Abstract
Triggering receptor expressed on myeloid cells 2 (TREM2) is an orphan immune receptor expressed on cells of myeloid lineage such as macrophages and microglia. The rare variant R47H TREM2 is associated with an increased risk for Alzheimer's disease, supporting the hypothesis that TREM2 loss of function may exacerbate disease progression. However, a complete knockout of the TREM2 gene in different genetic models of neurodegenerative diseases has been reported to result in both protective and deleterious effects on disease-related end points and myeloid cell function. Here, we describe a Trem2R47H transgenic mouse model and report that even in the absence of additional genetic perturbations, this variant clearly confers a loss of function on myeloid cells. The Trem2R47H variant–containing myeloid cells exhibited subtle defects in survival and migration and displayed an unexpected dysregulation of cytokine responses in a lipopolysaccharide challenge environment. These subtle phenotypic defects with a gradation in severity across genotypes were confirmed in whole-genome RNA-Seq analyses of WT, Trem2−/−, and Trem2R47H myeloid cells under challenge conditions. Of note, TREM2-activating antibodies that boost proximal signaling abrogated survival defects conferred by the variant and also modulated migration and cytokine responses in an antibody-, ligand-, and challenge-dependent manner. In some instances, these antibodies also boosted WT myeloid cell function. Our studies provide a first glimpse into the boost in myeloid cell function that can be achieved by pharmacological modulation of TREM2 activity that can potentially be ameliorative in neurodegenerative diseases such as Alzheimer's disease.
Keywords: neuroinflammation, neurodegeneration, Alzheimer's disease, microglia, antibody, myeloid cell, genetics, gene knockout, R47H TREM2
Introduction
Triggering receptor expressed on myeloid cells (TREM2) is expressed on cells of myeloid lineage, including macrophages and microglia (1, 2). TREM2 is an orphan immune receptor with a short intracellular domain and functions by signaling through its adaptor partner, DAP12 (3). Mutations in both TREM2 and DAP12 have been linked to the autosomal recessive disorder Nasu–Hakola disease, which is characterized by bone cysts, muscle wasting, and demyelination (2). More recently, variants in the TREM2 gene have been linked to increased risk for Alzheimer's disease (AD)5 and other forms of dementia, including frontotemporal dementia (4–6). In particular, the R47H variant has been identified in genome-wide studies as being associated with increased risk for late-onset AD with an overall adjusted odds ratio (for populations of all ages) of 2.3, second only to the strong genetic association of ApoE with Alzheimer's. The R47H mutation resides on the extracellular Ig V-set domain of the TREM2 protein and has been shown to impact lipid binding and uptake of Aβ (7–10), suggestive of a loss of function linked to disease. Two recent publications have demonstrated defects in the autophagy/lysosomal pathway (11) and decreased capacity of R47H microglia to compact plaques (12). In general, however, the effect of the variant on TREM2 expression and myeloid cell functioning and the ability to modulate the gene to have a beneficial effect in disease are emerging areas of TREM2 biology. Most preclinical models that have been designed to study TREM2's role in modulating myeloid cell function and disease progression in AD have employed complete gene knockouts that are probably not representative of a physiologically relevant state. Different knockout mice crossed with traditional AD mouse models (APP/PS1 and 5×FAD) have resulted in conflicting data with respect to the role of TREM2 in Alzheimer's disease; Wang et al. (8) have proposed a loss of function for the gene in disease based on decreased microglia survival and reduced ability to cluster around Aβ plaques and clear them (APP/PS1 and 5×FAD crossed with Colonna knockouts). Jay et al. (13) have provided evidence supportive of an amelioration of the neurodegenerative phenotype upon Trem2 deletion in their models, including an increase in phagocytosis and anti-inflammatory cytokines and an overall decrease in amyloid burden (APP/PS1 crossed with NIH KOMP knockouts, hereafter referred to as Trem2KOMP−/−). The effects were subsequently shown to be potentially temporally regulated based on disease progression (14). Similarly, conflicting results have been reported in tauopathy models also with an exacerbation in disease end points in one model (15) and an improvement in another (16) upon gene deletion. At the time of submission of this manuscript, Song et al. (17), described for the first time a deleterious effect of human R47H TREM2 when introduced into 5×FAD mice, noting subtle cell intrinsic and extrinsic TREM2-dependent effects. The authors noted differences in plaque load and microglial function between KO-5×FAD mice and human R47H TREM2–5×FAD mice. A more comprehensive understanding of the effect of the variant on myeloid cell function in general, especially in the absence of additional genetic perturbations that are used to generate neurodegenerative disease models, may help clarify some of the contradictory data pertaining to the role of the protein in progressive, neurodegenerative diseases. We describe below an R47H transgenic model and comprehensively characterize the loss-of-myeloid cell function conferred by the R47H variant. We also demonstrate that antibodies that activate proximal TREM2 signaling rescue some of these cumulative defects and even boost WT TREM2 activity. Our studies provide the first glimpse into pharmacological modulation of TREM2 as a means to modulate myeloid cell function in neurodegenerative diseases.
Results
Gene-edited Trem2R47H and Trem2−/− mice are specifically modified in the TREM2 gene with no off-target effects on other TREM genes in the locus
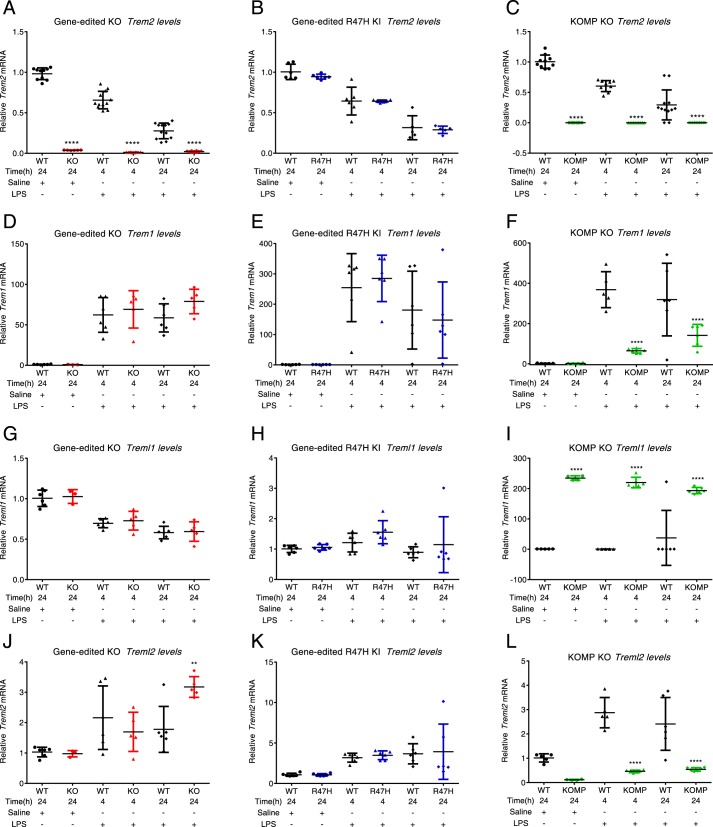
To address some of these fundamental questions linked to TREM2 biology and the effect of the R47H variant on TREM2 functioning, we have generated Trem2−/− and Trem2R47H knockin mice. The TREM gene locus in humans and mice includes multiple TREM and TREM-like genes with both ITAM- and ITIM-associated functions (18) (Fig. S1A). Regulatory elements located within the TREM2 gene can impact the expression of other TREM genes. To specifically target the TREM2 gene without perturbing additional regulatory elements, we used a gene editing–based approach to generate Trem2−/− or Trem2R47H mice. The Trem2−/− strain was generated by engineering a deletion in exon 1 of the Trem2 gene, and the Trem2R47H strain was generated by engineering a point mutation at residue 47 in the mouse Trem2 gene (Fig. S1, B and C), analogous to the human variant. Detailed qPCR analyses of brain homogenates from the gene-edited Trem2−/−, Trem2R47H, and commercial NIH KOMP mice confirmed a comparable loss of the gene in the knockouts and Trem2 expression comparable with WT age-matched controls for the Trem2R47H mice (Fig. 1, A–C). We further confirmed that upon LPS stimulation, R47H Trem2 was down-regulated in mouse brains comparable with age-matched WT controls, whereas no detectable levels of Trem2 mRNA were observed in the Trem2−/− animals under any of the conditions tested (Fig. 1, A–C). Similar and expected trends in expression patterns were observed in WT, gene-edited Trem2−/−, and Trem2R47H mice with respect to other Trem genes in the locus, including Trem1 (Fig. 1, D and E), Treml1 (Fig. 1, G and H), and Treml2 (Fig. 1, J and K) under basal and LPS-stimulated conditions. Of note, we and others (19) have observed unexpected changes in other Trem genes in the Trem locus under basal and LPS-stimulated conditions in the Trem2KOMP−/− mice; this includes a less significant Trem1 response compared with age-matched WT littermates (Fig. 1F, LPS stimulation, 4- and 24-h time points), a highly significant increase in Treml1 (Fig. 1I, steady state and LPS), and a significant decrease in Treml2 (Fig. 1L, steady state and LPS).
