S-acylation of the beet severe curly top virus C4 protein is essential for its membrane association, interaction with CLV1, and symptom determination during geminivirus infection.
Keywords: C4, CLAVATA, geminivirus, membrane association, S-acylation, symptom determination
Abstract
Geminiviruses, such as beet severe curly top virus (BSCTV), are a group of DNA viruses that cause severe plant diseases and agricultural losses. The C4 protein is a major symptom determinant in several geminiviruses; however, its regulatory mechanism and molecular function in plant cells remain unclear. Here, we show that BSCTV C4 is S-acylated in planta, and that this post-translational lipid modification is necessary for its membrane localization and functions, especially its regulation of shoot development of host plants. Furthermore, the S-acylated form of C4 interacts with CLAVATA 1 (CLV1), an important receptor kinase in meristem maintenance, and consequentially affects the expression of WUSCHEL, a major target of CLV1. The abnormal development of siliques in Arabidopsis thaliana infected with BSCTV is also dependent on the S-acylation of C4, implying a potential role of CLAVATA signaling in this process. Collectively, our results show that S-acylation is essential for BSCTV C4 function, including the regulation of the CLAVATA pathway, during geminivirus infection.
Introduction
Geminiviruses are a family of plant viruses with circular single-stranded DNA genomes that cause plant diseases worldwide. Geminivirus-encoded proteins regulate host cellular processes, including DNA replication, gene silencing, and hormone signaling, to complete their infection cycle (Ramesh et al., 2017). The cell cycle is an important process targeted by geminiviruses (Gutierrez, 1999); for instance, the geminivirus Rep protein binds to RETINOBLASTOMA-RELATED, a critical plant cell cycle regulator, and thereby releases the transcription factor E2F for virus replication (Arguello-Astorga et al., 2004). Although the Rep-related proteins are essential for virus replication, the abnormal cell division of host plants infected with many geminiviruses is induced by symptom determinants (Hanley-Bowdoin et al., 2013). Understanding the regulatory mechanisms of symptom determinants is therefore important for developing strategies to protect plants from geminiviruses.
The conserved C4 (or L4) proteins in monopartite geminiviruses and AC4 (or AL4) proteins in bipartite geminiviruses have important functions (Deom and Mills-Lujan, 2015). The disruption of C4 reduces symptom development in many geminivirus infections (Stanley and Latham, 1992; Teng et al., 2010), suggesting it is a major determinant of pathogenesis. The expression of C4 genes from several geminiviruses, including beet curly top virus (BCTV) and beet severe curly top virus (BSCTV), results in ectopic cell division in Arabidopsis thaliana and Nicotiana benthamiana (Latham, 1997; Piroux et al., 2007; Mills-Lujan and Deom, 2010), supporting the idea that the leaf-curling and vein-swelling symptoms are caused by an abnormal cell cycle. Previous studies have indicated that BSCTV C4 induces the expression of RKP for cell cycle regulation (Lai et al., 2009) and ATHB7/12 for infected cell development (Park et al., 2011), consistent with its effects on cell division. The C4 proteins have been shown to interact with SHAGGY-like protein kinases (Piroux et al., 2007) and affect the brassinosteroid (BR) pathways (Mills-Lujan and Deom, 2010; Bi et al., 2017). In addition, the C4 proteins of some geminiviruses are involved in virus movement and gene silencing (Ramesh et al., 2017). Despite these recent discoveries, the precise molecular mechanisms by which C4 contributes to symptom determination, as well as how C4 is regulated in the host cells, remain unclear.
Phosphorylation and myristoylation have been reported to regulate the function of C4 proteins in pathogenesis (Fondong et al., 2007; Piroux et al., 2007; Mills-Lujan et al., 2015); however, it is unclear whether other regulatory mechanisms contribute to the function of C4 during geminivirus infection. Because BSCTV is able to efficiently infect model plants such as Arabidopsis and N. benthamiana, causing severe symptoms (Park et al., 2002), it provides a powerful system to study the interactions between geminiviruses and hosts. Here, we determined that the C4 protein from BSCTV is S-acylated in plant cells. S-acylation is a type of lipid modification that transfers a long-chain fatty acid such as palmitate to the cysteine residues of proteins (Linder and Deschenes, 2007). Unlike myristoylation, which is irreversible, S-acylation is a reversible modification that regulates the subcellular localization, stability, and activity of the substrates, and is important for many biological processes in a variety of plant species (Li and Qi, 2017); for example, S-acylation regulates the membrane association of the RHO family proteins (Lavy et al., 2002) and calcineurin B-like proteins (CBLs) (Batistič et al., 2012) for signaling transduction. Recently, S-acylation was reported to be involved in the regulation of leaf senescence, stress tolerance, and root hair development (Hemsley et al., 2005; Zhou et al., 2013; Lai et al., 2015), but its functions in plant–virus interactions remain unknown.
Materials and methods
Plant material and growth conditions
Seeds of Arabidopsis thaliana Columbia-0 (wild-type, WT) were surface-sterilized for 2 min in 75% ethanol followed by 5 min in 1% NaClO solution, rinsed five times with sterile water and plated on Murashige and Skoog (MS) medium with 1.5% sucrose and 0.8% agar. They were then stratified at 4 °C in the dark for 2 d and subsequently grown under long-day conditions (16/8 h of light/dark) at 22 °C.
Protoplast transformation and confocal microscopy
For transient expression in protoplasts, the coding sequence (CDS) of C4 was cloned into a pBluscript-based vector under the 35S promoter, fused with yellow fluorescent protein (YFP), 3× FLAG, and 6× histidines (Liu et al., 2016). The C8S mutation version of this plasmid was generated by PCR using a 5′-primer harboring the mutation site. The plasmids expressing different versions of C4 proteins were transformed into Arabidopsis protoplasts as previously described (Yoo et al., 2007). The transformed protoplasts were incubated for 18 h at 22 °C, and the subcellular localization of YFP-fused proteins was observed under a Zeiss LSM 710 laser-scanning microscope (514 nm for excitation and 530–600 nm for emission).
