's t-test. Any difference with a p-value less than 0.05 were considered statistically significant.
3 | RESULTS
3.1 | Activation of Notch signaling inhibited expression of matrix anabolic genes in AF cells
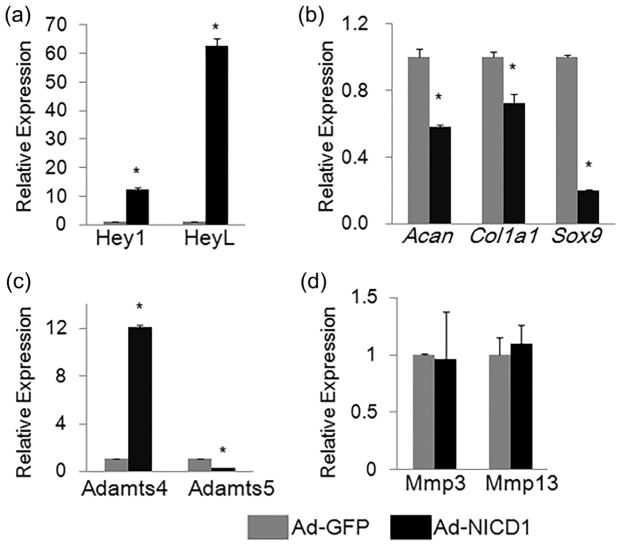
To determine the effects of Notch activation on AF cells, we utilized a rat AF cell line (rAF cells, hereafter) (Oh et al., 2016), and infected these cells with adenoviruses expressing Notch1 intracellular domain (NICD1) (Ad-NICD1) or adenoviruses expressing GFP (Ad-GFP) Total RNA was isolated from rAF cells after 3 days of adenoviral infection. Quantitative PCR (qPCR) analysis showed that the expression of Notch signaling target genes Hey1 and HeyL was increased (by 11- and 62-folds, respectively) in Ad-NICD1-infected cells (Figure 1a), confirming that overexpression of NICD1 activated Notch signaling in rAF cells.
FIGURE 1.
Effects of Notch signaling activation on expression of matrix catabolic and anabolic genes in rAF cells. qPCR analyses of Notch signaling target genes (a), matrix anabolic genes (b), and matrix catabolic genes (c,d) in rAF cells infected with adenoviruses expressing Notch1 intracellular domain (NICD1) (Ad-NICD1) or adenoviruses expressing GFP (Ad-GFP) for 3 days. Relative gene expression was calculated by 2−ΔΔCt method after normalizing first to house-keeping Gapdh, and then to the control groups (Ad-GFP-infected cells). Data are means ± SD. *p <0.05, n = 3
Outer and inner AF cells express high levels of collagen I (encoded by Col1a1) and aggrecan (encoded by Acan), respectively (Bron, Helder, Meisel, Van Royen, & Smit, 2009; Gruber, Leslie, Ingram, Norton, & Hanley, 2004; Oh et al., 2016; Singh, Masuda, Thonar, An, & Cs-Szabo, 2009). To investigate the effects of Notch signaling activation on matrix anabolism of rAF cells, we performed qPCR analyses to examine the mRNA levels of these two genes encoding extracellular matrix proteins, and found that expression of Acan and Col1a1 were significantly decreased (by 42% and 27%, respectively) in Ad-NICD1-infected cells compared to control cells infected with Ad-GFP (Figure 1b). In addition, we also analyzed expression of Sox9, a transcriptional factor known to regulate aggrecan expression. Similarly, the mRNA level of Sox9 was reduced by 80% in Ad-NICD1-infected rAF cells.
MMPs (matrix metalloproteinases) and ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) are key matrix-degrading enzymes that are involved in both osteoarthritis and intervertebral disc degeneration (Chen et al., 2017; Heinegard & Saxne, 2011; Risbud & Shapiro, 2014; Wang et al., 2011, 2012) Therefore, we examined the effects of Notch activation on the expression of Mmps (Mmp3 and Mmp13) and Adamts (Adamts4 and Adamts5). Results of qPCR showed that activation of NICD1 up-regulated the Adamts4 expression (11-folds increase), but not Adamts5 in rAF cells (Figure 1c). In contrast, NICD1 overexpression has no significant effect on mRNA levels of Mmp3 and Mmp13 (Figure 1d). Our data demonstrated that activation of Notch signaling in rAF cells inhibited expression of matrix anabolic genes.
