Abstract
RNA-binding proteins (RBP) are critical regulators of gene expression. Recent studies have uncovered hundreds of mRNA-binding proteins that do not contain annotated RNA-binding domains and have well-established roles in other cellular processes. Investigation of these nonconventional RBPs is critical for revealing novel RNA-binding domains and may disclose connections between RNA regulation and other aspects of cell biology. The endosomal sorting complex required for transport II (ESCRT-II) is a nonconventional RNA-binding complex that has a canonical role in multivesicular body formation. ESCRT-II was identified previously as an RNA-binding complex in Drosophila oocytes, but whether its RNA-binding properties extend beyond Drosophila is unknown. In this study, we found that the RNA-binding properties of ESCRT-II are conserved in Xenopus eggs, where ESCRT-II interacted with hundreds of mRNAs. Using a UV cross-linking approach, we demonstrated that ESCRT-II binds directly to RNA through its subunit, Vps25. UV cross-linking and immunoprecipitation (CLIP)-Seq revealed that Vps25 specifically recognizes a polypurine (i.e. GA-rich) motif in RNA. Using purified components, we could reconstitute the selective Vps25-mediated binding of the polypurine motif in vitro. Our results provide insight into the mechanism by which ESCRT-II selectively binds to mRNA and also suggest an unexpected link between endosome biology and RNA regulation.
Keywords: RNA-binding protein, RNA–protein interaction, RNA, endocytosis, RNA metabolism, RNA localization, ESCRT-II, polypurine motif, endosome maturation, GA-rich sequence, membrane trafficking
Introduction
Posttranscriptional control of gene expression is achieved through the regulation of mRNA at every stage of their life cycle, including splicing, nuclear export, localization in the cytoplasm, translation initiation, stability, and degradation. RNA-binding proteins (RBP)2 are key regulators of these processes, and eukaryotic genomes encode hundreds of RBPs to maintain tight regulation of mRNA (1). However, among the RBPs that are well-characterized, only a limited number of RNA-binding domains have been described (2, 3), and it has recently become clear that hundreds of proteins lacking conventional RNA-binding domains interact with and regulate mRNA (4–9). Many of these nonconventional mRNA-binding proteins have well-established functions in other processes within the cell, thus potentially providing a mechanism through which mRNA regulation can be linked to other aspects of cell biology (10, 11). Further studies are needed to better understand the mechanisms through which these nonconventional mRNA-binding proteins mediate mRNA regulation and coordinate their multiple roles within the cell.
Proteins that regulate mRNA localization within the cytoplasm are an important class of RBPs. Recent work has demonstrated that a large portion of all mRNAs exhibit a specific localization pattern (12–17) and that mRNA localization is a fundamental aspect of the posttranscriptional control of gene expression (18). mRNA localization is important for the control of cell fate patterning during early development and for local protein translation in many different cell types (15). Although many RBPs involved in the subcellular localization of mRNAs are well-characterized, the mechanisms through which the majority of localized mRNAs are selectively positioned in the cell are poorly understood. In an effort to identify novel factors involved in RNA localization, work in Drosophila has identified a highly conserved protein complex, ESCRT-II, as being involved in the trafficking of the bicoid mRNA during oogenesis (19). However, ESCRT-II lacks conventional RNA-binding domains and it is unclear how ESCRT-II interacts with mRNA. To date, no other mRNA partners of ESCRT-II have been identified, and whether the RNA-binding properties of ESCRT-II extend beyond Drosophila has not been explored.
ESCRT-II is a highly conserved protein complex composed of two copies of Vps25 and one copy each of Vps36 and Vps22 (20–22). ESCRT-II is part of the protein machinery that sorts activated cell surface receptors into intraluminal vesicles within the endosome, thereby maturing endosomes into multivesicular bodies (MVB) (23). This process is mediated by the sequential recruitment of ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III to the endosome. The role of ESCRT-II in this process is to deform the endosomal membrane through interactions between phospholipids and the Vps36-GLUE and Vps22-H0 domains (21, 24–26), to sort ubiquitinated cargo into membrane buds through interactions between the GLUE domain and ubiquitin (24, 26–28), and to recruit ESCRT-III through Vps25 (21, 29). Interestingly, mutational analysis has revealed that of all the ESCRT complexes, ESCRT-II is uniquely required for bicoid localization, suggesting that ESCRT-II's role in bicoid RNA trafficking is independent of its role in MVB formation (19).
To understand how the nonconventional RNA-binding complex, ESCRT-II, interacts with mRNA, we undertook a detailed characterization of the RNA-binding properties of ESCRT-II using both Xenopus laevis egg extract as well as purified components. We found that the RNA-binding properties of ESCRT-II are conserved across multiple species and that ESCRT-II has hundreds of mRNA binding partners. We determined that Vps25 is the RNA-binding subunit of ESCRT-II and identified a purine-rich RNA sequence motif recognized by Vps25. Furthermore, we established that the interaction between Vps25 and purine-rich sequences is direct using a UV cross-linking approach in egg extract and in vitro. Our results demonstrate that the RNA-binding functions of ESCRT-II are widespread and offer a mechanism for how a nonconventional RBP can selectively recognize and interact with RNA.
Results
ESCRT-II is an RNA-binding complex in Xenopus laevis eggs
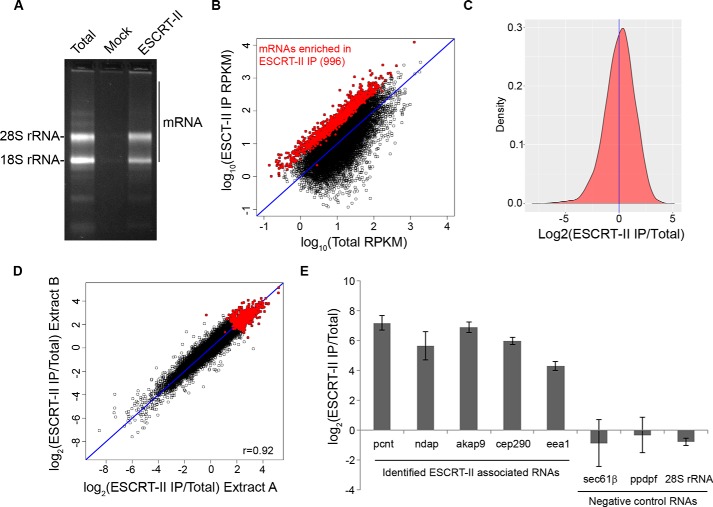
To determine whether ESCRT-II associates with transcripts in X. laevis eggs, we performed an RNA immunoprecipitation (RIP) from egg extract using antibodies specific to ESCRT-II. ESCRT-II immunoprecipitations consistently contained a complex mixture of RNA that differed subtly from total RNA, whereas a mock immunoprecipitation performed with nonspecific rabbit IgG co-immunoprecipitated very little RNA (Fig. 1A). We sequenced ESCRT-II-associated RNAs and identified 996 mRNAs that are significantly enriched in ESCRT-II immunoprecipitations (2-fold enriched over total RNA with a p value of <0.01 as determined by edgeR) (30) (Fig. 1, B and C, and Table S1). Two biological replicates of ESCRT-II RNA immunoprecipitations showed a strong correlation in co-precipitating mRNAs, demonstrating that these data are highly reproducible (r = 0.92) (Fig. 1D). We validated the high-throughput sequencing results by reverse transcription quantitative PCR (RT-qPCR) and found that ESCRT-II reproducibly interacts with top candidates identified by RIP-Seq but not with RNAs that are under-represented in the RIP-Seq library (Fig. 1E). Gene ontology analysis revealed that ESCRT-II–associated transcripts encode proteins involved with cell adhesion, membranes, the cytoskeleton/centrosome, and cell motility (31, 32) (Table 1). We concluded that mRNA binding by ESCRT-II is a conserved property of the complex and that ESCRT-II interacts with a wide variety of cellular mRNA.
Figure 1.
