Abstract
The inflammasome is a major component of the innate immune system, and its main function is to activate caspase-1, a cysteine protease that promotes inflammation by inducing interleukin-1β (IL-1β) maturation and release into the extracellular milieu. To prevent uncontrolled inflammation, this complex is highly regulated. When it is assembled, the inflammasome is insoluble, which has long precluded the analysis of its interactions with other proteins. Here we used the proximity-dependent biotinylation assay (BioID) to identify proteins associated with caspase-1 during inflammasome activation. Using the BioID in a cell-free system in which the inflammasome had been activated, we found that a caspase-1–biotin ligase fusion protein selectively labeled 111 candidates, including the p62/sequestosome-1 protein (p62). Using co-immunoprecipitation experiments, we demonstrated that p62 interacts with caspase-1. This interaction promoted caspase-1–mediated cleavage of p62 at Asp-329. Mechanistic and functional analyses revealed that caspase-1–mediated cleavage of p62 leads to loss of its interaction with the autophagosomal protein microtubule-associated protein 1 light chain 3 β (LC3B). Strikingly, overexpression of a p62 N-terminal fragment generated upon caspase-1 cleavage decreased IL-1β release, whereas overexpression of p62's C-terminal portion enhanced IL-1β release, by regulating pro-IL1β levels. Overall, the overexpression of both fragments together decreased IL-1β release. Taken together, our results indicate that caspase-1–mediated p62 cleavage plays a complex role in balancing caspase-1–induced inflammation.
Keywords: inflammasome, inflammation, autophagy, caspase-1 (CASP1), proteomics, proteolytic enzyme
Introduction
Caspase-1 is essential for processing and maturing pro-inflammatory cytokines; namely, interleukin-1β (IL-1β)3 and IL-18 (1). Caspase-1 is found in cells as an inactive precursor and is activated in response to inflammatory signals, including pathogen-associated molecular patterns and damage-associated molecular patterns. Following the recognition of pathogen-associated molecular patterns or damage-associated molecular patterns, a subset of intracellular pattern recognition receptors assembles into a macromolecular complex known as the inflammasome (2, 3). Most inflammasome pattern recognition receptors belong to the family of nucleotide-binding domain and leucine-rich repeat–containing receptors (NLRs). Caspase-1 activation takes place within inflammasome complexes. In addition to triggering IL-1β/IL-18 activation and release, active caspase-1 initiates an inflammatory form of cell death termed pyroptosis (4). Caspase-1 and caspase-11 (the latter being the murine ortholog of human caspases 4 and 5) cleave gasdermin D (GSDMD), leading to the release of the GSDMD N terminus polypeptide that oligomerizes in the plasma membrane, leading to pore formation and rupture of the plasma membrane (5, 6). The resulting disruption of plasma membrane integrity leads to osmotic swelling and release of inflammatory cytokines and culminates in pyroptotic cell death.
The inflammasome is a well-controlled machinery. Various mechanisms preventing its activation are known, and some competitive inhibitors have also been described, such as pyrin-only proteins or card-only proteins (7). Moreover, inflammasome activation does not always end in inflammatory cell death, and some processes may terminate its activation (8–11).
Autophagy is an intracellular degradation system associated with maintenance of cellular homeostasis. Autophagy plays a critical role in inflammasome regulation (8, 11). First, autophagy suppresses TLR4-dependent production of IL-1β and IL-18 by inhibiting TRIF-dependent reactive oxygen species production (12). Second, autophagy suppresses activation of the NLRP3 inflammasome by eliminating damaged mitochondria, thus preventing excess production of reactive oxygen species and mitochondrial DNA, in a process called mitophagy (13). Mitophagy depends on the adaptor p62/SQSTM1 (hereafter called p62) (14). Finally, autophagy selectively eliminates ubiquitinated ASC and IL-1β. Indeed, p62 targets these inflammasome components to the LC3B-positive cytosolic compartment, allowing their engulfment into autophagosomes and their elimination in autophagolysosomes.
In this study, we took advantage of the proximity-dependent biotinylation assay coupled with MS (BioID) to identify caspase-1–neighboring proteins during inflammasome activation (15). This BioID–caspase-1 assay identified 111 proteins, indicating that these candidates were in close proximity of caspase-1 but not necessary interactors.
Of these potential candidates, 28 (25%) had previously been linked to the inflammasome complex or autoinflammation. Among the hits, p62 was confirmed to interact with caspase-1 and identified as a new caspase-1 substrate. We demonstrate that caspase-1–mediated p62 cleavage at Asp-329 prevents its interaction with LC3B. Moreover, overexpression of p62 cleaved fragments regulates IL-1β release, suggesting that p62 is a complex regulator of the inflammasome.
Results
Expression of BirA*–caspase-1 in caspase-1−/− macrophages
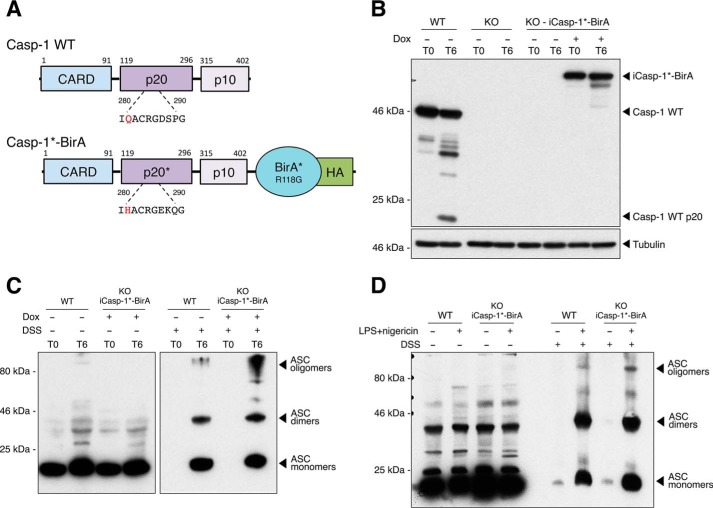
The analysis of inflammasome component interactions with other proteins has long been challenged by the insoluble nature of the complex. To overcome this technical issue, we used BioID, a method based on proximity-dependent biotinylation of proteins that are near neighbors of a fusion protein directly within cells or cell lysates. Biotinylated proteins can then be identified using streptavidin affinity capture and MS. The advantage of this approach is that the affinity capture can be performed under stringent conditions that disrupt native complexes and limit pulldown contaminants. In this system, the targeting protein is fused to a 35-kDa DNA-binding biotin protein ligase derived from Escherichia coli and called BirA (16). A BirA mutant (R118G, hereafter called BirA*) is defective in both self-association and DNA binding and has enhanced capacity for proximity-dependent biotin ligation (17). In addition, biotinylation is an exceptional endogenous modification in mammalian cells (18). To identify proteins in the vicinity of caspase-1 during inflammasome activation, we expressed a BirA*–caspase-1 fusion protein under an inducible promoter in immortalized bone marrow-derived macrophages (iBMDMs) derived from caspase-1−/− mice (Fig. 1). Full caspase-1 activation is associated with its cleavage within the inflammasome complex, and the half-life of processed caspase-1 is very short (2, 19). To increase the sensitivity of the system, we fused the BirA* ligase to a mutated caspase-1 containing a Q281H substitution within the catalytic site and observed no degradation of BirA*–caspase-1 after 6 h of inflammasome activation in a cell-free inflammasome-activating assay (Fig. 1, A and B). In this cell-free system, disruption of the cell membrane, along with hypotonic and low-potassium conditions, spontaneously activates the inflammasome (20). We then verified that inflammasome oligomerization was not altered by the BirA* fusion protein by assaying apoptosis-associated speck-like protein containing a CARD (ASC) oligomerization in the cell-free system. As expected, ASC oligomerization was preserved under inflammasome-activating conditions in reconstituted cells using the cell-free system (Fig. 1C) or in intact cells treated with the inflammasome activator nigericin (Fig. 1D)
Figure 1.