3.2 | Activation of Notch signaling promoted expression of matrix catabolic genes in ATDC5 cells
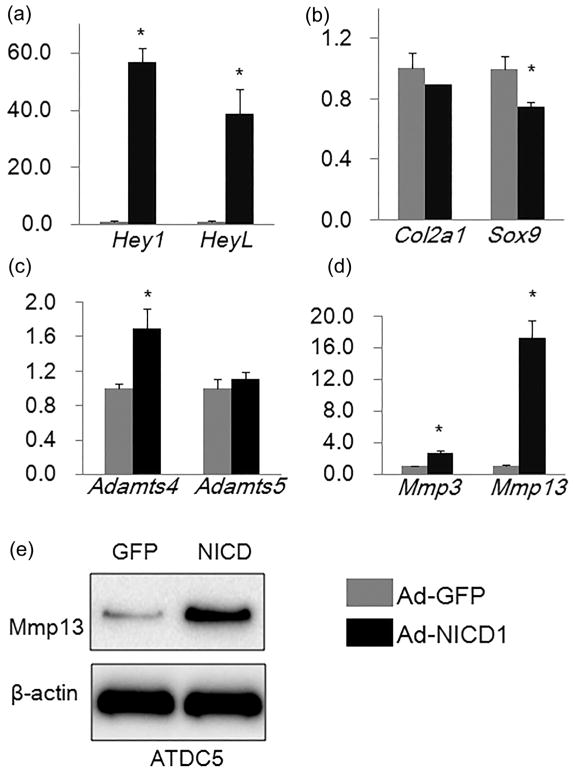
Since the endplate and growth plate are important cartilaginous components of IVDs, we next determine the effects of Notch activation on chondrogenic cells. To this end, we utilized the chondrogenic ATDC5 cell line, and infected these cells with adenoviruses expressing Notch1 intracellular domain (NICD1) (Ad-NICD1) or adenoviruses expressing GFP (Ad-GFP). Total RNA was isolated from ATDC5 cells 3 days after adenoviral infection. Results of qPCR analysis showed that the expression of Notch signaling target genes Hey1 and HeyL was significantly increased (by 57- and 39-folds, respectively) after 3 days of Ad-NICD1 infection (Figure 2a), indicating that overexpression of NICD1 activated Notch signaling in ATDC5 cells.
FIGURE 2.
Effects of Notch signaling activation on expression of matrix catabolic and anabolic genes in chondrogenic ATDC5 cells. (a–d) qPCR analyses of Notch signaling target genes (a), matrix anabolic genes (b), and matrix catabolic genes (c,d) in ATDC5 cells infected with Ad-GFP or Ad-NICD1 for 3 days. Relative gene expression was calculated by 2−ΔΔCt method after normalizing first to house-keeping Gapdh, and then to the control groups (Ad-GFP-infected cells). Data are means ± SD. *p <0.05, n = 3. (e) Western blot analysis of Mmp13 in ATDC5 cells infected with Ad-GFP or Ad-NICD1 for 3 days
To investigate the effects of Notch signaling activation on matrix anabolic genes in ATDC5 cells, we performed qPCR analyses to examine the mRNA levels of Col2a1 and Sox9. As shown in Figure 2b, the expression of Sox9 was mildly, but significantly decreased in Ad-NICD1 infected cells compared to Ad-GFP-infected cells. Similarly, the expression level of Col2a1 was lower in Ad-NICD1-infected cells (Figure 2b), but the difference did not reach the statistical significance (p = 0.14, n = 3).