ESCRT-II is an RNA-binding complex in Xenopus laevis eggs. A, ethidium bromide–stained agarose gel demonstrating the composition of total RNA and RNA that co-immunoprecipitates with anti-ESCRT-II antibodies or nonspecific rabbit IgG (Mock). Equal masses of total RNA and ESCRT-II RNA samples and equal volumes of ESCRT-II and mock RNA samples were loaded. B, scatter plot demonstrating the normalized sequencing reads of RNA isolated from an ESCRT-II IP or total RNA from one replicate. Points highlighted in red are 2-fold–enriched in the ESCRT-II IP over total egg extract RNA with a p value of <0.01 (calculated using two biological replicates and the edgeR exact test, based on the quantile-adjusted conditional maximum likelihood model). The blue line depicts equal values in both samples. C, histogram of ESCRT-II enrichment. The blue line indicates the mean. D, correlation of ESCRT-II enrichment (RPKM (reads per kilo base per million) in ESCRT-II IP/Total) across two biological replicates. The Pearson correlation coefficient is 0.92. The red points represent mRNAs that are significantly enriched in ESCRT-II IPs. The blue line depicts equal values in both samples. E, RT-qPCR validation of ESCRT-II enrichment (abundance in ESCRT-II immunoprecipitations/abundance in total RNA) of transcripts identified by RIP-Seq as associated with ESCRT-II. 28S rRNA or transcripts predicted by RIP-Seq to be under-represented in ESCRT-II immunoprecipitations were used as negative controls. Error bars represent S.E. from three biological replicates.
Table 1.
Gene ontology analysis of the ESCRT-II RIP-Seq library
Shown is a summary of the top four annotation clusters in overrepresented and underrepresented transcripts in the ESCRT-II RIP-Seq library identified by NCBI DAVID (31, 32).
| Annotation cluster | Enrichment score |
|---|---|
| Over-represented clusters | |
| Cell adhesion | 6.85 |
| Membranes | 3.86 |
| Cytoskeleton/Centrosome | 3.63 |
| Cell motility | 3.18 |
| Under-represented clusters | |
| Ribosomal/cytosolic proteins | 25.15 |
| Mitochondria | 3.51 |
| Glutathione S-transferase | 2.37 |
| rRNA/ncRNA processing | 2.24 |
ESCRT-II directly interacts with RNA through Vps25 in egg extract
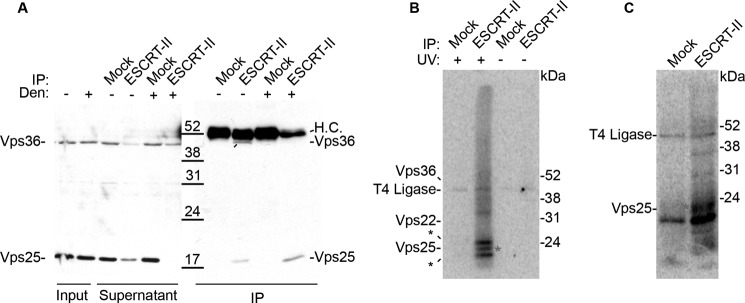
To determine whether ESCRT-II interacts directly with RNA targets and to determine which subunit of ESCRT-II is responsible for RNA binding, we performed UV cross-linking and immunoprecipitation (CLIP) (33, 34) in Xenopus egg extract. In this assay, RNA–protein complexes were cross-linked by UV irradiation, and then ESCRT-II immunoprecipitations were performed under native conditions. The immunoprecipitations were subsequently treated with RNase A, and then any remaining cross-linked and protein-protected nucleic acids were radiolabeled. The isolated RNA–protein complexes were separated by SDS-PAGE, and proteins bound directly to RNA were identified by the presence of the radiolabel. As a result, this assay not only reveals whether a protein of interest is directly bound to RNA, but in the case of an entire complex, may be used to identify the subunits of the complex that are responsible for RNA-binding. Our polyclonal ESCRT-II antibodies were raised against the entire ESCRT-II complex, but our ESCRT-II antibodies primarily recognize Vps25 and more weakly recognize Vps36 by Western blotting (Fig. 2A). However, immunoprecipitation for any ESCRT-II subunit co-immunoprecipitates the entire complex; therefore, any subunit of ESCRT-II that directly interacts with RNA will be detectable by this assay.
Figure 2.
ESCRT-II binds directly to RNA through Vps25 in Xenopus egg extracts. A, Western blotting of ESCRT-II IPs or mock IPs (nonspecific rabbit IgG) performed under native (−) or denaturing (+) conditions. Vps22 was not detectable by Western blotting with our ESCRT-II polyclonal antibody. H.C., heavy chain. B, autoradiograph of a CLIP experiment from Xenopus egg extract under high RNase conditions (0.1 mg/ml). A radioactive band consistent with the molecular mass of Vps25 (denoted by the red asterisk) is observed, but no bands at the molecular mass of Vps22 or Vps36 are apparent. The expected migrations of the ESCRT-II subunits are indicated to the left of the gel. T4 RNA ligase forms a covalent intermediate with pCp (used to radiolabel the RNA fragments) and appears in every lane. The bands above and below the Vps25 band (denoted by black asterisks) are nonspecific, as they appeared in the IgG control in some replicates of this experiment. C, autoradiograph of a CLIP experiment from Xenopus egg extract performed as described in B, except under denaturing immunoprecipitation conditions. The same polyclonal ESCRT-II antibody was used for A–C, but under denaturing conditions this antibody only immunoprecipitates Vps25.
Using this approach, we observed a radiolabeled band at 20 kDa, which is consistent with the molecular mass of Vps25 cross-linked to a short piece of RNA (3 kDa larger than Vps25 alone) (Fig. 2B). This band was absent in the mock immunoprecipitation as well as in a control performed without UV cross-linking. We did not observe any signals consistent with RNA binding to Vps22 or Vps36, although the presence of faint signals at molecular masses that are inconsistent with ESCRT-II subunits suggests that additional RNA-binding proteins associate with the complex. Together these results suggest that Vps25 is the RNA-binding subunit of ESCRT-II in Xenopus egg extract.
To confirm that the radiolabeled 20-kDa band observed in the ESCRT-II immunoprecipitation was in fact Vps25 and not a co-immunoprecipitating protein of the same size, we repeated the UV cross-linking experiment under denaturing immunoprecipitation conditions. As our ESCRT-II antibodies preferentially recognize Vps25, under denaturing conditions only Vps25 was isolated (Fig. 2A). Consistently, the 20-kDa radiolabeled band remained when Vps25 was immunoprecipitated under denaturing conditions, providing further evidence that Vps25 mediates RNA binding by ESCRT-II in Xenopus eggs (Fig. 2C).
Vps25 directly binds to RNAs that contain a GA-rich motif
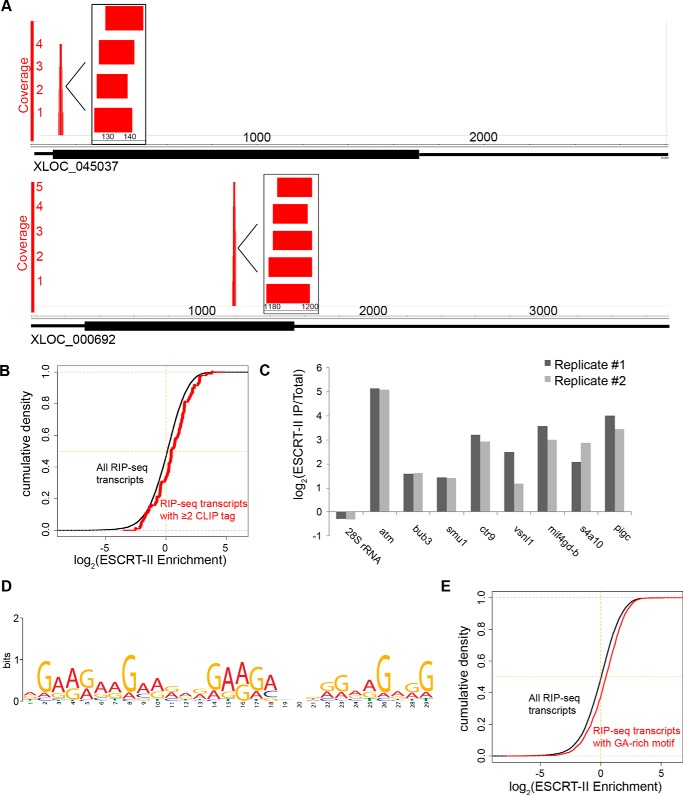
To determine whether ESCRT-II recognizes specific sequence motifs, we performed CLIP-Seq using the UV–cross-linked material that immunoprecipitated with Vps25 under denaturing conditions. Using the Illumina MiSeq platform, we obtained 11,167 unique, non-rRNA reads, which we aligned to a custom transcriptome derived from the Xenopus laevis 7.0 genome and our own sequencing data (35). The unique reads aligned to 1640 transcripts (Table S1 and S3), of which 196 transcripts had more than one aligning read. In addition, 114 transcripts had multiple CLIP tags (uniquely aligned reads) that overlapped at a particular region of the transcript (pileups), suggesting a selective binding site of ESCRT-II on the mRNA (Fig. 3A). The presence of multiple CLIP tags correlated with greater enrichment in native ESCRT-II immunoprecipitations as revealed by high-throughput sequencing (Fig. 3B) and RT-qPCR (Fig. 3C), providing validation that Vps25 CLIP tags arise from RNAs that co-immunoprecipitate with the entire ESCRT-II complex.
