Abstract
The histone H3 variant H3.3 is a highly conserved and dynamic regulator of chromatin organization. Therefore, fully elucidating its nucleosome incorporation mechanisms is essential to understanding its functions in epigenetic inheritance. We previously identified the RNase P protein subunit, Rpp29, as a repressor of H3.3 chromatin assembly. Here, we use a biochemical assay to show that Rpp29 interacts with H3.3 through a sequence element in its own N terminus, and we identify a novel interaction with histone H2B at an adjacent site. The fact that archaeal Rpp29 does not include this N-terminal region suggests that it evolved to regulate eukaryote-specific functions. Oncogenic H3.3 mutations alter the H3.3–Rpp29 interaction, which suggests that they could dysregulate Rpp29 function in chromatin assembly. We also used KNS42 cells, an H3.3(G34V) pediatric high-grade glioma cell line, to show that Rpp29 1) represses H3.3 incorporation into transcriptionally active protein-coding, rRNA, and tRNA genes; 2) represses mRNA, protein expression, and antisense RNA; and 3) represses euchromatic post-translational modifications (PTMs) and promotes heterochromatic PTM deposition (i.e. histone H3 Lys-9 trimethylation (H3K9me3) and H3.1/2/3K27me3). Notably, we also found that K27me2 is increased and K36me1 decreased on H3.3(G34V), which suggests that Gly-34 mutations dysregulate Lys-27 and Lys-36 methylation in cis. The fact that Rpp29 represses H3.3 chromatin assembly and sense and antisense RNA and promotes H3K9me3 and H3K27me3 suggests that Rpp29 regulates H3.3-mediated epigenetic mechanisms by processing a transcribed signal that recruits H3.3 to its incorporation sites.
Keywords: chromatin regulation, histone modification, gene regulation, glioblastoma, transcription, epigenetics, antisense RNA, histone H3.3, RNase P, Rpp29, histone post-translational modifications (PTMs)
Introduction
The nucleosome, composed of ∼147 bp of DNA wrapped around an octamer of histone proteins (H3, H4, H2A, and H2B), is the fundamental unit of chromatin (1). The N-terminal tails of the histones protrude from the nucleosome core and are the targets of numerous post-translational modifications (PTMs),4 which modulate nucleosome structure and recruit chromatin-regulatory factors (2). The bulk of the histones are expressed during S phase to package newly replicated DNA. However, histone variants are expressed throughout the cell cycle and incorporated into chromatin through replication-independent (RI) mechanisms. Histone H3.3 is a variant of the H3 histones that differs from canonical H3.1 and H3.2 by 5 and 4 and amino acids, respectively. H3.3 has emerged as a dynamic and integral regulator of transcriptional activation (3–6) and silencing (7–9), DNA replication and repair (10, 11), and differentiation and nuclear reprogramming (12–14). Therefore, fully elucidating H3.3-regulatory mechanisms will provide essential insight into how chromatin organization regulates cellular identity and genome integrity.
Distinct chromatin assembly factors regulate H3.3 incorporation and histone PTM deposition at different genomic sites (1, 15). The H3.3-specific chaperone, DAXX, together with the chromatin-remodeling factor, ATRX, are required for H3.3 deposition and H3 lysine 9 trimethylation (H3K9me3) at heterochromatin (7, 9, 16, 17). In the absence of ATRX-DAXX–mediated H3.3 chromatin assembly, heterochromatin organization is impaired, and endogenous retroviruses and imprinted genes are derepressed (15, 18, 19). HIRA, which is a component of the HUCA complex that also includes CABIN1, UBN1, and Asf1a, is required for H3.3 incorporation at genic sites (7, 20). Although the promoters and enhancers of the most highly transcribed genes have the highest H3.3 turnover rates (21, 22), H3.3 incorporation is better correlated with RNA pol II engagement and/or sequence-specific transcription factor binding at these sites than transcription (23). Transcription is, however, required for H3.3 incorporation into gene bodies (21). HIRA is also required for Polycomb repressive complex 2 (PRC2)-mediated deposition of H3 lysine 27 trimethylation (H3K27me3) at bivalent genes in embryonic stem cells (24). Although HIRA deletion does not impact basal transcription, it dysregulates differentiation, which suggests that H3.3 incorporation is required for developmental signals to be converted into epigenetically inherited gene expression programs (24).
H3.3's essential role in development is also highlighted by the finding that point mutations drive pediatric high-grade gliomas (HGGs) and skeletal cancers (2, 25–27). These mutations are located at or near PTM sites in the H3.3 N-terminal tail. In ∼80% of diffuse intrinsic pontine gliomas, lysine 27, in either H3.3 or the canonical H3.1, is converted to methionine (K27M). H3(K27M) inhibits EZH2, the H3 Lys-27 methyltransferase in PRC2, through a direct interaction with its active site, which globally decreases H3K27me3 (28–31). Similarly, Lys-36 is mutated to K36M in chondroblastoma (32) and blocks Lys-36 methyltransferases through a similar mechanism (33, 34). H3.3 glycine 34 is also mutated to either arginine or valine in hemispheric pediatric HGGs and to tryptophan or leucine in giant cell tumor of the bone (GCTB). Less is known about how the Gly-34 mutations promote tumorigenesis, but alterations in H3 Lys-36 methylation have been reported (30, 35–37). These results suggest that oncohistone H3.3s drive tumorigenesis by dysregulating the chromatin landscape during differentiation.
Although the identification of H3.3-specific chaperones has provided enormous insight into chromatin assembly and histone PTM deposition, H3.3 functions remain incompletely understood due in part to the difficulty of elucidating events that initiate nucleosome incorporation. To address this, we investigated H3.3 recruitment dynamics at an activated transgene array and determined that H3.3 accumulates with RNA and RNA-regulatory factors (38, 39). Specifically, we showed that H3.3 co-localizes with AS RNA (39) and the following RNA factors: RNase P protein subunits (Rpp29, Rpp21, and POP1); fibrillarin, an RNA methyltransferase; and RPL23a (ribosomal protein–like) (38). We also reported that Rpp29 depletion increases H3.3 chromatin deposition and sense (S) and AS RNA levels, which suggests that an RNase P variant represses H3.3 chromatin assembly and transcription. Consistent with our results, a recent study identified RNA proteins, including RPLs and splicing factors, as H3.3-interacting factors (40). This study also showed that H3.3(G34W) increases chromatin compaction and causes aberrant RNA processing. Taken together, these results support the hypothesis that H3.3 chromatin assembly is functionally integrated with RNA processing and suggest that dysregulation of these events contributes to tumorigenesis.
Here, we continue our investigation of Rpp29 function in H3.3 chromatin assembly. RNase P is an endoribonuclease found in all three domains of life as either a ribozyme-centered ribonucleoprotein (RNP) or a protein-only enzyme, which is essential for cleaving the 5′ leader sequence from precursor tRNAs (41, 42). In its RNP form, RNase P is composed of a single catalytic RNA and up to nine protein subunits. The bacterial RNP includes one protein. Archaea have up to five proteins, two of which are homologous to Rpp29 and Rpp21. Eukaryotes also have an RNase P variant, RNase MRP, which shares many of the same protein subunits but has a distinct catalytic RNA (i.e. RMRP). It is thought that the catalytic RNA and subunit complexity of RNase P have been preserved through evolution because they have essential roles in processes other than tRNA cleavage (41). In support of this hypothesis, RNase P variants and subunits have been shown to regulate RNA pol I and III transcription sites (43–45); to degrade noncoding RNA, including AS transcripts, in yeast (46); to regulate piRNA and tRNA gene chromatin (47); and to promote homology-directed DNA double-strand break repair (i.e. catalytic RNA, Rpp21, and Rpp29) (48). It is not known whether H3.3 is involved in any of these noncanonical RNase P functions. However, the findings that Rpp29 and H3.3 regulate DNA repair (10, 11, 49–51) suggest the possibility of functional overlap.
Here, we report that Rpp29 interacts with H3.3 through its eukaryote-specific N terminus. We also identify a novel interaction between Rpp29 and histone H2B and show that the oncogenic H3.3 mutations alter the H3.3–Rpp29 interaction, which suggests that they could dysregulate Rpp29 function in chromatin assembly. We also use shRNA knockdowns to show that Rpp29 represses H3.3 deposition and AS RNA expression from RNA pol I, pol II, and pol III genes. Using quantitative MS (qMS), we show that Rpp29 impacts histone PTMs. Specifically, Rpp29 represses euchromatic PTMs and promotes heterochromatic PTMs (i.e. H3K9me3 and H3.1/2/3K27me3). Taken together, these results suggest that Rpp29 regulates H3.3-mediated epigenetic mechanisms by processing a transcribed signal that recruits H3.3 to its incorporation sites.
Results
H3.3 interacts with the eukaryote-specific N terminus of Rpp29
We previously reported that Rpp29 co-localizes with histone H3.3 at an activated transgene array (38). We also showed that Rpp29 interacts with the 63 N-terminal amino acids of H3.3 (H3.3-N-tail-αN, Fig. 1B). In this assay, we incubate bacterially expressed and purified GST-tagged H3.3 constructs with lysates from cells expressing YFP-Rpp29 (Fig. 1A). Rpp29 associates with WT H3.3 (H3.3-WT) but not GST (Fig. 1C), as detected by immunoblotting with a GFP antibody (Fig. 1C). Additionally, the H3.3–Rpp29 interaction is diminished by the conversion of one lysine and three arginine residues, in and around the αN helix, to alanine (H3.3–4PTM; Fig. 1B, red asterisks indicate the locations of the point mutations). This result suggests that the H3.3 αN helix plays an important role in mediating the Rpp29 interaction.
Figure 1.
