Summary
Accurate pathological diagnosis is crucial for optimal management of cancer patients. For the ~100 known central nervous system (CNS) tumour entities, standardization of the diagnostic process has been shown to be particularly challenging - with substantial inter-observer variability in the histopathological diagnosis of many tumour types. We herein present the development of a comprehensive approach for DNA methylation-based CNS tumour classification across all entities and age groups, and demonstrate its application in a routine diagnostic setting. We show that availability of this method may have substantial impact on diagnostic precision compared with standard methods, resulting in a change of diagnosis in up to 12% of prospective cases. For broader accessibility we have designed a free online classifier tool (www.molecularneuropathology.org) requiring no additional onsite data processing. Our results provide a blueprint for the generation of machine learning-based tumour classifiers across other cancer entities, with the potential to fundamentally transform tumour pathology.
The developmental complexity of the brain is reflected in the vast array of distinct brain tumour entities defined in the current WHO classification of central nervous system (CNS) tumours 1. These tumours are clinically and biologically highly diverse, encompassing a wide spectrum from benign neoplasms that can frequently be cured by surgery alone (e.g. pilocytic astrocytoma), to highly malignant tumours responding poorly to any therapy (e.g. glioblastoma). Previous studies reported substantial inter-observer variability in the histopathological diagnosis of many CNS tumours, e.g., in diffuse gliomas 2, ependymomas 3 and supratentorial PNETs 4. To address this, some molecular grouping has been introduced into the update of the WHO classification, but only for selected entities such as medulloblastoma. Furthermore, several single-gene tests based on DNA methylation analysis (e.g., MGMT promoter methylation status), FISH (e.g., 1p/19q, EGFR, MYC, MYCN, PDGFRA, 19q13.42, etc.) or immunohistochemistry (CTNNB1, LIN28A, etc.) that are required to cover the most important differential diagnoses have been shown to be difficult to standardize. Such diagnostic discordance and uncertainty may confound decision-making in clinical practice as well as the interpretation and validity of clinical trial results.
The cancer methylome is a combination of both somatically acquired DNA methylation changes and characteristics reflecting the cell of origin 5,6. The latter property allows, for example, tracing of the primary site of highly dedifferentiated metastases of cancers of unknown origin 7. It has been convincingly shown that DNA methylation profiling is highly robust and reproducible even from small samples and poor quality material 8, and such profiles have been widely used to subclassify CNS tumours that were previously considered homogeneous diseases 4,9–16. Based on this preliminary work within single entities, we herein present a comprehensive approach for DNA methylation-based classification of all CNS tumour entities across age groups.
CNS tumour reference cohort
To establish a comprehensive CNS tumour reference cohort, we generated genome-wide DNA methylation profiles (minimum of eight cases per group) representing almost all WHO defined neuroectodermal and sellar region tumours 1. We further profiled mesenchymal tumours, melanoma, diffuse large B-cell lymphoma, plasmacytoma and six types of pituitary adenomas, in total comprising 76 histopathological entities and seven entity variants occurring in the CNS. All histopathological entities and variants were analysed by unsupervised clustering both within each entity and across histologically similar tumour entities, aiming to identify (i) distinct DNA methylation classes within one histopathological entity and (ii) DNA methylation classes comprising tumours displaying a varied histological phenotype. This iterative process led to the designation of 82 CNS tumour classes characterised by distinct DNA methylation profiles (Figure 1a). Twenty-nine of these were equivalent to a single WHO entity (category 1), 29 represented subclasses within a WHO entity (category 2), in eight the WHO grading was not fully recapitulated (category 3) and in 11 the boundaries of methylation classes were not identical to the entity boundaries of WHO (category 4) (Figure 1a). The remaining five represented DNA methylation classes not defined by the WHO classification (category 5), three of which were recently described 4 as well as the not yet well-defined class of anaplastic pilocytic astrocytoma and one new subclass of infantile hemispheric glioma. There was evidence for several additional classes of rare tumours, with too few cases to be included at present. In consideration of the impact of the tumour microenvironment on the methylation profile, we included 47 tumour samples with a pronounced inflammatory or reactive tumour microenvironment, respectively, both demonstrating distinct methylation profiles. We additionally selected 72 samples representing seven non-neoplastic CNS regions, resulting in a combined reference cohort of 2,801 samples from 91 classes (Figure 1a) that was visualized using t-SNE dimensionality reduction 17 (Figure 1b). This analysis further supported the separation of samples into the defined DNA methylation classes (see also Extended Data Figure 1a, b; unprocessed .idat files can be downloaded at NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo), accession number GSE90496). Supplementary Table 1 gives an overview of methylation class characteristics and Supplementary Table 2 shows case-by-case information of the reference samples.
Figure 1 |. Establishing of the DNA methylation-based CNS tumour reference cohort.
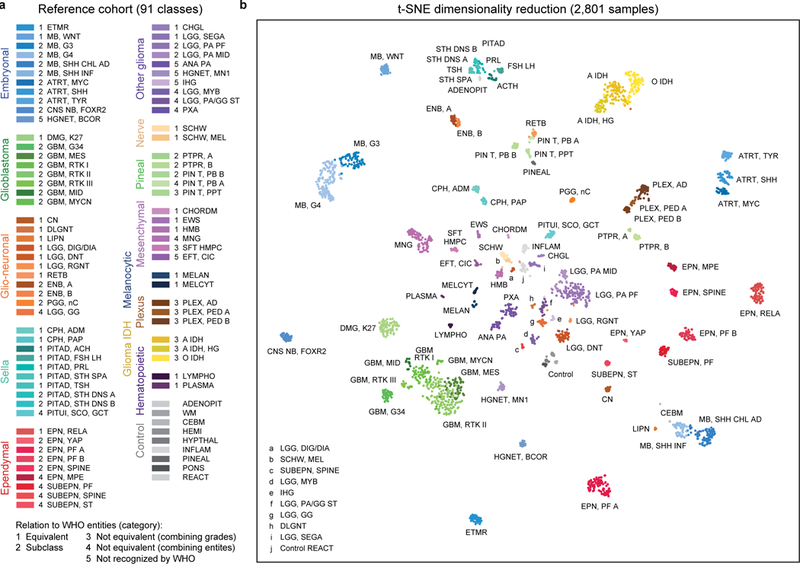
a, Overview of the 82 CNS tumour methylation classes and nine control tissue methylation classes of the reference cohort. The methylation classes are grouped by histology and color-coded. Category 1 methylation classes are equivalent to a WHO entity, category 2 methylation classes are a subgroup of a WHO entity, category 3 methylation classes are not equivalent to a unique WHO entity with combining of WHO grades, category 4 methylation classes are not equivalent to a unique WHO entity with combining of WHO entities, and category 5 methylation classes are not recognized as a WHO entity. Full names and further details of the abbreviated 91 classes are given in Supplementary Table 1. Embryonal tumours: shades of blue; Glioblastomas: shades of green; Other gliomas: shades of violet; Ependymomas: shades of red; Glio-neuronal tumours: shades of orange; IDH-mutated gliomas: shades of yellow; Choroid plexus tumours: shades of brown; Pineal region tumours: shades of mint green; Melanocytic tumours: shades of dark blue; Sellar region tumours: shades of cyan; Mesenchymal tumours: shades of pink; Nerve tumours: shades of beige; Haematopoietic tumours: shades of dark purple; Control tissues: shades of grey. b, Unsupervised clustering of reference cohort samples (n=2,801) using t-SNE dimensionality reduction. Individual samples are colour-coded in the respective class colour (n=91) and labelled with the class abbreviation. The colour code and abbreviations are identical to Figure 1a.
The stability of separation of methylation classes by t-SNE was analysed by iterative random downsampling of the reference cohort and indicated a high stability of the groups (Extended Data Figure 1c, d). Testing for confounding batch effects within our reference cohort did not reveal unexpected confounding factors (Extended Data Figure 2, Extended Data Figure 3a-c). For reference astrocytomas, oligodendrogliomas and glioblastomas we performed additional classification according to the TCGA pan glioma DNA methylation model18 indicating a strong association of the TCGA classes LGm1–6 with specific classes defined in our reference cohort (Extended Data Figure 3d, Supplementary Table 2).
Classifier development
Application in routine diagnostics requires fast and reproducible classification of samples as well as a measure of confidence for the specific call. To this end, we employed the Random Forest (RF) algorithm that is a so called ensemble method that combines the predictions of several ‘weak’ classifiers to achieve improved prediction accuracy19. Using this algorithm, we generated 10,000 binary decision trees, incorporating genome-wide information from all 2,801 reference samples and 91 methylation classes (Extended Data Figure 4). Each of these trees assigns a given diagnostic sample to one of the 91 classes, resulting in an aggregate raw score (Figure 2a). To obtain class probability estimates that can be used to guide diagnostic decision-making, we fitted a multinomial logistic regression calibration model that transforms the raw score into a probability that measures the confidence in the class assignment (‘calibrated score’). The calibration allows a comparison of classifier results between classes despite a different raw score distribution (Extended Data Figure 5a, b). Cross-validation of the RF classifier resulted in an estimated error rate of 4.89% for raw and 4.28% for calibrated scores and an area under receiver operating characteristic curve (AUC) of 0.99, indicating a high discriminating power (Figure 2b, Extended Data Figure 5c). The vast majority of cross-validation misclassifications occurred within eight groups of histologically and biologically closely related tumour classes, distinction of which is currently without clinical impact (with the possible exception of choroid plexus tumours 13; Figure 2b). We therefore defined eight ‘methylation class families’ (MCF), for which calibrated scores are summed up to a single score. This reduced the cross-validated error rate for the clinically relevant groupings to 1.14% (Figure 2b, Extended Data Figure 5c). Taking the maximum score for class assignment and using a multiclass approach 20, overall sensitivity and specificity was 0.989 and 0.999, respectively (Extended Data Figure 5c).
Figure 2 |. Development and cross-validation of the DNA methylation-based CNS tumour classifier.