Figure 3.
CLIP analysis of ESCRT-II–bound RNAs. A, genome browser views of CLIP read clusters for two transcripts. Inset illustrates a higher resolution view of aligned CLIP reads. 114 transcripts in the ESCRT-II CLIP-Seq library had overlapping unique reads. B, cumulative distribution plot demonstrating the fraction of RIP-Seq transcripts with ≥2 CLIP tags (red) or the entire ESCRT-II RIP-Seq library as a function of their enrichment in the ESCRT-II RIP-Seq library relative to total RNA. The p value for the difference between the two distributions is <0.0014 using a two-sided, unpaired t test. C, RT-qPCR validation of ESCRT-II enrichment (abundance in ESCRT-II immunoprecipitation/abundance in total RNA) of transcripts containing CLIP tags compared with a negative control RNA (28S rRNA). Two biological replicates are shown. D, MEME motif analysis (36) was used to analyze the sequence motifs present in ESCRT-II CLIP tags. The top-scoring motif is shown. E, cumulative distribution plot demonstrating the fraction of RIP-Seq transcripts containing a GA-rich motif (a 30-nt stretch of at least 90% purines) (red) or the entire ESCRT-II RIP-Seq library (black) as a function of their enrichment in the ESCRT-II RIP-Seq library. The p value for the difference between the two distributions is <1 × 10−12 using a two-sided unpaired t test.
Our RIP-Seq data demonstrated that ESCRT-II selectively interacts with specific mRNA targets. This selective interaction could be achieved through the recognition of a sequence or structural element on the RNA target or through additional trans-acting factors associated with the transcripts, or it could be influenced by the local population of transcripts in the vicinity of ESCRT-II in vivo. To determine whether ESCRT-II exhibited sequence-specific RNA binding, we performed a motif analysis of the CLIP-Seq library using MEME (36). The top motif was a stretch of G and A nucleotides that appeared in 113 of 1640 of the CLIP-Seq transcripts and 14 of 114 of the CLIP-Seq transcripts that had multiple reads (Fig. 3D). Additionally, a poly-C motif was present in 96 of 1640 CLIP-Seq transcripts and 46 of 114 CLIP-Seq transcripts with multiple reads. In support of the possibility that the GA-rich motif promotes binding to ESCRT-II, we found that GA-rich sequences (having a 30-nucleotide stretch of at least 90% purines) were preferentially enriched in the ESCRT-II RIP-Seq library compared with other ESCRT-II–bound transcripts (Fig. 3E and Table 2) (p < 1 × 10−12 using a two-sided, unpaired t test). No other dinucleotide or single nucleotide motif displayed preferential enrichment in ESCRT-II immunoprecipitations, with the exception of stretches of cytosines, the second motif identified in the CLIP-Seq library (Table 2). We concluded that ESCRT-II binds preferentially to mRNAs containing specific sequence motifs in Xenopus egg extracts but that ESCRT-II is also likely to recognize multiple sequence motifs.
Table 2.
Transcripts containing GA- or C-rich motifs are more enriched in ESCRT-II immunoprecipitations compared with the total RIP-Seq library
Shown are the means of the distributions of RIP-Seq transcripts containing 30-nt stretches of the indicated nucleotides or dinucleotides (see “Experimental procedures” regarding allowance of mismatches) compared with the mean of the distribution of all ESCRT-II RIP-Seq transcripts. The p values were determined using a two-sided, unpaired t test comparing the distributions of the indicated RIP-Seq transcripts with the distribution of all ESCRT-II RIP-Seq transcripts.
| Transcripts | Mean enrichment value | p value |
|---|---|---|
| All RIP-Seq transcripts | 1.59 | |
| GA | 1.97 | 9.19 × 10−13 |
| AC | 1.78 | 0.06 |
| AU | 1.61 | 0.55 |
| CU | 1.6 | 0.88 |
| GU | 1.75 | 0.02 |
| CG | 1.67 | 0.2 |
| A | 1.65 | 0.24 |
| U | 1.68 | 0.21 |
| G | 1.6 | 0.88 |
| C | 1.83 | 7.76 × 10−5 |
ESCRT-II interacts nonspecifically with RNA in vitro through the membrane-binding domain of Vps22
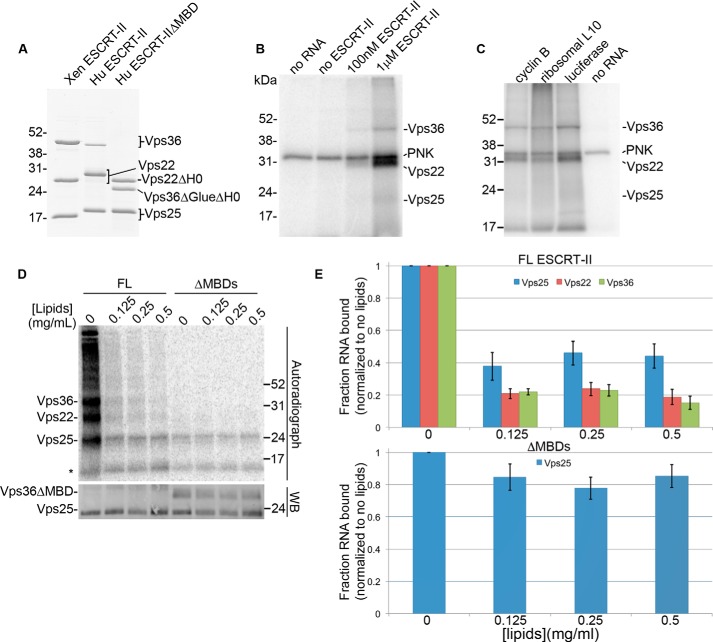
Our results indicated that ESCRT-II interacts directly with RNA through the Vps25 subunit in Xenopus egg extracts and that the selective interaction between ESCRT-II and a portion of its RNA targets could be determined by recognition of a purine-rich motif. We next asked whether we could recapitulate sequence-specific RNA binding in vitro using purified components (Fig. 4A). We first determined whether recombinant ESCRT-II interacts selectively with purified RNAs through Vps25 in vitro. To address this question, we performed binding reactions between recombinant ESCRT-II and purified RNA from Xenopus egg extracts. We then performed in vitro UV cross-linking on the binding reactions using an approach similar to the CLIP protocol to identify the RNA-binding subunit of recombinant ESCRT-II. Surprisingly, we found that binding to total egg RNA in vitro occurred primarily through Vps22, with some detectable binding through Vps36 and Vps25 (Fig. 4B), which contrasts with our observation of RNA binding primarily through Vps25 in egg extract. Similar results were obtained using single in vitro transcribed RNAs corresponding to transcripts under-represented in an ESCRT-II immunoprecipitation, suggesting that this in vitro interaction is nonspecific (Fig. 4C).
Figure 4.
