Abstract
Triggering receptor expressed on myeloid cells 2 (TREM2) is an immune receptor expressed on the surface of microglia, macrophages, dendritic cells, and osteoclasts. The R47H TREM2 variant is a significant risk factor for late-onset Alzheimer's disease (AD), and the molecular basis of R47H TREM2 loss of function is an emerging area of TREM2 biology. Here, we report three high-resolution structures of the extracellular ligand-binding domains (ECDs) of R47H TREM2, apo-WT, and phosphatidylserine (PS)-bound WT TREM2 at 1.8, 2.2, and 2.2 Å, respectively. The structures reveal that Arg47 plays a critical role in maintaining the structural features of the complementarity-determining region 2 (CDR2) loop and the putative positive ligand–interacting surface (PLIS), stabilizing conformations capable of ligand interaction. This is exemplified in the PS-bound structure, in which the CDR2 loop and PLIS drive critical interactions with PS via surfaces that are disrupted in the variant. Together with in vitro and in vivo characterization, our structural findings elucidate the molecular mechanism underlying loss of ligand binding, putative oligomerization, and functional activity of R47H TREM2. They also help unravel how decreased in vitro and in vivo stability of TREM2 contribute to loss of function in disease.
Keywords: Alzheimer disease, neuroinflammation, X-ray crystallography, lipid-binding protein, scavenger receptor, TREM2, R47H, neurodegeneration, microglia, antibody, myeloid cell, structure, triggering receptor expressed on myeloid cells 2
Introduction
TREM2 (triggering receptor expressed on myeloid cells 2) is an immune receptor expressed on the surface of microglia, macrophages, dendritic cells, and osteoclasts. Mutations in TREM2 have been linked to various neurodegenerative diseases, including Alzheimer's disease (AD),8 frontotemporal dementia, and Nasu–Hakola disease (NHD) (1). TREM2 is reported to bind to several putative ligands, including apolipoprotein E (apoE) and apolipoprotein J (apoJ) and is activated by anionic lipids (2–5). It also has been shown to play a role in the clearance of β-amyloid plaques (5), apoptotic cells (6), myelin debris (7), and bacterial beads (8). The ability to interact with multiple endogenous and surrogate ligands in various physiological and challenge environments by potentially engaging different molecular interaction surfaces is consistent with the promiscuous nature of this class of receptors (9).
Studies on the R47H variant (linked to late-onset AD) have revealed impaired NFAT reporter gene signaling in R47H TREM2/DAP12 cell lines, subtle but consistent differences in the secondary structure of the variant protein, and small differences in glycosylation and potential trafficking compared with WT protein (5, 10). Furthermore, the variant has been shown to bind more weakly to apoE isoforms (2, 3), which was confirmed in an unbiased library screening in which several proteins, including apoE and apoJ, showed reduced binding to R47H TREM2 compared with WT (4). Finally, immunofluorescence studies on brain sections from R47H carriers with AD also suggested loss of a novel microglial barrier function (11).
At the molecular level, the complete loss of function associated with some mutations linked to NHD and frontotemporal dementia, including Q33X and Y38C, can be explained by truncation (Q33X) or misfolding of the protein due to aberrant disulfide bond formation (Y38C). The effects of other mutations like R47H, R62H, T66M, and D87N, however, appear to be more subtle. The analysis of a 3.1 Å structure of WT TREM2 ECD revealed that mutations in NHD are buried, whereas AD risk variants are found on the surface, providing valuable preliminary insight into structural features that impact TREM2 function (12, 13).
An understanding of the structural and biophysical properties of WT and variant ECD is also important for better understanding the properties of the shed or soluble form of TREM2 (sTREM2). sTREM2 is composed of the extracellular putative ligand binding domain of TREM2, where the Ig domain is followed by a short stalk terminating at His157 (14, 15). Several of the disease-associated variants (most notably T66M) present with reduced surface TREM2 expression and a concomitant reduction in sTREM2 levels (16). Recently, Song et al. (17) noted that R47H TREM2 5x FAD mice (AD genetic mouse models carrying the R47H variant) appeared to have lower levels of sTREM2 in the brain compared with CV (common variant) TREM2 5x FAD mice. However, similar transcript and surface TREM2 levels have been reported (17), and the differences could not be explained by differences in cleavage propensity by known TREM2 proteases like ADAM17. Key outstanding questions include how the R47H variant affects levels of sTREM2 in vivo; what link exists between sTREM2 levels, TREM2 receptor activity, and microglia activity; and how sTREM2 interferes with therapeutics designed to target the receptor.
To elucidate the molecular mechanism of the loss of function conferred by the R47H variant and to inform effective therapeutic strategies targeting TREM2 in neurodegenerative diseases, we have carried out structural, biochemical, functional, and biophysical studies on the ECDs of both R47H and WT TREM2. Here we report high-resolution crystal structures of R47H TREM2 and WT TREM2 at 1.8 and 2.2 Å, respectively. We also describe the co-crystal structure of TREM2 complexed with a putative ligand, phosphatidylserine (PS), at 2.2 Å. Furthermore, we describe the effect of the variant on both in vitro and in vivo stability of sTREM2. Together, these data advance our understanding of the molecular interactions underpinning the effects of the R47H TREM2 variant on the fold, stability, functional activity, and putative positive ligand–interacting surface (PLIS) impacting ligand binding.
Results
Crystal structures of R47H and WT TREM2 ECD
Human TREM2 is a polypeptide chain of 230 amino acids that consists of a single 156-amino acid N-terminal mature ECD (residues 19–174, with residues 1–18 constituting signal peptide), a membrane-spanning region (residues 175–195), and a C-terminal cytosolic tail (residues 196–230). Here, we focused our structural and biophysical efforts on the ECD of WT and R47H TREM2 proteins for the reasons discussed above. To generate soluble and homogenous proteins, we engineered various forms of WT and R47H TREM2 ECDs, including truncations and glycosylation-site mutations (Asn20 and Asn79) and also screened different expression systems (see “Experimental procedures” and Table S4). We were ultimately successful in obtaining diffracting crystals for R47H TREM2 produced by Escherichia coli expression and WT TREM2 with an N20D mutation produced by mammalian expression.
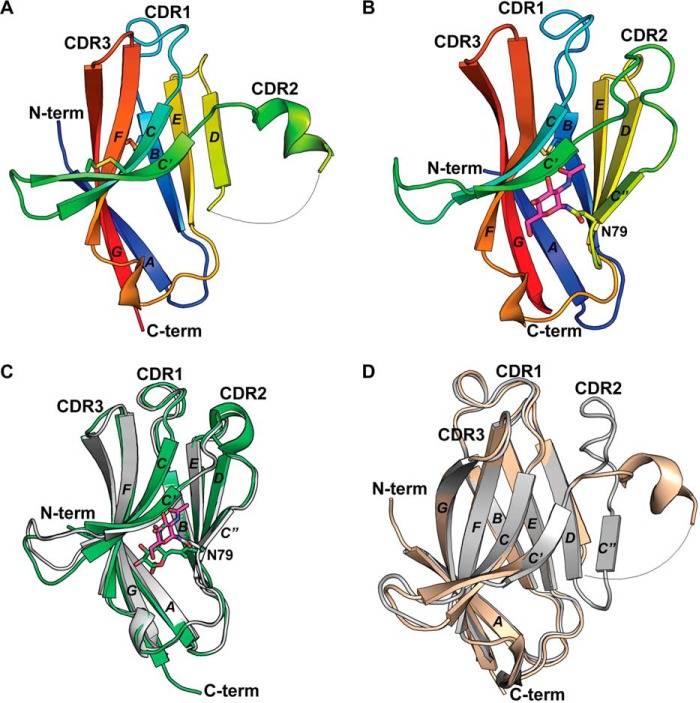
The crystal structure of R47H TREM2 (residues 19–131) was solved to 1.8 Å resolution in the P21212 space group with two molecules per asymmetric unit (asu) arranged in a dimeric “tail-to-tail” configuration (see “Experimental procedures,” Table S1, and Fig. S1A). The total buried solvent-accessible surface area (SASA) between the two molecules in the asu is 605 Å2 with a shape complementarity (Sc) score of 0.49, suggesting that the protein is unlikely to exist as a dimer in solution, consistent with size-exclusion chromatography data. Analysis of the three antibody-equivalent complementarity-determining region (CDR) loops reveals a distinct and nonstandard 2-turn helix CDR2 loop conformation between β-strands C′ and C″ (Fig. 1A).
Figure 1.