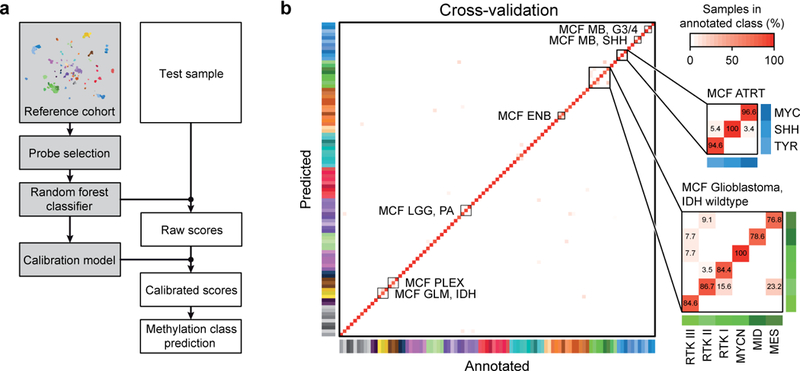
a, Schematic of principal classifier components (grey) and processing steps for individual test samples (white). The most informative probes are selected for training of the Random Forest classifier. The classifier produces raw scores representing the number of decision trees assigning a test sample to a specific methylation class. To enable inter-class-comparability a calibration model is used, which transforms raw into calibrated scores. Calibrated scores represent an estimated probability measure of methylation class assignment. b, Heatmap showing results of a three-fold cross-validation of the Random Forest classifier incorporating information of n=2801 biologically independent samples allotted to 91 methylation classes. Deviations from the bisecting line represent misclassification errors (using the maximum calibrated score for class prediction). Methylation class families (MCF) are indicated by black squares. The colour code and abbreviations are identical to Figure 1a.
For application to diagnostic tumour samples, a threshold value for the prediction of a matching class is required. Using Receiver Operating Characteristic (ROC) curve analysis of the maximum calibrated scores we devised an optimal “common” calibrated score threshold of ≥0.9 (Extended Data Figure 5d, e). For subclasses within methylation class families, we defined a threshold value of ≥0.5 as sufficient for a valid prediction, as long as all family member scores add up to a total score of ≥0.9. Single class specificity and sensitivity for the ≥0.9 threshold are provided in Supplementary Table 3.
Clinical implementation
For evaluation of clinical utility, we prospectively analysed a series of 1,155 diagnostic CNS tumours in parallel with standard histopathological workup (Figure 3a, b). For 51 cases (4%) the material was not suitable for methylation profiling, mostly because of too low tumour cell content or limited total material. Methylation profiling was performed for the remaining 1,104 samples and the cases were assigned as either ‘matching to a defined DNA methylation class’ (calibrated score ≥0.9) or as ‘no match’ cases (highest score <0.9) (for a case-by-case list see Supplementary Table 4). The investigated cases comprised 64 different histopathological entities from both adult (71%) and paediatric patients (29%). The spectrum of entities was enriched for rare and difficult to diagnose cases received for referral, and therefore did not exactly match the distribution seen in daily routine diagnostic practice. Histopathological evaluation was performed blinded to DNA methylation profiling results and included standard molecular testing.
Figure 3 |. Implementation of the classifier in diagnostic practice.
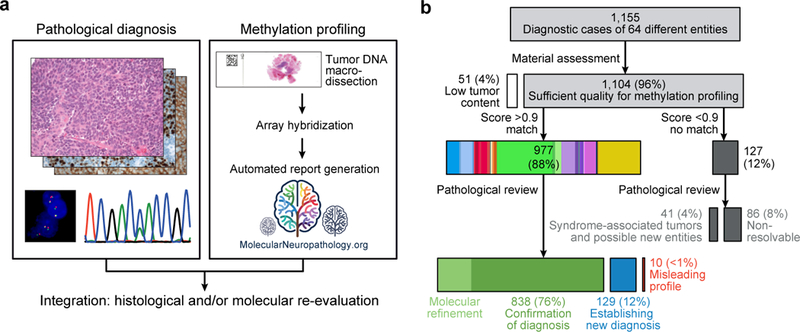
a, Classifier validation by an independent prospective cohort of diagnostic samples. Pathological diagnosis was established by current pathological standard according to the 2016 version of the WHO classification of CNS tumours and compared to classification by methylation profiling. Cases were categorized as “confirmation of diagnosis”, “establishing new diagnosis”, “misleading profile”, or “no match to defined class”. b, Overview of methylation profiling result from 1,155 diagnostic samples and integration with pathological diagnosis.
In total, 88% of profiled samples (n=977/1,104) matched to an established DNA methylation class with a calibrated classifier score ≥0.9 (Figure 3b). For 838 of these (838/1,104; 76%), results obtained by pathology and DNA methylation profiling were concordant. In 171 of the cases, an unambiguous molecular subgroup could be assigned, which would not have been available based on histopathology evaluation only (e.g., molecular subgroups of medulloblastoma and ependymoma, many of which were included in the latest version of the WHO classification of CNS tumours 1).
For the remaining 139 samples with a calibrated classifier score ≥0.9, the DNA methylation class was discordant from the pathological diagnosis. These cases were histologically and molecularly re-evaluated, including additional molecular diagnostics (DNA copy-number profiling, targeted gene sequencing, gene panel sequencing21, and gene fusion analysis of a subset of cases, see Supplementary Table 5). This resulted in a revision of the initial histopathological diagnosis in 129 of the 139 cases (12% of all cases, Figure 4) in favour of the predicted methylation class. In agreement with several recent reports 16,22,23, several of these were IDH-wildtype astrocytomas and anaplastic astrocytomas reclassified as IDH-wildtype glioblastomas. Establishing a new diagnosis had a profound clinical impact: a change in WHO grading was observed in 71% of these cases (92/129), with both upgrading (41%, 53/129) and downgrading (30%, 39/129; Figure 4). Discrepant results could not be resolved in only 10 cases (<1% of profiled cases), and the histopathological diagnosis was retained.
Figure 4 |. Reassessment of discrepant cases and establishment of new diagnosis.
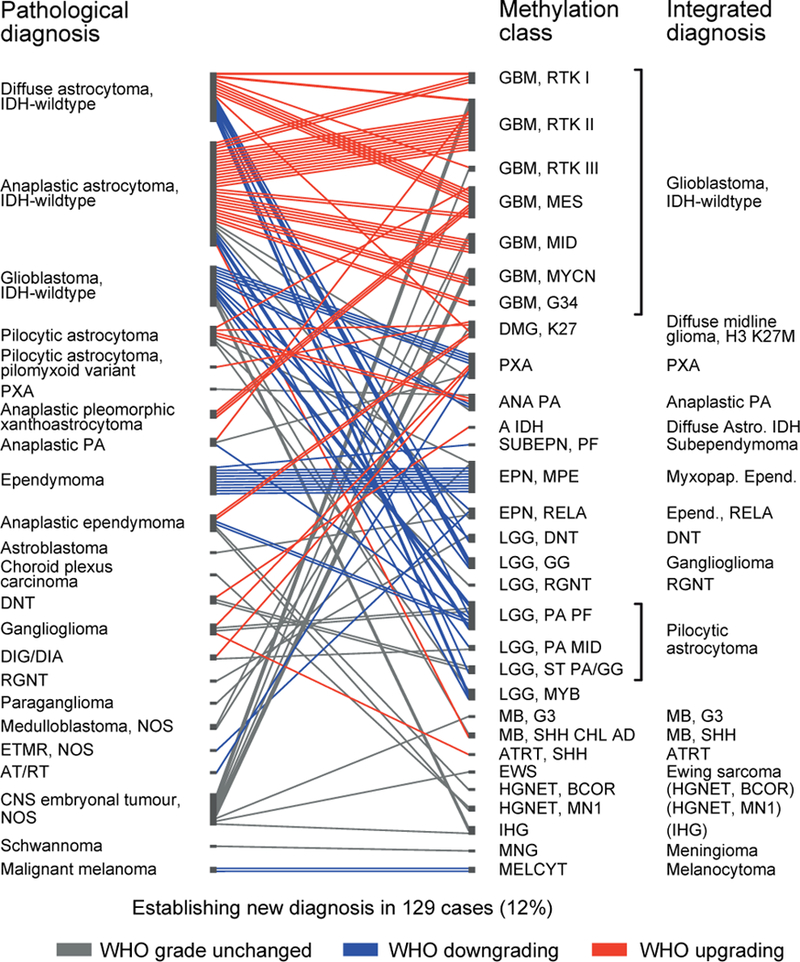
Discrepancy between pathological diagnosis (left) and methylation profiling (middle) was observed for 139 cases. For 129 cases histological and molecular reassessment (Supplementary Table 5) resulted in change of the initial diagnosis with formulation of a new integrated diagnosis (right). For 92 cases this involved change of WHO grading, with both down- (blue) and upgrading (red). Integrated diagnoses in brackets are not recognized as a WHO entity. For methylation class abbreviations see Supplementary Table 1.
To substantiate the impact in clinical practice we contacted five external centres that have started to implement methylation profiling for diagnostic cases using our algorithm. In total, these centres analysed 401 diagnostic cases and in 50 cases (12%) a new diagnosis was established after methylation profiling, very closely recapitulating our rate of reclassification (Extended Data Figure 6a, Supplementary Table 6). For individual centres the rate of reclassification varied between 6% and 25%, most likely due to differences of the spectrum of investigated cases and more upfront molecular testing by some centres (Extended Data Figure 6b, Supplementary Table 6).
Twelve percent of tumours from the prospective cohort (127/1,104) could not be assigned to a DNA methylation class using the rigid calibrated classifier score cutoff of ≥0.9 (Figure 3b). To further clarify the role of these non-classifiable cases we performed an unsupervised t-SNE analysis of the reference cohort together with the diagnostic cohort (Figure 5a). This demonstrated a high overlap of the classifiable cases with the reference cohort, whereas non-classifiable cases frequently fell in the periphery of the reference classes or even completely separate from these and frequently grouped with other non-classifiable cases (Figure 5a). This may indicate that such cases represent rare novel molecular entities that have not been previously recognized. An example for a likely novel CNS tumour entity is exemplified in Figure 5b, c.
Figure 5 |. DNA methylation-based identification of potential new CNS tumour entities.
