Abstract
Omega-3 polyunsaturated fatty acids (PUFA) are produced in some unicellular organisms, such as marine gammaproteobacteria, myxobacteria, and thraustochytrids, by large enzyme complexes called PUFA synthases. These enzymatic complexes resemble bacterial antibiotic-producing proteins known as polyketide synthases (PKS). One of the PUFA synthase subunits is a conserved large protein (PfaA in marine proteobacteria) that contains three to nine tandem acyl carrier protein (ACP) domains as well as condensation and modification domains. In this work, a study of the PfaA architecture and its ability to initiate the synthesis by selecting malonyl units has been carried out. As a result, we have observed a self-acylation ability in tandem ACPs whose biochemical mechanism differ from the previously described for type II PKS. The acyltransferase domain of PfaA showed a high selectivity for malonyl-CoA that efficiently loads onto the ACPs domains. These results, together with the structural organization predicted for PfaA, suggest that this protein plays a key role at early stages of the anaerobic pathway of PUFA synthesis.
Keywords: fatty acid synthase (FAS), polyketide, polyunsaturated fatty acid (PUFA), acyl carrier protein (ACP), acyltransferase
Introduction
Polyunsaturated fatty acids (PUFA)2 are molecules with beneficial properties for human health synthesized among eukaryotic organisms by desaturation and elongation of shorter fatty acids (1, 2). However, it has been reported that the heterotrophic fungus-like thraustochytrids, some myxobacteria, and some marine gammaproteobacteria are able to de novo synthesize PUFAs from acyl-CoA precursors (3). These organisms accumulate PUFAs like eicosapentaenoic acid (EPA; 20:5) or docosahexaenoic acid (DHA; 22:6) by using modular proteins of the Pfa family, which show a modular organization similar to that of polyketide synthases (PKS) and fatty acid synthases (FAS) (4). Thus, current knowledge of PKS and FAS domains could help in the understanding of the mechanism by which PUFA synthases work. Pfa systems are formed by three (thraustochytrids and myxobacteria) or four polypeptides (gammaproteobacteria). In marine bacteria like Moritella marina, which we used as a model, these proteins are named PfaA, PfaB, PfaC, and PfaD (5). PfaD is predicted to contain a singular enoyl reductase (ER) domain (6), whereas PfaA, PfaB, and PfaC are multimodular proteins, each of them formed by a series of domains that show sequence and structural homology with the ones present in PKS and FAS systems. These domains can be classified into condensing and modifier motifs according to their predicted biochemical function (7). The condensing domains are responsible for extending the fatty acid chain. They comprise acyltransferases (AT) that select the acyl building blocks, and keto synthases (KS) that polymerize these extender units by a condensation reaction. The fatty acid intermediates are held by the acyl carrier protein domains (ACP) that transfer them to the different catalytic domains. Thereupon the modifier domains, keto reductases (KR), dehydratases (DH) and enoyl reductases (ER), reduce intermediate keto groups to form partially or fully saturated fatty acid moieties. Unlike FAS systems, Pfa proteins have redundant domains (KS, AT, ACP, and DH), whose roles are not currently clear, so there is a growing interest in understanding its biological function.
One of the first steps in FAS and PKS synthesis is the selection of extender blocks through the coordinated action of ACP and AT. For this process, ACP has to be previously activated with a 4′-phosphopantetheine prosthetic group (PPT) that acts as a linkage between the ACP active serine and the acyl group (8). ACP activation in PUFA synthases is performed by a specific 4′-phosphopantetheine transferase (PfaE in M. marina). Transfer of extender units to the terminal sulfhydryl group of PPT is carried out by an AT domain, at least in FAS and modular PKS (9). However, it has been reported the self-acylation ability of ACP in iterative PKS systems, being ACP capable of selecting the extender blocks in the absence of AT (10).
The main striking feature of Pfa systems is the presence of ACP domains in tandem, which can have from 3 to 9 repetitions with sequence identity ranging from 85 to 96% (11). A higher number of ACP domains within this tandem arrangement is directly related with improvements in the productivity of the system (12, 13). These tandem ACP domains are located in the PfaA polypeptide in all omega-3 PUFA synthases. PfaA shows a KS–AT condensing module at their N-terminal half, followed by the tandem ACPs and a KR–DH C-terminal domain (3).
The critical process for the polymerization of a polyketide or a fatty acid is the correct selection of the extender units through a collaborative process that involves AT and ACPs. Different extension units have been described in PKS synthesis (14), however, FAS systems show less complexity, being primarily malonyl the extender unit of choice (15). For the selection of these building blocks the presence of specific AT is necessary. Pfa systems have two different AT domains, one in PfaA, and a second one in PfaB. This domain was reported to be involved in the determination of the final product of the omega-3 PUFA synthesis (16), but its specific biochemical activity has never been proven, neither has there been clarify of the differential activity of both AT domains.
In the present article we studied the loading of extender blocks onto the tandem ACPs of PfaA. The presence of two AT domains (PfaA AT and PfaB AT) that had not been fully characterized suggested to us the need of a more comprehensive study. Here, we researched the AT substrate preference, mechanism of action, and interaction with the PfaA tandem ACPs, which are critical aspects to understand the omega-3 PUFA synthesis process by the Pfa family.
Results and discussion
PfaA domain organization is conserved in all omega-3 PUFA synthases
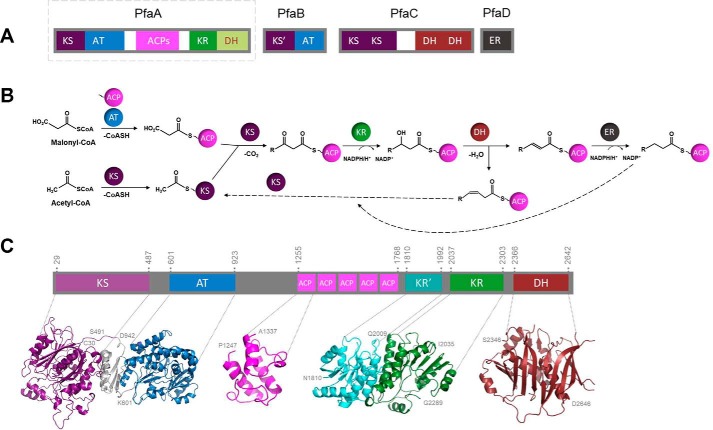
Pfa gene clusters of marine bacteria like M. marina (Fig. 1A) are formed by 3 genes that code for multimodular proteins (PfaA, PfaB, and PfaC) and a fourth gene that codes for a single protein domain (PfaD). Like in PKS or FAS, the basic domain architecture contains KS, AT, ACP, KR, and DH domains. Despite the general lack of biochemical evidences to assign a role to each redundant domain of the Pfa system, a general outline is normally assumed (4). An AT domain should load the malonyl extender units onto the ACPs, a KS domain adds these blocks to the growing chain by successive condensations and the KR, DH, and ER domains modify the keto groups formed in each condensation to form the fatty acid-unsaturation pattern (Fig. 1B). Besides the redundancy of KS and DH domains in omega-3 PUFA synthases, the main difference between its biochemical pathways and that of PKS are the acyl precursors used during the synthesis as well as the characteristic cyclization patterns of PKS that involves specific enzymes.
Figure 1.
PfaA analysis and general biochemical route. A, general scheme of the pfa genetic cluster of M. marina. Each domain is represented with a specific color. Domain regions correspond to KS, AT, acyl carrier proteins (5ACP), KR, DH, and ER. PfaA that will be further analyzed is squared with a dashed line. B, overview of the biochemical route for omega-3 PUFA synthesis. The dashed arrows represent the beginning of a new synthesis cycle. C, PfaA domain organization. Prediction of folding domains within the PfaA protein sequence made with InterPro. Each domain is represented with a specific color. Structural predictions made with Phyre2 of each domain are shown in the lower panel using the same color code. The first and last amino acid of each domain prediction is indicated in the figure. Only the structure of one of the five ACPs has been represented as they are similar.