H3.3- and H2B-binding regions are located in the eukaryote-specific N terminus of Rpp29. A, diagram of the human Rpp29 constructs expressed as YFP fusion proteins and evaluated for their ability to interact with the GST-histone constructs. The C terminus (green) is conserved in both archaea and eukaryotes, and the N terminus (red) is eukaryote-specific. Locations of H3.3-BR and H2B-BR are shown at the top of the diagram as gray bars. B, diagram of the H3.3-N-tail-αN (63 amino acids) expressed as GST fusion proteins. The αN helix and the H3.3-specific serine 31 (S31) are marked. Red asterisks, point mutations (K37A, R42A, R49A, and R52A) in the H3.3 four-point mutant (H3.3–4PTM) construct. C, in the binding assay, GST, GST-H3.3-WT, and GST-H3.3–4PTM (stained with colloidal blue, bottom panel) were incubated with protein lysates from cells expressing YFP-tagged Rpp29 constructs. Bound YFP-Rpp29 proteins were detected by immunoblotting with a GFP antibody. D, diagram of the N-terminal regions of H2A (40 amino acids), H2B (53 amino acids), and H4 (47 amino acids) expressed as GST fusion proteins. The α1 helices are marked. E, in the binding assay, GST and GST-H3.3, -H2A, -H2B, and -H4 (stained with colloidal blue; bottom) were incubated with full-length YFP-Rpp29. Bound YFP-Rpp29 was detected by immunoblotting with a GFP antibody. F, in the binding assay, GST and GST-H2B (stained with colloidal blue; bottom) were incubated with protein lysates from cells expressing YFP-tagged Rpp29 constructs. Bound YFP-Rpp29 proteins were detected by immunoblotting with a GFP antibody. G, diagram of H3.3-N-tail-αN showing the location of the αN helix, H3.3-specific Ser-31, and the oncogenic point mutations, K27M, G34R, G34V, G34L, and G34W. H, levels of YFP-Rpp29 pulled down by GST-H3.3-N-tail-αN proteins in three independent binding assays. Graphs represent mean ± S.D. (error bars). p values were calculated using an unpaired t test (n = 3 independent experiments). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns, not significant.
Here, we used this assay to identify the region of Rpp29 that interacts with H3.3-N-tail-αN (Fig. 1). Human Rpp29 is composed of 220 amino acids (Fig. 1A). The C terminus (green) is conserved in both archaea and eukaryotes, and the N terminus (red) is specific to eukaryotes. Deletion of the 58 C-terminal amino acids (Rpp29(1–162)) did not eliminate the interaction with H3.3, which indicates that this region is not required (Fig. 1C). Deletion of the 50 N-terminal amino acids of Rpp29 (Rpp29(50–220)) also did not eliminate the interaction. Deletion of 74 and 98 N-terminal amino acids (Rpp29(76–220)) did, however, prevent the association, which indicates that the H3.3-binding region (H3.3-BR) is between amino acids 51 and 73 (Fig. 1A). The location of the H3.3-BR in the eukaryote-specific N terminus of Rpp29 suggests that this region may have evolved to regulate functions other than pre-tRNA processing. We also note that Rpp29(50–220) interacts better with H3.3–4PTM, compared with full-length Rpp29 (Fig. 1C). It is possible that deleting the 50 amino acids N-terminally adjacent to the H3.3-BR alters its conformation in a way that increases its affinity for H3.3–4PTM.
Rpp29 interacts with histone H2B
We also used this binding assay to determine whether Rpp29 interacts with any of the other core histones. To do this, we expressed and purified the N-terminal tail α1-helices of H2A (40 aa), H2B (53 aa), and H4 (47 aa) as GST proteins (Fig. 1D) and incubated them with lysates from cells expressing YFP-Rpp29. This analysis revealed that Rpp29 also interacts with H2B but not H2A and H4 (Fig. 1E). To identify the H2B-binding region (H2B-BR) in Rpp29, we incubated GST-H2B with the Rpp29 deletion constructs (Fig. 1F). Deletion of the first 98 but not the first 75 acids of Rpp29 eliminated their interaction. This result indicates that the H2B-BR is within amino acids 76–98. The location of the H2B-BR region C-terminally adjacent to the H3.3-BR suggests that H3.3 and H2B do not compete for binding to Rpp29 (Fig. 1A). Although we do not know the significance of the Rpp29–H2B interaction, it further supports a functional connection between Rpp29 and chromatin.
The oncogenic H3.3 mutations impact the Rpp29 interaction
Because the oncogenic H3.3 mutations are within the region that interacts with Rpp29 (Fig. 1G), we also wanted to evaluate their impact on the association. Therefore, we introduced them into the GST-H3.3-N-tail-αN construct and used the binding assay to evaluate their effects on the interaction with YFP-Rpp29 (Fig. 1H). Compared with WT H3.3, the K27M, G34W, and G34L mutations significantly increased the association, whereas G34R decreased it. Although G34V did not have a significant effect, valine is smaller and has different chemical properties compared with arginine, phenylalanine, and leucine. Therefore, its effect on the Rpp29 interaction may not be detectable in this assay. Taken together, these results suggest that the oncogenic mutations significantly impact the H3.3–Rpp29 interaction, which could affect Rpp29 function in H3.3 chromatin assembly.
Rpp29 represses H3.3 incorporation into transcriptionally active genes
We also previously reported that Rpp29 represses H3.3 incorporation into chromatin using both ChIP, at a DAXX-ATRX–regulated transgene array, and a high-salt extraction assay (38). To evaluate the effects of Rpp29 depletion on H3.3 deposition at endogenous genes, we knocked Rpp29 down in KNS42 cells, a pediatric HGG cell line that harbors the H3.3(G34V) mutation, and used ChIP to measure H3.3 incorporation into several genes previously reported to be highly up-regulated (i.e. DLX-5, DLX-6, and MYCN) (Fig. 2D) and down-regulated (i.e. MYT1 and DLK1) (Fig. S1G) (27, 35). As a negative control, we knocked down Rpp20, which is one of the RNase P subunits that did not co-localize with H3.3 at the activated transgene array (38). As a positive control, we knocked down HIRA, which is required for H3.3 incorporation into single-copy genes (7). Fig. 2A shows that all of the shRNAs significantly decreased their respective mRNAs compared with the nonsilencing control (pLKO). We also measured H3.3 (i.e. hH3F3A and hH3F3B) mRNA (Fig. 2B) and evaluated H3.3 and H3 (i.e. H3.1, H3.2, and H3.3) protein levels using immunoblotting (Fig. 2C) to confirm that they are not affected by the knockdowns. The H3.3 antibody specifically recognizes H3.3 but not H3.1 and H3.2 (Fig. S1A). It also recognizes H3.3(G34V) (Fig. S1B), which indicates that H3.3(G34V) will also be measured in the ChIP analyses in KNS42 cells.
Figure 2.
Rpp29 represses H3.3 chromatin incorporation and mRNA and antisense RNA at single-copy genes. A, qRT-PCR analysis of Rpp29, Rpp20, and HIRA mRNA levels in KNS42 cells 144 h after knockdowns compared with the nonsilencing control, pLKO. p values were calculated using unpaired t test (n = 3 independent experiments with two technical replicates each). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns, not significant. B, qRT-PCR analysis of H3F3A and H3F3B mRNA levels in KNS42 cells 144 h after knockdowns (n = 3 independent experiments). C, immunoblot of H3.3, H3, and HIRA levels in knockdown cells. GAPDH is used as a loading control. D, diagrams of DLX-5, DLX-6, and MYCN genes showing the location of the transcription start sites (arrows) and exons (thick bars). Red bars show the location of the primer pairs used for ChIP analysis. E, ChIP qRT-PCR analysis of H3.3 incorporation into DLX-5, DLX-6, and MYCN in KNS42 cells following knockdowns (n = 3 independent experiments). F, qRT-PCR analysis of DLX-5, DLX-6, and MYCN mRNA levels in KNS42 cells following knockdowns (n = 3 independent experiments). G, immunoblot analysis of MYCN levels in knockdown cells detected using a MYCN-specific antibody. GAPDH is used as a loading control. H, strand-specific qRT-PCR analysis of DLX-5, DLX-6, and MYCN antisense RNA levels in KNS42 cells following knockdowns (n = 3 independent experiments). Error bars, S.D.
Rpp29 depletion significantly increased H3.3 incorporation at the promoters and gene bodies of DLX-5, DLX-6, and MYCN (Fig. 2E), which indicates that it represses H3.3 deposition at transcriptionally active genes. In contrast, Rpp20 depletion did not significantly alter H3.3 incorporation, with the exception of an increase at exon 3 of DLX-6 (Fig. 2E). Because Rpp20 is a component of canonical RNase P, its depletion could indirectly increase H3.3 incorporation by decreasing the stability of the subunits that regulate H3.3 chromatin assembly. In contrast, HIRA depletion significantly decreased H3.3 levels at five of the six promoter and exon sites (Fig. 2E), which is consistent with reports that HIRA is required for H3.3 deposition at single-copy genes (7). Additionally, Rpp29 depletion did not alter HIRA levels (Fig. 2C), which indicates that the increase in H3.3 deposition (Fig. 2E) is not due to an increase in HIRA.
Rpp29 depletion in the ovarian cancer cell line OVCAR-3 (Fig. S1C) also increases H3.3 incorporation into active genes (i.e. DLX-5 and PAX-8) (Fig. S1, D and E), which further supports the conclusion that Rpp29 represses H3.3 chromatin assembly at endogenous genes. Notably, Rpp29 depletion did not affect H3.3 deposition at down-regulated and inactive genes in both KNS42 (i.e. MYT1 and DLK1) (27, 35) (Fig. S1, G and H) and OVCAR-3 cells (i.e. MYO-D) (Fig. S1, D and E). These results suggest that Rpp29 represses H3.3 chromatin assembly at active genes.
Because H3.3 is also incorporated into tRNA and rRNA genes (21, 52–54) and RNase P variants bind to RNA pol I and pol III transcription sites (43–45), we also evaluated the effects of Rpp29 depletion on H3.3 incorporation at rDNA repeats and a tRNA-leucine gene (Fig. S1, J and K). Indeed, Rpp29 knockdown significantly increased H3.3 incorporation at these sits (Fig. S1K). Taken together, these results suggest that a subcomplex of RNase P, which includes Rpp29, regulates H3.3 chromatin assembly at transcriptionally active RNA pol I, pol II, and pol III genes.
Rpp29 represses mRNA and protein expression
To determine whether the increases in H3.3 incorporation induced by Rpp29 depletion correlate with changes in gene expression, we measured DLX-5, DLX-6, and MYCN mRNA in Rpp29-knockdown cells (Fig. 2F). Compared with the nonsilencing control (pLKO), Rpp29 depletion increased mRNA levels ∼2-fold. In contrast, Rpp20 and HIRA depletion did not significantly affect them (Fig. 2F). This suggests that Rpp29 represses transcription, which is consistent with our previous report that Rpp29 depletion increases expression of the S RNA from the transgene and that Rpp29 overexpression accelerates transgene chromatin compaction (38). The finding that HIRA depletion did not significantly alter mRNA levels is consistent with previous reports that HIRA and H3.3 are not essential for basal transcription in mouse ES cells (7, 19).
