A basic helix-loop-helix transcription factor PbbHLH4 induces the biosynthesis of monoterpenes for floral scent in Phalaenopsis orchids.
Keywords: Floral scent, geranyl diphosphate synthase, monoterpenes, orchids, Phalaenopsis, transcription factors
Abstract
Floral scent is an important factor in attracting pollinators and repelling florivores. In Phalaenopsis bellina (Orchidaceae), the major floral scent components are monoterpenoids. Previously, we determined that expression of GERANYL DIPHOSPHATE SYNTHASE (PbGDPS) is highly correlated with monoterpene biosynthesis in Phalaenosis orchids. Here, we found that both cis- and trans-regulation were present on the GDPS promoters, with trans-regulation playing a key role. To investigate the regulation of biosynthesis of floral scent, we compared the transcriptomic data of two Phalaenopsis orchids with contrasting scent phenotypes. Eight transcription factors (TFs) that exhibited sequential elevation in abundance through floral development in P. bellina were identified, and their transcript levels were higher in the scented orchid than the scentless one. Five of these TFs transactivated several structural genes involved in monoterpene biosynthesis including PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNAC1. Ectopic transient expression of each of these TFs in scentless orchids resulted in stimulation of terpenoid biosynthesis. PbbHLH4 most profoundly induced monoterpene biosynthesis, with a 950-fold increase of monoterpenoid production in the scentless orchid. In conclusion, we determined that biosynthesis of orchid floral monoterpenes was sequentially regulated, with PbbHLH4 playing a crucial role for monoterpene biosynthesis.
Introduction
Terpenoids, or terpenes, represent the largest group of plant floral volatiles (Tholl, 2015). They play important roles in attracting pollinators for successful reproduction (Blight et al., 1997; Byers et al., 2014) and in defense against pathogens and florivores (Junker et al., 2011; Huang et al., 2012). Apart from their natural roles, terpenoids are widely used in the cosmetics and perfume industry and as food additives because of their unique aromas and flavors (Schwab et al., 2008; Caputi and Aprea, 2011).
The biosynthesis of terpene starts from the production of basic C5 units, isopentenyl diphosphate (IDP) and its isomer, dimethylallyl diphosphate (DMADP). Both C5 units are synthesized from the mevalonate (MVA) pathway in the cytosol or the methylerythritol phosphate (MEP) pathway in plastids. The C5 precursors derived from the MVA pathway are preferentially used for biosynthesis of sesquiterpenoids, and those generated by the MEP pathway are predominately used for monoterpenoids and diterpenoids. A group of enzymes called short-chain prenyltransferases are responsible for the successive condensation of IDP with DMADP to produce intermediates for terpene synthases (TPSs), including geranyl diphosphate synthase (GDPS) that produces geranyl diphosphate (GDP, C10) for monoterpenes, farnesyl diphosphate synthase that generates farnesyl diphosphate (FDP, C15) for sesquiterpenes, and geranylgeranyl diphosphate synthase that supplies geranylgeranyl diphosphate (C20) for diterpenes (Dudareva et al., 2004).
Phalaenopsis bellina has a pleasant scent and is widely used as a breeding parent for its scent phenotype. The main floral scent volatiles in P. bellina are monoterpenoids, including linalool, geraniol, and their derivatives (Hsiao et al., 2006). A combination of bioinformatics and genomics has highlighted PbGDPS as being enriched in the scented P. bellina but not in the scentless P. equestris (Hsiao et al., 2006). Recombinant PbGDPS has dual prenyltransferase activities that produce GDP and FDP by using IDP/DMADP and IDP/GDP as substrates, respectively. PbGDPS shows flower-specific expression, and its highest expression is concurrent with maximal emissions of monoterpenoids on day 5 post-anthesis (D+5), which suggests a crucial role of PbGDPS in floral scent production in P. bellina (Hsiao et al., 2008). Recently, we found that the emission of monoterpenes is under the control of light and a circadian rhythm in Phalaenopsis orchids (Chuang et al., 2017).
Although our knowledge of the biosynthesis of floral terpenoids is increasing, little is known about its regulation. To date, the types of transcription factors (TFs) involved in terpene biosynthesis have been found to vary, and most studies on the regulation of terpene biosynthesis have been performed in vegetative tissues and fruit. Examples include AabHLH1, AabZIP1, AaERF1, AaERF2, AaNAC1, AaORA, and AaWRKY1 in Artemisia annua (Ma et al., 2009; Yu et al., 2012; Lu et al., 2013; Ji et al., 2014; Zhang et al., 2015; Lv et al., 2016), CitAP2.10 and CitERF71 in sweet orange (Shen et al., 2016; Li et al., 2017), CrBIS1, CrBIS2, CrBPF1, CrMYC2, CrORCA2-5, CrWRKY1, and CrZCT1-3 in Catharanthus roseus (van der Fits and Memelink, 2000; Pauw et al., 2004; Suttipanta et al., 2011; Zhang et al., 2011; Li et al., 2013, 2015a; Van Moerkercke et al., 2015, 2016; Paul et al., 2017), GaWRKY1 in cotton (Xu et al., 2004), MsYABBY5 and MsMYB in spearmint (Wang et al., 2016; Reddy et al., 2017), NACs and EILs in kiwifruit (Nieuwenhuizen et al., 2015), OsbZIP79, OsDPF, and OsTGAP1 in rice (Okada et al., 2009; Miyamoto et al., 2015; Yamamura et al., 2015), SlMYC1, SlEOT1, and SlWRKY73 in tomato (Spyropoulou et al., 2014a, 2014b), and ZmEREB58 in maize (Li et al., 2015b). In contrast, regulation of floral terpene biosynthesis has been less well studied. To date, there has been only one study showing that AtMYC2 promotes inflorescence sesquiterpene production in Arabidopsis (Hong et al., 2012). A summary of TFs known to regulate terpene biosynthesis is presented in Table S1 available at the Dryad Digital Repository, (https://doi.org/10.5061/dryad.kt056q7, Chuang et al., 2018).