To determine the effects of Notch activation on expression of matrix catabolic genes in ATDC5 cells, we examined the expression of Adamts4, Adamts5, Mmp3, and Mmp13, genes encoding for the key matrix-degrading enzymes that are implicated in cartilage degradation (Liu et al., 2015). Results of qPCR analysis showed that the mRNA levels of Adamts4, Mmp3, and Mmp13 were significantly induced by Notch signaling activation in ATDC5 cells (Figures 2c and d). In particular, the mRNA level of Mmp13 was increased by 16-folds (Figure 2d). Consistent with the qPCR results, Western blot analysis showed that protein level of Mmp13 was remarkably increased in Ad-NICD1-infected cells compared to Ad-GFP-infected cells (Figure 2e). In contrast, overexpression of NICD1 did not significantly affect mRNA level of Adamts5 in ATDC5 cells (Figure 2c). These data indicated that the major effect of activation of Notch signaling in chondrogenic ATDC5 cells is to promote matrix catabolism.
3.3 | Activation of Notch signaling suppressed expression of key matrix-degrading enzymes in NP cells
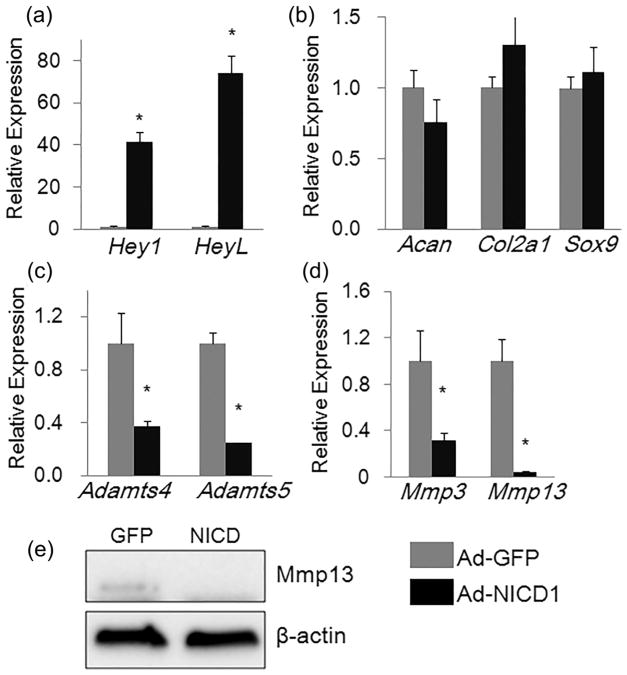
To determine the effects of Notch signaling activation on NP cells, we utilized a rat NP line (rNP cells hereafter)(Oh et al.,2016),and infected these cells with adenoviruses expressing Notch1 intracellular domain (NICD1) (Ad-NICD1) or adenoviruses expressing GFP (Ad-GFP). Total RNA was isolated from rNP cells 3 days after adenoviral infection. Results of qPCR analysis showed that expression of the Notch signaling target genes, such as Hey1 and HeyL, was significantly increased (by 40- and 73- folds, respectively)inAd-NICD1-infected rNP cells, indicating that overexpression of NICD1 robustly activated Notch signaling in these cells (Figure 3a).
FIGURE 3.
Activation of Notch signaling suppressed expression of genes encoding key matrix-degrading enzymes in rNP cells. (a–d) qPCR analyses of Notch signaling target genes (a), matrix anabolic genes (b), and matrix catabolic genes (c,d) in rNP cells infected with Ad-GFP or Ad-NICD1 for 3 days. Relative gene expression was calculated by 2−ΔΔCt method after normalizing first to house-keeping Gapdh, and then to the control groups (Ad-GFP-infected cells). Data are means ± SD. *p <0.05, n = 3. (e) Western blot analysis of Mmp13 in rNP cells infected with Ad-GFP or Ad-NICD1 for 3 days
NP cells synthesize and produce high levels of extracellular matrix collagen II and aggrecan (Millward-Sadler et al., 2009; Oh et al., 2016; Sive et al., 2002). To examine the effects of Notch activation on matrix anabolism in rNP cells, we performed qPCR to analyze the expression of Acan and Col2a1 as well as Sox9, the transcriptional regulator of these genes. As shown in Figure 3b, mRNA levels of Sox9, Acan, and Col2a1 genes were not significantly changed after 3 days of NICD1 over-expression, indicating that Notch activation did not have a major effect on expression of extracellular matrix-related anabolic genes in rNP cells.
