Abstract
Protoporphyrinogen IX oxidase (PPO), the last enzyme that is common to both chlorophyll and heme biosynthesis pathways, catalyzes the oxidation of protoporphyrinogen IX to protoporphyrin IX. PPO has several isoforms, including the oxygen-dependent HemY and an oxygen-independent enzyme, HemG. However, most cyanobacteria encode HemJ, the least characterized PPO form. We have characterized HemJ from the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis 6803) as a bona fide PPO; HemJ down-regulation resulted in accumulation of tetrapyrrole precursors and in the depletion of chlorophyll precursors. The expression of FLAG-tagged Synechocystis 6803 HemJ protein (HemJ.f) and affinity isolation of HemJ.f under native conditions revealed that it binds heme b. The most stable HemJ.f form was a dimer, and higher oligomeric forms were also observed. Using both oxygen and artificial electron acceptors, we detected no enzymatic activity with the purified HemJ.f, consistent with the hypothesis that the enzymatic mechanism for HemJ is distinct from those of other PPO isoforms. The heme absorption spectra and distant HemJ homology to several membrane oxidases indicated that the heme in HemJ is redox-active and involved in electron transfer. HemJ was conditionally complemented by another PPO, HemG from Escherichia coli. If grown photoautotrophically, the complemented strain accumulated tripropionic tetrapyrrole harderoporphyrin, suggesting a defect in enzymatic conversion of coproporphyrinogen III to protoporphyrinogen IX, catalyzed by coproporphyrinogen III oxidase (CPO). This observation supports the hypothesis that HemJ is functionally coupled with CPO and that this coupling is disrupted after replacement of HemJ by HemG.
Keywords: photosynthesis, heme, cyanobacteria, enzyme purification, membrane protein, coproporphyrinogen III oxidase, HemJ, protoporphyrinogen IX oxidase, Synechocystis sp. PCC 6803
Introduction
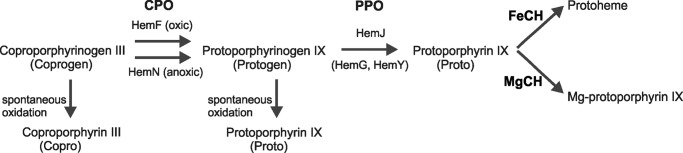
The last common step for heme and chlorophyll biosynthesis (1), conversion of protoporphyrinogen IX (Protogen)2 into protoporphyrin IX (Proto), is catalyzed by the enzyme protoporphyrinogen IX oxidase (Fig. 1). Although the six-electron oxidation of Protogen occurs also spontaneously, its enzymatic conversion is apparently necessary for the correct channeling of Proto to chelatases for metal insertion (Fig. 1). Three analogous enzymes HemY, HemG, and lastly HemJ (recently reannotated as PgoX, PgdH1, and PgdH2 (2)), exhibiting no mutual homology, have been found to serve as a protoporphyrinogen oxidase (3–5). HemY is an oxygen-dependent enzyme of ∼55 kDa, creating membrane bound dimers and using FAD as a co-factor (6). HemY occurs in most of bacteria phyla and in almost all eukaryotes (7).
Figure 1.
Late steps of tetrapyrrole biosynthesis in cyanobacteria (Synechocystis 6803). Conversion of Coprogen to Protogen is catalyzed by CPO. In Synechocystis 6803, HemF works as the sole CPO under oxic conditions, and HemN works under anoxic conditions. The CPO product Protogen is oxidized to Proto by PPO. In most cyanobacteria, this reaction is performed by HemJ PPO isoform, in others by HemG or HemY. Proto is than channeled to ferrochelatase (FeCH) leading to Protoheme or to magnesium-chelatase (MgCH), first enzyme of the dedicated chlorophyll pathway. Coprogen and Protogen can be spontaneously oxidized to Copro and Proto, respectively.
HemG (21 kDa) is mostly found in γ-proteobacteria (7). The protein forms membrane-associated oligomers (8, 9) and uses noncovalently bound flavin mononucleotide as a co-factor (9). HemG is functional in oxic as well as anoxic conditions (9). Electrons from the Proto oxidation catalyzed by HemG are withdrawn via ubiquinone, cytochrome bo, and cytochrome bd oxidases to oxygen or under anoxic conditions to the terminal electron acceptors fumarate and nitrate by corresponding reductases (8).
Most recently, the slr1790 gene from the cyanobacterium Synechocystis 6803 has been described as the third PPO and named hemJ (3). Although HemJ is the most common PPO variant in cyanobacteria, in cyanobacterial GenBank genomes, we found 435 hemJ genes and only 47 hemY genes, and its origin and spread within cyanobacteria is not clear. It is assumed that HemJ evolved within α-proteobacteria and spread to cyanobacteria and various proteobacteria through multiple horizontal gene transfers (7). In contrast, the phylogenetic tree of HemY for cyanobacteria is consistent with the cyanobacterial phylogeny, and HemY was probably the ancestral PPO in cyanobacteria (7). For completeness, eight genomes of the Synechococcus–Prochlorococcus lineage contain the HemG PPO variant probably obtained by horizontal gene transfer.
As with other PPOs, hemJ appears to be essential and could not be inactivated in Synechocystis 6803 (3). A partially segregated Synechocystis 6803 strain contained less than half of the chlorophyll content compared with WT, and although it accumulated Proto, it probably originated from nonenzymatic oxidation of Protogen (Fig. 1). Accordingly, the hemJ deletion strain from Acinetobacter baylyi exhibited auxotrophy for hemin and accumulated porphyrins when supplemented with 5-aminolevulinic acid (10).
To demonstrate that HemJ is a bona fide PPO, HemJ mutants were complemented with other PPO isoforms: several hemY variants and hemG rescued hemin auxotrophy in A. baylyi (10), and expression of Arabidopsis thaliana hemY in Synechocystis 6803 enabled hemJ inactivation. This mutant strain accumulated Proto when treated with HemY inhibitor acifluorfen (3). This accumulation of intermediates is consistent with substrate channeling between the terminal enzymes of heme synthesis. Formation of the multienzyme heme biosynthetic complex, containing the three terminal enzymes CPO, PPO, and ferrochelatase (FeCH) is predicted (11) and partly documented (12, 13). For prokaryotes, identification of such complexes is further complicated by the existence of multiple enzyme isoforms and different heme biosynthetic pathways (2). In addition to PPOs there are two forms of CPOs in Synechocystis 6803 as in most organisms performing oxygenic photosynthesis: HemN and HemF (14). Under micro-oxic conditions, the conversion of coproporphyrinogen III (Coprogen) to Protogen is catalyzed by an oxygen-independent CPO encoded by hemN, whereas oxidative decarboxylation of the substrate requires oxygen and is catalyzed by HemF, an enzyme structurally and functionally unrelated to HemN (14, 15) (Fig. 1). HemN is a monomeric, iron–sulfur cluster–containing protein (16), HemF is a dimeric protein with two independent active sites (17). Herein, we report the biochemical characterization of HemJ homologously expressed in Synechocystis 6803 and the results of complementation of hemJ mutant by expression of hemG from Escherichia coli. We also show that there is a functional coupling between Synechocystis 6803 CPO and HemJ.