In this study, we aimed to elucidate the transcriptional regulation of floral scent biosynthesis by carrying out a comparative transcriptome analysis between scented and scentless orchids. Five TFs with higher expression in the scented orchid were identified, namely PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNAC1. They showed a sequential expression pattern during floral development, which indicates developmental regulation. Ectopic transient expression of these TFs revealed that terpenoid biosynthesis in general was increased, with PbbHLH4 strongly inducing monoterpene production in the scentless orchid. Taken together, the results broaden our knowledge of the regulating mechanisms controlling floral monoterpene biosynthesis. Many Phalaenopsis varieties and other modern floricultural cultivars have completely lost their scented phenotype as a result of traditional breeding methods, due to negative correlations between floral scent and other favorable traits (Vainstein et al., 2001; Hsiao et al., 2011). This study therefore provides a starting point for molecular breeding to introduce the scented trait in Phalaenopsis orchids.
Materials and methods
Plant material and growth conditions
Six Phalaenopsis orchids including two scented and four scentless ones that are commonly utilized as breeding parents were used in this study. The two scented orchids, P. bellina and P. Meidarland Bellina Age ‘LM128’, were purchased from Ming-Hui Orchids Nursery (Yunlin, Taiwan) and Meidarland Orchids (Tainan, Taiwan), respectively. The four scentless orchids were P. javanica (from Mi-Tuo Orchids, Kaohsiung, Taiwan), P. mannii (from Ji An Guang Feng, Hualien, Taiwan), and P. aphrodite subsp. formosana (hereafter abbreviated to P. aphrodite) and P. Sogo Yukidian ‘V3’ (from Taiwan Sugar Corp., Tainan, Taiwan). The scented orchid (P. bellina) has a much bigger genome than the scentless P. aphrodite (15.03 pg/2C versus 2.80 pg/2C) (Lin et al., 2001). All plants were kept in a greenhouse at the National Cheng Kung University (NCKU, Tainan, Taiwan) under natural conditions.
Chromatographic analysis of floral volatiles
The floral metabolites of the Phalaenopsis orchids were collected on day 5 post-anthesis (D+5) for 6 h (from 10.00 h to 16.00 h) as this represents their maximum emission interval (Chuang et al., 2017). The flowers on the plants were placed in a scent-extracting apparatus as described by Chuang et al. (2017). To analyse the scent composition, metabolites were sampled from a single flower, with three biological replicates. The scent emission pattern of P. bellina was measured on a single flower from day 1 prior to anthesis (D–1) to D+23, with three biological replicates (senescence of P. bellina flowers usually occurs from D+25 to D+28). As a negative control, metabolites originating from the scent-extracting apparatus were analysed for background. The volatiles collected were eluted by hexane and identified by gas chromatography/high-resolution mass spectrometry (GC/HRMS) at the NCKU Instrument Center, as described by Hsiao et al. (2006, 2008).
Transcriptome construction, assembly, and annotation
Transcriptomes were constructed for four floral stages of P. bellina: anthesis day (D0), D+3, D+5, and D+7. Total RNA was extracted from the entire flowers with duplicate biological repeats as described previously (Hsiao et al., 2006, 2008). Quality control of RNA was performed using the RNA 6000 Nano Assay supplied with the Agilent 2100 bioanalyser. Preparation of four cDNA libraries and subsequent sequencing was carried out using an Illumina HiSeq 2000 at the Beijing Genomics Institute. De novo assembly for whole-transcriptome construction was carried out using the clean reads from the four libraries by using Trinity (release-20130225) (Grabherr et al., 2011), and the resulting sequences were the unigenes. The expression level of each unigene in each sample was calculated as fragments per kilobase of transcript per million mapped reads (FPKM) based on the number of fragments uniquely aligned to the unigene in each library.
To gain insight into the function of the unigenes, the non-redundant (Nr) protein database at NCBI (https://www.ncbi.nlm.nih.gov/) was used to annotate them by using blastx with an E-value cut-off of 1.0 × 10–5. The floral transcriptome of P. aphrodite and its annotation were downloaded from Orchidstra (Su et al., 2011, 2013a, 2013b), upgraded to Orchidstra 2.0 (http://orchidstra2.abrc.sinica.edu.tw/orchidstra2/index.php), in which the relative expression of unigenes is derived from microarray analyses at both the floral bud and full-blossom stages. Since two different methods were used to generate the Phalaenopsis transcriptome data, the most significant results were validated experimentally by quantitative real-time PCR.
Identification of structural genes and TFs related to terpene biosynthesis
Three reference genes widely used in Phalaenopsis orchids were identified by using local blastn, namely Actin4, Actin9, and Ubiquitin10. We identified genes encoding enzymes related to monoterpene biosynthesis according to the KEGG annotation (https://www.genome.jp/kegg/annotation/). The genes isolated from the P. bellina floral transcriptome were confirmed by Nr annotation with an E-value cut-off of 1.0 × 10–5. The related genes in the P. aphrodite transcriptomes were identified by using local tblastx with an E-value cut-off of 1.0 × 10–50 and confirmed using the Nr annotation available on Orchidstra.
With the 181 TPS sequences collected from other plants as queries, TPS sequences of the two transcriptomes were isolated by using tblastx with an E-value cut-off of 1.0 × 10–5. The two transcripts of PbTPS5 and PbTPS10 were analysed. Since most monoterpene synthases (MTPSs) identified from other plants had an average peptide length of 600 residues, we focused on those with lengths close to this side, namely PbTPS5-1 (605 amino acids) and PbTPS10-2 (595 amino acids). The putative protein sequences of GDPS and four TPSs from both P. bellina and P. aphrodite are shown in Fig. S1 available at Dryad.