Figure 1.
Gene-edited Trem2R47H and Trem2−/− mice are specifically modified in the TREM2 gene with no off-target effects on other TREM genes in the locus unlike the Trem2KOMP mice. A, no detectable Trem2 mRNA in the gene-edited Trem2−/− brains under any treatment conditions; B, Trem2 mRNA is significantly down-regulated upon LPS treatment in WT and R47H Trem2 brains; C, no detectable Trem2 mRNA in the gene-edited Trem2KOMP brains under any treatment conditions; D and E, Trem1 is up-regulated upon LPS stimulation in gene-edited Trem2−/− and Trem2R47H brains, comparable with corresponding WT control brains; F, Trem1 up-regulation in the Trem2KOMP−/− mice post-stimulation is lower compared with the in-house mice and age-matched littermate WT controls; G and H, no significant changes were noted in Treml1 levels in WT, gene-edited Trem2−/−, and Trem2R47H brain samples with saline treatment and LPS stimulations; I, Treml1 is highly up-regulated in the Trem2KOMP−/− mice under basal conditions (saline treatment) and remains high upon LPS treatment; J and K, Treml2 is up-regulated 2–4-fold upon LPS treatment in the in-house Trem2−/− mice and the Trem2R47H mice; L, Treml2 mRNA is significantly reduced in the Trem2KOMP mice under both basal and LPS-stimulated conditions. All results are presented as -fold change over saline-treated WT controls. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; data represented as mean ± S.D.; statistical analysis: one-way ANOVA with Sidak's correction for multiple comparison.
Trem2R47H macrophages and microglia reveal a cell survival defect with defects noted primarily in apoptosis
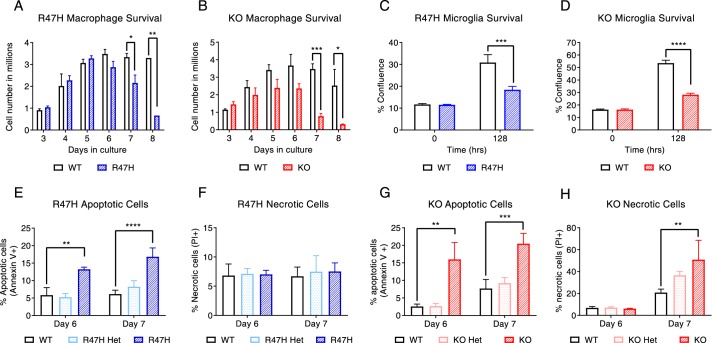
Next, we compared the phenotypes of Trem2R47H, Trem2−/−, and WT bone marrow–derived macrophages (BMDMs) and microglia. We noted reduced proliferation and survival of the gene-edited Trem2R47H and Trem2−/− BMDMs (Fig. 2, A and B) and microglia (Fig. 2, C and D) under limiting conditions of CSF-1, consistent with previously published data on knockout alone (9, 20). As well, we noted gene dosage–dependent effects on proliferation and survival of both Trem2R47H (Fig. S2A) and Trem2−/− (Fig. S2B) myeloid cells. A more detailed analysis reveals that the variant macrophages have a primarily apoptotic and negligible necrotic response (Fig. 2, E and F) compared with the knockout macrophages, which have significant increases in both apoptotic and necrotic events (Fig. 2, G and H). Also, a breakdown of the apoptotic and necrotic events by gene dosage revealed clearly that in the case of the variant, the effects are only apparent in the homozygous R47H macrophages and not in the heterozygous macrophages, again supportive of the more subtle nature of the mutation. In the case of the heterozygous knockouts, whereas the necrotic events did not achieve statistical significance (p = 0.08), they consistently demonstrated an increasing trend compared with WT.
Figure 2.
Trem2R47H macrophages and microglia reveal a cell survival defect that is similar to but less severe than the Trem2−/− population. A, Trem2R47H mouse BMDMs exhibit a survival defect when cultured under limiting conditions of CSF-1. B, Trem2−/− BMDMs had a more pronounced survival defect in similar culture conditions; C and D, Trem2R47H (blue) and Trem2−/− (red) mouse adult microglia exhibit a survival defect in culture; E, a significant increase in annexin V–stained BMDM population was detected at 6 and 7 days in culture in the case of the Trem2R47H BMDMs, supportive of increased apoptotic events; F, the total numbers of necrotic events 6 and 7 days in culture in the Trem2R47H BMDMs were comparable with WT macrophages; G, increased annexin V+ populations were noted at days 6 and 7 in Trem2−/− BMDMs; H, Trem2−/− BMDMs had significantly increased PI+ populations at day 7 compared with Trem2+/+ BMDMs, supportive of increased necrotic events in these populations. Data are presented as mean ± S.D. (error bars) and are from a representative experiment. The experiment was conducted twice independently. *, p < 0.05, **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Statistical analysis: two-way ANOVA with Sidak's correction for multiple comparisons (A–D) or Dunnett's correction for multiple comparisons (E–H).
RNA-Seq analysis provides genome-wide molecular corroboration for the more subtle effect of the variant, including effects on several genes linked to AD
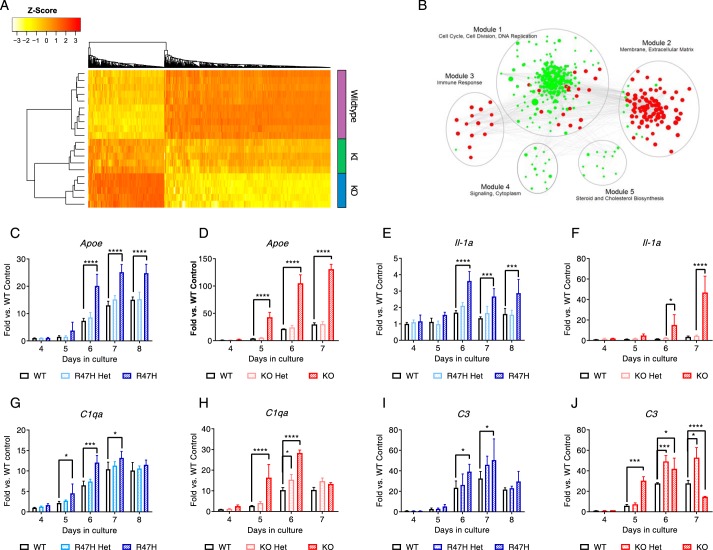
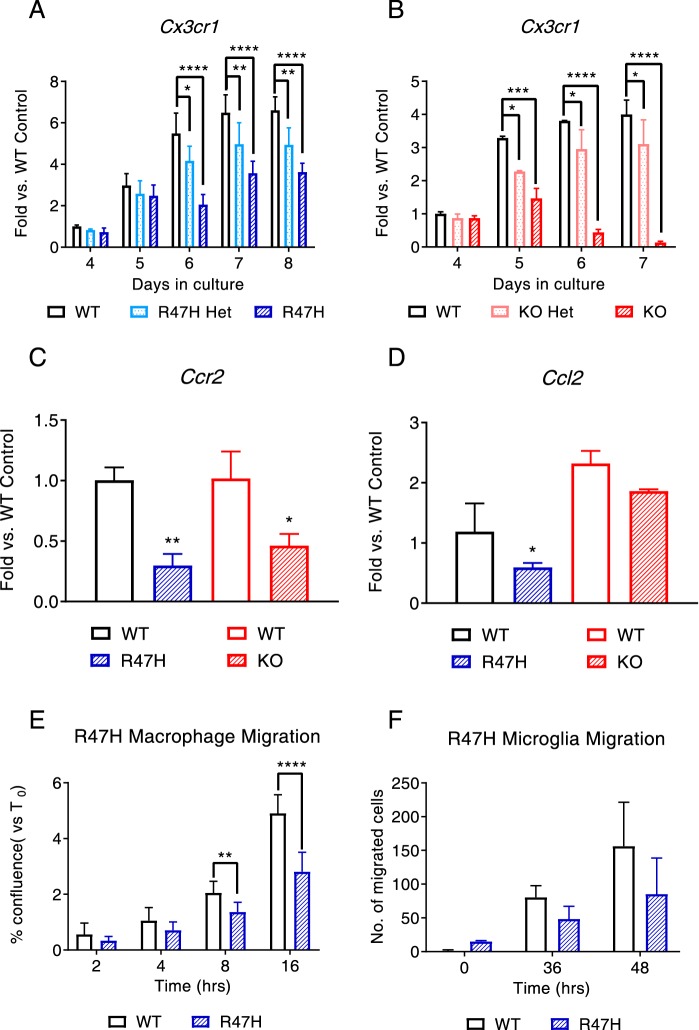
To determine whether the parallels and differences between the variant and knockout macrophages can be explained at the molecular level, we performed RNA-Seq analyses comparing WT, Trem2−/−, and Trem2R47H macrophages. At day 7 under limiting conditions of CSF-1, consistent with the gradation in severity of phenotypes observed, we observed the greatest differential transcript expression in the Trem2−/− macrophages with an intermediate effect in Trem2R47H macrophages (heat map in Fig. 3A). Pathway analyses point to a role for TREM2 in cell cycle/proliferation and survival, immune response and migration, and lipid and cholesterol homeostasis (Fig. 3B). Whereas the total number of genes involved in proliferation, necrosis, and apoptosis was higher in the knockout group, the R47H group had a larger number of apoptotic genes. Overall, the total number of genes that were differentially expressed was higher in the knockout group compared with the R47H variant group. We confirmed differences in several genes in each of the modules, including genes linked to the proliferation/survival module as well as some known genetic factors linked to Alzheimer's like ApoE (Fig. 3, C and D), pro-inflammatory cytokines that are up-regulated in AD like Il-1α (Fig. 3, E and F), and complement genes C1qa (Fig. 3, G and H) and C3 (Fig. 3, I and J). A temporal component is associated