Results
Synechocystis 6803 HemJ forms a heme-binding oligomer
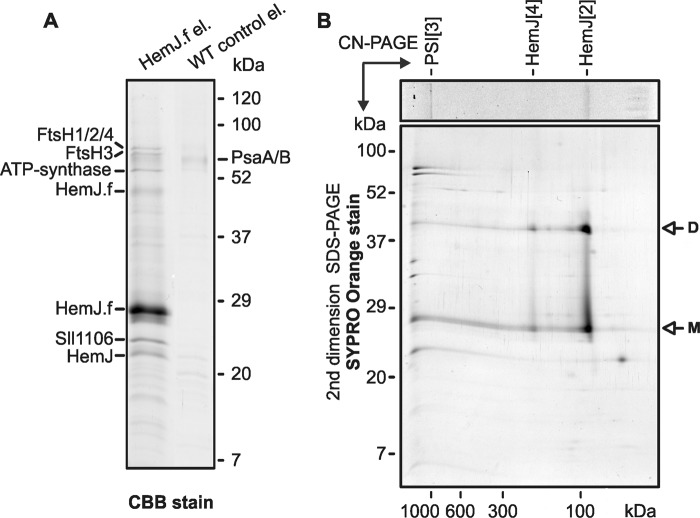
Because heterologous expression of HemJ proteins from various organisms frequently resulted in no or poor expression (3, 10), we expressed the HemJ enzyme fused on its C terminus with 3× FLAG tag (HemJ.f) homologously in Synechocystis 6803 under psbAII promoter. After full segregation of the hemJ.f strain, it was possible to delete the WT copy of the hemJ gene, demonstrating that the HemJ.f protein is functional (Fig. S1). However, for the HemJ isolation, a strain containing both WT and tagged variants of HemJ was used to potentially co-isolate HemJ.f, together with the native form of the protein (see below): oligomeric states have previously been described for other PPO variants (8). After affinity chromatography of hemJ.f membrane fraction solubilized by nonionic detergent, the purified HemJ.f eluate was markedly reddish. Native separation of the eluate on clear native gel (CN–PAGE) resulted in two reddish bands: CN1 and only slightly visible CN2 (Fig. 2). Photosystem I (PSI) was shown to bind nonspecifically to the anti-FLAG affinity resin (18). Hence, HemJ.f was also isolated from a strain lacking PSI (ΔPSI) to achieve maximum purity (Fig. 2). Absorption spectra of the eluted HemJ.f protein showed an absorption maximum at 412 nm (Fig. 3A), which was presumed to be the Soret band of a bound tetrapyrrole. After deletion of PSI only a small amounts of chlorophyll (absorbing at 671 nm) and variable amount of carotenoids (absorbance 450–520 nm) co-eluted with HemJ.f (Fig. 3A). The bound tetrapyrrole was extracted by acetone and was identified as protoheme by HPLC (Fig. S2) (protoheme is referred as heme b, when it is bound to a protein). To spectroscopically characterize this bound heme further, absorption spectra of the HemJ.f eluate from the ΔPSI strain were measured after oxidation by air or after reduction by dithionite (Fig. 3B). The oxidized spectrum indicates a high spin heme b (λmax = 411 nm), and the reduced spectrum (λmax = 424 nm) is typical for a ferrous six-coordinate b-type heme. To assess the stoichiometry of heme binding to HemJ.f, protein isolated by affinity chromatography was further purified by size-exclusion chromatography (Fig. 4A). Fractions with the highest absorption at 415 nm were collected and used for heme and protein quantification. The protein concentration was estimated from the absorbance at 280 nm using calculated extinction coefficient for HemJ.f. The estimated molar ratio of heme to HemJ.f monomer was 0.85 ± 0.05. Eluted fractions did not display any sign of chlorophyll or carotenoids (Fig. 4B). Size-exclusion chromatography was also used to estimate the size of the native HemJ complex (Fig. 4A). The main elution peak corresponds to a protein of ∼150 kDa, indicating a higher oligomeric state of HemJ.f (27 kDa monomer) even when taking into account its association with detergent. Analysis of the HemJ.f eluate by SDS–PAGE revealed that several proteins specifically co-eluted from the anti-FLAG resin with HemJ.f (Fig. 5A). These protein bands were identified by MS as Sll1106, FtsH proteases, and ATP synthase subunits (Table S1). Sll1106, together with HemJ, was also identified by MS in the CN1 band from CN–PAGE (Fig. 2). Nevertheless, the reddish CN1 band was present even when HemJ.f was purified from a Δsll1106 background (Fig. S3), indicating that heme is associated with HemJ.f. Furthermore, when a gel strip from CN–PAGE (Fig. 2) was separated in a second dimension by SDS–PAGE (Fig. 5B), colored CN1 and CN2 bands dissociated into two spots, apparently representing monomer and dimer of the HemJ.f (27 kDa). The presence of the HemJ.f dimer, even on the denaturing SDS gel, suggests its remarkable stability. The upper CN2 band contains the HemJ.f oligomer, most probably a tetramer, which is in agreement with the size of the complex determined by the size-exclusion chromatography (Fig. 4A).
Figure 2.
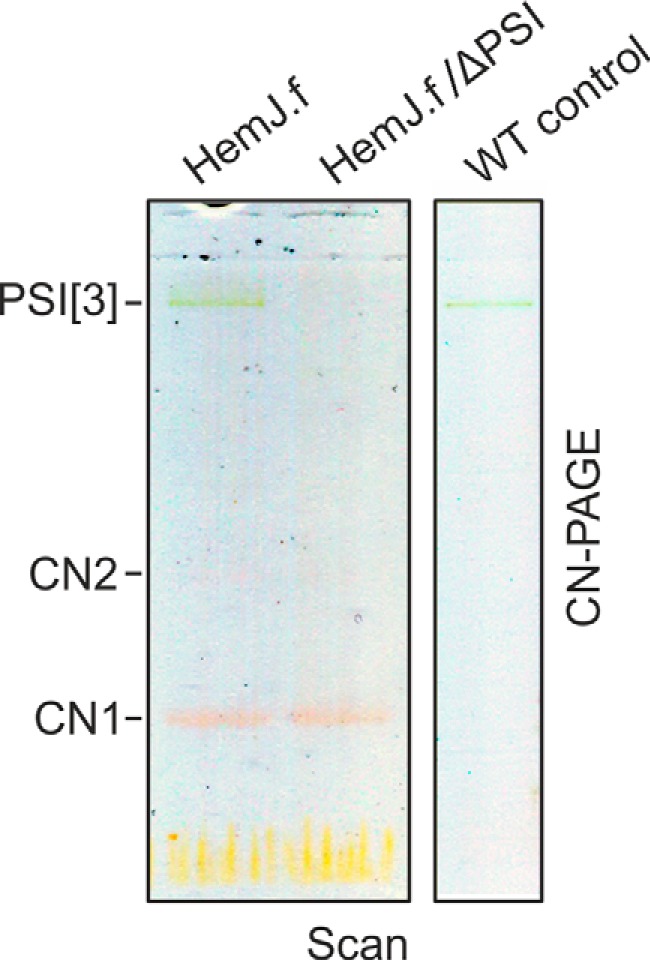
Separation of the purified HemJ.f by clear native gel electrophoresis. Native isolations of HemJ.f from WT and ΔPSI backgrounds that resulted in a reddish eluate were further separated by 4–14% clear native gel electrophoresis (CN–PAGE). Reddish bands CN1 and CN2 (poorly visible) were identified as HemJ.f. As already reported (18), a small amount of trimeric PSI (PSI[3]) is a typical contamination of FLAG eluates obtained from Synechocystis 6803. The gel was scanned in transmittance mode (Scan) using an LAS 4000 Imager (Fuji).
Figure 3.
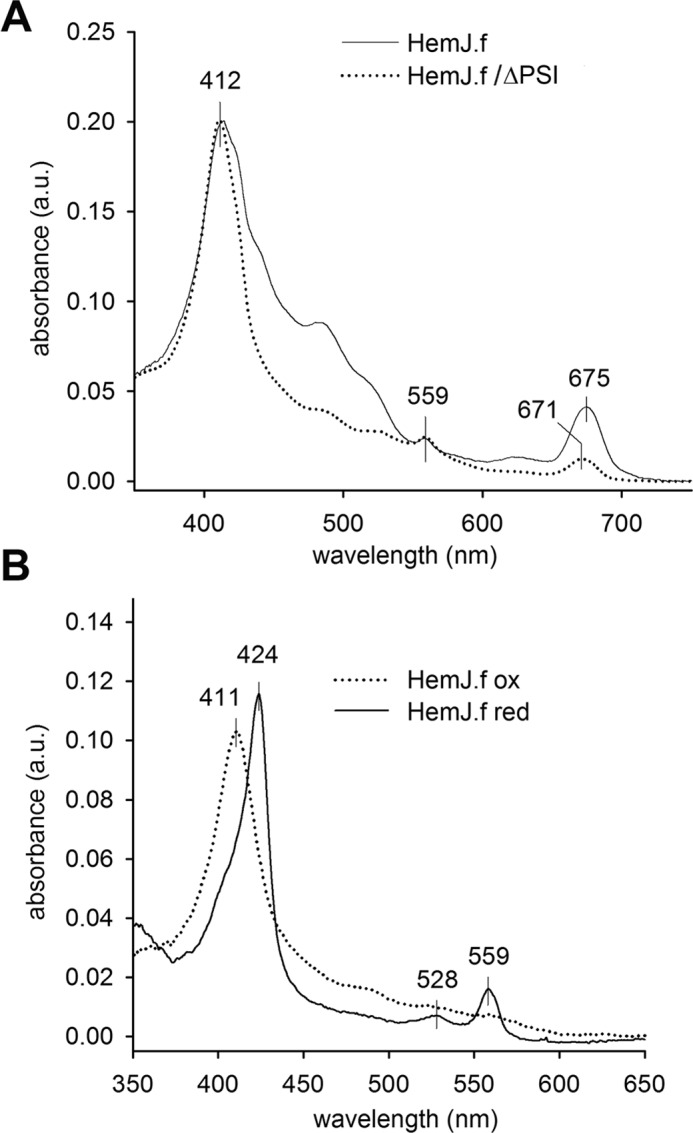
Spectroscopy analysis of the purified HemJ.f. A, absorption spectra of HemJ.f eluate. The peak at 675 nm derives from chlorophyll, whereas the peaks at 412 and 559 nm are characteristic for heme. The eluate with lower carotenoid content (absorption at 450–520 nm) was used for spectroscopy analysis. B, absorption spectra of oxidized and reduced eluate obtained from the ΔPSI genetic background. a.u., absorbance units.
