Abstract
Defining the etiology of inflammatory bowel disease (IBD) continues to elude researchers, in part due to the possibility that there may be different triggers for a spectrum of disease phenotypes that are currently classified as either Crohn’s disease (CD) or ulcerative colitis (UC). What is clear is that genetic susceptibility plays an important role in the development of IBD, and large genome-wide association studies using case-control approaches have identified more than 230 risk alleles. Many of these identified risk alleles are located in a variety of genes important in host-microbiome interactions. In spite of these major advances, the mechanisms behind the genetic influence on disease development remain unknown. In addition, the identified genetic risks have thus far failed to fully define the hereditability of IBD. Host genetics influence host interactions with the gut microbiota in maintaining health through a balance of regulated immune responses and coordinated microbial composition and function. What remains to be defined is how alterations in these interactions can lead to disease. The nature and cause of changes in the microbiota in patients with IBD are poorly understood. In spite of the large catalog of alterations in the microbiota of IBD patients, inflammation itself can alter the microbiota, leaving open the question of which is cause or effect. The composition and function of the gut microbiota are influenced by many factors, including environmental factors, dietary factors, and, as recent studies have shown, host genetic makeup. More than 200 loci have shown potential to influence the microbiota, but replication and larger studies are still required to validate these findings. It would seem reasonable to consider the combination of both host genetic makeup and the inheritance of the microbiota as interdependent heritable forces that could explain the nature of an individual’s susceptibility to IBD or indeed the actual cause of IBD. In this review, we will consider the contribution of the host genetics, the microbiome, and the influence of host genetics on the microbiota to the heritability of IBD.
Keywords: human microbiota, dysbiosis, inflammation, epigenetics
INTRODUCTION
The factors that trigger the onset of inflammatory bowel disease (IBD) are unknown; however, the current hypothesis proposes that genetically susceptible individuals experience an environmental trigger resulting in an inappropriate immune response potentially against certain gut microbes.1, 2 The term “heritability” is commonly used to refer to the observed differences in a trait among individuals of a population that are due to genetic differences or inheritance from previous generations. In addition to genetic inheritance, factors including environment and diet can contribute to the variation between individuals and may even contribute to clustering of traits within families, and therefore suggest a hereditable influence. It is clear from human studies that there is a strong heritable component to IBD, but the genetic basis for this does not seem to fully explain the observed heritability.3, 4 As we begin to appreciate the host interactions with the microbes that occupy the gut,4–6 we have also seen elements of hereditability influencing the composition of gut microbiota. In mouse work, this appreciation has led to the importance of using littermate controls to assess genetic contribution to a phenotype.7 In this review, we will discuss the notion that hereditability of IBD is a function of not only of host genetics, but also of the gut microbiome and other factors that influence their interaction.
GENETICS OF IBD
It is well established that IBD susceptibility has a strong genetic component,8 with up to 12% of IBD patients having a family history of IBD.9 Family and twins studies have shown that the degree of heritability of IBD depends on the disease phenotype, with host genetics playing a smaller contribution in ulcerative colitis (UC) as compared with Crohn’s disease (CD).4 First-degree relatives (FDRs) of IBD patients have increased risk of IBD, and this risk is higher in CD relatives compared with UC relatives.9 The incidence rate ratio in CD FDRs is 7.77 (95% confidence interval [CI], 7.05–8.56), whereas the incidence rate ratio in UC FDRs is 4.08 (95% CI, 3.81–4.38).9 Furthermore, the concordance rate in monozygotic twins for UC is 10%–15%, whereas it reaches 30%–35% in CD.10, 11
To date, genome-wide association studies (GWAS) have identified more than 230 single nucleotide polymorphisms (SNPs) associated with IBD.3, 12, 13 For many of these SNPs, the risk ascribed (as expressed by the observed odd ratios [ORs]) remains relatively low, especially for most of the newly discovered SNPs.3 For instance, the first risk variant for IBD was located in the nucleotide oligomerization domain containing protein 2 gene (NOD2), which has the highest OR of 3.1 in CD. Additionally, the interleukin 23 receptor (IL23R) risk allele has an OR of 2.0 in IBD, whereas recently identified rs1748195 located in the DOCK7 gene only has an OR of 1.07 in European descent.3, 4
In order to assess the combined effect of all the risk alleles, a genetic risk score (GRS) can be calculated using the observed odds ratios for each SNP and disease phenotype. Healthy FDRs of CD have a higher CD-GRS (calculated using only SNPs associated with CD) than the general healthy population, but their CD-GRS is lower than individuals diagnosed with CD.14 Similarly, FDRs of UC have a higher UC-GRS than the general healthy population. It is also important to acknowledge that there are many instances in which a patient with CD may have a family member with UC and vice versa.9 In 1 study, 20.5% of families with 2 or more affected members were pure CD families and 43.4% were pure UC families, whereas 36.1% were mixed CD and UC families.9 Perhaps this is not surprising given that there are many genetic loci that are shared between CD and UC.4 Therefore, there is a clear genetic basis to the observed increase of IBD in families; however, the IBD risk–associated SNPs seem to account for only a portion of the observed heritability of IBD. Conversely, many healthy individuals who carry these risk alleles never develop disease.4
Influence of Ethnicity on Genetic Associations of IBD
Ethnic groups for which IBD incidence has historically been rare have recently been experiencing a sudden rise in the number of new IBD cases.15 Several GWAS were performed in defined ethnic populations,16–18 including Japanese, Koreans, Ashkenazi Jews, etc. These studies describe small but noticeable differences in the list of SNPs that are associated with IBD in different ethnicities as compared with Caucasians of European decent (the most widely studied group affected by IBD). Some loci such as NOD2 seem to be more significantly associated with IBD in certain cohorts/populations. For example, the NOD2 locus has a smaller effect size in East Asian population3 as compared with the European population. In more recent work looking at the Japanese population, 2 East Asia–specific IBD susceptibility loci were identified,19 and in a Korean cohort, 3 novel CD risk loci were identified.17 In 2012, Jostins et al. identified 163 SNPs associated with IBD in cohorts of European ancestry, of which 30 were specific to CD, 23 to UC, and 110 were shared between both.4 Among those SNPs, 41 were not replicated in a separate non-European cohort.3 In another independent GWAS study with 25,305 individuals of European ancestry, almost all genetic risk loci were shared across different ethnic populations.12 Lastly, in a large study of Ashkenazi Jews, the most common IBD-associated variants conserved the directionality of their effects, as observed in the European ancestry cohort.16 Overall, these GWAS studies suggest that a large common core of IBD risk–associated SNPs exists, which may help explain similar pathophysiology across ethnicities.
Genetically Defined Pathways Associated With IBD
More than 230 IBD risk alleles have been identified, suggesting a number of genetic pathways involved in the observed risk. The strongest genetic risk locus in IBD is NOD2,20–22 which codes for an important cytosolic pattern recognition receptor in the host-microbe immune response. NOD2 is expressed in gut epithelial cells (including Paneth cells) and lamina propria lymphocytes (including T cells) but is most strongly expressed in monocytes and macrophages.23–26 This receptor recognizes and binds to muramyl dipeptide (MDP), a component of peptidoglycan derived from the bacterial cell wall.27 Binding of MDP to NOD2 results in oligomerization and induces activation of NK-κB and MAPK and increases transcription of pro-inflammatory cytokines.7, 28 The 3 common NOD2 mutations observed in IBD are located within the leucine-rich repeat (LRR) domain responsible for binding MDP29; suggesting that the absence of bacterial sensing via NOD2 could explain the increased risk.30 Paneth cell antimicrobial response abnormalities have also been associated with NOD2 mutations.31, 32 Paneth cells secrete antimicrobial peptides such as α-defensins, which are reduced in IBD patients,33 perhaps related to the abnormal Paneth cell morphology that has been observed in CD patients with NOD2 mutations.34 Furthermore, abnormal Paneth cells have been associated with an altered ileal microbiota in pediatric CD patients.35 Taken together, this suggests that the role NOD2 plays in IBD pathogenesis is via influencing responses to gut microbiota, although the exact mechanism(s) by which this occurs has not yet been defined.7
Autophagy, a degradation system used by cells for clearance of cytosolic debris and dysfunctional organelles, and possibly in the intracellular responses to pathogens, has also been implicated in IBD. Genetic variants identified in several genes involved in the pathway, including ATG16L1, LRRK2, and IRGM have been identified as risk-associated loci for IBD.36–38 The function of the autophagy pathway in general cell maintenance and its role in response to intracellular microbes suggest diverse mechanisms that can lead to IBD, although it remains unclear what direct role autophagy plays in IBD pathogenesis.39, 40
The gut mucosa is an important physical, chemical, and immunological interface between the gut microbiota and the host; thus any dysregulation or breakdown in this barrier could contribute to disease.41, 42 Altered physical epithelial barrier function, thinner mucus layer, and altered responses to endoplasmic reticulum (ER) stress (via mutations in MUC19, ITLN1, FUT2, and XBP1) have all been identified as risk-associated loci for IBD.43–46
The observation that many of the identified genetic risk loci in IBD implicate genes involved in the complex interplay between a host and gut microbes points to a central role for host-microbe interactions at the root of IBD pathogenesis.