To determine whether the increase in mRNA induced by Rpp29 depletion correlates with an increase in protein expression, we immunoblotted lysates for MYCN (Fig. 2G). Indeed, compared with the controls, MYCN is increased in Rpp29-knockdown cells. Although it is possible that Rpp29 depletion indirectly increases H3.3 chromatin assembly and transcription by disrupting the canonical functions of RNase P and RNase MRP in tRNA and rRNA processing, respectively, the increase in MYCN suggests that translation is not significantly impaired in Rpp29-knockdown cells.
Rpp29 represses antisense RNA
Our finding that Rpp29 depletion increases H3.3 incorporation at transcriptionally active genes suggests that Rpp29 represses H3.3 chromatin assembly by regulating a transcriptional event. It was previously reported that noncoding RNA, including AS transcripts, accumulate in yeast with a mutation in the catalytic RNA subunit of RNase P (46). Additionally, we previously showed that H3.3 co-localizes with Rpp29 and AS RNA at an activated transgene array and that Rpp29 depletion increases both H3.3 accumulation and transgene AS RNA levels (38, 39). To determine the impact of Rpp29 depletion on AS RNA expressed from endogenous genes, we measured it using strand-specific qRT-PCR. Rpp29 knockdown elevated AS RNA levels at the transcriptionally active but not the down-regulated genes in both KNS42 and OVCAR-3 cells (Fig. 2H and Fig. S1, F, I, and L). Therefore, the increase in H3.3 incorporation induced by Rpp29 depletion correlates with increases in both S and AS RNA.
Rpp20 depletion did not affect AS RNA (Fig. 2H), consistent with the finding that it does not impact H3.3 chromatin incorporation (Fig. 2E) or mRNA levels (Fig. 2F). Notably, however, HIRA depletion increased AS RNA levels (Fig. 2H), which is consistent with reports that when components of the HIR complex are deleted in yeast, cryptic transcription increases (55, 56). Although we have not shown a direct interaction between AS RNA and Rpp29 in this study, our previous report that Rpp29 and H3.3 colocalize with AS RNA at the activated transgene array (38, 39) and our current finding that H3.3 deposition correlates with increased S and AS RNA expression in Rpp29-knockdown cells suggest that 1) H3.3 is recruited to its incorporation sites by RNA, and 2) Rpp29 represses H3.3 chromatin assembly through a currently unknown RNA-processing mechanism.
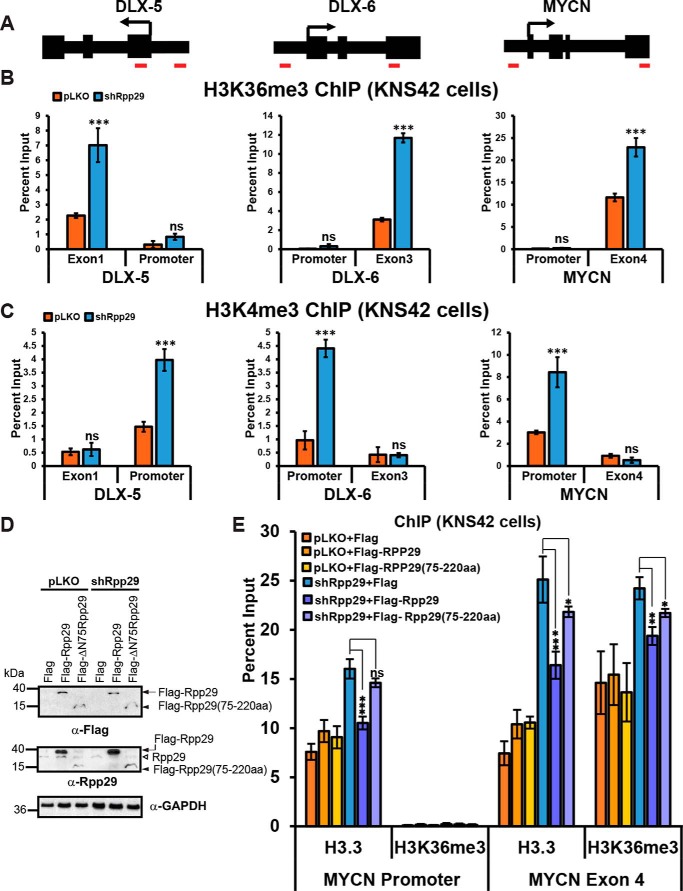
Rpp29 represses H3K36me3 and H3K4me3 at active genes
We also used ChIP to evaluate the effect of Rpp29 depletion on the deposition of H3K36me3, which is enriched in gene bodies, because a previous analysis in KNS42 cells indicated that it is elevated at DLX-5, DLX-6, and MYCN (35) (Fig. 3A). We also included H3K4me3 in this analysis because it is enriched at the promoters of active genes (57). Rpp29 depletion significantly increased both H3K36me3 (Fig. 3B) and H3K4me3 (Fig. 3C) levels. These results suggest that, in Rpp29-knockdown cells, the increases in H3.3 incorporation (Fig. 2E) and transcription (Fig. 2F) are accompanied by increased deposition of euchromatic PTMs.
Figure 3.
Rpp29 represses H3K36me3 and H3K4me3 at active genes. A, diagrams of DLX-5, DLX-6, and MYCN genes showing the location of the transcription start sites (arrows) and exons (thick bars). Red bars show the location of the primer pairs used for ChIP analysis. B, ChIP qRT-PCR analysis of H3K36me3 levels at DLX-5, DLX-6, and MYCN in KNS42 cells following knockdowns. p values were calculated using unpaired t test (n = 3 independent experiments with two technical replicates each) (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns, not significant). C, ChIP qRT-PCR analysis of H3K4me3 levels at DLX-5, DLX-6, and MYCN in KNS42 cells following knockdowns. D, immunoblot analysis of shRNA-resistant FLAG-tagged Rpp29 and Rpp29(75–220) expressed in knockdown cells using FLAG- and Rpp29-specific antibodies. Arrows, FLAG-tagged constructs and endogenous Rpp29. GAPDH is used as a loading control. E, ChIP qRT-PCR analysis of H3.3 and H3K36me3 levels at MYCN in KNS42 cells expressing shRNA-resistant FLAG-tagged constructs following knockdowns. Error bars, S.D.
Rpp29, but not Rpp29(75–220), restores repression of H3.3 chromatin assembly in Rpp29-knockdown cells
To confirm that the increase in H3.3 deposition in Rpp29-knockdown cells is due, directly, to Rpp29 depletion and that it is not an off-target effect of the shRNA, we measured H3.3 incorporation at the MYCN promoter and gene body after expressing shRNA-resistant FLAG-tagged Rpp29 constructs (Fig. 3, D and E). Immunoblots using α-FLAG and α-Rpp29 antibodies show expression of Rpp29 and Rpp29(75–220), which is deleted of the H3.3-BR (Fig. 1A), in knockdown cells (Fig. 3D). FLAG-Rpp29 significantly reduced H3.3 incorporation at both the promoter and gene body of MYCN (Fig. 3E). Similar trends were seen for H3K36me3. These results confirm that the increases in H3.3 and H3K36me3 deposition in Rpp29-knockdown cells are caused by Rpp29 depletion. The finding that Rpp29(75–220) cannot repress H3.3 and H3K36me3 deposition to the same extent as full-length Rpp29 (Fig. 3E) suggests that the N terminus, which includes the H3.3-BR, is essential for this function.
Rpp29 depletion alters global histone PTM levels
Evaluating histone PTM levels at specific genes by ChIP requires selection of a limited number of antibodies. Therefore, to determine the effects of Rpp29 depletion on all histone PTMs and variants (about 250 uniquely modified peptides from canonical and variant histones) in KNS42 cells, we used “one-pot shotgun” qMS, which identifies changes in both single and combinatorial PTMs with high sensitivity (58) (Fig. 4). The volcano plot shows the histone peptides with PTMs that are significantly decreased (left of y axis) and increased (right of y axis) in Rpp29-knockdown cells (Fig. 4A). Strikingly, Rpp29 depletion significantly increased the levels of unmodified peptides from several histones (i.e. H3, H4, and H2A) (Fig. 4B), which suggests that Rpp29 promotes PTM deposition and/or prevents PTM removal.
Figure 4.
Rpp29 depletion alters global histone PTM levels. A, volcano plot showing histone PTMs significantly altered following Rpp29 depletion as detected by quantitative MS. B, comparison of changes in unmodified histone peptide levels in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells. Error bars, S.D. p values were calculated using two-tailed unpaired t test: n = 5 experimental replicates using the means of triplicate measurements for each replicate (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001). C, comparison of changes in PTM levels on 9–17 aa peptides of H3 histones (i.e. H3.1, H3.2, and H3.3) in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells.
Rpp29 depletion changes histone H3 PTMs
Of interest, Rpp29 depletion significantly increased the levels of unmodified H3 peptides that include PTM sites regulated by H3.3-chromatin assembly (i.e. H3 Lys-9 and H3 Lys-27) (7, 15) (Fig. 4B). Therefore, we focused on analyzing the H3 histones. For these qMS studies, histones are digested with trypsin after first being propionylated, which restricts cleavage to sites C-terminal of arginine and produces the following H3 N-terminal tail peptides: 3–8 aa, 9–17 aa, 18–26 aa, and 27–40 aa. For the first three, it is not possible to distinguish H3.3 from H3.1 and H3.2 because their amino acid sequences are exactly the same. However, amino acid differences in the 27–40 aa peptides make it possible to evaluate the variants. We did not detect significant changes in the PTMs on the 3–8 aa peptide, including H3K4me3 (Fig. S2A), despite the increases detected by ChIP at selected genes (Fig. 3C). Because H3K4me3 is mainly enriched in the 5′ regions of active genes (57), it may not be possible to detect Rpp29-induced changes when analyzing the entire histone pool. A significant increase in H3K18me1 was detected on the 18–26 aa peptide in Rpp29-knockdown cells (Fig. S2B). However, the function of H3K18 methylation is not well-understood, making it difficult to speculate about its impact.
Rpp29 depletion decreases H3K9me3
Coincident with the increase in unmodified H3 9–17 aa (Fig. 4B), Rpp29 depletion induced a significant increase in H3K14ac and decrease in H3K9me3 (Fig. 4C). The H3K14ac increase is consistent with our finding that Rpp29 depletion increases PTMs associated with active chromatin (i.e. H3K4me3 and H3K36me3; Fig. 3). The decrease in H3K9me3 suggests that Rpp29 promotes heterochromatic histone PTM deposition and/or maintenance.