with the changes observed in some of the genes with an increase in gene expression followed by a decline, as in the case of C1qa and C3 (Fig. 3, G–J). We also noted changes in a host of chemokines/chemokine receptors in both the R47H and knockout groups, including a down-regulation of genes like Cx3cr1 (Fig. 4, A and B), Ccr2 (Fig. 4C), and Ccl2 (Fig. 4D). In a few instances, we also noted up-regulation of genes, including Ccl5 and Ccl22 (Fig. S3, A–D). The effect on Cx3cr1 was the most profound, with a significant gene dosage–dependent effect at multiple time points that was observed in both genotypes. The effects on Ccr2 and Ccl2 were more subtle and noted only at specific time points. For the first time, we noted a reduction in Flt1 in the TREM2 knockouts but no obvious effect in the variants (Fig. S3, E and F). Overall, in the majority of genes, the differential effect was significantly more pronounced in the knockouts compared with the R47H cells. The modulation of multiple migratory chemokines/chemokine receptors translated to a net reduction in migration/motility of R47H and knockout macrophages (Fig. 4E and Fig. S3G) and to a lesser extent Arg-47 microglia (Fig. 4F, trend but not statistically significant).
Figure 3.
RNA-Seq analysis provides molecular corroboration for the more subtle effect of the variant and modulation of several genes linked to AD. A, transcript heat map of differentially regulated genes between various treatment groups (n = 5 animals/genotype) is supportive of an intermediate perturbation in the Trem2R47H BMDMs compared with Trem2−/− and WT BMDMs. B, WGCNA analysis resulted in the identification of five modules/gene networks that are differentially regulated in the knockout and variant macrophages compared with WT. C–J, a subset of genes, including Apoe (C and D), Il-1α (E and F), C1qa (G and H), and C3 (I and J), confirmed as differentially regulated in R47H macrophages (C, E, G, and I; blue graphs) and knockout macrophages (D, F, H, and J; red graphs) by qPCR in a time course study. Data are presented as mean ± S.D. (error bars) and are from a representative experiment. The experiment was conducted twice independently. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001.
Figure 4.
RNA-Seq analysis also reveals dysregulation of multiple chemokines/chemokine receptors and accompanying migration defects in variant macrophages and microglia. A and B, Cx3cr1 was significantly down-regulated in R47H macrophages (A) and knockout macrophages (B) with a clear gene dosage effect observed at different time points. C and D, Ccr2 (C) and Ccl2 (D) were also confirmed as down-regulated in both R47H and knockout macrophages (trend for Ccl2) at different time points. Data are presented as mean ± S.D. (error bars) and are from a representative experiment. The experiment was conducted twice independently. E, migration defects were observed for the R47H macrophages in a plug area assay. Data are presented as mean ± S.D. and are from a representative experiment. The experiment was conducted three times independently. F, a trend of migration defects was also noted for R47H microglia in a transwell assay with C5a as the chemoattractant (data from three animals). *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; data represented as mean ± S.D.
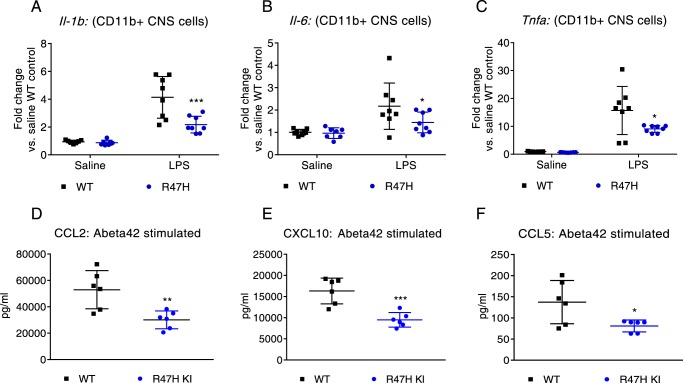
In vivo LPS and in vitro Aβ challenge reveal a surprising decrease in pro-inflammatory cytokines in microglia and macrophages
We next determined the effect of an acute in vivo challenge on variant and knockout microglia. Animals were administered LPS (5.0 mg/kg intraperitoneally (i.p.)), and CD11b+ microglia cells were isolated following overnight treatment according to the manufacturer's protocols. No significant differences were observed in the total number of microglial cells isolated under homeostatic and challenge conditions (data not shown). Contrary to our expectation of increased pro-inflammatory cytokines upon LPS challenge in the variant microglia based on the proposed functional role for TREM2, we observed statistically significant decreases in pro-inflammatory cytokines, including Il-1β, Il-6, and Tnf-α (Fig. 5, A–C). In vitro Aβ(1–42) (Anaspec AggreSureTM) challenge of macrophages revealed a similar trend, with statistically significant decreases in CCL2, CXCL10, and CCL5 protein levels (Fig. 5, D–F) in R47H macrophages compared with WT. A decreasing trend in CCL3 and CCL4 was also noted over and above what was observed with control scrambled Aβ(1–42) peptide (data not shown). Similar but less significant trends were observed when microglia were treated with Aβ(1–42), with the most significant changes observed for CCL3 and CCL4 (Fig. S4, A and B). In these instances where we measured protein levels in the conditioned media, an equal number of cells were plated at the start of the experiment, and the cells were grown in complete medium, where similar proliferation curves have been observed for WT and R47H microglia. Hence, it is unlikely that the differences observed can be attributed solely to reduced cell numbers. Further, in at least a few instances, we observed opposite trends for chemokines/cytokines, depending on the challenge administered, supportive of a generally dysregulated inflammatory state conferred by the variant/knockout that manifests differently based on the challenge. Of note, Ccl2 was one of the only chemokines tested that maintained a similar trend with most challenges. Ccl2 was reduced in Trem2−/− and Trem2R47H day 5 BMDM cultures compared with WT, although only the R47H data reached statistical significance (Fig. 4D). A small reduction of CCL2 was also noted in lavage fluid of the peritoneal cavity of knockout mice treated with zymosan (Fig. S4C) and to a lower extent (p > 0.05) in the brains of LPS-treated knockout mice (Fig. S4D).
Figure 5.