Acyl-RAC assays
Acyl-resin-assisted capture (Acyl-RAC) assays were performed following the protocol previously described by Lai et al. (2015). In particular, the plasmids expressing C4WT-YFP-FLAG3His6 or C4C8S-YFP-FLAG3His6 were transformed into protoplasts as described by Yoo et al. (2007). After incubation for 48 h (or other specified times), the cells were collected and lysed in lysis buffer (25 mM HEPES, 25 mM NaCl, 1 mM EDTA, pH 7.5) containing a protease inhibitor cocktail. Equal amounts of proteins were diluted in blocking buffer (100 mM HEPES, 1.0 mM EDTA, 2.5% SDS, 0.5% MMTS, pH 7.5) and incubated at 40 °C for 10 min with frequent vortexing. Three volumes of cold acetone were added, and the sample was allowed to precipitate at −20 °C for 20 min. The precipitated proteins were collected by centrifugation at 5000 g for 10 min, and the pellet was washed with 70% acetone, re-suspended in 300 µl of binding buffer (100 mM HEPES, 1.0 mM EDTA, 1% SDS, pH 7.5) and mixed with 40 µl of pre-washed Thiopropyl Sepharose 6B (Sigma). Then, 40 µl of either 2 M NH2OH (pH 7.5) or 2 M NaCl (control) was added into this mixture. The mixtures were rotated at room temperature for 2 h and 20 µl of each supernatant was saved as the total input. Resins were washed five times with binding buffer. Elution was performed using 60 µl of binding buffer containing 50 mM DTT at room temperature for 20 min. Supernatants were removed and mixed with protein sample buffer, incubated at 95 °C for 5 min, and then used for SDS-PAGE and immunological detection.
Cell fraction assays
The wild-type or mutant versions of C4-YFP-FLAG3His6 were transiently expressed in protoplasts. At 48 h after transformation, the protoplasts were collected for cell fraction as previously described (Mei et al., 2018) with minor modification. The cells were re-suspended in homogenization buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 13% sucrose with protease inhibitor). The samples were centrifugated at 3000 g for 20 min at 4 °C to remove nuclei and large cellular debris, and the supernatant was ultra-centrifuged at 50000 g for 1 h at 4 °C to generate soluble and pellet fractions. The pellet fraction was re-suspended in homogenization buffer. All fractions were then used for immunoblot analysis.
Generation of transgenic plants
For inducible expression of BSCTV C4 in Arabidopsis, the pER8-C4 plasmid described previously was used (Lai et al., 2009). To generate the C8S mutation versions of this plasmid, a 5′-primer harboring the mutation site was used in PCR amplification. The constructs were transformed into Agrobacterium EHA105, which was then used to transform wild-type Arabidopsis (Columbia) by the floral-dip method (Clough and Bent, 1998). The seeds were germinated on media including DMSO or 2 µM estradiol (Sigma), or germinated on regular MS medium for 7d and transferred onto media with or without 2 µM estradiol for an additional 3 weeks.
Transient expression of proteins in Nicotiana benthamiana leaves
For transient expression in N. benthamiana, the wild-type or mutant version of C4 was cloned into the pCanG-MYC vector and fused with a MYC tag at its C terminus under the 35S promoter. Leaves of N. benthamiana were infiltrated via A. tumefaciens as described by Liu et al. (2010). At 4 d after infiltration, the leaves were imaged and total protein was extracted for immunological blotting.
Virus inoculation
For construction of the BSCTV plasmids, the wild-type virus with 1.8 copies of the BSCTV genome in the pCAMBIA1300 vector was used, as previously described (Lai et al., 2009). To generate the virus carrying the C8S mutant on C4 (which did not alter the amino acid sequence of the overlapping Rep protein), the 0.8 copy and 1 copy of the virus genome were separately cloned into pET28a as an intermediate vector, and the mutation was introduced by site-directed mutagenesis. These mutant fragments were then used to replace the wild-type version of the BSCTV genome sequentially in pCAMBIA1300. The insertion direction and sequence information were confirmed by digestion and DNA sequencing. The constructs were transformed into A. tumefaciens EHA105. The agrobacteria were resuspended and adjusted to OD600=1, and then serial dilutions were used for inoculation on the leaves of N. benthamiana or Arabidopsis plants (Teng et al., 2010). Symptoms occurred around 9 d after inoculation.
DNA gel blot analysis
For the DNA gel blot analysis, total DNA was extracted using the CTAB method from protoplasts or N. benthamiana tissues after BSCTV infection. Total genomic DNA was separated by electrophoresis in 0.8% agarose gels and transferred to a Hybond N+ membrane. A whole-genome fragment of BSCTV, digested from pCambia-1300-BSCTV with EcoRI, was used as a probe (Teng et al., 2010). Probe labeling and signal detection were performed with a DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche).
Yeast two-hybrid assays
The CDS of C4 was cloned into pGBKT7 and the intracellular catalytic domain of CLV1 (704AA-967AA) was cloned into pGADT7. Yeast two-hybrid (Y2H) assays were performed using the Matchmaker GAL4-based Two-Hybrid System 3 (Clontech) according to the manufacturer’s instructions. Interactions were tested by stringent selection (SD/–Leu/–Trp/–His) supplied with 1 mM 3-amino-1,2,4-triazole (Sigma). Yeast cells with pGBKT7-p53 and pGADT7-T (Clontech) were used as positive controls; the yeast cells with pGBKT7-Lam and pGADT7-T (Clontech) were used as negative controls.
BiFC assays
The wild-type or mutant version of C4 was cloned into the pSPYCE-35 vector, while CLV1 was cloned into the pSPYNE-35S vector (Walter et al., 2004). The plasmids were co-transformed into Arabidopsis protoplasts. Combinations with empty vectors were used as negative controls. Bimolecular fluorescence complementation (BiFC) assays were performed as previously described (Walter et al., 2004). YFP signals were detected under a Zeiss LSM 710 laser-scanning microscope.
Co-Immunoprecipitation assays
For expression of CLV1-MYC in protoplasts, the full length CDS of CLV1 was cloned into pCanG-MYC by homologous recombination. Using pCanG-MYC alone as a control, pCanG-CLV1-MYC was co-transformed with the wild-type or mutant version of 35S::C4-YFP-FLAG3His6 into protoplasts (Yoo et al., 2007). At 48 h after transformation, the protoplasts were collected for co-immunoprecipitation (Co-IP) assays. Proteins were extracted in extraction buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10% glycerol, and 1% DDM) containing a protease inhibitor cocktail (Roche). After centrifugation at 13000 g for 10 min, the supernatant was incubated with anti-MYC (Cell Signaling Technology) antibody at 4 °C overnight, and rotated with the protein A resin for another 3 h. Then the resins were centrifuged and washed three times with washing buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10% glycerol, and 0.1% DDM). Proteins were eluted with SDS sample buffer and analysed by immunoblotting using anti-GFP (green fluorescent protein; Abcam) or anti-MYC (Cell Signaling Technology) antibodies. For Co-IP between CLV1-GFP and C4-MYC, the reagents and protocol were similar except that the GFP-Trap resin (Chromotek) was used instead.
GUS staining
For detection of the expression of WUS and CLV3, the WUSpro::GUS-GFP or CLV3pro::GUS-–GFP construct (previously described by Cui et al., 2015) was introduced into pER8-C4WT or pER8-C4C8S by genetic crossing. After growth on medium containing 2 µM estradiol for a specified time, the seedlings were sampled for GUS (β-glucuronidase) staining. The GUS activity analysis was performed according to the method described by De Schutter et al., 2007.