THE HUMAN GUT MICROBIOME
The human gut microbiota consists of a complex consortium of microorganisms that include members of the superkingdom of Archaea, Bacteria, and Eukarya.47 It remains possible that any member or consortia of the microbiota might be associated with or even cause IBD. To date, much of the work has been focused on the bacterial community within the gut microbiota due in large part to the evolution of technology using next-generation sequencing to study the composition and function of the bacterial population within the gut microbiota. Focusing on the bacteria will help us understand the technological and bioinformatic challenges that will need to be applied to the study of other members of the gut microbiota, including fungi and viruses.
The composition of the gut microbiota and its functional relationship with the host is an important determinant of health and disease. With the advent of the Human Microbiota Project (HMP), defining the composition and function of the microbiota in healthy subjects, we are beginning to understand the nature of this ecological niche as it relates to the human host.48, 49 The determinants of the composition of the microbiota include environmental factors such as exposure to pollution, diet, and exposure to drugs, especially antimicrobials.50–53 Most importantly, the inoculum delivered to the newborn infant during childbirth seems to dictate the microbiota composition during the critical period of development in early life, during which the immune system and gut microbes develop a bidirectional relationship.54–57 A stable gut microbial composition is achieved between 1 and 3 years of age, and it remains stable unless there is a major perturbation such as an illness, use of antibiotics, or major changes in diet.54, 58–60 Although the environment is a major influence on the microbiota, it is also apparent that the gut microbial composition is influenced by host genetic factors.51, 61, 62
Heritability of the Microbiota
The gut microbiome seems to be influenced by hereditable factors, as is evident from twin studies and studies of large cohorts of healthy subjects. Monozygotic twins have a more similar microbiome than dizygotic twins, with dizygotic twins being more similar than unrelated individuals.59, 63–65 Several studies have now shown that elements of the microbiome, including specific taxa, are heritable. The largest heritability study on the gut microbiome was performed in a cohort of 416 mostly female twin pairs from the United Kingdom, with a mean age of 60 years, thus reducing the potential confounding effect of a shared environment on the microbiome.66 This study identified 26 taxa to be heritable (using a nominal P < 0.05), with Christensenella being the taxa with the highest heritability score. These 26 heritable taxa were mostly replicated, and new heritable taxa were identified in 2 separate large cohorts.67, 68 In a third study of 271 related healthy individuals from 123 pedigrees, we showed that almost a third of the bacterial taxa in stool were heritable.51 The finding that there are familial influences on the bacterial composition raises the possibility that specific host genetic variants may account for the individual variability in microbial profiles.
Association of IBD Risk–Associated SNPs and Gut Microbiota in Healthy Subjects
To assess possible genetic associations with microbiota composition, several approaches have been taken. One study focused on the IBD risk–associated SNPs and their associations with microbiome composition in 582 healthy individuals.69 It found that a high genetic risk for IBD was associated with a decrease in the relative abundance of the acetate-to-butyrate converter Roseburia.69, 70
To our knowledge, no study has been able to identify a robust association of NOD2 genetic polymorphism with microbial taxa in healthy human subjects. Nonetheless, a suggestive association (4.6 × 10–6 < P < 1.3 × 10–4) was identified in a cohort of 1514 healthy individuals.62 In that study, the NOD2 locus was associated with the enterobactin biosynthesis pathway, which is highly correlated with Escherichia coli abundance. The authors suggested that this pathway could help E. coli to evade the host innate immune responses in inflammatory gut disease.62, 71
Among the genes involved in barrier function, the protein encoded by the alpha1, 2-fucosyltransferase 2 gene (FUT2), which is responsible for secretion of the ABO histo-blood group antigens in the mucosa, may be involved in alterations of the gut microbiota. The minor allele (A) confers a nonsecretor phenotype that is associated with CD susceptibility, with an odds ratio of 1.1 for CD (95% CI, 1.071–1.143; P < 10–15).4 Early studies have shown that FUT2 polymorphisms were associated with the microbiome.72–74 Similar results were confirmed more recently in a cohort of 33 healthy individuals75; however, a larger study of 1503 healthy twins76 could not replicate these findings. Therefore, replication studies with larger cohorts are necessary to define the effects of any associations between host genotype and microbial composition.
GWAS of Microbiota In Healthy Individuals
In an effort to define the genetic basis of the hereditability of the gut microbiome, GWAS between a host’s genetics and their microbiome composition have been performed. The first such GWAS used the Human Microbiome Project metagenomics data.77 Here, the high sequencing depth of fecal DNA allowed for identification of host genetic polymorphisms.52 Despite the small sample size (93 individuals), they identified several associations between host genetics and microbial taxa. The most convincing was the association of rs56064699 located in the LCT gene, which encodes the lactase gene responsible for lactose hydrolysis. The minor allele of rs56064699 was associated with an increase in the relative abundance of Bifidobacteria.52 This association was later replicated in a small cohort of healthy Hutterites.78 It is possible that these initial studies have identified associations with higher effect size than are likely to be seen in subsequent studies, due in part to the phenomenon referred to as the winners-curse effect.79
The gut microbiome can also be assessed for diversity, such as alpha diversity, which measures the presence/absence of unique taxa within a sample, whereas beta diversity assesses the presence and abundance of taxa between samples.80–83 Alpha diversity of the microbiome is decreased in both UC and CD patients compared with healthy controls.80–83 As measured by the Shannon index, this parameter was shown to be heritable; however, to our knowledge, no SNPs have been associated with microbial alpha diversity,51, 78 suggesting that environmental exposures might have a larger effect on this parameter compared with host genetics.
Another proxy for an “IBD-like microbiota” is the microbial dysbiosis index (MD-index) described by Gevers et al.84 This index is defined based on measures of altered microbial composition in the ileum and was suggested to be specific to the diagnosis of CD.84 Even though this index showed a moderate score for heritability, a GWAS in healthy subjects failed to identify any significant association between SNPs and the MD-index.51 Several SNPs did show a nominal association with the MD-index, for example, rs2138126, rs2138125, and rs2589132 (P < 1.4 × 10–6), all located within the regulatory associated protein of MTOR complex 1 (RPTOR), which is involved in immunity and obesity.85, 86 In addition, among all of the suggestive associations (ie, nominal associations with 0.05 > P > 10–8), rs11575056 (P = 1.9 × 10–6) were also associated with the MD-index.4 This is potentially interesting because rs11575056 is located close to the chemokine (C-C motif) ligand 8 (CCL8), an important inflammatory response chemokine,87 and next to a known IBD risk region (17q12).