To determine the effects of Notch activation on expression of matrix catabolic genes, we examined the expression of matrix-degrading Adamts (Adamts4, Adamts5) and MMPs (MMP3 and MMP13). Interestingly, mRNA levels of all above catabolic enzymes were significantly suppressed by Notch signaling activation in rNP cells (Figures 3c and d). In particular, the mRNA level of Mmp13 was reduced by more than 90% (Figure 3d). Consistent with the qPCR results, Western blot analysis showed that the basal level of Mmp13 protein was completely blocked by Notch activation in rNP cells (Figure 3e).
Taken together, these data indicate that Notch signaling activation in rNP cells suppressed expression of key matrix-degrading enzymes (Adamts4, Adamts5, Mmp3, and Mmp13) without obvious effect on expression of matrix anabolic genes.
3.4 | Activation of Notch signaling in NP cells prevented TNF-α- and inflammatory macrophage-induced Mmp13 expression
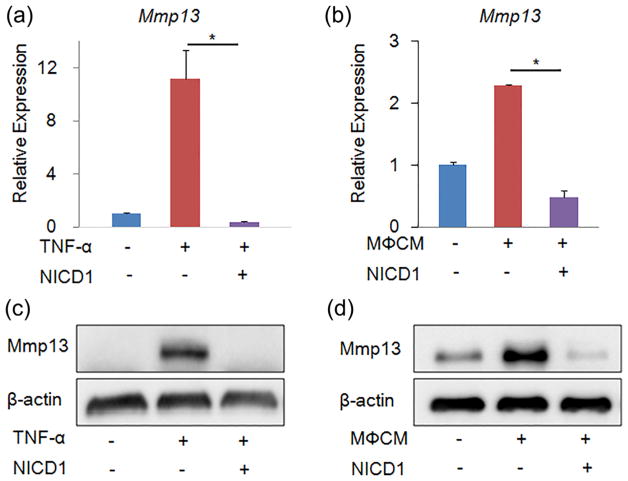
After confirming that activation of Notch signaling suppressed expression of matrix catabolic genes (particularly Mmp13) in rNP cells, we further investigated whether overexpression of NICD1 could block or attenuate inflammatory cytokines- or inflammatory cells-induced Mmp13 expression in NP cells. Since TNF-α is one of the major cytokines that are implicated in pathogenesis of IVD degeneration (Risbud & Shapiro, 2014), we first tested whether NICD1 overexpression protected TNF-α-induced Mmp13 expression in rNP cells. To this end, we infected rNP cells with Ad-NICD1 or Ad-GFP for 1 day, and then treated these cells with or without 20 ng/ml TNF-α for additional 2 days. Consistent with the literature (Millward-Sadler et al., 2009), results of qPCR analysis revealed that TNF-α induced Mmp13 expression by more than 10-folds in rNP cells (Figure 4a). Interestingly, overexpressing NICD1 in rNP cells prevented TNF-α-induced mRNA expression of Mmp13 gene (Figure 4a). Consistent with the qPCR results, results of Western blot analysis showed that overexpressing NICD1 completely abolished TNF-α-induced Mmp13 upregulation (Figure 4b). Then, we tested whether NICD1 overexpression prevented inflammatory macrophage-induced Mmp13 expression in rNP cells. We infected rNP cells with Ad-NICD1 or Ad-GFP for 1 day, and then treated these cells with 30% conditional medium collected from inflammatory macrophages (M3CM). Results of qPCR analysis revealed that M3CM significantly induced mRNA levels of Mmp13 in rNP cells, which was blocked by NICD1 overexpression (Figure 4c). Consistently, results of Western blot analysis showed that induction of Mmp13 protein by M3CM was completely inhibited by NICD1 overexpression (Figure 4d).
FIGURE 4.
Activation of Notch signaling prevented TNF-α- and inflammatory macrophage-induced mRNA and protein expression of Mmp13 in nucleus pulposus cells. (a–b) qPCR analyses of catabolic Mmp13 in rat nucleus pulposus cells that were infected with Ad-GFP or Ad-NICD1 for 1 day, and then treated with 20 ng/ml TNF-α (a) or 30% conditional medium collected from inflammatory macrophages (M3CM) (b) for 2 days. Relative gene expression was calculated by 2−ΔΔCt method after normalizing first to house-keeping Gapdh, and then to the control groups (Ad-GFP infected cells). Data are means ± SD. *p < 0.05, n = 3. (a–b) Western blot analyses of Mmp13 in rat nucleus pulposus cells. rNP cells were treated as described above
Taken together, these data suggest that activation of Notch signaling may be beneficial to alleviate inflammatory cytokines-mediated pro-catabolic effects on NP cells.