Rpp29 depletion alters H3 Lys-27 and Lys-36 methylation
Because serine is substituted at amino acid 31 in H3.3 in place of alanine in H3.1/H3.2 (Fig. 5A), it is possible to differentiate the 27–40 aa peptides of these variants using MS (Fig. 5). Similarly, H3.3(G34V) can be distinguished from WT H3.3. Fig. 5B shows that H3.1/H3.2 are ∼10 times more abundant than H3.3 and H3.3(G34V). Of note, Rpp29 depletion decreased H3.1/H3.2 K27me3, a PTM enriched at the promoters of inactive genes (59) (Fig. 5C). It also increased K27me1 and K27ac, which are associated with active chromatin (60). These results are consistent with our finding that Rpp29 depletion decreases the heterochromatic PTM, H3K9me3, and increases euchromatic PTMs (Figs. 3 and 4C). Because PRC2 is responsible for depositing all H3K27 methylation states (60), these results suggest that Rpp29 regulates PRC2 activity.
Figure 5.
Rpp29 depletion alters PTM levels on H3 27–40 aa peptides. A, sequence diagrams of the histone H3.1/H3.2, H3.3, and H3.3(G34V) 27–40 aa peptides. B, comparison of relative amounts of H3.1, H3.2, and H3.3 27–40 aa peptides in KNS42 cells calculated using chromatographic peak areas of peptides detected by qMS. Error bars, S.D. p values were calculated using two-tailed unpaired t test: n = 5 experimental replicates using the means of triplicate measurements for each replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. C, comparison of changes in PTM levels at Lys-27 on the 27–40 aa peptide of H3.1/H3.2 in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells. D, comparison of changes in PTM levels at Lys-36 on the 27–40 aa peptide of H3.1/H3.2 in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells. E, comparison of changes in PTM levels at Lys-27 on the 27–40 aa peptide of H3.3 in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells. F, comparison of changes in PTM levels at Lys-36 on the 27–40 aa peptide of H3.3 in Rpp29 knockdown (shRpp29) and nonsilencing control (pLKO) cells.
Rpp29 depletion also significantly increased K36me1 and K36me2 on H3.1/H3.2 (Fig. 5D). The fact that H3K36me2 and H3K36me3 antagonize PRC2-mediated H3K27me3 but still permit K27me1 (61, 62) suggests that Rpp29 promotes H3K27me3 by repressing H3K36me2 (Fig. 5C). The increase in K36me2 also suggests that the activity of the H3K36-specific dimethylase, Ash1 (61) (Fig. 5D), which is a Trithorax group protein that antagonizes Polycomb silencing (63), is up-regulated in Rpp29-knockdown cells. Although significant changes in H3.1/H3.2 K36me3 were not detected (Fig. 5D), the increase in K27me1 (Fig. 5C) suggests that the activity of the H3 Lys-36 trimethylase, SETD2, is also up-regulated because it is known to promote H3K27me1 by inhibiting its conversion to H3K27me2 (60).
Although similar trends in Lys-27 and Lys-36 methylation were seen for H3.3 (Fig. 5, E and F), all of these changes were not significant, due, perhaps, to its lower levels in KNS42 cells (Fig. 5B). However, similar to H3.1/2, Rpp29 depletion significantly decreased H3.3K27me3 (Fig. 5E) and increased H3.3K36me1 (Fig. 5F). It also decreased the combinatorial PTMs, H3.3K27me3/K36me1 and H3.3K27me1/K36me3 (Fig. S2C). It was previously reported that K27me1 and K36me3 only coexist on H3.3 (60). Therefore, the decrease in H3.3K27me3 induced by Rpp29 depletion further supports the hypothesis that Rpp29 regulates H3.3-PRC2–mediated epigenetic mechanisms.
H3.3(G34V) dysregulates Lys-27 and Lys-36 methylation in cis
Rpp29 depletion did not induce significant changes in PTM deposition on the H3.3(G34V) 27–40 aa peptide (Fig. 6A) (data not shown). However, notably, H3.3(G34V) has significantly higher K27me1 and K27me2 and lower K36me1 compared with H3.3 (Fig. 6, B and C). This suggests that G34V directly impacts the modification of the adjacent Lys-27 and Lys-36 residues. This is consistent with a recent report that used immunoblotting to show that G34L and G34W increase K27me3 and decrease K36me2 and K36me3 in cis (37). G34V could increase K27me2 in cis by inhibiting its removal or preventing its conversion to K27me3. The decrease in K36me1 on the G34V peptide suggests that the mutation inhibits Lys-36 monomethylation. Because Lys-36 methylation antagonizes Lys-27 methylation (61, 62), G34V could also promote K27me2 by inhibiting Lys-36 methylation. This result is consistent with reports that the Gly-34 mutations impair Lys-36 methylation (30, 35, 36). Taken together, these results suggest that the H3.3 Gly-34 mutations dysregulate the post-translational modification of their adjacent Lys-27 and Lys-36 residues.
Figure 6.
G34V increases K27me2 and decreases K36me1 in cis. A, sequence diagrams of the histone H3.3 and H3.3(G34V) 27–40 aa peptides. B, comparison of changes in PTM levels at Lys-27 on the 27–40 aa peptides of H3.3 and H3.3(G34V). Error bars, S.D. p values were calculated using two-tailed unpaired t test: n = 5 experimental replicates using the means of triplicate measurements for each replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. C, comparison of PTM level differences at Lys-36 on the 27–40 aa peptides of H3.3 and H3.3(G34V). D, proposed model of the Rpp29 function in H3.3 chromatin assembly and epigenetic regulation.
Discussion
The presence of RNase P in all three domains of life and the preservation of its RNP form attest to both its ancient origins and the theory that RNA served as both enzyme and genome in the “RNA world” (41, 64). Due to its presence throughout cellular evolution, it is possible that RNase P played a foundational role in genome evolution. As such, our finding that the RNase P protein subunit, Rpp29, regulates H3.3 chromatin assembly provides an important opportunity to gain insight into the genomic functions of RNase P.
To investigate Rpp29 function in H3.3-mediated chromatin regulation, we first determined that Rpp29 interacts with the H3.3 N terminus through a sequence element in its own N terminus. The fact that this N-terminal region is not present in archaeal Rpp29 suggests that it evolved to regulate a eukaryote-specific function. Of interest, most archaeal histones are only composed of the histone fold region; they lack the N-terminal extensions of the eukaryotic histones, where almost all of the PTMs are deposited (65). Therefore, it is possible that the N termini of Rpp29 and H3.3 co-evolved during the development of more complex gene-regulatory mechanisms. Our finding that Rpp29 depletion impacts histone PTM deposition supports this hypothesis. It will be interesting to determine whether the Rpp29 C terminus interacts with the H3.3 histone fold region because it would suggest that RNase P regulated genomic mechanisms before the evolution of the eukaryotic nucleosome.
Additional studies are also needed to define the mechanism through which the H3.3 and Rpp29 N termini interact. Because we incubated bacterially expressed H3.3 with lysates from cells expressing YFP-Rpp29, we do not know whether their interaction is direct. As both proteins are basic, we hypothesize that a negatively charged factor, such as the RNase P or RNase MRP catalytic RNA (RPPH1 and RMRP, respectively), mediates their association. Previously, we were unable to co-localize either of these RNAs with Rpp29 and H3.3 at the transgene array using RNA FISH or to disrupt the H3.3–Rpp29 interaction using RNases that degrade single and dsRNA (38). However, these negative results do not rule out the possibility that RPPH1 or RMRP is a component of the Rpp29–H3.3 complex, as their conformations could make them inaccessible to RNA FISH probes and RNases. In the future, it will be necessary to evaluate these interactions using purified components.
Our finding that Rpp29 interacts with the N terminus of histone H2B through a region that is C-terminally adjacent to the H3.3-BR also supports our conclusion that an RNase P variant regulates chromatin. Future studies will be needed to determine the functional significance of their association and whether the H2B–Rpp29 interaction is direct or mediated by RNA. It will also be important to determine whether Rpp29 interacts with H3.3 and H2B in the context of the nucleosome or during the process of chromatin assembly. Our previous finding that the H3.3 N terminus (H3.3-N-tail-αN) co-localizes with Rpp29, RNA, and RNA proteins at the activated transgene array suggests that they interact upstream of H3.3 nucleosome deposition (38). This idea is also supported by our finding that the αN helix, which contacts DNA in the nucleosome (66), is important for the H3.3–Rpp29 interaction (38). Understanding the timing of the Rpp29–H3.3 and –H2B interactions could also provide insight into whether Rpp29 regulates the establishment or maintenance of histone PTM patterns.
We also determined that Rpp29 represses H3.3 incorporation into the promoters and gene bodies of endogenous transcriptionally active genes, specifically single-copy protein–coding as well as rRNA and tRNA genes, but not inactive genes. The finding that the increase in H3.3 incorporation at active genes correlates with an increase in AS RNA suggests a regulatory relationship between noncoding RNA and H3.3 chromatin assembly. It was previously reported that in yeast, cryptic transcription increases when RI chromatin assembly factors, including HIRA homologs, are deleted (55, 56). It is thought that loss of chromatin structure grants the transcription machinery access to cryptic promoters (67, 68). Our finding that HIRA depletion both decreases H3.3 incorporation and increases AS RNA supports this idea. However, our finding that Rpp29 depletion increases both H3.3 incorporation and AS RNA suggests that RNA-processing events are also required to repress cryptic transcripts. This idea is supported by reports that RNA exosome factors repress cryptic transcription (69). Based on our findings that H3.3 interacts with Rpp29 (38) and that H3.3 depletion increases AS RNA (39), it is also possible that H3.3, itself, functions as an RNA-processing factor before nucleosome deposition. Indeed, there may be a regulatory dependence between RNA processing and H3.3 nucleosome assembly. Therefore, it is possible that chromatin assembly factor deletion up-regulates cryptic transcription by dysregulating both chromatin structure and RNA processing.