R47H microglia produce lower levels of pro-inflammatory cytokines upon an acute in vivo challenge. A–C, CD11b+ cells isolated from brains of Trem2R47H animals treated with LPS for 24 h show a significant reduction in Il-1β (A), Il-6 (B), and Tnf-α (C) mRNA levels relative to Trem2+/+ animals. D–F, BMDMs from Trem2R47H animals show significant reductions in CCL2 (D), CXCL10 (E), and CCL5 (F) levels following treatment with Aβ(1–42) compared with WT littermates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; data represented as mean ± S.D. Statistical analysis: one-way ANOVA with Sidak's correction for multiple comparison.
Antibodies that boost TREM2 signaling also modulate myeloid cell functioning
Whereas the Trem2R47H macrophages phenocopy (albeit to a more subtle degree) the Trem2−/− macrophages, the phenotypes cannot be explained simply by a reduction in cell-surface expression of R47H TREM2. WT and Trem2R47H BMDMs appeared to have comparable levels of surface TREM2 expression (Fig. S5, A and C). However, we and others have shown that the R47H variant has impaired ligand sensing/signaling (8, 21).
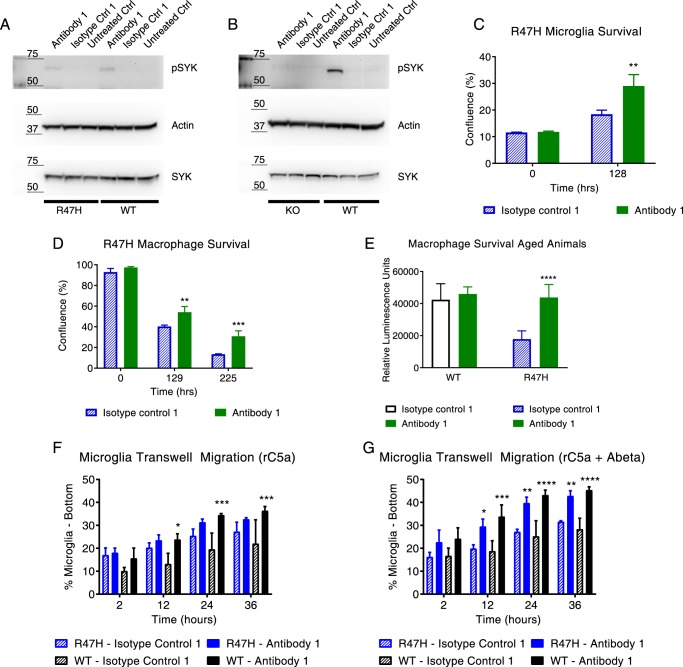
To determine whether the defects in proximal signaling and myeloid cell function can be rescued by a pharmacologic agent, we tested commercial and internally generated mouse antibodies in pSyk activation assays and on more distal myeloid cell functioning. We first confirmed the specificity of a commercially available TREM2 antibody (henceforth denoted by antibody 1) by demonstrating a lack of FACS shift compared with isotype control on Trem2−/− BMDMs (Fig. S5B). Next, we demonstrated that the antibody increased pSyk levels in both R47H and WT BMDMs, with the effect being more pronounced in the WT BMDMs (Fig. 6A). The antibody had no effect on the Trem2−/− macrophages, further supportive of specific activation of TREM2 (Fig. 6B).
Figure 6.
An antibody that boosts TREM2 signaling also modulates myeloid cell function. A and B, antibody 1 activates TREM2/DAP12-mediated Syk signaling in Trem2+/+ and Trem2R47H BMDMs. The effect is less pronounced in the Trem2R47H BMDMs compared with WT BMDMs; the TREM2 agonist antibody does not increase pSyk levels in the Trem2−/− BMDMs, confirming a TREM2-specific effect. No significant differences were observed in total Syk levels or in the loading control (actin). Data are shown from a representative set of animals. C, the antibody also restores the survival defect in Trem2R47H microglia, as demonstrated in a real-time cell confluence assay. Isotype control antibody (blue) is not able to achieve the same boost in survival. Data are plotted as mean ± S.D. (error bars) and are from a single representative experiment. The experiment was conducted twice independently. D, improved survival was also noted in Trem2R47H BMDMs with antibody 1 treatment (green) relative to isotype control (blue hash). E, BMDMs obtained from aged (18-month-old) R47H animals also demonstrate a similar survival defect that can be rescued with agonist antibody treatment (green), whereas the isotype control has no effect (blue hash). F, antibody 1 treatment significantly increased migration toward recombinant C5a as a chemoattractant in Trem2+/+ microglia relative to isotype control. G, antibody 1 treatment significantly increased migration toward recombinant C5a + Aβ(1–42) as a chemoattractant in Trem2+/+ and Trem2R47H microglia relative to isotype control. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; data represented as mean ± S.D.
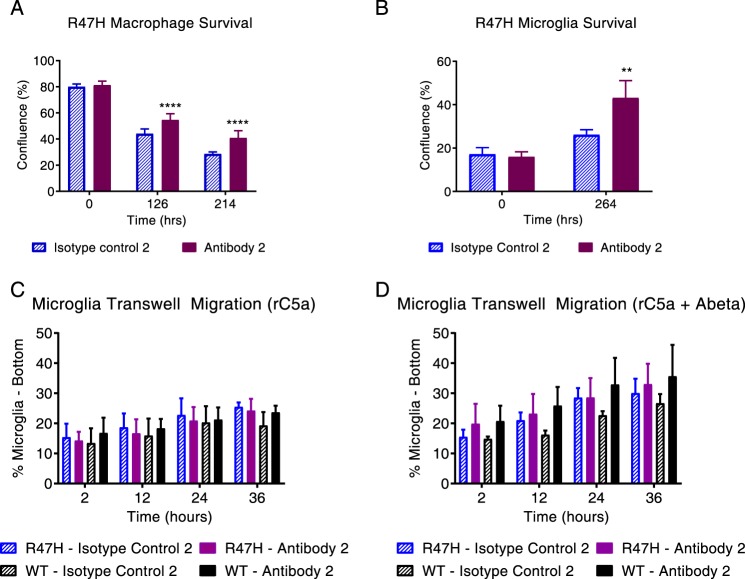
We then determined that the antibody improved survival of Trem2R47H microglia and macrophages (Fig. 6, C and D, green), whereas an equivalent rescue in cell survival was not observed when treated with isotype control antibodies (Fig. 6, C and D, blue). The effect was equally robust when we treated macrophages from aged animals (Fig. 6E). In fact, the antibody was also able to boost the function of WT macrophages and microglia (Fig. S6, A and B) but had no effect on knockout macrophage survival (Fig. S6C). Additionally, the antibody increased migration of microglia toward recombinant C5a (a classic microglial chemoattractant) both with and without aggregated Aβ(1–42) (a putative endogenous ligand) (Fig. 6, F and G). Aβ(1–42) alone did not serve as a chemoattractant in the transwell migration assay (data not shown). It should be noted that the boost in migration reached statistical significance for the R47H microglia only in the presence of Aβ (Fig. 6G, blue bars), whereas the WT microglia showed significantly increased migration under both treatment paradigms (Fig. 6, F and G, black bars). It is also noteworthy that CCL2, a myeloid chemoattractant and the one chemokine whose level was reduced in different treatment conditions, actually increased upon antibody treatment in the Trem2R47H macrophages (Fig. S7, A and B). Additionally, antibody 1 also significantly modified expression levels of genes in module 1 (i.e. the proliferation and survival module) (>50 based on their annotation in the Gene Ontology network) in macrophages. Representative data are shown for Melk, Nek2, and Mmp14 (Fig. S7, C–E). The modulated gene signature is consistent with the antibody suppressing cell death events and promoting proliferation. To determine whether the ability to modulate different aspects of myeloid cell function was unique to antibody 1, we tested another internally generated mouse TREM2 antibody (henceforth referred to as antibody 2) that did not compete with antibody 1 for TREM2 binding and also activated Syk signaling. In this instance, antibody 2 recognized only mouse TREM2. We confirmed by FACS (Fig. S5, C and D) and immunoblotting (Fig. S5E) that antibody 2 did not have any signal in the knockout BMDMs but was able to recognize TREM2 in WT and R47H BMDMs. Antibody 2 was also able to boost survival of both R47H macrophages and microglia (Fig. 7, A and B). Further, as in the case of antibody 1, antibody 2 was also able to boost survival of WT cells (Fig. S6, D and E) but did not impact the survival or rescue survival defects observed in Trem2−/− cells (Fig. S6F). Also, the antibody did not affect migration in any of the genotypes (Fig. 7, C and D). Thus, even a comparison of just two antibodies with putatively different TREM2-binding regions and different properties resulted in different myeloid cell function modulation profiles.