To isolate TFs and regulators, 28193 proteins in the PlnTFDB database (ver. 3.0; Pérez-Rodríguez et al., 2010) were downloaded as queries to search the P. bellina transcriptome by using tblastn with an E-value cut-off of 1.0 × 10–50. The resulting sequences were then classified by using iTAK (Zheng et al., 2016). Among them, 335 genes annotated as bHLH, bZIP, ERF, NAC, MYB, and WRKY were isolated, and 165 genes with FPKM>1 were imported into the Short Time-series Expression Miner (STEM) software (Ernst and Bar-Joseph, 2006) for classification of their expression profiles. For STEM analysis, the temporal expression profiles were transformed to start at 0 by subtracting the FPKM levels of the four floral stages (D0, D+3, D+5, and D+7) by the value for the first stage. The STEM Clustering Method provided by the software was used to cluster the factors into 10 profiles according to their expression patterns.
Phylogenetic analysis
The full-length amino acid sequences of the putative TPSs isolated from the P. bellina and P. aphrodite transcriptomes (Su et al., 2013a) were analysed phylogenetically with TPS sequences of other plant species. The MTPSs were predicted by the known TPSs in the same clade depending on the phylogenetic analysis (Fig. S2 at Dryad). For TF analysis, we downloaded basic helix-loop-helix (bHLH) TF families of Arabidopsis from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/index.jsp). The subgroup classification was analysed according to Heim et al. (2003). The MEGA6 software was used for alignment via ClustalW and the phylogenetic tree was built using the neighbor-joining method with 1000 bootstrap trials.
Transactivation assay of the GDPS promoter in three Phalaenopsis orchids
Promoter fragments of PbGDPS were isolated by using a Universal GenomeWalker kit (Clontech, USA) and the primer design and PCR conditions for genome walking followed the manufacturer’s instructions (sequences of all primers used in this study are listed in Table S2 at Dryad). The 2.7-kb promoter fragment of PbGDPS was used for isolation of the PaGDPS promoter. We isolated a promoter fragment of GDPS from P. bellina and P. aphrodite, designated PbGDPSp for P. bellina and PaGDPSpA and PaGDPSpB for P. aphrodite because the sequencing results revealed that the promoter fragments of PaGDPS were polymorphic with two types based on the deletion region. The 1076-, 1065-, and 1028-bp fragments of PbGDPSp, PaGDPSpA, and PaGDPSpB, respectively, were cloned into pJD301(f) to drive the firefly luciferase gene. In addition, an internal control pJD301(R) was included, which contained the Renilla luciferase gene driven by the cauliflower mosaic virus (CaMV) 35S promoter. The plasmids harboring promoter fragments and the internal control were then co-bombarded into Phalaenopsis floral tissues at a ratio of 10 to 0.1 μg as described previously (Hsu et al., 2014). Bombardment of flowers of P. bellina and P. aphrodite at the D+5 stage was carried out with three replicates. The luciferase activity of each sample was measured after 20 h of incubation. The tissues were ground in liquid nitrogen using a pestle and mortar, and the resulting fine powder was dissolved in the PLB solution provided with the Dual-Luciferase Reporter (DLR) Assay System (Promega). The luciferase activity was determined according to the manufacturer’s instructions. Pairwise comparisons between groups were performed using Tukey’s honestly significant difference test at α=0.05.
Quantitative real-time PCR
Total RNA was extracted as described previously (Hsiao et al., 2006, 2008). For P. Meidarland Bellina Age ‘LM128’, P. aphrodite, P. javanica, and P. mannii, total RNA was extracted at the floral D+5 stage. For P. bellina total RNA was extracted at five floral stages: D–1, D0, D+3, D+5, and D+7. After removal of DNA contamination by DNase (NEB, UK), the RNA samples were reverse-transcribed to cDNA using SuperScript III (ThermoFisher Scientific). Primers were designed to detect transcripts of P. bellina and P. aphrodite simultaneously based on the corresponding transcripts in the two orchid transcriptomes. Quantitative real-time PCR was carried out using a StepOnePlus Quantitative Real-Time PCR System and a SYBR Green kit (Applied Biosystems) as described previously (Hsu et al., 2015). The expression of all genes was normalized to the reference gene, PbActin1. Three biological replicates were used, and pairwise comparisons between groups were performed using Tukey’s honestly significant difference test at α=0.05.
Examination of the transactivation of TFs on the promoters of structural genes
Promoter fragments of PbGDPS2, PbTPS5, and PbTPS10 were isolated from genomic DNA of P. bellina by using primers designed according to the P. equestris draft genome (Cai et al., 2015). After sequence confirmation, 1175-, 887-, and 1016-bp of promoter fragments for PbGDPS2, PbTPS5, and PbTPS10, respectively, were cloned into pJD301(f) to drive the firefly luciferase gene. The coding sequences for PbbHLH2, PbbHLH4, PbbHLH5, PbbHLH6, PbbZIP4, PbERF1, PbMYB22, and PbNAC1 were amplified from full-bloom flowers of P. bellina using gene-specific primers and cloned into pBI221 to replace the GUS (β-glucuronidase) gene and were driven by the CaMV 35S promoter. Three separate plasmids—pBI221 containing TFs, pJD301(f) containing promoter fragments (four sequences, together with the PbGDPSp), and the internal control pJD301(R)—were co-bombarded into P. aphrodite floral tissues at a ratio of 1.5:1.5:0.15 (total 3.15 µg) as described previously (Hsu et al., 2014). The luciferase activity of each sample was measured after 20 h of incubation. The relative fold-change in activity was calculated by comparison with the control assay for GUS in pBI221. The assays with TFs were performed with three biological repeats. The numbers of replicates for native promoter activity without TFs (with GUS instead) were as follows: PbGDPSp, n=27; PbGDPS2p, n=12; PbTPS5p, n=15; and PbTPS10p, n=18. Several combinations showing variance among replicates were performed with six replicates to obtain more reliable results: PbbHLH2, PbbHLH6, and PbNAC1 for PbGDPSp. Pairwise comparisons between groups were performed by using Tukey’s honestly significant difference test at α=0.05.