R47H TREM2 and human WT TREM2 ECD crystal structures. A, cartoon depiction of R47H TREM2 variant Ig-like V-domain (in rainbow colors) with a conserved disulfide bond between Cys36–Cys110 and Cys51–Cys60. B, cartoon depiction of WT TREM2 Ig-like V-domain (in rainbow colors) with a conserved disulfide bond between Cys36–Cys110 and Asn79-linked NAG depicted in fuchsia sticks. C, cartoon depiction of previously published human WT TREM2 Ig-like V-domain and Asn79-linked NAG at 3.1 Å (in lime green) (12) overlaid with current 2.2-Å human WT TREM2 Ig-like V-domain (in gray) and Asn79-linked NAG depicted in fuchsia sticks. Mammalian cell expression constructs for the 3.1-Å Ig-like domain encompassed amino acids 19–134; residues 20–131 were visualized for each noncrystallographic symmetry (NCS) molecule in the asu. D, cartoon depiction of R47H TREM2 variant Ig-like V-domain (in beige) overlaid with WT TREM2 in gray.
The structure of WT TREM2 ECD (residues 19–141) was determined to 2.2 Å resolution (see “Experimental procedures” and Table S1) in the P41212 space group with six molecules per asu (Fig. 1B and Fig. S1, B–D). Three of the six molecules in the asu assumed a core trimeric arrangement (molecules A, B, and C), with the other three molecules packed in the interfaces between each pair of neighboring molecules in the trimer core (molecules D, E, and F) (Fig. S1, B–F). The total buried SASAs between the molecule pairs within the core trimer (A-B, A-C, and B-C) are between 672 and 687 Å2, with the Sc score ranging from 0.68 to 0.73 (Tables S2 and S3). In contrast, the total buried SASAs of the exterior TREM2 molecules with the neighboring trimer exhibit a broader range from 472 to 672 Å2. Correspondingly, the shape complementarities of these surfaces also exhibit a broader range from 0.60 to 0.81. Here the Sc scores are borderline to typical Sc scores of 0.70–0.76 for protein oligomeric interfaces and the buried SASAs are approaching the cutoff of 850 Å2 for monomeric proteins (18, 19). These scores, along with solution state characterization by size exclusion and dynamic light scattering, indicate that the 6-mer observed in the crystal packing of WT TREM2 is less likely to be the biologically relevant form. However, the interface analysis also indicates that WT TREM2 has a higher propensity to oligomerize than R47H TREM2, and the trimeric interfaces provide one example of putative interaction interfaces. In addition, at 2.2 Å resolution, most residues in our WT TREM2 structure are well defined in the electron density maps, enabling us to clearly elucidate atomic interactions.
CDR2 loop remodeling in R47H TREM2 can disrupt ligand interactions
Whereas the R47H variant and WT structures are highly similar in the core fold, there is a remarkable structural change in the CDR2 loop region, with CDR2 loop in the R47H structure adopting a completely different conformation compared with the WT structure (Fig. 1D). The downstream β-strand C″ and C″-D loop are disordered in the R47H variant as well (Fig. 1D). Superposition of the R47H and WT structures results in an r.m.s. deviation of 1.1 Å over all of the backbone Cα atoms (residues 19–130). When residues 67–81 from the CDR2 loop, β-strand C″, and loop C″-D are excluded, however, the r.m.s. deviation drops to 0.52 Å, indicating that the variant structure differs from WT mainly in the region of the CDR2 loop to loop C″-D.
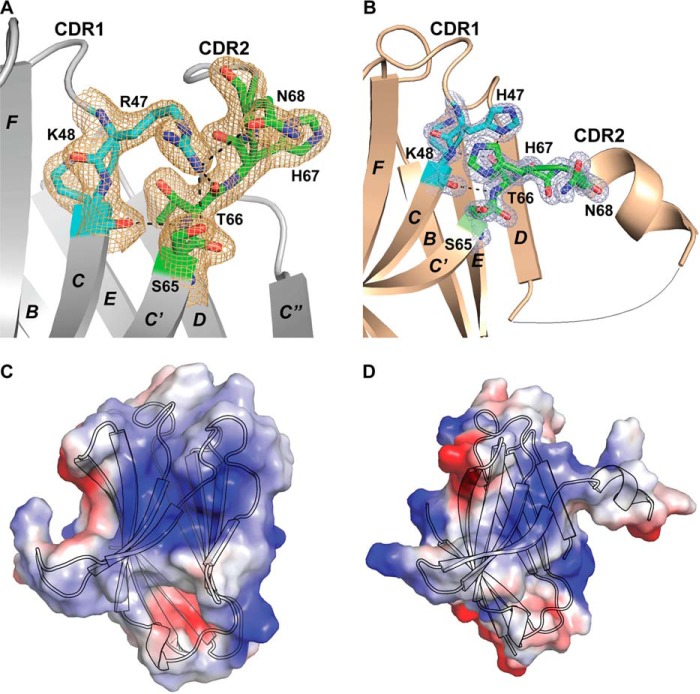
At the molecular level, the CDR2 loop remodels as a short helix in the R47H variant. This occurrence can be described by an entirely different set of interactions in the region surrounding the R47H mutation, which resides in the C-terminal region of the CDR1 loop, compared with the WT structure. In the WT structure, the Arg47 side chain projects into the N-terminal region of the CDR2 loop, participating in extensive hydrogen bond networks with residues in the CDR2 loop, including the side chain of Ser65, the backbone carbonyls of Thr66 and His67, and the side chain of Asn68 (Fig. 2A). As a result, Arg47 plays a critical structural role in maintaining the fold of the N-terminal area of the CDR2 loop. Other hydrogen bond interactions in the region extend from the carbonyl of Lys48 to the amide of Thr66 and from the amide of Lys48 to the side-chain hydroxyl of Thr66 (Fig. 2A).
Figure 2.
WT and R47H TREM2 putative ligand-interacting interfaces and molecular interactions. The 2mFo − DFc electron density map corresponding to the depicted protein side chains is demonstrated for both structures in A and B (contoured to 1.0σ). A, molecular interactions between CDR1 and CDR2 in WT TREM2, including hydrogen bonds from Arg47 NH2 to Ser65 OH, Oδ1 Asn68, and Thr66 OH and Arg47 Nϵ to His67 carbonyl oxygen; Thr66 OH to Lys48 backbone amide; and Thr66 backbone amide to Lys48 carbonyl oxygen. B, molecular interactions between CDR1 and CDR2 in R47H TREM2, including a hydrogen bond from the His47 imidazole to Thr66 OH and His47 imidazole π–π stacking interactions with the His67 imidazole from the CDR2 loop. C, electrostatic interface diagram depicting a large positively charged patch composed of Arg47, Arg62, His67, and Arg77 on the external side of the A-G-F-C-C′ sheet. A cartoon representation depicts the WT TREM2 Ig-like V domain in black outline. D, electrostatic interface diagram depicting minimization of a positively charged patch composed of His47, Arg62, His67, and Arg77 on the external side of the A-G-F-C-C′ sheet. A cartoon shows the R47H TREM2 variant Ig-like V-domain in black outline.
Whereas the R47H variant maintains the two hydrogen bond interactions between Lys48 in the C terminus of the CDR1 loop and Thr66 in the N terminus of CDR2 loop, the hydrogen bond network interactions from Arg47 to Ser65, Thr66, His67, and Asn68 of the CDR2 loop noted in the WT structure are all absent in the R47H variant structure. The imidazole group of the His47 side chain in the R47H variant of CDR1 makes no direct hydrogen-bond interactions with the backbone of the CDR2 loop. Instead, the His47 imidazole moiety directly hydrogen-bonds with the side-chain hydroxyl of Thr66 and also forms π–π stacking interactions with the His67 imidazole ring from the CDR2 loop (Fig. 2B). His67 undergoes a dramatic conformational change to swing ∼180° toward the area of the C terminus of CDR1, facilitating interactions with His47 (Fig. 2, A and B). By contrast, in the WT structure, residue His67 from the CDR2 loop adopts an extended conformation toward the exterior of the protein (Fig. 2A).
As a result of numerous intramolecular interactions from Arg47 to CDR2, the CDR2 loop in the WT structure is very stable, as indicated by its low B-factors (Fig. S2A). In the R47H variant structure, however, whereas the N-terminal part of the CDR2 loop that interacts with the CDR1 loop remains stable, the rest of the CDR2 loop (residues Leu69–Leu75 in the short helix) presents high B-factors due to lack of intramolecular interactions, followed by a disordered β-strand C″ compared with the rest of the protein (Fig. S2B).
Consequently, Arg47 plays a critical structural role in maintaining the tertiary architecture in the region around the C terminus of the CDR1 loop and the N terminus of the CDR2 loop. The arginine-to-histidine mutation in the R47H variant abolishes the extensive interactions specific for the arginine residue, reducing the stability of the CDR2 loop and its downstream β-strand C″. In this manner, CDR2 loop remodeling in the R47H variant can be traced back readily to the single amino acid mutation of R47H in the CDR1 loop, probably causing the rest of the CDR2 loop to be moved out of register with its partner strands.