Figure 7.
In vitro profiling of a second TREM2-activating antibody with a different myeloid function modulation profile compared with antibody 1. A and B, an internally generated agonist antibody (antibody 2) also showed a similar boost in survival of Trem2R47H macrophages (A, purple bar) and microglia (B, purple bar) as antibody 1 relative to isotype control (blue hash, A and B). C and D, no increase in migration was observed in Trem2+/+ (black) and Trem2R47H microglia (purple) following treatment with antibody 2 relative to isotype control (blue) both in the presence and absence of rC5a alone or rC5a + Aβ as a chemoattractant. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; data represented as mean ± S.D.
Discussion and conclusions
In summary, we have generated powerful and specific TREM2 animal models to elucidate the effect of the R47H variant on myeloid cell function and show that pharmacologic agents like antibodies can rescue the loss of function conferred by the variant and even boost WT function. We demonstrate that targeting the Trem2 gene using gene editing allows for specific perturbation of the gene without disrupting other genes in the locus. Both TREM1 and TREM2 signal via DAP12 with probably opposing effects with respect to modulating inflammatory response (pro-inflammatory versus anti-inflammatory) (22); similarly, TREML1 and TREML2 also modulate inflammatory response; in fact, TREML2 is expressed on similar cells as TREM2, including microglia, and recently has been proposed to play an opposing role to TREM2 (23, 24). Hence, we expect that perturbation of multiple genes in the Trem locus as in the Trem2KOMP−/− mice can confound some of the results from mice generated by crossing these knockouts with disease models. Our TREM models will probably not suffer from these complications.
R47H variant macrophages and microglia reveal survival and migration defects in culture similar to but less severe than knockout cells (8). We note that the difference in severity in the phenotype (under challenge conditions) between a complete deletion of the gene versus the occurrence of the variant is recapitulated at the transcript level with similar but more significant changes in the Trem2−/− macrophages compared with R47H macrophages. The less pronounced effect of the mutation with most effects being noted primarily in the homozygous variant cells/animals is also consistent with the identification of R47H TREM2 as a rare variant associated with a polygenic disease like AD. Our data reveal how the variant can exert a subtle but cumulative effect on myeloid cell functioning over the lifetime of the individual. The occurrence of the variant alone without any other genetic risk factors confers a loss of myeloid function for the Trem2 gene that is probably exacerbated in the context of AD. Additional risk factors, notably age as well as genetic and environmental risk factors, then probably serve as epidemiological challenges and predispose heterozygous carriers to an increased risk of disease. The subtle nature of this particular variant and a comparison with heterozygous and homozygous knockouts also suggest that in the context of the larger late-onset Alzheimer's disease (LOAD) population (patients who do not carry this mutation), a small but cumulative loss of function for WT TREM2 with age can similarly contribute to disease. The phenotype associated with the R47H variant is also in stark contrast to the more overt phenotypes associated with a complete loss of function as in Nasu-Hakola disease or other forms of more aggressive neurodegenerative diseases (Q33X, Y38C, and T66M).
Pathway analyses of the differentially regulated genes provide mechanistic insight into the observed phenotypic defects. We note a dysregulation in different aspects of myeloid cell functioning, including DNA replication, cell cycle regulation, proliferation, cell death, chemokine/cytokine modulation, and complement pathway. We do not observe significant changes in genes directly linked to phagocytosis. Our data support the hypothesis that a loss of function of the TREM2 gene contributes directly to a fundamental proliferation/survival deficit that can then translate indirectly to functional effects like reduced phagocytic capacity. The role of TREM2 as a more fundamental regulator of the homeostatic state of microglia is still unclear. Recent work from Keren-Shaul et al. (25), employing single-cell RNA-Seq techniques, suggests that microglia surrounding plaques are modulated in two waves, a primary TREM2-independent wave with an up-regulation of ApoE and a down-regulation of genes like Cx3Cr1 and P2ry12 and a second TREM2-dependent wave that modulates lipid sensing and phagocytosis. However, some studies have reported a TREM2 depletion–linked reduction in ApoE and locking of microglia in a homeostatic state (26). Based on our gene networks and gene analysis, we find that some of the key wave 1 genes are also significantly regulated by the variant (increased ApoE, decreased Cx3Cr1) under some challenge conditions. Our data suggest that TREM2 can more fundamentally regulate the transition out of a homeostatic state for myeloid cells in a challenge-dependent manner. With respect to regulation of the chemokine environment, the variant clearly contributes to a dysregulation of the chemokine environment in vitro. However, the observed decreasing trends in traditional pro-inflammatory cytokines upon LPS/zymosan challenge in vivo in variant microglia is reflective of the highly complex, temporally regulated, and ligand/challenge-sensitive nature of the TREM2-dependent response (27–30). In fact, recent data pointing to opposing roles for TREM2 in tauopathy can potentially be attributed to the age of the animals and the different genetic models of tauopathy utilized in the two studies and further support the complex nature of TREM2-mediated modulation of neuroinflammation (15, 16).
The ability of antibodies that boost proximal signaling to rescue the viability, proliferation, and migration defects elegantly demonstrates the potential application of pharmacologic agents in rescuing cumulative genetic defects and boosting microglia activity in a TREM2-dependent manner. In fact, the ability of antibodies 1 and 2 to boost WT microglia function under challenge conditions in vitro is supportive of the putative therapeutic potential of a TREM2 agonist antibody for the >99% of LOAD patients who do not carry the R47H mutation. However, the different effects on migration, depending on which antibody is used, are supportive of more divergent and complex distal biology. The next steps will entail in vivo administration of antibodies with different in vitro profiles in neurodegenerative disease mouse models to more rigorously define the properties of an efficacious TREM2 therapeutic for the treatment of AD or other neurodegenerative diseases. Finally, most efforts on TREM2 have been focused on its functioning in microglia. Recent studies on the CNS immune environment have focused on the cross-talk between activated microglia and reactive astrocytes (31). The increase in complement genes and factors like Il-1α in the TREM2 variants and knockouts that have been proposed to play a role in mediating microglia–astrocyte cross-talk also points to potentially novel non-cell-autonomous roles for TREM2 in modulating the CNS immune environment that will need further investigation.
Materials and methods
All animal procedures were approved by the Amgen Institutional Animal Care and Use Committee.
Gene-edited in-house mouse construction
A pair of mRNAs targeting exon 2 of the TREM2 gene was synthesized. The target sequences were as follows: left, 5′-TCCTTGAGGGTGTCATGTAC-3′; right, 5′-TGCGTCTCCCCCAGTGCTTC-3′. The binding sites were separated by a 12-bp spacer region. A 143-mer R47H single-stranded oligonucleotide (ssODN), which was silently mutated with code modification in gene-editing binding sites to prevent cutting and also to create an MluI enzyme cutting site for the genotype, was synthesized by Integrated DNA Technologies. The ssODN sequence was 5′-CAA GCC CTC AAC ACC ACG GTG CTG CAG GGC ATG GCC GGC CAG TCG TTA AGG GTA TCC TGC ACT TAT GAC GCG TTG AAA CAT TGG GGC AGA CAT AAG GCC TGG TGT CGG CAG CTG GGT GAG GAG GGC CCA TGC CAG CGT GTG GT-3′.
Microinjection
Two TREM2 gene-edited mRNAs were injected into the pronuclei of fertilized oocytes obtained from superovulated females of the C57BL/6 strain for TREM2 knockout. Two TREM2 gene-edited mRNAs and a 143-mer ssODN with an R47H mutation were injected into the pronuclei of fertilized oocytes obtained from superovulated females of the C57BL/6 strain for TREM2 R47H mutation. Genotyping for TREM2 KO and R47H KI mice was confirmed by PCR from tail genomic DNA.