Another study used 2 population-based cohorts, 1 from Schleswig-Holstein (Germany) comprised of the 914 healthy individuals recruited from the PopGen study and a second with 1115 individuals recruited from the FoCus study, comprised of 371 obese subjects from the FoCus obesity cohort, to identify SNPs associated with microbial composition.61 Their data suggested that host genetics contributes to overall bacterial composition,61 with 42 loci associated with microbiome composition, as measured by the Bray-Curtis beta diversity index. Here rs7974353, which is located within the vitamin D receptor (Vdr) locus, accounted for a small fraction (0.75%) of the microbiota variation in the combined cohort. Carriers of homozygote TT of the rs7974353 Vdr variant had a decrease in the relative abundance of Parabacteroides.61 Vitamin D receptor has been associated with an increased risk of IBD,88–90 particularly the homozygote minor genotype for the Vdr (TaqI polymorphism at codon 352 of exon 8), which is more frequent in CD patients compared with UC patients or healthy controls.88 Altogether, these results suggest that genetic variants of Vdr modulate the Parabacteroides relative abundance and that this interaction might influence IBD risk.91 It is of note that they also showed an upregulation of VDR expression in biopsies of CD and UC patients with acute inflammation as compared with healthy controls. This upregulation was accompanied by a lower relative abundance of Parabacteroides, possibly supporting such an interaction.61
In a study by Bonder et al., 42 SNPs were associated with bacterial function and composition using metagenomic shotgun sequencing of 1514 healthy subjects.62 In this study, 9 loci were associated with specific bacterial taxa and 33 were associated with bacterial function using a conventional P value threshold for GWAS (P < 5 × 10–8). Other associations were related to IBD loci, but with nonsignificant P values (<5 × 10–4). Among these, the rs2155219 located in the C11orf30–LRRC32 locus is part of a cell-cell signaling pathway (gene ontology [GO] term: 0007267) and was previously associated with IBD risk with an OR of 1.15.4 Interestingly, using the same data set, this GO term was correlated with the abundance of 2 potentially IBD-associated bacteria, Coprococcus comes and Proteobacteria.62 Other IBD-associated genes were identified as associated with the gut microbiome function: CCL2 associated with phosphopantothenate biosynthesis III, DAP associated with sucrose degradation IV (sucrose phosphorylase), and IL23R associated with 4-chlorobenzoate degradation.62
In a large discovery cohort comprised of healthy FDRs of CD patients, we identified 58 SNPs that were associated with 33 bacterial taxa, and 4 of these associations were replicated in a second mixed cohort of European ancestry comprised of Canadian, American, and Israeli.51 When IBD risk–associated SNPs were analyzed in this cohort, no statistical association was observed after correction for multiple testing.51 This cohort is enriched in IBD risk–associated variants as the cohort was composed of healthy FDRs of CD patients; therefore, one would expect that the power to detect associations of IBD risk–associated SNPs would be greater than in the general healthy population.14 Given that none of the recent GWAS papers identified IBD risk loci associated with bacterial taxa, it is tempting to conclude that genetic influence on the microbiome composition is independent of IBD genetic risk. Interestingly, our study identified that the relative abundance of Faecalibacterium, a taxa frequently decreased in IBD as compared with healthy controls, is influenced by rs1394174 located within the CNTN6 gene. This suggests that the relative abundance of Faecalibacterium may be driven both by host genetics and environment factors.51, 67 Moreover, the association of rs1394174 with Faecalibacterium was replicated in an independent cohort of 696 healthy individuals.92
The analysis of host genetic influence on the gut microbiota is still in its infancy, and the findings described highlight the complexity of this analysis and the potential pitfalls of working with relatively small cohorts. Important covariates have frequently not been included in GWAS analysis of the microbiota. For example, it is known that stool frequency and consistency are associated with microbiota composition.93, 94 Many other factors such as diet, antibiotic use, and disease diagnosis also influence microbiota.95–97 Further, it should be highlighted that the presence of inflammation in patients with established disease will further confound the apparent associations. Larger-scale analysis or meta-analysis is required to fully define the effect of host genetics on the gut microbiome. Toward this end, the MiBioGen consortium will combine data from multiple centers currently comprising more than ~19,000 individuals (Wang et al., unpublished data). Whether these SNP-microbiome associations identified in healthy subjects can be implicated in the triggering of IBD onset remains to be shown. The finding that there are associations between host genetic makeup and gut microbiota leads us to speculate that the gut microbiota may serve as a heritable vector influencing the onset of IBD.
Associations of Host Genetics and the Gut Microbiome in IBD Patients
Few studies have investigated the effect of host genetics on the gut microbiome in individuals diagnosed with IBD. This is probably because inflammation itself is a major confounder and strongly influences the composition of the microbiota.1 Thus, it is much more challenging to identify any specific effect of host genetics (especially if it has a small effect size) on the microbiota of individuals with an inflamed gut. Despite this fact, in 474 IBD patients, there was an association between NOD2 risk allele dosage, comprised of rs104895431, rs104895467, rs2066844, rs2066845, rs5743277, rs5743293, and the microbiota from intestinal biopsies, particularly with Enterobacteriaceae.98 As this association is not observed in healthy subjects, this finding suggests that NOD2 polymorphisms may be important in defining host-microbiome interactions along the gastrointestinal tract during inflammation.51, 69
In a small study of 9 CD patients homozygous for the T300A risk allele in ATG16L1, the patients had impaired clearance of potential pathobionts, including Bacteroidaceae, Enterobacteriaceae, and Fusobacteriaceae.99 This implies that the ATG16L1 polymorphism associated with CD could alter the microbiota composition through alterations in secretion of antibacterial peptides39, 40; however, in a study of 313 patients with IBD, the IBD-GRS (based on 200 IBD risk–associated SNPs) showed no association with microbial composition.69 These results suggest that host genetics can influence the microbiome composition but that inflammation can mask or alter this association.
Microbiome Alterations in IBD
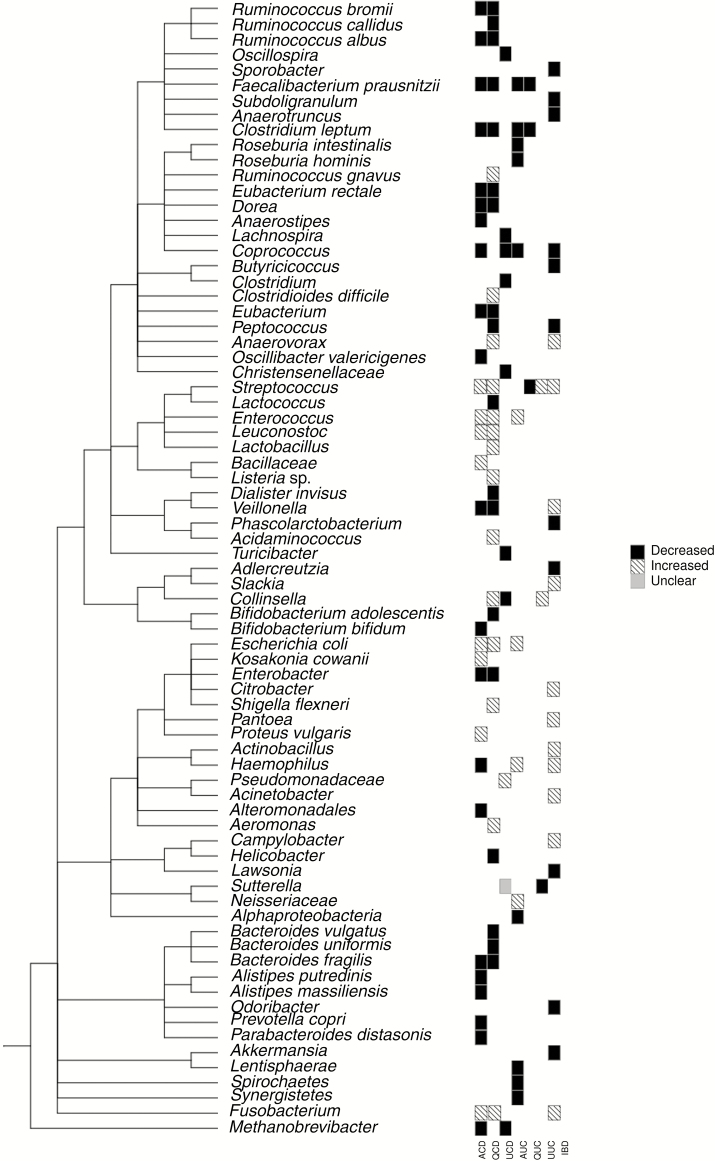
Given the observation that fecal diversions in CD patients can decrease the inflammatory process and that inflammation in mouse models of colitis is dependent on the presence of a gut microbiota, it has long been proposed that the gut microbiome plays a significant role in IBD. Identifying alterations in the gut microbiota of IBD patients could provide insight into the influence of the gut microbiome on the etiology of IBD.100 In an attempt to define the microbiota in patients with IBD, most studies have compared IBD cases with healthy controls (Fig. 1, Table 1). Studies attempting to identify a single bacterium or a group of bacteria as causal have failed. Nonetheless, the attempts to identify specific changes in the microbial composition in IBD patients using case-control studies have led to a number of interesting findings. IBD patients tend to have reduced abundance of bacteria belonging to the phyla of Firmicutes and Bacteroidetes, while being enriched for bacteria from the phyla of Proteobacteria and Actinobacteria.101, 102 A decreased relative abundance of Bacteroides, Faecalibacterium, Roseburia, Blautia, Ruminococcus, and Coprococcus, in addition to other taxa within the families of Ruminococcaceae and Lachnospiraceae, was reported in several studies in CD.82, 84, 103 On the other hand, the family of Enterobacteriaceae is increased in IBD.82, 84, 104 Two members of this family, Enterococcus and Escheriachia coli, are increased in CD105, 106 and UC.107 Other changes that are often reported include a decrease in the genera Bifidobacterium,45, 108Prevotella,109 and Coprococcus,102, 106 as compared with healthy controls. How these described alterations relate to the underlying nature of IBD, either CD or UC, remains to be determined. In order to conclude whether any of these changes occurs in IBD patients independent of the inflammatory process, or before the development of intestinal inflammation, will require a prospective cohort study of high-risk individuals before disease onset.
FIGURE 1.