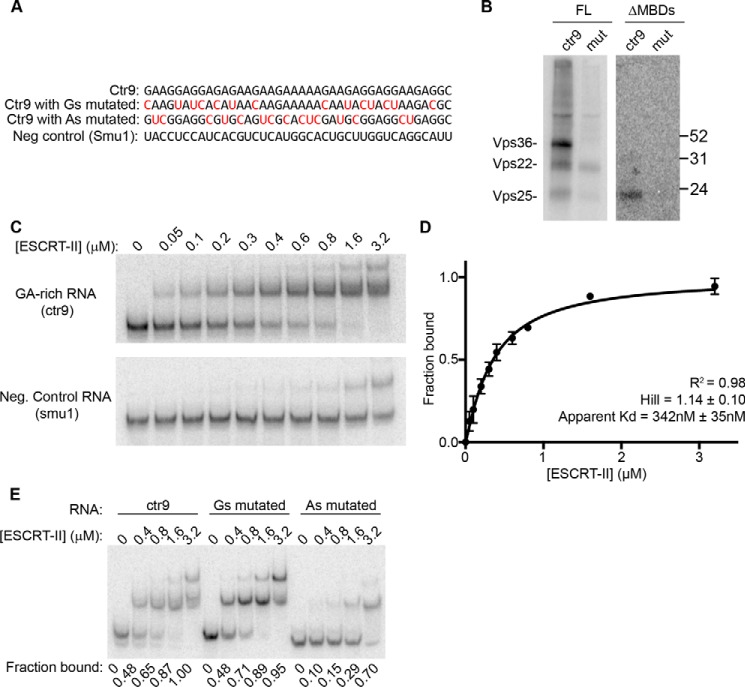
Analysis of ESCRT-II/RNA binding in vitro. A, Coomassie Blue-stained gel of the recombinant Xenopus (Xen) and human (Hu) ESCRT-II complexes used in the in vitro RNA-binding assays. ΔMBD lacks the membrane-binding domains of human ESCRT-II. B–D, autoradiographs of UV–cross-linked in vitro binding reactions with: B, Xenopus ESCRT-II and 5′-end-labeled total egg RNA; C, Xenopus ESCRT-II and individual 5′-end-labeled in vitro transcribed RNAs that are under-represented in ESCRT-II immunoprecipitations; and D, full-length HuESCRT-II (FL) or HuESCRT-IIΔMBD (ΔMBD) and a body-labeled, in vitro transcribed GA-rich CLIP tag (a region of the ctr9 mRNA). B and C, a covalent intermediate of PNK and [γ-32P]ATP (used to radiolabel the RNA fragments) is indicated. D, Folch fraction liposomes were included in the binding reactions at the indicated concentrations. A fluorescent Western blotting (WB) of the same nitrocellulose membrane shown in the autoradiograph is shown as a loading control. The asterisk represents a nonspecific band. A–D, the expected migrations of the ESCRT-II subunits are indicated. E, quantification of the autoradiograph shown in D and two additional, independent replicates depicting the fraction of RNA bound by each ESCRT-II subunit at the indicated concentrations of Folch fraction liposomes relative to binding with no liposomes present. Error bars are S.E.
Vps22 and Vps36 both contain membrane-binding domains; Vps22 binds to membranes through its H0 domain (21) and Vps36 binds to membranes through its GLUE domain (21, 24, 25). Because these positively charged domains bind to negatively charged phospholipids in cells, we reasoned that in vitro, these domains may be inappropriately exposed and could bind nonspecifically to the negatively charged RNA backbone. We therefore addressed the influence of membranes on the RNA-binding properties of the ESCRT-II subunits. Using the in vitro UV cross-linking assay, we examined the interaction of ESCRT-II with a GA-rich ESCRT-II CLIP tag (a region of the CTR9 homolog, Paf1/RNA polymerase II complex component (ctr9) mRNA) in the presence or absence of Folch fraction liposomes, used previously to investigate interactions between ESCRT-II and membranes in vitro (25, 29). In support of the hypothesis that RNA binding by Vps22 and Vps36 in vitro occurs through their membrane-binding domains, we found that binding of the ctr9 CLIP tag to Vps22 and Vps36 is reduced in the presence of Folch fraction liposomes in a dose-dependent manner, whereas RNA binding through Vps25 shows an initial decrease in the presence of lipids but is unaffected by increasing lipid concentrations (Fig. 4, D and E). Deletion of the membrane-binding domains in Vps22 and Vps36 (ESCRT-II ΔMBD) (21) (Fig. 4A) abrogated the binding of these subunits to RNA and rendered the affinity of the ESCRT-II complex for RNA largely insensitive to the presence of membranes (Fig. 4, D and E). The initial decrease in RNA binding by Vps25 in response to lipids in the context of full-length ESCRT-II but not the ΔMBD construct suggests that nonspecific RNA binding by the Vps22 and Vps36 membrane-binding domains may promote the overall affinity of ESCRT-II for RNA, whereas lipid binding by Vps22 and Vps36 may negatively regulate ESCRT-II/RNA interactions. Of note, the full-length and ΔMBD ESCRT-II complexes used in this assay were human ESCRT-II. Thus, our results suggest that RNA binding by ESCRT-II is additionally conserved in humans. We conclude that the membrane-binding domains of ESCRT-II interact nonspecifically with RNA in vitro and that membranes and RNA compete for the same binding site in Vps22 and Vps36.
Vps25 has specificity for GA-rich sequences in vitro
Our ESCRT-II/RNA binding data in egg extract suggest that Vps25 has specificity for GA-rich sequences. To address whether ESCRT-II binds specifically to this sequence motif in vitro through Vps25, we compared the binding of full-length human ESCRT-II or ESCRT-II ΔMBD to the ctr9 GA-rich CLIP tag or a mutated version of ctr9 that lacks the majority of the adenosines in the motif (Fig. 5A). As described above, full-length ESCRT-II bound to ctr9 through each of its subunits, and Vps22 bound to the mutated ctr9 RNA to a similar degree (Fig. 5B). In contrast, ESCRT-II ΔMBD cross-linked only to ctr9 in vitro, and consistent with our previous results, this binding occurred only through Vps25 (Fig. 5B). Thus, the mechanism and specificity of the ESCRT-II/RNA interaction that we observed in egg extract can be recapitulated in vitro using purified components.
Figure 5.
ESCRT-II specifically binds GA-rich RNAs in vitro. A, sequences of transcripts used for in vitro binding reactions. Nucleotides highlighted in red are mutations made to the ctr9 CLIP tag. B, autoradiograph of UV–cross-linked in vitro binding reactions as described in Fig. 4, performed with recombinant full-length HuESCRT-II (FL) or HuESCRT-II ΔMBD (ΔMBD) and the in vitro transcribed, body-labeled ctr9 CLIP tag or the ctr9 CLIP tag with the adenosines mutated. C, RNA EMSA with ESCRT-II ΔMBD at the indicated concentrations and the ctr9 CLIP tag or a negative control sequence of the same length (a region of the smu1 transcript). D, quantification of three independent RNA EMSA experiments using ESCRT-II ΔMBD and the ctr9 CLIP tag. Error bars are S.E. E, RNA EMSA reactions with ESCRT-II ΔMBD at the indicated concentrations and the ctr9 CLIP tag containing or lacking mutations in the majority of guanosines or adenosines. The fraction of RNA bound by ESCRT-II ΔMBD is indicated below each lane.
We additionally observed that Vps36 bound specifically to the GA-rich CLIP tag in vitro in the context of full-length ESCRT-II (Fig. 5B). This interaction depends on its membrane-binding domain and is inhibited by Folch fraction liposomes (Fig. 4D), suggesting that membranes and the GA-rich CLIP tag bind to the same region of Vps36 in vitro. Vps36-mediated RNA binding was not observed in egg extracts and will require further detailed study to investigate its in vivo significance. Of note, the interaction between ESCRT-II and bicoid mRNA was suggested to occur through the membrane-binding domain of Vps36 based on a similar in vitro UV cross-linking assay (19).
To further investigate the interaction between Vps25 and GA-rich sequences in vitro, we performed an RNA electrophoretic mobility shift assay (EMSA) with the ctr9 GA-rich CLIP tag and ESCRT-II ΔMBD, which was chosen to avoid the contribution of nonspecific binding. We found that ESCRT-II ΔMBD binds to the ctr9 CLIP tag with an apparent Kd of ∼342 nm and has a much lower affinity for a negative control RNA (Fig. 5, C and D) or a version of the ctr9 CLIP tag that harbors mutations to its adenosines (Fig. 5E). In contrast, replacement of the guanosines in the ctr9 fragment with other nucleotides did not impact the affinity of ESCRT-II for the RNA (Fig. 5E), suggesting that the adenosines of the motif make a greater contribution to ESCRT-II binding.