Genomic analyses indicate that most genes are overlapped by long noncoding RNAs that are expressed from either an upstream or an antisense promoter (70). These noncoding RNAs are thought to be alternative genetic information that regulates cellular responses to environmental and developmental stimuli (56, 68). This suggests that the signaling events that activate genes also make the cryptic promoters, contained within them, accessible to the transcription machinery. If noncoding RNA recruits H3.3 to these transcription units for deposition, it suggests a mechanism through which the information that they encode could be converted into epigenetically heritable chromatin structures. The finding that histone PTM enzymes, including the H3K36me3 methyltransferase Set2 (71, 72), also regulate cryptic transcription supports this hypothesis (70). This idea also suggests that the recruitment of H3.3 and Rpp29 to genes is a consequence of transcriptional activation and not an initiating event, which is consistent with our previous reports that H3.3 and Rpp29 begin to accumulate with RNA at the transgene array ∼50 min after activation (38, 39). Importantly, noncoding RNAs also regulate PRC2 recruitment to its target genes (73). Therefore, elucidating the mechanisms through which noncoding RNA regulates H3.3 chromatin assembly could also provide new insight into PRC2 regulation and H3(K27M)-driven tumorigenesis (28–31).
Overall, our ChIP and qMS data suggest that Rpp29 represses euchromatic PTMs and promotes heterochromatic PTM deposition. Specifically, by ChIP, we detected increases in the euchromatic PTMs, H3K36me3 and H3K4me3, and by qMS, we detected increases in the following H3 PTMs: K14ac, K27ac, K27me1, K36me2. The fact that Hira deletion does not affect H3K4me3 and H3K36me3 levels in mouse ESCs suggests that RI H3.3 chromatin assembly is not required for their deposition (7). Therefore, Rpp29 may indirectly repress euchromatic PTMs by repressing transcription. The finding that Rpp29 depletion decreases both H3K9me3 and H3K27me3 suggests that Rpp29 may also repress euchromatic PTMs by promoting heterochromatic PTM deposition.
Currently, we do not know the mechanism through which Rpp29 promotes H3K9me3 and H3K27me3. Because Rpp29 depletion increases K36me2, which is known to antagonize K27me3 (61, 62), Rpp29 could promote K27me3 by repressing H3 Lys-36 methylation. It has also been reported that RNA prevents PRC2 from binding to chromatin (74, 75). Therefore, as a component of an RNase P variant that regulates H3.3 deposition, Rpp29 could promote H3K27me3 by degrading RNAs that inhibit PRC2 targeting. This idea is consistent with our finding that the expression of exogenous Rpp29 accelerates RNA degradation and chromatin compaction at the activated transgene array (38). Alternatively, Rpp29 could process the noncoding RNAs that recruit PRC2 to its target sites through an unknown mechanism (73). At this time, we also do not know whether the effects of Rpp29 depletion on histone PTMs precede or follow changes in transcription.
The finding that Rpp29 levels impact histone PTM regulation and gene expression has important implications for disease processes. Rpp29 (i.e. POP4) is overexpressed in ovarian and breast tumors with 19q12 amplifications (76, 77) and depleted in oligodendrogliomas with 1p/19q co-deletions (78). Therefore, Rpp29 copy number changes could promote tumorigenesis by altering the epigenetic inheritance of transcriptional programs. Of interest, the RNase MRP catalytic RNA, RMRP, is required for differentiation of T helper 17 (T17) lymphocytes, which indicates that it plays a role in tissue-specific transcriptional regulation (79). Like Rpp29, RMRP is also amplified in cancers, including glioma (80), breast (81), lung (82), prostate (83), and gastric tumors (84). If RMRP is a component of an RNase P variant that regulates H3.3 deposition, its overexpression could drive tumorigenesis by dysregulating epigenetic mechanisms.
We also found that the oncogenic H3.3 mutations impact the Rpp29–H3.3 interaction. Several, including the GCTB mutation, G34W, increase the affinity for Rpp29. Of interest, H3.3(G34W) has been reported to down-regulate transcription and increase chromatin compaction (40). Because we show that Rpp29 promotes heterochromatic PTM deposition, it is possible that H3.3(G34W) promotes tumorigenesis by enhancing the gene-silencing function of Rpp29. It is also interesting to note that RNase P and RNase MRP subunits, as well as H3.3 in GCTB, are mutated in diseases that originate in the metaphyseal region of bones (85). The recessive inheritance of mutations in RMRP causes cartilage hair hypoplasia, which is characterized by short stature due to impaired bone growth (86). The protein subunit, POP1, is also mutated in skeletal dysplasias (87–89). Therefore, it will be interesting to determine whether H3.3 and these RNase P factors regulate the same signaling events during bone development.
Finally, our analysis of PTM deposition on H3.3(G34V) revealed that this point mutation dysregulates the intramolecular modification of the Lys-27 and Lys-36 residues, consistent with a recent report examining the impact of G34L and G34W on these sites in cis (37). Specifically, we show that K27me1 and K27me2 are increased and K36me1 is decreased on H3.3(G34V) compared with WT. The requirement of PRC2 for all forms of H3K27 methylation (73) suggests that G34V, like K27M, may drive tumorigenesis by dysregulating PRC2 function (28–31). Importantly, H3K27me2 prevents the accumulation of H3K27me1 and H3K27ac, which are required for enhancer firing (90). Because enhancer activity is important for differentiation, it is possible that the persistence of K27me2 on H3.3(G34V) promotes tumorigenesis by preventing enhancer activation. Therefore, genome-wide analyses of K27me2 in tumor lines harboring the oncogenic Gly-34 mutations could identify dysregulated enhancers and genic elements that cause differentiation to be blocked.
Taken together, our current and previous investigations of Rpp29- and H3.3-regulatory dynamics (38, 91) suggest that 1) H3.3 is recruited to its incorporation sites at transcriptionally active genes by noncoding RNA, and 2) Rpp29 represses transcription and H3.3 deposition and promotes H3.3-meditated epigenetic mechanisms by processing RNA transcribed from these sites through a currently unknown mechanism (Fig. 6D). Importantly, our results indicate that Rpp29 impacts the deposition of PTMs on the region of the H3.3 N-terminal tail that is a hot spot for oncogenic mutations (i.e. Lys-27, G34L/R/V/W, and Lys-36) (25). Although Rpp29 depletion did not alter Lys-27 and Lys-36 methylation on H3.3(G34V) in KNS42 cells, our finding that Rpp29 affects PTM deposition at Lys-27 and Lys-36, which surround it, suggests that G34V could impact Rpp29 function through a currently unknown mechanism. The requirement of H3.3 deposition for differentiation but not for basal transcription in mouse ESCs (7, 19, 24) suggests that the essential function of H3.3 in differentiation could be to convert the alternate genetic information encoded in the noncoding transcriptome into heritable gene expression programs through histone PTM deposition in partnership with a variant of RNase P that includes Rpp29.
Experimental procedures
Plasmids
H3.3-YFP, GST-H3.3-N-tail-αN-WT, GST-H3.3-N-tail-αN 4-PTM, YFP-Rpp29-C3, and pLKO were described previously (39, 92). The K27M, G34R, G34V, G34W, and G34L mutations were introduced into GST-H3.3-N-tail-αN and H3.3-YFP using the QuikChange site-directed mutagenesis kit (Agilent Technologies). The GST-H4 (1–48 aa), H2A (1–41 aa), and H2B (1–53 aa) constructs were created by cloning PCR products into pGEX-4T3 vector (SalI/NotI). YFP-Rpp29 deletion constructs (1–162, 50–220, 75–220, and 99–220 aa) were made by cloning PCR products into YFP-C3 (XhoI/EcoRI). Rpp29 was made shRNA-resistant using the QuikChange site-directed mutagenesis kit (Agilent Technologies), and full-length and Rpp29(75–220) were subjected to PCR and cloned into pCMV-FLAG vector (HindIII/BamHI).
Cell culture and transfections
KNS42 cells (35) (gift of Lynn Bjerke and Chris Jones, Institute of Cancer Research, Sutton, Surrey, UK) were grown in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% FBS (Atlanta Biologics Ltd.). U2OS cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1 mm penicillin/streptomycin (Thermo Fisher Scientific). Transfections for binding assays in U2OS cells were carried out by electroporation or lentiviral infection. OVCAR-3 cells were grown in RPMI 1640 medium supplemented with 10% FBS and 1 mm penicillin/streptomycin.
Knockdown analyses
Knockdowns were done using the following shRNAs (Sigma): shPop4 (Rpp29) (TRCN0000049878), shRpp20 (TRCN0000049859), and shHIRA (TRCN0000020518). Lentiviruses were prepared in 293-T cells as described previously (93). Briefly, 1 × 106 cells were plated in 10-cm dishes, and cells were infected the next day with virus. At 24 h postinfection, cells were split into 10-cm dishes (1:3). At 48 h, puromycin was added (0.5 μg/ml). At 120 h, cells were harvested for RNA analysis or ChIP.
GST-binding assay
The assay was performed as described previously (38). Briefly, bacterially expressed GST-tagged histones were bound to GSH-Superflow Resin (Clontech), equilibrated with resuspension buffer (25 mm Tris, pH 7.5, 0.5 m KCl, 10% glycerol, 5 mm β-mercaptoethanol, phenylmethylsulfonyl fluoride), and incubated with precleared lysates of U2OS cells expressing YFP-Rpp29 constructs overnight (4 °C) with rotation. The next day, beads were washed three times with resuspension buffer, heated (95 °C) for 3 min in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1) and 3× loading dye (150 mm Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, bromphenol blue, 3% β-mercaptoethanol), and immunoblotted.
Immunoblotting and densitometry analysis
The following antibodies were used: GFP (which also recognizes YFP) (1:1000; catalog no. 11814460001, Roche Applied Science); histone H3.3 (1:1000; catalog no. 09-838, EMD Millipore); histone H3 (1:3000; catalog no. ab1791, Abcam); MYCN (1:1000; catalog no. 9405S, Cell Signaling Technology, Danvers, MA); HIRA antibody (1:1000; catalog no. 12463, Cell Signaling Technology); Rpp29 antibody (1:500; catalog no. NBP1-92281, Novus Biologicals); FLAG antibody (1:10,000; M2; Sigma-Aldrich); GAPDH (1:5000; catalog no. 2118S, Cell Signaling Technology). Measurements of band intensities from scanned images of immunoblots and colloidal blue (Thermo Fisher Scientific) stained gels were done using ImageJ software (version 1.48, National Institutes of Health, Bethesda, MD).