To analyze the M. marina PfaA (mmPfaA) protein sequence and make a preliminary prediction of its domains, the InterPro online server (17) was used. The predicted domain organization of mmPfaA (Fig. 1C) shows five different folding motifs along its 2,652 amino acids. From the N-terminal to the C-terminal domain the predictions were as follows: a KS domain (aa 29–487), an AT domain (aa 601–923), five ACP domains in a tandem arrangement with an overall sequence identity of 82% (aa 1255–1768), two KR domains (aa 1810–1992 and 2037–2303), and a DH domain (aa 2366–2642).
Structural predictions made with Phyre2 (18) by using closely related solved structures as templates (listed under “Experimental procedures”) also support the identity of the defined domains (Fig. 1C). We have found a duplication of the thiolase-fold, typical of KS domains (19) represented in purple, and a ferredoxin-like subdomain (in gray) followed by an α/β hydrolase-fold that corresponds to the AT domain (20, 21) represented in blue. Five repetitions of the four α-helices in an up-down-down topological arrangement that correspond to the ACPs (22) are represented in pink. Two repetitions of the Rossman-fold (23) with a twisted, parallel β-sheet composed of seven β-strands flanked in both sides by eight α-helices that correspond to two KR domains are represented in cyan and green and a repetition of the hotdog-fold (24) that corresponds to the DH domain represented in red.
Comparisons with characterized similar domains helped us to identify the residues presumably involved in catalysis. The KS active site is formed by the Cys229–His364–His404 triad involved in the condensation reaction, whereas the AT active residue responsible of the extender unit binding was identified at Ser703 (Fig. S1). The five serine active residues of the ACPs responsible for PPT binding were localized at Ser1293, Ser1395, Ser1496, Ser1597, and Ser1703, respectively (Fig. S2). KR catalytic triad is formed by the Lys2195–Ser2219–Tyr2232 residues, whereas Asn2236 is involved in the formation of the proton-wire (Fig. S3). We have predicted the existence of two differentiated KR-like structures identified as KR′–KR, where KR′ is a pseudo-KR domain that lacks sequence homology with any described catalytic KR, but shows structural homology with KR domains of PKS type I and FAS systems. KR′ does not contain any of the catalytic residues conserved in KR domains (25, 26), indicating that this KR′ domain is a structural element of PfaA. This full KR′–KR domain is present in the marine bacteria analyzed but partially absent in myxobacteria and thraustochytrids. Within the KR catalytic domain, the Asn/Lys catalytic dyad involved in proton replenishment (Asn2236 and Lys2195 in PfaA) is swapped with respect to KR domains from PKS type II (27). This swapping also occurs in mammalian FAS and PKS type I, which indicates an evolutionary relationship of the PfaA protein with FAS and PKS type I systems (Fig. S4). Finally, His2379–Asp2563 (Fig. S5) are predicted to be the active residues of the C-terminal DH domain involved in the reduction of the hydroxyl group generated at the keto reduction step. This C-terminal DH domain was not described in most of the previous publications that assume the existence of a larger KR domain (5, 7, 12, 16, 28). We have proved here the conservation of the residues conforming the active sites of all predicted domains by doing sequence alignments with distant homologs of FAS and PKS systems and also with homologous proteins of gammaproteobacteria, myxobacteria, and thraustochytrids (Figs. S1–S5), all of them previously reported omega-3 PUFA producers (29).
Thus, our bioinformatic analysis predicted a KS–AT–ACP(x5)–KR′–KR–DH organization in PfaA. This organization is conserved among PfaA-like proteins, also named PFA1 in thraustochytrids, or Pfa2 in myxobacteria (3, 4) although the number of ACP domains is variable and the KR′ structural domain is absent in some systems. In fact, ACP repetitions can vary from three in Shewanella pealeana to nine in some thraustochytrids (11, 30). The conservation of this scheme indicates that this particular structural conformation may be essential for the proper function of the PUFA synthase protein complex.
Tandem ACPs of PfaA are self-acylated with malonyl-CoA
An essential role in polyketide or FA synthesis is performed by ACP domains. These domains determine the final product by selecting and carrying the extender units that will be condensed and modified by other enzymatic Pfa domains. PfaA-like proteins show the peculiarity of having a variable number of tandem ACP domains. Thus, we were interested in studying the specificity of these ACP domains within PUFA synthases to understand the synthesis process.
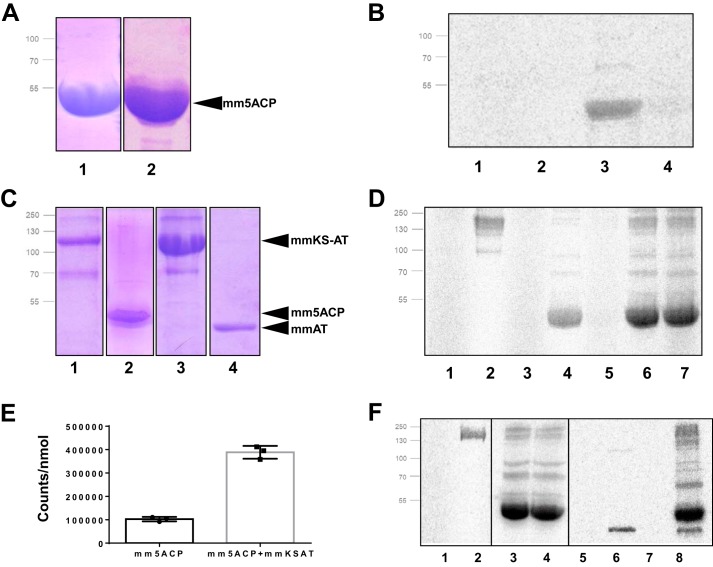
Based on the information obtained by in silico analysis, we have cloned into the expression vector pET29c a DNA fragment coding for the predicted 5 tandem ACP domains of M. marina PfaA, as described under “Experimental procedures.” Using this construction, pET29c::5ACP, we have purified these 5 tandem ACP domains in their apo form. The tandem ACPs construction was also co-expressed with mmPfaE, the phosphopantetheinyl transferase (PPTase) of M. marina. mmPfaE was cloned into an expression vector with a different antibiotic resistance (pET3a vector), so that we could directly purify the active ACP holo-form by coexpression of pET29c:5ACP and pET3a:PfaE plasmids (Fig. 2A).
Figure 2.