Discussion
We have demonstrated that ESCRT-II is a conserved RNA-binding protein that interacts with hundreds of different mRNAs, validating the novel observation that ESCRT-II binds to bicoid mRNA and is required for its localization in Drosophila (19). Our data indicate that ESCRT-II interacts with mRNA through the Vps25 subunit in Xenopus egg extracts and in vitro. Additionally, we demonstrated that Vps25 exhibits sequence-specific RNA-binding properties. Our results provide molecular insight into the mechanism of RNA interaction by ESCRT-II and set the stage for further study of the impact that ESCRT-II has on RNA regulation.
Mechanism of the ESCRT-II/RNA interaction
Several recent studies have demonstrated that hundreds of proteins directly interact with mRNA, AND yet the majority of these mRNA-binding proteins do not contain annotated RNA-binding domains (4, 5, 8, 9). Our work highlights a critical need to understand how noncanonical RNA-binding proteins interact with mRNA. In this work we used UV cross-linking in Xenopus egg extract to demonstrate that ESCRT-II interacts directly with RNA through the Vps25 subunit, which is composed primarily of winged helix domains (20, 21, 29). Winged helix domains are well-characterized nucleic acid binding domains but are primarily found on DNA-binding proteins and are also involved in many protein/protein interactions (37). The structure of ESCRT-II is such that each subunit of Vps25 contains one solvent-exposed winged helix domain (20) with the ability to participate in nucleic acid interactions. The exposed surface of Vps25 that participates in interactions with RNA is also the same surface that is used to interact with the Vps20 subunit of ESCRT-III during MVB cargo sorting (38, 39). In the future it will be important to determine which amino acids participate in the interaction of Vps25 with RNA and if Vps25/RNA interactions are competitive with Vsp25/Vps20 interactions.
Initial studies of the interaction between ESCRT-II and bicoid mRNA have concluded, based on an in vitro assay, that ESCRT-II interacts with bicoid through the Vps36-GLUE domain (19). In our study, we found that the Vps36-GLUE domain could interact with RNA in vitro, but we failed to observe a Vps36/RNA interaction by performing CLIP in Xenopus egg extract. We additionally observed that the Vps22-H0 domain exhibits nonspecific RNA binding in vitro. The interaction of Vps36 and Vps22 with RNA was competitive with membrane binding, whereas the interaction of Vps25 with RNA was largely unaffected by binding to membranes. Our results suggest that Vps25, and possibly Vps36, drives sequence-specific mRNA interactions that may be aided by promiscuous interactions between RNA and the Vps22-H0 domain. Upon membrane binding, the interaction of ESCRT-II with RNA would occur primarily through the Vps25 subunit. In the future it will be important to examine the precise coordination of RNA binding by ESCRT-II upon recruitment to the endosome.
Implications of RNA binding by ESCRT-II
ESCRT-II was first demonstrated to interact with RNA based on the observation that ESCRT-II is required for the normal localization of bicoid mRNA. Additionally, a recent study demonstrated that ESCRT-II deletion mutants impact the translation of deleted in colorectal cancer (DCC) mRNA, a transcript that is localized to growth cones during neuronal development in Xenopus (40). In this study, we did not test the role of ESCRT-II in mRNA localization in Xenopus eggs because of a lack of the genetic resources necessary to address such questions. However, many of the ESCRT-II–associated mRNAs display distinct localization patterns in the cell. ESCRT-II is associated with a number of cytoskeletal mRNAs, which are enriched on spindle microtubules (12, 41), but it does not dramatically associate with mRNAs that localize to the vegetal cortex in Xenopus oocytes. Many ESCRT-II–enriched mRNAs encode membrane-binding proteins that are translated on endoplasmic reticulum–associated ribosomes. Given that endosomes undergo transient interactions with different membrane compartments in the cell, it will be interesting to examine the relationship between ESCRT-II/RNA binding and membrane interactions.
Why the cell would repurpose an endosomal sorting complex as an RNA-binding factor is a major outstanding question. One possibility is that ESCRT-II might play completely separate roles in RNA regulation and membrane trafficking. Cases of such “moonlighting” proteins are widespread throughout biology and may arise to reduce the number of genes required in an organism or as a natural process in evolution to adapt existing proteins for new functions (reviewed in Ref. 42). However, a more exciting possibility is that ESCRT-II could provide a link between RNA regulation and endosome biology. In particular, it will be interesting to investigate whether ESCRT-II is involved in the selective sorting of RNAs into exosomes (43) or whether ESCRT-II interacts with mRNAs undergoing miRNA-mediated silencing on endosomes (44, 45). Our finding that a core component of the ESCRT machinery is a conserved RNA-binding factor warrants further investigation into the interconnection of RNA regulation and endosome biology.
Experimental procedures
Recombinant proteins
Two expression constructs of the ESCRT-II complex were generated by cloning the open reading frames of X. laevis Vps22 (IMAGE clone: 7197421), Vps36 (IMAGE clone: 6641897), and Vps25 (pMB 404) into the polycistronic pST39 vector (46). The first contained a His6 tag on the N terminus of Vps22 (pMB 449) and the second contained a GST tag at the N terminus of Vps36 (pMB 432). Xenopus IMAGE Consortium cDNA clones were purchased from Open Biosystems. The coding region of Vps25 was cloned from egg extract. Full-length human ESCRT-II and the human ESCRT-II deletion construct lacking membrane-binding domains (HuESCRT-II ΔMBD) with His6 tags on the N terminus of Vps22 were a gift from James Hurley (21). HuESCRT-II ΔMBD is as follows: Vps25(1–176), Vps22(25–258), and Vps36(170–386). His6-tagged proteins were expressed in BL21 Rosetta, Rosetta(DE3), or Rosetta(DE3)pLysS-competent cells. Cultures grown to an OD of 0.4–0.6 were then induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 16 °C or were induced at OD 1.0 and grown for 1 h at 37 °C. Cells were collected by centrifugation and resuspended in PBS plus 10 mm imidazole and protease inhibitors (leupeptin, pepstatin, and chymostatin). Cells were lysed by French press, treated with RNase A, type XIIa (Sigma) and RQ1 DNase (Promega), and then cleared by centrifugation at 12,000 × g for 20 min. Proteins were bound to nickel-nitrilotriacetic acid–agarose (Qiagen), washed with PBS plus 10 mm imidazole, eluted with PBS plus 500 mm imidazole, and then dialyzed to PBS. Recombinant protein used for RNA EMSA was further purified by gel filtration using a Superdex 200 column and eluted in 1× RNA-binding buffer (see “RNA EMSA” below). GST-ESCRT-II, expressed and induced similarly, was purified with GSH-agarose beads (GE Healthcare Life Sciences) according to the manufacturer's instructions. Single-use aliquots of purified proteins were snap-frozen in liquid nitrogen and stored at −80 °C.
Antibodies and Western blotting
Two different rabbit polyclonal Xenopus ESCRT-II antibodies were generated by immunizing rabbits with either GST- or His6-tagged ESCRT-II by Covance. Serum was affinity-purified by incubation overnight at 4 °C with His6–ESCRT-II coupled to Affi-Gel 10–agarose beads (Bio-Rad Laboratories). Following washes with PBS, the antibody was eluted from the beads with 0.2 m glycine, 500 mm NaCl, pH 2.0. The pH of the eluted antibody was neutralized with 1.5 m Tris-Cl, pH 8.8, and then the eluted antibody was dialyzed into PBS plus 50% glycerol. Whole-molecule ChromPure rabbit IgG was purchased from Jackson ImmunoResearch. Chicken polyclonal antibodies against human ESCRT-II were generated against HuESCRT-II ΔMBD by Aves Labs, Inc., and purified against full-length HuESCRT-II. In general, antibodies were used at 0.2 μg/ml for Western blotting. All Western blotting was detected with horseradish peroxidase–conjugated secondary antibodies from Jackson ImmunoResearch at 1:10,000. 12% Tris-glycine–SDS-polyacrylamide gels were used for Western blotting.