This phylogenetic tree presents a summary of the described variation of a selection of taxa from publications assessing the fecal microbiome from active CD (ACD), quiescent CD (QCD), unspecified CD (UCD), active UC (AUC), quiescent UC (QUC), unspecified UC (UCD), and IBD patients. In the heat map, black case indicates the taxa that are described as significantly decreased in most (>50%) of the assessed studies in diseased patients, striped cases signifies that the taxa are significantly increased, and the grey means that variations are contradicting between the assessed studies (50% of studies have contradictory results). The plot was generated as follows: Taxa names were first collected from publications and searched for on the NCBI Taxonomy database. The Taxonomy IDs were then imported to the PhyloT web interface (http://phylot.biobyte.de/) to generate a phylogenetic tree (for better visibility, the internal nodes were collapsed). The final plot was then generated in R using the ggtree and the gheatmap commands from the ggtree package.
TABLE 1:
Fecal Bacterial Taxa Associated With IBD
| Taxa | Increase | Decreased | Similar | Disease Activity |
|---|---|---|---|---|
| Total diversity | 103, 105, 106, 108–115 | 109 C | Active CD | |
| 80, 105, 108, 110, 112, 114, 116 | 109 | Quiescent CD | ||
| 117 | Unspecified CD | |||
| 103, 106, 111, 112, 118 | 113 | Active UC | ||
| 112 | 114 | Quiescent UC | ||
| Aeromonas | 82 I | Quiescent CD | ||
| Acidaminococcaceae | 106 | Active CD | ||
| Acidaminococcus | 82 I | Quiescent CD | ||
| Acinetobacter | 103 | IBD | ||
| Actinobacteria | 118 | Active UC | ||
| Actinobacillus | 103 | IBD | ||
| Adlercreutzia | 103 | IBD | ||
| Akkermensia | 115 | IBD | ||
| Alistipes | 115 | IBD | ||
| Alistipes putredinis | 119 | Active CD | ||
| Alistipes massiliensis | 109 | Active CD | ||
| Alphaproteobacteria | 118 | Active UC | ||
| Alteromonadales [Chromatiaceae] | 109 | Active CD | ||
| Anaerosptipes | 114 | Unspecified CD | ||
| Anaerovorax | 115 | IBD | ||
| 82 I | Quiescent CD | |||
| Anaerotruncus | 115 | IBD | ||
| Bacillaceae | 106 | Active CD | ||
| Bacteroidaceae | 117 | Unspecified CD | ||
| 106 | Active UC | |||
| Bacteroides | 108 | 105 | 111 | Active CD |
| 108 | 80, 108 | Quiescent CD | ||
| 106 | 111 | Active UC | ||
| Bacteroides fragilis | 110 | Active CD | ||
| 110, 120 | Quiescent CD | |||
| Bacteroides uniformis | 116 a | Quiescent CD | ||
| Bacteroides vulgatus | 120 | Quiescent CD | ||
| Bacteroidetes | 119 | Active CD | ||
| 118 | Active UC | |||
| Betaproteobacteria | 118 | Active UC | ||
| Bifidobacteria | 105 | 110, 111 | Active CD | |
| 82 | 110 | Quiescent CD | ||
| 111 | Active UC | |||
| Bifidobacterium | 108 | 121 | Active CD | |
| 108 | Quiescent CD | |||
| 121 | Active UC | |||
| Bifidobacterium adolescentis | 122 | Quiescent CD | ||
| Bifidobacterium bifidum | 119 | Active CD | ||
| Blautia | 106 | 84 | Active CD | |
| 114 | Unspecified CD | |||
| Butyricicoccus | 115 | IBD | ||
| Camplylobacter | 103 | IBD | ||
| Citrobacter | 103 | IBD | ||
| Clostridiaceae | 114 | Unspecified CD | ||
| Clostridiales | 109 R | Active CD | ||
| 114 | Unspecified CD | |||
| Clostridium | 114 | Unspecified CD | ||
| Clostridium coccoides group | 80 | Quiescent CD | ||
| 111, 123 | Active UC | |||
| 123 | Quiescent UC | |||
| Clostridium cluster XIVa | 108, 117 | Active CD | ||
| 108, 122 | Quiescent CD | |||
| Clostridium difficile | 120 | Quiescent CD | ||
| Clostridium leptum subgroup | 110–112, 119, 121 | Active CD | ||
| 80, 110, 112, 121 | Quiescent CD | |||
| 112, 121, 123 | Active UC | |||
| 112 | 121 | Quiescent UC | ||
| Collinsella | 82 | Quiescent CD | ||
| 114 | Unspecified CD | |||
| 114 | Unspecified UC | |||
| Coprococcus | 106, 109 | Active CD | ||
| 106 | Active UC | |||
| 114 | Unspecified CD | |||
| 103 | IBD | |||
| Coriobacteriaceae | 84 | Active CD | ||
| Christensenellaceae | 114 | |||
| Deltaproteobacteria | 118 | Active UC | ||
| Dialister | 114 | Unspecified CD | ||
| Dialister invisus | 122 | Quiescent CD | ||
| Dorea | 84, 117 | Active CD | ||
| 117 | Quiescent CD | |||
| Enterobacter cowanii | 119 | Active CD | ||
| Enterobacteriaceae | 82 I | Quiescent CD | ||
| 117 | Unspecified CD | |||
| 106 | Active UC | |||
| 103, 115 | IBD | |||
| Escherichia | 117, 124 | 108 | Active CD | |
| 117 | 108 | Quiescent CD | ||
| 114 | Unspecified CD | |||
| 106 | 113 | Active UC | ||
| 103, 115 | IBD | |||
| Escherichia coli | 119, 120 | 119 | Active CD | |
| 120 | Quiescent CD | |||
| 121 | Active UC | |||
| Enterobacter | 108 | Active CD | ||
| 108 | Quiescent CD | |||
| Epsilonproteobacteria | 118 | Active UC | ||
| Erysipelotrichaceae | 117 | 114 | Unspecified CD | |
| 103 | IBD | |||
| Eubacteriaceae | 106 | Active CD | ||
| Eubacterium | 108 | Active CD | ||
| 108 | Quiescent CD | |||
| Eubacterium rectale | 119 | Active CD | ||
| 120 | Quiescent CD | |||
| Enterobacteria | 105 | 111 | Active CD | |
| 105 | Quiescent CD | |||
| 111 | Active UC | |||
| Enterococcus | 106 | Active CD | ||
| 120 | Quiescent CD | |||
| 106 | Active UC | |||
| Enterococcacae | 106 | Active CD | ||
| 106 | Active UC | |||
| 103 | IBD | |||
| Faecalibacterium | 108, 109, 117, 124 R, 106 | Active CD | ||
| 82 C | 82 I | Quiescent CD | ||
| 114 | Unspecified CD | |||
| Faecalibacterium prausnitzii | 112, 121, 125 | Active CD | ||
| 112, 119, 120, 122, 125 | Quiescent CD | |||
| 112, 121, 123, 126 | Active UC | |||
| 112, 127 | 123 | Quiescent UC | ||
| Firmicutes | 80 | Quiescent CD | ||
| 118 | Active UC | |||
| Fusobacteria | 106, 117 | Active CD | ||
| 118 | Active UC | |||
| Fusobacterium | 117 | Active CD | ||
| 82 I, 114 | 82 C | Quiescent CD | ||
| 103 | IBD | |||
| Gammaproteobacteria | 117 | Active CD | ||
| 117 | Unspecified CD | |||
| 118 | Active UC | |||
| Haemophilus | 113 | Active CD | ||
| 113 | Active UC | |||
| 103 | IBD | |||
| Helicobacter | 128 | Quiescent CD | ||
| Lachnospiraceae | 109 R, 117 | Active CD | ||
| 117 | Unspecified CD | |||
| Lachnospira | 114 | Unspecified CD | ||
| Lactobacillaceae | 106 | Active CD | ||
| Lactobacillales | 84 | Active CD | ||
| 114 | Unspecified CD | |||
| 103 | IBD | |||
| Lactobacillus | 121 | Active CD | ||
| 82 I | Quiescent CD | |||
| 121 | Active UC | |||
| Lactococcus | 82 C | Quiescent CD | ||
| Lawsonia | 115 | IBD | ||
| Lentisphaerae | 118 | Active UC | ||
| Listeria sp. | 120 | Quiescent CD | ||
| Leuconostoc | 108 | Active CD | ||
| 108 | Quiescent CD | |||
| Methanobrevibacter | 109 | Active CD | ||
| 114 | Unspecified CD | |||
| Mogibacteriaceae | 114 | Unspecified CD | ||
| Neisseriaceae | 106 | Active UC | ||
| Odoribacter | 115 | IBD | ||
| Oscillibacter | 115 | IBD | ||
| Oscillibacter valericigenes | 119 | Active CD | ||
| Oscillospira | 114 | Unspecified CD | ||
| Pantoea | 103 | IBD | ||
| Pasteurellaceae | 106 | Active UC | ||
| Parabacteroides | 115 | IBD | ||
| 114 | Unspecified CD | |||
| Parabacteroides distasonis | 119 | Active CD | ||
| Peptococcus | 117 | IBD | ||
| 82 I | Quiescent CD | |||
| Peptostreptococcaceae | 114 | Unspecified CD | ||
| Phascolarctobacterium | 115 | IBD | ||
| Prevotellaceae | 117 | Unspecified CD | ||
| Prevotella | 108 | 117 | Active CD | |
| 117 | 108 | Quiescent CD | ||
| 82 | Quiescent UC | |||
| Prevotella copri | 109 | Active CD | ||
| Proteus | 108 | Active CD | ||
| 108 | Quiescent CD | |||
| Proteus vulgaris | 119 | Active CD | ||
| Proteobacteria | 106 | Active CD | ||
| 106, 118 | Active UC | |||
| 117 | Unspecified CD | |||
| Pseudomonadaceae | 117 | Unspecified CD | ||
| Rikenellaceae | 115 | IBD | ||
| 106 | Active CD | |||
| 114 | Unspecified CD | |||
| Roseburia | 106, 117 | Active CD | ||
| 82, 117 | Quiescent CD | |||
| 114 | Unspecified CD | |||
| Roseburia hominis | 126 | Active UC | ||
| Ruminococcaceae | 106, 109 | Active CD | ||
| 114, 117 | Unspecified CD | |||
| Ruminococcus | 108, 109 R, 84, 106 | Active CD | ||
| 82 | 108 | Quiescent CD | ||
| 114 | 114 | Unspecified CD | ||
| 103, 115 | IBD | |||
| Ruminococcus albus | 119 | Active CD | ||
| 120 | Quiescent CD | |||
| Ruminococcus bromii | 119 | Active CD | ||
| 120 | Quiescent CD | |||
| Ruminococcus callidus | 120 | Quiescent CD | ||
| Ruminococcus gnavus | 122 | Quiescent CD | ||
| Ruminococcus intestinalis | 123 | Active UC | ||
| 123 | Quiescent UC | |||
| Shigella | 117, 124 | Active CD | ||
| 106, 117 | Quiescent CD | |||