Generation of HA-tagged BirA*–caspase-1–overexpressing macrophages. A, schematic showing caspase-1 WT (casp-1 WT) and the engineered Q281H-mutated caspase-1 fused to HA-tagged BirA* (casp-1*-BirA). B, immortalized bone marrow-derived macrophages from Caspase-1−/− mice were reconstituted with a doxycycline (Dox)-inducible casp-1*-BirA (iCasp-1*-BirA). Reconstituted cells (KO-iCasp-1*-BirA) and their WT or caspase-1−/− (KO) controls were plated in the presence or absence of doxycycline (1 μg/ml) and incubated for 24 h. Cells were harvested, centrifuged, and resuspended in a hypotonic low-potassium buffer at 4 °C. Cells were mechanically lysed, and nuclei and plasma membranes were removed. The cell-free preparation was then incubated at 37 °C for 6 h (T6) or not (T0). Proteins were analyzed by Western blotting after boiling and fractionating on a 10% SDS-polyacrylamide gel. C, ASC oligomerization was analyzed after cross-linking or not with DSS and analyzed by Western blotting under nonreducing conditions. D, ASC oligomerization was also verified in cells previously plated in the presence of doxycycline upon activation of the inflammasome by LPS (1 μg/liter, 3 h) + nigericin (5 μm, 45 min). ASC oligomers were cross-linked or not with DSS and analyzed by Western blotting under nonreducing conditions.
Identification of proteins in the vicinity of caspase-1 during inflammasome activation in a cell-free assay
Studying inflammasome activation in cytosolic extracts provides multiple advantages, as it is synchronized, rapid, and strong. We thus decided to use this system to identify the repertoire of proteins coming in close proximity with caspase-1 during inflammasome activation. Doxycycline was added to the cells to induce BirA*–caspase-1 expression 24 h before the experiment. Cells were then mechanically lysed in a hypotonic, low-potassium buffer at 4 °C and incubated at 37 °C for 6 h in the presence of biotin (Fig. 2A). Control cells were left at 4 °C to prevent inflammasome activation. Biotinylated proteins were subsequently captured with streptavidin-coupled beads and extensively washed, and bound proteins were analyzed by MS. Because ASC is the main caspase-1–interacting protein during inflammasome activation, streptavidin pulldown of ASC was selected as the readout for a successful procedure before MS analysis (Fig. 2B).
Figure 2.
Identification of caspase-1–neighboring proteins during inflammasome activation in a cell-free assay. A, schematic presenting the BioID approach adapted to the cell-free assay for inflammasome activation. B, reconstituted cells (KO-iCasp-1*-BirA) and their WT or Caspase-1−/− (KO) controls were plated in presence or absence of doxycycline (Dox, 1 μg/ml) and incubated for 24 h. Cells were harvested, centrifuged, and resuspended in a hypotonic low-potassium buffer at 4 °C. Cells were mechanically lysed, and nuclei and plasma membranes were removed. The cell-free soup was then incubated at 37 °C for 6 h (T6) or not (T0). Supernatants were then incubated with streptavidin-coated beads overnight at 4 °C. Beads were collected and rigorously washed. Streptavidin-bound proteins were then analyzed by Western blotting after boiling and fractionating on a 15% SDS-polyacrylamide gel. ASC pulldown was selected as the readout for successful procedure before MS analysis. IP, immunoprecipitation. C, sorting process of the proteins identified by the caspase-1 BioID assay (1108 proteins were identified by MS, 628 were present in the uninduced sample, 112 proteins did not pass the 2× enrichment threshold between the 37 °C and the 4 °C samples, and 146 proteins were excluded based on CRAPome analysis, resulting in 111 proteins identified with confidence in the caspase-1 BioID assay). These candidates were analyzed for gene ontology (GO) enrichment according to biological process (PANTHER classification system).
Proteins unique to the BioID–caspase-1 (BirA*–caspase-1) pulldown and not detected with identical pulldowns from control cells (no doxycycline) were then selected based on an enrichment factor of >2 during inflammasome activation. As expected, MS identified ASC and caspase-1. Furthermore, caspase-1 was the most abundantly identified protein, detected at similar levels before and after inflammasome activation. Self-biotinylation of the bait protein explains this result. Among the 1108 proteins identified by MS, 480 were not detected in control cells (without doxycycline). 368 of 480 (77%) had an enrichment factor of ≥2 (Fig. 2C, left panel) upon inflammasome activation (37 °C versus 4 °C). We then removed 146 proteins that were contained in the Contaminant Repository for Affinity Purification (www.crapome.org;4 47) because they are known as usual contaminants of streptavidin-based affinity capture. This selection resulted in a final number of 111 proteins. Importantly, 28 proteins (25%) had previously been linked to the inflammasome complex or autoinflammation (Table S1), validating our screen to identify inflammasome-related proteins. Gene ontology enrichment analysis revealed significant enrichment in proteins involved in the regulation of NF-κB signaling (fold enrichment, 9.23; p < 0.01), in the regulation of vesicle-mediated transport (fold enrichment, 6.59; p < 0.001), in the regulation of protein transport (fold enrichment, 3.87; p < 0.001), and in metabolic processes (fold enrichment, 2.33; p < 0,01) (Fig. 2C, right panel).
Caspase-1 interacts with p62
In our BioID–caspase-1 screen, we identified caspase-1 proximity biotinylation of p62 (three unique peptides), a major adaptor protein involved in selective autophagy. p62 mainly serves as a cargo to drive ubiquitinated proteins to the LC3B-positive autophagosome before selective degradation within the autophagolysosome (21, 22). The function of the protein requires its complete integrity (23). p62 is also implicated in cell survival, inflammation, and osteoclastogenesis through its interactions with TRAF6/RANK/NF-κB and in apoptosis by scaffolding ubiquitinated caspase-8 (22).
We demonstrated the interaction between caspase-1 and p62 by performing a co-immunoprecipitation in HEK293T transfected with plasmids expressing FLAG-tagged caspase-1 and HA-tagged p62 (Fig. 3A). In addition, inflammasome activation in primary human macrophages using LPS + nigericin treatment led to the formation of caspase-1 specks corresponding to the inflammasome complex that colocalized with p62 (Fig. 3B). In contrast, upon LPS priming alone, p62 and caspase-1 had cytosolic localization but did not show any obvious colocalization, suggesting that p62 interacts specifically with caspase-1 upon inflammasome assembly.
Figure 3.
p62 interacts with caspase-1 upon inflammasome activation. A, HEK293T cells were transfected with plasmids encoding FLAG-tagged WT caspase-1 and HA-tagged p62 and incubated overnight. Cells were detached, lysed, and centrifuged. Supernatants were then incubated for 3 h at 4 °C with anti-FLAG– or anti-HA–agarose beads. Beads were collected and washed. Bound proteins were analyzed by Western blotting after boiling and fractionating on a 10% SDS-polyacrylamide gel. IP, immunoprecipitation. B, primary human monocyte-derived macrophages were treated as indicated. Cells were fixed, permeabilized, and incubated with the indicated antibodies, and images were collected using a confocal microscope.