Figure 4.
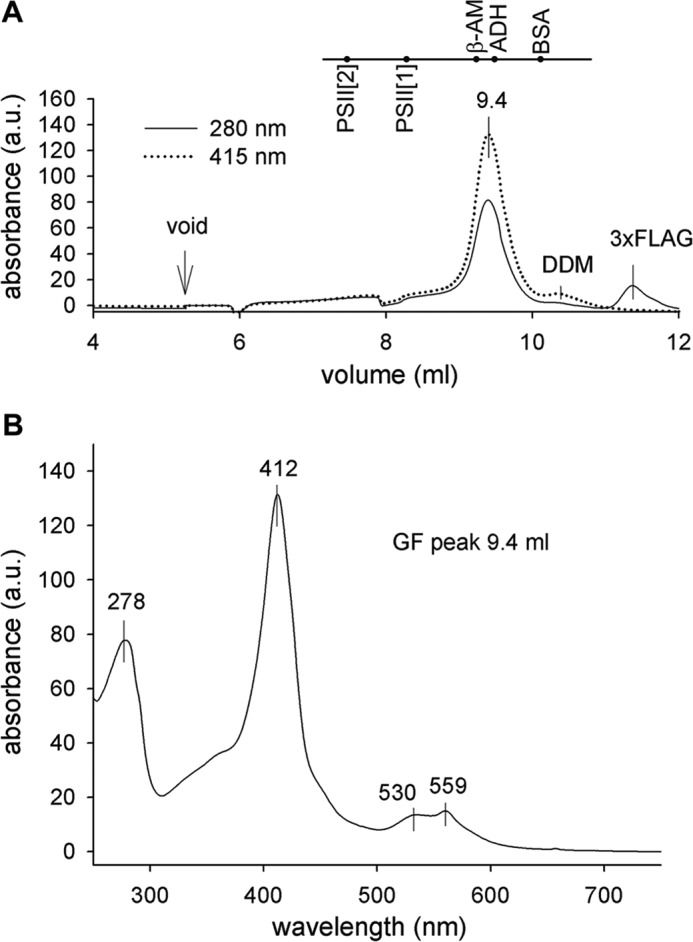
Separation of the purified HemJ.f protein by size-exclusion chromatography. A, the HemJ.f pulldown obtained from the ΔPSI genetic background was loaded on a size-exclusion chromatography column, and eluted proteins/complexes were detected by absorbance 280 and 415 nm. Positions of standards are shown at the top of the graph: PSII[2], PSII dimer (600 kDa); PSII[1], PSII monomer (300 kDa); β-AM, β-amylase (200 kDa); ADH, alcohol dehydrogenase (150 kDa); BSA, BSA (66 kDa); DDM, micelle of dodecyl-β-maltoside; 3xFLAG, 3× FLAG peptide used to elute HemJ.f from the anti-FLAG-M2 agarose resin. B, absorption spectrum of 9.4 ml size-exclusion chromatography fraction was recorded by a HPLC diode-array detector.
Figure 5.
One-dimensional SDS–PAGE and two-dimensional CN/SDS–PAGE separation of HemJ.f eluate. A, proteins isolated by affinity chromatography from HemJ.f strain and from WT control cells were separated by 12 to 20% SDS–PAGE and stained with CBB, and the individual proteins bands were identified by MS (Table S1). B, the gel strip from CN–PAGE (see Fig. 1) was further separated in a second dimension by 12–20% SDS–PAGE and stained with SYPRO Orange. HemJ.f bands (marked as CN1 and CN2 in Fig. 1) were tentatively assigned as dimeric (HemJ[2]) and tetrameric (HemJ[4]) HemJ.f oligomers, respectively.
In vitro PPO activity of the purified HemJ.f
Because previous studies provided conflicting results regarding the ability of the isolated HemJ to perform Protogen oxidation/dehydrogenation, we attempted to measure PPO activity of the eluted HemJ.f. However, no measurable PPO activity in the eluate was detected with or without artificial electron acceptors menadione or benzoquinone. Marginal activity comparable with the spontaneous oxidation of Protogen was detected in Synechocystis 6803 crude extracts and in the membrane fraction, although this activity was probably an artifact, because it was present also in these fractions after heat treatment (90 °C, 3 min). Recombinant human HemY (19) was used as a positive control for the PPO activity measurement.
The HemJ is not fully replaceable by HemG
Because we were not able to detect HemJ activity, we decided to confirm the function of HemJ via complementation with the PPO analog HemG. Two strains were engineered: one with the Synechocystis 6803 hemJ gene placed under a copper-regulated promoter (petJ) and the second expressing hemG from E. coli under the psbAII promoter. In both strains it was possible to delete the WT hemJ gene, indicating that both constructs were functional. In the P(petJ)::hemJ strain, it was possible to decrease the amount of hemJ by adding copper to the growth medium. This led to lower amounts of enzymatically produced Proto, followed by decreases in the levels of phycobilins and chlorophyll and lower levels of major chlorophyll-binding photosynthetic complexes, especially trimeric PSI (Fig. S4, A and B). To identify how the repression of hemJ expression affects tetrapyrrole biosynthesis, the accumulation of chlorophyll/heme intermediates was analyzed in the P(petJ)::hemJ strain suppressed by copper (Fig. 6). The suppressed strain exhibited significantly decreased levels of the chlorophyll precursors magnesium–protoporphyrin IX and monovinyl protochlorophyllide along with an accumulation of coproporphyrin III (Copro) and Proto; the latter tetrapyrrole was also visibly excreted into the medium. Detection of both Copro and Proto was likely to result from the accumulation and nonenzymatic oxidation of their reduced precursors in the cell (3, 20).
Figure 6.
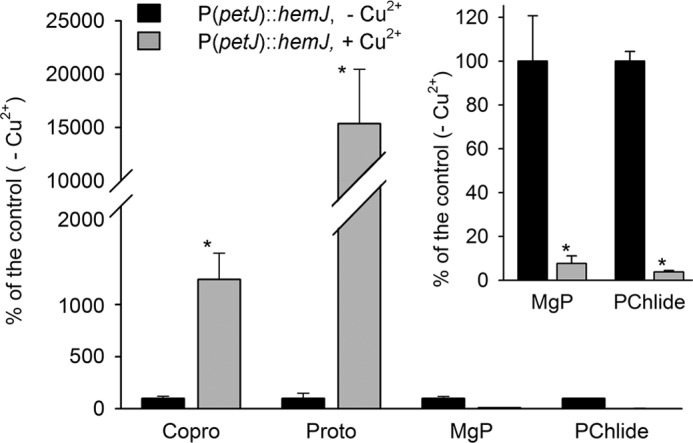
Analysis of heme/chlorophyll precursors in the P(petJ)::hemJ strain grown photoautotrophically in medium with or without copper. Heme/chlorophyll precursors were extracted with 70% methanol from the P(petJ)::hemJ cells at OD730 = 0.3–0.4 and separated on a HPLC equipped with two fluorescence detectors (48). The amounts of later chlorophyll precursors magnesium-protoporphyrin IX (MgP) and monovinyl protochlorophyllide (PChlide) were markedly reduced in cells cultivated with the repressed hemJ expression when compared with the same mutant cells grown without copper. On the contrary, Proto and Copro massively accumulated in the repressed cells. The inset shows a different scale for the less abundant precursors. *, significance difference tested using a paired t test (p = 0.05).
The HemG complementation strain ΔhemJ/hemG grew on glucose slightly slower than WT (Fig. S5) and accumulated lower amounts of chlorophyll and phycobilins (Fig. S4C). However, the strain did not grow autotrophically (Fig. S5). When the ΔhemJ/hemG cells were transferred to glucose-free medium, the cellular level of chlorophyll-binding photosynthetic complexes gradually decreased (Fig. S4D). Also chlorophyll precursors were almost undetectable in the cells, except for monovinyl chlorophyllide (Fig. 7), most probably originating from chlorophyll a recycling from pigment–protein complexes by its dephytylation (21). In addition, a large quantity of an unidentified tetrapyrrole with absorption spectrum resembling Copro eluted at 13.4 min on the HPLC profile of the extract from the ΔhemJ/hemG strain incubated without glucose (Fig. 7). This compound was completely missing in the WT strain (Fig. 7A), as well as in the ΔhemJ/hemG mutant grown on glucose (Fig. S6).
Figure 7.