3.5 | Sustained Notch signaling in postnatal IVD severely disrupted growth plate and endplate cartilage
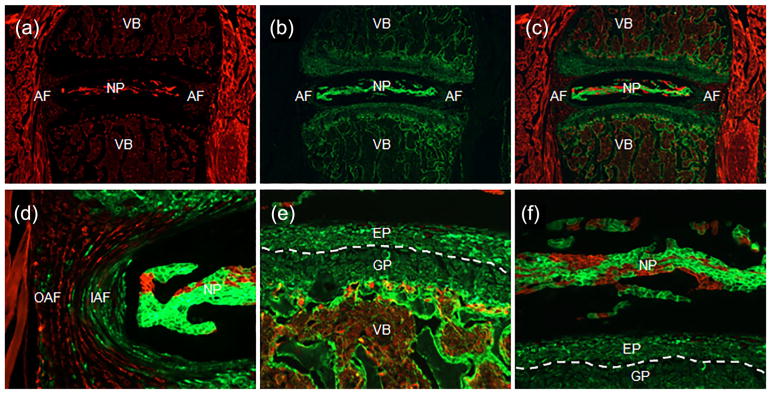
The above in vitro experiments have showed that Notch signaling activation could cause detrimental or beneficial effects on intervertebral discs depending on cell types. To further investigate the effects of Notch activation on intervertebral discs in vivo, we generated a tissue-specific and temporally controlled Notch1 gain-of-function (NICD1 GOF) mouse model using the Cre/LoxP technology coupled with tetracycline-on (Tet-On) system: Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+. In this system, reverse tetracycline trans-activator (rtTA) is expressed only in Col2a1-expressing cells and their progenies, where rtTA binds to the tetO promoter and subsequently activates overexpression of NICD1 only in the presence of the tetracycline or its analog doxycycline (Dox). Since Col2a1Cre determines the tissue-specificity, we first characterized the cell types targeted by Col2a1Cre in postnatal IVD using the R26-mT/mG reporter mice. The R26-mT/mG mice ubiquitously express mTomato (a membrane-localized red fluorescent protein) prior to Cre-mediated recombination, and turn on expression of a membrane-targeted green fluorescent protein (mGFP) in cells with Cre-mediated recombination. Therefore, cells with GFP expression represent cells targeted by Col2a1Cre. Using this approach, we found that, Col2a1Cre targeted inner AF, most of NP, and the entire growth plate and endplate in intervertebral disc tissues (Figure 5).
FIGURE 5.
Col2a1Cre targeted the majority cells in the postnatal intervertebral discs. (a–c) Fluorescence images of mGFP (a), mTomato (b), and mGFP/mTomato (c) on intervertebral disc sections from 2 month-old Col2a1Cre; R26-mT/mG mice. (d–f) Higher magnification mGFP/ mTomato images for AF (d), endplate/growth plate (e), and NP (f). NP, nucleus pulposus; IAF, inner annulus fibrosus; OAF, outer annulus fibrosus; EP, endplate; GP, growth plate; VB, verterbral body
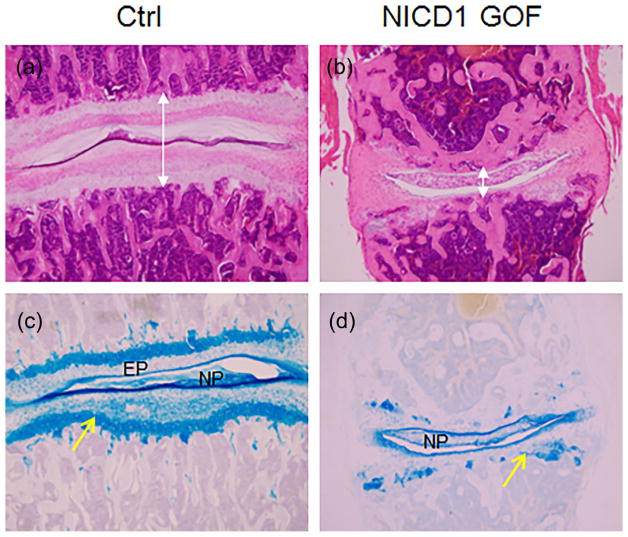
We then set to induce NICD1 overexpression in postnatal IVDs. In previous studies, we have determined proper Dox concentrations and found that 100 μg/g Dox (3 times/week, 4 weeks) could reach sustained Notch activation in cartilages (Liu et al., 2015), In the present study, we used the same approach to induce Notch activation in IVDs of 1-month-old NICD1 GOF (Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+) mice. After 12 times of Dox injection, we harvested disc tissues from NICD1 GOF mice and their littermate controls, and performed histological analysis. As shown in Figure 6, the total height of IVD was significantly decreased in NICD1 GOF mice. More strikingly, results of H&E and Alcian blue staining showed that the growth plate and the endplate cartilage tissues were severely disorganized with a significant loss of proteoglycan in NICD1 GOF mice. In contrast, the proteoglycan content in NP cells was comparable between NICD1 GOF and control mice.