Results
S-acylation of C4 in plant cells is essential for its membrane localization
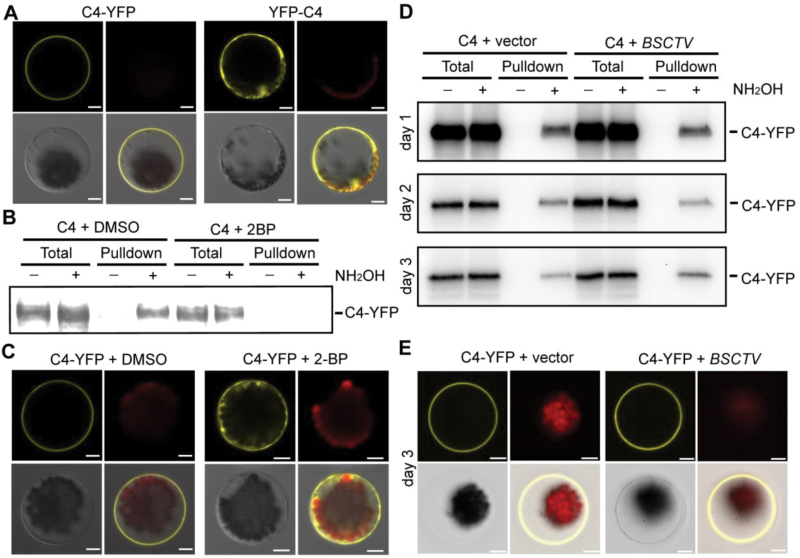
The C4 or AC4 proteins of most geminiviruses are localized to the membrane, although our previous study had shown that the N-terminal GFP/YFP fusion of the C4 protein from BSCTV was distributed in the cytosol of plant cells (Teng et al., 2010). When YFP was fused to the C terminus of BSCTV C4, the fluorescence signals were predominantly observed on the plasma membrane of the plant protoplasts (Fig. 1A), implying that the N-terminal fusion interferes with the membrane localization of the protein. The bioinformatics software CSS-Palm (Ren et al., 2008) predicted that the eighth residue on BSCTV C4 is a cysteine (C8) that is highly available for S-acylation. Cysteine residues are also found in the N termini of other geminiviruses (see Supplementary Fig. S1 at JXB online), suggesting a potential conserved mechanism among these virus species. An acyl-RAC assay (Lai et al., 2015) was used to provide biochemical evidence for the S-acylation of BSCTV C4. The C4 protein was pulled-down on Thiopropyl Sepharose in the presence of NH2OH (Fig. 1B), suggesting that it was S-acylated in plant cells. Because 2-bromopalmitate (2-BP) is an inhibitor of S-acylation (Jennings et al., 2009), it was used to determine the specificity of this assay. The modification of C4 was found to be sensitive to 2-BP, providing further support for the S-acylation of C4. Consistently, the membrane localization of C4-YFP was disrupted in the presence of 2-BP (Fig. 1C), suggesting that S-acylation was essential for the membrane association of C4. The S-acylation levels of C4 were also monitored during the BSCTV infection of plant cells. Compared with the vector control, BSCTV infection did not alter the S-acylation or subcellular localization of C4 (Fig. 1D, E).
Fig. 1.
S-acylation of BSCTV C4 in plant cells is essential for its membrane localization. (A) C4-YFP-FLAG3His6 or YFP-C4 were expressed in protoplasts and fluorescence was detected by confocal microscopy 18 h after transformation. The YFP signal (yellow), chloroplast autofluorescence (red), bright field (gray), and merged images are shown. Scale bars are 10 µm. (B) Detection of C4 S-acylation in plant cells. C4-YFP-FLAG3His6 was expressed in protoplasts (treated overnight with DMSO or 20 µM S-acylation inhibitor 2-BP) for acyl-resin-assisted capture assays. The signals from the total lysates are shown as the input control, and pulldown indicates the S-acylation proteins captured on the Thiopropyl Sepharose. S-acylation pulldown is dependent on NH2OH. The signals were detected using immunological blotting with an anti-FLAG antibody. (C) C4-YFP-FLAG3His6 was expressed in protoplasts with or without a treatment with 20 µM 2-BP. The fluorescence signals (yellow) were detected using confocal microscopy. Scale bars are 10 µm. (D) S-acylation of C4-YFP-FLAG3His6 was detected in protoplasts without (vector) or with BSCTV infection. The levels of S-acylation were measured 1, 2, and 3 d after infection. (E) Representative images of localization of C4-YFP-FLAG3His6 3 d after BSCTV infection. Scale bars are 10 µm. All results in this figure are representative of three independent experiments.