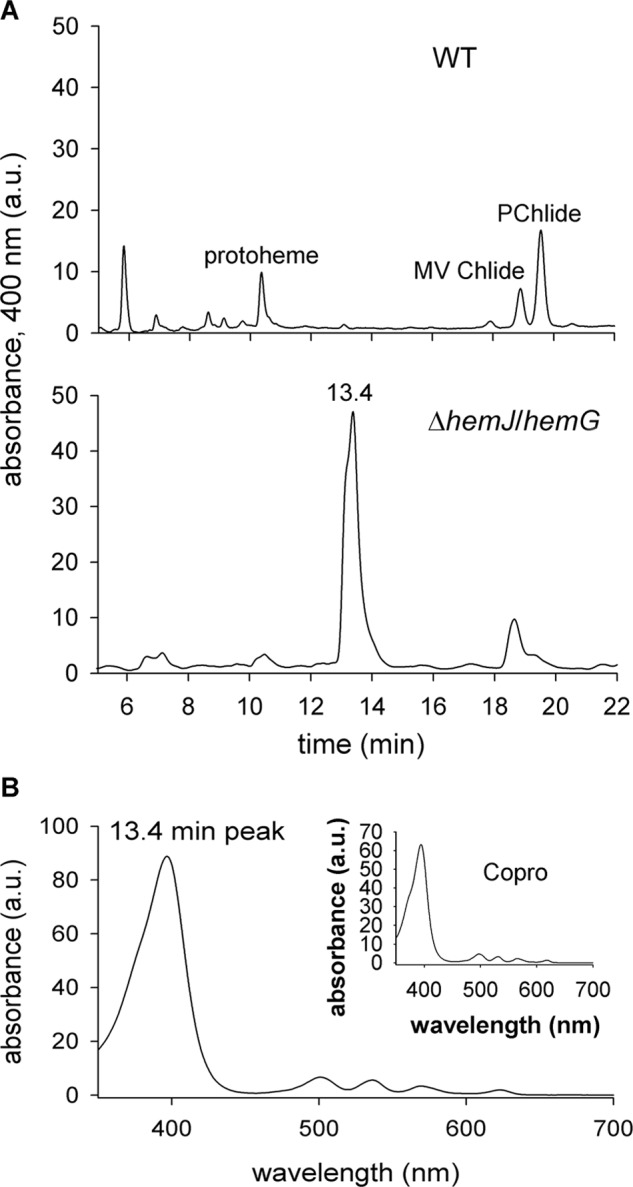
Detection of an unusual tetrapyrrole in the ΔhemJ/hemG strain incubated without glucose. A, polar tetrapyrroles were extracted with 70% methanol from 50 ml of WT and ΔhemJ/hemG cells grown photoautotrophically for 3 days and harvested at OD730 = 0. 3–0.4. The obtained extract was separated on a HPLC (see “Experimental procedures”), and eluted pigments were detected by a diode-array detector at 400 nm. MV Chlide, monovinyl chlorophyllide; Pchlide, monovinyl protochlorophyllide. B, absorption spectrum of the compound eluting at 13.4 min. The inset shows absorption spectrum of the Copro standard.
Harderoporphyrin accumulation in the ΔhemJ/hemG strain
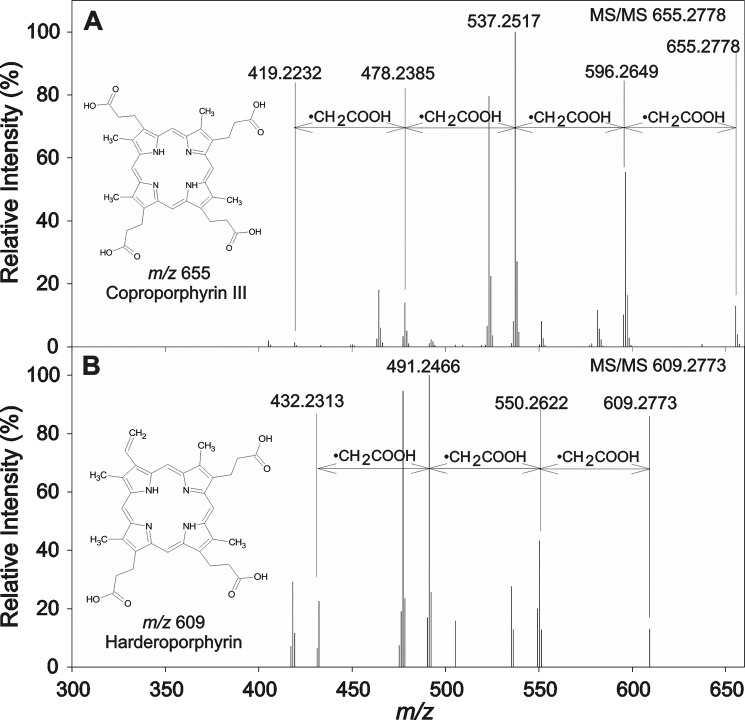
The unknown tetrapyrrole observed in ΔhemJ/hemG mutant was isolated and analyzed using HPLC coupled to high resolution tandem MS (HPLC–HRMS/MS). The compound exhibited a m/z value of 609.2708 corresponding to the elemental composition C35H37N4O6. Based on the similarity of this elemental composition to Copro (C36H39N4O8), the fragmentation spectra of these two molecules were compared (Fig. 8A and Table S2). Fragmentation of the Copro molecular ion led to successive benzylic cleavages with the loss of a •CH2COOH radical (Δ 59 Da) from all four propionic acid residues (m/z 655 → 596 → 537 → 478 → 419) as described previously (22). The queried compound exhibited only three consecutive losses of •CH2COOH radicals, suggesting the presence of only three propionic groups. From the difference in the molecular weight of both compounds, it can be assumed that the fourth propionic acid group of the tetrapyrrole ring is substituted by a vinyl group. Such substitution is consistent with that described for harderoporphyrin (23), although the exact position of the vinyl group on the tetrapyrrole ring cannot be determined via MS/MS experiments. Hence, the compound accumulating in the ΔhemJ/hemG strain with a molecular weight of 609 is likely to be harderoporphyrin, a spontaneously oxidized intermediate of the CPO reaction, which is normally generating Protogen.
Figure 8.
HRMS/MS fragmentation of the Copro (A) and harderoporphyrin (B). A, analytical standard of Copro was subjected to HPLC–HRMS/MS analysis to provide background data for identification of the unknown tetrapyrrole found in ΔhemJ/hemG cells. It provided molecular ion at m/z 655 when subjected to HPLC–HRMS/MS, and its subsequent fragmentation led to formation of ions corresponding to four consecutive losses of propionic radical. B, harderoporphyrin was isolated from ΔhemJ/hemG cells grown without glucose. The HPLC–HRMS/MS analysis provided molecular ion at m/z 609 with only three consecutive losses detected in its MS/MS spectrum. The fourth propionic group is substituted by a vinyl group as inferred from differences between Copro and harderoporphyrin m/z values.
Discussion
We have isolated tagged HemJ protein from solubilized Synechocystis 6803 membranes and found that it forms an oligomeric, most likely tetrameric complex (Figs. 4A and 5B). Because of the hydrophobic nature of this enzyme, it is unclear whether the tetramer is the native form or results from nonphysiological aggregation. However, the HemJ dimer is very stable even after SDS electrophoresis (Fig. 5B). Distantly related HemJ homologs with known three-dimensional structure (see below) have previously been shown to form dimers (24, 25).
The natively isolated HemJ complex was reddish in color (Fig. 2) because of the presence of heme b (Fig. 3). Interestingly, Gomelsky and Kaplan (26) previously overexpressed a Rhodobacter sphaeroides hemJ gene (orf1) located upstream of their gene of interest and noted that the E. coli strain overproducing this protein turned pink. Our rough estimate of heme stoichiometry indicated a ratio of 0.85 heme b per subunit. It seems plausible that HemJ binds a single heme b with partial heme loss occurring during isolation. Even though HemJ.f eluate contained also chlorophyll and variable amounts of carotenoids (Fig. 3A), neither was detected in HemJ.f fraction from size-exclusion chromatography (Fig. 4B); thus we expect these pigments to be bound unspecifically.
To predict the localization of the observed heme b within HemJ, we have modeled the three-dimensional structure of HemJ protein (Fig. 9). The modeling was performed on the HemJ peptide from R. sphaeroides (WP_023003745), which contains only four helices, as is found for most of the proteins of the HemJ family. The Synechocystis 6803 fifth helix, which is present only in cyanobacteria, did not provide enough coverage for co-evolutionary modeling, and also the overall homology in this helix was much lower (see supplemental information for details). The HemJ protein forms a very common structural motif in membrane proteins, a four–α-helix bundle. The overlay of the predicted structures from R. sphaeroides and Synechocystis 6803 showed very good agreement within this four–α-helix bundle (Fig. S8E), indicating that this domain is sufficient for basic HemJ function. Advanced alignment and secondary structure prediction methods (27) revealed distant homology of HemJ to other heme-binding membrane redox proteins. In several of our models, the invariant His-16 of HemJ (His-12 in Fig. S7) was aligned with heme-binding His residues of such distantly related templates. Because His is the most common residue providing at least one of the usual two heme ligands (28) and because His-16 is the only invariant His in the PF03653 Pfam family, we believe that His-16 of HemJ also ligates heme b. This would bury the observed heme b in the middle of the membrane, oriented perpendicularly to the membrane surface (Fig. 9). Please note that we are using the term “remote homolog” for HemJ without showing that it has common ancestry with other heme-binding redox membrane proteins. Observed structural similarity and one similarly placed His residue can simply be the result of convergent evolution of a heme-binding membrane protein. Absorbance spectra of reduced HemJ (Fig. 3B) indicate the presence of a ferrous six-coordinate heme b with the second ligand probably provided by another amino acid side chain. Interestingly, from five amino acids (His, Met, Cys, Tyr, and Lys) that can act as axial heme ligands of hemoproteins (28), there is an invariant Lys-94 (Lys-91 in Fig. S7) located on the distal side of the proposed heme location. This Lys could provide the sixth ligand of HemJ. Moreover, conserved Trp-90 (Trp-87 in Fig. S7), rarely substituted by Phe in other members of the Pfam family, may stabilize the heme pocket, as aromatic residues (phenylalanine, tyrosine, and tryptophan) play important roles in protein–heme interactions through stacking interactions with the porphyrin (28). The distant homology to several membrane oxidases together with heme absorption spectra indicate that the heme b in HemJ is redox-active and is involved in electron transfer.