Ligand-binding assays of mm5ACP, mmKS-AT, and mmAT domains. A, 12% SDS-PAGE gel containing 20 μg of purified mm5ACP in its holo- (lane 1) and apo- (lane 2) forms. B, 20 μm mm5ACP incubated with [14C]malonyl-CoA and [14C]acetyl-CoA and analyzed by 12% radio-SDS-PAGE. Lanes 1, apo-5ACP + malonyl-CoA; 2, apo-5ACP + acetyl-CoA; 3, holo-5ACP + malonyl-CoA; 4, holo-5ACP + acetyl-CoA. C, 12% SDS-PAGE gel containing purified mmKS-AT (lane 1), holo-mm5ACP (lane 2), mmKS*(C229A)-AT (lane 3), and mmAT (lane 4). D, same domains incubated with [14C]malonyl-CoA and [14C]acetyl-CoA, and analyzed by 12% radio-SDS-PAGE. Lanes 1, mmKS-AT + acetyl-CoA; 2, mmKS-AT + malonyl-CoA; 3, holo-5ACP + acetyl-CoA; 4, holo-5ACP + malonyl-CoA; 5, holo-mm5ACP + mmKS-AT + acetyl-CoA; 6, holo-mm5ACP + mmKS-AT + malonyl-CoA; 7, holo-mm5ACP + mmKS-AT + acetyl-CoA + malonyl-CoA; E, quantification of the protein band intensities of holo-mm5ACP + malonyl-CoA (D, lane 4) and holo-mm5ACP + mmKS-AT + malonyl-CoA (D, lane 6) by densitometric scanning, normalized with respect to the molarity of ACP in each sample. F, mmKS(C229A)-AT mutant and mmAT isolated domain were incubated with [14C]malonyl-CoA and [14C]acetyl-CoA, and analyzed by 12% radio-SDS-PAGE. Lanes 1, mmKS(C229A)-AT + acetyl-CoA; 2, mmKS(C229A)-AT + malonyl-CoA; 3, holo-mm5ACP + mmKS(C229A)-AT + malonyl-CoA; 4, holo-mm5ACP + mmKS(C229A)-AT + malonyl-CoA + acetyl-CoA; 5, mmAT + acetyl-CoA; 6, mmAT + malonyl-CoA; 7, holo-mm5ACP + mmAT + acetyl-CoA; 8, holo-mm5ACP + mmAT + malonyl-CoA. Protein mapping positions are indicated in A and C by black arrows.
To investigate the mmACP substrate preference and test their possible ability to transfer and incorporate acyl groups, we incubated 20 μm of purified apo- or holo-mm5ACP in 20-μl reactions with 14C-radiolabeled malonyl-CoA or 14C-radiolabeled acetyl-CoA. Results (Fig. 2B) indicate that the holo-form of mm5ACP was able to perform self-acylation with malonyl-CoA (lane 3) but did not show activity against acetyl-CoA (lane 4). This catalytic activity associated with holo-ACPs has been extensively studied in the literature, being normally linked to PKS systems but not widespread in FAS (10, 31–34). A couple of examples of FAS proteins with self-acylation activity have also been described (10), specifically from Plasmodium falciparum and Brassica napus, being thus ACP self-acylation not exclusive from PKS systems. The self-malonylation ability of mm5ACP from PUFA synthases pointed out for the first time a functional homology of these domains with the ones present in iterative PKS and other self-acylating systems. mm5ACP purified in the absence of PfaE, apo-mm5ACP, did not react with any of the two substrates analyzed (Fig. 2, lanes 1 and 2). Thus, mmPfaE seems to be essential for mm5ACP PPT binding and cannot be substituted by the E. coli homolog 4′-phosphopantetheinyl transferase (accession number P37623).
We have demonstrated the ACP ability to be self-malonylated in its natural tandem arrangement form. This activity has also been reported for ACP domains of PKS II and some FAS systems. Unlike FAS systems, auto-malonylation activity is necessary in PKS II due to the absence of an associated AT domain (32). However, Pfa systems include one or two AT domains in their architecture, so, the presence of independent self-acylating ACPs in PUFA synthases was not foreseen. The ACPs self-malonylation ability proved here provides additional evidence of the evolutionary position of PUFA synthases between FAS and PKS systems. Besides that, the demonstrated ACP relative independence raises questions about the actual role of the cis- and trans-AT domains (PfaA AT and PfaB AT domains, respectively) that may not be essential to the synthesis process.
PfaA AT domain promotes malonyl-CoA binding to ACPs
The core components required to produce a regular fatty acid condensation contain three domains, KS, AT, and ACP, that perform one cycle of chain extension. An acetyl group or starter unit is attached to the cysteine thiol of the KS domain and the chain extender unit is selected by the AT and bound to a thiol residue of the ACPs PPT where is ready for condensation. Thus, AT is the domain responsible for selecting the molecules that will be polymerized to form the backbone of the fatty acid (Fig. 1B). The extender unit is malonate in FAS, but can be malonate, methylmalonate, ethylmalonate, or other extender units in PKSs. Although we have demonstrated that mmPfaA ACPs are able to perform self-malonylation we also checked if the mmPfaA AT domain stimulates this malonylation.
Because the KS–AT didomain usually functions as a tightly coordinated protein (25), both mmPfaA domains (mmKSAT) were initially included in this experiment as a single polypeptide. The concentration of proteins and substrates was experimentally established to improve visualization of the bands. As described under “Experimental procedures,” 5 μm mmKSAT, 20 μm mm5ACP, and 20 μm 14C-radiolabeled malonyl-CoA or [14C]acetyl-CoA were incubated in a 20-μl reaction volume. mmKSAT was able to bind covalently 14C-radiolabeled malonyl (Fig. 2D, lane 2). However, under similar conditions [14C]acetyl binding to mmKSAT was not observed (Fig. 2D, lane 1). The mmKSAT didomain was subsequently incubated with both substrates in the presence of mm5ACP. When mmKSAT was mixed with mm5ACP and malonyl-CoA (Fig. 2D, lane 6), a stronger radioactive signal was detected, bound to mm5ACP, demonstrating that the mmKSAT didomain transfers the malonyl extender units to the mmACPs. Quantification of the mm5ACP band intensity showed a significant increase of 3.5 ± 0.68 times when mmKS-AT is added to the experiment (Fig. 2E). 14C-Radiolabeled acetyl was not transferred to ACP (Fig. 2D, lane 5) and the presence of both 14C-radiolabeled acetyl and 14C-radiolabeled malonyl did not change the binding activity (lane 7).
To prove that this overloading effect was due to the activity of the AT domain within the mmKSAT didomain, an inactive mutant for the KS active site (C229A) was used. We observed that this mutant was perfectly able to bind malonyl by itself (Fig. 2F, lane 2) and transfer this malonyl to ACP, when present (lane 3). As expected, acetyl-CoA does not have any effect on the reaction (Fig. 2D, lanes 1 and 4). As a complement to this experiment, we designed a construction carrying only the AT domain. The isolated AT domain (mmAT) was also able to bind malonyl but not acetyl (Fig. 2D, lanes 6 and 5, respectively). Moreover, mmAT was effectively loaded only the malonyl groups units, not the acetyl groups, onto mm5ACP (lanes 8 and 7, respectively), demonstrating structural and functional independence from its KS partner.
PfaA AT domain was highly specific for malonyl-CoA and able to load these extender units onto its adjacent ACPs. Despite the ACP self-malonylation activity, PfaA AT domain was needed to reach a higher level of malonyl-ACP (Fig. 2D, lanes 4 and 6). This fact highlights that ACPs are not fully independent, thus the adjacent AT domain could be able to modulate the ACPs acylation state during the omega-3 PUFA synthesis. On the other hand, as the ACP domain is present from three to nine times in PfaA-like proteins, its stoichiometry in relationship with the AT domain must be from 3:1 to 9:1, so the ACPs self-acylating ability may have emerged to drive high reaction rates, as was previously suggested in the literature in relationship with its tandem organization (11–13).
Surprisingly, and as a remarkable difference with FAS systems, we were not able to demonstrate acetyl binding with any of the domains analyzed. In all previously described models for FAS it is always necessary to prime the system with acetyl units to perform the first decarboxylative condensation (35). Biosynthesis in bacterial aromatic PKS is initiated by priming acetate as starter unit, but some aromatic PKSs can use different starter units (36) or prepare their starter units through decarboxylation mechanisms (37). Because Pfa are hybrid systems located diffusely between FAS and PKS, it is difficult to speculate about the nature of this initiator system. Despite having defined the ability of mmAT to activate the ACPs with malonyl, a more comprehensive characterization of the system is needed to elucidate how the acetyl seed is incorporated to the nascent chain (in case such starter molecule is necessary).