FIGURE 6.
Sustained Notch signaling in postnatal IVD severely disrupted growth plate and endplate cartilage. (a–b) H&E staining of intervertebral disc sections from 1-month-old wild-type (Ctrl) (a) or NICD1 GOF (Col2a1Cre; tetO-NICD1; Rosa-rtTAf/+) mice (b) 4 weeks after they were injected with 100 μg/g Dox (three times per week). (c–d) Alcian blue staining of intervertebral disc sections from 1-month-old Ctrl (c) or NICD1 GOF mice (d) 4 weeks after they were injected with 100 μg/g Dox (three times per week). Yellow arrows point to the growth plate of IVD; EP, the end plate of IVD. Lines with double arrow indicate the height of IVDs
4 | DISCUSSION
Prior studies have demonstrated that Notch signaling was increased in AF and NP tissues of degenerated IVDs, and induced by IL1-β and TNF-α in NP cells (Millward-Sadler et al., 2009; Wang et al., 2013). However, the effects of Notch activation on disc cells are still unclear. In this study, we overexpressed the Notch1 intracellular domain (NICD1) in AF, NP, and ATDC5 cells via adenoviral infection, and examined the effects of Notch signaling activation on catabolic and anabolic gene expression in these cells. We also generated a mouse model with sustained activation of Notch signaling in postnatal IVD. Our studies revealed that activation of Notch signaling in the intervertebral discs could lead to opposite effects on IVD homeostasis depending on disc cell types in which Notch signaling is activated. The present study, for the first time to our knowledge, established that Notch activation in either AF or chondrogenic cells in IVDs inhibited the expression of matrix anabolic genes but stimulated the expression of matrix catabolic genes (MMPs and ADAMTSs); whereas Notch activation in NP cells resulted in decreased expression of matrix-degrading proteases (Mmp3, Mmp13, Adamts4, Adamts5) and attenuated TNF-α- and inflammatory macrophage-induced Mmp13 expression. Our findings suggested that Notch signaling could be a potential therapeutic target for treating IVD degeneration, but whether stimulating or inhibiting Notch signaling should be strictly based on cell-types targeted.
The role of Notch signaling in disc homeostasis and degeneration is likely complicated. Besides the pathological role of hyperactive Notch signaling in disc homeostasis and degeneration, the proper level of endogenous Notch signaling appears to be required for IVD maintenance and homeostasis. Indeed, Notch signaling components and target genes were expressed in both AF and NP tissues of IVD tissues (Hiyama et al., 2011). Consistent with the hypoxic environment where inner AF and NP cells reside in vivo, hypoxia activated the Notch signaling activity in these cells (Hiyama et al., 2011). Moreover, suppression of Notch signaling significantly decreased proliferation of both AF and NP cells in vitro. However, it is still unclear whether Notch signaling plays a similar physiological role in vivo. Furthermore, it is important to determine whether the physiological role of Notch signaling in discs is cell type-specific. Future studies are warranted in these directions.