Xenopus laevis egg extracts and RNA immunoprecipitations
Xenopus egg extract was prepared as described previously (47). All experiments performed using X. laevis were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee (Office of Laboratory Welfare (OLAW) Assurance A3596-01). Anti-ESCRT-II or Rabbit IgG was coupled to protein A–Dynabeads (Life Technologies) according to the manufacturer's instructions. In general, 40 μg of antibody and 160 μl of Dynabeads were used per 100 μl of egg extract for immunoprecipitations. In some cases, the extract was diluted 1:4 in PBS prior to the immunoprecipitation; we confirmed that this dilution did not affect the enrichment of candidate RNAs in an ESCRT-II immunoprecipitation. The RNA immunoprecipitations presented in Fig. 3C were performed from extract diluted in PBS plus 0.5% Triton. Extract was incubated with antibody-coupled beads at 4 °C for 1–2 h. Following incubation, the beads were washed twice with PBS plus 0.5% Triton and twice with PBS alone; then the beads were transferred to a fresh tube and resuspended in TRIzol® (Life Technologies). 2 μl of the input extract was also resuspended in TRIzol, and RNA was purified from all samples following the manufacturer's instructions. Purified RNA was resuspended in water and analyzed on a 0.8% agarose gel with ethidium bromide, used as input for library construction, or used as input for reverse transcription followed by qPCR.
RIP-Seq library construction and sequencing
RIP-Seq libraries were prepared using the TruSeq stranded mRNA sample prep kit (Illumina). 200 ng of input and ESCRT-II–immunoprecipitated RNA samples prepared from biological replicates were first enriched for mRNA by LNA oligo(dT) selection (Exiqon) and then were used as input for library construction. Barcoded libraries were pooled and sequenced by SR-50 on the Illumina HiSeq platform.
Reverse transcription quantitative PCR
200 ng of RNA samples was used for reverse transcription with random hexamers and SuperScript III® reverse transcriptase (Life Technologies) according to the manufacturer's instructions. Samples were treated with RNase H (New England Biolabs (NEB)) for 20 min at 37 °C following the reverse transcription reaction. The cDNA was used as input for qPCR with iQTM SYBR® Green Supermix (Bio-Rad Laboratories); primer sets are listed in Table S2. qPCRs for each biological replicate were performed in technical triplicates.
UV cross-linking experiments and CLIP-Seq
Native UV cross-linking experiments in egg extract, modeled after previously described CLIP protocols (33, 48), were performed by UV cross-linking of egg extract diluted 1:10 in PBS for 10 min on ice in a Stratalinker 2400 (Stratagene) equipped with UVC bulbs. Samples without UV cross-linking were performed in parallel. Samples were treated with 0.1 mg/ml RNase A type XIIa (Sigma) for 30 min at room temperature and then cleared by centrifugation at 10,200 × g for 10 min at 4 °C. Samples were precleared with protein A–Dynabeads for 1 h at 4 °C at a ratio of 100 μl of beads/1 ml of diluted extract. Samples were then immunoprecipitated with anti-ESCRT-II or mock immunoprecipitated with rabbit IgG and washed as described above for RNA immunoprecipitations. Samples resuspended in 1× PNK buffer (NEB) with 1 unit/μl RNase inhibitor (Roche Applied Science) and 0.75 unit/μl T4 PNK (NEB) were incubated for 45 min at 37 °C to dephosphorylate the 3′-ends of the RNA. The beads were then washed three times in PBS, transferred to a fresh tube, and then resuspended in 1× T4 RNA ligase buffer (50 mm Tris-HCl, 10 mm MgCl2, and 10 mm DTT, pH 7.5) with 1 mm ATP, 0.02 mg/ml BSA, and 1 unit/μl RNase inhibitor (Roche Applied Science) and 3′-end–labeled by the addition of 1 μl of pCp (cytidine 3′-5′-(bis)phosphate, 5′-32P, 3000 Ci/mmol, PerkinElmer) and 0.5 μl of T4 RNA ligase (Fermentas) per 20-μl reaction. The reactions proceeded for 2 h at room temperature and were then washed three times in PBS, transferred to a fresh tube, resuspended in protein loading buffer, run on SDS-PAGE, transferred to a nitrocellulose membrane, and analyzed by autoradiography on a Typhoon Trio scanner (GE Healthcare).
The denaturing UV cross-linking experiments followed a similar protocol, except the extracts were initially diluted 1:4 in PBS for UV cross-linking, and then following RNase A treatment, SDS and EDTA were added to a final concentration of 0.5% and 1 mm, respectively. The samples were heated at 70 °C for 10 min and then diluted 1:2 with PBS plus 1% Triton to reduce the SDS concentration for immunoprecipitation.
To construct the CLIP-Seq library, UV cross-linking and denaturing immunoprecipitations were performed as described above except with a low RNase A concentration (1 μg/ml); and instead of 3′-end labeling with pCp, the samples were resuspended in a 20-μl mix containing 1× T4 RNA ligase 2, truncated buffer (NEB), 6 μl of PEG 8000, 1 μl of RNase inhibitor (Roche Applied Science), 1 μl of truncated T4 RNA ligase 2 (NEB), and 30 pmol of adenylated DNA oligo (Blower lab oligo database number oMB1863: 5′-rAppCGGCCGCCACCATCAAT-3ddC-3′). The ligation reaction was incubated overnight at 16 °C with agitation in a ThermoMixer. The samples were washed, electrophoresed, and transferred as described above, and then a region of the nitrocellulose between 31 and 38 kDa was excised and RNA extracted as described previously (48), except with preincubation of the proteinase K solution at 37 °C to inactivate the RNase. Following the proteinase K treatment, the RNA was phenol-chloroform extracted.
SDS-PAGE used for UV cross-linking experiments in egg extract included: 4–12% NuPage Bis-Tris (for native UV CLIP experiment), 12% NuPage Bis-Tris (for low RNase denaturing immunoprecipitations (IP) used for library construction) run in MOPS running buffer (50 mm MOPS, 50 mm Tris, 1 mm EDTA, and 0.1% SDS), or 12% Tris-glycine (for high RNase denaturing IP). Western blotting of all samples was performed in parallel to monitor the immunoprecipitation of ESCRT-II.
CLIP-Seq library construction and sequencing
A custom library protocol was developed using primers and adapters similar to previously described protocols (49), except with a NotI site inserted into the 3′-adapter and the reverse transcription primer for enzymatic removal of empty adapters. The isolated ESCRT-II CLIP RNA was used as input for reverse transcription with 20 pg of the oMB1861 primer (5′-(Phos)-CAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGT CGC-(iSp18)-CACTCA-(iSp18)-TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGGCGGCC G-3′) in a 10-μl reverse transcription reaction using SuperScript III® reverse transcriptase (Life Technologies) as described below. The sample was then precipitated with isopropanol and resuspended in 1× CircLigase buffer with 50 nm ATP, 2.5 mm MnCl2, and 0.3 μl of CircLigase (Epicentre) in a 10-μl reaction. The sample was incubated for 1 h at 60 °C, then for 10 min at 80 °C, and then cleaned up with the QIAquick PCR purification kit (Qiagen). The sample was PCR-amplified using oMB1522 (5′-AATGATACGGCGACCACCGAGATCTACAC-3′) and oMB1904 (5′-TTCAGACGTGTGCTCTTCCG-3′), isopropanol-precipitated, and then run on an 8% TBE–acrylamide gel. The bands containing an insert were gel-purified away from unused empty adapters and then digested with NotI (NEB) to further remove empty adapters. The sample was purified with the QIAquick PCR purification kit and then PCR-amplified with oMB1519 (5′-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGCTCTTCCG-3′) and oMB1522 to incorporate Illumina adapter sequences. After purification with the QIAquick PCR purification kit, the sample was sequenced by SR-50 on the Illumina MiSeq platform.
In vitro UV cross-linking experiments
1 μg of purified total egg RNA or 15 nm unlabeled in vitro transcribed RNA was incubated with the indicated concentrations of ESCRT-II in 100 μl of PBS for 30 min at room temperature. The samples were then UV–cross-linked for 5 min on ice in a Stratalinker 2400 (Stratagene) equipped with UVC bulbs and then treated with RNase A (Sigma type XIIA) at 0.1 mg/ml for 30 min at room temperature. ESCRT-II was then isolated by immunoprecipitation with anti-ESCRT-II coupled to protein A–Dynabeads, and after washing, the beads were resuspended in PNK buffer and treated with PNK (NEB) and [γ-32P]ATP (PerkinElmer) for 45 min at 37 °C. The beads were washed with PBS, resuspended in protein loading buffer, and analyzed by SDS-PAGE and autoradiography.