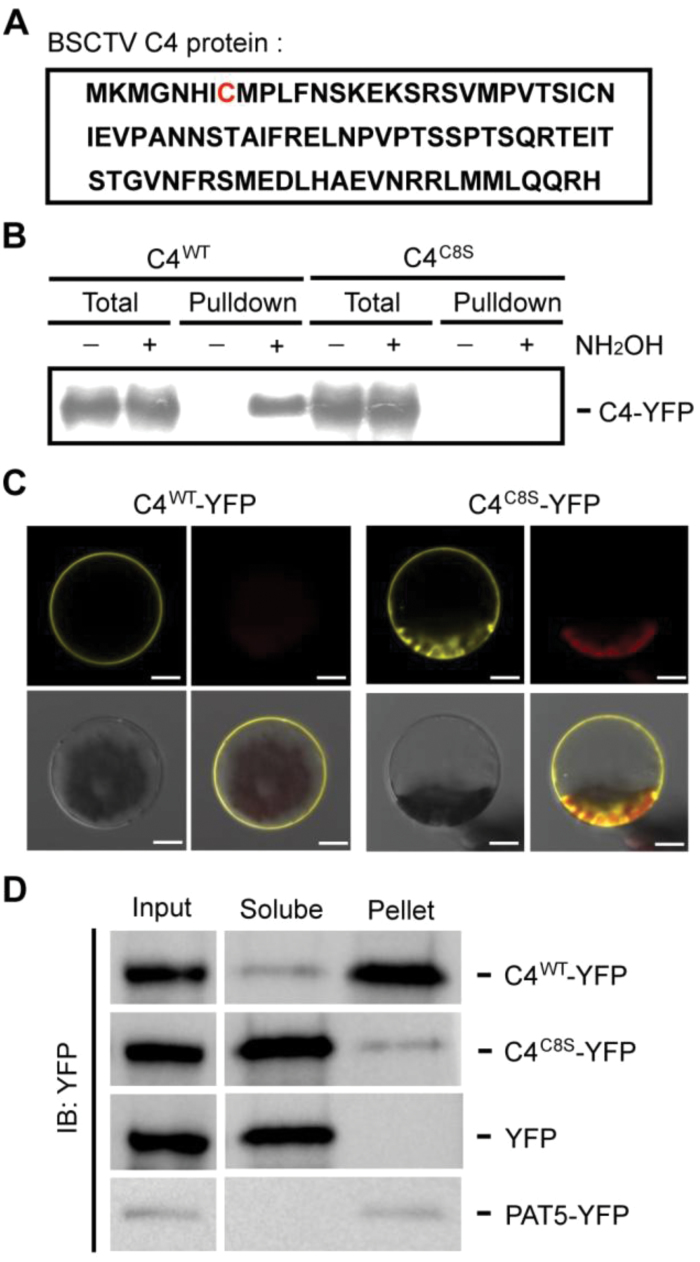
The putative residue for S-acylation, C8, was mutated from a cysteine to a serine (C8S) (Fig. 2A). An acyl-RAC assay was used to determine that this mutation resulted in the complete loss of BSCTV C4 S-acylation (Fig. 2B), indicating that this residue is the major S-acylation site on the viral protein. To confirm the requirement for S-acylation in the localization of C4, the fluorescence signals from plant cells expressing the wild-type or mutated versions of C4-YFP were detected. The results showed that mutating the C8 S-acylation site impaired the membrane association of C4 (Fig. 2C), consistent with our results from the 2-BP treatment. The S-acylation-mediated regulation of the subcellular localization of C4 was confirmed using a cell fraction assay. Similar to the transmembrane protein PAT5 (Protein S-Acyltransferase 5) (Batistič, 2012), the wild-type C4 was predominantly localized in the membrane fraction. When the S-acylation site was mutated, most C4 proteins were detected in the soluble fraction, with only a small amount of mutated protein associated with the membrane (Fig. 2D), supporting the results of the microscopy observations. The mutation of the other C4 cysteine residue, C28, did not affect the S-acylation or localization of the protein (Supplementary Fig. S2), confirming that only the eighth residue was S-acylated on C4.
Fig. 2.
Identification of the S-acylation site in BSCTV C4. (A) The predicted S-acylation site in BSCTV C4. The amino acid sequence of C4 is shown and the potential S-acylation site (C8) is highlighted in red. (B) Characterization of the S-acylation site on the C4 protein using acyl-resin-assisted capture assays. The wild-type (WT) or C8S (cysteine to serine) mutant version of C4-YFP-FLAG3His6 was expressed in protoplasts. The total lysate is shown as the input control, and pulldown indicates the S-acylation proteins captured on Thiopropyl Sepharose. The signals were detected using immunological blotting with an anti-FLAG antibody. (C) The localization of the WT or C8S version of C4-YFP-FLAG3His6, detected using confocal microscopy. The YFP signal (yellow), chloroplast autofluorescence (red), bright field (gray), and merged images are shown. Scale bars are 10 µm. (D) Protoplasts expressing the WT or C8S version of C4-YFP-FLAG3His6 were fractionated into soluble and pellet fractions. YFP and PAT5-YFP (a transmembrane protein) were used as controls. The signals were measured from an immunological blot using an anti-GFP antibody. All results in this figure are representative of three independent experiments.
Because myristoylation at the N terminus of proteins may regulate their S-acylation, the potential myristoylation site (G4) on BSCTV C4 was also mutated from a glycine to an alanine residue, which disrupted both the S-acylation and membrane association of C4 (Supplementary Fig. S3). The N terminus of BSCTV C4 contains two more residues than the C4 proteins of other geminiviruses (Supplementary Fig. S1). To exclude their effect on S-acylation, the first two residues of C4 were deleted (C4D2); however, the patterns of S-acylation and localization of C4D2 were similar to those of the full-length protein (Supplementary Fig. S4), suggesting that the S-acylation of BSCTV C4 was not dependent on the extra residues at its N terminus.
S-acylation of C4 is critical for its function as a symptom determinant
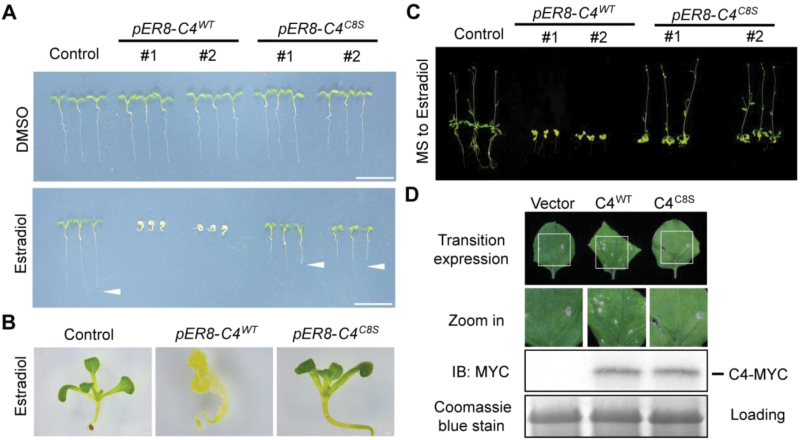
Because C4 is a symptom determinant in BSCTV, the wild-type or C8S versions of C4 were expressed in Arabidopsis to elucidate the effect of S-acylation on C4 function in the host plants. Transgenic plants constitutively expressing BSCTV C4 could not be obtained because of their ectopic cell division (Lai et al., 2009); therefore, the C4 gene was cloned into the pER8 vector and its expression was controlled using an inducible promoter (Zuo et al., 2000). In the presence of estradiol, the expression of both the wild-type and C8S versions of C4 were induced at similar levels in their respective transgenic lines (Supplementary Fig. S5). Seeds were germinated on media with or without the inducer to compare the effects of C4 on plant growth (Fig. 3A). The development of the transgenic plants without the induced C4 expression was normal, but in the presence of estradiol, plant growth was completely inhibited by the wild-type C4, consistent with previous results (Lai et al., 2009). The mutation of the S-acylation site dramatically attenuated the abnormal host development caused by C4, demonstrating the importance of S-acylation on the function of the viral protein.
Fig. 3.