A comparative study reveals evolutionary divergences between ACPs
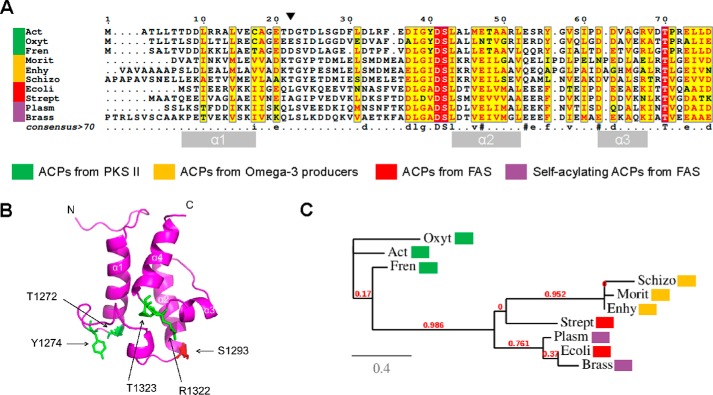
To study the mmACP conserved residues that may be involved in the protein function, we have performed a multiple sequence alignment (Fig. S2), which includes representative single ACP domains of PfaA-like proteins from gammaproteobacteria, myxobacteria, and thraustochytrids. Three conserved motifs were found in the alignment. The active site motif contains the conserved serine involved in PPT binding, the “TGY” conserved motif, and the “RT” dyad. The structure of each of the five mmACP domains is predicted to be a four-helix bundle (Fig. 1C) like all the ACP structures solved so far. The three conserved motifs are located at the same side of the bundle with TGY and the active site between α1 and α2 and RT between α3 and α4 (Fig. 3B). Based on this preliminary analysis we have compared the first ACP domain of M. marina (mmACP1; aa Ile2155–Ala1348) and other ACPs from marine bacteria with domains of homologous PKS and FAS systems, including the two exceptional examples with self-acylation activity (P. falciparum and B. napus) (Fig. 3A). The results of this analysis revealed that mmACP1 Thr1272 of the TGY conserved motif maps at the position where PKS II homologs conserve a negatively charged Asp/Glu residue and FAS proteins show hydrophobic residues. We did not find any conservation pattern at the position of mmACP1 residue Tyr1274 of the TGY conserved motif. RT dyad residue Thr1323 of mmACP1 is fully conserved across all domains analyzed and the conservation of Arg1322 found in omega-3 PUFA producers was not observed in FAS nor PKS, having indistinctly neutral or charged residues in that position.
Figure 3.
Comparative analysis of representative ACP domains from different organisms. A, multiple sequence alignment of representative ACPs from PKSII, omega-3 PUFA synthases, and FAS. Alignment names correspond to actinorhodin PKSII ACP (Act) from S. coelicolor (UniProt accession number Q02054), oxytetracycline PKSII ACP (Oxyt) from Streptomyces rimosus (accession number P43677), frenolicin PKSII ACP (Fren) from Streptomyces roseofulvus (accession number Q54996), PfaA ACP1 (Morit) from M. marina (accession number Q9RA21), Pfa ACP1 (Enhy) from Enhygromyxa salina (accession number A0A0C2CTM9), Pfa1 ACP1 (Schizo) from Schizochytrium ATCC 20888 (accession number AAK72879), ACP (E. coli) from E. coli FAS (accession number P0A6A8), and ACP (Strept) from Streptomyces coelicolor (accession number P72393), ACP (Plasm) from P. falciparum (accession number O77077), and ACP (Brass) from B. napus (accession no. P17650). Identical residues are shown in white on a red background, whereas similar residues are shown in red. The position of the “D” motif, characteristic of self-acylating ACPs, has been marked with a black arrow. Left panel indicates with a color code the type of organism in each sequence. Position of α-helices is represented with gray boxes at the bottom of the alignment. B, predicted structure of mmACP1 from M. marina. The four residues selected for the alanine screening are highlighted in green and the active serine in red. C, phylogram representation of all the previous sequences, showing evolutive divergence between them.
As a result of the previous analysis, the threonine and tyrosine of the TGY conserved motif and the RT dyad (Fig. S2) were taken into consideration as possible residues involved in the acylation of the ACPs. WT holo-mmACP1 as well as four holo-mmACP1 constructions with alanine substitutions in these four residues (Thr1272, Tyr1274, Arg1322, and Thr1323) completely conserved among omega-3 PUFA producers (Fig. 3B) were designed and purified to test their possible implication in the ACP malonylation process.
First the self-malonylation capacity of the mmACP1 protein was analyzed. As shown in Fig. S6A, mmACP1 was malonylated in the presence of 14C-radiolabeled malonyl-CoA as described for mm5ACP. Thus, mmACP self-malonylation activity does not depend on the tandem organization of mm5ACP, rather it seems to be an intrinsic property of each domain. As expected, apo-mmACP1 did not react with malonyl-CoA. We have then assayed the self-malonylation of the four mmACP1 mutants (Fig. S6A, lanes 3–6). All four mutants showed a similar ability to covalently bind malonyl by themselves with the same enzymatic efficiency (Fig. S6B). Finally, this single mmACP1 domain and its four mutants were tested as substrates of the mmKSAT didomain in binding assays with 14C-radiolabeled malonyl and purified mmKSAT (Fig. S6C). We have observed that mmACP1 and each of the four mutants were identically loaded with malonyl by mmKSAT (Fig. S6D).
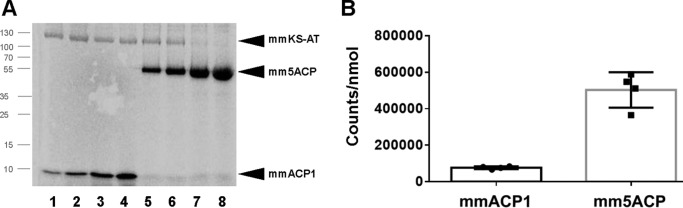
When the binding activity of mmACP1 and mm5ACP was compared in the presence of mmKSAT (Fig. 4A), the mm5ACP-bound radioactive malonyl-CoA/mol was 6.6 ± 1.7 times higher than in mmACP1 (Fig. 4B). This result is consistent with a simultaneous occupation of the 5 ACP active sites, each being of the five ACP domains of mm5ACP able to be malonylated by PfaA AT.
Figure 4.
Ligand-binding assays of mmACP1 and mm5ACP with mmKS-AT. A, 12% radio-SDS-PAGE gel containing 2.2 μg of mmKS-AT and 0.32, 0.64, 1.6, or 3.2 μg (lanes 1–4) of mmACP1 or 0.8, 0.8, 2.0, or 4.0 μg (lanes 5–8) of mm5ACP incubated 1 h with [14C]malonyl-CoA. Protein mapping positions are indicated by black arrows. B, quantification of the protein band intensities was by densitometric scanning, normalized with respect to the molarity of ACP in each sample.
The autoacylation ability of ACPs was originally linked to the presence of several conserved arginine residues that are poorly conserved among non-self-acylating FAS (33). Moreover, Tyr56 and Arg72 of the actinorhodin PKS were further described to be not only directly related to self-acylation capacity but with the ability to transfer the malonyl groups to other ACPs (32, 38). Finally, it was demonstrated that at least in PKS type II the self-malonylation was promoted by an acidic Asp/Glu residue and helped by some Thr residues (10). All these amino acids are located at the putative binding clefts of the ACPs and more specifically within the disordered loops connecting the α helices, which are presumably involved in the protein–protein and protein–substrate recognition processes (10). Despite our efforts to find homologous residues conserved among omega-3 PUFA producers, we could not identify any of them involved in catalysis. These results lead us to think that we are in front of a substantially different mechanism from the one described for PKS or FAS proteins, so there should exist evolutionary divergences between Pfa self-acylation mechanisms and other self-acylating systems. To analyze this divergent process, a phylogenetic tree was made (Fig. 3C), including the sequences used in the multiple sequence alignment of Fig. 3A. It can be noticed that an evolutionary proximity between all the analyzed FAS ACPs, which are closely related to the ones from the Pfa systems, are both distant from PKSII. These results evidence either a loss of the self-acylation ability for most of the FAS systems during evolution or an alternative emergence of this capacity within PUFA systems.