Transient ectopic expression of TFs in orchids
Plasmids were constructed as described previously (Hsu et al., 2015). The coding sequences of PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNAC1 were amplified from full-bloom flowers of P. bellina using gene-specific primers and transferred to the vector p1304NhXb under a duplicated CaMV 35S promoter. Agrobacterium tumefaciens EHA105 carrying the resulting clones was infiltrated into scentless flowers of P. aphrodite and P. Sogo Yukidian ‘V3’ on the day of anthesis (D0) with three replicates as described previously (Hsu et al., 2015). The promoter containing GUS only was used as the negative control. The metabolites emitted from infiltrated flowers were collected at 4 d post-infiltration (DPI) for 8 h (from 8.00 h to 16.00 h) as described above, and the compounds were identified by GC/HRMS as described by Hsiao et al. (2006, 2008). Total RNA was extracted at 5 DPI as described above, and gene expression analysis was performed by quantitative real-time PCR. Statistical analysis was performed using Student’s t-test at α=0.05.
Accession numbers
Sequence data supporting the findings of this study have been deposited in GenBank under accession numbers PbGDPS (EU023907) and PbbHLH4 (KY979199).
Results
Profiles of floral volatile in scented P. bellina and scentless P. aphrodite
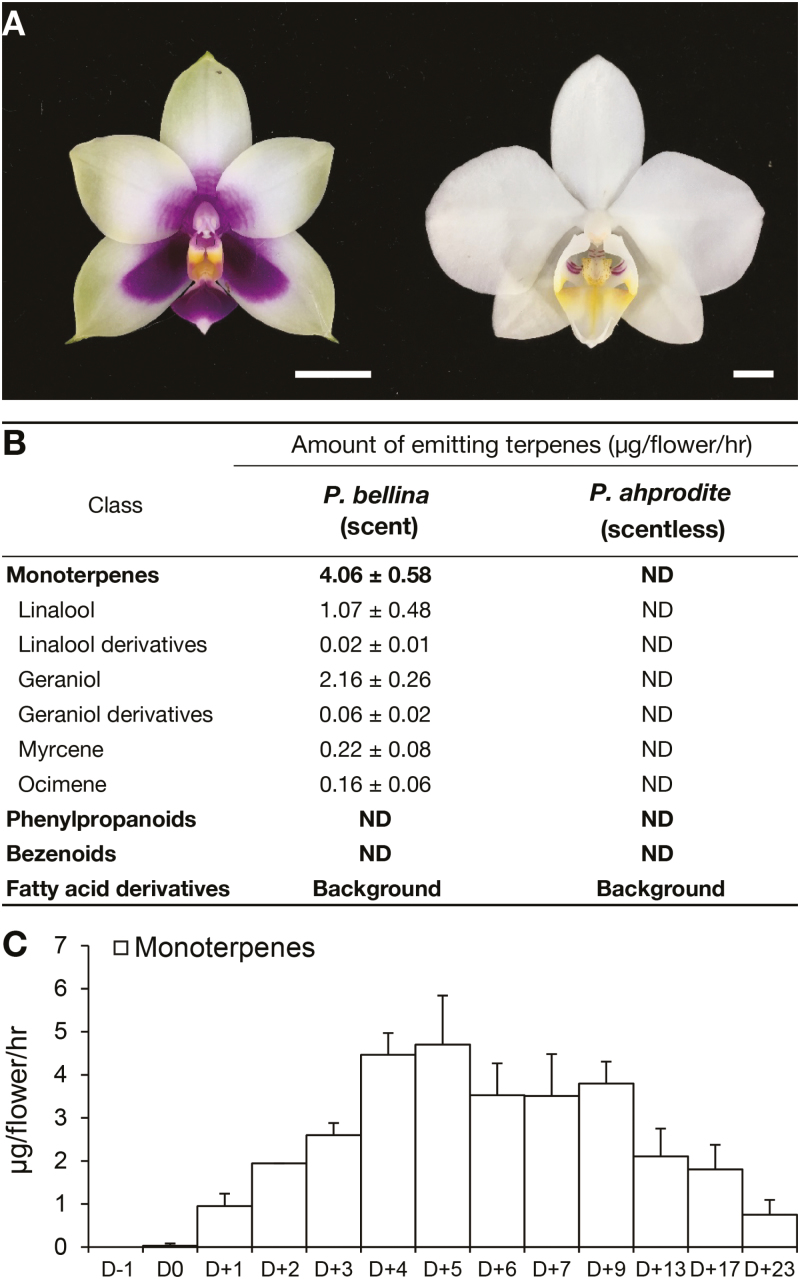
To study the regulation mechanism of floral scent biosynthesis in Phalaenopsis orchids, we selected two native species with contrasting scent profiles, P. bellina (scented) and P. aphrodite (Fig. 1A), which are important parents in breeding programs for scent and white-color traits, respectively. In addition, P. aphrodite is a commonly cultivated and commercialized Phalaenopsis species. We first checked the volatiles emitted from a single flower of the two species (Fig. 1B). Two monoterpenoids, linalool and geraniol and their derivatives, accounted for 25% and 50%, respectively, of the total amount of floral volatiles in P. bellina. In contrast, no scent compounds were detected in P. aphrodite. The emission pattern of monoterpenes throughout the floral developmental stages was then further examined in P. bellina. Monoterpenoids were absent in floral buds (D–1), were initiated on the day of anthesis (D0), rapidly increased in emissions during flower maturation, peaked at the full-bloom stages (D+4 and D+5), and gradually decreased thereafter with flower age (Fig. 1C). Thus, production of monoterpenoids was developmentally co-ordinated during the flowering of P. bellina.