S-acylation is important for the effect of C4 on Arabidopsis development. (A) Seedlings of estradiol-inducible pER8-C4WT (wild-type) and pER8-C4C8S (C8 mutated from cysteine to serine) plants germinated on media with (bottom panel) or without (top panel) 2 µM estradiol for 6 d. Scale bars are 1 cm. (B) Shoot phenotypes of control, pER8-C4WT, and pER8-C4C8S plants germinated on a medium containing 2 µM estradiol. (C) Seedlings were germinated and grown on regular medium for 7 d, then transferred onto a medium containing 2 µM estradiol for 3 weeks. The images are from independent experiments using individual transgenic lines, which produced similar patterns. (D) Agrobacteria carrying pCanG-MYC (Vector), pCanG-C4WT-MYC, or pCanG-C4C8S-MYC were injected into Nicotiana benthamiana leaves. After 4 d, their phenotypes were observed and the levels of C4 proteins were detected with using immunological blotting with an anti-MYC antibody. Coomassie Blue staining of total proteins was used as a loading control. WT, wild-type. All results in this figure are representative of at least three independent experiments.
In the pER8-C4C8S plants, root development was reduced and the phenotypes of the hypocotyls and cotyledons were also slightly altered, while the development of the shoot was normal (Fig. 3B). To further explore the effects of C4 on shoot development, 1-week-old seedlings were transferred from regular medium to inducing medium for C4 expression. In comparison with the pER8-C4WT plants, which had reduced shoot growth, no abnormal effects on shoot development were observed in the pER8-C4C8S plants (Fig. 3C), indicating that S-acylation was critical for the function of C4 in the regulation of the shoot meristem. The function of S-acylation in the symptom development mediated by C4 was also supported by a transient expression assay in N. benthamiana leaves. When similar levels of proteins were expressed, the wild-type C4 induced leaf curling and cell death, but this effect was impaired when the S-acylation site was mutated (Fig. 3D, Supplementary Fig. S6).
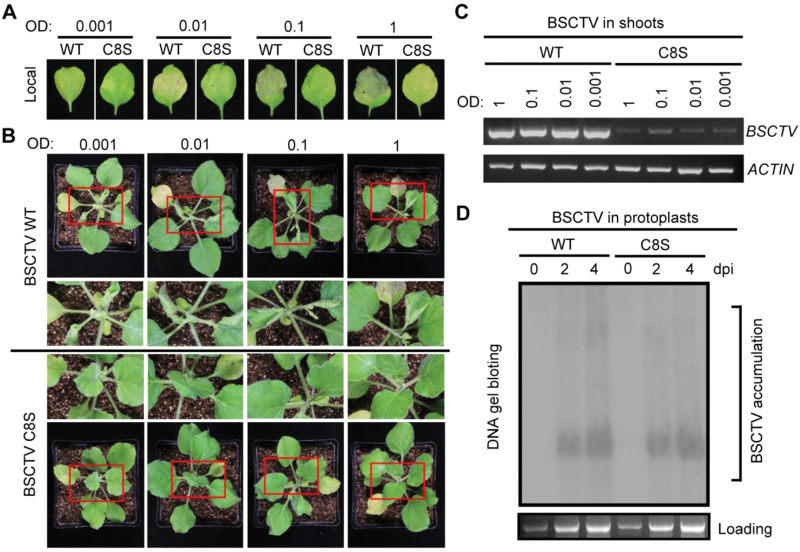
Next, we investigated the role of C4 S-acylation in the alteration of host shoot development during BSCTV infection. BSCTV carrying wild-type or C8S C4 (which did not alter the amino acid sequence of the overlapping Rep protein) was used to inoculate N. benthamiana leaves (Teng et al., 2010). Host cell death was induced by the wild-type virus, resulting in a top-curling phenotype, but not by the virus containing the mutated C4 (Fig. 4A, B). Compared with the wild-type virus, much lower levels of the C4C8S virus were detected in the newly emerged shoots (Fig. 4C, Supplementary Fig. S7); however, the accumulation of both types of viruses were similar in the protoplasts and the leaves local to the inoculation site (Fig. 4D, Supplementary S7). These results suggested that S-acylation of C4 may regulate the movement but not the replication of the virus, consistent with previous results from the depletion of BSCTV C4 (Teng et al., 2010). Although the accumulation of the mutated virus was detectable in the shoot region, the plants showed no symptoms even a long time after inoculation (Fig. S8), supporting the conclusion that the S-acylation of C4 was critical for symptom determination during BSCTV infection.
Fig. 4.
S-acylation is necessary for the function of C4 in symptom determination during BSCTV infection. (A) Nicotiana benthamiana plants were inoculated with pCAMBIA1300-BSCTV-C4WT-1.8copy (wild-type) or pCAMBIA1300-BSCTV-C4C8S-1.8copy (C8 mutated from a cysteine to a serine) using different concentrations of Agrobacterium relative to OD600. The leaves local to the inoculation site were photographed after 9 d. (B) The top-curling phenotype of N. benthamiana induced by infection with BSCTV. The plants were photographed 9 d after inoculation. Enlarged images of the shoot tips are shown for a better comparison. The top-curling symptom did not appear on the plants infected with BSCTV-C4C8S even a long time after inoculation. (C) Levels of BSCTV in the newly emerged shoots of N. benthamiana plants were detected using PCR 9 d after inoculation. ACTIN (N. benthamiana) was used as a control. (D) Accumulation of BSCTV in Arabidopsis protoplasts was measured 0, 2, and 4 d after infection. The DNA was extracted and subjected to a DNA gel blot using the BSCTV genome as a probe. The plant genomic DNA was used as the loading control. dpi, days post-infection; WT, wild-type. The results in this figure are representative of at least three independent experiments.
S-acylation of C4 is necessary for its interaction with CLV1
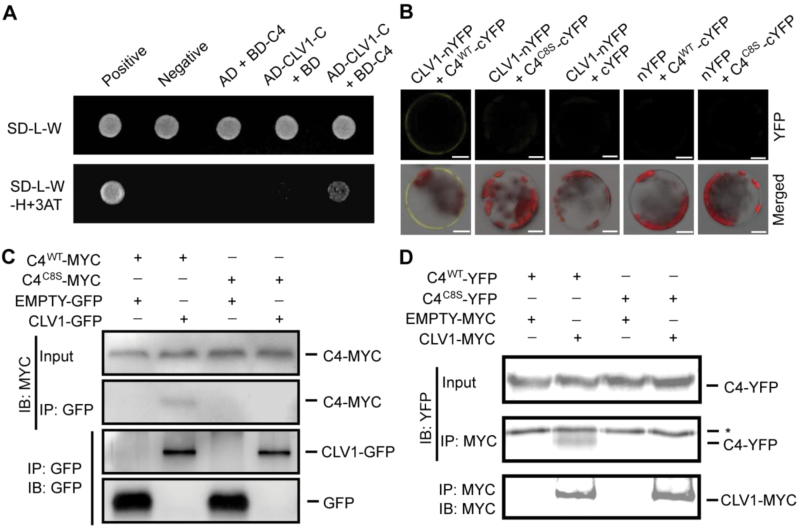
Our data showed that S-acylation was important for the effect of C4 on shoot development; therefore, we next attempted to uncover the potential mechanism regulating this process. We hypothesized that the target of C4 may be associated with the plasma membrane and may play a role in regulating shoot development. A previous study showed that the intracellular catalytic domain of a leucine-rich repeat receptor-like kinase (At3g24240) interacts with BCTV C4 in a Y2H assay (Piroux et al., 2007). Using a bioinformatics analysis, we found that the amino-acid sequence of the interaction domain of this protein was highly similar to that of CLAVATA 1 (CLV1), a receptor kinase involved in the regulation of the shoot meristem (Clark et al., 1997; Hazak and Hardtke, 2016). A Y2H assay revealed that BSCTV C4 interacted with the intracellular catalytic domain of CLV1 in yeast cells (Fig. 5A).