ESCRT-II human deletion recombinant proteins were UV–cross-linked to body-labeled RNA to avoid obscuring bands with the PNK-[γ-32P]ATP intermediate. Body-labeled RNA fragments were in vitro transcribed with T7 RNA polymerase using PCR products amplified from the pCR2.1 vector (Life Technologies) containing the indicated sequences (see “RNA EMSA”) as a template in the presence of [α-32P]GTP at a ratio of unlabeled GTP to labeled GTP of 160:1. The fragments were then purified by a G25 Sephadex column (GE Healthcare) and phenol-chloroform extraction. Binding reactions were performed with 100 nm RNA and 1 μm ESCRT-II recombinant protein in the presence of RNase inhibitor. The UV cross-linking experiment was performed as described above except without 5′-end labeling and with the anti-HuESCRT-II antibody used for the immunoprecipitation. Rabbit anti-chicken (IgY) Fc fragment (Jackson ImmunoResearch) was used to couple the chicken anti-HuESCRT-II to protein A–Dynabeads.
For UV cross-linking experiments including liposomes, brain extract lipids (Folch fraction type I, Sigma) were resuspended in a buffer containing 20 mm HEPES, pH 7.4, and 150 mm NaCl; then vesicles were formed by sonication and cleared by centrifugation at maximum speed for 5 min. The supernatant containing the liposomes was included at the indicated concentrations in in vitro binding reactions with human ESCRT-II recombinant proteins and body-labeled RNA as described above. All other steps were performed similarly, except the immunoprecipitations were washed twice with PBS plus 1% Triton X-100, twice with PBS, and then transferred to a fresh tube prior to SDS-PAGE. Autoradiograph bands were quantified using ImageJ software.
SDS-PAGE for UV cross-linking experiments were performed with 12% Tris-glycine gels or 12% NuPage Bis-Tris gels. Western blotting was performed in parallel to monitor the immunoprecipitation of ESCRT-II. In the experiments in which body-labeled RNA was used, the gels were transferred to nitrocellulose for autoradiography to remove free RNA. Western blotting to monitor immunoprecipitation of ESCRT-II was performed using an Alexa Fluor® 647 anti-chicken secondary antibody (Invitrogen, Molecular Probes) and imaged on a Gel DocTM XR+ gel documentation system (Bio-Rad).
RNA EMSA
To generate RNA probes for EMSA, the indicated sequences were cloned into the NotI and BamHI sites of pCR2.1 (Life Technologies), amplified with T7 and M13 primers, and in vitro transcribed with T7 RNA polymerase. In vitro transcription reactions were supplemented with [α-32P]ATP at a ratio of unlabeled to labeled ATP of 160:1. After completion, the in vitro transcription reactions were DNase-treated, gel-purified, then phenol-chloroform–extracted. 200 pm radioactively labeled RNA probe was used for the EMSA. RNA probes were first heated to 95 °C for 1 min and then cooled to 4 °C to remove secondary structure. Gel shifts were performed in 1× RNA-binding buffer (10 mm HEPES, 150 mm KCl, 1 mm MgCl2, 1 mm EDTA, 5% glycerol, 1 mm DTT, and 0.05% Nonidet P-40, pH 7.7) in the presence of 0.1 mg/ml yeast tRNA and 0.8 unit/μl RNase inhibitor (Roche Applied Science). HuESCRT-II ΔMBD was used for gel shifts at the indicated concentrations. Binding reactions proceeded for 30 min at 20 °C and then were loaded onto a prerun native 8% TBE–acrylamide gel (with 19:1 acrylamide:bis). Gels were run at 200 V for 55 min in 0.5× TBE at 4 °C and then dried and analyzed by autoradiography. Bands were quantified using ImageJ software, and curves were fit to the Hill equation using GraphPad Prism allowing the Hill coefficient and Kd value to float.
Analysis of Xenopus cDNA libraries
Sequencing reads from the Illumina HiSeq were collapsed into unique reads using a custom Perl script (50). Reads aligning to X. laevis rRNA sequences were subtracted using a custom Perl script and then were aligned to a draft version of the X. laevis genome (version 7.0, downloaded from http://www.xenbase.org/) (53)3 using TopHat and Cufflinks (51). Reads were counted against a custom gene model (35) using the Cuffdiff program. Only transcripts having more than 100 combined normalized alignment counts were included in further analyses.
All statistical tests of sequencing data were performed in R (individual tests are listed in each figure legend), except the statistical comparison of ESCRT-II–enriched RNAs versus total RNA was performed using edgeR (30). All graphs of sequencing data were prepared using R. All sequencing data generated in this study were deposited in the NCBI Sequence Read Archive (SRA) under accession number Bioproject PRJNA336357.
To perform gene ontology analysis, we used BlastX to search the X. laevis transcripts generated by Cufflinks to the Human Uniprot database and retained all hits with an e-value of <1 × 10−20. We then used the Human Uniprot accession numbers as input for gene ontology enrichment analysis using NCBI DAVID (31, 52). For background in this analysis, we used all expressed transcripts in which all replicates of the experiment had greater than 100 combined reads.
Analysis of CLIP-Seq cDNA library
FastQ sequences were filtered to remove linker sequences then were collapsed into unique reads using a custom Perl script. Reads with >90% of a single base were removed using a custom Perl script. The remaining reads were aligned to X. laevis rRNA genes using Bowtie allowing for up to three mismatches, and the aligning reads were removed. The remaining reads were aligned to sequences derived from a custom gene model (35) allowing up to three mismatches and retaining the best strata of alignments using the parameters –m 3 –best –strata. Reads aligning to more than two transcripts or to antisense strands were removed. Reads of equal length aligning to the same position of the transcriptome were removed under the assumption that they are PCR duplicates. The alignments were counted with a weighted counting strategy using a custom Perl script, such that if a read aligned twice, each transcript received a count of 0.5. We used the SAMtools mpileup command to create coverage files and a custom Perl script to analyze transcripts with multiple aligning reads. To perform motif analysis, we collapsed overlapping reads into CLIP tags and extended the sequences with 25 nt of flanking sequence from the aligned transcript on either side of the CLIP tag. The resulting sequences were used as input for motif prediction using MEME (36), with all expressed transcripts from our RIP-Seq datasets with greater than 100 reads used as the background model.
Analysis of dinucleotide or mononucleotide motifs
We used a 30-nt sliding window to scan all mRNAs in the Xenopus transcriptome for mono- and dinucleotide motifs. We calculated the window with the highest percentage of the bases of interest. To compare the motifs to mRNA enrichment in ESCRT-II immunoprecipitations, we first examined histograms of the percent identity distributions for each motif. We then generated mismatch cutoffs that resulted in approximately equal numbers of mRNAs in the top-scoring bin. For example, to obtain approximately equal occurrences of each motif, we allowed up to three mismatches in a 30-nt window for AG, whereas we allowed up to 12 mismatches for strings of G, which occurs less frequently in the transcriptome. We next identified transcripts that contain each motif (allowing the appropriate number of mismatches) and compared these subsets of transcripts to the total ESCRT-II RIP-Seq library for their enrichment in an ESCRT-II immunoprecipitation.
Author contributions
A. B. E. and M. D. B. conceptualization; A. B. E. and M. D. B. data curation; A. B. E. and M. D. B. formal analysis; A. B. E. validation; A. B. E. and M. D. B. investigation; A. B. E. visualization; A. B. E. and M. D. B. methodology; A. B. E. and M. D. B. writing-original draft; A. B. E. and M. D. B. writing-review and editing; M. D. B. supervision; M. D. B. funding acquisition; M. D. B. project administration.
Supplementary Material
Acknowledgments
We thank Ashwini Jambhekar and Caterina Schweidenback for critical comments on this manuscript and members of the Blower laboratory for helpful suggestions. We thank Josh Plant for preliminary work investigating the interaction of ESCRT-II with RNA in Xenopus eggs and Elijah Carrier for assistance with generating ESCRT-II antibodies. We thank Song Tan for kindly donating the polycistronic pST39 expression vector and James Hurley for generously donating the human ESCRT-II and ESCRT-II ΔMBD expression constructs.