Apo-mmACP1 was also purified to analyze the efficiency of the phosphopantetheinylation by PfaE. Using high definition mass spectrometry, as described under “Experimental procedures,” we observed in the sample containing the purified apo-mmACP1 a main peak of 11,352 Da corresponding to the theoretical molecular weight of mmACP1 (Fig. S7). A main peak of 11,692 Da, a theoretical molecular mass of mmACP1 plus PPT, was found in the sample containing holo-mmACP1. The presence of apo-ACP was not detected in the holo-ACP sample (Fig. S7), thus confirming that mmPfaE is the phosphopantetheinylase (PPTase) of mmACP and that the reaction is 100% effective.
mmPfaE modeling by Phyre2 predicted a typical PPTase structure like that of the Bacillus subtillis PPTase named Spf (PDB accession number 1QR0) (Fig. S8). In fact, Trujillo et al. (30) showed that the Photobacterium profundum mmACP homolog is effectively phosphopantetheinylated by B. subtillis Spf. In that work they also detected some in vivo phosphopantetheinylation by the Escherichia coli PPTase, a reaction that was not detected in our experiments. Spf interacts with the second helix of ACP by residues around its first (α4 in Fig. S8) and third helix (α6) (39). Then the Mg2+ ion coordinated by residues Asp107, Glu109, and Glu151 catalyzes the PPTase reaction. As shown in Fig. S8, whereas the catalytic residues are conserved in mmPfaE as well as in E. coli, residues involved in ACP interaction are not 100% conserved. These differences could be responsible for the necessity of a specific PfaE protein in PUFA synthases.
PfaA AT is the main malonyltransferase domain within the Pfa cluster
Together with the AT domain of PfaA, there is another putative AT domain in the pfa cluster of M. marina, the AT domain of PfaB. PfaB is a 883-aa protein that determines the final product in the EPA/DHA biosynthesis (16), although the molecular mechanism is unknown. Its sequence contains a structural pseudo-KS motif in DHA producers like M. marina that is absent in EPA producers like Shewanella baltica, and an AT domain at the C-terminal position (Fig. S9).
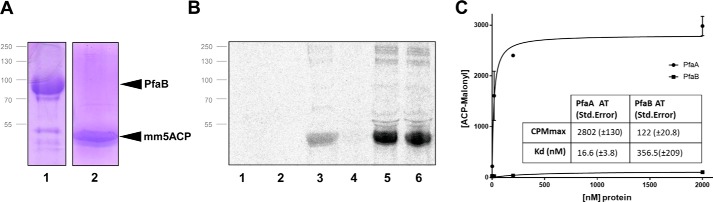
PfaB activity and specificity was studied using the same assay than the one used for the PfaA KS–AT didomain. When PfaB was incubated with 14C-radiolabeled acetyl-CoA or malonyl-CoA (Fig. 5B, lanes 1 and 2, respectively), the presence of a radioactive acyl-PfaB covalent complex was not detected. When PfaB was incubated with [14C]malonyl-CoA in the presence of ACPs, an increase in the intensity of the malonyl-ACP band was detected (lane 5). However, the PfaB concentration used for this experiment was 50 μm; 5 times higher than that used for KSAT PfaA in the previous experiment, suggesting that PfaB may show a lower affinity for malonyl-CoA. As with PfaA, no activity against acetyl-CoA was detected (lane 4) and no changes in the binding pattern in the presence of both substrates either (lane 6). Self-malonylation of ACPs was again included in the experiment as a positive control (lane 3).
Figure 5.
Binding assays of mmPfaB and mm5ACP. A, 12% SDS-PAGE containing the purified mmPfaB (lane 1) and holo-mm5ACPs proteins (lane 2). Protein mapping positions are indicated with black arrows. B, mmPfaB and holo-mm5ACPs were incubated with radiolabeled substrates [14C]malonyl-CoA and [14C]acetyl-CoA and analyzed by 12% radio-SDS-PAGE. Lanes 1, mmPfaB + acetyl-CoA; 2, mmPfaB + malonyl-CoA; 3, holo-mm5ACP + malonyl-CoA; 4, holo-mm5ACP + mmPfaB + acetyl-CoA; 5, holo-mm5ACP + mmPfaB + malonyl-CoA; 6, holo-mm5ACP + mmPfaB + acetyl-CoA + malonyl-CoA. C, determination of ATs dissociation constants. The AT concentration dependence of [14C]malonyl-ACP formation was measured by titration experiments. Concentration of protein (PfaA or PfaB AT domains) was progressively increased and the reaction product (malonyl-mm5ACP) was measured with a scintillation counter.
To measure the different activity shown by the AT domains of PfaA and PfaB against malonyl-CoA, a titration experiment was performed. Concentration of 14C-radiolabeled malonyl-CoA was fixed to 20 μm, whereas the concentration of the AT domain was progressively increased from 2 nm to 2 μm. The amount of malonyl-ACP was measured with a scintillation counter (Fig. 5C). This resulted in a dissociation constant (Kd) of 16.6 ± 3.8 nm for PfaA and 365.6 ± 209 nm for PfaB and a maximum reaction rate of 2802 ± 130 cpm for PfaA and 122 ± 20.8 cpm for PfaB (Fig. 5C). Thus, the Kd is significantly lower for PfaA so its affinity for malonyl-CoA is higher, pointing out its role in the loading of the malonyl extender units.
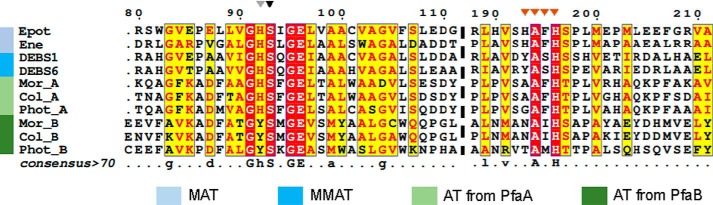
The XAXH motif within acyltransferases, close to the active site, has been identified as the main determinant of substrate specificity, HAFH and YASH being the most common variations, showing preference for malonyl (MAT) or methylmalonyl (MMAT), respectively (9, 40, 41). A multiple sequence alignment was performed comparing representative MAT and MMAT domains from PKS systems with PfaA and PfaB proteins from omega-3 PUFA synthases (Fig. 6). Conservation of a [AG]AFH motif characteristic of MAT domains was observed in PfaA proteins, whereas a [NT]A[IM]H[ST] motif was found in PfaB proteins, indicating a sequence divergence that may promote the specificity for other substrates different from malonyl-CoA. In addition, the histidine residue of the GHSXGE motif, characteristic of MAT proteins and involved in the stabilization of the malonyl–enzyme intermediate (42) is conserved in PfaA, but is replaced by tyrosine in PfaB. These motifs are in agreement with the different in vitro affinity for malonyl of KSAT PfaA and PfaB.
Figure 6.