Fig. 5.
S-acylation of C4 is important for its interaction with CLV1. (A) Detection of the interaction between C4 (fused to the binding domain, BD) and the intracellular domain (704AA-967AA) of CLV1 (CLV1C; fused to the activation domain, AD) in a yeast two-hybrid assay. Protein interactions were tested using a stringent (SD/–Leu/–Trp/–His) selection medium containing 3-amino-1,2,4-triazole (3AT). (B) The in vivo interaction between C4 and CLV1 was measured using bimolecular fluorescence complementation assays. The combinations with empty vectors were used as negative controls. The YFP signals and merged signals (yellow for YFP, red for chloroplast autofluorescence, and gray for bright field) are shown. Scale bars are 10 µm. (C) Detection of the interaction between C4-MYC and CLV1-GFP using co-immunoprecipitation (Co-IP). The wild-type (WT) or mutant form of pCanG-C4-MYC (C8 mutated from a cysteine to a serine) was co-transformed with 35S::CLV1-GFP or 35S::GFP (control). After 48 h, the transformed protoplasts were collected for Co-IP using GFP-Trap resin. (D) Measurement of the interaction between C4-YFP-FLAG3His6 and CLV1-MYC using Co-IP. The wild-type or mutant form of C4-YFP-FLAG3His6 was co-expressed with pCanG-CLV1-MYC or pCanG-MYC (control). Co-IP was performed using an anti-MYC antibody with Protein A resins. * indicates the unspecific signal from the antibody. The results in this figure are representative of independent experiments.
Next, we assessed this interaction in plant cells and investigated whether it was dependent on the S-acylation of C4. First, the results from a BiFC assay showed that full-length CLV1 and C4 interacted on the plasma membrane in vivo, but that the depletion of S-acylation disrupted this association (Fig. 5B). This interaction was further confirmed by performing Co-IP assays in plant cells, which showed that CLV1 specifically interacted with wild-type C4 but not with the mutated C4 (Fig. 5C, D), providing direct evidence that their interaction was mediated by S-acylation in plant cells.
C4-induced misexpression of WUS is dependent on S-acylation
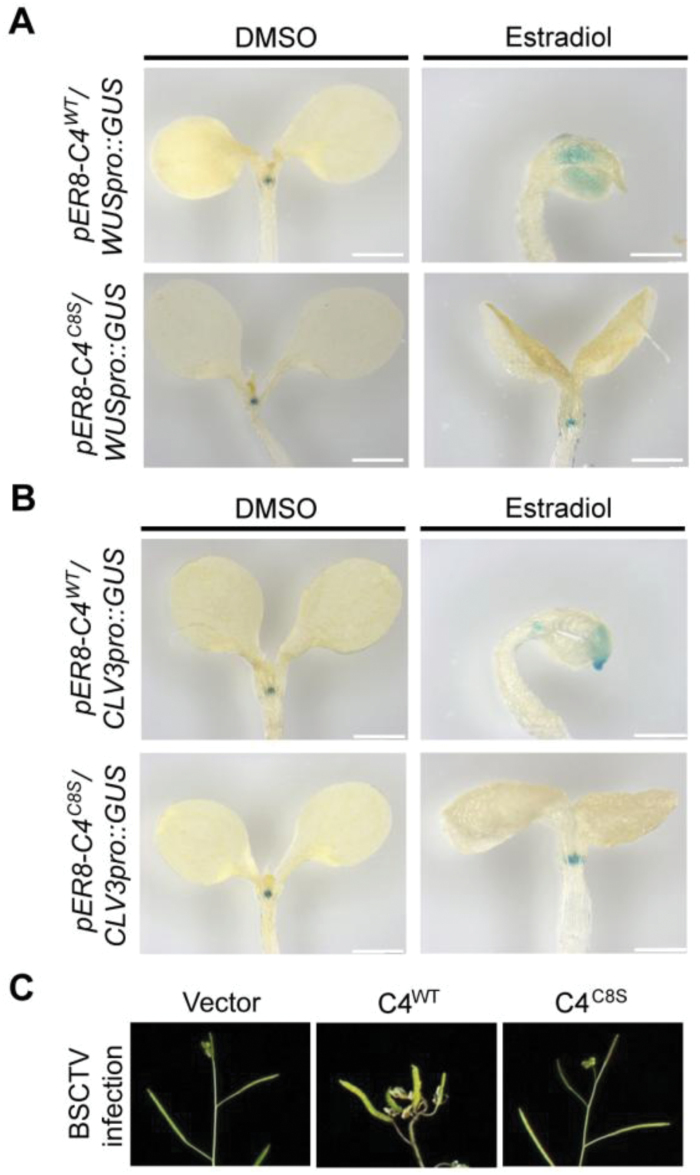
CLV1 is a receptor-like kinase that perceives the secreted peptide CLV3 on the plasma membrane, then negatively regulates the expression of the homeodomain transcription factor WUSCHEL (WUS) to modulate cell differentiation in the shoot and floral meristems (Brand et al., 2000). The interaction between C4 and CLV1 may therefore interfere with this signaling transduction. To detect the expression of WUS, a GUS gene under the control of the WUS promoter was crossed into pER8-C4WT and pER8-C4C8S plants. WUS was expressed in the shoot-meristem region of the seedlings in the absence of C4 expression (Fig. 6A). The pattern of GUS staining was dramatically changed in the presence of C4 expression and showed that the expression of WUS was inhibited in the meristem but enhanced in the cotyledons, suggesting the abnormal regulation of this pathway by the viral protein. By contrast, the expression of WUS was not affected by the C4C8S mutant protein, supporting the role of S-acylation in this process.
Fig. 6.
