What is transvection?
There is something magical about transvection. It conveys the power and elegance of classical Drosophila genetics, and has attracted and perplexed many distinguished scientists since its discovery by Ed Lewis in 1954. Transvection refers to a special class of genetic complementation of mutant alleles on homologous chromosomes. The prevailing view is that regulatory DNAs located on one homolog can regulate the transcription unit on the other homolog in trans. In some cases, enhancers appear to trans-activate genes located on the other homolog, but transvection can also lead to trans-repression of gene expression across homologous chromosomes.
What is the evidence for transvection?
Lewis described genetic complementation between mutant alleles of the Hox gene Ultrabithorax (Ubx) in Drosophila. Both bx34e and Ubx1 mutants exhibit abnormalities in the patterning of the thorax, including partial transformations of halteres into wings: however, bx34e/Ubx1 trans-heterozygotes display less severe transformations than predicted from the phenotypes produced by the individual alleles. Only after the molecular cloning and mapping of Ubx mutant alleles did the mechanistic basis for this trans-complementation process come into focus.
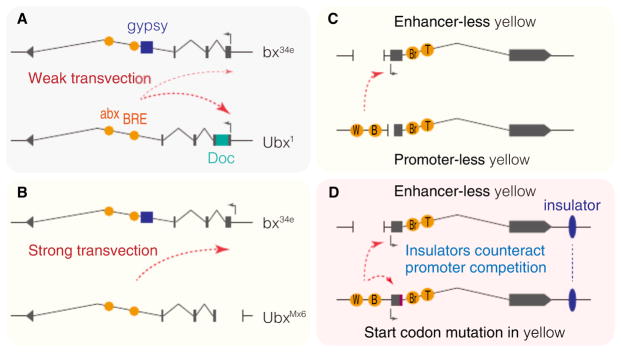
Ubx is regulated by multiple enhancers, including the intronic enhancers abx and BRE (Figure 1A). The bx34e mutant allele is caused by the insertion of a gypsy transposable element between these intronic enhancers and the Ubx promoter. The gypsy element contains twelve binding sites for the Zn-finger protein Su(Hw), which functions as an insulator to block enhancer–promoter interactions. Ubx1 is a protein transposable element within the 5′ untranslated region (UTR) of the first exon. The favored interpretation of this partial genetic complementation is that the intronic abx and BRE enhancers in the Ubx1 allele are able to activate transcription from the bx34e allele in trans to produce functional Ubx transcripts (Figure 1A). Chromosome rearrangements disrupt this transvection effect, suggesting that trans enhancer–promoter interactions require physical association of the two alleles. Indeed, visualization of the endogenous Ubx locus by DNA fluorescence in situ hybridization (FISH) revealed substantial allele pairing during Drosophila development.
Figure 1. Transvection in Drosophila.
(A) Transvection in bx34e/Ubx1 trans-heterozygotes. Intronic abx and BRE enhancers on the Ubx1al-lele activate transcription from the bx34e allele in trans. (B) Loss of cis-linked promoter augments transvection. UbxMx6 mutation consists of a 3.4 kb deletion including the Ubx promoter. (C) 5′ wing and body enhancers on the promoter-less allele activate transcription from the enhancer-less allele in trans. W, wing enhancer; B, body enhancer; Br, bristle enhancer; T, tarsal claw enhancer. (D) Insulator DNAs facilitate transvection at yellow.
How common is transvection?
Since the discovery of transvection at the Ubx locus, there have been numerous reports of trans-homolog interactions in Drosophila. For example, a mutant allele of yellow lacking 5′ enhancers can be complemented by a null mutation lacking the promoter region (Figure 1C). Synthetic promoter-less/enhancer-less yellow mutant alleles were integrated into different locations of the genome and found to recapitulate transvection. These results suggest that somatic pairing of homologous chromosomes may be pervasive in the Drosophila genome. Supporting this view, transvection has been observed at many different endogenous loci, including Abd-B, Scr, dpp, w, ap, eya and vg. DNA FISH assays have revealed frequent pairing events at many chromosomal locations. In mammalian systems, trans-homolog enhancer–promoter interactions have not been so clearly demonstrated, but homolog pairing is thought to take place during X-chromosome inactivation, B-cell maturation and ES cell differentiation.