| 115 | Active UC | |||
| Shigella flexneri | 120 | Quiescent CD | ||
| Slackia | 103 | IBD | ||
| Spirochaetes | 118 | Active UC | ||
| Sporobacater | 115 | IBD | ||
| Streptococcaceae | 84, 106 | Active CD | ||
| 117 | Unspecified CD | |||
| Streptococcus | 84, 106, 108, 117 | Active CD | ||
| 108, 117 | Quiescent CD | |||
| 82 | Quiescent UC | |||
| 114 | Unspecified UC | |||
| 103 | IBD | |||
| Sutterella | 114 | 117 | Unspecified CD | |
| 114 | Unspecified UC | |||
| Subdoligranulum | 115 | IBD | ||
| Synergistetes | 106 | Active UC | ||
| Turicibacter | 114 | Unspecified CD | ||
| Verrucomicrobiale | 115 | IBD | ||
| 118 | Active UC | |||
| Veillonellaceae | 117 | Unspecified CD | ||
| Veillonella | 108, 117 | Active CD | ||
| 82 I | 82 C, 108 | Quiescent CD | ||
| 103 | IBD |
Abbreviations: I, ileal CD; C, colonic CD; R, resection.
aSignificant for ileal CD only. Microbiota was analyzed using different methods including dot blot hybridization,105 DGGE,110 fluorescence in situ hybridization (FISH) adapted to flow cytometry,111 T-RFLP,108 quantitative polymerase chain reaction (qPCR) and TTGE,112 cloning and full-length 16S rRNA gene sequencing,113 V4 sequencing of the bacterial 16S rRNA,103, 109, 114 Sanger sequencing,106, 115 454 pyrosequencing,82, 106, 116 macroarray,80 terminal restriction fragment length polymorphism (T-RFLP)80, 108, 116 microarray,118 qPCR and microarray,119 microarray,120 qPCR,121, 125, 127, 128 DGGE,122 V4 sequencing of the bacterial 16S rRNA and shotgun sequencing,84 FISH,123 16S sequencing,124 qPCR and DGGE.126 Articles were selected based the reviews published by Li et al.129 and Ni et al.130
Nevertheless, the association between adherent-invasive Escherichia coli (AIEC) and the development of CD in the terminal ileum is worth discussing. AIEC is a member of the Enterobacteriaceae family, and this family has been reported to be increased in the majority of the studies assessing microbial variations in CD patients compared with healthy controls.131, 132 On the other hand, a decrease in the relative abundance of Faecalibacterium prausnitzii has also been described in CD and UC.102, 121, 133 This taxon, along with Roseburia hominis (another “protective” taxon decreased in CD patients), can produce butyrate, which has anti-inflammatory properties.83, 121, 123, 126, 134
Studies that have looked more broadly at changes in the absolute abundance of mucosa-associated bacteria (as assessed by the total number of 16S rDNA gene copies) have shown an increase in subjects with IBD as compared with non-IBD, suggesting that IBD patients may have increased bacterial translocation into the gut mucosa.104, 135 On a larger scale, studies of the composition of the gut microbiota in IBD patients have suggested that there is a generalized imbalance in microbiome composition as compared with healthy controls, sometimes referred to as “dysbiosis” in IBD. Again, It is difficult to know if dysbiosis is a cause or a result of the disease process, as the inflammatory process itself can alter the composition of the microbiota.1 Aerobic microbes in IBD are increased as compared with nondisease conditions,45, 136 perhaps due to the increase in the reactive oxygen species generated during inflammation that react with endogenous luminal sulphur compounds to form new respiratory electron acceptors. This can, for example, favor the growth of species resistant to reactive oxygen species similar to Salmonella typhimurium.137 This same phenomenon could be implicated in the alteration of the other aerobic microbes in the gut of IBD patients.
ENVIRONMENTAL DETERMINANTS OF RISK FOR IBD
There may be other factors influencing IBD that seem to have a hereditable influence. Environmental factors and demographic factors, such as smoking and exposure to second-hand smoke, urban vs rural life,138 air pollution,139 and cultural influences on diet,140 all represent shared exposures that may constitute familial risk for IBD.141 Many of these factors may explain the apparent association between “Westernisation” and the risk of IBD development, as has been described in China and offspring of South Asian immigrants to North America.142 These observations again raise the issue of how cultural influences, such as diet, play a role in modulating the risk of IBD in certain ethnic communities and in certain geographic locations. It will be important to identify the role of these environmental influences and factors in IBD.143
Smoking has been associated with IBD risk, specifically with the risk of CD,144, 145 and is associated with increased intestinal permeability,146 but what remains unclear is whether the impact is mediated through the gut microbiome.147 It also remains unclear whether second-hand smoke exposure can increase risk of IBD onset. A meta-analysis failed to identify a relationship between childhood passive smoke exposure and CD148; however, more recent studies have contradictory results, thus the influence of second-hand smoke on IBD onset requires further investigation.149–151
Diet is a major factor capable of modifying gut microbiota composition.95, 152 Indeed, as part of the IBD European Prospective Cohort Study (IBD-EPIC study), many dietary factors have been associated with IBD onset.153–156 Milk consumption was shown to be potentially associated with a decreased risk of developing CD (P = 0.23 for CD) but not with UC (P = 0.60),154 whereas a role of flavones and resveratrol in the risk of developing CD was also observed.155 The IBD-EPIC study findings also support a role for dietary linoleic acid in the etiology of UC.155 In this cohort, obesity, as measured by body mass index (BMI), was not associated with the development of UC or CD.156 Overall, total fiber intake from fruit, vegetables, or cereals, and the subsequent development of either CD or UC were not associated.153 However, it is important to acknowledge that most of these analyses are based on food frequency questionnaire data that may have important limitations.158 As meals and diet are usually shared between parents and their children from the same household, the influence of diet on the gut microbiota profile can seem heritable.
Epigenetics
The term “epigenetics” refers to heritable changes in phenotype that occur independently of changes to the DNA sequence. Epigenetic modifications include DNA methylation and histone modifications.159 The concept that IBD can be influenced by epigenetically induced changes in gene expression is supported by the fact that GWAS have identified many IBD-associated variants adjacent to genes encoding DNA methyltransferases and their interacting proteins.4 For example, the mutation or differential expression of the uhrf1 gene has been linked to DNA methylation changes in mammals and zebrafish.160 As a result of inactivation or expression modulation of this gene in zebrafish, the DNA methylation changes lead to an IBD-like phenotype.161–164
Epigenetic modifications are known to be influenced by environmental factors such as diet and the microbiota,165 particularly in colorectal cancer.166 Indeed, the gut microbiota was shown to alter host histone acetylation and methylation in human colon tissues167 and in gene promoters linked to inflammatory responses such as IL23 or IgA.168 Moreover, fermentation end products, especially short-chain fatty acids such as acetate, butyrate, and propionate, produced by microbial fermentation of fiber, may be important for the epigenetic regulation of inflammatory reactions.167
Epigenetics is likely to play a role in IBD pathogenesis; however, the changes in inflammatory diseases remain to be defined.159 There are several challenges in studying epigenetics in IBD. Epigenetic signatures are cell type specific165; however, a standardized database of methylated genes in IBD have been recently released169 and will help research into the epigenetic basis of disease. Determining whether an IBD epigenetic signature exists would require that future studies consider several crucial aspects such as isolation of disease-relevant cell types obtained from carefully selected cohorts of patients with homogenous disease location and activity.165 Currently, most of the studies on epigenetics in IBD focus on UC patients and are mainly focused on methylation of DNA from biopsies or DNA derived from peripheral blood.170, 171 Studies on CD patients are only starting to emerge.