The mechanism underlying differential effects of Notch activation on different cell types in IVDs is still unknown. We and others showed that Notch activation increased IL-6 and other inflammatory cytokines in chondrogenic ATDC5 and primary chondrocytes (Liu et al., 2015; Zanotti & Canalis, 2013), and that neutralization of IL-6 alleviated induction of Mmp13 by Notch activation in these cells (Liu et al., 2015; Zanotti & Canalis, 2013). Interestingly, our RNA-seq analyses indicated that IL-6 and several other inflammatory cytokines were down-regulated in rNP cells infected with Ad-NICD1 (data not shown). Moreover, this study showed that Notch activation in rNP cells alleviated TNF-α- and inflammatory macrophage-induced expression of Mmp13. Therefore, it is possible that Notch activation exerted differential effects on expression of catabolic genes in different types of disc cells through modulating expression of IL-6 and other inflammatory cytokines. Another possibility is that Notch/RBPjk binds directly to the promoter/enhancer of catabolic genes (such as Mmp13) and activates their transcription in ATDC5 and rAF cells, but represses the expression of catabolic genes indirectly through its downstream effectors (i.e., Hes/Hey transcriptional repressors) in rNP cells. Future studies will be directed at elucidating the precise mechanism(s) underlying the cell type-specific effects of Notch signaling activation on intervertebral discs, which will contribute to identifying the potential targets for developing therapies to treat IVD degeneration and back pain.
Acknowledgments
Funding information
The National Key R&D Program of China, Grant number: 2016YFC1100203; the Priority Academic Program Development of Jiangsu High Education Institutions (PAPD); Natural Science Foundation of Jiangsu Province, Grant number: BK20140289; National Natural Science Foundation of China, Grant numbers: 81401768, 81772294; NIH/NIAMS, Grant number: AR063071
This work was funded by the National Key R&D Program of China (2016YFC1100203) (JC), the National Natural Science Foundation of China (81772294 to JC, 81401768 to JL), the Priority Academic Program Development of Jiangsu High Education Institutions (PAPD) (JC), the Natural Science Foundation of Jiangsu Province (BK20140289) (JL), and NIH/NIAMS grant AR063071 (MJH).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflict of interest to declare.
References
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, … Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Research. 2005;33(5):e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron JL, Helder MN, Meisel HJ, Van Royen BJ, Smit TH. Repair, regenerative and supportive therapies of the annulus fibrosus: Achievements and challenges. European spine journal: Official Publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2009;18(3):301–313. doi: 10.1007/s00586-008-0856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shen J, Zhao W, Wang T, Han L, Hamilton JL, Im HJ. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Research. 2017;5:16044. doi: 10.1038/boneres.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber HE, Leslie K, Ingram J, Norton HJ, Hanley EN. Cell-based tissue engineering for the intervertebral disc: In vitro studies of human disc cell gene expression and matrix production within selected cell carriers. The Spine Journal: Official Journal of the North American Spine Society. 2004;4(1):44–55. doi: 10.1016/s1529-9430(03)00425-x. [DOI] [PubMed] [Google Scholar]
- Gu X, Fu X, Lu J, Saijilafu, Li B, Luo ZP, Chen J. Pharmacological inhibition of S6K1 impairs self-renewal and osteogenic differentiation of bone marrow stromal cells. Journal of Cellular Biochemistry. 2018;119(1):1041–1049. doi: 10.1002/jcb.26272. [DOI] [PubMed] [Google Scholar]
- Heinegard D, Saxne T. The role of the cartilage matrix in osteoarthritis. Nature Reviews Rheumatology. 2011;7(1):50–56. doi: 10.1038/nrrheum.2010.198. [DOI] [PubMed] [Google Scholar]