Caspase-1 cleaves p62 at Asp-329
We then found that the inflammasome activation in a THP-1–derived cell-free system resulted in p62 cleavage and appearance of a 30-kDa migrating fragment (Fig. 4A). To further assess whether the cleavage of p62 depended on caspase-1, we used another model of inflammasome activation in the presence or absence of specific inhibitors. Inflammasome activation in primary human monocytes resulted in the cleavage of p62, and this cleavage was prevented by addition of Z-YVAD, a caspase-1 inhibitor (Fig. 4B). In contrast, the addition of specific inhibitors of caspase-8 (Z-IETD) or calpain, two proteases known for their ability to cleave p62 (24), did not prevent this cleavage. We further confirmed that caspase-1 was sufficient to cleave p62 by cotransfecting HEK293T cells with plasmids encoding p62 and various caspases (Fig. 4C). HEK293T cells lack the inflammasome machinery (25), but the ectopic expression of caspases is sufficient to obtain their self-activation. In this experiment, we confirmed that caspase-1 was very efficient in cleaving p62 compared with other caspases. In addition, we also detected cleavage of p62 by the pro-inflammatory caspase-4, an ortholog of the murine caspase-11. In agreement with a previous report, we observed that caspase-8 also induced degradation of p62 through multiple cleavage sites (24).
Figure 4.
Caspase-1 cleaves p62 at Asp-329. A, p62 cleavage in THP-1 cells was assayed using the cell-free system. The inflammasome was activated by incubating the cell-free soup at 37 °C during 1 h or 2 h in the presence or absence of pancaspase inhibitor (Z-VAD) or caspase-1 inhibitor (Z-YVAD). Caspase-1 (casp-1) and p62 were then analyzed by Western blotting after boiling and fractionating on a 10% SDS-polyacrylamide gel. Casp-1 p20 is the cleavage fragment, observed when caspase-1 is activated. B, primary human monocytes, obtained from whole blood of healthy donors, were incubated in the presence of the inflammasome activator (LPS and nigericin) with or without caspase-8 inhibitor (Z-IETD), calpain inhibitor, and caspase-1 inhibitor (Z-YVAD). Cells were then washed and lysed, and p62 cleavage was analyzed by Western blotting after boiling and fractionating on a 10% SDS-polyacrylamide gel. Ratios of cleaved/uncleaved p62 were calculated using ImageJ. C, HEK293T cells were co-transfected with plasmids encoding p62 and various caspases (casp-1 to 9) and incubated overnight. Cells were then washed and lysed, and p62 and IL-1β were analyzed by Western blotting after boiling and fractionating on a 10% SDS-polyacrylamide gel. D, HEK293T cells were co-transfected with plasmids encoding WT caspase-1 and WT or mutated p62 containing point mutations at the indicated amino acids and incubated overnight. Cells were then washed and lysed, and p62 and caspase-1 expression and cleavage were analyzed by Western blotting. E, HEK293T cells were co-transfected with plasmids encoding FLAG-tagged WT caspase-1 and WT or mutated p62. Cells were then washed, lysed, and incubated with anti-FLAG–agarose during 3 h at 4 °C. Beads were then rigorously washed, and bound proteins were analyzed by Western blotting. IP, immunoprecipitation.
According to the molecular weight of the fragments and the amino acid sequence, caspase-1–mediated cleavage of human p62 was predicted to be at Asp-329 or Asp-347. We thus generated p62 expression constructs harboring the point mutations p.[D329A] and p.[D347A] to interrogate the contribution of the corresponding residues to caspase-1–mediated p62 processing. HEK293T cells were cotransfected with caspase-1 and the WT or the mutated p62 constructs. The cleavage of p62 by caspase-1 was no longer detected in D329A-mutated p62 but unaffected in the mutant carrying the p.[D347A] mutation (Fig. 4D). The p.[D329A] mutation did not prevent the interaction between caspase-1 and p62, suggesting that the residue at Asp-329 is directly targeted by caspase-1 proteolytic activity and does not contribute to p62 interaction with caspase-1 (Fig. 4E).
Caspase-1 cleavage disrupts p62 interaction with LC3B and modulates IL-1β secretion
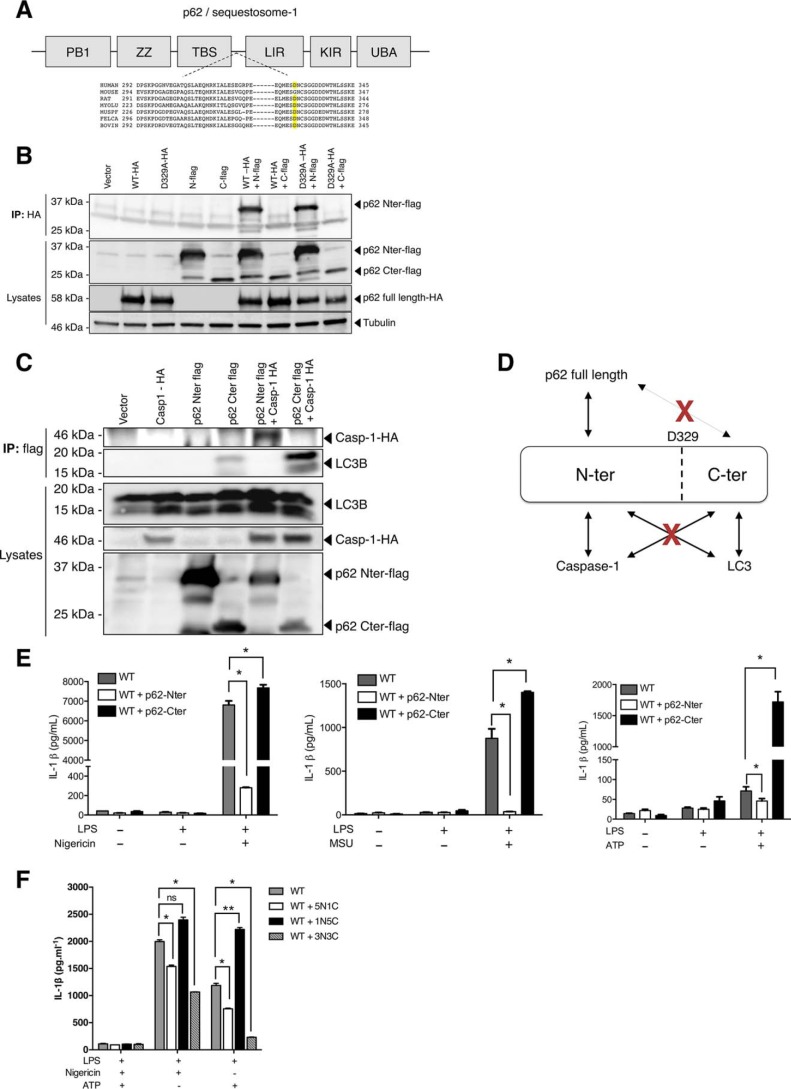
Autophagy is known to regulate the inflammasome, and p62 has been shown to deliver ASC and IL-1β to the autophagosome, resulting in suppression of inflammasome activation (8, 26). In addition, mice and cells deficient for p62 show higher IL-1β release upon inflammasome activation (8, 27). We sought to determine the role of p62 cleavage by caspase-1 on its interaction with LC3B and on IL-1β secretion. p62 is composed of six domains, including an N-terminal PB1 domain for oligomerization, a ubiquitin-binding domain (UBA), and an LC3B-interacting domain (LIR) (21, 22) (Fig. 5A). p62 interacts with the autophagosome docking protein LC3B via its LIR domain to shuttle ubiquitinated proteins into newly formed autophagosomes (21). Furthermore, the cleavage of p62 at Gly-241 by a protease from Coxsackievirus results in the disruption of p62 function in selective autophagy (23, 28). More precisely, the C-terminal fragment retained the binding activity to LC3B and exhibited a dominant-negative effect against native p62, probably by competing for LC3B binding. Because p62 self-oligomerization is required for ubiquitinated protein engulfment in autophagosomes (22, 29), we tested whether this ability was retained in both N-terminal and C-terminal fragments. Although cotransfection of plasmids encoding full-length p62 and N-terminal p62 showed that the oligomerization ability was retained for the N-terminal fragment, we did not observe any interaction between full-length p62 and the p62 C-terminal fragment (Fig. 5B), suggesting that this fragment had lost the ability to promote p62 oligomerization. In addition, we found that the N-terminal fragment mediated the interaction of caspase-1 with p62, whereas the LC3B-binding ability was limited to the C-terminal fragment (Fig. 5C). Altogether, these results suggest that cleavage of p62 by caspase-1 may disrupt p62 cargo function (Fig. 5D). To further characterize the functional consequences of the cleavage, we transduced WT iBMDMs with an inducible lentiviral vector encoding either of the two cleaved fragments of p62. We then activated the NLRP3 inflammasome with nigericin, MSU, or ATP. We demonstrated that ectopic expression of the N-terminal fragment of p62 decreased the release of IL-1β upon inflammasome activation, whereas overexpression of the C-terminal fragment significantly increased IL-1β release (Fig. 5E).