RT-PCR and quantitative PCR (qPCR) analyses
To collect RNA, cells were trypsinized, pelleted, and lysed in TRIzol (Life Technologies, Inc.). RNA was purified using the Direct-zol RNA mini prep kit (Zymo Research, Irvine CA). For qRT-PCR analysis of mRNA, reverse transcription of total RNA was done with random hexamer primer (50 μm; IDT DNA Technologies) using Omniscript (Qiagen) followed by qPCR with SYBR Green (Sigma) using a 7500 Fast real-time PCR machine (Applied Biosystems, Invitrogen). Antisense RNA was measured using strand-specific RT-PCR as described previously (39). qPCR data were analyzed using the 2(−ΔCt) method according to Applied Biosystems guidance, using GAPDH for normalization.
The following primer pairs were used to measure mRNA levels: Rpp29/POP4, 5′-GGAGCTGCGGCTCTTTGA-3′ and 5′-GGAGAGGGAGGAAAAGGCTGTA-3′; Rpp20, 5′-ACCGTGGAGCTTGTTGATGAG-3′ and 5′-GTCAGTGGCTCCCGTGTGT-3′; HIRA, 5′-CAGGAGGATGACGAGAAGGA-3′ and 5′-ACTGTTTGACCACCGCACAC-3′; GAPDH, 5′-ATGGAAATCCCATCACCATCTT-3′ and 5′-CGCCCCACTTGATTTTGG-3′; MYCN, 5′-GGCGTTCCTCCTCCAACA-3′ and 5′-TCTTGGGACGCACAGTGATG-3′; DLX-5, 5′-CCGGCGACTTCCAAGCT-3′ and 5′-TGAGACGGATGGTGCATAGC-3′; DLX-6, 5′-TGGCTGACGGCTTGGAA-3′and 5′-GCTGCCCGAACTCCATGA-3′; PAX-8, 5′-TTTGCTTGGCTCTTTCTACACCTC-3′ and 5′-GAATGTCTGTTTTAAGCTCCCTGG-3′; MYO-D, 5′-CCGCCTGAGCAAAGTAAATGA-3′ and 5′-GGCAACCGCTGGTTTGG-3′; DLK1, 5′-TGCAACCCCCAAAATGGAT-3′ and 5′-GGAGCCACTCTATTACCTGCAAA-3′; MYT1, 5′-GGCAAGGAGGGCTATGCA-3′ and 5′-AAAACACAAGCACCCCCAAA-3′; rDNA, 5′-ACCTGGCGCTAAACCATTCGT-3′ and 5′-GGACAAACCCTTGTGTCGAGG-3′; tRNA Leu, 5′-GAGGACAACGGGGACAGTAA-3′ and 5′-TCCACCAGAAAAACTCCAGC-3′.
ChIP
Cross-linked ChIP was done using the following antibodies: histone H3.3 (5 μg/reaction; catalog no. 09-838, EMD Millipore); H3K36me3 (2.5 μg/reaction; catalog no. ab9050, Abcam); H3K4me3 (2.5 μg/reaction; catalog no. ab8580, Abcam). Cells were incubated with 1% formaldehyde at room temperature for 20 min, followed by treatment with 0.125 m glycine for 5 min. Cells were washed with 1× PBS, pelleted, snap-frozen in liquid nitrogen, and stored (−80 °C). The cell pellets were lysed in Buffer 1 (50 mm HEPES-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, and 0.25% Triton X-100) for 10 min (4 °C) and pelleted at 1350 × g for 5 min. Cells were next incubated in Buffer 2 (10 mm Tris-HCl, pH 8, 200 mm NaCl, 1 mm EDTA, and 0.5 mm EGTA) for 10 min at 4 °C. The pellet was resuspended in Buffer 3 (10 mm Tris-HCl, pH 8, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, and 0.5% sodium laurylsarcosine), sonicated, checked for fragmentation efficiency, and spun at 20,000 × g for 10 min (4 °C). The supernatants were precleared for 2 h in protein A+G-agarose (Invitrogen) and bacterial DNA. Inputs were collected, and the remaining supernatant was incubated with antibodies overnight (4 °C) with rotation. The next day, the samples were incubated with protein A+G-agarose and bacterial DNA for 2 h (4 °C) with rotation. The beads were pelleted at 2000 rpm for 2 min (4 °C) and washed four times with radioimmune precipitation assay wash buffer (50 mm HEPES-KOH, pH 7.5, 500 mm lithium chloride, 1 mm EDTA, 1% Nonidet P-40, and 0.7% sodium deoxycholate). DNA was eluted in 150 μl of elution buffer (50 mm Tris-HCl, pH 8.0, 1% SDS, and 10 mm EDTA), decross-linked overnight, and purified using the Qiaquick PCR purification kit (Qiagen).
The following primer pairs were used: MYCN promoter, 5′-CATCCAGAGGTCTTGTTCCTAAGG-3′ and 5′-GAGGTTGCTCCTGTGTAATTACGA-3′; MYCN exon 4, 5′-GGAAAAATTGCAGGCAAGACA-3′ and 5′-AATGTGCAAAGTGGCAGTGACT-3′; DLX-5 promoter, 5′-GCGTCCGGGAGATACTCATC-3′; 5′-TGGAGTGTTGGGAAGCAAGTT-3′; DLX-5 exon 1, 5′-GAGCGATGACAGGAGTGTTTGA-3′ and 5′-TGAGACGGATGGTGCATAGC-3′; DLX-6 promoter, 5′-GCAACCTACAAGGGTGCAAAG3′ and 5′-GGAGCCCCAGTTGCCAATAT-3′; DLX-6 exon 3, 5′-AGTATGCCCCCCAACAGCTA-3′ and 5′-CTTGGGCAACTCACATCATCTG-3′; rRNA promoter, 5′-CTGCGATGGTGGCGTTTTTG-3′ and 5′-ACAGCGTGTCAGCAATAACC-3′ (94, 95); 28S rRNA, 5′-ACCTGGCGCTAAACCATTCGT-3′ and 5′-GGACAAACCCTTGTGTCGAGG-3′; tRNA Leu, 5′-GAGGACAACGGGGACAGTAA-3′ and 5′-TCCACCAGAAAAACTCCAGC-3′ (96); PAX-8 exon 4, 5′-CCTGGGAACAGGCCAGACT-3′ and 5′-TCACAAACCCATGTGTAGACTGAGT-3′; MYO-D exon 1, 5′-CCGCCTGAGCAAAGTAAATGA-3′ and 5′-GGCAACCGCTGGTTTGG-3′; DLK1 exon 4, 5′-TGCAACCCCCAAAATGGAT-3′ and 5′-GGAGCCACTCTATTACCTGCAAA-3′; MYT1 exon 4, 5′-GGCAAGGAGGGCTATGCA-3′ and 5′-AAAACACAAGCACCCCCAAA-3′.
Histone sample preparation for qMS
Histone samples for qMS were prepared as described previously with minor modifications (97). Cells were resuspended in nuclear isolation buffer (15 mm Tris, pH 7.5, 15 mm NaCl, 60 mm KCl, 5 mm MgCl2, 1 mm CaCl2, 250 mm sucrose, supplemented with 1 mm DTT, 500 μm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 5 nm microcystin, and 10 mm sodium butyrate immediately before use) with 0.2% Nonidet P-40 and incubated on ice for 10 min. Nuclei were pelleted at 3400 × g for 5 min at 4 °C and washed twice in nuclear isolation buffer without detergent. Histones were extracted with 0.4 n H2SO4 for 2–4 h at 4 °C, separated from insoluble debris by centrifugation as above, and precipitated overnight at 4 °C by the addition of tricholoracetic acid to a final concentration of 25%. Histones were pelleted by centrifugation as above, washed sequentially in acetone plus 0.1% HCl and acetone, air-dried, and resuspended in 0.1 m ammonium bicarbonate for derivatization with propionic anhydride. To propionylate free lysine residues, 1 volume of propionic anhydride reagent (25% propionic anhydride in 2-propanol) was added to 2 volumes of histone sample (generally containing 5–20 μg of protein) and incubated at 37 °C for 15 min. The sample was then dried in a SpeedVac and derivatized once more before overnight digestion with trypsin (1:20 ratio by mass) at room temperature in 0.1 m ammonium bicarbonate. After digestion, histones were derivatized twice more to propionylate free N termini and then desalted with C18 Stage Tips before analysis by LC-MS/MS.
Mass spectrometry and data analysis
Histone peptides were analyzed by LC-MS/MS as follows. Peptides were separated on fused silica capillary columns (75-μm inner diameter × 15 cm), packed with C18 resin (3 μm ReproSil-Pur 120 C18-AQ, Dr. Maisch GmbH), via an EasyLC 1000 nano-LC system (Thermo Fisher Scientific). Solvents A and B were composed of 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. The chromatography gradient consisted of 2–28% solvent B over 45 min, 28–85% solvent B over 5 min, and 85% solvent B for 10 min at a flow rate of 300 nl/min. Peptides eluted directly into an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) running in data-independent acquisition (DIA) mode. Each instrument cycle began with a full MS precursor scan in the Orbitrap from 300 to 1100 m/z in positive profile mode at a resolution of 120,000, followed by eight DIA MS/MS scans in the ion trap (positive centroid mode with collision-induced dissociation fragmentation at 35% normalized collision energy) with isolation windows of 50 m/z increasing from 325 m/z to 675 m/z, a second full MS scan, and eight more DIA MS/MS scans centered on 725–1075 m/z. The mass range of the DIA scans varied slightly, beginning with 120–1500 m/z for the first DIA scan and ending with 295–1500 m/z for the final DIA scan. EpiProfile, a custom software algorithm written in Matlab, was used to extract chromatographic peak areas of the masses corresponding to the uniquely modified peptide species (98). For each peptide sequence, the relative abundance of a uniquely modified form was obtained by calculating its peak area as a proportion of the sum of the peak areas for all modified and unmodified forms of that peptide sequence.
Author contributions
P. K. S. and S. M. J. conceptualization; P. K. S., P. J. L., and S. M. J. data curation; P. K. S. and P. J. L. formal analysis; P. K. S. and P. J. L. validation; P. K. S. and P. J. L. investigation; P. K. S., P. J. L., and B. A. G. methodology; P. K. S., P. J. L., and B. A. G. writing-review and editing; B. A. G. resources; B. A. G. and S. M. J. supervision; B. A. G. funding acquisition; B. A. G. and S. M. J. project administration; S. M. J. writing-original draft.