The drastic conformational change in the CDR2 loop in the R47H variant also results in substantial modifications in the putative PLIS on the A-G-F-C-C′ sheet of WT TREM2 composed mainly of a few basic residues, including Arg47 (CDR1 loop), Arg62 (β-strand C′), His67 (CDR2 loop), and Arg77 (β-strand C″) (Fig. 2, C and D). Kober et al. (12) also reported a similar PLIS. The altered interactions in the R47H variant discussed above, especially the swing of His67 to make His47–His67 stacking, result in a disruption in the close interactions of the CDR2 loop with CDR1 and β-strand C″, subsequently resulting in a net reduction in the PLIS of the variant and the appearance of a significantly altered surface (Fig. 2D).
With regard to the influence of crystal contacts on the altered conformation of the CDR loops, His47 in the R47H variant completely remodels the fold encompassing CDR2, which is not involved in crystal packing in the asymmetric unit or between symmetry-related molecules. As well, examination of CDR1 and CDR3 reveals that these loops are not involved in crystal contacts involving CDR2, again within the asymmetric unit or between symmetry-related molecules. The WT TREM2 CDR2 loop does have crystal contacts with both the CDR1 and D-E loops of neighboring molecules. However, the CDR2 loop has a fold highly similar to the fold observed in the recently reported WT TREM2 crystal structure (12), although the two structures are determined in two different space groups with different crystal packing(s) (Fig. 1C). Due to the numerous interactions of Arg47 with the CDR2 loop, it is more likely that the extensiveness of this interaction makes intramolecular contact more dominant and thus less likely to be impacted by crystal packing.
Conformation and stability change driven by R47H variant are independent of glycosylation and correlate well with lower levels of sTREM2 in vivo
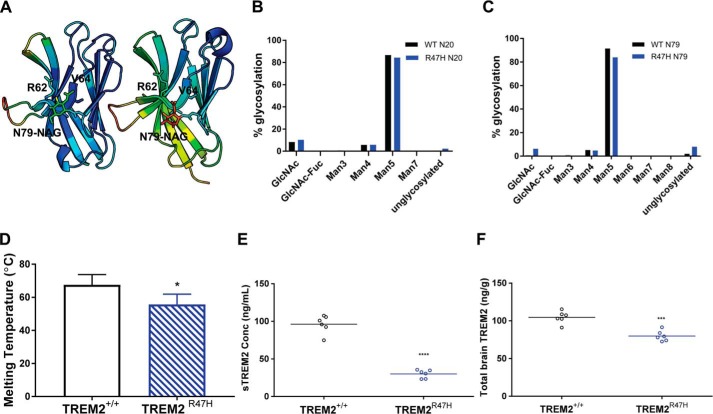
Comparison of the R47H and WT structures clearly reveals the impact of an arginine-to-histidine mutation at residue 47 on the TREM2 ECD structural fold. The protein sources for each structure differ, however, and this difference must be addressed. The WT structure was derived from mammalian protein, whereas the R47H variant structure was derived from E. coli expressed protein and lacks the glycosylation observed at Asn79 in the WT structure. To address the possible contribution of Asn79 glycosylation to the observed differences between the WT and R47H TREM2 structures, we examined the interactions of the NAG glycan with other residues in the WT structure in greater detail. In the WT structures presented here and the previously reported structure (Protein Data Bank code 5ELI) (12), the Asn79-NAG shares the same hydrogen bond interaction through the amide of Asn79 to the backbone carbonyl of Val63 of β-strand C′, and the GlcNAc moieties have only weak van der Waals contacts with Arg62, Val63, and Val64 of β-strand C′ (Fig. 3A). In addition, the NAG moieties in the two WT structures have different orientations (Fig. 1C) and much higher B-factors in both instances compared with TREM2 residues in the vicinity of the NAGs (Fig. 3A), suggesting dynamic, nonspecific NAG conformations. As a result, the presence or absence of glycan is unlikely to play a significant role in the structural fold of the TREM2 protein. Whereas the lack of a glycan on Asn79 in the R47H variant might contribute to some extent to the disordering of the more distal β-strand C″, it is unlikely to result in the dramatic N-terminal remodeling of the CDR2 loop. Instead, the dramatic conformational change of His67 is well supported by the altered atomic interactions resulting from the single arginine-to-histidine mutation at residue 47.
Figure 3.
Stability characterizations of WT and R47H TREM2. A, cartoon diagrams of current 2.2-Å WT TREM2 Ig-like V-domain (left) and previously solved 3.1-Å WT TREM2 Ig-like V-domain (right), with related B-factors colored in rainbow gradient. Low values trend toward cool coloring, and high values trend toward warm coloring. B, percent glycosylation of Asn20 in human WT TREM2 ECD (black) versus R47H TREM2 variant ECD (blue), indicating highly equivalent glycan content. C, percent glycosylation of Asn79 in human WT TREM2 ECD (black) versus R47H TREM2 variant ECD (blue), indicating highly equivalent glycan content. D, melting temperature measurements for WT TREM2 ECD (black diagonal lines) and R47H TREM2 variant ECD (blue diagonal lines) indicate a lower average Tm = 55.7 °C for R47H TREM2 variant ECD compared with an average Tm = 67.6 °C for WT TREM2 ECD; data are presented as mean ± S.D.; *, p < 0.05 unpaired t test with Welch's correction. E and F, levels of sTREM2 (E) in plasma and total TREM2 (F) in brains of R47H transgenic mice (blue circles) are significantly reduced compared with WT littermate controls (black circles); n = 6 animals/genotype; ***, p < 0.001; ****, p < 0.0001, unpaired t test with Welch's correction.
To further probe the effect of glycosylation on R47H TREM2, we conducted structural and biophysical studies on mammalian R47H TREM2. Despite intensive effort, we were unable to obtain crystals of mammalian expressed R47H TREM2 or E. coli expressed WT TREM2 (Table S4). We were, however, able to obtain pure, homogeneous protein for mammalian expressed WT and R47H TREM2 ECD (residues 19–174) for biophysical characterization. An analysis of glycan content for these two protein samples of WT and R47H variant revealed nearly equivalent glycan type and content at both the Asn20 and Asn79 positions (Fig. 3, B and C). Thermal stability measurements (melting temperature (Tm)), however, revealed a 10 °C difference between constructs with a Tm = 55.7 °C for the R47H variant compared with Tm = 67.5 °C for WT TREM2 (Fig. 3D). These data clearly support the profound effect of the R47H mutation in driving conformational changes that result in stability loss. Both WT and R47H variant proteins utilized in these experiments have the same glycan type and content, and hence the observed differences in stability can be explained by the variant alone. Finally, our structural observations, including the remodeled short α-helix and disordered β-strand (C″) in the R47H variant compared with WT TREM2, are consistent with and provide detailed and unprecedented molecular insight into the differences in WT and R47H TREM2 observed by lower-resolution CD spectroscopy (12).
To determine whether the in vitro differences in stability and structure of WT and R47H ECDs translate to differences in in vivo levels, we developed a homogeneous ELISA to determine steady-state, homeostatic levels of soluble and total TREM2 in plasma and brains of WT and R47H TREM2 mice (for a brief description of mice, see “Experimental procedures”; for a detailed description, see Cheng et al. (33)). The assay developed specifically recognizes TREM2, as evidenced by no detectable levels in knockout animal plasma and brains (data not shown). The standard curves using recombinant TREM2 protein confirm that the assay recognizes WT and R47H TREM2 equivalently. R47H variant mice revealed significantly reduced sTREM2 (plasma, Fig. 3E) and total TREM2 (brain homogenate, sTREM2 + total TREM2, Fig. 3F) as compared with WT animals (Fig. 3, E and F), correlating well with the reduced stability of the variant. It should be noted that these differences in sTREM2 protein were noted despite similar levels of transcript and surface protein expression in microglia and macrophages isolated from these mice compared with WT age-matched littermate controls (33). Song et al. (17) have also reported lower levels of sTREM2 in the R47H TREM2 5x FAD mice.