Unlike GWAS studies, which require large cohorts to detect genetic variations, previous studies on DNA methylation in IBD patients were able to detect several methylated positions using much smaller cohorts.165 This might indicate that epigenetics may provide much stronger signals as compared with GWAS. If epigenetics proves to be important in IBD pathogenesis, future studies on focused subsets or phenotypes of IBD patients might get around the issue of disease heterogeneity. Nonetheless, early studies using only 18 patients with CD compared with 25 healthy controls identified as many as 4287 differentially methylated positions in DNA derived from peripheral blood cells, indicating that CD patients display a specific methylation “landscape.”172 Another study found that 3196 probes were differentially methylated in DNA derived from peripheral blood mononuclear cells (PBMCs) from 149 IBD cases and 39 controls. After enrichment analysis, the authors showed that the differentially methylated probes were significantly associated with 104 GO terms. Interestingly, in CD, the most enriched pathways identified involve immune responses, regulation of T-cell activation, and cellular response to molecules of bacterial origin,173 suggesting that epigenetic modification is involved in microbial and immune response dysregulation in IBD. Another study has compared intestinal epithelial cells purified from 66 IBD patients to 30 non-IBD controls and showed that there were distinct and stable patterns of DNA methylation in the 2 groups.171 An alternative approach to study epigenetic mechanisms is the development of ex vivo intestinal organoid–based analysis, which has already shown promising results despite limited sample size.171
Altogether, multidimensional analysis of the complex immune cell infiltrates in intestinal tissue of IBD patients harbors the key to understanding disease pathogenesis and how this contributes to the perceived hereditability of these diseases.174 Despite technical difficulties in obtaining homogenous cohorts and biopsy samples, epigenetics has the potential to provide an explanation to the missing heritability of IBD.
CONCLUSION
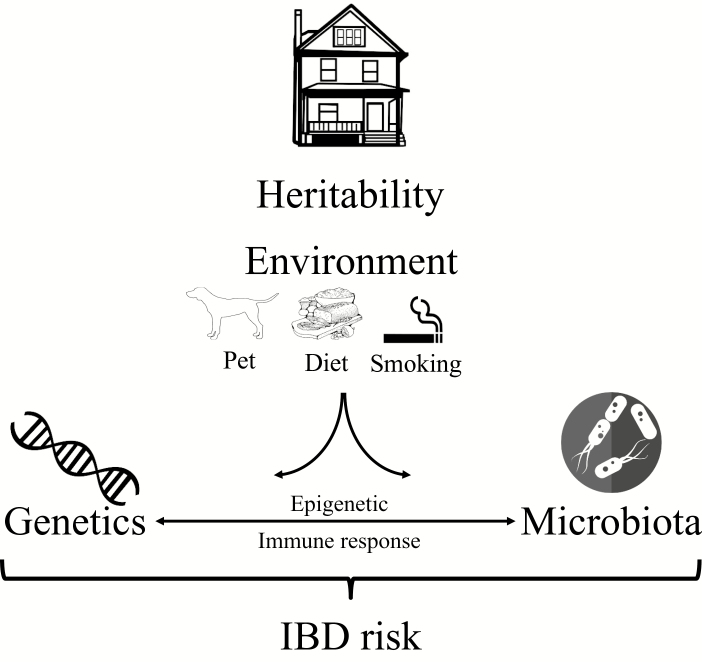
What can be defined as heritability of IBD can be explained by the fact that host genetics and the microbiome contribute to IBD. It is reasonable to suggest that the missing heritability, that is, the occurrence of IBD in families not explained by genetic factors, could be explained by the interaction of host genetics with the microbiota under certain environmental conditions. GWAS have identified several genetic variants linked to IBD that are shared across different ethnic groups, and that the list of risk-associated SNPs will probably keep increasing. In addition to defined genetic risk, the microbiota, which can be influenced by host genetics and environmental factors, plays a role. Thus, genetic risk and microbiome composition certainly contribute to the heritability of IBD. In addition, nongenetic components such as shared environmental factors in individuals from the same household, for example, smoking or pet exposure, could contribute to the onset of IBD (Fig. 2). Overall, the search for a “cure” for IBD requires appreciating the complex interplay between host genetics, gut microbes, environmental triggers, and the mucosal immune response.
FIGURE 2.
Factors that combine to explain the heritability of IBD. In addition to host genetics, the microbiome might contribute to heritability of IBD. Nongenetic components that are shared in families of relatives of IBD patients include dietary and cultural habits; pet exposure can also influence the IBD risk through influencing the microbiota.
Conflicts of interest: All authors disclose no potential conflicts (financial, professional, or personal) that are relevant to the manuscript.
Supported by: This study was supported by grants from Crohn’s and Colitis Canada, Canadian Institutes of Health Research (CIHR; grant No. CMF108031), and the Helmsley Charitable Trust. Williams Turpin is a recipient of a Postdoctoral Fellowship Research Award from the CIHR Fellowship/Canadian Association of Gastroenterology (CAG)/Ferring Pharmaceuticals Inc. and a fellowship from the Department of Medicine, Mount Sinai Hospital, Toronto.
REFERENCES
- 1. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. [DOI] [PubMed] [Google Scholar]
- 2. Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. [DOI] [PubMed] [Google Scholar]
- 3. Liu JZ, van Sommeren S, Huang H et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jostins L, Ripke S, Weersma RK et al. ; International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chu H. Host gene-microbiome interactions: molecular mechanisms in inflammatory bowel disease. Genome Med. 2017;9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Philpott DJ, Sorbara MT, Robertson SJ et al. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. [DOI] [PubMed] [Google Scholar]
- 8. Liu JZ, Anderson CA. Genetic studies of Crohn’s disease: past, present and future. Best Pract Res Clin Gastroenterol. 2014;28:373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moller FT, Andersen V, Wohlfahrt J et al. Familial risk of inflammatory bowel disease: a population-based cohort study 1977-2011. Am J Gastroenterol. 2015;110:564–71. [DOI] [PubMed] [Google Scholar]
- 10. Spehlmann ME, Begun AZ, Burghardt J et al. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–76. [DOI] [PubMed] [Google Scholar]
- 11. Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011;17:1–5. [DOI] [PubMed] [Google Scholar]
- 12. de Lange KM, Moutsianas L, Lee JC et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo Y, de Lange KM, Jostins L et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat Genet. 2017;49:186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kevans D, Silverberg MS, Borowski K et al. ; GEM Project IBD genetic risk profile in healthy first-degree relatives of Crohn’s disease patients. J Crohns Colitis. 2016;10:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gearry RB. IBD and environment: are there differences between East and West. Dig Dis. 2016;34:84–9. [DOI] [PubMed] [Google Scholar]
- 16. Kenny EE, Pe’er I, Karban A et al. A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet. 2012;8:e1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang SK, Hong M, Zhao W et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–7. [DOI] [PubMed] [Google Scholar]
- 18. Yang SK, Hong M, Zhao W et al. Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm Bowel Dis. 2013;19:954–66. [DOI] [PubMed] [Google Scholar]
- 19. Fuyuno Y, Yamazaki K, Takahashi A et al. Genetic characteristics of inflammatory bowel disease in a Japanese population. J Gastroenterol. 2016;51:672–81. [DOI] [PubMed] [Google Scholar]
- 20. Hugot JP, Chamaillard M, Zouali H et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 21. Ogura Y, Bonen DK, Inohara N et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 22. Radford-Smith G, Pandeya N. Associations between NOD2/CARD15 genotype and phenotype in Crohn’s disease—are we there yet?World J Gastroenterol. 2006;12:7097–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogura Y, Inohara N, Benito A et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–8. [DOI] [PubMed] [Google Scholar]
- 24. Gutierrez O, Pipaon C, Inohara N et al. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J Biol Chem. 2002;277:41701–5. [DOI] [PubMed] [Google Scholar]
- 25. Rosenstiel P, Fantini M, Bräutigam K et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124:1001–9. [DOI] [PubMed] [Google Scholar]
- 26. Zanello G, Goethel A, Forster K et al. Nod2 activates NF-kB in CD4+ T cells but its expression is dispensable for T cell-induced colitis. PLoS One. 2013;8: e82623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Girardin SE, Boneca IG, Viala J et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–72. [DOI] [PubMed] [Google Scholar]
- 28. Barnich N, Aguirre JE, Reinecker HC et al. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-{kappa}B activation in muramyl dipeptide recognition. J Cell Biol. 2005;170:21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lesage S, Zouali H, Cézard JP et al. ; EPWG-IBD Group; EPIMAD Group; GETAID Group CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eckmann L, Karin M. NOD2 and Crohn’s disease: loss or gain of function?Immunity. 2005;22:661–7. [DOI] [PubMed] [Google Scholar]
- 31. Lala S, Ogura Y, Osborne C et al. Crohn’s disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47–57. [DOI] [PubMed] [Google Scholar]
- 32. Ogura Y, Lala S, Xin W et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wehkamp J, Harder J, Weichenthal M et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. VanDussen KL, Liu TC, Li D et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu TC, Gurram B, Baldridge MT et al. Paneth cell defects in Crohn’s disease patients promote dysbiosis. JCI Insight. 2016;1:e86907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hampe J, Franke A, Rosenstiel P et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. [DOI] [PubMed] [Google Scholar]
- 37. McCarroll SA, Huett A, Kuballa P et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Q, Pan Y, Yan R et al. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat Immunol. 2015;16:918–26. [DOI] [PubMed] [Google Scholar]
- 39. Cadwell K, Liu JY, Brown SL et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cadwell K, Stappenbeck TS, Virgin HW. Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol. 2009;335:141–67. [DOI] [PubMed] [Google Scholar]