Figure 9.
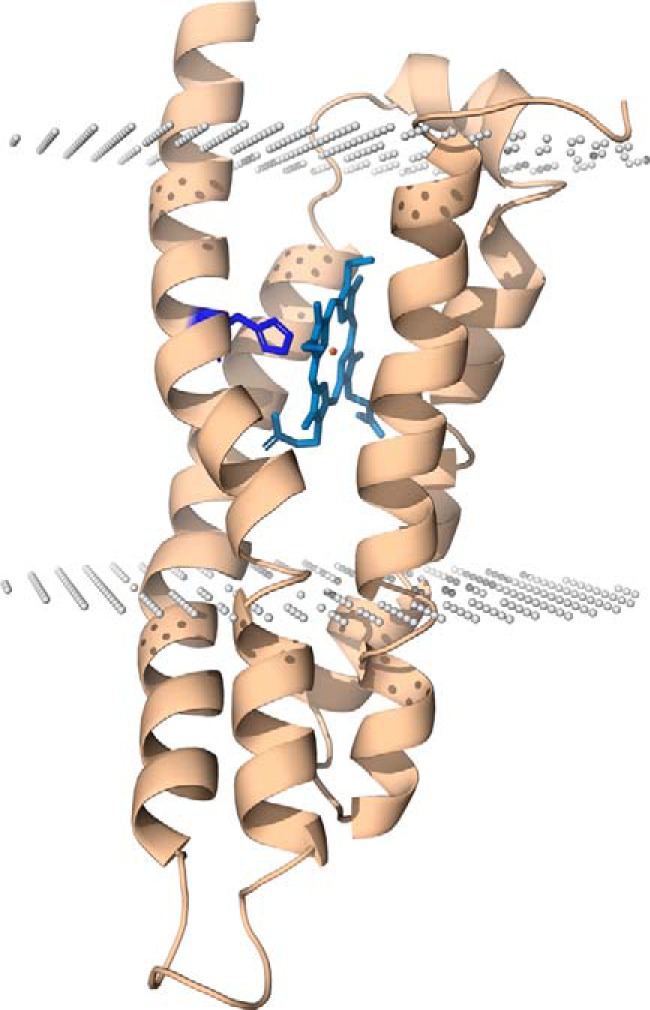
Model structure of R. sphaeroides HemJ monomer. The HemJ model was predicted by automatic structure prediction server RaptorX (53). The His-16 proposed to bind heme is shown in blue. For prediction of heme-binding site, the server COFACTOR was used (55). PPM server (56) was used to predict position of the protein structure model within membrane. Both the N and C termini of the protein are on the periplasmic/lumenal side of the membrane.
This is in agreement with the expected oxidase/dehydrogenase activity of HemJ, which is still poorly understood. Before the enzyme identity was known, PPO activity belonging to HemJ was characterized in the membrane fraction of R. sphaeroides (29). This activity was inhibited by respiratory inhibitors and also by extraction of quinones from membranes with pentane, and no enzyme activity was detected after membrane solubilization (29). This indicated that PPO activity in R. sphaeroides is closely linked with components of the respiratory electron transport chain. In A. baylyi, both membrane and soluble fractions were necessary to detect HemJ activity, suggesting that a soluble factor may be required. In another study, authors documented oxygen-dependent PPO activity of purified HemJ from R. sphaeroides (3). However, detected specific PPO activity was much lower than that for the combined fractions in A. baylyi mentioned above (10), and the authors also did not note the presence of a colored co-factor in the purified enzyme. We were not able to detect any oxygen-dependent PPO activity of the HemJ protein isolated from solubilized membranes. No activity was detected even in the presence of menadione, a soluble analog of menaquinone probably serving as the electron acceptor for HemG (9). This is in line with the report that HemJ does not complement an E. coli ΔhemG mutant, suggesting that the connection of HemJ to the respiratory chain is distinct from that of HemG (8).
Even though we were not able to measure PPO activity in our eluate, the data presented herein (Fig. 6 and Fig. S4, A and B) further document that HemJ is indeed a bona fide PPO as indicated previously (3, 10). As would be expected for PPO, HemJ down-regulation resulted in accumulation of tetrapyrrole precursors Copro and Proto at the same time as depletion of chlorophyll precursors occurred (Fig. 6). The HemJ protein is indispensable, and complete inactivation of the encoding gene was possible only after expression of another copy of HemJ or HemG. Although we were able to complement missing HemJ with HemG as shown before (10), this replacement was not without consequences. The resulting ΔhemJ/hemG strain did not grow autotrophically (Fig. S5), and the tetrapyrrole biosynthetic pathway was disturbed (Fig. 7A).
Reactive protoporphyrin intermediates generally do not accumulate in strains with an undisturbed tetrapyrrole biosynthetic pathway. Mutation of genes encoding tetrapyrrole biosynthetic enzymes or treatment of cells with enzyme inhibitors frequently results in accumulation of high levels of tetrapyrrole intermediates (4), probably leading to increased formation of reactive oxygen species. PPO-deficient mutants commonly accumulate earlier heme intermediates: uroporphyrin, Copro, and also Proto (30). Accumulation of oxidized forms is caused by nonspecific oxidation of Coprogen and Protogen in the cell or during isolation (3, 20) (Fig. 1). Repression of HemJ expression in the P(petJ)::hemJ strain also led to Copro and Proto accumulation (Fig. 6). The complemented ΔhemJ/hemG strain expressing HemG accumulated less Copro and Proto when grown on glucose compared with the WT (not shown), indicating that the pathway is functional and provides enough Proto for normal chlorophyll accumulation and almost normal growth (Figs. S4, C and D, and S5). After transfer to medium without glucose, the cells of the ΔhemJ/hemG strain stopped growing. Chlorophyll levels also quickly decreased, suggesting that not enough Proto accessible to magnesium-chelatase is made (Fig. S4C). Interestingly, at the same moment, the cells started to accumulate an unusual porphyrin identified as harderoporphyrin (Figs. 7 and 8), a probable auto-oxidation product of harderoporphyrinogen, which is a tricarboxylic intermediate in a two-step decarboxylation of the heme precursor Coprogen (31), catalyzed by CPO (Fig. 1). This indicates problems with the ΔhemJ/hemG mutant at this stage of tetrapyrrole biosynthesis. Even though there are two CPO enzymes (HemN and HemF) in Synechocystis 6803, HemF is expected to serve as the sole CPO under prevailing oxic conditions (15). We speculate that HemF is functionally coupled with HemJ and that this coupling is disturbed in the ΔhemJ/hemG strain. Surprisingly, the improper coupling was not demonstrated by accumulation of Protogen (Proto) but by accumulation of harderoporphyrinogen. This indicates a reduced ability of Synechocystis 6803 HemF to convert harderoporphyrinogen to Protogen after replacement of downstream PPO of HemJ-type by that of the HemG type. The only other examples of harderoporphyrin accumulation are for specific mutants of human CPO, which is an oxygen-dependent HemF (32). The reduced ability to convert harderoporphyrinogen to Protogen in the abnormal CPO was explained by reduced affinity for harderoporphyrinogen, which may leave the enzyme more easily (33). This would suggest that replacement of HemJ by HemG also destabilizes the active site of HemF, implying that a physical contact between HemJ and HemF leads to conformational changes of the HemF active site. Because both Coprogen and harderoporphyrinogen are substrates for CPO, the release of harderoporphyrinogen could also occur when an excess of Coprogen is accumulated in the cell (34). However, Coprogen also accumulated in the P(petJ)::hemJ strain (Fig. 6) without concomitant accumulation of harderoporphyrin. We do not know exactly why we see harderoporphyrin accumulation particularly under autotrophic growth. It is possible that glucose, similarly to oxygen, regulates the accumulation of both CPO forms HemF and HemN. The red microalga Galdieria partita, also containing both forms of CPO, excretes Copro in the medium when growing mixotrophically on glucose (35). This was explained by inactivation of the oxygen-dependent CPO in microoxic conditions when grown on glucose (36). In Synechocystis 6803, the two-component regulatory histidine kinase Hik31 is involved in the response to glucose and in switching between photoautotrophic and photoheterotrophic growth (37). At the same time, Hik31 has an additional role in the transition between oxic and microoxic growth (38).