R47H variant differentiates from WT TREM2 in lipid binding, oligomerization, and signaling
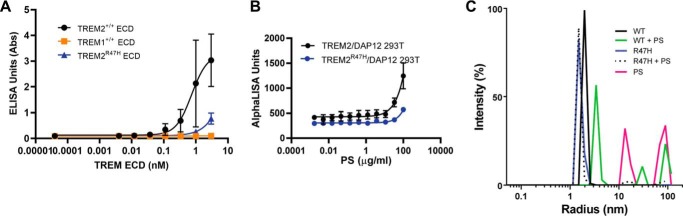
Although the R47H variant signals poorly in an NFAT reporter gene assay, Kober et al. (12) have reported that both WT and R47H TREM2 bind comparably to phospholipids. We carried out lipid binding studies using PS Lipid Snooper® strips (Avanti Lipids) and performed dose-response titrations with Fc-tagged forms of WT, R47H TREM2 (generated internally), and control TREM1 (R&D Systems) proteins. An ELISA detecting the Fc part of the protein was utilized as the detection format. We determined that the R47H variant TREM2 ECD binds to PS less efficiently compared with WT TREM2 ECD (Fig. 4A). We also observed differences between WT and R47H TREM2 binding to other lipids, including oxidized PS, phosphatidylethanolamine, and sphingomyelin (Fig. S3, A–I), consistent with the reported defect in NFAT-mediated signaling for the R47H variant (5). We did not observe binding to lipids like phosphatidic acid and lysophosphatidic acid and observed poor binding to phosphatidylcholine and cardiolipin (Fig. S3, A–I). Overall, WT TREM2 binds to a broad panel of phospholipids with a preference for anionic lipids. Lower binding efficiency is noted for R47H TREM2, and no binding is observed for TREM1. Finally, we also compared the functional activity of WT and R47H TREM2. Consistent with the NFAT reporter gene assay, we also noted reduced pSyk signaling after PS stimulation in R47H TREM2/DAP12-overexpressing 293T lines compared with WT TREM2/DAP12 lines (Fig. 4B).
Figure 4.
In vitro characterization of WT and R47H TREM2. A, reduced binding of R47H TREM2 ECD compared with WT ECD as measured by ELISA. TREM1 ECD is used as a nonbinding control. B, PS-mediated activation of pSyk in HEK cells expressing R47H TREM2 is reduced compared with cells expressing WT TREM2. C, dynamic light scattering diagram of hydrodynamic radius (nm) versus intensity (%) of WT TREM2 and WT TREM2 incubated with PS and of R47H TREM2 variant incubated with PS revealed a hydrodynamic radius of 20 Å (black) for WT and 16 Å (blue) for the variant. The addition of PS shifts WT TREM2 ECD to a high-order oligomer with a radius of 35 Å (green); no shift is observed for the R47H variant (black dots). PS-only control is depicted in pink.
Next, we examined whether PS functions to drive or stabilize TREM2's oligomerization state. The structural analysis of the crystal packing of WT and R47H TREM2 suggests that the WT protein may oligomerize more readily compared with R47H TREM2. We utilized dynamic light scattering to assess the solution state oligomerization of WT and R47H TREM2 variant in the presence of PS. Both WT and R47H TREM2 were monomeric in solution, with a smaller hydrodynamic radius (Rh) of 16 Å for the R47H variant compared with 20 Å for WT (Fig. 4C). The particle size increased to 35 Å for WT protein in the presence of PS, indicating an increased propensity of the WT protein to oligomerize in solution, which is enhanced by ligands (Fig. 4C). In contrast, R47H variant showed little change in particle size upon incubation with PS in solution (Rh = 15 Å), consistent with its poor binding to the PS phospholipid.
PS stabilizes oligomeric packing of WT TREM2
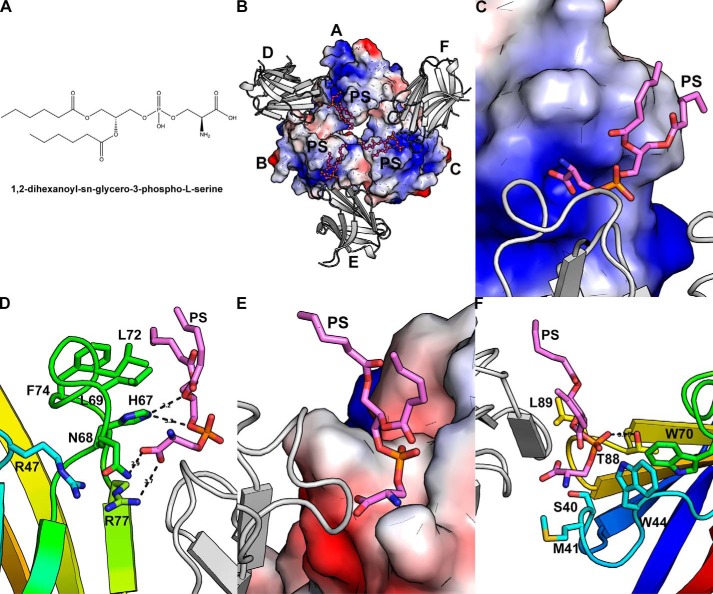
To investigate PS binding to WT TREM2 and its impact on oligomerization and activity, we determined the crystal structure of PS-bound WT TREM2 to 2.2 Å through a soaking experiment with a phospholipid of 1,2-dihexanoyl-sn-3-phospho-l-serine (Fig. 5A). The PS-bound structure (see “Experimental procedures” and Table S1) retained the same space group as the WT apo-structure with a slight change in the unit cell dimensions (shrinking by 0.33 Å along the x and y axes and 0.37 Å along the z axis). The oligomeric arrangement in the asu was also retained, and all six molecules were highly similar, with an average r.m.s. deviation of 0.27 Å for the backbone Cα atoms between the six molecules. Residues 19–21 and 130–140 were also disordered in the PS-soaked structure. As in the WT structure, glycosylation at Asn79 was modeled as NAG.
Figure 5.
PS binding to WT TREM2. A, chemical structure of 1,2-dihexanoyl-sn-glycero-3-phospho-l-serine. B, PS-bound WT TREM2 structure, including inner trimer (electrostatic surface), outer TREM2 protomers of higher-order complex (gray cartoon), and PS (fuchsia sticks). C, zoomed-in depiction of best-modeled PS molecule (fuchsia sticks) bound between inner trimer (electrostatic surface) and outer TREM2 protomer (gray cartoon). D, molecular interactions of best-modeled PS molecule (fuchsia sticks) with TREM2 of inner trimer (rainbow cartoon and sticks). Neighboring outer TREM2 protomer is depicted as a gray cartoon. E, zoomed-in depiction of best-modeled PS molecule (fuchsia sticks) bound between inner trimer (gray cartoon) and outer TREM2 protomer (electrostatic surface). F, adjacent residues to best-modeled PS molecule (fuchsia sticks) from TREM2 of neighboring outer TREM2 (rainbow cartoon and sticks). TREM2 of the inner trimer is depicted as a gray cartoon.
The PS-soaked trimer maintained the same general trefoil-like configuration as the apo-WT trimer but with an increase in total buried surface area to 690 Å2 (2.7%), 715 Å2 (5.9%), and 708 Å2 (3%) for the inner trimer molecules (Table S2). The complementarity of the molecular surfaces of the trimer was maintained with PS binding, with overall (Sc) scores between the molecules in the trimer core (0.67, 0.66, and 0.74) remaining comparable with those of apo-WT (Table S3) (18). On the other hand, the total SASAs of the exterior TREM2 molecules with the neighboring trimer decreased in all instances except one (molecules D-B), along with a decrease in shape complementarity in all instances except one (molecules E-C) (Tables S2 and S3). Three PS molecules were modeled into the extra electron density observed in the “pore” of the TREM2 trimer (Fig. 5B and Fig. S4A). The electron density was clearly observed for the phosphoserine head, but variable electron density was observed for the hexanoyl tails for three PS molecules. In general, ligand density fit was of lower quality, indicating lower ordering for these molecules, and therefore any binding interactions noted should be considered in a general sense. The electron density for one PS molecule, however, was observed in its entirety (Fig. S4B). Each PS molecule bound between two TREM2 molecules: one from the trimer core (molecule A, B, or C) and the other one from the outer layer (molecule D, E, or F) (Fig. 5B). Contact analysis of the PS molecule with two contacting TREM2 molecules shows that PS utilized more interactions with the TREM2 molecule in the trimer core than the neighboring molecule in the outer layer.
Molecular interaction analysis for the PS molecule with TREM2 in the trimer core showed that the negatively charged headgroup of the PS molecule fit snugly into a positively charged surface depression formed by His67 (CDR2 loop), Asn68 (CDR2 loop), and Arg77 (N terminus of β-strand C″ following the CDR2 loop) (Fig. 5C). The carboxylate moiety of PS formed a few hydrogen bond interactions with both amide N and Nδ of Asn68 and NH1 of Arg77 as well as stacking interactions with the imidazole moiety of His67 (Fig. 5D). Additional hydrogen-bond interactions were also observed from Nϵ of His67 to O2 of the phosphoryl moiety and O11 of the hexanoyl. Furthermore, the hydrophobic hexanoyl tails of the PS molecule made van der Waals interactions with the hydrophobic residues Leu69, Leu74, and Leu76 of the CDR2 loop (Fig. 5D).