Supplementary Material
Acknowledgments
We acknowledge the Wistar Cancer Center Genomics and Molecular Screening Core Facilities (supported by Wistar Cancer Center Core Facilities Grant P30 CA10815). We thank Sylvie Shaffer for artistic contributions to the model.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- PTM
- post-translational modification
- AS
- antisense
- S
- sense
- GCTB
- giant cell tumor of the bone
- HGG
- high-grade glioma
- H2B-BR
- H2B-binding region
- H3.3-BR
- H3.3-binding region
- PRC2
- Polycomb repressive complex 2
- qMS
- quantitative MS
- RI
- replication-independent
- RMRP
- RNase MRP catalytic RNA
- pol
- polymerase
- RPL
- ribosomal protein–like
- RNP
- ribonucleoprotein
- GST
- glutathione S-transferase
- aa
- amino acids
- YFP
- yellow fluorescent protein
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR
- DIA
- data-independent acquisition
- H3
- histone H3
- K
- lysine
- me1
- me2, and me3, mono-, di-, and trimethylation
- ac
- acetylation.
References
- 1. Talbert P. B., and Henikoff S. (2017) Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 10.1038/nrm.2016.148 [DOI] [PubMed] [Google Scholar]
- 2. Weinberg D. N., Allis C. D., and Lu C. (2017) Oncogenic mechanisms of histone H3 mutations. Cold Spring Harb. Perspect. Med. 7, a026443 10.1101/cshperspect.a026443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., and Felsenfeld G. (2009) H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 10.1038/ng.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mito Y., Henikoff J. G., and Henikoff S. (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 10.1038/ng1637 [DOI] [PubMed] [Google Scholar]
- 5. Wirbelauer C., Bell O., and Schübeler D. (2005) Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19, 1761–1766 10.1101/gad.347705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pchelintsev N. A., McBryan T., Rai T. S., van Tuyn J., Ray-Gallet D., Almouzni G., and Adams P. D. (2013) Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019 10.1016/j.celrep.2013.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong L. H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M. A., Mann J., and Choo K. H. (2009) Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drané P., Ouararhni K., Depaux A., Shuaib M., and Hamiche A. (2010) The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 10.1101/gad.566910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luijsterburg M. S., de Krijger I., Wiegant W. W., Shah R. G., Smeenk G., de Groot A. J. L., Pines A., Vertegaal A. C. O., Jacobs J. J. L., Shah G. M., and van Attikum H. (2016) PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 10.1016/j.molcel.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frey A., Listovsky T., Guilbaud G., Sarkies P., and Sale J. E. (2014) Histone H3.3 is required to maintain replication fork progression after UV damage. Curr. Biol. 24, 2195–2201 10.1016/j.cub.2014.07.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van der Heijden G. W., Derijck A. A., Pósfai E., Giele M., Pelczar P., Ramos L., Wansink D. G., van der Vlag J., Peters A. H., and de Boer P. (2007) Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 39, 251–258 10.1038/ng1949 [DOI] [PubMed] [Google Scholar]
- 13. Torres-Padilla M. E., Bannister A. J., Hurd P. J., Kouzarides T., and Zernicka-Goetz M. (2006) Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 [DOI] [PubMed] [Google Scholar]
- 14. Loppin B., Bonnefoy E., Anselme C., Laurençon A., Karr T. L., and Couble P. (2005) The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 10.1038/nature04059 [DOI] [PubMed] [Google Scholar]
- 15. Voon H. P., and Wong L. H. (2016) New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic acids Res. 44, 1496–1501 10.1093/nar/gkw012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong L. H., McGhie J. D., Sim M., Anderson M. A., Ahn S., Hannan R. D., George A. J., Morgan K. A., Mann J. R., and Choo K. H. (2010) ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 10.1101/gr.101477.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., and Allis C. D. (2010) Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 107, 14075–14080 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elsässer S. J., Noh K. M., Diaz N., Allis C. D., and Banaszynski L. A. (2015) Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 10.1038/nature14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jang C. W., Shibata Y., Starmer J., Yee D., and Magnuson T. (2015) Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–1392 10.1101/gad.264150.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tagami H., Ray-Gallet D., Almouzni G., and Nakatani Y. (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- 21. Kraushaar D. C., Jin W., Maunakea A., Abraham B., Ha M., and Zhao K. (2013) Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 10.1186/gb-2013-14-10-r121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang C., Zhang Z., Xu M., Li Y., Li Z., Ma Y., Cai T., and Zhu B. (2013) H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 9, e1003558 10.1371/journal.pgen.1003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C., and Zhu B. (2014) H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin? BioEssays 36, 579–584 10.1002/bies.201400005 [DOI] [PubMed] [Google Scholar]
- 24. Banaszynski L. A., Wen D., Dewell S., Whitcomb S. J., Lin M., Diaz N., Elsässer S. J., Chapgier A., Goldberg A. D., Canaani E., Rafii S., Zheng D., and Allis C. D. (2013) Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 10.1016/j.cell.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindroth A. M., and Plass C. (2013) Recurrent H3.3 alterations in childhood tumors. Nat. Genet. 45, 1413–1414 10.1038/ng.2832 [DOI] [PubMed] [Google Scholar]
- 26. Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., Becksfort J., Qu C., Ding L., Huether R., Parker M., Zhang J., Gajjar A., Dyer M. A., Mullighan C. G., Gilbertson R. J., et al. (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M., Hovestadt V., Albrecht S., Kool M., Nantel A., Konermann C., et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 28. Bender S., Tang Y., Lindroth A. M., Hovestadt V., Jones D. T., Kool M., Zapatka M., Northcott P. A., Sturm D., Wang W., Radlwimmer B., Højfeldt J. W., Truffaux N., Castel D., Schubert S., et al. (2013) Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 10.1016/j.ccr.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 29. Chan K. M., Fang D., Gan H., Hashizume R., Yu C., Schroeder M., Gupta N., Mueller S., James C. D., Jenkins R., Sarkaria J., and Zhang Z. (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 10.1101/gad.217778.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis P. W., Müller M. M., Koletsky M. S., Cordero F., Lin S., Banaszynski L. A., Garcia B. A., Muir T. W., Becher O. J., and Allis C. D. (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venneti S., Garimella M. T., Sullivan L. M., Martinez D., Huse J. T., Heguy A., Santi M., Thompson C. B., and Judkins A. R. (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 23, 558–564 10.1111/bpa.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Behjati S., Tarpey P. S., Presneau N., Scheipl S., Pillay N., Van Loo P., Wedge D. C., Cooke S. L., Gundem G., Davies H., Nik-Zainal S., Martin S., McLaren S., Goodie V., Robinson B., et al. (2013) Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 10.1038/ng.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fang D., Gan H., Lee J. H., Han J., Wang Z., Riester S. M., Jin L., Chen J., Zhou H., Wang J., Zhang H., Yang N., Bradley E. W., Ho T. H., Rubin B. P., et al. (2016) The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 10.1126/science.aae0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu C., Jain S. U., Hoelper D., Bechet D., Molden R. C., Ran L., Murphy D., Venneti S., Hameed M., Pawel B. R., Wunder J. S., Dickson B. C., Lundgren S. M., Jani K. S., De Jay N., et al. (2016) Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 10.1126/science.aac7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bjerke L., Mackay A., Nandhabalan M., Burford A., Jury A., Popov S., Bax D. A., Carvalho D., Taylor K. R., Vinci M., Bajrami I., McGonnell I. M., Lord C. J., Reis R. M., Hargrave D., et al. (2013) Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 3, 512–519 10.1158/2159-8290.CD-12-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yadav R. K., Jablonowski C. M., Fernandez A. G., Lowe B. R., Henry R. A., Finkelstein D., Barnum K. J., Pidoux A. L., Kuo Y. M., Huang J., O'Connell M. J., Andrews A. J., Onar-Thomas A., Allshire R. C., and Partridge J. F. (2017) Histone H3G34R mutation causes replication stress, homologous recombination defects and genomic instability in S. pombe. Elife 6, e27406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi L., Shi J., Shi X., Li W., and Wen H. (2018) Histone H3.3 G34 Mutations alter histone H3K36 and H3K27 methylation in cis. J. Mol. Biol. 430, 1562–1565 10.1016/j.jmb.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newhart A., Powers S. L., Shastrula P. K., Sierra I., Joo L. M., Hayden J. E., Cohen A. R., and Janicki S. M. (2016) RNase P protein subunit, Rpp29, represses histone H3.3 nucleosome deposition. Mol. Biol. Cell 27, 1154–1169 10.1091/mbc.e15-02-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newhart A., Rafalska-Metcalf I. U., Yang T., Joo L. M., Powers S. L., Kossenkov A. V., Lopez-Jones M., Singer R. H., Showe L. C., Skordalakes E., and Janicki S. M. (2013) Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site. J. Biol. Chem. 288, 19882–19899 10.1074/jbc.M113.473181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lim J., Park J. H., Baude A., Yoo Y., Lee Y. K., Schmidt C. R., Park J. B., Fellenberg J., Zustin J., Haller F., Krücken I., Kang H. G., Park Y. J., Plass C., and Lindroth A. M. (2017) The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing. Sci. Rep. 7, 13459 10.1038/s41598-017-13887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gopalan V., Jarrous N., and Krasilnikov A. S. (2018) Chance and necessity in the evolution of RNase P. RNA 24, 1–5 10.1261/rna.063107.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jarrous N. (2017) Roles of RNase P and its subunits. Trends Genet. 33, 594–603 10.1016/j.tig.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 43. Reiner R., Ben-Asouli Y., Krilovetzky I., and Jarrous N. (2006) A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 20, 1621–1635 10.1101/gad.386706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reiner R., Krasnov-Yoeli N., Dehtiar Y., and Jarrous N. (2008) Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One 3, e4072 10.1371/journal.pone.0004072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Serruya R., Orlovetskie N., Reiner R., Dehtiar-Zilber Y., Wesolowski D., Altman S., and Jarrous N. (2015) Human RNase P ribonucleoprotein is required for formation of initiation complexes of RNA polymerase III. Nucleic Acids Res. 43, 5442–5450 10.1093/nar/gkv447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marvin M. C., Clauder-Münster S., Walker S. C., Sarkeshik A., Yates J. R. 3rd, Steinmetz L. M., and Engelke D. R. (2011) Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 17, 1441–1450 10.1261/rna.2737511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molla-Herman A., Vallés A. M., Ganem-Elbaz C., Antoniewski C., and Huynh J. R. (2015) tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J. 34, 3009–3027 10.15252/embj.201591006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abu-Zhayia E. R., Khoury-Haddad H., Guttmann-Raviv N., Serruya R., Jarrous N., and Ayoub N. (2017) A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Sci. Rep. 7, 1002 10.1038/s41598-017-01185-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., and Tyler J. K. (2016) Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife 5, e15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adam S., Polo S. E., and Almouzni G. (2013) Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155, 94–106 10.1016/j.cell.2013.08.029 [DOI] [PubMed] [Google Scholar]