Fig. 1.
Floral volatile profiles of Phalaenopsis bellina and P. aphrodite. (A) Single flowers of P. bellina (left) and P. aphrodite (right). Scale bars are 1 cm. (B) Floral volatile profiles of P. bellina and P. aphrodite at the full-bloom stage, 5 d after anthesis (D+5). Data are means (±SE) of three replicates. ND, not detected. ‘Background’ indicates that the fatty-acid derivatives detected were emitted from the scent-extracting apparatus. (C) The emission pattern of total monoterpenes from the day before anthesis (D–1) to D+23 in P. bellina. Data are means (±SE) of three replicates.
Key steps in monoterpene biosynthesis in Phalaenopsis orchids
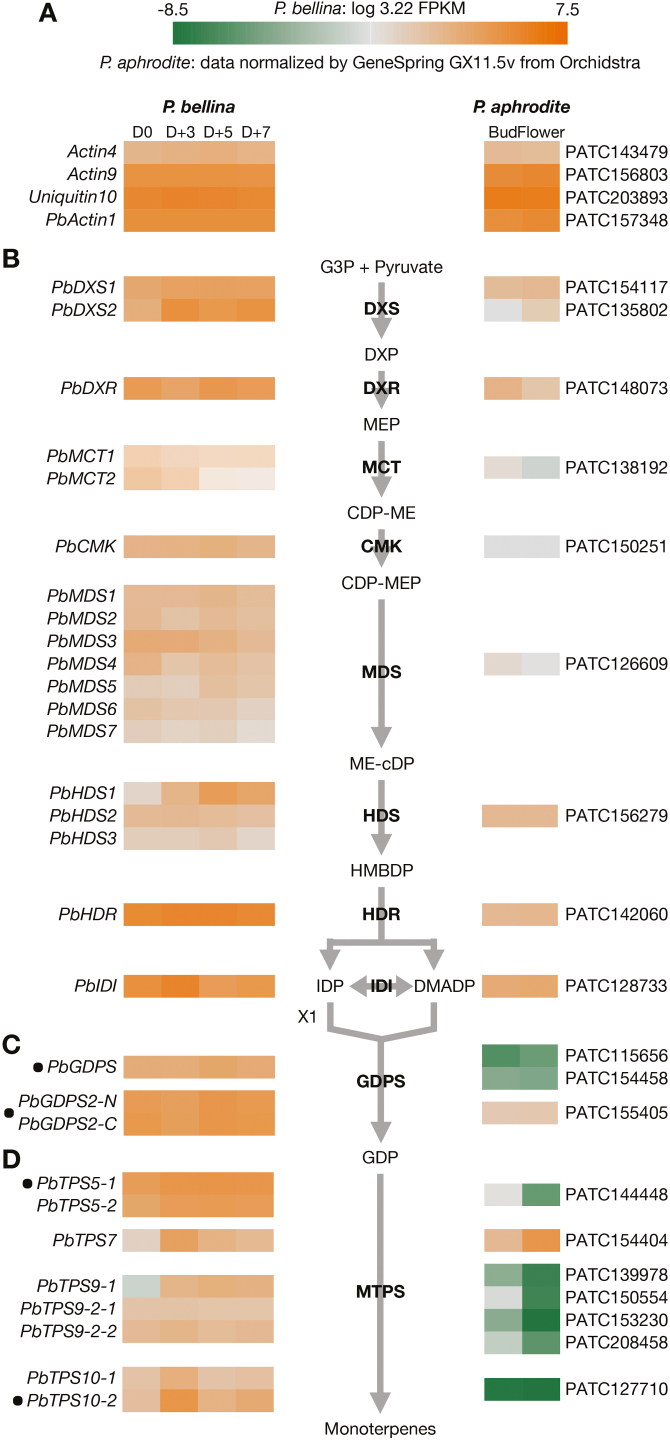
To explore the mechanisms underlying the contrasting monoterpene biosynthesis of the two orchids, the floral transcriptomes of P. bellina and P. aphrodite were compared. The transcriptomic data of P. bellina were constructed using floral material at four developmental stages (D0, D+3, D+5, and D+7) that represented the four phases of the monoterpene emission pattern: onset, increase, peak, and decline (Fig. 1C). In addition, floral transcriptomic data for P. aphrodite were downloaded from Orchidstra (Su et al., 2011, 2013a), which contains information for both the floral bud and full-blossom stages. The transcript abundance of the P. bellina transcriptome was determined by using FPKM, and the gene expression of P. aphrodite was analysed by microarray assays (Su et al., 2013b). To compare the transcriptomic profiling between the two different platforms, the FPKM values of the P. bellina transcriptome were transformed by log3.22. After this transformation, the expression of three reference genes that are typically analysed in Phalaenopsis orchids were found to exhibit equivalent levels between the RNA-seq and microarray assay data: Actin4 (Chen et al., 2005; Hsieh et al., 2013; Pan et al., 2014; Hsu et al., 2015), Actin9 (Hsiao et al., 2008; Pan et al., 2011, 2014; Hsu et al., 2015), and Ubiquitin10 (Lu et al., 2007; Hsiao et al., 2008) (Fig. 2A). In addition, Actin1, which had identical expression between the two transcriptomes (Fig. 2A), was used as an internal calibrator for further analysis. The expression levels of these reference genes in both transcriptomes are available at Dryad (see Dataset 1, ‘Reference’ spreadsheet, https://doi.org/10.5061/dryad.kt056q7).