LPS administration
4–5-month-old male KOMP and gene-edited mice were all maintained in C57BL/6 background. They were housed at a constant ambient temperature of 21 °C with a 12-h/12-h light dark cycle and were given access to food and water ad libitum. Equal numbers of animals from each genotype were randomly assigned to different groups. Treatment groups were given Escherichia coli LPS (Sigma-Aldrich) prepared in sterile saline and administered i.p. by single injection at a dose of 5 mg/kg, whereas control groups received saline i.p. Animals were dosed by a trained technician who was blinded with respect to animals' identifications, including genotypes. Animals with clinical signs of pain or distress after treatments, including hunched posture, rough hair coat, increased respiration, and lethargy were excluded by humane euthanasia. Tissue samples were collected at the 4- and 24-h time points. Mice were euthanized with CO2 inhalation for 2 min, and blood was withdrawn by cardiac puncture. Brains were removed, divided in half, and stored at −80 °C until use. The treatment group sizes were as follows: Trem2−/− saline-treated, n = 3; WT littermates, n = 6; Trem2−/− LPS-treated, n = 3; WT littermates, n = 6; Trem2R47H saline-treated, n = 5; WT littermates, n = 6; Trem2R47H LPS-treated, n = 6; WT littermates, n = 6; Trem2KOMP saline-treated, n = 4; WT littermates, n = 5. Post hoc analysis showed that a sample size of n = 5 was adequate to detect 20% difference between groups with respect to -fold change of transcripts. A second experiment was run with Trem2−/− and Trem2KOMP mice (n = 6 animals in each treatment group), confirming the results shown (data not shown).
Brain qPCR
Frozen brain halves were homogenized in RNA lysis buffer, and RNA was isolated by using the RNeasy minikit (Qiagen, Redwood City, CA). Total RNA was quantified on a Nanodrop Spectrophotometer (Thermo Fisher Scientific), and the cDNA was synthesized using the Cells-to-CT Bulk RT Reagents (Applied Biosystems). qPCR was done in a 384-well format using TaqMan® Universal PCR Master Mix (Applied Biosystems) on a Viia7 real-time PCR system (Thermo Fisher Scientific) with the following cycle conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, 1 min at 60 °C.
Antibodies
Rat monoclonal anti-human/mouse TREM2 (rat IgG2b clone 237920, R&D Systems) and internally generated mouse monoclonal anti-mouse TREM2 were used to activate TREM2 signaling along with the respective isotype controls (monoclonal rat IgG2B clone 141945, R&D Systems; internally generated isotype control).
Differentiation of mouse BMDMs
Bone marrow cells from Trem2−/−, Trem2−/+, Trem2R47H,Trem2R47H/+, and Trem2+/+ mice were obtained from femurs and tibiae using standard protocols (32) and seeded in DMEM supplemented with 10% heat-inactivated FBS and 50 ng/ml mouse CSF-1 (R&D Systems) in non-TC-treated sterile Petri dishes (Thermo Fisher Scientific). Nonadherent cells were removed by washing with cold PBS (Gibco) on day 5 or day 6 in culture (unless otherwise indicated), and the adherent macrophages were harvested by gentle scraping, counted, and used for further downstream analysis.
BMDM survival studies and flow cytometry analysis
Equal numbers of freshly harvested bone marrow cells from Trem2−/−, Trem2−/+, Trem2R47H, Trem2R47H/+, and Trem2+/+ mice (n = 3 animals/genotype with the exception of WT age-matched littermate controls for day 6 samples in the knockout experiment) were differentiated and harvested on the indicated days, and cell numbers were determined using a ViCell Cell Counter (BD Biosciences). Day 6 and day 7 BMDMs were harvested and stained with FITC-annexin V and PI and analyzed on an LSR II flow cytometer (BD Biosciences). TREM2 surface expression was also measured in the macrophages using APC-conjugated mouse anti-human TREM2 antibody (clone 23790, R&D Systems).
Microglia isolation and survival study
Young mice (n = 3/genotype) were euthanized via CO2 asphyxiation, and brains were dissected out. Single-cell suspensions were isolated using Miltenyi Biotec's adult mouse brain dissociation kit, followed by microglia isolation using Miltenyi Biotec's CD11b microbeads. Microglia were plated on a poly-d-lysine–coated 96-well plate in 200 μl of complete medium (DMEM/F-12 medium supplemented with 10% heat-inactivated FBS, 5% L929 conditioned medium, 1% pencillin/streptomycin, 1× Glutamax, 20 ng/ml granulocyte/macrophage colony–stimulating factor, 20 ng/ml M-CSF, 5 ng/ml TGF-β1). On the third day after plating and every 2–3 days thereafter, medium was replaced with basal medium (DMEM/F-12 with 10% heat-inactivated FBS). Confluence was measured using Incucyte Zoom (Essen Bioscience). For antibody treatment, basal medium was supplemented with 300 nm TREM2-activating antibodies or respective isotype control.
RNA-Seq
Day 6 BMDMs were harvested from Trem2−/−, Trem2R47H, and respective WT littermates (n = 5 animals/genotype), and total RNA was isolated using an RNeasy minikit (Qiagen) according to the manufacturer's protocol.
cDNA library preparation and NSG
1–2 μg of total RNA purified from BM-derived ex vivo macrophages was used for cDNA library preparation by using a modified protocol based on the Illumina Truseq RNA sample preparation kit (Illumina, San Diego, CA) and the published methods for strand-specific RNA-Seq (33, 34). After poly(A) selection, fragmentation, and priming, reverse transcription was carried out for first strand cDNA synthesis in the presence of RNaseOut (Life Technologies, Inc., Carlsbad, CA) and actinomycin D (MP Biomedicals, Santa Ana, CA). The synthesized cDNA was further purified by using AMPure RNAClean beads (Beckman Coulter, Pasadena, CA) following the commercial instruction. A modified method by incorporation of dUTP instead of dTTP was prepared and used for the second strand synthesis (33, 34). After AMPure XP bead purification (Beckman Coulter), following the standard protocol recommended by Illumina, end-repairing, A-tailing, and ligation of index adaptors were sequentially performed for generation of cDNA libraries. After size selection of libraries using Pippen Prep (SAGE Biosciences, Beverly, MA), the dUTP-containing cDNA strands were destroyed by digestion of USER enzymes (New England Biolabs, Ipswich, MA) followed by a step of PCR enrichment for introduction of strand specificity. After cleaning up, the enriched cDNA libraries were analyzed in an Agilent Bioanalyzer and quantified by Quant-iTTM Pico-Green assays (Life Technologies) before being sequenced onto the Illumina HiSeq platform. Each library generated at least 35 million 75-bp pair-end reads for downstream analysis.
RNA-Seq data analysis
RNA-Seq sequencing reads were aligned using OSA aligner (35) embedded in the Omicsoft ArrayStudio pipeline (Omicsoft Inc.). Mouse genome version GRCm38 and UCSC gene annotation were used in the alignment and quantification. Quantification was performed to the gene and transcript level based on RSEM (36). Normalized gene expression level was calculated by fragments per kilobase per million reads (FPKM) and then quantile-normalized at 70 percentile to 10 (FPKQ). Only genes with at least one sample expressed at FPKQ ≥ 1 were used in the following statistical analysis. Raw read counts from the selected genes were compared using the R Bioconductor package DESeq2 following negative binomial distribution (37). Genes with Benjamini-Hochberg–corrected p value <0.05 and -fold change ≥1.5 or ≤⅔ were selected as significantly differentially expressed genes. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA; Qiagen). Differentially expressed genes in both Trem2−/− versus WT and Trem2R47H were used to construct a gene co-expression network. Weighted gene co-expression network analysis (WGCNA) was performed using the Bioconductor package WGCNA. Modules in the network were defined by dynamic tree cutting in WGCNA. Genes with an absolute correlation coefficient of >0.95 with other genes were shown on the network. The red nodes are genes up-regulated in both Trem2−/− and Trem2R47H, whereas the green nodes are genes down-regulated in both Trem2−/− versus WT and Trem2R47H.
qPCR confirmation of subset of differentially regulated genes
BMDMs from Trem2R47H, Trem2−/− and respective WT littermates were harvested daily between day 4 and day 8. Microglia were isolated from Trem2R47H and WT littermates treated with saline or LPS, as described previously. Total RNA was isolated using the RNeasy minikit (Qiagen) according to the manufacturer's protocol. cDNA was generated using the RT Reagents (Thermo Fisher Scientific). Primers specific to Apoe, Il-1a, Cx3cr1, C1qa, Ccl5, Ccl22, Ccr2, Flt-1, and C3 (macrophages) and Il-1b, Il-6, and Tnf-a (microglia) were ordered from Thermo Fisher Scientific, and quantitative RT-PCR was performed in a Viia7 real-time PCR machine (Thermo Fisher Scientific) using the Taqman gene expression master mix.