Multiple sequence alignment of AT protein regions implicated in substrate recognition. Alignment of representative AT protein regions from PKS and omega-3 PUFA synthases is shown. Sequences correspond to epothilone PKS AT2 domain (Epot) from Sorangium cellulosum (accession number Q9KIZ8), dynemicin PKS DynE8 AT domain (Ene) from Micromonospora chersina (accession number Q84HI8), DEBS ATs 1 and 6 (DEBS1 and DEBS6) from Saccharopolyspora erythraea (accession number Q03131 and Q03132), PfaA AT domains (Mor_A, Col_A, and Phot_A) from M. marina, Colwellia psychrerythraea, and Photobacterium profundum (accession numbers Q9RA21, Q47ZG8, and Q93CG8, respectively), PfaB AT domains (Mor_B, Col_B, and Phot_B) from M. marina, Colwellia psychrerythraea, and Photobacterium profundum (accession numbers Q9RA20, Q47ZG9, and Q93CG7, respectively). Identical residues are shown in white on a red background, whereas similar residues are shown in red. The position of the “S” catalytic motif has been marked with a black arrow, the XAXH motif, the determinant of specificity was marked with orange arrows, and the histidine of the GHSXGE motif was marked with a gray arrow. The color legend indicates the selectivity against malonyl (MAT) or methylmalonyl (MMAT) of the previously known domains.
We have demonstrated here that PfaB, when highly concentrated, showed a detectable capacity to select and load malonyl units onto the ACPs. Considering the trans-AT nature of PfaB, the AT redundancy, its reduced ability to load malonyl, and its previous description as final product determinant (16), we think that the natural substrate of PfaB should be another molecule, probably involved at the last stages of the PUFA synthesis.
In summary, through a bioinformatic analysis we have completed the characterization of mmPfaA, defining an KS–AT–ACP(x5)–KR′–KR–DH organization that seems conserved in all Pfa systems. Due to the presence of ACPs with the ability to be malonylated in parallel, the existence of an AT domain selector of malonyl units, the presence of a KS domain (whose catalysis has not been demonstrated yet) and a KR–DH modifier domain, we propose PfaA as a preparative protein for the omega-3 PUFA synthesis. The PfaA role would be the generation of mature extender units that will eventually be polymerized by the rest of Pfa proteins.
Experimental procedures
Domain prediction and structural modeling
The InterPro online tool (43) was used to predict the signatures of the domains by scanning a database of previously known protein signatures with our query proteins. Jpred (44) was used to predict the secondary structure elements of the entire polypeptides. The 3D structures of the different domains were predicted by homology modeling using the Phyre2 server (18). The crystal structures used as templates for each domain were as follows: the mammalian fatty acid synthase (mFAS) (PDB accession number 2VZ8) for the mmKS-AT didomain; the ACP of Mycobacterium tuberculosis (1KLP) for the mmACP1 domain; the KR from Saccharopolyspora spinose (4IMP) for the mmKR domain; and the dehydratase domain from CurF module of curacin polyketide synthase (3KG6) for the mmDH domain. Images of the resulting models were generated using the program PyMol (45).
Sequence analysis software and public gene expression data sets
All protein and nucleic acid sequences were obtained from the public databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Protein database searches were performed using BLASTP (46), available on the NCBI web site. Multiple sequence alignments were made using the software T-coffee (47) (http://www.ebi.ac.uk/Tools/msa/tcoffee/)3 and plotted using the program ESPript 3.0 (48) (http://espript.ibcp.fr/ESPript/ESPript/).3
Phylogenetic analysis of the protein sequences was performed in phylogeny.fr (49). Sequences were aligned using MUSCLE (version 3.8.31). Gaps and not well-aligned regions were removed by using Gblocks (c0.91b). Dendrograms were constructed by using the maximum likelihood method included in PhylML (version 3.1/3.0 aLRT). Reliability for the internal branch was set using the bootstrapping method (100 replicates). The dendrogram was drawn with TreeDyn (version 198.3).
Strains and culture conditions
The E. coli strain DH5α was employed for cloning procedures and E. coli strain BL21 (DE3) for protein expression. Liquid cultures were prepared in flasks containing ¼ volume LB medium (Pronadisa, Spain) supplemented with either kanamycin sulfate (Sigma) at a final concentration of 50 μg/ml and/or ampicillin (Sigma) at 100 μg/ml and incubated at 37 °C and 180 rpm. For solid media culture LA was used (LB medium supplemented with 1.5% (w/v) agar; Pronadisa, Spain).
DNA manipulation and plasmid construction
The pDHA plasmid DNA containing pfa genes from M. marina, used as template to clone the different constructions, was kindly provided by Orikasa's lab (50). All DNA fragments were amplified by polymerase chain reaction (PCR) with oligonucleotides purchased from Sigma-Genosys (Sigma) and the high-fidelity DNA polymerase Phusion (Thermo Fisher, EEUU). All constructions were carried out using the isothermal assembly Gibson method (51).
DNA was analyzed by agarose gel electrophoresis (1% (w/v) agarose in TBE buffer) in a horizontal running system (Bio-Rad), using Hyperladder I (Biolabs, UK) as molecular weight marker and 0.05 mg/ml of SYBR Safe (Invitrogen, Life Technologies) for staining. Quantity One software (Bio-Rad) was used to visualize the images taken with a Gel Doc 2000 UV system. Plasmid DNA isolation was performed by using the QIAprep Spin Miniprep kit (Qiagen, Germany). PCR products were purified with the GenElute PCR Clean-Up kit (Sigma) or extracted from the agarose gels using the GenElute Gel Extraction kit (Sigma). The DNA concentration was determined at 260 nm with a Nano-Drop ND-1000 spectrophotometer (Thermo Scientific). Microdialized (Millipore GS, 0.05 μm pore size) isothermal assembly reactions were transformed into competent E. coli DH5α or BL21 (DE3) cells by electroporation using 0.2-cm Gene Pulser cuvettes (Bio-Rad) in a MicroPulserTM electroporator (Bio-Rad). The polymerase Biotaq (Bioline, London, UK) was used for PCR verification of the genetic constructions.
Expression and purification
E. coli BL21 (DE3) competent aliquots were individually transformed with plasmid constructions pET29c::KS-AT, pET29c::KS*-AT, pET29c::AT, pET29c::PfaB, pET29c::5ACP, or pET29c::1ACP. Plasmids pET29c::5ACP or pET29c::1ACP and their mutants were also co-transformed with pET3a::PfaE to produce the holo-ACP proteins. Individual transformant colonies were grown in 2-liter flasks containing 1 liter of LB supplemented with kanamycin (and ampicillin when coexpressing pET3a::PfaE) at 37 °C and induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside when A600 was ∼0.6. After induction, the incubation temperature was decreased to 15 °C in all cases but the ACPs constructions were expressed at 18 °C. Cell cultures were collected 18 h post-induction by centrifugation at 5,500 × g for 15 min at 4 °C and pellets were stored at −80 °C until analyzed. For purification, pellets were resuspended in buffer A (300 mm NaCl, 50 mm Tris-HCl, pH 7.5, 20 mm imidazole, and 1 mm PMSF), sonicated, and clarified by ultracentrifugation at 100,000 × g for 15 min at 4 °C. Supernatants were loaded onto a HisTrap HP column (GE Healthcare, 5 ml) previously equilibrated with buffer A. His-tagged bound proteins were eluted by a concentration gradient (0–500 mm) of imidazole (Sigma). To improve the purification, the eluted protein was concentrated using Amicon Ultra 30k Centrifugal filters (Millipore, Ireland) and loaded onto a size-exclusion chromatography Superdex 75 GL10_30 column (Amersham Biosciences, UK) previously equilibrated with buffer B (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1 mm EDTA). Purified proteins were stored at −80 °C in 5% glycerol for further experiments.