Figure 5.
Caspase-1 cleavage disrupts p62 interaction with LC3B and modulates IL-1β secretion. A, representation of the p62 structure. The PB1 (Phox and Bem1) N-terminal domain allows p62 oligomerization and/or interactions with other autophagy adaptors. The LIR domain allows interaction with the LC3B-coated compartment (autophagosome), and the UBA domain binds ubiquitinated proteins. ZZ, ZZ-type zinc finger domain; TBS, TRAF6-binding domain; KIR, KEAP1-interacting region. Alignment of the protein sequence surrounding Asp-329 demonstrates conservation in numerous mammalian species (human, rat, bat (Myotis lucifugus), ferret (Mustela furo), cat (Felis catus), bovine) but not in mice. B, HEK293T cells were cotransfected with plasmids encoding HA-tagged WT p62 (WT-HA) or its uncleavable mutant (D329A-HA) along with the FLAG-tagged p62 N-terminal (N-FLAG) or C-terminal (C-FLAG) fragments and incubated overnight. Cells were then washed, lysed, and incubated with anti-HA–agarose during 3 h at 4 °C. Beads were then rigorously washed, and bound proteins were analyzed by Western blotting. IP, immunoprecipitation. C, HEK293T cells were cotransfected with plasmids encoding HA-tagged caspase-1 (casp1-HA) and FLAG-tagged p62 N-terminal or C-terminal fragments and incubated overnight. Cells were then washed, lysed, and incubated with anti-FLAG–agarose during 3 h at 4 °C. Beads were rigorously washed, and bound proteins were analyzed by Western blotting. D, schematic presenting the interactions between p62 fragments and p62 full-length, caspase-1, and LC3B. E and F, WT iBMDMs were transduced stably with (E) the pINDUCER21 doxycycline-inducible lentiviral vector encoding human p62 N-terminal or C-terminal fragments or transiently (F) with various ratios (1:1, 5:1, and 1:5) of the pINDUCER21 doxycycline-inducible lentiviral vector encoding human p62 N-terminal (N) and C-terminal (C) fragments as indicated. Cells were incubated with doxycycline for 24 h (1 μg/ml) to induce ectopic expression of p62 fragments. The inflammasome was then activated by addition of a priming signal (LPS, 1 μg/liter during 3 h) followed by either nigericin (5 μm, 45 min), MSU (50 μg/ml, 6 h), or ATP (1 mm, 60 min) as indicated. The supernatants were collected. IL-1β concentrations were measured by ELISA. *, p < 0.05; **, p < 0.001; ns, not significant.
Because ectopic expression of the N- and C-terminal fragments of p62 has inverse effects on IL-1β release, we assessed the functional impact of co-expression of the two fragments. We thus transduced WT iBMDMs with different ratios (1:1, 5:1, and 1:5) of lentiviral vectors encoding the N- or C-terminal fragments of p62. Upon activation of the inflammasome by ATP or nigericin, we observed a decrease in IL-1β secretion in cells expressing high p62 N-terminal fragment levels, whereas IL-1β secretion was increased in cells expressing high p62 C-terminal fragment levels (Fig. 5F and Fig. S1). These results thus recapitulated the above findings. Nevertheless, when cells expressed both the N- and C-terminal fragments with an identical ratio (1:1), the overall effect was inhibition of IL-1β secretion, suggesting a negative dominant effect of the N-terminal over the C-terminal fragment.
Autophagy has been reported to control IL-1β secretion by targeting pro-IL-1β for degradation (26). We thus measured the levels of pro-IL-1β and two other inflammasome components, ASC and caspase-1, in the lysates of transduced cells. We observed increased levels of pro-IL-1β in p62 C-terminal fragment–expressing cells. Conversely, the levels of ASC and pro-caspase-1 in untreated cells, LPS-treated cells, and LPS+ATP-activated cells were not affected by p62 fragments expression (Fig. S2). The increase in pro-IL-1β level was not observed in WT and p62 N-terminal fragment–expressing cells. This may, at least in part, explain the increased levels of IL-1β secreted in C-terminal fragment–expressing cells after inflammasome activation. Altogether, these findings indicate that p62 is a complex regulator of the inflammasome and that the two fragments cleaved by caspase-1 may differentially regulate IL-1β release.
Discussion
In this study, we took advantage of the recently described BioID assay to identify caspase-1–neighboring proteins during inflammasome activation. The BioID assay consists of MS identification of proteins that have been biotinylated by the BirA proximity-dependent biotin ligase fused to a “bait” protein following streptavidin affinity capture (15). One limitation of the BioID assay is the problem of scale when dealing with low-abundance proteins. Such proteins may be difficult to identify, and a sufficient start material is necessary. Indeed, in preliminary setup experiments, we were not able to detect robust ASC biotinylation upon inflammasome stimulation in whole cells. We have thus chosen to use the previously described cell-free assay, where inflammasome assembly is very robust and occurs spontaneously upon disruption of cellular integrity in a low-potassium buffer (20). We assume that this method precludes the identification of various proteins usually contained in cell compartments that have been removed during the cell-free preparation, such as the cell membrane or nuclear proteins. Nevertheless, using the cell-free assay offers number advantages: strong, rapid, and synchronized inflammasome activation along with a large quantity of proteins in a reduced volume.
The analysis of inflammasome component interactions with other proteins has long been prevented by the insoluble nature of the inflammasome when in its mature complex. The use of the BioID assay sidesteps issues associated with bait protein solubility because biotinylation occurs before protein solubilization (15). Neighboring proteins are covalently modified with biotin, and robust lysis conditions can be used to solubilize polypeptides localized to poorly soluble cellular compartments.