- 51. Jones J. M., Bhattacharyya A., Simkus C., Vallieres B., Veenstra T. D., and Zhou M. (2011) The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol. Lett. 136, 156–162 10.1016/j.imlet.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 52. Lin C. J., Koh F. M., Wong P., Conti M., and Ramalho-Santos M. (2014) Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell 30, 268–279 10.1016/j.devcel.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jullien J., Astrand C., Szenker E., Garrett N., Almouzni G., and Gurdon J. B. (2012) HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 5, 17 10.1186/1756-8935-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ahmad K., and Henikoff S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- 55. Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., Mennella T. A., and Keogh M. C. (2012) The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287, 1709–1718 10.1074/jbc.M111.316489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., Hughes T. R., and Winston F. (2008) Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6, e277 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ruthenburg A. J., Allis C. D., and Wysocka J. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 10.1016/j.molcel.2006.12.014 [DOI] [PubMed] [Google Scholar]
- 58. Lin S., and Garcia B. A. (2012) Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 512, 3–28 10.1016/B978-0-12-391940-3.00001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., and Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- 60. Ferrari K. J., Scelfo A., Jammula S., Cuomo A., Barozzi I., Stützer A., Fischle W., Bonaldi T., and Pasini D. (2014) Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53, 49–62 10.1016/j.molcel.2013.10.030 [DOI] [PubMed] [Google Scholar]
- 61. Yuan W., Xu M., Huang C., Liu N., Chen S., and Zhu B. (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 10.1074/jbc.M110.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 63. Klymenko T., and Müller J. (2004) The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377 10.1038/sj.embor.7400111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maizels N., and Weiner A. M. (1994) Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. U.S.A. 91, 6729–6734 10.1073/pnas.91.15.6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sandman K., and Reeve J. N. (2006) Archaeal histones and the origin of the histone fold. Curr. Opin. Microbiol. 9, 520–525 10.1016/j.mib.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 66. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 67. Hennig B. P., and Fischer T. (2013) The great repression: chromatin and cryptic transcription. Transcription 4, 97–101 10.4161/trns.24884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Venkatesh S., Workman J. L., and Smolle M. (2013) UpSETing chromatin during non-coding RNA production. Epigenetics Chromatin 6, 16 10.1186/1756-8935-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rougemaille M., and Libri D. (2010) Control of cryptic transcription in eukaryotes. Adv. Exp. Med. Biol. 702, 122–131 10.1007/978-1-4419-7841-7_10 [DOI] [PubMed] [Google Scholar]
- 70. Kopp F., and Mendell J. T. (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim J. H., Lee B. B., Oh Y. M., Zhu C., Steinmetz L. M., Lee Y., Kim W. K., Lee S. B., Buratowski S., and Kim T. (2017) Erratum: Modulation of mRNA and lncRNA expression dynamics by the Set2-Rpd3S pathway. Nat. Commun. 8, 16122 10.1038/ncomms16122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., Kumar S., Natarajan K., and Workman J. L. (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 10.1038/nature11326 [DOI] [PubMed] [Google Scholar]
- 73. Davidovich C., and Cech T. R. (2015) The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022 10.1261/rna.053918.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang X., Paucek R. D., Gooding A. R., Brown Z. Z., Ge E. J., Muir T. W., and Cech T. R. (2017) Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 24, 1028–1038 10.1038/nsmb.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Beltran M., Yates C. M., Skalska L., Dawson M., Reis F. P., Viiri K., Fisher C. L., Sibley C. R., Foster B. M., Bartke T., Ule J., and Jenner R. G. (2016) The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 26, 896–907 10.1101/gr.197632.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Natrajan R., Mackay A., Wilkerson P. M., Lambros M. B., Wetterskog D., Arnedos M., Shiu K. K., Geyer F. C., Langerød A., Kreike B., Reyal F., Horlings H. M., van de Vijver M. J., Palacios J., Weigelt B., and Reis-Filho J. S. (2012) Functional characterization of the 19q12 amplicon in grade III breast cancers. Breast Cancer Res. 14, R53 10.1186/bcr3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wrzeszczynski K. O., Varadan V., Byrnes J., Lum E., Kamalakaran S., Levine D. A., Dimitrova N., Zhang M. Q., and Lucito R. (2011) Identification of tumor suppressors and oncogenes from genomic and epigenetic features in ovarian cancer. PLoS One 6, e28503 10.1371/journal.pone.0028503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ceccarelli M., Barthel F. P., Malta T. M., Sabedot T. S., Salama S. R., Murray B. A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S. M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., et al. (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 10.1016/j.cell.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang W., Thomas B., Flynn R. A., Gavzy S. J., Wu L., Kim S. V., Hall J. A., Miraldi E. R., Ng C. P., Rigo F. W., Meadows S., Montoya N. R., Herrera N. G., Domingos A. I., Rastinejad F., et al. (2015) DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528, 517–522 10.1038/nature16193 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Feng W., Li L., Xu X., Jiao Y., and Du W. (2017) Up-regulation of the long non-coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch. Med. Sci. 13, 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rheinbay E., Parasuraman P., Grimsby J., Tiao G., Engreitz J. M., Kim J., Lawrence M. S., Taylor-Weiner A., Rodriguez-Cuevas S., Rosenberg M., Hess J., Stewart C., Maruvka Y. E., Stojanov P., Cortes M. L., et al. (2017) Recurrent and functional regulatory mutations in breast cancer. Nature 547, 55–60 10.1038/nature22992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Meng Q., Ren M., Li Y., and Song X. (2016) LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One 11, e0164845 10.1371/journal.pone.0164845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Park J., and Jeong S. (2015) Wnt activated β-catenin and YAP proteins enhance the expression of non-coding RNA component of RNase MRP in colon cancer cells. Oncotarget 6, 34658–34668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Song H., Sun W., Ye G., Ding X., Liu Z., Zhang S., Xia T., Xiao B., Xi Y., and Guo J. (2013) Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 11, 225 10.1186/1479-5876-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Futamura N., Urakawa H., Tsukushi S., Arai E., Kozawa E., Ishiguro N., and Nishida Y. (2016) Giant cell tumor of bone arising in long bones possibly originates from the metaphyseal region. Oncol. Lett. 11, 2629–2634 10.3892/ol.2016.4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ridanpää M., van Eenennaam H., Pelin K., Chadwick R., Johnson C., Yuan B., vanVenrooij W., Pruijn G., Salmela R., Rockas S., Mäkitie O., Kaitila I., and de la Chapelle A. (2001) Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104, 195–203 10.1016/S0092-8674(01)00205-7 [DOI] [PubMed] [Google Scholar]
- 87. Barraza-García J., Rivera-Pedroza C. I., Hisado-Oliva A., Belinchón-Martínez A., Sentchordi-Montane L., Duncan E. L., Clark G. R., Del Pozo A., Ibáñez-Garikano K., Offiah A., Prieto-Matos P., Cormier-Daire V., and Heath K. E. (2017) Broadening the phenotypic spectrum of POP1-skeletal dysplasias: identification of POP1 mutations in a mild and severe skeletal dysplasia. Clin. Genet. 92, 91–98 10.1111/cge.12964 [DOI] [PubMed] [Google Scholar]
- 88. Elalaoui S. C., Laarabi F. Z., Mansouri M., Mrani N. A., Nishimura G., and Sefiani A. (2016) Further evidence of POP1 mutations as the cause of anauxetic dysplasia. Am. J. Med. Genet. A 170, 2462–2465 10.1002/ajmg.a.37839 [DOI] [PubMed] [Google Scholar]
- 89. Glazov E. A., Zankl A., Donskoi M., Kenna T. J., Thomas G. P., Clark G. R., Duncan E. L., and Brown M. A. (2011) Whole-exome re-sequencing in a family quartet identifies POP1 mutations as the cause of a novel skeletal dysplasia. PLoS Genet. 7, e1002027 10.1371/journal.pgen.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ferenczy M. W., Ranayhossaini D. J., and Deluca N. A. (2011) Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J. Virol. 85, 4993–5002 10.1128/JVI.02265-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Newhart A., Negorev D. G., Rafalska-Metcalf I. U., Yang T., Maul G. G., and Janicki S. M. (2013) Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site. Mol. Biol. Cell 24, 1454–1468 10.1091/mbc.e12-09-0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Newhart A., Rafalska-Metcalf I. U., Yang T., Negorev D. G., and Janicki S. M. (2012) Single-cell analysis of Daxx and ATRX-dependent transcriptional repression. J. Cell Sci. 125, 5489–5501 10.1242/jcs.110148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Everett R. D., Parada C., Gripon P., Sirma H., and Orr A. (2008) Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82, 2661–2672 10.1128/JVI.02308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ghoshal K., Majumder S., Datta J., Motiwala T., Bai S., Sharma S. M., Frankel W., and Jacob S. T. (2004) Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 279, 6783–6793 10.1074/jbc.M309393200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tanaka Y., Okamoto K., Teye K., Umata T., Yamagiwa N., Suto Y., Zhang Y., and Tsuneoka M. (2010) JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 29, 1510–1522 10.1038/emboj.2010.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gomez-Roman N., Grandori C., Eisenman R. N., and White R. J. (2003) Direct activation of RNA polymerase III transcription by c-Myc. Nature 421, 290–294 10.1038/nature01327 [DOI] [PubMed] [Google Scholar]
- 97. Sidoli S., and Garcia B. A. (2017) Characterization of individual histone posttranslational modifications and their combinatorial patterns by mass spectrometry-based proteomics strategies. Methods Mol. Biol. 1528, 121–148 10.1007/978-1-4939-6630-1_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yuan Z. F., Lin S., Molden R. C., Cao X. J., Bhanu N. V., Wang X., Sidoli S., Liu S., and Garcia B. A. (2015) EpiProfile quantifies histone peptides with modifications by extracting retention time and intensity in high-resolution mass spectra. Mol. Cell. Proteomics 14, 1696–1707 10.1074/mcp.M114.046011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.