This work was supported by NIGMS, National Institutes of Health Grant RO1 GM086434 (to M. D. B.) and a grant from the National Science Foundation Graduate Research Fellowships Program (GRFP) (to A. B. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2.
All sequencing data generated in this work were deposited in the NCBI SRA under accession no. Bioproject PRJNA336357.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- RBP
- RNA-binding protein
- ESCRT
- endosomal sorting complex required for transport
- MVB
- multivesicular body
- RIP
- RNA immunoprecipitation
- qPCR
- quantitative PCR
- CLIP
- cross-linking and immunoprecipitation
- MBD
- membrane-binding domain
- EMSA
- electrophoretic mobility shift assay
- PNK
- polynucleotide kinase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation
- TBE
- Tris borate–EDTA
- nt
- nucleotide(s).
References
- 1. Anantharaman V., Koonin E. V., and Aravind L. (2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30, 1427–1464 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glisovic T., Bachorik J. L., Yong J., and Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lunde B. M., Moore C., and Varani G. (2007) RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., Wyler E., Bonneau R., Selbach M., Dieterich C., and Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- 5. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., and Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 6. Kwon S. C., Yi H., Eichelbaum K., Fohr S., Fischer B., You K. T., Castello A., Krijgsveld J., Hentze M. W., and Kim V. N. (2013) The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122–1130 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- 7. Mitchell S. F., Jain S., She M., and Parker R. (2013) Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castello A., Fischer B., Frese C. K., Horos R., Alleaume A. M., Foehr S., Curk T., Krijgsveld J., and Hentze M. W. (2016) Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He C., Sidoli S., Warneford-Thomson R., Tatomer D. C., Wilusz J. E., Garcia B. A., and Bonasio R. (2016) High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beckmann B. M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A. M., Schwarzl T., Curk T., Foehr S., Huber W., Krijgsveld J., and Hentze M. W. (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127 10.1038/ncomms10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castello A., Hentze M. W., and Preiss T. (2015) Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 26, 746–757 10.1016/j.tem.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blower M. D., Feric E., Weis K., and Heald R. (2007) Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J. Cell Biol. 179, 1365–1373 10.1083/jcb.200705163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buxbaum A. R., Haimovich G., and Singer R. H. (2015) In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16, 95–109 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P., and Krause H. M. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 15. Martin K. C., and Ephrussi A. (2009) mRNA localization: Gene expression in the spatial dimension. Cell 136, 719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mili S., Moissoglu K., and Macara I. G. (2008) Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453, 115–119 10.1038/nature06888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Medioni C., Mowry K., and Besse F. (2012) Principles and roles of mRNA localization in animal development. Development 139, 3263–3276 10.1242/dev.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mardakheh F. K., Paul A., Kümper S., Sadok A., Paterson H., Mccarthy A., Yuan Y., and Marshall C. J. (2015) Global analysis of mRNA, translation, and protein localization: Local translation is a key regulator of cell protrusions. Dev. Cell 35, 344–357 10.1016/j.devcel.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Irion U., and St Johnston D. (2007) bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445, 554–558 10.1038/nature05503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hierro A., Sun J., Rusnak A. S., Kim J., Prag G., Emr S. D., and Hurley J. H. (2004) Structure of the ESCRT-II endosomal trafficking complex. Nature 431, 221–225 10.1038/nature02914 [DOI] [PubMed] [Google Scholar]
- 21. Im Y. J., and Hurley J. H. (2008) Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell 14, 902–913 10.1016/j.devcel.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teo H., Perisic O., González B., and Williams R. L. (2004) ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell 7, 559–569 10.1016/j.devcel.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 23. Hurley J. H. (2008) ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20, 4–11 10.1016/j.ceb.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slagsvold T., Aasland R., Hirano S., Bache K. G., Raiborg C., Trambaiolo D., Wakatsuki S., and Stenmark H. (2005) Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 280, 19600–19606 10.1074/jbc.M501510200 [DOI] [PubMed] [Google Scholar]
- 25. Teo H., Gill D. J., Sun J., Perisic O., Veprintsev D. B., Vallis Y., Emr S. D., and Williams R. L. (2006) ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125, 99–111 10.1016/j.cell.2006.01.047 [DOI] [PubMed] [Google Scholar]
- 26. Wollert T., and Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alam S. L., Langelier C., Whitby F. G., Koirala S., Robinson H., Hill C. P., and Sundquist W. I. (2006) Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat. Struct. Mol. Biol. 13, 1029–1030 10.1038/nsmb1160 [DOI] [PubMed] [Google Scholar]
- 28. Hirano S., Suzuki N., Slagsvold T., Kawasaki M., Trambaiolo D., Kato R., Stenmark H., and Wakatsuki S. (2006) Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat. Struct. Mol. Biol. 13, 1031–1032 10.1038/nsmb1163 [DOI] [PubMed] [Google Scholar]
- 29. Teo H., Veprintsev D. B., and Williams R. L. (2004) Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 279, 28689–28696 10.1074/jbc.M400023200 [DOI] [PubMed] [Google Scholar]
- 30. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ule J., Jensen K., Mele A., and Darnell R. B. (2005) CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 10.1016/j.ymeth.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 34. Ule J., Jensen K. B., Ruggiu M., Mele A., Ule A., and Darnell R. B. (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- 35. Schweidenback C. T., Emerman A. B., Jambhekar A., and Blower M. D. (2015) Evidence for multiple, distinct ADAR-containing complexes in Xenopus laevis. RNA 21, 279–295 10.1261/rna.047787.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., and Noble W. S. (2009) MEME suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harami G. M., Gyimesi M., and Kovács M. (2013) From keys to bulldozers: Expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem. Sci. 38, 364–371 10.1016/j.tibs.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 38. Im Y. J., Wollert T., Boura E., and Hurley J. H. (2009) Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell 17, 234–243 10.1016/j.devcel.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langelier C., von Schwedler U. K., Fisher R. D., De Domenico I., White P. L., Hill C. P., Kaplan J., Ward D., and Sundquist W. I. (2006) Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80, 9465–9480 10.1128/JVI.01049-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Konopacki F. A., Wong H. H., Dwivedy A., Bellon A., Blower M. D., and Holt C. E. (2016) ESCRT-II controls retinal axon growth by regulating DCC receptor levels and local protein synthesis. Open Biol. 6, 150218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharp J. A., Plant J. J., Ohsumi T. K., Borowsky M., and Blower M. D. (2011) Functional analysis of the microtubule-interacting transcriptome. Mol. Biol. Cell 22, 4312–4323 10.1091/mbc.e11-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huberts D. H., and van der Klei I. J. (2010) Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta 1803, 520–525 10.1016/j.bbamcr.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 43. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNA and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 44. Gibbings D. J., Ciaudo C., Erhardt M., and Voinnet O. (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–1149 10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- 45. Lee Y. S., Pressman S., Andress A. P., Kim K., White J. L., Cassidy J. J., Li X., Lubell K., Lim D. H., Cho I. S., Nakahara K., Preall J. B., Bellare P., Sontheimer E. J., and Carthew R. W. (2009) Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 11, 1150–1156 10.1038/ncb1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan S. (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21, 224–234 10.1006/prep.2000.1363 [DOI] [PubMed] [Google Scholar]
- 47. Hannak E., and Heald R. (2006) Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1, 2305–2314 10.1038/nprot.2006.396 [DOI] [PubMed] [Google Scholar]
- 48. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D. J., Luscombe N. M., and Ule J. (2011) iCLIP: Transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 30, pii: 2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., and Weissman J. S. (2012) The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwarz D. S., and Blower M. D. (2014) The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J. Cell Biol. 207, 41–57 10.1083/jcb.201406037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang da W., Sherman B. T., Zheng X., Yang J., Imamichi T., Stephens R., and Lempicki R. A. (2009) Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.11 [DOI] [PubMed] [Google Scholar]
- 53. James-Zorn C., Ponferrada V. G., Burns K. A., Fortriede J. D., Lotay V. S., Liu Y., Brad Karpinka J., Karimi K., Zorn A. M., and Vize P. D. (2015) Xenbase: Core features, data acquisition, and data processing. Genesis 53, 486–497 10.1002/dvg.22873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.