Substrate channeling and the presence of stable or transient multienzyme complexes is expected to be an advantageous mechanism for pathways with reactive intermediates to protect the cell from oxidative damage. In tetrapyrrole biosynthesis, most information supporting the presence of multienzyme complexes for at least some enzymes comes from immunoprecipitation followed by immunoblotting and/or MS (12, 13). In developing erythroid cells, the presence of a mitochondrial heme metabolism complex minimally consisting of FeCH, PPO, and aminolevulinic acid synthase was documented (12). In bacteria, co-immunoprecipitation and immunogold labeling supported a complex involving HemY type of PPO and FeCH in the thermophilic cyanobacterium Thermosynechococcus elongatus (13).
In pulldown experiments we aimed to co-purify HemJ with its partners in tetrapyrrole biosynthesis. The most prominent protein co-purified with HemJ was Sll1106 (Fig. 5A), a protein with unknown function containing an extended glycine zipper motif, which can be found in a number of channel forming proteins (39). However, it is unlikely that Sll1106 is required for PPO function, because in a set of 40 cyanobacterial genomes, HemJ was coded in 32 genomes, whereas Sll1106 homologs were present in only 20 (data not shown). A Sll1106 homolog was found also in some cyanobacteria coding for HemY-type of PPO. Thus, we do not expect Sll1106 to be directly involved in HemJ activity, and its function remains to be elucidated. Interestingly, all four FtsH protease homologs encoded in Synechocystis 6803 were present in significant quantities in the HemJ.f eluate (Fig. 5A). FtsHs are universally conserved transmembrane metalloproteases responsible for quality control of membrane and membrane-associated proteins. They form heterooligomeric complexes with distinct functions (40–42). For example, the FtsH2/3 heterooligomeric complex responsible for PSII repair and biogenesis was the most abundant in the eluate. This may reflect a functional connection to PSII or may just reflect the membrane region in the cell where the PPO reaction is performed.
Experimental procedures
Construction of Synechocystis 6803 strains
The strains used in this study were derived from nonmotile, glucose-tolerant Synechocystis 6803 strain obtained from the laboratory of Peter J. Nixon (Imperial College, London, UK). To prepare a strain expressing HemJ (WP_020861394.1) with a 3× FLAG tag at the C terminus (HemJ.f), the hemJ gene (slr1790) was cloned into the pPD-CFLAG plasmid containing the Synechocystis 6803 psbAII promoter, a sequence encoding the tag, kanamycin resistance cassette, and flanking sequences for homologous recombination that allow insertion of hemJ tagged on its C terminus into the Synechocystis 6803 genome in place of the psbAII gene (43). Because pPD-CFLAG plasmid containing hemJ.f (pPD-hemJ.f) isolated from E. coli frequently contained frameshift mutations within hemJ, the fragment for transformation was obtained by PCR using pPD-hemJ.f ligation mixture as a template. The resultant PCR fragment was used for transformation of WT and ΔPSI strains (44). The vector used for disruption of hemJ by a chloramphenicol resistance cassette was constructed using the megaprimer PCR method (45). First, upstream and downstream regions of hemJ were amplified from WT genomic DNA with flanking sequences of chloramphenicol resistance cassette from pACYC184 vector. Then the chloramphenicol resistance cassette and regions upstream and downstream of hemJ were mixed in one PCR with the primer annealing temperature at 50 °C. 5 μl of PCR mixture was used for transformation into the hemJ.f Synechocystis 6803 strain. To construct the P(petJ)::hemJ strain expressing HemJ under the control of copper-repressed promoter, pPsbAIpetJ-FLAG plasmid (46) was used with the hemJ gene inserted into specific restriction sites, leaving out the 3× FLAG tag. The resulting plasmid was used for WT transformation, and the WT copy of hemJ was deleted afterward. To prepare Synechocystis 6803 strain expressing HemG from E. coli under the Synechocystis 6803 psbAII promoter, the hemG gene (47) was again cloned into the pPD-CFLAG plasmid, leaving out the 3× FLAG tag. The WT strain was transformed with the resultant plasmid, and the WT copy was subsequently deleted. In all cases, Synechocystis 6803 transformants were selected on BG11 agar plates with increasing levels of corresponding antibiotic. Full segregation was confirmed by PCR.
Growth conditions
Synechocystis 6803 strains were grown photoautotrophically in BG11 medium in shaken conical flasks at 29 °C and irradiance of 40 μmol photons m−2 s−1. The ΔhemJ/hemG strain was grown in the medium supplemented with 5 mm glucose. P(petJ)::hemJ strain was grown in the medium without copper, and the addition of 1 μm CuSO4 was used for repression of hemJ expression. For the purification of HemJ.f protein, 4 liters of cells were grown mixotrophically to an OD730 of 0.5–0.7 in the medium supplemented with 5 mm glucose.
Analysis of cellular tetrapyrroles
Whole-cell absorption spectra were measured with Shimadzu UV-3000 spectrophotometer. Heme/chlorophyll precursors were measured from 2 ml of culture OD730 of ∼0.4 using HPLC according to procedure described by Pilný et al. (48). For the detection of porphyrins accumulated in the ΔhemJ/hemG strain, 50 ml of cells were grown without glucose for 3 days and harvested at an OD730 of ∼0.4. Pigments were extracted by an excess of 70% methanol and separated by a HPLC method described in Pilný et al. (48), and their absorbance was detected by a diode-array detector.
Preparation of thylakoid membranes and protein complex purification
Harvested cells were washed and resuspended in thylakoid buffer containing 25 mm MES/NaOH, pH 6.5, 25% glycerol, 10 mm MgCl2, 10 mm CaCl2 and then broken in a Mini-Beadbeater-16 (Biospec). For protein purification, EDTA-free protease inhibitor (Roche) was added into the buffer prior to breaking the cells. The membranes were pelleted by centrifugation (55,000 × g, 20 min, 4 °C) and resuspended in excess of the thylakoid buffer, and the centrifugation step was repeated.
For the isolation of HemJ.f protein, the membrane fraction was resuspended in the thylakoid buffer and solubilized for 1 h at 10 °C with 1.5% n-dodecyl-β-d-maltoside (DDM; Enzo Life Sciences). After centrifugation (55,000 × g, 20 min, 4 °C), the solubilized proteins were purified using anti-FLAG-M2 agarose resin (Sigma–Aldrich). The resin was intensively washed with 20 resin volumes of the thylakoid buffer containing 0.04% DDM, and the HemJ.f was finally eluted with two resin volumes of the same buffer containing in addition 300 μg/ml of 3× FLAG peptide (Sigma–Aldrich).
Electrophoresis and size-exclusion chromatography
The protein composition of purified complexes was analyzed by SDS–PAGE in a denaturing 12–20% polyacrylamide gel containing 7 m urea (49). For native electrophoresis, solubilized membrane proteins or isolated complexes were separated on 4–14% CN–PAGE (50). To resolve individual components of protein complexes, the gel strip from the CN–PAGE was first incubated in 2% SDS and 1% DTT for 30 min at room temperature, and then proteins were separated along the second dimension by SDS–PAGE in a denaturing 12–20% polyacrylamide gel containing 7 m urea (49). Proteins separated by SDS–PAGE were stained with Coomassie Brilliant Blue (CBB) or SYPRO Orange afterward. Mass spectrometry analysis of CBB-stained protein bands/spots from the SDS–PAGE gels was accomplished essentially as described by Bučinská et al. (18).
For size-exclusion chromatography, the HemJ.f eluate prepared from ΔPSI genetic background was injected onto an Agilent-1200 HPLC and separated on a Yarra 3000 column (Phenomenex) using 25 mm HEPES buffer, pH 7.5, containing 0.25% DDM at a flow rate of 0.2 ml min−1 at 10 °C. Fractions corresponding to HemJ.f were pooled and concentrated on a 100-kDa cutoff microconcentrator (Millipore).
Identification and quantification of the HemJ.f-bound heme
For the analysis of heme, the HemJ.f protein was isolated by the affinity chromatography and subsequently further purified by size-exclusion chromatography (see previous paragraph). Heme was extracted from the isolated and concentrated protein by 90% of acetone, 2% HCl; passed through a 0.22-μm filter; and separated by HPLC on Nova-Pak C18 column (Waters) using a 25–100% linear gradient with H2O (A)/acetonitrile (B) both containing 0.1% TFA as a mobile phase at a flow rate of 1 ml min−1 at 40 °C. Protoheme was detected using a diode-array detector (Agilent-1200). The protein concentration was estimated in two fractions from the size-exclusion chromatography by sample absorbance at 280 nm using the ProtParam tool (http://web.expasy.org/protparam) and a calculated extinction coefficient for the HemJ.f protein of 41,000 m−1 cm−1. Full reduction of heme in HemJ.f eluate from the ΔPSI strain was achieved by adding a few grains of sodium dithionite. UV-visible spectra were measured at room temperature with a Shimadzu UV-3000 spectrophotometer.