Protein intact mass determination
40 μg of purified apo- or holo-mmACP1 proteins were desalted using C4 and C18 Micro SpinColumnTM (Harvard Apparatus). Eluted samples were dried in a SpeedVac (Thermo Scientific) and resuspended in 25 μl of 50% acetonitrile, 0.25% formic acid. The protein was directly injected into a SYNAPT HDMS mass spectrometer (Waters) and MS spectra were manually acquired in the m/z 500–1700 range. Protein intact mass was determined by MaxEnt1 software (Waters) and default deconvolution parameters were used. Mass ranges were selected based on available protein sequence information and software was set to iterate to convergence. Experimentally obtained masses were matched to mmACP protein amino acid sequence, with or without 4′-phosphopantetheine, using the BioLynx tool embedded in MassLynx 4.1 software (Waters).
Radioactive binding assays
To investigate the enzymatic activity of the different AT and ACP domains in vitro the selected protein combinations were incubated with radiolabeled substrates and analyzed by SDS-PAGE and autoradiography (radio-SDS-PAGE). The reaction mixture (20 μl) contained 20 μm [14C]malonyl-CoA (55 Ci/mol) or 20 μm [14C]acetyl-CoA (29 Ci/mol), 20 μm of the ACPs and, according to the experiment, 5–50 μm of the AT domain in a buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, and 0.5 mm EDTA. Following incubation at room temperature for 30 min, the reaction was quenched with 10 μl of protein loading buffer (0.1% bromphenol blue, 1% SDS, 40% glycerol), and loaded onto a 12% acrylamide SDS gel. After electrophoresis the gels were stained with Coomassie to visualize the proteins, dried, and subjected to autoradiography to detect the radioactive substrates. The procedure was repeated in triplicate for each reaction.
Radioactive titration experiments
Each titration assay vial contained 20 μm [14C]malonyl-CoA, 20 μm ACP, and 2–2000 mm of the AT domain analyzed in a buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, and 0.5 mm EDTA in a final volume of 20 μl. The reaction mixture was incubated at room temperature for 5 min and stopped by the addition of 160 μl of ice-cold 10% trichloroacetic acid (TCA). The samples were incubated on ice for 10 min, and centrifuged at 10,000 rpm for 10 min. The pellet was washed with 10% TCA twice and finally resuspended in 60 μl of 4% SDS in 20 mm NaOH. The suspension was combined with 5 ml of scintillation mixture and analyzed with a Packard Tri-Carb liquid scintillation counter. The experiments were performed in triplicate.
Author contributions
O. S. and G. M. formal analysis; O. S. investigation; O. S. and G. M. methodology; O. S. and G. M. writing-original draft; G. M. conceptualization; G. M. supervision; G. M. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Dr. Yoshitake Orikasa, Obihiro University of Agriculture and Veterinary Medicine (Japan), for the generous gift of plasmid pDHA4. We are also grateful to Mapi Garcillan for critical reading of the manuscript and to anonymous reviewers for their valuable comments. Mass spectrometry analysis was performed in the Proteomics Core Facility-SGIKER (member of ProteoRed-ISCIII) at the University of the Basque Country, UPV/EHU.
This work was supported by Grant BFU2014-55534-C2-2-P from the Spanish Ministry of Economy and Competitiveness (to G. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PUFA
- polyunsaturated fatty acid
- EPA
- eicosapentaenoic acid
- DHA
- docosahexaenoic acid
- PKS
- polyketide synthases
- FAS
- fatty acid synthases
- ER
- enoyl reductase
- AT
- acyltransferase
- KS
- keto synthases
- ACP
- acyl carrier protein
- KR
- keto reductases
- DH
- dehydratases
- PPT
- 4′-phosphopantetheine
- PPTase
- phosphopantetheinyl transferase
- aa
- amino acid(s).
References
- 1. Lee J. M., Lee H., Kang S., and Park W. J. (2016) Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients 8, E23 10.3390/nu8010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tapiero H., Ba G. N., Couvreur P., and Tew K. D. (2002) Polyunsaturated fatty acids (PUFA) and eicosanoids in human health and pathologies. Biomed. Pharmacother. 56, 215–222 10.1016/S0753-3322(02)00193-2 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida K., Hashimoto M., Hori R., Adachi T., Okuyama H., Orikasa Y., Nagamine T., Shimizu S., Ueno A., and Morita N. (2016) Bacterial long-chain polyunsaturated fatty acids: their biosynthetic genes, functions, and practical use. Mar. Drugs 14, E94 10.3390/md14050094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gemperlein K., Rachid S., Garcia R. O., Wenzel S. C., and Müller R. (2014) Polyunsaturated fatty acid biosynthesis in myxobacteria: different PUFA synthases and their product diversity. Chem. Sci. 5, 1733–1741 10.1039/c3sc53163e [DOI] [Google Scholar]
- 5. Okuyama H., Orikasa Y., Nishida T., Watanabe K., and Morita N. (2007) Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol. 73, 665–670 10.1128/AEM.02270-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bumpus S. B., Magarvey N. A., Kelleher N. L., Walsh C. T., and Calderone C. T. (2008) Polyunsaturated fatty acid-like trans-enoyl reductases utilized in polyketide biosynthesis. J. Am. Chem. Soc. 130, 11614–11616 10.1021/ja8040042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaulmann U., and Hertweck C. (2002) Biosynthesis of polyunsaturated fatty acids by polyketide synthases. Angew. Chem. Int. Ed. 41, 1866–1869 10.1002/1521-3773(20020603)41:11%3C1866::AID-ANIE1866%3E3.0.CO%3B2-3 [DOI] [PubMed] [Google Scholar]
- 8. Orikasa Y., Nishida T., Hase A., Watanabe K., Morita N., and Okuyama H. (2006) A phosphopantetheinyl transferase gene essential for biosynthesis of n - 3 polyunsaturated fatty acids from Moritella marina strain MP-1. FEBS Lett. 580, 4423–4429 10.1016/j.febslet.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 9. Dunn B. J., and Khosla C. (2013) Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J. R. Soc. Interface. 10, 2013.0297 10.1098/rsif.2013.0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Misra A., Sharma S. K., Surolia N., and Surolia A. (2007) Self-acylation properties of type II fatty acid biosynthesis acyl carrier protein. Chem. Biol. 14, 775–783 10.1016/j.chembiol.2007.05.013 [DOI] [PubMed] [Google Scholar]
- 11. Jiang H., Zirkle R., Metz J. G., Braun L., Richter L., Van Lanen S. G., and Shen B. (2008) The role of tandem acyl carrier protein domains in polyunsaturated fatty acid biosynthesis. J. Am. Chem. Soc. 130, 6336–6337 10.1021/ja801911t [DOI] [PubMed] [Google Scholar]
- 12. Hayashi S., Satoh Y., Ujihara T., Takata Y., and Dairi T. (2016) Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci. Rep. 6, 35441 10.1038/srep35441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang H., Rajski S. R., and Shen B. (2009) Tandem acyl carrier protein domains in polyunsaturated fatty acid synthases. Methods Enzymol. 459, 79–96 10.1016/S0076-6879(09)04604-7 [DOI] [PubMed] [Google Scholar]
- 14. Chan Y. A., Podevels A. M., Kevany B. M., and Thomas M. G. (2009) Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26, 90–114 10.1039/B801658P [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beld J., Lee D. J., and Burkart M. D. (2015) Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering. Mol. BioSyst. 11, 38–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orikasa Y., Tanaka M., Sugihara S., Hori R., Nishida T., Ueno A., Morita N., Yano Y., Yamamoto K., Shibahara A., Hayashi H., Yamada Y., Yamada A., Yu R., Watanabe K., and Okuyama H. (2009) pfaB products determine the molecular species produced in bacterial polyunsaturated fatty acid biosynthesis. FEMS Microbiol. Lett. 295, 170–176 10.1111/j.1574-6968.2009.01582.x [DOI] [PubMed] [Google Scholar]