- Hiyama A, Skubutyte R, Markova D, Anderson DG, Yadla S, Sakai D, … Risbud MV. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: Implications in degenerative disc disease. Arthritis and Rheumatism. 2011;63(5):1355–1364. doi: 10.1002/art.30246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. Journal of Cell Science. 2013;126(Pt 10):2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Fu X, Yang H, Long F, Chen J. MTORC1 signaling promotes limb bud cell growth and chondrogenesis. Journal of Cellular Biochemistry. 2017;118(4):748–753. doi: 10.1002/jcb.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Ellman MB, An HS, Yan D, van Wijnen AJ, Murphy G, … Im HJ. Lactoferricin mediates anabolic and anti-catabolic effects in the intervertebral disc. Journal of Cellular Physiology. 2012;227(4):1512–1520. doi: 10.1002/jcp.22867. [DOI] [PubMed] [Google Scholar]
- Kim JS, Ellman MB, Yan D, An HS, Kc R, Li X, … Im HJ. Lactoferricin mediates anti-inflammatory and anti-catabolic effects via inhibition of IL-1 and LPS activity in the intervertebral disc. Journal of Cellular Physiology. 2013;228(9):1884–1896. doi: 10.1002/jcp.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen J, Mirando AJ, Wang C, Zuscik MJ, O'Keefe RJ, Hilton MJ. A dual role for NOTCH signaling in joint cartilage maintenance and osteoarthritis. Science Signaling. 2015;8(386):ra71. doi: 10.1126/scisignal.aaa3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ren Y, Mirando AJ, Wang C, Zuscik MJ, O'Keefe RJ, Hilton MJ. Notch signaling in postnatal joint chondrocytes, but not subchondral osteoblasts, is required for articular cartilage and joint maintenance. Osteoarthritis and Cartilage. 2016;24(4):740–751. doi: 10.1016/j.joca.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128(24):5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord-derived cells using a Noto-cre mouse: Implications for intervertebral disc development. Disease Models & Mechanisms. 2012;5(1):73–82. doi: 10.1242/dmm.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward-Sadler SJ, Costello PW, Freemont AJ, Hoyland JA. Regulation of catabolic gene expression in normal and degenerate human intervertebral disc cells: Implications for the pathogenesis of intervertebral disc degeneration. Arthritis Research & Therapy. 2009;11(3):R65. doi: 10.1186/ar2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirando AJ, Liu Z, Moore T, Lang A, Kohn A, Osinski AM, … Hilton MJ. RBP-Jkappa-dependent Notch signaling is required for murine articular cartilage and joint maintenance. Arthritis and Rheumatism. 2013;65(10):2623–2633. doi: 10.1002/art.38076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Oh CD, Im HJ, Suh J, Chee A, An H, Chen D. Rho-Associated kinase inhibitor immortalizes rat nucleus pulposus and annulus fibrosus cells: Establishment of intervertebral disc cell lines with novel approaches. Spine. 2016;41(5):E255–E261. doi: 10.1097/BRS.0000000000001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nature Reviews Rheumatology. 2014;10(1):44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Masuda K, Thonar EJ, An HS, Cs-Szabo G. Age-related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine. 2009;34(1):10–16. doi: 10.1097/BRS.0b013e31818e5ddd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive JI, Baird P, Jeziorsk M, Watkins A, Hoyland JA, Freemont AJ. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Molecular Pathology: MP. 2002;55(2):91–97. doi: 10.1136/mp.55.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BF. The prevalence of low back pain: A systematic review of the literature from 1966 to 1998. Journal of Spinal Disorders. 2000;13(3):205–217. doi: 10.1097/00002517-200006000-00003. [DOI] [PubMed] [Google Scholar]
- Wang H, Tian Y, Wang J, Phillips KL, Binch AL, Dunn S, … Risbud MV. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: Implications in intervertebral disc degeneration. The Journal of Biological Chemistry. 2013;288(23):16761–16774. doi: 10.1074/jbc.M112.446633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Shen J, Jin H, Im HJ, Sandy J, Chen D. Recent progress in understanding molecular mechanisms of cartilage degeneration during osteoarthritis. Annals of the New York Academy of Sciences. 2011;1240:61–69. doi: 10.1111/j.1749-6632.2011.06258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tang D, Shu B, Wang B, Jin H, Hao S, … Chen D. Conditional activation of beta-catenin signaling in mice leads to severe defects in intervertebral disc tissue. Arthritis and Rheumatism. 2012;64(8):2611–2623. doi: 10.1002/art.34469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Interleukin 6 mediates selected effects of Notch in chondrocytes. Osteoarthritis and Cartilage. 2013;21(11):1766–1773. doi: 10.1016/j.joca.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]