As well, each PS molecule was bound by a TREM2 molecule originating from the outer layer. The binding surface was mainly to the polar PS headgroup and composed of the polar and hydrophobic residues Thr88 and Leu89 from the D-E loop and Ser40, Met41, and Trp44 from the CDR1 loop (Fig. 5E). These interactions were mainly van der Waals in nature with one hydrogen-bond interaction from O3 of phosphate to the hydroxyl of Thr88 (Fig. 5F). Finally, we observed that the hydrophobic hexanoyl tails did not interact with this outer layer TREM2 molecule.
Discussion
In summary, we present extensive structural analysis elucidating the impact of the Alzheimer's risk variant R47H on TREM2 structure, in vitro stability, in vivo homeostatic levels, and ligand interactions. The global fold of the extracellular domains of WT TREM2 described here and the previously reported lower-resolution WT structure (Protein Data Bank code 5ELI) (12) are highly similar. However, the detailed molecular interaction analysis and comparison of the PLIS of R47H versus WT TREM2 and apo-WT versus PS-bound WT TREM2 structures provide the first molecular insight into the profound effects of the R47H variant on TREM2 ligand–interacting surfaces, stability, and function. These effects could not have been predicted based on an analysis of the WT structure alone, especially the dramatic conformational changes involving the key residue, His67.
This key residue, His67, when considered in combination with the R47H mutation, probably alters the pH dependence of the R47H variant. The R47H variant is apt to be more sensitive to pH change as a result of the mutation, with subsequent reshaping of the PLIS. The R47H crystal structure was obtained with a buffer of pH 8.0. Thus, it is likely that the imidazole ring of His47 and His67, with a pKa of 6.0, assumes a greater state of neutrality, giving rise to a larger potential for π–π stacking interactions, as shown by observed His47–His67 π–π stacking in the crystal structure. At pH lower than 6.0, increased protonation of His47 probably creates a repulsive effect with neighboring cationic groups, including His67, possibly causing conformational change of His67. The intracellular trafficking of WT and R47H TREM2 variant, however, indicates that both are predominantly localized in the Golgi apparatus (pH 6.7 to 6.0) (20) before moving to secretory granules (pH 5.5) and the cell surface (pH 7.4). Under these varying pH conditions, the presence of this type of histidine switch would be far more detrimental to the maintenance of a PLIS than arginine, which would remain protonated under all pH variations and retain hydrogen bonding with neighboring residues to maintain the CDR2 loop conformation.
We also provide experimental evidence that the absence of glycosylation at Asn79 in the E. coli expression–derived R47H structure probably has little to no effect on the observed conformational changes between WT and Arg47 TREM2 variant structures. A careful analysis of the glycan interactions at Asn79 in both WT structures as well as the detailed glycan analysis of mammalian R47H and WT TREM2 revealed similar glycan composition. This lends credence to the hypothesis that much of the ligand-interacting surface remodeling observed in the R47H variant structure can be attributed mainly to the single point mutation. This is further supported by the stability differences noted between mammalian R47H and WT TREM2 (with comparable glycan composition) as well as secondary structure differences between the two proteins reported previously (12).
The CDR2 loop remodeling and loss of the proposed PLIS in the R47H structure provide the first indications of the manner in which ligand interaction may be reduced or abolished in the variant. The PS-bound WT TREM2 structure offers even further insight into additional ways in which the variant can disrupt ligand interaction. Whereas clustering is a common activation mechanism for pattern recognition receptors in general and TREM2 specifically, the biochemical data and interface analysis of the previously solved WT structure suggested that the protein is an inactive monomer in solution, providing little insight into ligand engagement or the molecular basis for clustering of the receptor. Analyses of our current WT TREM2 structure, including crystal packing, Sc scores, and total SASAs of the oligomeric interfaces, suggest a propensity for WT TREM2 to oligomerize by engaging interfaces that are not accessible in the variant structure.
A potential increase in oligomerization upon lipid binding and activation of signaling are also supported by our data. Moreover, the molecular interactions of PS with TREM2 clearly reveal multiple ways in which Arg47 maintains the molecular architecture conducive to ligand binding, whereby CDR2 loops and PLISs across multiple protomers of the oligomeric arrangement come together to drive ligand binding and activation. The electrostatic and hydrogen-bond interactions between the PS headgroup and the surface formed by the CDR2 loop of the molecule from the WT trimer core probably account for much of the affinity driving the binding. Additionally, interactions provided by the surface cavity formed by the CDR1 loop and D-E loop of the TREM2 molecule from the outer trimer layer also undoubtedly tend to boost avidity.
Finally, the interactions between PS and WT TREM2 noted in the PS-bound structures are focused mainly in the surface area of the CDR2 loop by key residues His67, Asn68, and Arg77. In the R47H variant structure, these three residues, especially His67 and Asn68 from the CDR2 loop, undergo dramatic conformational changes following the R47H mutation. As a result, the PLIS binding to the negatively charged PS headgroup is abolished, and association with the outer layer molecules involved in the hexamer oligomerization is eliminated. Analysis of the PS-bound WT TREM2 structure thus allows a straightforward interpretation of the loss of PS binding and oligomerization in the case of the R47H AD variant, although Arg47 does not interact with PS directly.
This complex set of interactions between TREM2 and PS that involve multiple surfaces and interactions can to some extent explain the promiscuity of this class of receptors and the ability of TREM2 to bind to a broad range of phospholipids and other putative ligands. However, this is probably only the first, albeit very important, piece in a complex puzzle. It is plausible that TREM2 may interact with other putative ligands like apoE in addition to PS by engaging different ligand-interacting surfaces. It may even engage PS differently based on PS surface density (as proposed for TIM-4, discussed below) and if it is presented with other ligands. The ability of ligands with presumably differing affinities and avidities to interact with TREM2 by nonoverlapping molecular mechanisms also probably differentially impacts downstream signaling and activity. Precedence already exists for different affinities and avidities of disparate ligands resulting in diverse biological responses (inhibitory versus stimulatory) by differential recruitment of downstream components (21).
There are lessons to be learned from molecular studies on other PS-binding proteins that bind to other putative ligands as well and also engage PS in different ways. One example is C1q, the ligand recognition unit of the C1 complex of complement. C1q recognizes surface-exposed PS on early apoptotic cells and demonstrates specific, avidity-driven binding that is inhibited by annexin V in a dose-dependent manner (22). The 2.0 Å crystal structure of C1q globular region soaked with PS (22), like the PS-TREM2 structure, indicates that PS acts as a bridging molecule between two molecules of a C1q globular region trimer. Analysis of the structure points to the probability of induced formation of multiple charged surfaces where PS may bind in a promiscuous manner and promote binding and oligomerization of the C1q complex, in response to apoptotic cells. Additionally, the structures of the globular domain of C1q soaked with heparin sulfate and deoxy-d-ribose show that these molecules bind to nonoverlapping regions compared with phosphoserine, supporting the ability to form a large interaction area that can interact differently with diverse ligands. Further supportive of the putative multiple interaction interfaces for this class of proteins, the Ig binding region of C1q has been modeled on the outer side of the heterotrimer in a similar paradigm to TREM2 and its ligands discussed above.
Yet another example of engagement of multiple interaction interfaces for a PS binding protein is the TIM family of receptors that have high selectivity for PS (unlike TREM2) but also bind additional nonoverlapping ligands (galectin-9 for TIM-3). In this case, the primary PS-binding site chelates Ca2+ and interacts with the FG′ loop in the Ig V-domain. Recent data, however, suggest that TIM-4 contains additional basic residues/regions that serve as weak additional binding sites for PS (23). Thus, whereas the precedent exists for multiple interaction interfaces for PS-binding proteins that display ligand promiscuity like TREM2, we show for the first time at the molecular level how ligand promiscuity may be achieved for TREM2 and the profound effect of the variant on this.
Furthermore, a comparison of the in vitro stability of WT versus R47H TREM2 reveals a reduced stability of the variant protein compared with WT. Whereas this probably contributes to the reduced levels in vivo in our mice and the R47H-5x FAD mice (17), further studies aimed at measuring sTREM2 half-life in vivo are necessary to prove this unequivocally.
Our studies, while focusing on just one of TREM2's possible ligands, reveal the complexity surrounding TREM2–ligand interactions and emphasize the promiscuity of this receptor. Future work focused on multiple ligands interacting in concert with full-length TREM2-DAP12 complexes will provide a greater understanding of TREM2-DAP12 signaling biology and its dysregulation in disease.
Experimental procedures
Protein expression and purification of WT and R47H TREM2
For the WT TREM2 protein, the TREM2 gene encoding the human TREM2 signal peptide and extracellular domain (residues 1–174) with the N20D mutation to eliminate glycosylation and improve protein stability was subcloned into a pTT5 vector with a C-terminal tobacco etch virus cleavage site and His6 tag. The vector was co-transfected into HEK293S GnTI− cells (ATCC) along with EBNA plasmid and grown in 5-liter Thomson flasks with Freestyle293 medium (Invitrogen) at 37 °C and 120 rpm. Yeastolate was added at 20 ml/liter the day after transfection. Cells were harvested on day 7. The supernatant was loaded onto a nickel-nitrilotriacetic acid column and washed with 25 mm Tris-HCl, pH 7.6, 300 mm sodium chloride, and 15 mm imidazole and then eluted with 25 mm Tris-HCl, pH 7.6, 300 mm sodium chloride, and 300 mm imidazole. Both wash and elution buffers contained Complete, EDTA-free protease inhibitor mixture (Roche Applied Science).