For antibody treatment of macrophages before qPCR, day 5 BMDMs from Trem2−/− and Trem2R47H and respective WT littermates (n = 3 animals/genotype) were harvested and reseeded in 6-well plates at 2 million cells/well. The cells were treated with rat IgG2b or anti-Trem2 antibody (R&D MAB17291) overnight. The subsequent steps for performing qPCR were the same as described above, using primers specific to Melk, Nek2, Mmp14, and Ccl2.
Macrophage plug area migration assay
Day 5 BMDMs from Trem2+/+, Trem2R47H, and Trem2−/− mice were harvested and seeded into RadiusTM 96-well migration assay plates (Cell Biolabs) in complete RPMI medium supplemented with 50 ng/ml M-CSF (R&D Systems). The cells were treated with either anti-TREM2 antibody, isotype control, or vehicle for 24 h. The cells were washed the next day following the manufacturer's protocol to remove the Biocompatible Gel layer and expose the cell-free area for migration. The medium was replaced with fresh growth medium supplemented with 50 ng/ml M-CSF and antibody, isotype, or vehicle control as above. The cell confluence was monitored using the Incucyte Zoom imaging system, and data were plotted as percent confluence.
Peritoneal lavage
Zymosan A (Sigma-Aldrich Z-4250, Saccharomyces cerevisiae) powder was suspended in PBS (1 mg/ml), sonicated with heat, and vortexed every 10 min for 30 min. The Zymosan suspension was further diluted to 0.2 mg/ml in PBS, and 500 μl was injected i.p. into Trem2+/+ and Trem2−/− mice (n = 6/genotype). 3.5 and 6.5 h postinjection, mice were euthanized via CO2 asphyxiation. Peritoneal lavage was performed with 5 ml of ice-cold PBS. The peritoneal lavage fluid was centrifuged to pellet cells, and the supernatant was collected for further analysis.
Chemokine LuminexTM assay
Microglia was isolated as described previously and plated on a poly-d-lysine–coated 96-well plate in 200 μl of complete medium at 40,000 cells/well. After 4 days, medium was replaced with complete medium supplemented with TREM2 antibody or corresponding isotype control. After 6 h, cells were treated with AggreSureTM Aβ(1–42) or scrambled control (Anaspec). Final concentrations of antibody and Aβ were 300 nm and 10 μg/ml, respectively, in a 200-μl volume. 16 h after Aβ treatment, cell culture supernatant was collected and assayed for chemokine levels with Chemokine 9-Plex Mouse ProcartaPlexTM Panel 1 (Thermo Fisher) and analyzed with a Bio-PlexTM 200 analyzer (Bio-Rad).
Chemokine ELISAs
Day 5 BMDMs from Trem2R47H mice and WT littermates (n = 3 animals/genotype) were harvested, and an equal number of cells were plated in 96-well plates and allowed to adhere overnight. On the following day, cells were treated with 10 μg/ml AggreSureTM Aβ(1–42) (Anaspec) for 2 h. The conditioned medium was collected after 2 h, and chemokine levels were measured using Quantikine ELISA kits (R&D Systems). CCL2 levels in brain homogenates from LPS-treated animals (see “LPS administration” above) and peritoneal lavage from Zymosan-treated animals were also measured using the same kit. Samples were measured in triplicate. For measurement of CCL2 protein following antibody treatment, day 6 BMDMs from Trem2R47H and WT littermates (n = 2 animals/genotype) were treated with anti-TREM2 antibody or isotype control for 24 h in triplicate, and the CCL2 levels in the conditioned medium were measured using the same kit.
BMDM imaging
Day 6 BMDMs from Trem2−/− and Trem2R47H mice (n = 2 animals/genotype) were harvested, and equal numbers of cells were seeded in 96-well plates. 2 h post-plating, the cells were treated with anti-TREM2 antibodies or corresponding isotype controls, the cell confluence was monitored using the Incucyte Zoom Imaging System, and data were plotted as percent confluence. Each sample was measured in triplicate.
Cell viability
Day 6 BMDMs from 18-month-old Trem2R47H and age-matched WT littermates (R47H, n = 22 animals; WT, n = 24 animals) were harvested, and an equal number of cells were seeded in 96-well plates. 2 h post-plating, the cells were treated with R&D monoclonal antibody or isotype control for 14 days, and cell viability was measured using CellTiter Glo (Promega). Each treatment was done in triplicate.
Syk phosphorylation assay
For measurement of Syk phosphorylation, day 5 BMDMs from Trem2−/−, Trem2R47H, and respective WT littermates (n = 3 animals/genotype) were plated in 6-well tissue culture–treated dishes and allowed to adhere overnight. Cells were stimulated with anti-TREM2 antibodies or appropriate controls and incubated for 10 min at 37 °C. Each treatment was done in duplicate. At the end of the incubation time, the medium was removed, and cells were lysed with M-PER lysis buffer (Thermo Fisher Scientific) supplemented with HALT protease and phosphatase inhibitor (Thermo Fisher Scientific).
pSyk Western blotting
15 or 30 μg of total protein from the aforementioned BMDM cell lysates were separated by 4–12% BisTris SDS-PAGE (Thermo Fisher Scientific) and then transferred to polyvinylidene difluoride membranes. The membranes were probed with rabbit anti-pSYK (Cell Signaling) or rabbit anti-SYK (Cell Signaling) and mouse anti-actin (Sigma-Aldrich). Bands were visualized and quantified on the Bio-Rad imager.
Adult microglia transwell migration assay
Microglia were plated in complete medium at 5000 cells/well in transwell plates (Essen Bioscience) precoated with 20 μg/ml Protein G (Life Technologies) and 5 μg/ml I-CAM (Life Technologies). Transwells were placed in reservoir plates containing 200 μl of complete medium with 1 μg/ml recombinant C5a (R&D Systems). For migration assays with antibody treatment, microglia were plated at 3000 cells/well in complete medium supplemented with 300 nm TREM2 antibodies or corresponding isotype control. Transwells were initially placed in a reservoir with complete medium, and on day 5, medium in the transwell was replaced with 300 nm TREM2 antibodies or corresponding isotype controls in complete medium. Transwells were moved to a new reservoir plate containing 200 μl of complete medium with 1 μg/ml recombinant C5a or 100 ng/ml recombinant C5a and 10 μg/ml AggreSureTM Aβ 1–42 (Anaspec). Transwell migration was monitored and analyzed using Incucyte Zoom (Essen Bioscience).
Author contributions
Q. C., J. D., S. T., P. W., J. Y., N. S., D. C., D. T., H. C., and S. S. methodology; J. D., P. W., J. Y., N. S., D. C., D. T., and S. S. data curation; J. D. formal analysis; J. D., S. T., P. W., J. Y., N. S., D. C., D. T., H. C., and S. S. investigation; S. T. and S. S. writing-review and editing; C.-M. L., D. C., S. K., H. C., I. N. F., S. W., and S. S. conceptualization; C.-M. L., S. K., and S. S. supervision; S.S. writing-original draft.
Supplementary Material
This work was supported by Amgen Inc., and all authors were employees of Amgen at the time the research was conducted.
This article contains Figs. S1–S7.
- AD
- Alzheimer's disease
- LOAD
- late-onset Alzheimer's disease
- qPCR
- quantitative PCR
- LPS
- lipopolysaccharide
- BMDM
- bone marrow–derived macrophage
- i.p.
- intraperitoneally
- ssODN
- single-stranded oligonucleotide
- M-CSF
- macrophage colony–stimulating factor
- WGCNA
- weighted gene co-expression network analysis
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance
- Aβ
- amyloid β.