PPO assay
PPO activity was monitored using a continuous fluorometric assay as previously described (51). Production of the fluorescent Proto, from nonfluorescent Protogen was detected using a fluorescence plate leader at 25 °C. Reaction mixtures consisted of 50 mm MOPS, pH 8.0, 2 mm GSH, 20 μm Protogen, 200 nm HemJ.f, and 1 mm menadione or benzoquinone. Protogen oxidation by Human HemY was used as a positive control.
Isolation of harderoporphyrin and HPLC–HRMS/MS analysis
The unknown tetrapyrrole (peak, 13.4 min; see Fig. 7A) was prepurified from 600 ml of ΔhemJ/hemG cells grown without glucose for 3 days. Harvested cells were extracted by 50 ml of 70% methanol, the solvent was evaporated on a rotary evaporator, and the dried pigments were dissolved in 2 ml of methanol. This solution was separated on Agilent-1200 using the same solvents as described for the analysis of cellular tetrapyrroles but employing a semipreparative C8 column (Luna 5 μm, 250 × 10 mm, Phenomenex). The peak corresponding to the unknown tetrapyrrole was collected and dried in a vacuum concentrator. For HPLC–HRMS/MS analysis, the sample was analyzed on Thermo Scientific Dionex UltiMate 3000 UHPLC+ (Sunnyvale, CA) equipped with a diode-array detector. Separation of compounds was performed on reversed-phase Phenomenex Kinetex C18 column (150 × 4.6 mm, 2.6 μm; Torrance, CA) using H2O (A)/acetonitrile (B) both containing 0.1% HCOOH as a mobile phase with the flow rate of 0.5 ml min−1. For the separation, the following gradient was used: A/B 85/15 (0 min), 85/15 (in 1 min), 0/100 (in 25 min), 0/100 (in 30 min), and 85/15 (in 35 min). Analysis of mass spectra was performed on a Bruker Impact HD high resolution mass spectrometer (Billerica, MA) with electrospray ionization. The following settings were used: dry temperature, 200 °C; drying gas flow, 12 liters min−1; nebulizer, 3 bars; capillary voltage, 3800 V; and end-plate offset, 500. The spectra were collected in the range 20–2000 m/z with precursor ion selection set to 550–700 and automatic exclusion after five spectra. The analysis was calibrated using sodium formate at the beginning of the analysis. Collision energy for fragmentation was set to 60 eV.
Protein modeling
For modeling we have used HemJ polypeptide from R. sphaeroides (WP_023003745) with four transmembrane helices. The modeling was performed using several automatic structure prediction servers: Robetta (52), RaptorX (53), and RaptorX-Contact (54). The server COFACTOR was used for prediction of heme-binding site (55), and PPM server (56) was used to predict the position of the protein structure model within membrane. The protein models were aligned and visualized using PyMOL software (57).
Author contributions
P. S., R. S., and M. T. conceptualization; P. S., R. S., P. H., and M. T. formal analysis; P. S. validation; P. S., J. H., and M. T. investigation; P. S., R. S., J. H., P. H., and M. T. visualization; P. S., R. S., M. S., and M. T. methodology; P. S. and M. T. writing-original draft; P. S., R. S., M. S., P. H., and M. T. writing-review and editing; M. T. supervision; M. T. project administration.
Supplementary Material
Acknowledgments
We are grateful to the laboratory of Prof. Harry Dailey (University of Georgia) for supplying us with expression plasmid for HemY. We thank Eva Prachová and Jan Pilný for technical assistance and Peter Koník for performing the mass spectrometric analyses.
This work was supported by Project P501/12/G055 of the Czech Science Foundation and by Czech Ministry of Education Projects LO1416 and LM2015055. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains text, references, Tables S1 and S2, and Figs. S1–S8.
- Protogen
- protoporphyrinogen IX
- Proto
- protoporphyrin IX
- PPO
- protoporphyrinogen IX oxidase
- CPO
- coproporphyrinogen III oxidase
- Coprogen
- coproporphyrinogen III
- CN
- clear native
- PSI
- Photosystem I
- PSII
- Photosystem II
- Copro
- coproporphyrin III
- DDM
- n-dodecyl-β-d-maltoside
- CBB
- Coomassie Brilliant Blue
- FeCH
- ferrochelatase.
References
- 1. Czarnecki O., and Grimm B. (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63, 1675–1687 10.1093/jxb/err437 [DOI] [PubMed] [Google Scholar]
- 2. Dailey H. A., Dailey T. A., Gerdes S., Jahn D., Jahn M., O'Brian M. R., and Warren M. J. (2017) Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 81, e00048–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kato K., Tanaka R., Sano S., Tanaka A., and Hosaka H. (2010) Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 107, 16649–16654 10.1073/pnas.1000771107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishimura K., Nakayashiki T., and Inokuchi H. (1995) Cloning and identification of the hemG gene encoding protoporphyrinogen oxidase (PPO) of Escherichia coli K-12. DNA Res. 2, 1–8 10.1093/dnares/2.1.1 [DOI] [PubMed] [Google Scholar]
- 5. Narita S., Tanaka R., Ito T., Okada K., Taketani S., and Inokuchi H. (1996) Molecular cloning and characterization of a cDNA that encodes protoporphyrinogen oxidase of Arabidopsis thaliana. Gene 182, 169–175 10.1016/S0378-1119(96)00545-8 [DOI] [PubMed] [Google Scholar]
- 6. Dailey H. A., and Dailey T. A. (1996) Protoporphyrinogen oxidase of Myxococcus xanthus: expression, purification, and characterization of the cloned enzyme. J. Biol. Chem. 271, 8714–8718 10.1074/jbc.271.15.8714 [DOI] [PubMed] [Google Scholar]
- 7. Kobayashi K., Masuda T., Tajima N., Wada H., and Sato N. (2014) Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of protoporphyrinogen IX. Genome Biol. Evol. 6, 2141–2155 10.1093/gbe/evu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Möbius K., Arias-Cartin R., Breckau D., Hännig A. L., Riedmann K., Biedendieck R., Schröder S., Becher D., Magalon A., Moser J., Jahn M., and Jahn D. (2010) Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. U.S.A. 107, 10436–10441 10.1073/pnas.1000956107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boynton T. O., Daugherty L. E., Dailey T. A., and Dailey H. A. (2009) Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48, 6705–6711 10.1021/bi900850y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boynton T. O., Gerdes S., Craven S. H., Neidle E. L., Phillips J. D., and Dailey H. A. (2011) Discovery of a gene involved in a third bacterial protoporphyrinogen oxidase activity through comparative genomic analysis and functional complementation. Appl. Environ. Microbiol. 77, 4795–4801 10.1128/AEM.00171-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferreira G. C., Andrew T. L., Karr S. W., and Dailey H. A. (1988) Organization of the terminal two enzymes of the heme biosynthetic pathway: orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J. Biol. Chem. 263, 3835–3839 [PubMed] [Google Scholar]
- 12. Medlock A. E., Shiferaw M. T., Marcero J. R., Vashisht A. A., Wohlschlegel J. A., Phillips J. D., and Dailey H. A. (2015) Identification of the mitochondrial heme metabolism complex. PLoS One 10, e0135896 10.1371/journal.pone.0135896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masoumi A., Heinemann I. U., Rohde M., Koch M., Jahn M., and Jahn D. (2008) Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus. Microbiology 154, 3707–3714 10.1099/mic.0.2008/018705-0 [DOI] [PubMed] [Google Scholar]
- 14. Brzezowski P., Richter A. S., and Grimm B. (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985 10.1016/j.bbabio.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 15. Goto T., Aoki R., Minamizaki K., and Fujita Y. (2010) Functional differentiation of two analogous coproporphyrinogen III oxidases for heme and chlorophyll biosynthesis pathways in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 51, 650–663 10.1093/pcp/pcq023 [DOI] [PubMed] [Google Scholar]
- 16. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., and Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 10.1093/nar/29.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phillips J. D., Whitby F. G., Warby C. A., Labbe P., Yang C., Pflugrath J. W., Ferrara J. D., Robinson H., Kushner J. P., and Hill C. P. (2004) Crystal structure of the oxygen-depandant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. J. Biol. Chem. 279, 38960–38968 10.1074/jbc.M406050200 [DOI] [PubMed] [Google Scholar]
- 18. Bučinská L., Kiss E., Koník P., Knoppová J., Komenda J., and Sobotka R. (2018) The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of Photosystem II. Plant Physiol. 176, 2931–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hobbs C., Dailey H. A., and Shepherd M. (2016) The HemQ coprohaem decarboxylase generates reactive oxygen species: implications for the evolution of classical haem biosynthesis. Biochem. J. 473, 3997–4009 10.1042/BCJ20160696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lermontova I., and Grimm B. (2006) Reduced activity of plastid protoporphyrinogen oxidase causes attenuated photodynamic damage during high-light compared to low-light exposure. Plant J. 48, 499–510 10.1111/j.1365-313X.2006.02894.x [DOI] [PubMed] [Google Scholar]