The WT TREM2 protein sample was eluted from a HiLoad 16/600 Superdex 200pg (GE Healthcare) in 50 mm Hepes, pH 7.5, 150 mm NaCl, and 3% trehalose to remove aggregates. The major peak was then purified on a Mono Q 10/100GL (GE Healthcare), which then became cleaved at the C terminus as a result of a spontaneous degradative event. This truncated TREM2 eluted in the flow-through, which was subsequently crystallized.
For the R47H TREM2 protein, the TREM2 gene encoding the human TREM2 Ig-like domain (residues 19–131) with the R47H mutation was subcloned into a pET24 vector. The vector was transfected into BL-21-(DE3)-RIL Codon Plus (Agilent Technologies Inc.) and grown at 37 °C to A600 0.9 in TB-Kan-Cam-Tet medium. Expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. The pellet was lysed in 10 mm Tris-HCl and 5 mm EDTA, pH 8.0, and then spun down and washed with Milli-Q water. Membrane layers were sloughed off to recover inclusion bodies, which were then resuspended, spun down, and washed twice. The washed inclusion bodies were solubilized by adding a small amount of 50 mm Tris, pH 8.5, and enough DTT to yield a final concentration of 10 mm. 8 m guanidine HCl in 50 mm Tris was added to yield a final concentration of 6 m guanidine HCl. The inclusion bodies were stirred at room temperature for 1.5 h or until the mixture appeared uniformly solubilized.
The solubilized pool was then introduced in a 1:25 dilution to a refold buffer consisting of 2 m urea, 50 mm Tris-HCl, 20% glycerol, 160 mm l-arginine, 3 mm cysteine, and 1 mm cystamine at pH 8.9. The refold was allowed to react for 48 h until Ellman's test was negative. The resulting pool was sterile-filtered and concentrated 10-fold before being diluted 10-fold in 25 mm Hepes, pH 8.8, for loading onto a 20-ml Fractogel SO3 (EMD Millipore). Step elution was into 600 mm NaCl. The resulting elution peak was pooled and brought up to 200 mm l-arginine to enhance stability. This pool was then concentrated and loaded onto a Superdex 16/60 75 column (GE Healthcare) equilibrated with 1× PBS. The final pool was again brought up to 200 mm l-arginine.
Crystallization
For the WT TREM2 crystals, the sample (in 50 mm Hepes, pH 7.5, 50 mm NaCl) was concentrated to 10 mg/ml. Crystals grew in 0.2 m sodium iodide, 1.6 m ammonium sulfate at 20 °C via sitting-drop vapor diffusion.
For the R47H TREM2 crystals, 5 mm N-acetyl-d-lactosamine was added to the refolded sample (1× PBS, 200 mm l-arginine) to improve solubility during concentration and then concentrated to 4 mg/ml. The sample also contained 200 mm l-arginine to improve solubility. Crystals grew in 30% PEG 6000, 0.1 m Tris, pH 8.0, at 20 °C via sitting-drop vapor diffusion. No N-acetyl-d-lactosamine was observed in the crystal structure.
For the PS-soaked WT TREM2 crystals, PS liposomes were generated from 1,2-dihexanoyl-sn-glycero-3-phospho-l-serine suspended in chloroform (Avanti Polar Lipids, Inc.) by drying the mixture to remove solvent and yield a lipid film. The lipid film was subsequently hydrated by the addition of 50 mm Tris, pH 8.0, 150 mm NaCl and sonicated to produce a lipid suspension suitable for crystal-soaking experiments. WT TREM2 crystals were then soaked with 1 mm PS for 3 h before being frozen for data collection.
Data collection and structure determination
R47H TREM2, WT TREM2, and PS-soaked WT TREM2 diffraction data were collected at the Advanced Light Source (Berkeley, CA), beamline 5.0.2, at a temperature of 100 K and a wavelength of 1.0 Å. Data sets were processed and scaled with XDS and the CCP4 program suite (24, 25). R47H TREM2 was solved to 1.8 Å via molecular replacement using Molrep (25) and Phaser (26) using a solved structure of human TREM1 (Protein Data Bank code 1SMO). The model was rebuilt using Buccaneer. Iterative refinement cycles were performed using CCP4 (25) and COOT (27). WT TREM2 was solved to 2.2 Å via molecular replacement using Phaser (26) using the human R47H TREM2 mutant as a search model. Iterative refinement cycles were performed using Phenix.refine (28) and COOT (27). PS-soaked WT TREM2 diffraction data were solved to 2.2 Å via molecular replacement using Phaser (26) using human WT TREM2 as a search model. Density maps were improved using the density modification program Parrot (25). Buried surface areas for all were calculated using PISA (29), and shape complementarity was assessed with Sc (18).
Tm measurements
Two constructs were assessed: WT TREM2(19–179)-His-FLAG and R47H TREM2(19–179)-His-FLAG. Samples were concentrated to 1 mg/ml in 50 mm Hepes, pH 7.5, 150 mm NaCl and loaded into an Avacta Optim 1000 fluorescence spectrometer. Tm was assessed in linear fashion, from 15 to 90 °C at 0.3 °C/min and plate hold = 0 s, incubation time = 60 s. Measurements were processed with Optim Analytic software.
Glycan profiling
N-Glycan profiles of WT TREM2(19–179)-His-FLAG and R47H TREM2(19–179)-His-FLAG were determined by tryptic peptide mapping followed by LC-MS/MS analysis (30). Tryptic digestion was performed with a similar procedure described previously (30). About 25 μg of each protein was denatured and reduced with 4.0 m guanidine HCl (Sigma-Aldrich) and 1 mm DTT (Sigma-Aldrich) in 0.1 m Tris buffer (pH 7.5) at 37 °C for 30 min. Each denatured/reduced protein sample (at a concentration of ∼50 μg/ml) was alkylated with 2 mm iodoacetic acid (Sigma-Aldrich) at 25 °C for 20 min, followed by quenching of the reaction with 1 mm DTT. The reduced/alkylated protein samples were then exchanged and concentrated into a 0.1 m Tris buffer at pH 7.5 with a Vivaspin 500 10,000 MWCO filter (Sartorius Stedim, Bohemia, NY) according to the manufacturer's suggested procedure. Trypsin digestion was initiated by adding 5 μg of trypsin (Roche Diagnostics) to achieve an enzyme/substrate ratio of 1:5 and incubating at 37 °C for 60 min. After digestion, each sample (∼47 μl in volume) was quenched by adding 4.5 μl of 10% and 16 μl of 8 m guanidine HCl. About 5.5 μg (15 μl of solution) of each protein digest was injected for LC-MS/MS analysis.
LC-MS/MS peptide mapping was performed on a Thermo Scientific (San Jose, CA) Vanquish UHPLC system directly connected to a Thermo Scientific Orbitrap Fusion Tribrid mass spectrometer equipped with an electrospray ionization interface. Proteolytic peptides were separated on a Waters (Milford, MA) CSH column (2.1 × 150 mm, 1.7-μm particle) at 65 °C with a flow rate of 0.2 ml/min. Peptides were eluted with an acetonitrile gradient of 0.5–40% in 92 min, followed by column washing and re-equilibration, with 0.02% TFA in the mobile phase. The mass spectrometer was set up for a full scan in the Orbitrap with 120,000 resolution, followed by data-dependent collision-induced dissociation MS/MS scans in the linear trap with dynamic exclusion. Data analysis was performed on a MassAnalyzer (31) for automated identification and quantification of glycopeptides. Glycopeptides were identified by comparing experimental MS/MS with theoretically predicted MS/MS (30, 32). The detailed algorithm has been described previously (30, 31).
Gene-edited in-house mouse construction
All animal procedures were approved by the Amgen institutional animal care and use committee. A pair of mRNAs targeting exon 2 of the TREM2 gene was synthesized. The target sequences were as follows: left, 5′-TCCTTGAGGGTGTCATGTAC-3′; right, 5′-TGCGTCTCCCCCAGTGCTTC-3′. The binding sites were separated by a 12-bp spacer region. 143-mer R47H single-stranded oligonucleotide (ssODN), which was silently mutated with code modification in gene-editing binding sites to prevent cutting and also create an MluI enzyme-cutting site for genotype, was synthesized by Integrated DNA Technologies. The ssODN sequence was 5′-CAA GCC CTC AAC ACC ACG GTG CTG CAG GGC ATG GCC GGC CAG TCG TTA AGG GTA TCC TGC ACT TAT GAC GCG TTG AAA CAT TGG GGC AGA CAT AAG GCC TGG TGT CGG CAG CTG GGT GAG GAG GGC CCA TGC CAG CGT GTG GT-3′.