Fig. 2.
Comparative expression profiles of putative genes encoding enzymes for monoterpene biosynthesis from two Phalaenopsis transcriptomes. (A) The putative gene expression in P. bellina and P. aphrodite transcriptomes is represented by a color gradient from orange to green. The reference genes and their homologs in P. aphrodite were included. (B) Expression of genes encoding enzymes in the MEP pathway. The abbreviated names of enzymes in each catalytic step are in bold. Putative genes are organized according to their annotated function. Isopentenyl diphosphate isomerase (IDI) is the enzyme that catalyses the isomerization between IDP and DMADP. (C) Expression of GDPS and GDPS2 in both transcriptomes. PbGDPS2-N and PbGDPS2-C indicate the N and C termini of PbGDPS2, respectively. (D) Expression of putative genes annotated as MTPSs in both transcriptomes. Genes that were further analysed by transactivation assays in planta are labeled with black dots. The homologous genes between two Phalaenopsis orchids are aligned opposite each other. Abbreviations for enzymes or compounds not described in the article are as follows: CDP-ME, 4-diphosphocytidyl-2-C-methylerythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; G3P, glyceraldehyde-3-phosphate; IDI, isopentenyl diphosphate isomerase, HMBDP, 4-hydroxy-3-methyl-but-2-enyl pyrophosphate; ME-cDP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate.
The putative genes encoding each step of the MEP pathway were further identified in both orchids. Expression of these genes, which are responsible for the biosynthesis of IDP and DMAPP, did not differ much between the two orchids (Fig. 2B), although the putative genes encoding 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT), 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS), and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase (HDR) were slightly down-regulated in P. aphrodite. In addition, multigene families identified for several steps in the MEP pathway for P. bellina included 1-deoxy-D-xylulose 5-phosphate synthase (DXS), MCT, MDS, and 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS).
Intriguingly, in addition to the original PbGDPS (27 kDa) (Hsiao et al., 2008), another gene encoding geranyl diphosphate synthase (57 kDa) was also identified and designated as PbGDPS2. We detected significant differential expression of GDPS between P. bellina and P. aphrodite whereas little or no differences were observed for the expression of GDPS2 (Fig. 2C). In addition, among four putative genes encoding monoterpene synthases (MTPSs) (Fig. S2 at Dryad), three were more significantly up-regulated in P. bellina, namely PbTPS5, PbTPS9, and PbTPS10 (Fig. 2D), consistent with its monoterpene production. TPS7 was expressed at similar levels in both orchids. Therefore, we concluded that the enhanced expression of GDPS and MTPSs may have accounted for the monoterpene biosynthesis in the orchids. GDPS was the first enzyme in the pathway with significant differential expression, and it provides the precursors for further monoterpene biosynthesis. Hence, the absence of elevated GDPS expression in the scentless P. aphrodite might be responsible for the lack of monoterpene accumulation. The expression levels of these structural genes in both transcriptomes are available at Dryad (see Dataset 1, ‘Enzyme’ spreadsheet, https://doi.org/10.5061/dryad.kt056q7).
trans-factors are critical for GDPS expression
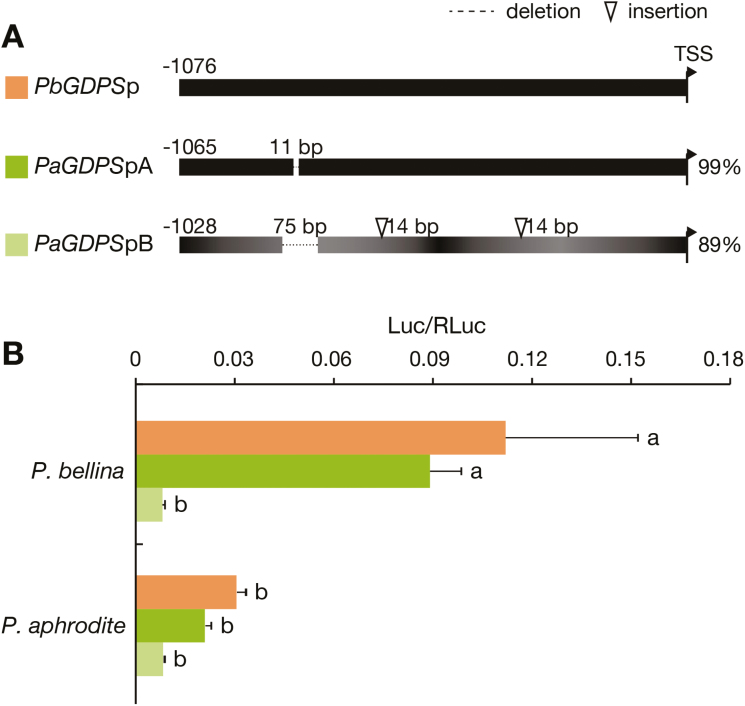
These results prompted us to investigate the mechanisms underlying the absence of GDPS expression in the scentless P. aphrodite. We isolated a 1-kb promoter fragment of GDPS from P. bellina and P. aphrodite, designated PbGDPSp for P. bellina and PaGDPSpA and PaGDPSpB for P. aphrodite because two promoter fragments were isolated. PaGDPSpA shared 99% identity with PbGDPSp, with an 11-bp deletion between nucleotides –836 to –823 (Fig. 3A). PaGDPSpB shared 89% identity with PbGDPSp, with a 75-bp deletion (nucleotides –859 to –785), two 14-bp insertions at nucleotides –763 and –355, and numerous nucleotide substitutions.