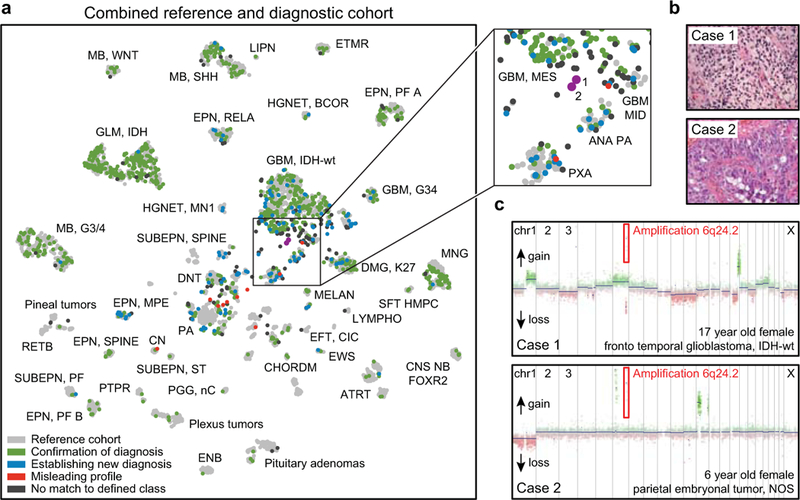
a, Unsupervised clustering of the combined reference (n=2,801, grey) and diagnostic cohort (n=1,104, coloured) using t-SNE dimensionality reduction. Abbreviated names indicate the reference cohort classes as in Figure 1. The diagnostic samples are colour coded as “confirmation of diagnosis” (n=838, green), “establishing new diagnosis” (n=129, blue), “misleading profile” (n=10, red) and “no match to defined class” (n=127, dark grey). The matching (green) and reclassified (blue) cases show high overlap with the reference cases. The non-classifiable (black) and the misleading (red) cases frequently fall in the periphery of the reference classes or are completely separate of these. The magnification (right) highlights two non-classifiable cases (here in magenta for easier identification) that group together in the t-SNE representation. b, Both highlighted non-classifiable cases occurred in female children, and had primitive neuroectodermal histology (glioblastoma- or embryonal tumour-like). Histology was assessed by three independent pathologists with similar results. c, Both cases shared a high-level amplification of chromosome 6q24.2 (common amplified region chr6:144,149,293–144,649,987). The common region includes only 5 protein coding genes: LTV1 (LTV1 ribosome biogenesis factor), ZC2HC1B (zinc finger C2HC-type containing 1B), PLAGL1 (PLAG1 like zinc finger 1), SF3B5 (splicing factor 3b subunit 5) and STX11 (syntaxin 11). This amplification was not observed in any of the other tumours from the reference or diagnostic cohort. Copy number analysis was performed once using copy number information deriving from the methylation array data.
Technical and inter-laboratory testing
Technical robustness of the RF classifier was investigated by inter-laboratory comparison. Results of two independent laboratories (starting from DNA extraction) were highly correlated, with only two of 53 samples (4%) showing a classifier score slightly lower than 0.9 in one of the centres whereas all other cases were classified identically (Extended Data Figure 7a). Calculation of copy number profiles was also stable across laboratories (Extended Data Figure 7b). To ascertain forward compatibility with developing technologies, we further used the RF classifier to interrogate newer EPIC DNA methylation arrays and high-coverage whole-genome bisulfite sequencing data. For all 16 samples from different CNS tumours profiled on both array platforms, raw scores (Extended Data Figure 7c) and calibrated scores (not shown) were highly correlated and running them through the classifying algorithm resulted in the same prediction for every case. Further, for all 50 high-coverage whole-genome bisulfite sequencing samples (11 different CNS tumour entities), the highest prediction score was for the same class as with the 450k array, suggesting that our approach is applicable to different DNA methylation profiling techniques with only slight adaptations (Extended Data Figure 7d).
Global dissemination of the platform
To ensure unrestricted community access to our classification system, we created a free web platform for data upload, automatic normalization, Random Forest classification, and PDF report generation (www.molecularneuropathology.org). DNA copy-number profiles24 and O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status25 are additionally provided, since they can be generated from the same data source – thus having the potential of replacing several time- and cost-intensive single-gene tests. A representative website report is shown in Extended Data Figure 8. During upload, the data provider can chose to give consent that the data may be used for further classifier development. We expect that this web platform can thereby act as a hub for a worldwide cooperative network to continuously identify and track rare tumour classes so that they can eventually be added to the catalogue of known human cancers. Since the launch of the website 14 months ago in December 2016, over 4,500 cases have been uploaded from over 15 participating centres. New biological insights are also likely to be gained based on the interrelationships of tumour classes, and by closer examination of how differential DNA methylation affects tumour biology.
Discussion
We here demonstrate that DNA methylation-based CNS tumour classification using a comprehensive machine learning approach is a valuable asset for clinical decision making. In particular, the high level of standardization has great promise to reduce the substantial inter-observer variability observed in current CNS tumor diagnostics. Further, in contrast to traditional pathology, whereby there is a pressure to assign all tumours to a described entity even for atypical or challenging cases, the objective measure that we provide here allows for ‘no match’ to a defined class. This information can also be of substantial value in highlighting that a tumour is not a typical example of a given differential diagnosis, and may rather belong to a rarer, yet undefined class. We defined 5 categories of methylation classes that have different clinical implications. Category 1 can be directly translated to WHO entities. Category 2 represents subclasses of WHO entities. For all but ependymal tumours, subclassification currently has little clinical consequence and a translation back to the WHO class may be appropriate for clinical purposes. Category 3 reflects the fact that WHO grading cannot be fully recapitulated by methylation profiling for several classes. Further data is required to assess if the methylation classes of this category may provide a more robust means of prognostication than histology alone, as has been demonstrated for several other classes 4,9,11. In category 4, the WHO entity boundaries are not identical to the boundaries of the methylation classes. Until additional data on the exact boundaries become available, this category should be critically discussed in the clinical context and orthogonal testing should be undertaken whenever possible. Category 5 represents putative new entities that are currently not recognized by the WHO, and while limited data on these cases is currently available, the biological rationale for a novel class was considered strong.
A study in which reference pathology and molecular diagnostics including DNA methylation profiling are blinded for each other´s results is currently ongoing for all childhood brain tumours diagnosed in Germany to objectivise the potential effect of re-classification on patient outcome (http://pediatric-neurooncology.dkfz.de/index.php/en/diagnostics/molecular-neuropathology), with results due over the next few years.
A uniform implementation of the classification algorithm holds great promise for standardization of tumour diagnostics across centres and across clinical trials. Further, the digital nature of methylation data facilitates easy exchange and will allow aggregation of extensive tumour libraries. This will likely result in the detection of exceptionally rare tumour classes and a continued refinement of classifiers. Inclusion of new classes will allow a prompt translation into diagnostic practice, almost certainly resulting in a more dynamic tumour classification. In our experience, adaptation of this technique in diagnostic laboratories is relatively straightforward. Extended Data Figure 9 summarizes a sample workflow for diagnostic implementation. We expect that the principle of using DNA methylation signatures as part of a combined histo-molecular tumour classification will improve diagnostic accuracy not only in neuropathology, but will serve as a blueprint in other fields of tumour pathology
Methods (online only)
Patient material
Patient material and clinical data of the retrospective reference cohort (total n=2,801) were obtained from the National Center for Tumour Diseases (NCT) in Heidelberg and supplemented with samples from additional centres (Supplementary Table 2) according to protocols approved by the institutional review boards with written consent obtained from each patient. Tumours were histopathologically re-assessed according to the current WHO classification1. Areas with highest tumour cell content (≥70%) were selected for DNA extraction. Subsets of the reference cohort have been previously published4,9–16,26–33. Additional patient characteristics are given in Supplementary Table 2. The prospectively assessed clinical cohort was analysed as part of the National Center for Tumour Diseases Precision Oncology Program according to procedures approved by the institutional review board at the Medical Faculty Heidelberg. All patients gave written consent for diagnostic procedures, comprising onward molecular testing including methylation profiling. Additional patient characteristics are given in Supplementary Table 4. Details of the online-analysed cohort of the five additional centres are given in Supplementary Table 6. Usage of the data was according to protocols approved by the institutional review boards of the University of Basel, Frankfurt am Main University Hospital, University Medical Center Utrecht and Princess Máxima Center for Pediatric Oncology Utrecht, Giessen University Hospital and University College London Hospitals. All patients gave written consent for diagnostic procedures, comprising onward molecular testing including methylation profiling. For all the above human research participants all relevant ethical regulations were followed.
Data generation, processing and Random Forest classifier generation
Samples were analysed using Illumina Infinium HumanMethylation450 BeadChip (450k) arrays according to the manufacturer’s instructions. To investigate stability across platforms a selection of samples were additionally assessed using the successor Methylation BeadChip (EPIC) array or whole-genome bisulfite sequencing (WGBS, generated and analysed as described6). Array data analysis was performed using R version 3.2.0 34, using a number of packages from Bioconductor35 and other repositories. A Random Forest19 classifier compatible with both 450k and EPIC platforms was trained, and a calibration model that calculates class probabilities from Random Forest scores was devised. A detailed description of all methods is provided below.
Methylation array processing
The 450k array was used to obtain genome-wide DNA methylation profiles for tumour samples and normal control tissues, according to the manufacturer’s instructions (Illumina, San Diego, USA). DNA methylation data was generated at the Genomics and Proteomics Core Facility of the DKFZ (Heidelberg, Germany) and the NYU Langone Medical Center (New York, USA). Data was generated from both fresh-frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples. For most fresh-frozen samples, >500 ng of DNA was used as input material. 250 ng of DNA was used for most FFPE tissues. On-chip quality metrics of all samples were carefully controlled. Copy-number variation (CNV) analysis from 450k methylation array data was performed using the conumee Bioconductor package version 1.3.0. Two sets of 50 control samples displaying a balanced copy-number profile from both male and female donors were used for normalization.
Raw signal intensities were obtained from IDAT-files using the minfi Bioconductor package version 1.14.0 36. Each sample was individually normalized by performing a background correction (shifting of the 5 % percentile of negative control probe intensities to 0) and a dye-bias correction (scaling of the mean of normalization control probe intensities to 10,000) for both colour channels. Subsequently, a correction for the type of material tissue (FFPE/frozen) was performed by fitting univariate, linear models to the log2-transformed intensity values (removeBatchEffect function, limma package version 3.24.15). The methylated and unmethylated signals were corrected individually. Estimated batch effects were also used to adjust diagnostic samples or test samples within the cross-validation. Beta-values were calculated from the retransformed intensities using an offset of 100 (as recommended by Illumina). To analyse for possible confounding batch effects within our pre-processed reference cohort dataset (after adjusting for FFPE versus frozen material) we applied the sva algorithm 37,38. We found no significant surrogate variable (data not shown).
The following filtering criteria were applied: Removal of probes targeting the X and Y chromosomes (n=11,551), removal of probes containing a single-nucleotide polymorphism (dbSNP132 Common) within five base pairs of and including the targeted CpG site (n=7,998), probes not mapping uniquely to the human reference genome (hg19) allowing for one mismatch (n=3,965), and probes not included on the Illumina EPIC array (n=32,260). In total, 428,799 probes targeting CpG sites were kept for further analysis.