- 41. Zeissig S, Bürgel N, Günzel D et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut. 2007;56:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. [DOI] [PubMed] [Google Scholar]
- 43. Schultsz C, Van Den Berg FM, Ten Kate FW et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089–97. [DOI] [PubMed] [Google Scholar]
- 44. Kaser A, Lee AH, Franke A et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fyderek K, Strus M, Kowalska-Duplaga K et al. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15:5287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–40. [DOI] [PubMed] [Google Scholar]
- 48. Peterson J, Garges S, Giovanni M et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lloyd-Price J, Mahurkar A, Rahnavard G et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012;33:459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turpin W, Espin-Garcia O, Xu W et al. ; GEM Project Research Consortium Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–7. [DOI] [PubMed] [Google Scholar]
- 52. Blekhman R, Goodrich JK, Huang K et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tilg H, Moschen AR. Food, immunity, and the microbiome. Gastroenterology. 2015;148:1107–19. [DOI] [PubMed] [Google Scholar]
- 54. Yassour M, Vatanen T, Siljander H et al. ; DIABIMMUNE Study Group Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bokulich NA, Chung J, Battaglia T et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martinez KA 2nd, Devlin JC, Lacher CR et al. Increased weight gain by C-section: functional significance of the primordial microbiome. Sci Adv. 2017;3:eaao1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pannaraj PS, Li F, Cerini C et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lozupone CA, Stombaugh JI, Gordon JI et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yatsunenko T, Rey FE, Manary MJ et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galloway-Peña JR, Smith DP, Sahasrabhojane P et al. Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Med. 2017;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang J, Thingholm LB, Skiecevičienė J et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bonder MJ, Kurilshikov A, Tigchelaar EF et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–12. [DOI] [PubMed] [Google Scholar]
- 63. Turnbaugh PJ, Hamady M, Yatsunenko T et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–42. [DOI] [PubMed] [Google Scholar]
- 65. Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM et al. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. 2001;13:129–34. [Google Scholar]
- 66. Goodrich JK, Waters JL, Poole AC et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goodrich JK, Davenport ER, Beaumont M et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goodrich JK, Davenport ER, Waters JL et al. Cross-species comparisons of host genetic associations with the microbiome. Science. 2016;352:532–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Imhann F, Vich Vila A, Bonder MJ et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Duncan SH, Holtrop G, Lobley GE et al. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2004;91:915–23. [DOI] [PubMed] [Google Scholar]
- 71. Singh V, Yeoh BS, Xiao X et al. Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat Commun. 2015;6:7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rausch P, Rehman A, Kunzel S et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wacklin P, Mäkivuokko H, Alakulppi N et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One. 2011;6:e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wacklin P, Tuimala J, Nikkilä J et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One. 2014;9:e94863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gampa A, Engen PA, Shobar R et al. Relationships between gastrointestinal microbiota and blood group antigens. Physiol Genomics. 2017;49:473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davenport ER, Goodrich JK, Bell JT et al. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1500 twins. BMC Genomics. 2016;17:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. The Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Davenport ER, Cusanovich DA, Michelini K et al. Genome-wide association studies of the human gut microbiota. PLoS One. 2015;10:e0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kraft P. Curses—winner’s and otherwise—in genetic epidemiology. Epidemiology. 2008;19:649–651; discussion 657. [DOI] [PubMed] [Google Scholar]
- 80. Manichanh C, Rigottier-Gois L, Bonnaud E et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Willing B, Halfvarson J, Dicksved J et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–60. [DOI] [PubMed] [Google Scholar]
- 82. Willing BP, Dicksved J, Halfvarson J et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854.e1. [DOI] [PubMed] [Google Scholar]
- 83. Lepage P, Häsler R, Spehlmann ME et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–36. [DOI] [PubMed] [Google Scholar]
- 84. Gevers D, Kugathasan S, Denson LA et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang K, Chi H. Tuning mTOR activity for immune balance. J Clin Invest. 2013;123:5001–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Morris BJ, Carnes BA, Chen R et al. Genetic variation in the raptor gene is associated with overweight but not hypertension in American men of Japanese ancestry. Am J Hypertens. 2015;28:508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Asano K, Takahashi N, Ushiki M et al. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat Commun. 2015;6:7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Simmons JD, Mullighan C, Welsh KI et al. Vitamin D receptor gene polymorphism: association with Crohn’s disease susceptibility. Gut. 2000;47:211–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ardesia M, Ferlazzo G, Fries W. Vitamin D and inflammatory bowel disease. Biomed Res Int. 2015;2015:470805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schoon EJ, Geerling BG, Van Dooren IM et al. Abnormal bone turnover in long-standing Crohn’s disease in remission. Aliment Pharmacol Ther. 2001;15:783–92. [DOI] [PubMed] [Google Scholar]
- 91. Limketkai BN, Mullin GE, Limsui D et al. Role of vitamin D in inflammatory bowel disease. Nutr Clin Pract. 2017;32:337–45. [DOI] [PubMed] [Google Scholar]
- 92. Rothschild D, Weissbrod O, Barkan E et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–5. [DOI] [PubMed] [Google Scholar]
- 93. Hadizadeh F, Walter S, Belheouane M et al. Stool frequency is associated with gut microbiota composition. Gut. 2017;66:559–60. [DOI] [PubMed] [Google Scholar]
- 94. Vandeputte D, Falony G, Vieira-Silva S et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. David LA, Maurice CF, Carmody RN et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Panda S, El khader I, Casellas F et al. Short-term effect of antibiotics on human gut microbiota. PLoS One. 2014;9:e95476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. De Filippo C, Cavalieri D, Di Paola M et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Knights D, Silverberg MS, Weersma RK et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sadaghian Sadabad M, Regeling A, de Goffau MC et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2015;64:1546–52. [DOI] [PubMed] [Google Scholar]
- 100. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Frank DN, St Amand AL, Feldman RA et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sokol H, Seksik P, Furet JP et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. [DOI] [PubMed] [Google Scholar]
- 103. Shaw KA, Bertha M, Hofmekler T et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kleessen B, Kroesen AJ, Buhr HJ et al. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034–41. [DOI] [PubMed] [Google Scholar]
- 105. Seksik P, Rigottier-Gois L, Gramet G et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen L, Wang W, Zhou R et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore). 2014;93:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pilarczyk-Zurek M, Chmielarczyk A, Gosiewski T et al. Possible role of Escherichia coli in propagation and perpetuation of chronic inflammation in ulcerative colitis. BMC Gastroenterol. 2013;13:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Andoh A, Kuzuoka H, Tsujikawa T et al. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J Gastroenterol. 2012;47:1298–307. [DOI] [PubMed] [Google Scholar]
- 109. Halfvarson J, Brislawn CJ, Lamendella R et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Scanlan PD, Shanahan F, O’Mahony C et al. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006;44:3980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sokol H, Seksik P, Rigottier-Gois L et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–11. [DOI] [PubMed] [Google Scholar]
- 112. Kabeerdoss J, Sankaran V, Pugazhendhi S et al. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol. 2013;13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nwosu FC, Thorkildsen LT, Avershina E et al. Age-dependent fecal bacterial correlation to inflammatory bowel disease for newly diagnosed untreated children. Gastroenterol Res Pract. 2013;2013:302398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Pascal V, Pozuelo M, Borruel N et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Papa E, Docktor M, Smillie C et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7:e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dicksved J, Halfvarson J, Rosenquist M et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. Isme J. 2008;2:716–27. [DOI] [PubMed] [Google Scholar]