Fig. 3.
GDPS promoter activities in planta. (A) Sequence differences of three GDPS promoter fragments isolated from P. bellina (PbGDPSp) and P. aphrodite (PaGDPSpA and PaGDPSpB). The numbers indicate base-pairs upstream of the translational start site (TSS). Dotted lines and triangles indicate large-fragment deletions and insertions, respectively, in the two promoter fragments of PaGDPS as compared to PbGDPSp. Comparative similarities are indicated on the right. The numerous substitutions of PaGDPSpB are indicated by the varying shading. (B) Comparative activities of three promoter fragments in the floral tissues of the two Phalaenopsis species as determined by dual-luciferase assays. The activation level is given by the ratio of Luc/RLuc. Data are means (±SE) of three biological replicates. Pairwise comparisons between groups were performed using Tukey’s honestly significant difference test, and different letters indicate significant differences at α=0.05.
To assess the activity of these three promoter fragments, we particle-bombarded live perianths of scented P. bellina and scentless P. aphrodite flowers with various constructs for dual luciferase assays (Fig. 3B). Both PbGDPSp and PaGDPSpA activities were substantially greater in scented than scentless flowers, so the activators for both promoters were up-regulated in the scented P. bellina but down-regulated in the scentless P. aphrodite. In contrast, we detected no differential luciferase activity for PaGDPSpB in scented or scentless plants, which suggested impaired promoter activity of PaGDPSpB associated with its deletions, insertions, or nucleotide substitutions. These results implied that both trans-factors and cis-elements are required for the GDPS promoter activity, although trans-factors played the major role.
Correlation analysis to identify eight TFs associated with PbGDPS expression
Several types of TFs for terpene biosynthesis have been detected in various plant species, including bHLH, bZIP, ERF, NAC, MYB, and WRKY. A total of 335 TFs categorized in these types were identified in the P. bellina transcriptome by a BLAST search of PlnTFDB (Pérez-Rodríguez et al., 2010) and classified by iTAK (Zheng et al., 2016). Because PbGDPS was highly expressed in the P. bellina transcriptome (FPKM>30), we selected 165 TF genes showing FPKM values >1 for further analysis. We applied two criteria to identify candidate TFs regulating GDPS for monoterpene biosynthesis: (1) the TFs had to appear either prior to or concurrent with the expression pattern of PbGDPS during flower development, and (2) the expression of the TFs had to be up-regulated in P. bellina but down-regulated in P. aphrodite.
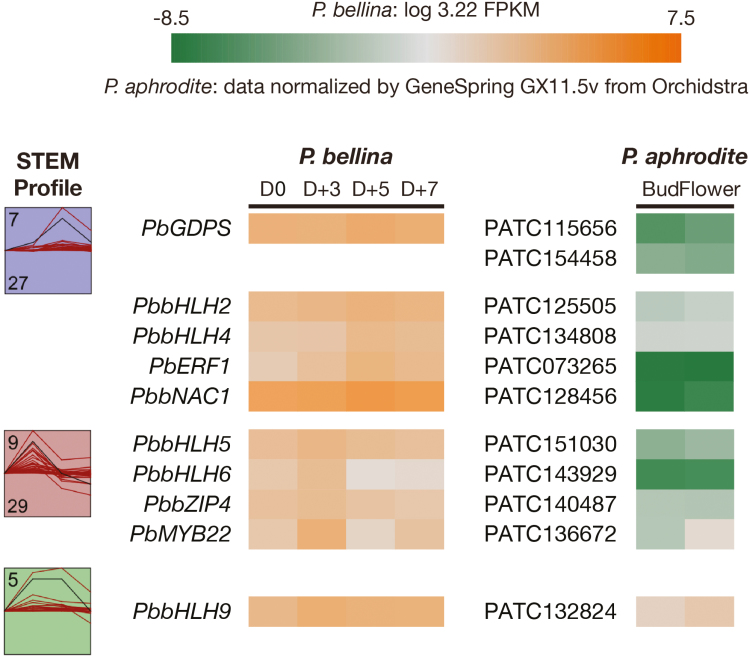
In the first screening, clustering by STEM (Ernst and Bar-Joseph, 2006) was performed to associate the expression patterns of the 165 TFs with that of PbGDPS, and 10 distinct STEM profiles (labelled 0 to 9) were generated (see Dataset 2 for full clustering results and see Fig. S3 at Dryad). PbGDPS showed maximal expression on D+5 and was classified into profile ‘7’, together with 26 TFs with concurrent expression (Fig. 4). In addition, 29 TFs in profile ‘9’ were selected for their expression prior to PbGDPS, which peaked on D+3. In the second screening, the expression of 55 TFs was compared to that of the scentless P. aphrodite. Among them, eight candidate TFs, namely PbbHLH2, PbbHLH4, PbHLH5, PbbHLH6, PbbZIP4, PbERF1, PbMYB22, and PbNAC1, showed enhanced differential expression between the two transcriptomes and were chosen for further analysis. The expression levels of these TF genes in both transcriptomes are available at Dryad (Dataset 1, ‘TF’ spreadsheet, https://doi.org/10.5061/dryad.kt056q7).
Fig. 4.
Bioinformatics analysis of transcription factors (TFs) to identify candidates. Two STEM profiles, No. 7 and No. 9, were selected for identifying candidate TFs in P. bellina transcriptomes (see Methods). Expression is represented by a color gradient from orange to green. The four time points represent the FPKM values at four floral developmental stages (day of anthesis D0, D+3, D+5, and D+7), which were normalized and transformed to set the time series to start at 0 (Fig. S2 at Dryad). The STEM profiles include the profile number at the top-left and the number of genes at the bottom-left. The model expression pattern of each profile is displayed as a black line, while the red lines are for individual genes. PbbHLH9, a bHLH TF belonging to STEM profile No. 5, is also included.