Another limitation of BioID relies on the expression of an exogenous protein that is fused to BirA* that may modify the protein structure, assembly properties, and targeting (15). In our study, we chose to fuse BirA* to a mutated caspase-1 (Q281H) to avoid protein degradation and a possible toxicity associated with overexpression of the catalytically active caspase-1 (Fig. 1, A and B). This strategy allowed the accumulation of caspase-1 without disrupting ASC oligomerization (Fig. 1, D and E) and ASC–caspase-1 interaction (Fig. 2B).
The BioID identified 1108 candidates, of which 480 were not detected with identical pulldowns from control cells. Upon inflammasome activation, 368 were enriched with a factor of ≥2. We made a final sort by using the Contaminant Repository for Affinity Purification (www.crapome.org;4 47) and identified 111 proteins interacting with/in the vicinity of caspase-1. This strategy is debatable because it could have excluded caspase-1 interactors at steady state and proteins that are usual contaminants but would still be relevant caspase-1 interactors. Indeed, many proteins involved in cell metabolism, such as glycolysis (30), have been excluded from the analysis based on their known tendency to contaminate streptavidin affinity capture assays (www.crapome.org;4 47). Because our strategy was an exploratory approach, and given that the BioID was not intended to be used as a quantitative approach, we voluntarily chose to be less specific to gain sensibility and to perform a second step of validation for candidate proteins identified in the BioID assay.
Interestingly, we identified 28 (25%) proteins that had previously been associated with either caspase-1 or the inflammasome or autoinflammatory phenotypes in mice or humans (30–34). For instance, we identified phospholipase-Cγ2 (PLCG2), which has been recently related to a novel human autoinflammatory disease, PLCG2-associated antibody deficiency and immune dysregulation (PLAID) (35). The pathophysiological mechanisms linking PLCG2 structural anomalies and the autoinflammatory phenotype are being uncovered (36), but a direct interaction between caspase-1 and PLCG2 has never been reported before. In our BioID screen, we also identified cytoplasmic phospholipase A2 (cPLA2), an enzyme that had been previously demonstrated to be a substrate of several caspases, including caspase-1 (37, 38). cPLA2 is believed to be involved in the “eicosanoid storm” accompanying inflammasome activation, a crucial link between the inflammasome and inflammatory lipids (39). As for now, most of the identified caspase-1 substrates have been related to caspase-1–induced cell death. Accumulating evidence suggests that caspase-1 may also play a role in other innate immunity-related pathways. For example, caspase-1 cleaves and dampens cGAS-STING–mediated interferon production (40). Thus, looking at caspase-1 interactors/substrates that are not involved in cell death may reveal additional functions for caspase-1.
Our screen did not identify known caspase-1 substrates, such as IL-1β, IL-18, or GSDMD. For IL-1β, this result was expected because we did not use any priming step, which is necessary to induce detectable pro-IL1β within the cytosol. BirA* may have precluded conformational changes necessary to caspase-1 interactions with some of its substrates (19). Finally, the cell-free assay process may have removed some proteins that associate with organelles and the cell membrane.
Our screen identified p62 as a caspase-1–neighboring protein, and subsequent analyses revealed that p62 was also one of its substrates. While going on with further analysis, we demonstrated that p62 was cleaved by caspase-1 at Asp-329. Interestingly, this aspartate residue is conserved among mammalian species, except in Mus musculus, where it is replaced by a glycine residue. This difference may suggest different regulatory functions for p62 between mice and humans or an alternative cleavage site in mice. Interestingly, a large number of human inflammasome regulators (e.g. card-only proteins and pyrin-only proteins) are absent in mice, indicating that inflammasome-regulatory mechanisms have been subjected to different selection pressures during co-evolution of the hosts with their pathogens.
In addition to caspase-1, in vitro experiments indicated that human caspase-4 might process p62. Although the relevance of this observation has not been tested in this study, it may suggest that p62 processing may also contribute to noncanonical inflammasome activation in humans.
Mechanistic analyses further revealed that the cleavage of human p62 by caspase-1 disrupts its normal interactions with LC3B. Selective autophagy plays a key role in inflammasome inactivation (8–11). p62 knockout cells and mice have an increased response to inflammasome activation, as demonstrated by increased release of IL-1β (8, 27). We sought to determine whether cleavage of p62 at Asp-329 decreased or enhanced its inhibitory role in inflammasome activation. We thus overexpressed N-terminal and C-terminal fragments in cells with various ratios and observed seemingly antagonist effects of both fragments. Although the N-terminal fragment strongly inhibited IL-1β release, the C-terminal fragment increased cytokine release. For the latter, we observed an accumulation of pro-IL-1β, likely explaining its effects. Mechanistic analyses demonstrated that only the N-terminal fragment was able to bind the full-length p62 protein and that oligomerization of p62 is required for its full adaptor function. We hypothesize that the N-terminal p62 fragment may enhance p62 oligomerization, culminating in increased inhibitory effects on the inflammasome complexes through autophagic degradation. On the other hand, the C-terminal fragment, in addition to precluding pro-IL-1β degradation by autophagy, is unable to bind the full-length p62 proteins but retains the ability to bind LC3B and may act as a dominant-negative fragment, competing with full-length p62 for LC3B binding to decrease p62-mediated inhibition of the inflammasome. Indeed, such an effect has already been reported for a longer p62 C-terminal fragment cleaved at Gly-241 by a protease from Coxsackievirus (23, 28).
Coexpression of similar levels of N- and C-terminal fragments led to decreased IL-1β release, suggesting a negative dominant effect of the N-terminal fragment. Overall, these results suggest that p62 is a complex regulator of the inflammasome with dual functions, either enhancing or quelling inflammation (Fig. 6).
Figure 6.

A proposed model for p62 regulation of the inflammasome. In the negative regulation model (left panel), caspase-1 cleavage of p62 and subsequent release of the N-terminal fragment (1) may favor full-length p62 self-oligomerization (2), interaction with the LC3B-coated compartment (3), and engulfment of the inflammasome into the autophagosome (4), allowing quelling of the inflammation. In the positive regulation model (right panel), caspase-1 cleavage of the entire p62 pool may alter p62 function and give rise to a dominant-negative C-terminal fragment (5) that competes with p62 for LC3B binding (6) to inhibit autophagic degradation of pro-IL-1β and the inflammasome (7), resulting in increased inflammasome activation, IL-1β release, and inflammatory cell death.
Conclusions
Using the BioID approach, we identified 111 proteins in close proximity of caspase-1 during inflammasome activation; 75% of them had not been related before to the inflammasome. p62, which has been reported as a negative regulator of the inflammasome, is cleaved by caspase-1. Our results provide another layer in the p62-mediated inflammasome regulation mechanisms by showing that p62 cleavage by caspase-1 could either result in inhibiting or increasing inflammasome activation.
Experimental procedures
Cell lines
iBMDMs from WT or caspase-1−/− mice (a gift from D. M. Monack and P. Broz, Stanford University, Stanford, CA) and the human monocytic cell lines THP-1 and U937 were cultured in RPMI medium (Gibco) supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin. THP-1 and U937 cells were grown in suspension in 75-cm2 flasks. HEK293T cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% (v/v) FBS, 4 mm l-glutamine, and 1% penicillin/streptomycin. All cells were grown at 37 °C with 5% CO2.