- 21. Kopečná J., Pilný J., Krynická V., Tomčala A., Kis M., Gombos Z., Komenda J., and Sobotka R. (2015) Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in the cyanobacterium Synechocystis 6803. Plant Physiol. 169, 1307–1317 10.1104/pp.15.01150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim C. K. (2010) High-performance Liquid Chromatography and Mass Spectrometry of Porphyrins, Chlorophylls and Bilins, World Scientific Publishing, Singapore [Google Scholar]
- 23. Kennedy G. Y. (1970) Harderoporphyrin: a new porphyrin from the Harderian glands of the rat. Comp. Biochem. Physiol. 36, 21–36 10.1016/0010-406X(70)90647-X [DOI] [PubMed] [Google Scholar]
- 24. Miller M. J., Hermodson M., and Gennis R. B. (1988) The active form of the cytochrome d terminal oxidase complex of Escherichia coli is a heterodimer containing one copy of each of the two subunits. J. Biol. Chem. 263, 5235–5240 [PubMed] [Google Scholar]
- 25. Lu P., Ma D., Yan C., Gong X., Du M., and Shi Y. (2014) Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 111, 1813–1818 10.1073/pnas.1323931111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomelsky M., and Kaplan S. (1996) The Rhodobacter sphaeroides 2.4.1 rho gene: expression and genetic analysis of structure and function. J. Bacteriol. 178, 1946–1954 10.1128/jb.178.7.1946-1954.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma J., Peng J., Wang S., and Xu J. (2012) A conditional neural fields model for protein threading. Bioinformatics 28, i59–i66 10.1093/bioinformatics/bts213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T., Bonkovsky H. L., and Guo J.-T. (2011) Structural analysis of heme proteins: implications for design and prediction. BMC Struct. Biol. 11, 13 10.1186/1472-6807-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacobs N. J., and Jacobs J. M. (1981) Protoporphyrinogen oxidation in Rhodopseudomonas spheroides, a step in heme and bacteriochlorophyll synthesis. Arch. Biochem. Biophys. 211, 305–311 10.1016/0003-9861(81)90458-6 [DOI] [PubMed] [Google Scholar]
- 30. Sasarman A., Chartrand P., Lavoie M., Tardif D., Proschek R., and Lapointe C. (1979) Mapping of a new hem gene in Escherichia coli K12. J. Gen. Microbiol. 113, 297–303 10.1099/00221287-113-2-297 [DOI] [PubMed] [Google Scholar]
- 31. Rand K., Noll C., Schiebel H. M., Kemken D., Dülcks T., Kalesse M., Heinz D. W., and Layer G. (2010) The oxygen-independent coproporphyrinogen III oxidase HemN utilizes harderoporphyrinogen as a reaction intermediate during conversion of coproporphyrinogen III to protoporphyrinogen IX. Biol. Chem. 391, 55–63 [DOI] [PubMed] [Google Scholar]
- 32. Schmitt C., Gouya L., Malonova E., Lamoril J., Camadro J. M., Flamme M., Rose C., Lyoumi S., Da Silva V., Boileau C., Grandchamp B., Beaumont C., Deybach J. C., and Puy H. (2005) Mutations in human CPO gene predict clinical expression of either hepatic hereditary coproporphyria or erythropoietic harderoporphyria. Hum. Mol. Genet. 14, 3089–3098 10.1093/hmg/ddi342 [DOI] [PubMed] [Google Scholar]
- 33. Nordmann Y., Grandchamp B., de Verneuil H., Phung L., Cartigny B., and Fontaine G. (1983) Harderoporphyria: a variant hereditary coproporphyria. J. Clin. Invest. 72, 1139–1149 10.1172/JCI111039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elder G. H., Evans J. O., Jackson J. R., and Jackson A. H. (1978) Factors determining the sequence of oxidative decarboxylation of the 2- and 4-propionate substituents of coproporphyrinogen III by coproporphyrinogen oxidase in rat liver. Biochem. J. 169, 215–223 10.1042/bj1690215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stadnichuk I., Rakhimberdieva M. G., Bolychevtseva Y. V., Yurina N. P., Karapetyan N. V., and Selyakh I. O. (1998) Inhibition by glucose of chlorophyll a and phycocyanobilin biosynthesis in the unicellular red alga Galdieria partita at the stage of coproporphyrinogen III formation. Plant Sci. 136, 11–23 10.1016/S0168-9452(98)00088-0 [DOI] [Google Scholar]
- 36. Sarian F. D., Rahman D. Y., Schepers O., and van der Maarel M. (2016) Effects of oxygen limitation on the biosynthesis of photo pigments in the red microalgae Galdieria sulphuraria strain 074G. PLoS One 11, e0148358 10.1371/journal.pone.0148358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kahlon S., Beeri K., Ohkawa H., Hihara Y., Murik O., Suzuki I., Ogawa T., and Kaplan A. (2006) A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC 6803 to the presence of glucose. Microbiology 152, 647–655 10.1099/mic.0.28510-0 [DOI] [PubMed] [Google Scholar]
- 38. Summerfield T. C., Nagarajan S., and Sherman L. A. (2011) Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 157, 301–312 10.1099/mic.0.041053-0 [DOI] [PubMed] [Google Scholar]
- 39. Kim S., Jeon T. J., Oberai A., Yang D., Schmidt J. J., and Bowie J. U. (2005) Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 14278–14283 10.1073/pnas.0501234102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mann N. H., Novac N., Mullineaux C. W., Newman J., Bailey S., and Robinson C. (2000) Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479, 72–77 10.1016/S0014-5793(00)01871-8 [DOI] [PubMed] [Google Scholar]
- 41. Krynická V., Tichý M., Krafl J., Yu J., Kaňa R., Boehm M., Nixon P. J., and Komenda J. (2014) Two essential FtsH proteases control the level of the Fur repressor during iron deficiency in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 94, 609–624 10.1111/mmi.12782 [DOI] [PubMed] [Google Scholar]
- 42. Komenda J., Knoppová J., Krynická V., Nixon P. J., and Tichý M. (2010) Role of FtsH2 in the repair of Photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochim. Biophys. Acta 1797, 566–575 10.1016/j.bbabio.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 43. Chidgey J. W., Linhartová M., Komenda J., Jackson P. J., Dickman M. J., Canniffe D. P., Koník P., Pilný J., Hunter C. N., and Sobotka R. (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26, 1267–1279 10.1105/tpc.114.124495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen G., Boussiba S., and Vermaas W. F. (1993) Synechocystis sp. PCC-6803 strains lacking Photosystem-I and phycobilisome function. Plant Cell 5, 1853–1863 10.2307/3869700,10.1105/tpc.5.12.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J., Lee H. J., Shin M. K., and Ryu W. S. (2004) Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques 36, 398–400 10.2144/04363BM04 [DOI] [PubMed] [Google Scholar]
- 46. Knoppová J., Sobotka R., Tichý M., Yu J., Konik P., Halada P., Nixon P. J., and Komenda J. (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26, 1200–1212 10.1105/tpc.114.123919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sasarman A., Letowski J., Czaika G., Ramirez V., Nead M. A., Jacobs J. M., and Morais R. (1993) Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can. J. Microbiol. 39, 1155–1161 10.1139/m93-174 [DOI] [PubMed] [Google Scholar]
- 48. Pilný J., Kopečná J., Noda J., and Sobotka R. (2015) Detection and quantification of heme and chlorophyll precursors using a High Performance Liquid Chromatography (HPLC) system equipped with two fluorescence detectors. Bio-protocol 5, e1390 [Google Scholar]
- 49. Dobáková M., Sobotka R., Tichý M., and Komenda J. (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wittig I., Karas M., and Schägger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell Proteomics 6, 1215–1225 10.1074/mcp.M700076-MCP200 [DOI] [PubMed] [Google Scholar]
- 51. Shepherd M., and Dailey H. A. (2005) A continuous fluorimetric assay for protoporphyrinogen oxidase by monitoring porphyrin accumulation. Anal. Biochem. 344, 115–121 10.1016/j.ab.2005.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim D. E., Chivian D., and Baker D. (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32, W526–W531 10.1093/nar/gkh468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., and Xu J. (2012) Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang S., Sun S., Li Z., Zhang R., and Xu J. (2017) Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 10.1371/journal.pcbi.1005324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C., Freddolino P. L., and Zhang Y. (2017) COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 45, W291–W299 10.1093/nar/gkx366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., and Lomize A. L. (2012) OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 10.1093/nar/gkr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 2.0, Schroedinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.