Microinjection
Two TREM2 gene-edited mRNAs and a 143-mer ssODN with R47H mutation were injected into the pronuclei of fertilized oocytes obtained from superovulated females of the C57BL/6 strain for the R47H TREM2 mutation. Genotyping for R47H KI mice was confirmed by PCR from tail genomic DNA.
Measurement of total and soluble TREM2
Equal numbers of 2–3-month-old R47H and WT mice were used in the study. Mice were euthanized by CO2 inhalation. Blood was withdrawn by cardiac puncture and collected in EDTA tubes. Plasma was obtained by centrifugation at 4 °C. Brains were removed and homogenized in 10 volumes (w/v) of 0.5% Triton X-100 in TBS with protease inhibitor mixtures. The homogenates were centrifuged at 100,000 rpm for 30 min at 4 °C. The supernatants were aliquoted and stored at −80 °C until further processing.
ELISA plates (Meso Scale Discovery GOLD 96-well small spot Streptavidin sector plate) were coated with 25 μl/well of a 5-μg/ml antibody (R&D biotinylated anti-TREM2, polyclonal goat IgG) solution in PBS. The plate was sealed and incubated overnight, at 4 °C with gentle shaking. The next day, plates were washed three times with TBS + 0.05% Tween 20. Excess liquid was gently removed by blotting plates on a paper towel. Plates were blocked with 150 μl/well of a 3% blocking solution (600 mg of MSD Blocker A in 20 ml of TBS + 0.05% Tween 20). Plates were sealed and incubated for 1 h at room temperature with gentle shaking. During the blocking step, samples and the standard curve were prepared. All samples were tested in triplicate, 25 μl/well. Plasma was diluted 1:10 and 1:15 into Tris lysis buffer (MesoScale Discovery). Brain lysates were diluted 1:3 and 1:5 in TBS + 0.5% Triton X-100 buffer. A 12-point standard curve was prepared in Tris lysis buffer for plasma or TBS + 0.5% Triton X-100 buffer for brain, with 3-fold serial dilutions using recombinant mouse soluble TREM2 protein (muTrem2V1(1–171)::GSS::FCN297G). After blocking, plates were washed as described previously. After the addition of a standard curve, samples, and blanks to plates, they were sealed and incubated overnight, at 4 °C with gentle shaking. The next day, 25 μl of detection antibody (R&D anti-TREM2, monoclonal rat IgG2B clone 237920, labeled with ruthenium (SULFO-TAG NHS-Ester as per MSD SOP)) was added to each well. The plate was sealed and incubated for 1 h at room temperature, protected from light, with gentle shaking. Plates were subjected to the wash protocol, and then plate orientation was flipped, and the wash protocol was repeated. Read Buffer (MSD Read Buffer T + surfactant 4× stock) was prepared by diluting to 2× with H2O and adding 150 μl/well. Plates were read immediately on an MSD Sector 6000 reader.
Dynamic light scattering
WT TREM2(19–140) used for structure solution was diluted to 0.1 mg/ml in crystallization buffer and incubated with and without 0.1 mg/ml PS liposomes (C18:0, Avanti) for 1 h at 4 °C. R47H TREM2(19–131) used for structure solution was diluted to 0.1 mg/ml in crystallization buffer and incubated with and without 0.1 mg/ml PS liposomes (C18:0, Avanti) for 1 h at 4 °C. Dynamic light scattering measurements were taken 50 times for each sample, using a Wyatt DynaPro Plate Reader II.
Preparation of PS liposome solution
1,2-Dioleoyl-sn-glycero-3-phospho-l-serine sodium salt (DOPS) was purchased from Avanti Polar Lipids, and chloroform and Dulbecco's PBS were purchased from Sigma-Aldrich. Solid DOPS (13 mg) was placed into a glass vial, chloroform (2 ml) was added, and the mixture was gently swirled until all solids dissolved. Solvent was then evaporated using a rotary evaporator (Laborota4001, Heidolf). PBS (13 ml) was filtered through a 0.45-μm nylon membrane filter and added to the vial containing DOPS film. The sample was briefly vortexed and then placed in a sonic bath for 15 min to yield an opaque solution. The sample was further dispersed using a digital sonifier (Branson), equipped with a 0.125-inch tip, for 60 s at 20% amplitude. Then 12 ml of the resulting opaque solution were extruded by passaging 1-ml samples 10 times through a mini-extruder (Avanti Polar Lipids) equipped with 100-nm polycarbonate membrane using two glass 1-ml syringes. The polycarbonate membrane was replaced after processing 6 ml of sample. The resulting translucent DOPS solution was collected into glass vials and stored at 2–8 °C until further use.
Cell line generation
Stable cells were generated in a HEK293T parental background using nucleofection. Briefly, the huTREM2 (or huR47H-TREM2) gene and the huDAP12 gene were cloned into the pBUDCE4.1 vector (Thermo Fisher Scientific) and transfected into HEK293T cells using the Amaxa 4D Nucleofector (Lonza Biosciences). Positive expressors were selected by adding 100 μg/ml Zeocin and screened for TREM2 surface expression by FACS analysis.
pSyk AlphaLISA
huTREM2–293T or huR47H-TREM2–293T was plated in 384-well white CulturPlates (PerkinElmer Life Sciences) overnight. On the following day, the cells were treated in quadruplicates with PS liposomes for 30 min at 37 °C. At the end of the treatment, the medium was removed, and cells were lysed in M-PER lysis buffer (Thermo Fisher Scientific) supplemented with HALT protease and phosphatase inhibitors (Thermo Fisher Scientific). The pSyk levels in the cell lysates were tested using a pSyk AlphaLISA developed in-house using a mouse biotinylated anti-Syk (BD Biosciences) and a rabbit anti-pSyk (Cell Signaling Technologies) antibody pair with anti-rabbit IgG acceptor beads (PerkinElmer Life Sciences) and Streptavidin donor beads (PerkinElmer Life Sciences). Briefly, 5 μl of lysate was incubated for 2 h with 15 μl of “acceptor mix” containing 1 nm anti-Syk, 1 nm anti-pSyk, and 10 μg/ml acceptor beads in AlphaLISA immunoassay buffer (PerkinElmer Life Sciences). At the end of the incubation time, the reaction was further incubated for 2 h with “donor mix” containing 40 μg/ml donor beads in AlphaLISA immunoassay buffer. Plates were read on an Envision 2103 multiLabel plate reader (PerkinElmer Life Science) using HTS AlphaScreen label settings. Raw data were designated as “AlphaLISA units” and analyzed using four-parameter log(dose) versus response nonlinear regression with GraphPad Prism version 6 (GraphPad Software, La Jolla, CA). Data are shown as mean ± S.E. from a representative experiment, which was repeated twice.
Lipid-binding assay
ELISA Lipid Snooper® strips were purchased from Avanti Polar Lipids. Strips were first blocked by incubating with 200 μl of blocking buffer (3% fatty acid–free BSA (Sigma) in PBS) for 1 h at room temperature with shaking. Strips were decanted and washed three times with 300 μl of PBS and then incubated with Fc-fused ECD of WT TREM2 (in-house), R47H TREM2 (in-house), or TREM1 (R&D Systems) from 0 to 300 nm in 100 μl of blocking buffer in duplicates. Strips were incubated for 2 h at room temperature with shaking. Strips were decanted and washed three times with 300 μl of PBS and then incubated with 100 μl of horseradish peroxidase–conjugated goat anti-human Fc-γ (Jackson ImmunoResearch) diluted 1:5000 in blocking buffer at room temperature with shaking for 1 h. Strips were decanted and washed three times with 300 μl of PBS and then incubated with 100 μl of TMB substrate (Thermo Fisher Scientific) at room temperature in the dark. Reaction was stopped with stop solution (Thermo Fisher Scientific), and absorbance at 450 nm was recorded using Envision (PerkinElmer Life Sciences). The absorbance readings versus concentrations were plotted and analyzed using four-parameter log(dose) versus response nonlinear regression with GraphPad Prism version 6 (GraphPad Software). The experiment was conducted twice independently.
Author contributions
A. S., E. M., H. C., I. N. F., S. S., and Z. W. conceptualization; A. S. validation; A. S., S. Talreja, J. D., E. B., R. K., J. R., N. S., S. Thibault, J. B., I. N. F., S. S., and Z. W. investigation; A. S. and X. M. visualization; A. S., S. Talreja, J. D., X. M., S. S., and Z. W. writing-original draft; S. Talreja, J. D., R. K., S. Thibault, J. B., I. N. F., S. S., and Z. W. methodology; Z. Z., S. Wood, A.-C. L., H. C., S. S., and Z. W. supervision; S. Wang resources; S. S. project administration.