- 117. Eun CS, Kwak MJ, Han DS et al. Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of Western patients?BMC Gastroenterol. 2016;16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Michail S, Durbin M, Turner D et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mondot S, Kang S, Furet JP et al. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm Bowel Dis. 2011;17:185–92. [DOI] [PubMed] [Google Scholar]
- 120. Kang S, Denman SE, Morrison M et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–42. [DOI] [PubMed] [Google Scholar]
- 121. Wang W, Chen L, Zhou R et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Joossens M, Huys G, Cnockaert M et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–7. [DOI] [PubMed] [Google Scholar]
- 123. Kumari R, Ahuja V, Paul J. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J Gastroenterol. 2013;19:3404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Thorkildsen LT, Nwosu FC, Avershina E et al. Dominant fecal microbiota in newly diagnosed untreated inflammatory bowel disease patients. Gastroenterol Res Pract. 2013;2013:636785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jia W, Whitehead RN, Griffiths L et al. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn’s disease?FEMS Microbiol Lett. 2010;310:138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Machiels K, Joossens M, Sabino J et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- 127. Varela E, Manichanh C, Gallart M et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther. 2013;38:151–61. [DOI] [PubMed] [Google Scholar]
- 128. Man SM, Zhang L, Day AS et al. Detection of enterohepatic and gastric helicobacter species in fecal specimens of children with Crohn’s disease. Helicobacter. 2008;13:234–8. [DOI] [PubMed] [Google Scholar]
- 129. Li J, Butcher J, Mack D et al. Functional impacts of the intestinal microbiome in the pathogenesis of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:139–53. [DOI] [PubMed] [Google Scholar]
- 130. Ni J, Wu GD, Albenberg L et al. Gut microbiota and IBD: causation or correlation?Nat Rev Gastroenterol Hepatol. 2017;14:573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Darfeuille-Michaud A, Boudeau J, Bulois P et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–21. [DOI] [PubMed] [Google Scholar]
- 132. Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Frank DN, Robertson CE, Hamm CM et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Scanlan PD, Shanahan F, O’Mahony C et al. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006;44:3980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Png CW, Lindén SK, Gilshenan KS et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–8. [DOI] [PubMed] [Google Scholar]
- 136. Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn’s disease. Inflamm Bowel Dis. 2004;10:824–33. [DOI] [PubMed] [Google Scholar]
- 137. Winter SE, Thiennimitr P, Winter MG et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Benchimol EI, Kaplan GG, Otley AR et al. Rural and urban residence during early life is associated with risk of inflammatory bowel disease: a population-based inception and birth cohort study. Am J Gastroenterol. 2017;112:1412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kaplan GG, Hubbard J, Korzenik J et al. The inflammatory bowel diseases and ambient air pollution: a novel association. Am J Gastroenterol. 2010;105:2412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lee D, Albenberg L, Compher C et al. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology. 2015;148:1087–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ananthakrishnan AN, Bernstein CN, Iliopoulos D et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15:39–49. [DOI] [PubMed] [Google Scholar]
- 142. Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–7. [DOI] [PubMed] [Google Scholar]
- 143. Rogler G, Vavricka S. Exposome in IBD: recent insights in environmental factors that influence the onset and course of IBD. Inflamm Bowel Dis. 2015;21:400–8. [DOI] [PubMed] [Google Scholar]
- 144. Lakatos PL, Vegh Z, Lovasz BD et al. Is current smoking still an important environmental factor in inflammatory bowel diseases? Results from a population-based incident cohort. Inflamm Bowel Dis. 2013;19:1010–7. [DOI] [PubMed] [Google Scholar]
- 145. Higuchi LM, Khalili H, Chan AT et al. A prospective study of cigarette smoking and the risk of inflammatory bowel disease in women. Am J Gastroenterol. 2012;107:1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kevans D, Turpin W, Madsen K et al. ; GEM Project Determinants of intestinal permeability in healthy first-degree relatives of individuals with Crohn’s disease. Inflamm Bowel Dis. 2015;21:879–87. [DOI] [PubMed] [Google Scholar]
- 147. Bernstein CN. Review article: changes in the epidemiology of inflammatory bowel disease-clues for aetiology. Aliment Pharmacol Ther. 2017;46:911–9. [DOI] [PubMed] [Google Scholar]
- 148. Jones DT, Osterman MT, Bewtra M et al. Passive smoking and inflammatory bowel disease: a meta-analysis. Am J Gastroenterol. 2008;103:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Chivese T, Esterhuizen TM, Basson AR et al. The influence of second-hand cigarette smoke exposure during childhood and active cigarette smoking on Crohn’s disease phenotype defined by the montreal classification scheme in a Western Cape population, South Africa. PLoS One. 2015;10:e0139597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Mahid SS, Minor KS, Stromberg AJ et al. Active and passive smoking in childhood is related to the development of inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:431–8. [DOI] [PubMed] [Google Scholar]
- 151. Guo AY, Stevens BW, Wilson RG et al. Early life environment and natural history of inflammatory bowel diseases. BMC Gastroenterol. 2014;14:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wu GD, Chen J, Hoffmann C et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Andersen V, Chan SS, Luben R et al. Fibre intake and the development of inflammatory bowel disease – a European prospective multi-centre cohort study (EPIC-IBD). J Crohn’s Colitis. 2018;12(2):129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Opstelten JL, Leenders M, Dik VK et al. Dairy products, dietary calcium, and risk of inflammatory bowel disease: results from a European prospective cohort investigation. Inflamm Bowel Dis. 2016;22:1403–11. [DOI] [PubMed] [Google Scholar]
- 155. Lu Y, Zamora-Ros R, Chan S et al. Dietary polyphenols in the aetiology of Crohn’s disease and ulcerative colitis—a multicenter European prospective cohort study (EPIC). Inflamm Bowel Dis. 2017;23:2072–82. [DOI] [PubMed] [Google Scholar]
- 156. Tjonneland A, Overvad K, Bergmann MM et al. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: a nested case-control study within a European prospective cohort study. Gut. 2009;58:1606–11. [DOI] [PubMed] [Google Scholar]
- 157. Chan SS, Luben R, Olsen A et al. Body mass index and the risk for Crohn’s disease and ulcerative colitis: data from a European prospective cohort study (the IBD in EPIC study). Am J Gastroenterol. 2013;108:575–82. [DOI] [PubMed] [Google Scholar]
- 158. Shim JS, Oh K, Kim HC. Dietary assessment methods in epidemiologic studies. Epidemiol Health. 2014;36:e2014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Stylianou E. Epigenetics: the fine-tuner in inflammatory bowel disease?Curr Opin Gastroenterol. 2013;29:370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Goll MG, Halpern ME. DNA methylation in zebrafish. Prog Mol Biol Transl Sci. 2011;101:193–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Obata Y, Furusawa Y, Endo TA et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15:571–9. [DOI] [PubMed] [Google Scholar]
- 162. Marjoram L, Alvers A, Deerhake ME et al. Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc Natl Acad Sci U S A. 2015;112:2770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Pack M. IBD. Fishing for missing heritability in IBD. Nat Rev Gastroenterol Hepatol. 2015;12:318–20. [DOI] [PubMed] [Google Scholar]
- 164. Mudbhary R, Hoshida Y, Chernyavskaya Y et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Jenke AC, Zilbauer M. Epigenetics in inflammatory bowel disease. Curr Opin Gastroenterol. 2012;28:577–84. [DOI] [PubMed] [Google Scholar]
- 166. Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. 2017;61: doi:10.1002/mnfr.201500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Krautkramer KA, Kreznar JH, Romano KA et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64:982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Kumar H, Lund R, Laiho A et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. mBio. 2014;5: doi:10.1128/mBio.02113-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Xiong Y, Wei Y, Gu Y et al. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017;45:D888–D95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Karatzas PS, Gazouli M, Safioleas M et al. DNA methylation changes in inflammatory bowel disease. Ann Gastroenterol. 2014;27:125–32. [PMC free article] [PubMed] [Google Scholar]
- 171. Howell KJ, Kraiczy J, Nayak KM et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154(3):585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Li Yim AYF, Duijvis NW, Zhao J et al. Peripheral blood methylation profiling of female Crohn’s disease patients. Clin Epigenetics. 2016;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. McDermott E, Ryan EJ, Tosetto M et al. DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J Crohns Colitis. 2016;10:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Cho JH. The promise of epigenetics. Has it delivered new insights?Dig Dis. 2016;34:12–19. [DOI] [PubMed] [Google Scholar]