Unsupervised analysis
Pairwise Pearson correlation was calculated for all 2,801 reference samples by selecting the 32,000 most variably methylated probes (s.d. > 0.228, Extended Data Figure 1a). The same probes were used for principal component analysis (PCA). For PCA, pairwise probe covariances of centred beta-values were calculated. Eigenvalue decomposition was performed using the eigs function of the RSpectra package version 0.12. The number of non-trivial components was determined by comparing eigenvalues to the maximum eigenvalue of a PCA using randomized beta-values (shuffling of sample labels per probe) (Extended Data Figure 1b). Principal component scores for all non-trivial components (n=94) were used for t-SNE analysis (t-Distributed Stochastic Neighbour Embedding17, Rtsne package version 0.11, Figure 1b). The following non-default parameters were used: theta=0, pca=F, max_iter=2500. A similar approach was used for the combined analysis of reference and diagnostic cases (Figure 5a).
The Random Forest algorithm
The Random Forest (RF) 19 algorithm is a so-called ensemble method that combines the predictions of several ‘weak’ classifiers to achieve improved prediction accuracy. The RF algorithm uses binary decision trees (Classification and Regression Trees, CART39) as ‘weak’ classifiers (Extended Data Fig. 4). Each of these trees is a sequence of binary splitting rules that are learned by recursive binary splitting. The CART algorithm starts with all samples assigned to a ‘root’ node and tries to find the variable, e.g., a measured CpG probe, and a corresponding cutoff that results in the purest split into the different classes. To measure this gain in class ‘purity’ the Gini index is used. To fit a tree, the CART algorithm iteratively repeats these steps until no further improvements can be made. To predict the class of a new diagnostic case the binary splitting rules are compared with the new data starting in the root node down to one of the leaf nodes. The tree then predicts or votes for the class of that leaf node. Decision trees have the advantage that they are non-parametric and do not rely on any distributional assumptions. The main disadvantages of decision trees is that they often tend to overfit the data and that they have a weak prediction performance. To improve the prediction accuracy the RF algorithm combines thousands of trees by bootstrap aggregation (bagging). In brief, each tree is fitted using training datasets that are generated by drawing bootstrap samples. In addition, at each node only a random subset of the available variables is used to find an optimal splitting rule. This additional source of randomization allows selecting variables with lower predictive value. This feature guarantees that the resulting trees are decorrelated, i.e., they use different variables to find an optimal prediction rule. Taking the majority vote over thousands of bootstrap aggregated and decorrelated trees greatly improves the prediction accuracy of the RF. The majority vote, i.e., the proportion of trees voting for a class, can be interpreted as empirical class probabilities.
Classifier development
To train the RF classifier, the randomForest R package 40 was used. First, the most important features (probes) were selected by applying the Random Forest algorithm to the beta-values of all filtered 428,799 probes. For efficient computation, the probes were split into 43 sets of approximately 10,000 probes. For each set, 100 trees were fitted using 654 randomly sampled candidate features at each split (mtry parameter, square root of 428,799, as would be used by default when not splitting into sets). To take the imbalanced methylation class sizes into account a downsampling strategy was followed that ensures an identical number of samples per class (parameter sampsize=rep(8, 91)), eight reflecting the minimum number of cases in the 91 classes) 41. For all other parameters the default settings were used. This procedure was repeated 100 times, essentially fitting 10,000 trees per probe. Finally, features are selected by the permutation-based variable importance measure as implemented in the randomForest R package40. The importance measure is the class-specific mean decrease in classification accuracy when the feature is permutated. We select features by ranking them using the minimal rank of the variable importance measures across all classes.
The final RF classifier was trained by fitting 10,000 trees with the parameter mtry=100 using beta-values of the 10,000 probes selected during feature selection. Imbalanced class sizes were accounted for by downsampling (as described above), and for all other parameters the default settings were used. An overview of the processes is given in Extended Data Fig. 4.
Classifier cross-validation
Overfitting of the training data is a typical problem expected when training classifiers on high-dimensional data. As it often cannot be avoided, the typical strategy to deal with this problem is to evaluate the model accuracy on an independent test dataset or apply cross-validation methods42. Because some of the newly defined methylation groups presented in this work cannot be diagnosed by classical histopathological methods or other established molecular assays, an independent test set to assess model accuracy is not available. Therefore, the accuracy of the presented RF model with the accompanying calibration model was evaluated by a three-fold, nested cross-validation (CV). For this, the reference dataset is split into three equally sized parts. In each CV iteration, two-thirds of the data were used to train a RF classifier in the same way as the RF classifier for the complete dataset was trained. Then, the remaining one-third of the data is predicted using this RF classifier. After the third iteration of the CV is completed, each of the 2,801 reference samples has been predicted by an independent RF classifier, i.e. where the sample was not used for estimating batch effects, performing variable selection, or training of the classifier.
Classifier score calibration
The classification scores generated by our multiclass RF (i.e. the proportion of trees voting for a class) perform well when they are used to assign the correct class labels, but they do not reflect class probabilities. Furthermore, the distribution of the RF scores varies between classes, which makes an inter-class comparison difficult. Moreover, to evaluate a diagnostic classification, the uncertainties associated with an individual prediction in terms of confidence scores or estimated class probabilities are needed.
To obtain scores that are comparable between classes and that are improved estimates of the certainty of individual predictions we performed a classification score recalibration by mapping the original scores to more accurate class probabilities43,44. To find such a mapping, a L2-penalized, multinomial, logistic regression-model was fitted, which takes the methylation class as response variable and the RF scores as explanatory variables. The R package glmnet45 was used to fit this model. In addition, the model was fitted by incorporating a small ridge-penalty (L2) on the likelihood to prevent from over fitting, as well as to stabilize estimation in situations where classes are perfectly separable. The amount of this regularization, i.e. the penalization parameter, is determined by running a ten-fold cross-validation and choosing the largest value that lies within one standard error of the minimum cross-validation error. Independent RF scores are needed to fit this model, i.e. the scores need to be generated by a RF classifier that was not trained using the same samples, otherwise the RF scores will be systematically biased and not comparable to scores of unseen cases. As such, RF scores generated by the three-fold CV are used.
To validate the class predictions generated by using the recalibrated scores of the calibration model, a nested three-fold CV loop is incorporated into the main three-fold CV that validates the RF classifier (Extended Data Fig. 4). Within each CV run this nested three-fold CV is applied to generate independent RF scores, which are then used to train a calibration model. The predicted RF scores resulting from predicting the one-third test data of the outer CV loop are then recalibrated by applying the calibration model that was fitted on the RF scores generated during the nested CV. A similar CV scheme was used by Appel et al.46 to validate estimated classification probabilities.
Classifier performance measures
Performances of the resulting classifier predictions and scores generated by the CV were assessed by the misclassification error, multiclass area under the curve (AUC) and the multiclass Brier score. The misclassification error measures the frequency of falsely assigned class labels when using the maximum of the RF scores or re-calibrated scores as a cutoff to determine the predicted class, i.e. the majority vote. To measure the AUC for our multiclass RF the generalization of the AUC for multiclass classification problems by Hand and Till47 was used. To measure how well the resulting RF scores and recalibrated scores perform when used as class probabilities, the multiclass Brier Score42,48,49 was used. The Brier score is the mean-squared difference between the actual and the predicted class probability and thus measures the same characteristic as the mean squared error (MSE) measures for a continuous forecast.
Methylation class families
We observed that the majority of misclassification errors occurred within eight groups of histologically and biologically closely related tumour classes. We therefore defined eight ‘methylation class families’ (MCF). Since calibrated scores represent class probabilities, it is possible to apply the addition rule of probabilities to sum up calibrated class scores within one MCF to get a class probability for the MCF.
Threshold analysis
Finding an optimal cutoff for diagnostic tests usually involves finding an optimal trade-off between sensitivity and specificity. If there are no preferences regarding specificity or sensitivity, the optimal cutoff is chosen by the upper left corner of the ROC curve or by maximizing the Youden index (specificity+sensitivty-1). In an application like the one described here, where the cost of false negative is that a tumour cannot be classified and the cost of a false positive is a falsely predicted methylation class, a threshold with high specificity is preferred. ROC analysis is typically defined for binary classification problems. Finding a threshold for multiclass classifiers either involves performing a ROC analysis for each class resulting in class-wise individual thresholds or finding some common threshold for all classes.
The calibrated MC/MCF scores (here referring to MCF and MC classes that are not assigned to a MCF) are already validated probability estimates for the methylation class with a direct interpretation, i.e. we expect among all samples with scores of approx. 0.9 that 10% are falsely predicted. Applying an additional threshold is not required from a statistical point of view, but desired in clinical practice. In addition, due to calibration, scores are comparable across classes and it is thus reasonable to define a common threshold for all classes instead of finding optimal cutoff for each individual methylation class.
To determine a common threshold for the calibrated MC/MCF scores, we performed a ROC analysis of the maximum calibrated MC/MCF scores calculated via cross-validation. For this ROC analysis we defined a new binary class, i.e. samples correctly classified during the CV using the maximum calibrated MC/MCF score for classification were considered as ‘classifiable’ and samples falsely classified by using this score were considered ‘non-classifiable’.
Following this ROC analysis approach, we determined a cutoff of 0.836 that maximises the Youden index with a specificity of 93.8% and sensitivity of 93.4% (Extended Data 5d and e). A maximum specificity of 100% with a sensitivity of 82.7% can be achieved with a threshold of 0.958. Bootstrapped 95% confidence intervals (grey area in Extended Data Figure 5d) demonstrate the uncertainty of sensitivity and specificity estimates, especially in the left upper corner of the ROC figure, where the considered thresholds are located.
Both thresholds have been determined by cross-validation on our training data of high quality, but real life diagnostic samples were found to achieve slightly lower scores, due to a number of factors we cannot control, such as lower overall sample quality and lower tumour purity compared to samples in our reference cohort. Therefore, we decided to lower the maximum specificity threshold to allow a wider spectrum of samples to become a match. For this, we chose a threshold of ≥0.9 that lies in the middle between the Youden index and the threshold for maximum specificity.
Comparison to TCGA pan-glioma methylation classes
To compare our methylation-based classification of CNS tumours with described methylation classes of brain tumours by the Cancer Genome Atlas (TCGA) project, we downloaded the pre-processed methylation dataset described in Ceccarelli et al. 201618 including methylation data of 418 low grade glioma and 377 glioblastoma samples analysed by using the Illumina 450k array or 27k array platforms. To classify our samples according to the TCGA pan-glioma DNA methylation classification, we trained a Random Forest classifier on this dataset using the 1,300 CpG probe signature provided by the authors and using the default settings of the Random Forest algorithms implemented in the R package randomForest. The results of this classification for astrocytomas, oligodendrogliomas and glioblastomas are shown in Extended Data Figure 3d and are given on a case-by-case basis in Supplementary Table 2 and 4.