References
- 1. Schmid C. D., Sautkulis L. N., Danielson P. E., Cooper J., Hasel K. W., Hilbush B. S., Sutcliffe J. G., and Carson M. J. (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J. Neurochem. 83, 1309–1320 10.1046/j.1471-4159.2002.01243.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kiialainen A., Hovanes K., Paloneva J., Kopra O., and Peltonen L. (2005) Dap12 and Trem2, molecules involved in innate immunity and neurodegeneration, are co-expressed in the CNS. Neurobiol. Dis. 18, 314–322 10.1016/j.nbd.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 3. Bouchon A., Hernández-Munain C., Cella M., and Colonna M. (2001) A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med. 194, 1111–1122 10.1084/jem.194.8.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J. S., Younkin S., Hazrati L., Collinge J., Pocock J., Lashley T., Williams J., et al. (2013) TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 368, 117–127 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., Rujescu D., Hampel H., Giegling I., Andreassen O. A., Engedal K., et al. (2013) Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guerreiro R. J., Lohmann E., Brás J. M., Gibbs J. R., Rohrer J. D., Gurunlian N., Dursun B., Bilgic B., Hanagasi H., Gurvit H., Emre M., Singleton A., and Hardy J. (2013) Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 70, 78–84 10.1001/jamaneurol.2013.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeh F. L., Wang Y., Tom I., Gonzalez L. C., and Sheng M. (2016) TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-β by microglia. Neuron 91, 328–340 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Cella M., Mallinson K., Ulrich J. D., Young K. L., Robinette M. L., Gilfillan S., Krishnan G. M., Sudhakar S., Zinselmeyer B. H., Holtzman D. M., Cirrito J. R., and Colonna M. (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160, 1061–1071 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atagi Y., Liu C. C., Painter M. M., Chen X. F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S. S., Xu H., Younkin S., Das P., Fryer J. D., and Bu G. (2015) Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem. 290, 26043–26050 10.1074/jbc.M115.679043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey C. C., DeVaux L. B., and Farzan M. (2015) The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J. Biol. Chem. 290, 26033–26042 10.1074/jbc.M115.677286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ulland T. K., Song W. M., Huang S. C., Ulrich J. D., Sergushichev A., Beatty W. L., Loboda A. A., Zhou Y., Cairns N. J., Kambal A., Loginicheva E., Gilfillan S., Cella M., Virgin H. W., Unanue E. R., et al. (2017) TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell 170, 649–663.e13 10.1016/j.cell.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan P., Condello C., Keene C. D., Wang Y., Bird T. D., Paul S. M., Luo W., Colonna M., Baddeley D., and Grutzendler J. (2016) TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 92, 252–264 10.1016/j.neuron.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 13. Jay T. R., Miller C. M., Cheng P. J., Graham L. C., Bemiller S., Broihier M. L., Xu G., Margevicius D., Karlo J. C., Sousa G. L., Cotleur A. C., Butovsky O., Bekris L., Staugaitis S. M., Leverenz J. B., et al. (2015) TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer's disease mouse models. J. Exp. Med. 212, 287–295 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jay T. R., Hirsch A. M., Broihier M. L., Miller C. M., Neilson L. E., Ransohoff R. M., Lamb B. T., and Landreth G. E. (2017) Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer's disease. J. Neurosci. 37, 637–647 10.1523/JNEUROSCI.2110-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bemiller S. M., McCray T. J., Allan K., Formica S. V., Xu G., Wilson G., Kokiko-Cochran O. N., Crish S. D., Lasagna-Reeves C. A., Ransohoff R. M., Landreth G. E., and Lamb B. T. (2017) TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol. Neurodegener. 12, 74 10.1186/s13024-017-0216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leyns C. E. G., Ulrich J. D., Finn M. B., Stewart F. R., Koscal L. J., Remolina Serrano J., Robinson G. O., Anderson E., Colonna M., and Holtzman D. M. (2017) TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. U.S.A. 114, 11524–11529 10.1073/pnas.1710311114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song W. M., Joshita S., Zhou Y., Ulland T. K., Gilfillan S., and Colonna M. (2018) Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215, 745–760 10.1084/jem.20171529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allcock R. J., Barrow A. D., Forbes S., Beck S., and Trowsdale J. (2003) The human TREM gene cluster at 6p21.1 encodes both activating and inhibitory single IgV domain receptors and includes NKp44. Eur. J. Immunol. 33, 567–577 10.1002/immu.200310033 [DOI] [PubMed] [Google Scholar]
- 19. Kang S. S., Kurti A., Baker K. E., Liu C. C., Colonna M., Ulrich J. D., Holtzman D. M., Bu G., and Fryer J. D. (2018) Behavioral and transcriptomic analysis of Trem2-null mice: not all knockout mice are created equal. Hum. Mol. Genet. 27, 211–223 10.1093/hmg/ddx366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otero K., Turnbull I. R., Poliani P. L., Vermi W., Cerutti E., Aoshi T., Tassi I., Takai T., Stanley S. L., Miller M., Shaw A. S., and Colonna M. (2009) Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 10, 734–743 10.1038/ni.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song W., Hooli B., Mullin K., Jin S. C., Cella M., Ulland T. K., Wang Y., Tanzi R. E., and Colonna M. (2017) Alzheimer's disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimers Dement. 13, 381–387 10.1016/j.jalz.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bouchon A., Dietrich J., and Colonna M. (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 164, 4991–4995 10.4049/jimmunol.164.10.4991 [DOI] [PubMed] [Google Scholar]
- 23. Benitez B. A., Jin S. C., Guerreiro R., Graham R., Lord J., Harold D., Sims R., Lambert J. C., Gibbs J. R., Bras J., Sassi C., Harari O., Bertelsen S., Lupton M. K., Powell J., et al. (2014) Missense variant in TREML2 protects against Alzheimer's disease. Neurobiol. Aging 35, 1510.e9–26 10.1016/j.neurobiolaging.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng H., Liu C. C., Atagi Y., Chen X. F., Jia L., Yang L., He W., Zhang X., Kang S. S., Rosenberry T. L., Fryer J. D., Zhang Y. W., Xu H., and Bu G. (2016) Opposing roles of the triggering receptor expressed on myeloid cells 2 and triggering receptor expressed on myeloid cells-like transcript 2 in microglia activation. Neurobiol. Aging 42, 132–141 10.1016/j.neurobiolaging.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., David E., Baruch K., Lara-Astaiso D., Toth B., Itzkovitz S., Colonna M., Schwartz M., and Amit I. (2017) A unique microglia type associated with restricting development of Alzheimer's disease. Cell 169, 1276–1290.e17 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 26. Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O'Loughlin E., Xu Y., Fanek Z., Greco D. J., Smith S. T., Tweet G., Humulock Z., Zrzavy T., et al. (2017) The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishizuka K., Kimura T., Igata-yi R., Katsuragi S., Takamatsu J., and Miyakawa T. (1997) Identification of monocyte chemoattractant protein-1 in senile plaques and reactive microglia of Alzheimer's disease. Psychiatry Clin. Neurosci. 51, 135–138 10.1111/j.1440-1819.1997.tb02375.x [DOI] [PubMed] [Google Scholar]
- 28. Westin K., Buchhave P., Nielsen H., Minthon L., Janciauskiene S., and Hansson O. (2012) CCL2 is associated with a faster rate of cognitive decline during early stages of Alzheimer's disease. PLoS One 7, e30525 10.1371/journal.pone.0030525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guedes J. R., Santana I., Cunha C., Duro D., Almeida M. R., Cardoso A. M., de Lima M. C., and Cardoso A. L. (2016) MicroRNA deregulation and chemotaxis and phagocytosis impairment in Alzheimer's disease. Alzheimers Dement. (Amst.) 3, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Khoury J., Toft M., Hickman S. E., Means T. K., Terada K., Geula C., and Luster A. D. (2007) Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 10.1038/nm1555 [DOI] [PubMed] [Google Scholar]
- 31. Liddelow S. A., Guttenplan K. A., Clarke L. E., Bennett F. C., Bohlen C. J., Schirmer L., Bennett M. L., Münch A. E., Chung W. S., Peterson T. C., Wilton D. K., Frouin A., Napier B. A., Panicker N., Kumar M., et al. (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zanoni I., Ostuni R., Capuano G., Collini M., Caccia M., Ronchi A. E., Rocchetti M., Mingozzi F., Foti M., Chirico G., Costa B., Zaza A., Ricciardi-Castagnoli P., and Granucci F. (2009) CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature 460, 264–268 10.1038/nature08118 [DOI] [PubMed] [Google Scholar]
- 33. Perkins T. T., Kingsley R. A., Fookes M. C., Gardner P. P., James K. D., Yu L., Assefa S. A., He M., Croucher N. J., Pickard D. J., Maskell D. J., Parkhill J., Choudhary J., Thomson N. R., and Dougan G. (2009) A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 5, e1000569 10.1371/journal.pgen.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., and Soldatov A. (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu J., Ge H., Newman M., and Liu K. (2012) OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics 28, 1933–1934 10.1093/bioinformatics/bts294 [DOI] [PubMed] [Google Scholar]
- 36. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.