Supplementary Material
Acknowledgments
We thank Fernando Garçes and Sheree Johnstone for critical reading of the manuscript. We also thank Roman Shimanovich for some liposome preparations. X-ray diffraction experiments were performed on beamline 5.0.2 at the Advanced Light Source with support from the United States Department of Energy under Contract DE-AC03–76SF00098 at the Lawrence Berkeley National Laboratory.
Note added in proof
Jason Richardson and Zhongqi Zhang were inadvertently omitted on the version of this article that was published as a Paper in Press on May 24, 2018. This error has now been corrected.
This work was funded by Amgen Inc., and all authors were employees of Amgen at the time the research was conducted.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S4 and Tables S1–S4.
The atomic coordinates and structure factors (codes 5UD7, 5UD8, and 6B8O) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AD
- Alzheimer's disease
- NHD
- Nasu–Hakola disease
- ECD
- extracellular ligand-binding domain
- sTREM2
- shed or soluble form of TREM2
- PS
- phosphatidylserine
- PLIS
- positive ligand–interacting surface
- asu
- asymmetric unit
- SASA
- solvent-accessible surface area
- Sc
- shape complementarity
- CDR
- complementarity-determining region
- r.m.s.
- root mean square
- NAG
- N-acetylglucosamine
- ssODN
- single-stranded oligonucleotide
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine sodium salt.
References
- 1. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., Rujescu D., Hampel H., Giegling I., Andreassen O. A., Engedal K., et al. (2013) Variant of TREM2 associated with the risk of AD. N. Engl. J. Med. 368, 107–116 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atagi Y., Liu C.-C., Painter M. M., Chen X.-F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S. S., Xu H., Younkin S., Das P., Fryer J. D., and Bu G. (2015) Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem. 290, 26043–26050 10.1074/jbc.M115.679043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey C. C., DeVaux L. B., and Farzan M. (2015) The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J. Biol. Chem. 290, 26033–26042 10.1074/jbc.M115.677286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeh F. L., Wang Y., Tom I., Gonzalez L. C., and Sheng M. (2016) TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-β by microglia. Neuron 91, 328–340 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y., Cella M., Mallinson K., Ulrich J. D., Young K. L., Robinette M. L., Gilfillan S., Krishnan G. M., Sudhakar S., Zinselmeyer B. H., Holtzman D. M., Cirrito J. R., and Colonna M. (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160, 1061–1071 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi K., Rochford C. D. P., and Neumann H. (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657 10.1084/jem.20041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poliani P. L., Wang Y., Fontana E., Robinette M. L., Yamanishi Y., Gilfillan S., and Colonna M. (2015) TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125, 2161–2170 10.1172/JCI77983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Q., Zhang K., Jin Y., Zhu T., Cheng B., Shu Q., and Fang X. (2013) Triggering receptor expressed on myeloid cells-2 protects against polymicrobial sepsis by enhancing bacterial clearance. Am. J. Respir. Crit. Care Med. 188, 201–212 10.1164/rccm.201211-1967OC [DOI] [PubMed] [Google Scholar]
- 9. Read C. B., Kuijper J. L., Hjorth S. A., Heipel M. D., Tang X., Fleetwood A. J., Dantzler J. L., Grell S. N., Kastrup J., Wang C., Brandt C. S., Hansen A. J., Wagtmann N. R., Xu W., and Stennicke V. W. (2015) Cutting edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J. Immunol. 194, 1417–1421 10.4049/jimmunol.1402303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kleinberger G., Yamanishi Y., Suarez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., Tahirovic S., Lleo A., Alcolea D., Fortea J., Willem M., et al. (2014) TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6, 243ra86 10.1126/scitranslmed.3009093 [DOI] [PubMed] [Google Scholar]
- 11. Yuan P., Condello C., Keene C. D., Wang Y., Bird T. D., Paul S. M., Luo W., Colonna M., Baddeley D., and Grutzendler J. (2016) TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 90, 724–739 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kober D. L., Alexander-Brett J. M., Karch C. M., Cruchaga C., Colonna M., Holtzman M. J., and Brett T. J. (2016) Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. eLife 5, e20391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kober D. L., Wanhainen K. M., Johnson B. M., Randolph D. T., Holtzman M. J., and Brett T. J. (2014) Preparation, crystallization, and preliminary crystallographic analysis of wild-type and mutant human TREM-2 ectodomains linked to neurodegenerative and inflammatory diseases. Protein Expr. Purif. 96, 32–38 10.1016/j.pep.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlepckow K., Kleinberger G., Fukumori A., Feederle R., Lichtenthaler S. F., Steiner H., and Haass C. (2017) An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol. Med. 9, 1356–1365 10.15252/emmm.201707672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thornton P., Sevalle J., Deery M. J., Fraser G., Zhou Y., Ståhl S., Franssen E. H., Dodd R. B., Qamar S., Gomez Perez-Nievas B., Nicol L. S., Eketjäll S., Revell J., Jones C., Billinton A., et al. (2017) TREM2 shedding by cleavage at the H157–S158 bond is accelerated for the Alzheimer's disease-associated H157Y variant. EMBO Mol. Med. 9, 1366–1378 10.15252/emmm.201707673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleinberger G., Brendel M., Mracsko E., Wefers B., Groeneweg L., Xiang X., Focke C., Deussing M., Suárez-Calvet M., Mazaheri F., Parhizkar S., Pettkus N., Wurst W., Feederle R., Bartenstein P., et al. (2017) The FTD-like syndrome causing TREM2 T66M mutation impairs microglia function, brain perfusion, and glucose metabolism. EMBO J. 36, 1837–1853 10.15252/embj.201796516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song W. M., Joshita S., Zhou Y., Ulland T. K., Gilfillan S., and Colonna M. (2018) Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215, 745–760 10.1084/jem.20171529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawrence M. C., and Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 10.1006/jmbi.1993.1648 [DOI] [PubMed] [Google Scholar]
- 19. Chothia C., and Janin J. (1975) Principles of protein–protein recognition. Nature 256, 705–708 10.1038/256705a0 [DOI] [PubMed] [Google Scholar]
- 20. Zhao Y., Li X., Huang T., Jiang L.-L., Tan Z., Zhang M., Cheng I. H.-J., Wang X., Bu G., Zhang Y.-W. Wang Q., and Xu H. (2017) Intracellular trafficking of TREM2 is regulated by presenilin 1. Exp. Mol. Med. 49, e405 10.1038/emm.2017.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iborra S., and Sancho D. (2015) Signalling versatility following self and non-self sensing by myeloid C-type lectin receptors. Immunobiology 220, 175–184 10.1016/j.imbio.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Païdassi H., Tacnet-Delorme P., Garlatti V., Darnault C., Ghebrehiwet B., Gaboriaud C., Arlaud G. J., and Frachet P. (2008) C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition 1. J. Immunol. 180, 2329–2338 10.4049/jimmunol.180.4.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tietjen G. T., Gong Z., Chen C.-H., Vargas E., Crooks J. E., Cao K. D., Heffern C. T. R., Henderson J. M., Meron M., Lin B., Roux B., Schlossman M. L., Steck T. L., Lee K. Y. C., and Adams E. J. (2014) Molecular mechanism for differential recognition of membrane phosphatidylserine by the immune regulatory receptor Tim4. Proc. Natl. Acad. Sci. U.S.A. 111, E1463–E1472 10.1073/pnas.1320174111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leslie A. G. W., and Powell H. R. (2007) Processing diffraction data with MOSFLM. Evol. Methods Macromol. Crystallogr. 245, 41–51 [Google Scholar]
- 25. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 30. Shah B., Jiang X. G., Chen L., and Zhang Z. (2014) LC-MS/MS peptide mapping with automated data processing for routine profiling of N-glycans in immunoglobulins. J. Am. Soc. Mass Spectrom. 25, 999–1011 10.1007/s13361-014-0858-3 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Z. (2009) Large-scale identification and quantification of covalent modifications in therapeutic proteins. Anal. Chem. 81, 8354–8364 10.1021/ac901193n [DOI] [PubMed] [Google Scholar]
- 32. Zhang Z., and Shah B. (2010) Prediction of collision-induced dissociation spectra of common N-glycopeptides for glycoform identification. Anal. Chem. 82, 10194–10202 10.1021/ac102359u [DOI] [PubMed] [Google Scholar]
- 33. Cheng Q., Danao J., Talreja S., Wen P., Yin J., Sun N, Li C.-M., Chui D., Tran D., Koirala S., Chen H., Foltz I. N., Wang S., and Sambashivan S. (2018) J. Biol. Chem. 293, 12620–12633 10.1074/jbc.RA118.001848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.