Estimating tumour purity from DNA methylation data
Due to the subjective nature of histological assessment of tumour purity, we additionally used the Ceccarelli et al. 2016 dataset18 to train a Random Forest regression (continuous response variable) model to predict tumour purity50. This Random Forest was trained on the 1,000 most important CpG probes for purity estimation selected also by a Random Forest (similar to the variable selection described for the Random Forest classifier). The out-of-bag (i.e. RF trees in which the respective sample, for which purity is predicted, was not used for training) mean squared error of the final model is 0.015, indicating that this model is able to yield reasonable predictions of tumour purity from methylation data (Extended Data Figure 3a-c). The estimated tumour purity for individual cases is given in Supplementary Table 2 and 4.
Extended Data
Extended Data Figure 1 |. Unsupervised clustering of the DNA methylation-based reference cohort.
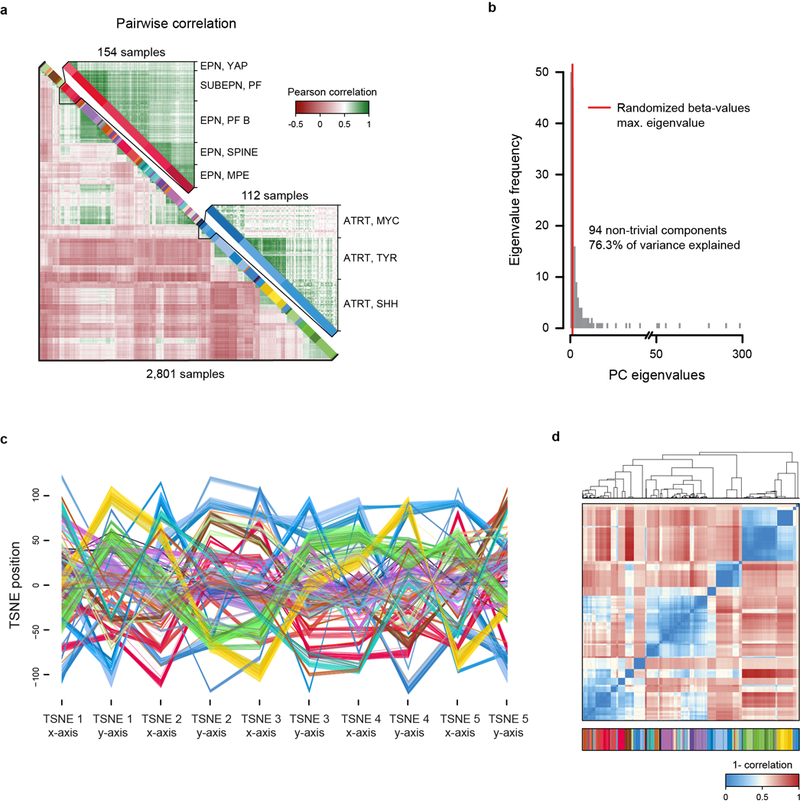
a, Heatmap showing the pairwise Pearson correlation (lower left) of the 32,000 most variably methylated CpG probes of all 2,801 biologically independent samples of the reference cohort. A detailed view on closely related ependymal classes (upper right) and the three subclasses identified in ATRT tumours (lower right) indicates higher correlation within classes. The colour code and abbreviations are identical to main Figure 1a. b, Barplot showing eigenvalue frequencies of a principal component analysis (PCA) using the same 32,000 most variably methylated CpG probes of all 2,801 biologically independent samples as in (a). The number of non-trivial components were determined by comparing eigenvalues to the maximum eigenvalue of a PCA using randomized beta-values (shuffling of sample labels per probe). c, X and Y coordinates of the first five of a total of 500 iterations of t-SNE dimensionality reduction generated by random downsampling to 90% of the 2,801 biologically independent samples to assess clustering stability. Axis positions of individual cases are connected by a line coloured according to the colour code of Figure 1a. The depiction illustrates the close proximity of cases of the same class across iterations, indicative of a high stability independent of the exact composition of the reference cohort. d, Pairwise correlation of X and Y coordinates between 2,801 biologically independent samples over all iterations of the downsampling analysis demonstrates a very high correlation within classes (average correlation 0.982), indicating a high stability of the t-SNE analysis.
Extended Data Figure 2 |. Unsupervised clustering is not biased by a range of possible confounding factors.
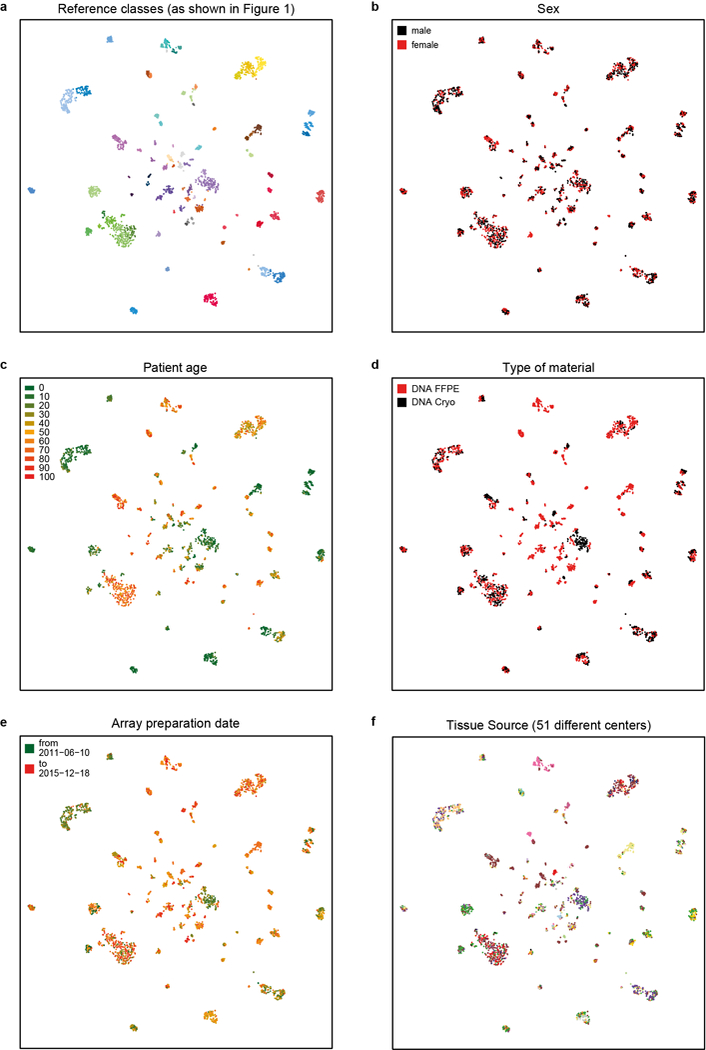
a, t-SNE representations of the 2,801 biologically independent samples constituting the reference cohort as shown in Figure 1b overlaid with potentially confounding factors (b-f). b, Distribution of patient sex among the classes illustrates equal or near equal distribution of many classes, but also an expected enrichment for one sex in some classes (e.g. female in meningioma or CNS high-grade neuroepithelial tumour with MN1 alteration). c, Patient age illustrates the expected age distribution of many tumour classes. d-f, The slightly uneven distribution of type of material (e.g. pilocytic astrocytoma or meningioma) (d), array preparation date (e), and tissue source (f) are related to the specifics of assembling the reference cohort and do not indicate an apparent confounding effect on the unsupervised clustering.
Extended Data Figure 3 |. Estimation of tumour purity and relation to TCGA pan-glioma methylation classes.
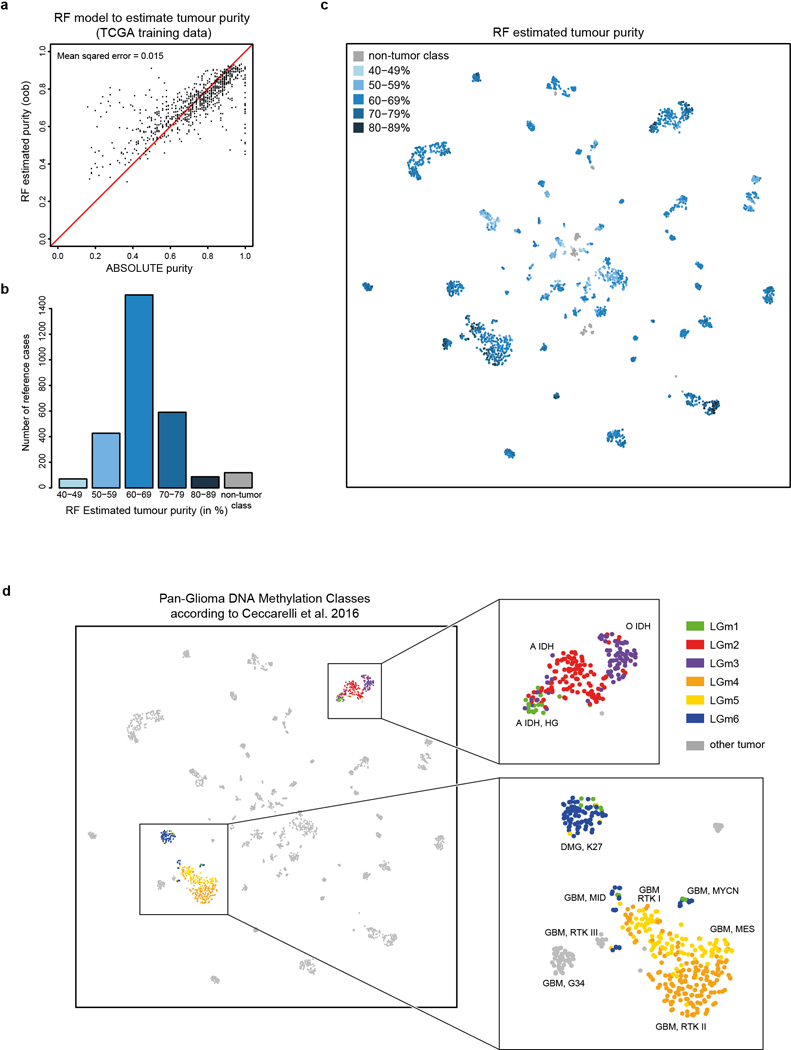
a, A Random Forest model was trained to predict ABSOLUTE tumour purity estimates50 using the TCGA pan-glioma dataset (795 biologically independent samples)18. The plot shows ABSOLUTE purity estimates and out-of-bag Random Forest tumour purity predictions (i.e. using only RF trees for which the respective sample was not involved in the training). The estimated mean squared error is 0.015, indicating that this model is able to yield reasonable predictions of tumour purity from methylation data. b, Bar plot showing the distribution of Random Forest predicted purity in the reference dataset (2,801 biologically independent samples). Purity estimates have been transformed into five categories indicated by different shades of blue. The exact case-by-case values are given in Supplementary Table 2. The median estimated purity in the reference cohort is 66% (range 42% to 87%) and 78% of samples have an estimated purity of at least 60%. c, t-SNE representation of the reference cohort (2,801 biologically independent samples) overlayed with Random Forest predicted purity categories. Methylation classes are generally composed of mixed tumour purity categories. Tumour purity shows some association with the WHO grade (WHO I median tumour purity 60%, range 39–77%; WHO II median 66%, range 43–80%; WHO III median 68% range 54–84%; WHO IV median 69% range 49–87%). A further association of tumour purity with the composition of classes in the unsupervised t-SNE analysis was not evident. d, t-SNE representation of the reference cohort (2,801 biologically independent samples) overlayed with predicted TCGA pan-glioma DNA methylation classes according to Ceccarelli et al. 2016. Pan-glioma methylation classes were predicted by training a Random Forest (RF) on the Ceccarelli et al. 2016 dataset including methylation data of 418 low grade glioma and 377 glioblastoma samples acquired using the Illumina 450k and 27k platforms. The RF was trained using the 1,300 CpG signature as described by the authors18 and using the default settings of the RF algorithm implemented in the R package randomForest. Pan-glioma class prediction was only performed for subsets of mostly adult astrocytomas, oligodendrogliomas and glioblastomas (magnified areas) included in the Ceccarelli et al. 2016 data set. LGm1, LGm2 and LGm3 show a high overlap with the methylation classes A IDH HG, A IDH and O IDH, respectively. LGm4 shows highest overlap with methylation class GBM RTK II. LGm5 shows highest overlap with methylation classes GBM MES and GBM RTK I. LGm6 show highest overlap with DMG K27, GBM MID and GBM MYCN.