Primary human monocytes and macrophages
Healthy donor blood was provided by the Etablissement Français du Sang in the Framework of Convention 14-1820. Peripheral blood mononuclear cells were isolated by density gradient centrifugation using lymphocyte separation medium (Eurobio). Monocytes were isolated from peripheral blood mononuclear cells by adhesion. Monocytes were differentiated into macrophages by culture for 6 days in RPMI 1640 medium, GlutaMAX medium (Thermo Fisher) supplemented with 10% (v/v) FBS, and 50 ng/ml macrophage colony-stimulating factor (Immunotools).
Reconstitution of caspase-1−/− cells and generation of HA-tagged BirA*–caspase-1–overexpressing cells
The lentiviral pINDUCER21 plasmid was obtained from Steve Elledge (Harvard Medical School, Boston, MA) (41). A human HA-tagged BirA*–caspase-1 construct was cloned into the pENTR1A dual selection vector (Invitrogen) and then cloned in the pINDUCER21, a GFP Tet-inducible lentiviral vector plasmid. Lentiviruses were produced as described previously (42). Caspase-1−/− and WT iBMDMs were infected with HA-tagged BirA*–caspase-1–expressing lentiviruses. GFP-positive cells were sorted by FACS 96 h post-infection. Cells were cultured in complete medium, and HA-tagged BirA*–caspase-1 expression was induced using tetracycline (doxycycline, 1 μg/ml) for 24 h.
Inflammasome activation
HA-tagged BirA*–caspase-1–expressing iBMDMs were cultured to a density of about 1.5 × 106 cells/ml in 150-cm2 plastic flasks or in 2-liter roller bottles. Inflammasome activation was induced in a cell-free system as described previously (20). When specified, biotin was added before inflammasome activation at a final concentration of 50 μm. For experiments using non-cell-free inflammasome activation, cells were washed twice with PBS, detached, enumerated, plated onto 12-well plates, and left to adhere overnight. The following day, culture media were changed before stimulation with Opti-MEM (Gibco) for Western blot analysis or with complete culture medium for ELISA analysis. Cells were primed with 1 μg/ml for 3 h and stimulated with 5 μm nigericin for 30 min, 50 μg/ml of MSU for 6 h, or 1 mm ATP for 60 min. After stimulation, supernatants were collected, and proteins were precipitated using the methanol/chloroform method. Proteins were then analyzed by Western blot analysis after boiling and fractionation on a 15% SDS-polyacrylamide gel.
Streptavidin pulldown and identification of biotinylated products by MS
Protein concentrations in supernatants from the cell-free assay were determined using the Bradford protein assay (Bio-Rad) and equalized in sample buffer. Supernatants were then incubated with 30 μl of Dynabeads (MyOne Streptavidin C1, Invitrogen) overnight at 4 °C. Beads were collected and washed twice for 8 min at RT in 1 ml of wash buffer (2% SDS in distilled H2O). This was repeated with a wash buffer containing 0.1% deoxycholate, 1% Triton X-100, 500 mm NaCl, 1 mm EDTA, and 50 mm Hopes; then with a wash buffer containing 250 mm LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mm EDTA, and 10 mm Tris (pH 8.1); and finally with a wash buffer containing 50 mm Tris (pH 7.4) and 50 mm NaCl. 10% of the sample was saved for Western blot analysis. Bound proteins were removed from the beads with 50 μl of Laemmli SDS sample buffer at 98 °C. Proteins eluted from the streptavidin beads by SDS sample buffer were reduced, alkylated, and separated by 1D SDS-PAGE. Mass spectrometry was performed as described previously (15, 43). The resulting .dat files were loaded into SCAFFOLD Q+ (Proteome Software). The acceptance level for proteins was two identified peptides with minimum 95% probability each.
ASC oligomerization assay
After stimulation, the supernatant was removed, and cells were detached with PBS containing 2 mm EDTA and centrifuged 5 min at 1500 × g. Cells were lysed on ice in 500 μl of buffer A (20) by mechanical lysis (30 passages through a 21-gauge needle). The cell lysates were centrifuged in 1.5-ml Eppendorf tubes at 1800 × g for 8 min to remove nuclei, and 30 μl of the supernatants were kept for Western blot analysis to test ASC expression in the lysates. The remaining supernatant was diluted twice with buffer A and centrifuged for 5 min at 2000 × g. After centrifugation, the supernatant was diluted with 1 volume of CHAPS buffer (20 mm Hepes-KOH (pH 7.5), 5 mm MgCl2, 0.5 mm EGTA, 0.1 mm phenylmethylsulfonyl fluoride, and 0.1% CHAPS) and again centrifuged at 5000 × g for 8 min to pellet the ASC pyroptosome. The supernatant was discarded, and the pellet was resuspended in 50 μl of CHAPS buffer with 4 mm of disuccinimidyl suberate (DSS, Thermo Scientific) for 30 min at room temperature (RT). Samples were centrifuged at 5000 × g for 8 min, and pellets were resuspended in 30 μl of 2× loading buffer under nonreducing conditions for Western blotting. For the cell-free assay, the ASC oligomerization assay was performed as described previously (20).
Immunofluorescence staining and confocal microscopy
For confocal microscopy, primary monocytes were sorted from healthy donor blood by sucrose gradient followed by CD14-positive selection (Miltenyi Biotec) and cultured for 6 days in complete RPMI medium supplemented with human macrophage colony-stimulating factor (100 ng/ml, Immunotools). Macrophages were plated the day before the experiment on 15-mm coverslips in 12-well plates. After treatment, the cells were fixed with 2% (v/v) paraformaldehyde (Applichem) for 15 min at room temperature. The cells were then permeabilized with 0.1% Triton X-100 in PBS for 10 min and washed once with PBS. The aspecific sites were blocked with 3% (w/v) BSA prepared in PBS for 30 min at RT. Antibodies against human p62 (Abcam) and human caspase-1 (Adipogen) were diluted 1:500 in 1% BSA and incubated on the coverslips overnight at 4 °C. The next day, the cells were washed five times with PBS, and the fluorescent secondary antibodies (anti-IgG (H+L), Alexa Fluor conjugates, Thermo Scientific), diluted 1:1000 in 1% BSA, were applied for 1 h. The coverslips were washed again with PBS and incubated for 5 min with Hoechst dye diluted in PBS at a final concentration of 4 μg/ml. The coverslips were mounted on slides using ProLong Gold Antifade (Invitrogen). Images were captured using an inverted confocal microscope (LSM 710 META, Zeiss) and processed with Zeiss Axiovision software.
Mutagenesis
Point mutations were performed using the HA-p62 plasmid (28027, Addgene) to obtain the p.[D329A] and p.[D347A] mutants, using the QuikChangeTM site-directed mutagenesis kit (Agilent Genomics), according to the manufacturer's instructions. The following primers (Microsynth AG) were used: 5′-GAA CAG ATG GAG TCG GCT AAC TGT TCA GGA-3′ and 5′-TCC TGA ACA GTT AGC CGA CTC CAT CTG TTC-3′ for p62 D329A and 5′-TCT TCA AAA GAA GTG GCC CCG TCT ACA GGT GAA-3′ and 5′-TTC ACC TGT AGA CGG GGC CAC TTC TTT TGA AGA-3′ for p62 D347A.