C4-induced misexpression of WUS is dependent on its S-acylation. (A) GUS staining levels of WUSpro::GUS in the estradiol-inducible pER8-C4WT (wild-type) and pER8-C4C8S (C8 mutated from a cysteine to a serine) plants 5 d after germination on media with (estradiol) or without (DMSO) induction by estradiol. Scale bars are 0.5 mm. (B) GUS staining levels of CLV3Pro::GUS in the pER8-C4WT and pER8-C4C8S plants 5 d after germination on media with or without estradiol. Scale bars are 0.5 mm. The results are representative of three independent experiments using individual transgenic lines, which had similar patterns. (C) Representative siliques of Arabidopsis infected with the geminivirus carrying the wild-type (WT) or C8S version of C4. The empty vector was used as an inoculation control.
The WUS protein migrates into adjacent cells to activate CLV3 expression and establish a feedback loop to balance the number of stem cells produced with the size of the meristem (Yadav et al., 2011); therefore, we also measured the transcription levels of CLV3 in the presence of C4. The expression of CLV3 was detectable but weaker in the meristems in the presence of wild-type C4, but it also abnormally occurred in the cotyledons (Fig. 6B). This result was consistent with feedback regulation and provided evidence that the inhibition of WUS expression by C4 was not a result of a misorganization of the shoot meristem; therefore, C4 may interact with CLV1 on the plasma membrane following its S-acylation, causing it to interfere with the CLAVATA pathway.
The mutation of components involved in the Arabidopsis CLAVATA pathway often causes phenotypic changes in the siliques (Somssich et al., 2016), so those on the BSCTV-infected plants may also have abnormal cell division. We found that infection with BSCTV carrying the wild-type but not the C8S mutant form of C4 induced abnormal development in the siliques in Arabidopsis (Fig. 6C), providing further evidence of a potential association between BSCTV infection and the CLAVATA pathway.
Discussion
Our current study revealed a novel mechanism for the role of S-acylation in the interaction between geminiviruses and plants. Certain viral proteins are regulated by S-acylation in mammalian host cells; for instance, S-acylation is essential for the association of the influenza virus haemagglutinin protein with the membrane microdomains of the animal cell (Veit et al., 2013). Similarly, the S-acylation of C4 regulates its localization to the plasma membrane of the plant cell and thus its consequential cellular effects. Our results showed that the S-acylation level of C4 was not altered during BSCTV infection (Fig. 1). When C4 is expressed in infected plant cells, the protein may be recognized and S-acylated by the host cell; however, this modification contributes to the efficiency of the viral infection, implying a functional interaction between pathogen and host during evolution.
It has been reported that myristoylation is also involved in the localization of geminivirus C4 proteins to the host membrane (Fondong et al., 2007; Piroux et al., 2007). Unlike myristoylation, S-acylation is reversible and causes the modified protein to have a higher affinity with the membrane, potentially providing a dynamic regulation of its membrane association (Linder and Deschenes, 2007). The mutation of the myristoylation site of C4 in BCTV resulted in the complete inactivation of this pathogenesis determinant (Piroux et al., 2007); here, we showed that S-acylation on BSCTV C4 was important for its function in regulation of shoot symptoms. Interestingly, in the current study, the mutation of the potential myristoylation site at the N terminus of BSCTV C4 disrupted its S-acylation and membrane localization (Supplementary Fig. S3), supporting the hypothesis that myristoylation may have consequential effects on S-acylation. The results from the cellular fraction showed that most C4C8S proteins were distributed in the cytosol, but a small amount of mutated proteins were retained at the membrane (Fig. 2), possibly mediated by myristoylation or physical interaction with other membrane proteins. This small proportion of C4 proteins may be sufficient to influence root development, which may be more sensitive to C4 (Fig. 3). Because S-acylation is reversibly regulated by S-acyltransferases and thioesterases (Lanyon-Hogg et al., 2017), the modification level of C4 may be dynamically controlled depending on the cellular conditions. There are 24 S-acyltransferases in Arabidopsis (Batistič, 2012) and many potential thioesterases, so it would be interesting to identify the specific enzymes involved in the regulation of C4 S-acylation. This would enhance our understanding of the precise mechanism controlling this symptom determinant.
Our results showed that the S-acylation of C4 did not have an effect on virus replication (Fig. 4), but may have contributed to virus movement, consistent with the previously described function of this protein (Teng et al., 2010). Although the movement of viruses to the newly emerged shoots was slower when the S-acylation site of C4 was mutated, BSCTV was still detectable in this region; however, even a long time after inoculation, the mutant BSCTV did not cause obvious symptoms in the shoots (Supplementary Fig. S8), supporting our conclusion that S-acylation is important for symptom development during BSCTV infection. The C4 proteins of several geminiviruses were previously identified as suppressors of gene silencing (Ramesh et al., 2017), so future research should investigate whether S-acylation is involved in the regulation of this process.
The identification of the interaction between C4 and CLV1 provided a potential mechanism to explain the effects of C4 on shoot development. Although the data from Y2H assays indicated that C4 interacted with the intracellular domain of CLV1, the Co-IP results using full-length proteins suggested that membrane localization of C4 mediated by S-acylation was critical for this interaction in plant cells (Fig. 5). Because CLV1 is a transmembrane protein, S-acylation of C4 may enhance its interaction with CLV1 in the membrane regions, affecting the perception of the CLV3 peptide for the downstream repression of WUS expression (Hazak and Hardtke, 2016). Our data showed that C4 inhibited WUS expression in the meristem regions but stimulated it in the cotyledons (Fig. 6), suggesting that this protein interferes with the CLAVATA signaling pathway. Membrane biochemistry is technologically difficult to study, but future investigations of the precise mechanisms by which C4 regulates CLV1 may yield interesting results. Geminivirus infections result in the abolishment of meristems and induce ectopic cell division in differentiated tissues (Park et al., 2002), which is consistent with our data showing abnormal WUS expression in the presence of C4. BSCTV induced cell division in the shoot tips of the plants, including the siliques, which is a similar phenotype to those of plants with mutations in components of the CLAVATA pathway; therefore, the CLAVATA signaling pathway may contribute to these symptoms. Previous studies indicated that BCTV C4 interacts with AtSKs and BIN2 (Mills-Lujan et al., 2015; Bi et al., 2017), interfering with BR signaling during infection. It will be also important to determine whether there is a C4-related connection between CLAVATA and BR signaling. In addition, C4 proteins from some geminiviruses are phosphorylated (Mills-Lujan et al., 2015). S-acylation is critical for the interaction between BSCTV C4 and CLV1, which is a receptor kinase; thus, it would be interesting to examine the relationship between the effects of S-acylation and phosphorylation on BSCTV C4.