Extended Data Figure 4 |. Development of the Random Forest classifier.
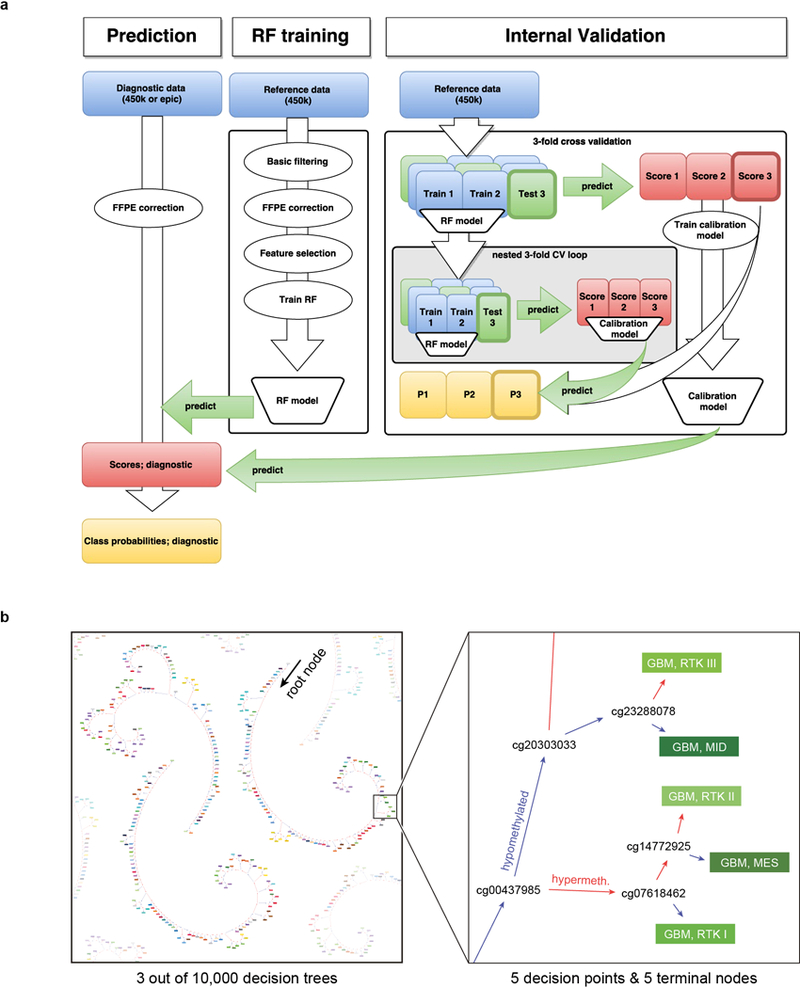
a, The RF training consists of four steps. First, a basic filtering for probes that are not included on the EPIC array, probes located on the X and Y- chromosomes, probes affected by SNPs, and probes not mapping uniquely to the genome is performed. In a second step, the probe-wise batch effects between samples from FFPE and frozen material are estimated and adjusted by a linear model approach. In a third step, feature selection is performed by training a RF using all probes and selecting the 10,000 probes with highest variable importance measure. In a last step, the final RF is trained using only the 10,000 selected probes. The validation of the RF classifier involves a three-fold nested cross-validation (CV). In the outer loop of the CV the complete RF training procedure described before is applied to the training data and the resulting RF is used to predict the test data to generate RF scores. In the inner loop of the CV a three-fold CV is applied to training data of the outer loop in order to generate RF scores independent of the test data in the outer loop. These scores are then used to fit a calibration model, i.e. a L2-penalized, multinomial, logistic regression that takes the RF scores of the test data in the outer CV loop to estimate tumour class probabilities (P1, P2, P3). To fit a calibration model to estimate class probabilities of diagnostic samples using all data in the reference set, the RF scores generated in the outer CV loop are used. b, Schematic depiction of three exemplary binary decision trees of the Random Forest classifier (left), and magnification on five exemplary decisions nodes relevant for glioblastoma classification (right). For prediction, a diagnostic sample enters the root node of each of the 10,000 trees. At every decision node, the decision path is determined on the methylation level of a single CpG, until reaching a terminal node that provides the class prediction. The joint class prediction of all trees represents the raw prediction score. The colour code and abbreviations are identical to Figure 1a.
Extended Data Figure 5 |. Comparison of raw and calibrated classifier scores and threshold definition.
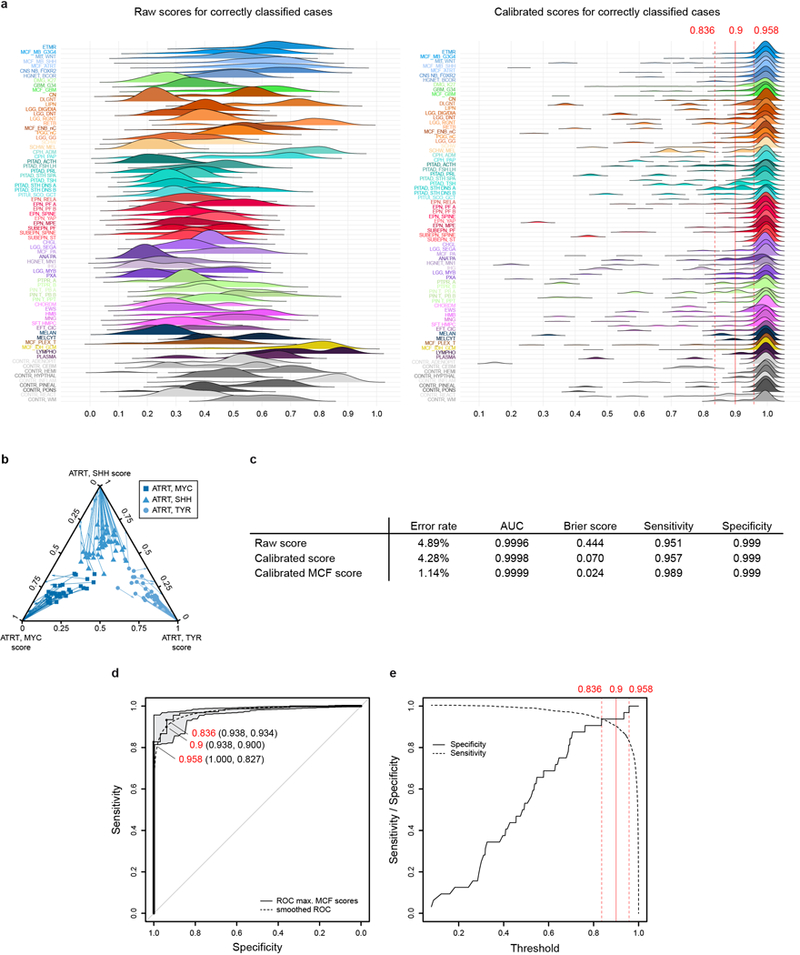
a, Density plots illustrating the distribution of raw and calibrated classifier scores for samples correctly classified during cross-validation (n=2,701 independent biological samples for raw and n=2769 independent biological samples for calibrated), depicted for each methylation class or methylation class family (MCF). Score calibration results in a harmonization of score distribution and allows the establishment of a shared classification threshold. Three thresholds for maximizing specificity (0.958), maximizing the Youden index (0.836), and the cutoff used in this study (0.9) are indicated by red lines (see also panels d and e). b, Multivariate score calibration exemplified in a ternary plot showing scores of the three ATRT subclasses (MYC, SHH, and TYR; together n=112 independent biological samples). Arrows indicate transformation of the scores for individual samples by the calibration model, which increases the discrimination between the three subclasses. c, The accuracy of prediction of the Random Forest classifier constructed of n=2801 biologically independent samples (measured by misclassification error, area under receiver operating characteristic curve (AUC), Brier score, multiclass Sensitivity and Specificity) is improved by score calibration and by combining classes into methylation class families (MCF). d, To determine a common threshold for the calibrated MCF scores, we performed a Receiver Operating Characteristic (ROC) analysis of the maximum calibrated MCF scores of all n=2801 biologically independent samples calculated via cross-validation. For this ROC analysis we defined a new binary class, i.e. samples correctly classified during the CV using the maximum calibrated MCF score for classification were considered as ‘classifiable’ (n=2769) and samples that got falsely classified by using this score were considered ‘non classifiable’ (n=32). Three thresholds for different sensitivity and specificity are highlighted in the ROC curve: A threshold of 0.958 achieving a maximum specificity of 1 with a sensitivity of 0.827, a threshold of 0.836 obtaining a maximum Youden index with Specificity 0.938 and sensitivity 0.934, and our recommended compromise threshold of 0.9 that results in a specificity of 0.938 and a sensitivity of 0.9. Bootstrapped 95% confidence intervals for estimated sensitivity and specificity are indicated in grey. e, Sensitivity and specificity for all possible thresholds applied to cross-validated maximum MCF classifier scores of all n=2801 biologically independent samples. Three thresholds for maximizing specificity (0.958), maximizing the Youden index (0.836) and 0.9 are highlighted by red lines.