Ectopic expression of p62 N-terminal and C-terminal fragments in cells
p62 N-terminal and p62 C-terminal constructs were obtained from the HA-p62 plasmid using the EcoR1 and NotI restriction enzymes (New England Biolabs) and the following primers (Microsynth AG): 5′-AAA GAA TTC ATG GCC ATG TCC TAC GTG-3′ and 5′-AAA GCG GCC GCC TAC GAC TCC ATC TGT TC-3′ for p62 Nter and 5′-AAA GAA TTC AAC TGT TCA GGA GGA-3′ and 5′-AAA GCG GCC GCC TAT CAC AAC GGC GGG-3′ for p62 Cter. p62 N-terminal and C-terminal constructs were then cloned into the pENTR1A dual selection vector and then cloned into pINDUCER21. WT iBMDMs were transduced with p62-expressing N-terminal or C-terminal lentiviruses. The lentiviral load was quantified, and different ratios were used to transduce the cells. GFP-positive cells were sorted by FACS 96 h post-infection. Cells were cultured in complete medium, and p62 fragment expression was induced using doxycycline (1 μg/ml) for 24 h. N- and C-terminal fragment ratios were verified by Western blot analysis.
HEK293T transfection and co-immunoprecipitation
HEK293T cells were plated the day before the experiment, transfected using polyethyleneimine, and incubated overnight at 37 °C. The plasmids used were from Tschopp and co-workers (2, 44, 45). N-terminal and C-terminal p62 constructs were cloned into a FLAG tag–expressing plasmid on a PCR3 backbone. The HA-p62 plasmid was from Addgene (28027). For co-transfection, the same amount of vector plasmid was transfected in control cells. Cells (1 × 106) were then washed with PBS, suspended in 200 μl of lysis buffer (0.1% NP-40, 20 mm Tris-HCl (pH 7.8), 150 mm NaCl, and 5 mm EDTA) supplemented with complete protease inhibitor (Roche), and incubated on ice for 15 min. Cells were then centrifuged at 1300 rpm at 4 °C for 5 min, and supernatant proteins were quantified. The supernatants were either collected in Laemmli SDS buffer and analyzed by Western blotting or incubated with 10 μl of anti-FLAG– or anti-HA–agarose (both from Sigma) at 4 °C overnight under agitation. After centrifugation at 6000 rpm for 30 min, supernatants were discarded, and beads were washed five times with lysis buffer. Immunoprecipitated material was eluted in SDS sample buffer for 5 min at 100 °C and separated with SDS-PAGE.
Western blotting
Proteins were denatured, separated by SDS-PAGE, and transferred to nitrocellulose. Membranes were blocked in 2.5% BSA in PBS with 0.4% Triton X-100 and incubated in the same buffer with horseradish peroxidase–conjugated secondary antibodies. Band intensities were quantified with ImageJ software using arbitrary pixel intensity units.
ELISA
Cytokine release was measured in supernatants from stimulated cells. Mouse IL-1β and human IL-1β ELISA kits (R&D Systems) were used according to the manufacturer's instructions.
Chemicals and antibodies
LPS, nigericin, ATP, calpain inhibitor, and doxycycline were from Sigma. MSU crystals were prepared as described previously described (46). Z-YVAD-fmk, Z-VAD-fmk, and Z-IETD-fmk were from Abcam. Anti-mouse caspase-1 p20, anti-mouse and anti-human ASC antibody, anti-human caspase-1, and anti-human tubulin were from Adipogen. Anti-mouse and anti-human LC3B antibodies were from Sigma-Aldrich. Anti-mouse and anti-human p62 and anti-HA tag antibodies were from Abcam. The anti-FLAG tag antibody was from GenScript.
Author contributions
Y. J., A. D. M., E. B., R. T., T. H., and F. M. conceptualization; Y. J., A. D. M., E. B., R. T., T. H., and F. M. data curation; Y. J. formal analysis; Y. J., B. L., A. D. M., E. B., A. P., R. T., T. H., and F. M. investigation; Y. J., A. D. M., E. B., R. T., T. H., and F. M. methodology; Y. J. writing-original draft; A. D. M., E. B., R. T., T. H., and F. M. supervision; A. D. M., E. B., R. T., T. H., and F. M. funding acquisition; A. D. M., E. B., R. T., T. H., and F. M. project administration; A. D. M., E. B., R. T., T. H., and F. M. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Manfredo Quadroni and the proteomics facility of the University of Lausanne for help with MS and analysis.
This work was supported by Fondation Innovations en Infectiologie, the Foundation for the Development of Internal Medicine in Europe, and the Groupama Foundation (to Y. J.), EC Seventh Framework Programme (FP7) Grants 311542 (to T. H.) and 281996 (to F. M.); the Société Nationale Française de Médecine Interne, and by a poste d'accueil from INSERM (to Y. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- IL
- interleukin
- NLR
- nucleotide-binding domain and leucine-rich repeat–containing receptor
- ASC
- apoptosis-associated speck-like protein containing a CARD
- iBMDM
- immortalized bone marrow-derived macrophage
- HEK
- human embryonic kidney
- HA
- hemagglutinin
- LPS
- lipopolysaccharide
- Z-VAD-fmk
- benzyloxycarbonyl-VAD-fluoromethyl ketone
- FBS
- fetal bovine serum
- RT
- room temperature
- MSU
- monosodium urate crystals.
References
- 1. Druilhe A., Srinivasula S. M., Razmara M., Ahmad M., and Alnemri E. S. (2001) Regulation of IL-1β generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 8, 649–657 10.1038/sj.cdd.4400881 [DOI] [PubMed] [Google Scholar]
- 2. Martinon F., Burns K., and Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 10, 417–426 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- 3. Martinon F., Mayor A., and Tschopp J. (2009) The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27, 229–265 10.1146/annurev.immunol.021908.132715 [DOI] [PubMed] [Google Scholar]
- 4. Bergsbaken T., Fink S. L., and Cookson B. T. (2009) Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 6. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 7. Latz E., Xiao T. S., and Stutz A. (2013) Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi C. S., Shenderov K., Huang N. N., Kabat J., Abu-Asab M., Fitzgerald K. A., Sher A., and Kehrl J. H. (2012) Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 13, 255–263 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu T., Tang Q., Liu K., Xie W., Liu X., Wang H., Wang R. F., and Cui J. (2016) TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep. 16, 1988–2002 10.1016/j.celrep.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 10. Saitoh T., and Akira S. (2016) Regulation of inflammasomes by autophagy. J. Allergy Clin. Immunol. 138, 28–36 10.1016/j.jaci.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 11. Harris J., Lang T., Thomas J. P. W., Sukkar M. B., Nabar N. R., and Kehrl J. H. (2017) Autophagy and inflammasomes. Mol. Immunol. 86, 10–15 10.1016/j.molimm.2017.02.013 [DOI] [PubMed] [Google Scholar]
- 12. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., and Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 10.1038/ni.1980,10.1038/nrm3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupfer C., Thomas P. G., Anand P. K., Vogel P., Milasta S., Martinez J., Huang G., Green M., Kundu M., Chi H., Xavier R. J., Green D. R., Lamkanfi M., Dinarello C. A., Doherty P. C., and Kanneganti T. D. (2013) Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 14, 480–488 10.1038/ni.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong Z., Umemura A., Sanchez-Lopez E., Liang S., Shalapour S., Wong J., He F., Boassa D., Perkins G., Ali S. R., McGeough M. D., Ellisman M. H., Seki E., Gustafsson A. B., Hoffman H. M., et al. (2016) NF-κB restricts inflammasome activation via elimination of damaged mitochondria. Cell 164, 896–910 10.1016/j.cell.2015.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cronan J. E. (2005) Targeted and proximity-dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. J. Nutr. Biochem. 16, 416–418 10.1016/j.jnutbio.2005.03.017 [DOI] [PubMed] [Google Scholar]