Although the protein sequences of geminivirus C4 proteins vary, cysteine residues are found in many members (Supplementary Fig. S1), suggesting a potential conserved mechanism mediated by S-acylation. The S-acylation of this pathogen determinant has not yet been biochemically characterized, however. Different geminiviruses cause differing symptoms, and their regulatory mechanisms may vary. In a previous study on myristoylation of AC4 from the East African cassava mosaic Cameroon virus (EACMCV), it was shown that a cysteine mutation did not apparently affect its function in pathogenesis (Fondong et al., 2007), implying that not all cysteines are essential for virus infection. In future studies, it would be interesting to investigate C4 conservation and variation following S-acylation in different types of geminiviruses, which could improve our understanding on the role of protein modifications in the interaction between viruses and plant cells.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Fig. S1. Sequence alignment of C4 proteins from different geminiviruses.
Fig. S2. Localization and S-acylation analysis of the C4C28S mutant.
Fig. S3. Localization and S-acylation analysis of the C4G4A mutant.
Fig. S4. Localization and S-acylation analysis of the C4D2 mutants.
Fig. S5. The expression levels of C4 in transgenic plants.
Fig. S6. Functional analysis of the C4C8A mutant in N. benthamiana leaves.
Fig. S7. The effect of C4 S-acylation on BSCTV accumulation in local inoculated leaves and newly emerged shoots.
Fig. S8. The effect of C4 S-acylation on symptoms at 2 weeks and 3 weeks after BSCTV inoculation.
Acknowledgments
We would like to thank Prof. Nam-Hai Chua (Rockefeller University) for kindly providing us with the pER8 vector. This work was supported by the National Natural Science Foundation of China (31771504, 31400314, 31670286, and 31390423), The University Innovation Program of the Department of Education of Guangdong Province (2014), the Guangdong Science and Technology Department (2015A020209155 and 2016A020208014), the Guangdong YangFan Innovative and Entrepreneurial Research Team Project (2015YT02H032), and the Guangzhou Scientific and Technological Program (201607010377).
References
- Arguello-Astorga G, Lopez-Ochoa L, Kong LJ, Orozco BM, Settlage SB, Hanley-Bowdoin L. 2004. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. Journal of Virology 78, 4817–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O. 2012. Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiology 160, 1597–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Rehers M, Akerman A, Schlücking K, Steinhorst L, Yalovsky S, Kudla J. 2012. S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Research 22, 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Fan W, Zhang P. 2017. C4 protein of sweet potato leaf curl virus regulates brassinosteroid signaling pathway through interaction with AtBIN2 and affects male fertility in Arabidopsis. Frontiers in Plant Science 8, 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cui Y, Rao S, Chang B, et al. 2015. AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of Arabidopsis in response to environmental hazards. Plant, Cell & Environment 38, 2098–2114. [DOI] [PubMed] [Google Scholar]
- De Schutter K, Joubès J, Cools T, et al. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. The Plant Cell 19, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom CM, Mills-Lujan K. 2015. Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signaling & Behavior 10, e1109758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondong VN, Reddy RV, Lu C, Hankoua B, Felton C, Czymmek K, Achenjang F. 2007. The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Molecular Plant-Microbe Interactions 20, 380–391. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. 1999. Geminivirus DNA replication. Cellular and Molecular Life Sciences 56, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nature Reviews Microbiology 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Hazak O, Hardtke CS. 2016. CLAVATA 1-type receptors in plant development. Journal of Experimental Botany 67, 4827–4833. [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Kemp AC, Grierson CS. 2005. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. The Plant Cell 17, 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2009. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. Journal of Lipid Research 50, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Chen H, Teng K, et al. 2009. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. The Plant Journal 57, 905–917. [DOI] [PubMed] [Google Scholar]
- Lai J, Yu B, Cao Z, Chen Y, Wu Q, Huang J, Yang C. 2015. Two homologous protein S-acyltransferases, PAT13 and PAT14, cooperatively regulate leaf senescence in Arabidopsis. Journal of Experimental Botany 66, 6345–6353. [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T, Faronato M, Serwa RA, Tate EW. 2017. Dynamic protein acylation: new substrates, mechanisms, and drug targets. Trends in Biochemical Sciences 42, 566–581. [DOI] [PubMed] [Google Scholar]
- Latham JR, Saunders K, Pinner MS, Stanley J. 1997. Induction of plant cell division by Beet curly top virus gene C4. The Plant Journal 11, 1273–1283. [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S. 2002. A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. The Plant Cell 14, 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qi B. 2017. Progress toward understanding protein S-acylation: prospective in plants. Frontiers in Plant Science 8, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. 2007. Palmitoylation: policing protein stability and traffic. Nature Reviews Molecular Cell Biology 8, 74–84. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q. 2010. An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. The Plant Journal 61, 893–903. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lai J, Yu M, et al. 2016. The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation. The Plant Cell 28, 2225–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Yang X, Huang C, Zhang X, Zhou X. 2018. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη-mediated phosphorylation in Nicotiana benthamiana. PLoS Pathogens 14, e1006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills-Lujan K, Andrews DL, Chou CW, Deom CM. 2015. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 10, e0122356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills-Lujan K, Deom CM. 2010. Geminivirus C4 protein alters Arabidopsis development. Protoplasma 239, 95–110. [DOI] [PubMed] [Google Scholar]
- Park J, Lee HJ, Cheon CI, Kim SH, Hur YS, Auh CK, Im KH, Yun DJ, Lee S, Davis KR. 2011. The Arabidopsis thaliana homeobox gene ATHB12 is involved in symptom development caused by geminivirus infection. PLoS ONE 6, e20054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Hur J, Park J, Lee S, Lee TK, Chang M, Davi KR, Kim J, Lee S. 2002. Identification of a tolerant locus on Arabidopsis thaliana to hypervirulent beet curly top virus CFH strain. Molecules and Cells 13, 252–258. [PubMed] [Google Scholar]
- Piroux N, Saunders K, Page A, Stanley J. 2007. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 362, 428–440. [DOI] [PubMed] [Google Scholar]
- Ramesh SV, Sahu PP, Prasad M, Praveen S, Pappu HR. 2017. Geminiviruses and plant hosts: a closer examination of the molecular arms race. Viruses 9, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. 2008. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Engineering, Design & Selection 21, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D. 2016. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. [DOI] [PubMed] [Google Scholar]
- Stanley J, Latham JR. 1992. A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190, 506–509. [DOI] [PubMed] [Google Scholar]
- Teng K, Chen H, Lai J, Zhang Z, Fang Y, Xia R, Zhou X, Guo H, Xie Q. 2010. Involvement of C4 protein of beet severe curly top virus (family Geminiviridae) in virus movement. PLoS ONE 5, e11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Serebryakova MV, Kordyukova LV. 2013. Palmitoylation of influenza virus proteins. Biochemical Society Transactions 41, 50–55. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes & Development 25, 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y. 2013. PROTEIN S-ACYL TRANSFERASE10 is critical for development and salt tolerance in Arabidopsis. The Plant Cell 25, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. 2000. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.