Extended Data Figure 6 |. Diagnostic utility of the DNA-methylation based classifier, assessed at different centres.
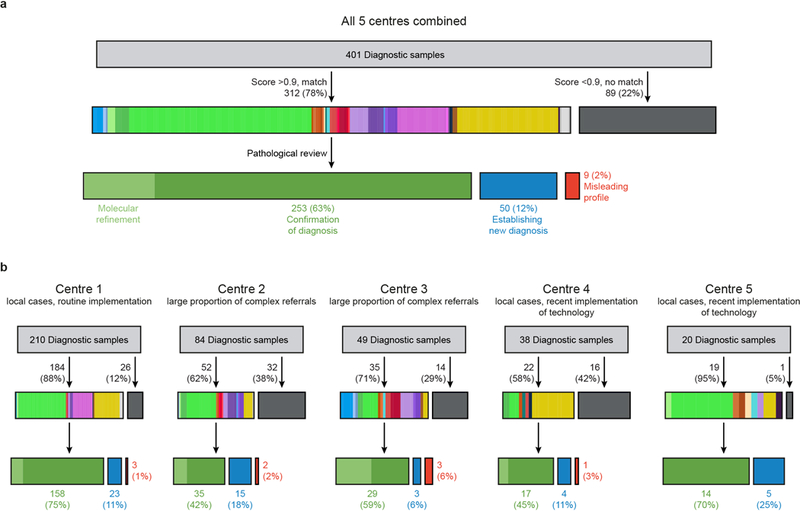
a, Implementation of the DNA methylation classifier by five external centres. In total, 401 independent biological samples were analysed. 78% matched to an established class with a cut-off score of ≥0.9 (class colours as in Figure 1a). A new diagnosis was established in 12% of cases. b, Depiction of individual centre results, illustrating the different composition of samples included in the analysis, variation in the rate of non-matching cases, and of cases where a new diagnosis was established. Case-by-case details are given in Supplementary Table 6.
Extended Data Figure 7 |. Inter-centre and inter-platform reproducibility of DNA methylation-based classification.
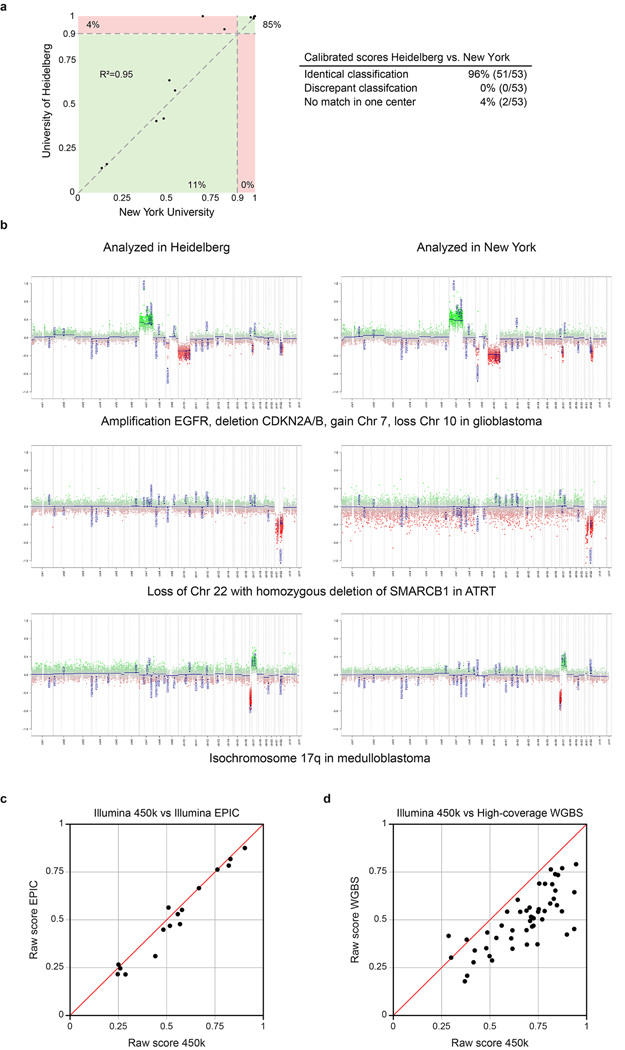
a, Calibrated scores of 53 independent biological samples representing diagnostic CNS tumour cases analysed at the University of Heidelberg and at the New York University pathology department. Both laboratories performed independent DNA extraction, array hybridization, and data analysis. Cases falling into green areas were classified identically in both centres (96%); cases in the red area were non-classifiable in one centre (4%). None of the 53 samples was assigned to a different methylation class by the two centres. b, Copy-number profiles calculated from the array data generated at both centres were highly comparable and allowed identification of chromosomal gains, losses, amplifications, and deletions. Calculations and interpretation were performed once at each centre. c, Plot of maximum raw classification scores of 16 different tumour samples generated using both 450k and EPIC arrays. All cases fall close to the bisecting line (red) indicating a high concordance of the scores. Further, the methylation class prediction was identical for all samples. d, The CNS tumour classifier also performs well with data generated by whole-genome bisulfite sequencing (WGBS). The plot shows classifier scores calculated from WGBS and 450k arrays of 50 cases comprising 11 different brain tumour entities (bisecting line in red). Methylation beta-values were calculated from high-coverage WGBS data (>10 fold average coverage) and run through the CNS tumour classifier and plotted against the same case analysed using 450k arrays. The highest class prediction score was identical in all cases.
Extended Data Figure 8 |.
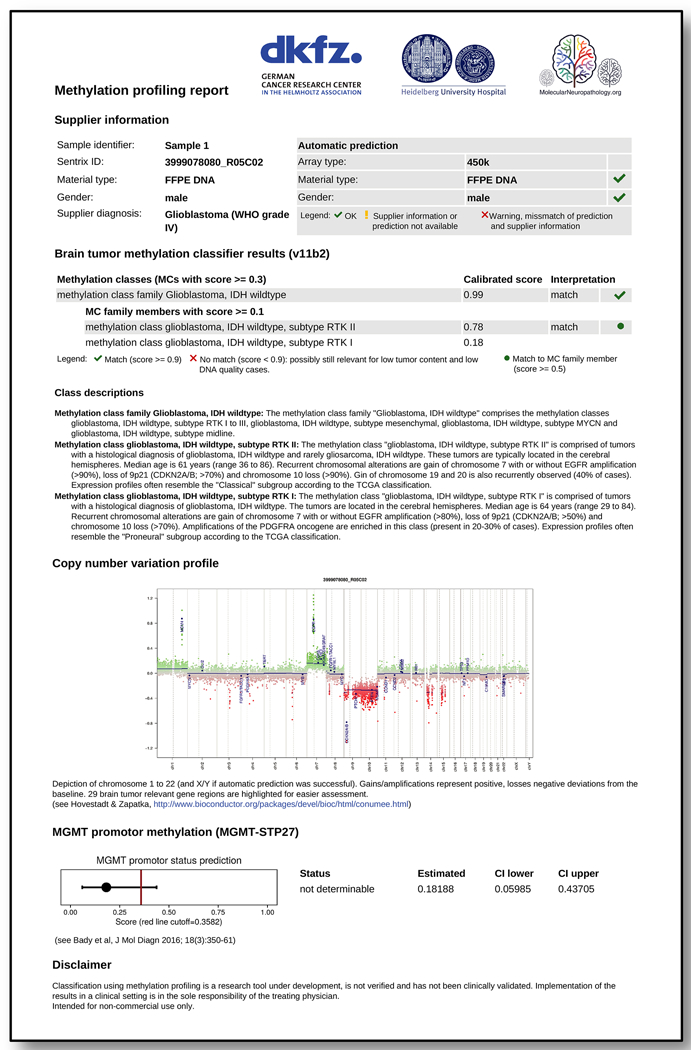
Sample website PDF report of a IDH wildtype glioblastoma sample.
Extended Data Figure 9 |.
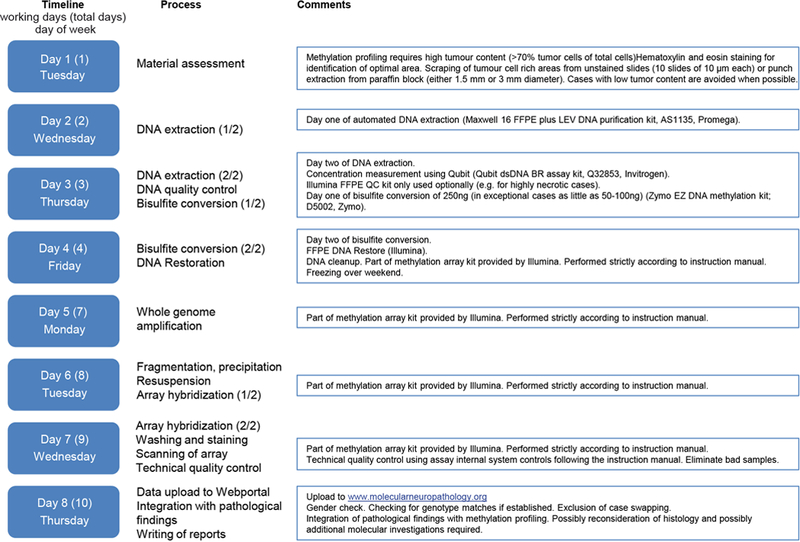
Exemplary workflow and timeline of diagnostic methylation profiling.
Supplementary Material
Acknowledgments
We thank U. Lass, A. Habel, I Oezen for technical and administrative support, the Microarray unit of the Genomics and Proteomics Core Facility (DKFZ) for methylation services, the German Glioma Network and the Neuroonkologische Arbeitsgemeinschaft for data sharing. This research was supported by the DKFZ-Heidelberg Center for Personalized Oncology (DKFZ-HIPO_036), the German Childhood Cancer Foundation (DKS 2015.01), an Illumina Medical Research Grant, the DKTK joint funding project ‘Next Generation Molecular Diagnostics of Malignant Gliomas’, the A Kids’ Brain Tumour Cure (PLGA) Foundation, the Brain Tumour Charity (UK) for the Everest Centre for Paediatric Low-Grade Brain Tumour Research, the Friedberg Charitable Foundation and the Sohn Conference Foundation (to M. Snuderl and M. Karajannis), the RKA-Förderpool (Project 37) and Stichting Kinderen Kankervrij and Stichting AMC Foundation (to E. Aronica), NIH/NCI 5T32CA163185 (to A. Olar), NIH/NCI Cancer Center Support Grant P30 CA008748 to MSKCC, the Luxembourg National Research Fond (FNR PEARL P16/BM/11192868 to M. Mittelbronn) and the National Institute of Health Research (NIHR) UCLH/UCL Biomedical Research Centre (S. Brandner).