- 17. Apweiler R., Attwood T. K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M. D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40 10.1093/nar/29.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nanson J. D., Himiari Z., Swarbrick C. M., and Forwood J. K. (2015) Structural characterisation of the β-ketoacyl-acyl carrier protein synthases, FabF and FabH, of Yersinia pestis. Sci. Rep. 5, 14797 10.1038/srep14797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holmquist M. (2000) αβ-Hydrolase fold enzymes structures, functions and mechanisms. Curr. Protein Pept. Sci. 1, 209–235 10.2174/1389203003381405 [DOI] [PubMed] [Google Scholar]
- 21. Li Z., Huang Y., Ge J., Fan H., Zhou X., Li S., Bartlam M., Wang H., and Rao Z. (2007) The crystal structure of MCAT from Mycobacterium tuberculosis reveals three new catalytic models. J. Mol. Biol. 371, 1075–1083 10.1016/j.jmb.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 22. Wong H. C., Liu G., Zhang Y.-M., Rock C. O., and Zheng J. (2002) The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277, 15874–15880 10.1074/jbc.M112300200 [DOI] [PubMed] [Google Scholar]
- 23. Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., and Jörnvall H. (2003) Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143, 247–253 [DOI] [PubMed] [Google Scholar]
- 24. Dillon S. C., and Bateman A. (2004) The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics 5, 109 10.1186/1471-2105-5-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maier T., Leibundgut M., and Ban N. (2008) The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 10.1126/science.1161269 [DOI] [PubMed] [Google Scholar]
- 26. Zheng J., and Keatinge-Clay A. T. (2011) Structural and functional analysis of C2-type ketoreductases from modular polyketide synthases. J. Mol. Biol. 410, 105–117 10.1016/j.jmb.2011.04.065 [DOI] [PubMed] [Google Scholar]
- 27. Keatinge-Clay A. T., and Stroud R. M. (2006) The structure of a ketoreductase determines the organization of the β-carbon processing enzymes of modular polyketide synthases. Structure 14, 737–748 10.1016/j.str.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 28. Jenke-Kodama H., Sandmann A., Müller R., and Dittmann E. (2005) Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22, 2027–2039 10.1093/molbev/msi193 [DOI] [PubMed] [Google Scholar]
- 29. Shulse C. N., and Allen E. E. (2011) Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PloS One 6, e20146 10.1371/journal.pone.0020146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trujillo U., Vázquez-Rosa E., Oyola-Robles D., Stagg L. J., Vassallo D. A., Vega I. E., Arold S. T., and Baerga-Ortiz A. (2013) Solution structure of the tandem acyl carrier protein domains from a polyunsaturated fatty acid synthase reveals beads-on-a-string configuration. PLOS ONE 8, e57859 10.1371/journal.pone.0057859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arthur C. J., Szafranska A., Evans S. E., Findlow S. C., Burston S. G., Owen P., Clark-Lewis I., Simpson T. J., Crosby J., and Crump M. P. (2005) Self-malonylation is an intrinsic property of a chemically synthesized type II polyketide synthase acyl carrier protein. Biochemistry 44, 15414–15421 10.1021/bi051499i [DOI] [PubMed] [Google Scholar]
- 32. Arthur C. J., Szafranska A. E., Long J., Mills J., Cox R. J., Findlow S. C., Simpson T. J., Crump M. P., and Crosby J. (2006) The malonyl transferase activity of type II polyketide synthase acyl carrier proteins. Chem. Biol. 13, 587–596 10.1016/j.chembiol.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 33. Hitchman T. S., Crosby J., Byrom K. J., Cox R. J., and Simpson T. J. (1998) Catalytic self-acylation of type II polyketide synthase acyl carrier proteins. Chem. Biol. 5, 35–47 10.1016/S1074-5521(98)90085-0 [DOI] [PubMed] [Google Scholar]
- 34. Matharu A. L., Cox R. J., Crosby J., Byrom K. J., and Simpson T. J. (1998) MCAT is not required for in vitro polyketide synthesis in a minimal actinorhodin polyketide synthase from Streptomyces coelicolor. Chem. Biol. 5, 699–711 10.1016/S1074-5521(98)90663-9 [DOI] [PubMed] [Google Scholar]
- 35. Wakil S. J., Stoops J. K., and Joshi V. C. (1983) Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 52, 537–579 10.1146/annurev.bi.52.070183.002541 [DOI] [PubMed] [Google Scholar]
- 36. Ray L., and Moore B. S. (2016) Recent advances in the biosynthesis of unusual polyketide synthase substrates. Nat. Prod. Rep. 33, 150–161 10.1039/C5NP00112A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szu P. H., Govindarajan S., Meehan M. J., Das A., Nguyen D. D., Dorrestein P. C., Minshull J., and Khosla C. (2011) Analysis of the ketosynthase-chain length factor heterodimer from the fredericamycin polyketide synthase. Chem. Biol. 18, 1021–1031 10.1016/j.chembiol.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Misra A., Surolia N., and Surolia A. (2009) Catalysis and mechanism of malonyl transferase activity in type II fatty acid biosynthesis acyl carrier proteins. Mol. Biosyst. 5, 651–659 10.1039/b820420a [DOI] [PubMed] [Google Scholar]
- 39. Tufar P., Rahighi S., Kraas F. I., Kirchner D. K., Löhr F., Henrich E., Köpke J., Dikic I., Güntert P., Marahiel M. A., and Dötsch V. (2014) Crystal structure of a PCP/Sfp complex reveals the structural basis for carrier protein posttranslational modification. Chem. Biol. 21, 552–562 10.1016/j.chembiol.2014.02.014 [DOI] [PubMed] [Google Scholar]
- 40. Liew C. W., Nilsson M., Chen M. W., Sun H., Cornvik T., Liang Z.-X., and Lescar J. (2012) Crystal structure of the acyltransferase domain of the iterative polyketide synthase in enediyne biosynthesis. J. Biol. Chem. 287, 23203–23215 10.1074/jbc.M112.362210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yadav G., Gokhale R. S., and Mohanty D. (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 10.1016/S0022-2836(03)00232-8 [DOI] [PubMed] [Google Scholar]
- 42. Poust S., Yoon I., Adams P. D., Katz L., Petzold C. J., and Keasling J. D. (2014) Understanding the role of histidine in the GHSxG acyltransferase active site motif: evidence for histidine stabilization of the malonyl-enzyme intermediate. PLOS ONE 9, e109421 10.1371/journal.pone.0109421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulder N., and Apweiler R. (2007) InterPro and InterProScan: tools for protein sequence classification and comparision. Methods Mol. Biol. 396, 59–70 10.1007/978-1-59745-515-2_5 [DOI] [PubMed] [Google Scholar]
- 44. Cuff J. A., Clamp M. E., Siddiqui A. S., Finlay M., and Barton G. J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14, 892–893 10.1093/bioinformatics/14.10.892 [DOI] [PubMed] [Google Scholar]
- 45. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 46. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., and Madden T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Notredame C., Higgins D. G., and Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 48. Gouet P., Courcelle E., Stuart D. I., and M√©toz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- 49. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–469 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orikasa Y., Nishida T., Yamada A., Yu R., Watanabe K., Hase A., Morita N., and Okuyama H. (2006) Recombinant production of docosahexaenoic acid in a polyketide biosynthesis mode in Escherichia coli. Biotechnol. Lett. 28, 1841–1847 10.1007/s10529-006-9168-6 [DOI] [PubMed] [Google Scholar]
- 51. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.