What is promoter competition?
Transvection is often attenuated by the presence of a linked promoter in cis. The first evidence for such promoter competition came from studies of the yellow locus. Similar results have been reported at other loci, including Ubx. The promoter-less mutant UbxMx6 produces more efficient suppression of the haltere-to-wing transformation than the promoter-containing Ubx1 mutant when placed in trans to the bx34e allele (Figure 1B). This observation suggests that the intronic enhancers located on the UbxMx6 allele are now able to effectively trans-activate the functional Ubx transcription unit located on the bx34e allele. Promoter competition has also been documented for the Hox gene Scr, suggesting that cis-preference may be a general property of transvection. This phenomenon might be related to prototypic examples of promoter competition seen at the β-globin locus, whereby the closest promoter displays preferential activation by a shared enhancer in cis.
What is the mechanism of transvection?
Insulator DNAs appear to be at the heart of trans-homolog enhancer–promoter interactions. As discussed above, enhancers preferentially activate neighboring promoters in cis. Trans-interactions are augmented by the loss of cis-promoters (Figure 1B and C). They are also augmented by insulator DNAs (Figure 1D). The analysis of the transvection mediating region of the Hox gene Abd-B suggests that insulators foster pairing of homologous chromosomes. Stable pairing might increase the opportunities for trans enhancer–promoter interactions. Importantly, insulators facilitate transvection even in the presence of a cis-linked promoter.
And future prospects?
Insulator DNAs are thought to be important agents of genome organization in vertebrates, leading to the formation of chromosomal loop domains, or TADs (topologically associating domains). It has been suggested that insulators also produce loop domains in Drosophila. For example, the depletion of CAP-H2, a key component of Condensin II, augments transvection at Ubx and yellow, possibly by antagonizing Cohesin. A major future goal is to determine the nature of putative chromosomal loop domains at transvecting loci. Do they serve as transcription ‘hubs’ for regulating homologous genes in cis and trans? With the advent of genome editing methods and quantitative live-imaging technologies, it won’t be long before these and other mysteries of transvection are revealed.
Where can I find out more?
- Chen JL, Huisinga KL, Viering MM, Ou SA, Wu CT, Geyer PK. Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci USA. 2002;99:3723–3728. doi: 10.1073/pnas.062447999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D, Fujioka M, Erokhin M, Georgiev P, Jaynes JB, Schedl P. Boundaries of loop domains (insulators): Determinants of chromosome form and function in multicellular eukaryotes. BioEssays. 2017;39 doi: 10.1002/bies.201600233. . Epub 2017 Jan 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ORB, Engel JD. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Mistry H, Schedl P, Jaynes JB. Determinants of chromosome architecture: insulator pairing in cis and in trans. PLoS Genet. 2016;12:e1005889. doi: 10.1371/journal.pgen.1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemkow MJ, Verveer PJ, Arndt-Jovin DJ. Homologous association of the Bithorax-Complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development. 1998;125:4541–4552. doi: 10.1242/dev.125.22.4541. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Green MM, Corces VG. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by Condensin II. Science. 2008;322:1384–1387. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- Hopmann R, Duncan D, Duncan I. Transvection in the iab-5,6,7 region of the bithorax complex of Drosophila: homology independent interactions in trans. Genetics. 1995;139:815–833. doi: 10.1093/genetics/139.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko E, Savitskaya E, Kravchuk O, Parshikov A, Georgiev P, Savitsky M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol Cell Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
- Martínez-Laborda A, González-Reyes A, Morata G. Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J. 1992;11:3645–3652. doi: 10.1002/j.1460-2075.1992.tb05449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, Levine M. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev Cell. 2004;7:925–932. doi: 10.1016/j.devcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Senaratne TN, Joyce EF, Nguyen SC, Wu CT. Investigating the Interplay between Sister Chromatid Cohesion and Homolog Pairing in Drosophila Nuclei. PLoS Genet. 2016;12:e1006169. doi: 10.1371/journal.pgen.1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth JW, Kennison JA. Transvection and silencing of the Scr homeotic gene of Drosophila melanogaster. Genetics. 2002;161:733–746. doi: 10.1093/genetics/161.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]