Footnotes
Code availability
The generated code is available from the corresponding author (S.M.P.) on reasonable request for non-commercial use.
Data Availability
The complete methylation values required for the construction of the classifier (reference set) as well as the prospective cohort (validation set) have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). The accession number is GSE109381. Supplementary Table 2 (reference cohort) and Supplementary Table 4 (prospective validation cohort) includes the IDAT-file names for assignment to patient characteristics. Source data for Figure 1b, 2b, 3b, 4, 5a,c and Extended Data Figure 1c, 2a-f, 3a-d, 5a,b,d,e, 6 and 7a,c,d are provided with the paper.
Author Contributions D.C. and D.T.W.J. composed the reference cohort and defined methylation classes; M.Sill and V.Hovestadt developed and technically validated the classifying algorithm; All four authors contributed equally to this study; D.Schrimpf developed the classification website; D.C., D.T.W.J., M.Sill, A.Benner, V.Hovestadt, D.Schrimpf, D.Stichel, M.Z., A.v.D., S.M.P developed additional methodology and software; D.C., D.T.W.J., M.Sill, D.Sturm, C.Koelsche, F.Sahm, L.C., D.E.R., A.Kratz, A.K.W., K.H., L.S., P.N.H., K.H.P., J.Schittenhelm, G.R., M.Prinz, W.B., F.Selt, H.Witt, T.M., O.W., S.Brehmer, M.Seiz-Rosenhagen, D.H., A.Kulozik, C.M.K., H.L.M., S.R., K.v.H., M.C.F., A.Gnekow, G.F., S.T., G.C., C.Monoranu, M.G., T.P., M.Bendszus, J.D., M.Platten, A.U., W.W., M.M., C.Hartmann, C.Herold-Mende, M.H., A.Korshunov, A.v.D., S.M.P. performed the prospective cohort analysis; P.N.H., K.H.P., H.D., B.K.G., J.H., S.F., P.W., Z.J., T.A., S.Brandner generated and collected the external centre data; K.W.P., A.O., N.W.E., A.K.B., R.C., A.Hölsken, E.H., R.Beschorner, J.Schittenhelm, O.S., K.W., K.W., V.P., M.Pages, P.T., D.L., E.A., F.G., E.R., W.S., C.G., F.J.R., A.Becker, M.Preusser, C.Haberler, R.Bjerkvig, J.C., M.F., M.D., S.Hofer, V.Hans, S.Heim, J.R.H., P.K., B.W.K., M.L., B.L., C.Mawrin, R.K., Z.K., F.H., A.Koch, A.Jouvet, C.Keohane, H.Mühleisen, W.M., U.P., M.Prinz, N.G., P.H., A.P., C.J., T.S.J., B.R., T.P., J.Schramm, G.S., M.Westphal, G.R., P.W., M.Weller, V.P.C., I.B., A.Huang, N.J., P.A.N., W.P., A.Gajjar, G.W.R., M.D.T., M.R., M.Karajannis, M.M., C.Hartmann, K.A., U.S., R.Buslei, P.L., M.Kool, C.Herold-Mende, D.W.E., M.H., S.Brandner, A.Korshunov, A.v.D., S.M.P. provided reference cohort material and data; K.L., M.Bewerunge-Hudler, M.Schick, R.F. performed methylation profiling; J.Serrano, K.K., A.T., M.Karajannis, M.Snuderl performed technical validation experiments; A.v.D. and S.M.P. supervised the project. The manuscript underwent an internal collaboration-wide review process. All authors approved the final version of the manuscript.
References
- 1.Louis DN, Ohgaki H, Wiestler OD & Cavenee WK WHO Classification of Tumours of the Central Nervous System (revised 4th edition). (IARC, 2016). [Google Scholar]
- 2.van den Bent MJ Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol . 120, 297–304, doi: 10.1007/s00401-010-0725-7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellison DW et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed 10, 7, doi: 10.1186/1477-5751-10-7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164, 1060–1072, doi: 10.1016/j.cell.2016.01.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez AF et al. A DNA methylation fingerprint of 1628 human samples. Genome Res . 22, 407–419, doi: 10.1101/gr.119867.110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hovestadt V et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541, doi: 10.1038/nature13268 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Moran S et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol . 17, 1386–1395, doi: 10.1016/S1470-2045(16)30297-2 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Hovestadt V et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol . 125, 913–916, doi: 10.1007/s00401-013-1126-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturm D et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437, doi: 10.1016/j.ccr.2012.08.024 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Reuss DE et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol . 130, 407–417, doi: 10.1007/s00401-015-1454-8 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Pajtler KW et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27, 728–743, doi: 10.1016/j.ccell.2015.04.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert SR et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol . 126, 291–301, doi: 10.1007/s00401-013-1124-7 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Thomas C et al. Methylation profiling of choroid plexus tumors reveals 3 clinically distinct subgroups. Neuro Oncol . 18, 790–796, doi: 10.1093/neuonc/nov322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mack SC et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450, doi: 10.1038/nature13108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johann PD et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 29, 379–393, doi: 10.1016/j.ccell.2016.02.001 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Wiestler B et al. Integrated DNA methylation and copy-number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathol . 128, 561–571, doi: 10.1007/s00401-014-1315-x (2014). [DOI] [PubMed] [Google Scholar]
- 17.van der Maaten L & Hinton G Visualizing data using t-SNE. The Journal of Machine Learning Research 9, 85 (2008). [Google Scholar]
- 18.Ceccarelli M et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–563, doi: 10.1016/j.cell.2015.12.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breiman L Random forests. Machine learning 45, 5–32 (2001). [Google Scholar]
- 20.Sokolova M & Lapalme G A systematic analysis of performance measures for classification tasks. Inf. Process. Manage . 45, 427–437, doi: 10.1016/j.ipm.2009.03.002 (2009). [DOI] [Google Scholar]
- 21.Sahm F et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol . 131, 903–910, doi: 10.1007/s00401-015-1519-8 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Weller M et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol . 129, 679–693, doi: 10.1007/s00401-015-1409-0 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research, N. et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med . 372, 2481–2498, doi: 10.1056/NEJMoa1402121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.conumee: Enhanced copy-number variation analysis using Illumina 450k methylation arrays. R package version 0.99.4, http://www.bioconductor.org/packages/release/bioc/html/conumee.html. v. 1.4.2 (2015). [Google Scholar]
- 25.Bady P, Delorenzi M & Hegi ME Sensitivity Analysis of the MGMT-STP27 Model and Impact of Genetic and Epigenetic Context to Predict the MGMT Methylation Status in Gliomas and Other Tumors. J. Mol. Diagn . 18, 350–361, doi: 10.1016/j.jmoldx.2015.11.009 (2016). [DOI] [PubMed] [Google Scholar]
Online Only References
- 26.Korshunov A et al. Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol . 131, 137–146, doi: 10.1007/s00401-015-1493-1 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Korshunov A et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol . 128, 279–289, doi: 10.1007/s00401-013-1228-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holsken A et al. Adamantinomatous and papillary craniopharyngiomas are characterized by distinct epigenomic as well as mutational and transcriptomic profiles. Acta Neuropathol Commun 4, 20, doi: 10.1186/s40478-016-0287-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim S et al. Papillary Tumor of the Pineal Region: A Distinct Molecular Entity. Brain Pathol . 26, 199–205, doi: 10.1111/bpa.12282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koelsche C et al. Melanotic tumors of the nervous system are characterized by distinct mutational, chromosomal and epigenomic profiles. Brain Pathol . 25, 202–208, doi: 10.1111/bpa.12228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat. Genet . 45, 927–932, doi: 10.1038/ng.2682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DT et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105, doi: 10.1038/nature11284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietsch T et al. Prognostic significance of clinical, histopathological, and molecular characteristics of medulloblastomas in the prospective HIT2000 multicenter clinical trial cohort. Acta Neuropathol . 128, 137–149, doi: 10.1007/s00401-014-1276-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2016).
- 35.Huber W et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12, 115–121, doi: 10.1038/nmeth.3252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aryee MJ et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369, doi: 10.1093/bioinformatics/btu049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leek JT & Storey JD Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS genetics 3, 1724–1735, doi: 10.1371/journal.pgen.0030161 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leek JT & Storey JD A general framework for multiple testing dependence. Proc. Natl. Acad. Sci. U. S. A . 105, 18718–18723, doi: 10.1073/pnas.0808709105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breiman L Classification and regression trees. (Chapman & Hall/CRC, 1984). [Google Scholar]
- 40.Liaw A & Wiener M Classification and Regression by randomForest. R News 2, 18–22 (2002). [Google Scholar]
- 41.Chen C, Liaw A & Breiman L Using random forest to learn imbalanced data. University of California, Berkeley, 1–12 (2004). [Google Scholar]
- 42.Kim KI & Simon R Overfitting, generalization, and MSE in class probability estimation with high-dimensional data. Biom J 56, 256–269, doi: 10.1002/bimj.201300083 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Boström H in Machine Learning and Applicati ons, 2008. ICMLA’08. Seventh International Conference on. 121–126 (IEEE). [Google Scholar]
- 44.Smola AJ Advances in large margin classifiers. (MIT press, 2000). [Google Scholar]
- 45.Friedman J, Hastie T & Tibshirani R Regularization paths for generalized linear models via coordinate descent. Journal of statistical software 33, 1 (2010). [PMC free article] [PubMed] [Google Scholar]
- 46.Appel IJ, Gronwald W & Spang R Estimating classification probabilities in high-dimensional diagnostic studies. Bioinformatics 27, 2563–2570 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Hand DJ & Till RJ A simple generalisation of the area under the ROC curve for multiple class classification problems. Machine learning 45, 171–186 (2001). [Google Scholar]
- 48.Simon R Class probability estimation for medical studies. Biom J 56, 597–600, doi: 10.1002/bimj.201300296 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Brier GW Verification of forecasts expressed in terms of probability. Monthly Weather Review 78, 1–3, doi: (1950). [DOI] [Google Scholar]
- 50.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol . 30, 41 3-421, doi: 10.1038/nbt.2203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