- 17. Kwon K., and Beckett D. (2000) Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 9, 1530–1539 10.1110/ps.9.8.1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapman-Smith A., and Cronan J. E. (1999) Molecular biology of biotin attachment to proteins. J. Nutr. 129, 477S–484S 10.1093/jn/129.2.477S [DOI] [PubMed] [Google Scholar]
- 19. Walsh J. G., Logue S. E., Lüthi A. U., and Martin S. J. (2011) Caspase-1 promiscuity is counterbalanced by rapid inactivation of processed enzyme. J. Biol. Chem. 286, 32513–32524 10.1074/jbc.M111.225862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jamilloux Y., and Martinon F. (2016) Cell-free assay for inflammasome activation. Methods Mol. Biol. 1417, 207–215 10.1007/978-1-4939-3566-6_14 [DOI] [PubMed] [Google Scholar]
- 21. Johansen T., and Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katsuragi Y., Ichimura Y., and Komatsu M. (2015) p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282, 4672–4678 10.1111/febs.13540 [DOI] [PubMed] [Google Scholar]
- 23. Shi J., Wong J., Piesik P., Fung G., Zhang J., Jagdeo J., Li X., Jan E., and Luo H. (2013) Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFκB signaling. Autophagy 9, 1591–1603 10.4161/auto.26059 [DOI] [PubMed] [Google Scholar]
- 24. Norman J. M., Cohen G. M., and Bampton E. T. (2010) The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 6, 1042–1056 10.4161/auto.6.8.13337 [DOI] [PubMed] [Google Scholar]
- 25. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., and Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris J., Hartman M., Roche C., Zeng S. G., O'Shea A., Sharp F. A., Lambe E. M., Creagh E. M., Golenbock D. T., Tschopp J., Kornfeld H., Fitzgerald K. A., and Lavelle E. C. (2011) Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J. Biol. Chem. 286, 9587–9597 10.1074/jbc.M110.202911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohtsuka S., Ishii Y., Matsuyama M., Ano S., Morishima Y., Yanagawa T., Warabi E., and Hizawa N. (2014) SQSTM1/p62/A170 regulates the severity of Legionella pneumophila pneumonia by modulating inflammasome activity. Eur. J. Immunol. 44, 1084–1092 10.1002/eji.201344091 [DOI] [PubMed] [Google Scholar]
- 28. Shi J., Fung G., Piesik P., Zhang J., and Luo H. (2014) Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ. 21, 1432–1441 10.1038/cdd.2014.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wurzer B., Zaffagnini G., Fracchiolla D., Turco E., Abert C., Romanov J., and Martens S. (2015) Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife 4, e08941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao W., Yeretssian G., Doiron K., Hussain S. N., and Saleh M. (2007) The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 282, 36321–36329 10.1074/jbc.M708182200 [DOI] [PubMed] [Google Scholar]
- 31. Wang L. J., Hsu C. W., Chen C. C., Liang Y., Chen L. C., Ojcius D. M., Tsang N. M., Hsueh C., Wu C. C., and Chang Y. S. (2012) Interactome-wide analysis identifies end-binding protein 1 as a crucial component for the speck-like particle formation of activated absence in melanoma 2 (AIM2) inflammasomes. Mol. Cell. Proteomics 11, 1230–1244 10.1074/mcp.M112.020594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., and Núñez G. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 10.1074/mcp.M800132-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agard N. J., Maltby D., and Wells J. A. (2010) Inflammatory stimuli regulate caspase substrate profiles. Mol. Cell Proteomics 9, 880–893 10.1074/mcp.M900528-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keller M., Rüegg A., Werner S., and Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 10.1016/j.cell.2007.12.040 [DOI] [PubMed] [Google Scholar]
- 35. Zhou Q., Lee G. S., Brady J., Datta S., Katan M., Sheikh A., Martins M. S., Bunney T. D., Santich B. H., Moir S., Kuhns D. B., Long Priel D. A., Ombrello A., Stone D., Ombrello M. J., et al. (2012) A hypermorphic missense mutation in PLCG2, encoding phospholipase Cγ2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am. J. Hum. Genet. 91, 713–720 10.1016/j.ajhg.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chae J. J., Park Y. H., Park C., Hwang I. Y., Hoffmann P., Kehrl J. H., Aksentijevich I., and Kastner D. L. (2015) Connecting two pathways through Ca2+ signaling: NLRP3 inflammasome activation induced by a hypermorphic PLCG2 mutation. Arthritis Rheumatol. 67, 563–567 10.1002/art.38961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lüschen S., Ussat S., Krönke M., and Adam-Klages S. (1998) Cleavage of human cytosolic phospholipase A2 by caspase-1 (ICE) and caspase-8 (FLICE). Biochem. Biophys. Res. Commun. 253, 92–98 10.1006/bbrc.1998.9754 [DOI] [PubMed] [Google Scholar]
- 38. Andrei C., Margiocco P., Poggi A., Lotti L. V., Torrisi M. R., and Rubartelli A. (2004) Phospholipases C and A2 control lysosome-mediated IL-1β secretion: implications for inflammatory processes. Proc. Natl. Acad. Sci. U.S.A. 101, 9745–9750 10.1073/pnas.0308558101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dennis E. A., and Norris P. C. (2015) Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 15, 511–523 10.1038/nri3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Ning X., Gao P., Wu S., Sha M., Lv M., Zhou X., Gao J., Fang R., Meng G., Su X., and Jiang Z. (2017) Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46, 393–404 10.1016/j.immuni.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 41. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A., et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bagnis C., Bailly P., and Chapel-Fernandes S. (2009) Using an EGFPmeter to evaluate the lentiviral vector production: tricks and traps. Methods Mol. Biol. 515, 151–163 10.1007/978-1-59745-559-6_10 [DOI] [PubMed] [Google Scholar]
- 43. Di Micco A., Frera G., Lugrin J., Jamilloux Y., Hsu E. T., Tardivel A., De Gassart A., Zaffalon L., Bujisic B., Siegert S., Quadroni M., Broz P., Henry T., Hrycyna C. A., and Martinon F. (2016) AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc. Natl. Acad. Sci. U.S.A. 113, E4671–E4680 10.1073/pnas.1602419113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thome M., Hofmann K., Burns K., Martinon F., Bodmer J. L., Mattmann C., and Tschopp J. (1998) Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr. Biol. 8, 885–888 10.1016/S0960-9822(07)00352-1 [DOI] [PubMed] [Google Scholar]
- 45. Thome M., Gaide O., Micheau O., Martinon F., Bonnet D., Gonzalez M., and Tschopp J. (2001) Equine herpesvirus protein E10 induces membrane recruitment and phosphorylation of its cellular homologue, bcl-10. J. Cell Biol. 152, 1115–1122 10.1083/jcb.152.5.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 10.1038/nature04516 [DOI] [PubMed] [Google Scholar]
- 47. Mellacheruvu D., Wright Z., Couzens A., Lambert J., St-Denis N., Li T., Miteva Y., Hauri S., Sardiu M., Low T., Halim V., Bagshaw R., Hubner N., Hakim A., Bouchard A., et al. (2013) The CRAPome: a Contaminant repository for affinity purification mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.