Confirmation of the transcript levels of structural genes and TFs by quantitative real-time PCR
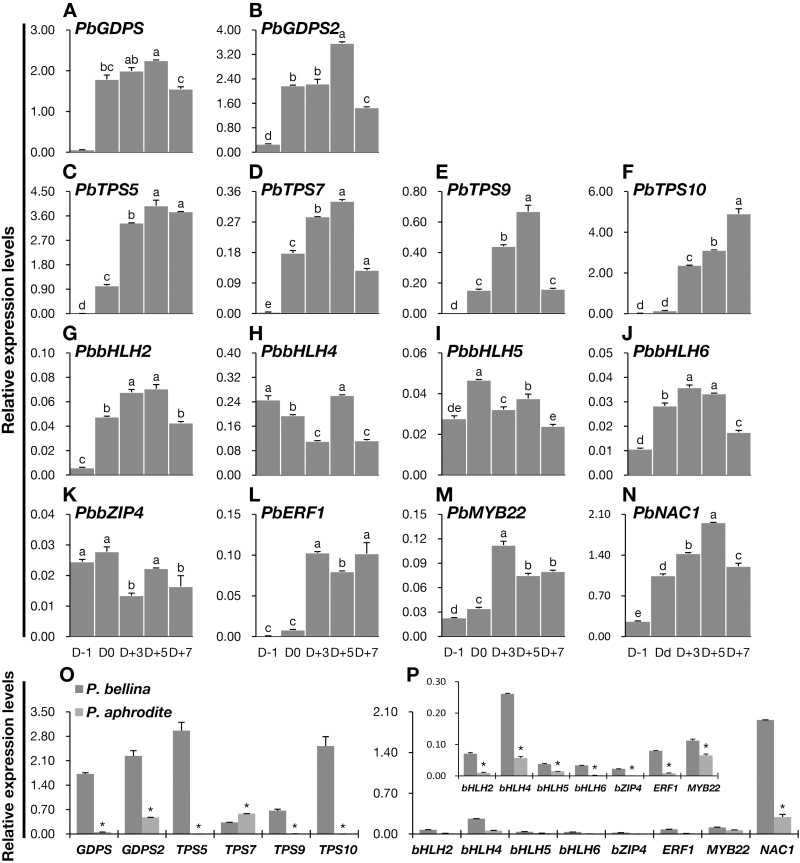
The transcript levels of GDPS, MTPSs, and the eight candidate TFs at the various floral developmental stages in the scented P. bellina were confirmed by quantitative real-time PCR (Fig. 5A–N). The expression of the structural genes involved in monoterpene biosynthesis sharply increased either upon anthesis (PbGDPS and PbGDPS2) (Fig. 5A, B) or at D+3 (PbTPS5, PbTPS7, PbTPS9, and PbTPS10) (Fig. 5C–F), concomitant with the emission pattern of monoterpenes (Fig. 1C). Among the four MTPSs, both PbTPS5 and PbTPS10 were highly expressed (Fig. 5C, F) and were responsible for the production of geraniol and linalool, respectively (Y-C Chuang, unpublished results). The expression of the eight TFs either peaked on D+5 (PbbHLH2, PbbHLH4, and PbNAC1) or peaked on D0 or D+3 prior to the expression of the structural genes (PbbHLH5, PbbHLH6, PbbZIP4, PbERF1, and PbMYB22).
Fig. 5.
Transcript levels of structural genes and transcription factors (TFs) in Phalaenopsis orchids. (A–N) Expression patterns during floral developmental stages in P. bellina as determined by quantitative real-time PCR for six structural genes for monoterpene biosynthesis, including PbGDPS (A), PbGDPS2 (B), PbTPS5 (C), PbTPS7 (D), PbTPS9 (E), PbTPS10 (F), and eight candidate TFs genes isolated by bioinformatics analysis, including PbbHLH2 (G), PbbHLH4 (H), PbbHLH5 (I), PbbHLH6 (J), PbbZIP4 (K), PbERF1 (L), PbMYB22 (M), and PbNAC1 (N). Expression was determined from the day before anthesis (D–1) to day 7 after anthesis (D+7). Pairwise comparisons between groups were performed by using Tukey’s honestly significant difference test, and different letters indicate significant differences at α=0.05. (O, P) Expression levels of six structural genes (O) and the eight candidate TFs (P) in the flowers of scentless P. aphrodite (at the D+5 stage) as determined by quantitative real-time PCR. Their expression levels in the scented P. bellina are also included for comparison. The insert in (P) shows the relatively lower expression levels of PbbHLH2, PbbHLH4, PbbHLH5, PbbHLH6, PbbZIP4, PbERF1, and PbMYB22 in P. aphrodite. Statistical analysis of the expression levels of Pa genes and Pb genes was performed by using Student’s t-test at α=0.05. Expression was normalized to that of Actin1. Data are means (±SE) from three replicates.
The expression levels of these TF genes in the scentless P. aphrodite were also verified and showed results that were consistent with the comparative transcriptomic analysis. GDPS, TPS5, TPS9, and TPS10 were highly expressed in scented P. bellina but were barely detectable in scentless P. aphrodite (Fig. 5O). The differential expression of GDPS2 was to slightly lesser extent than for GDPS, while TPS7 showed slightly higher expression in scentless P. aphrodite than in scented P. bellina. The differential expression of the eight TFs were confirmed, with all showing higher expression in P. bellina (Fig. 5P).
Transactivation of TFs on the promoters of the structural genes
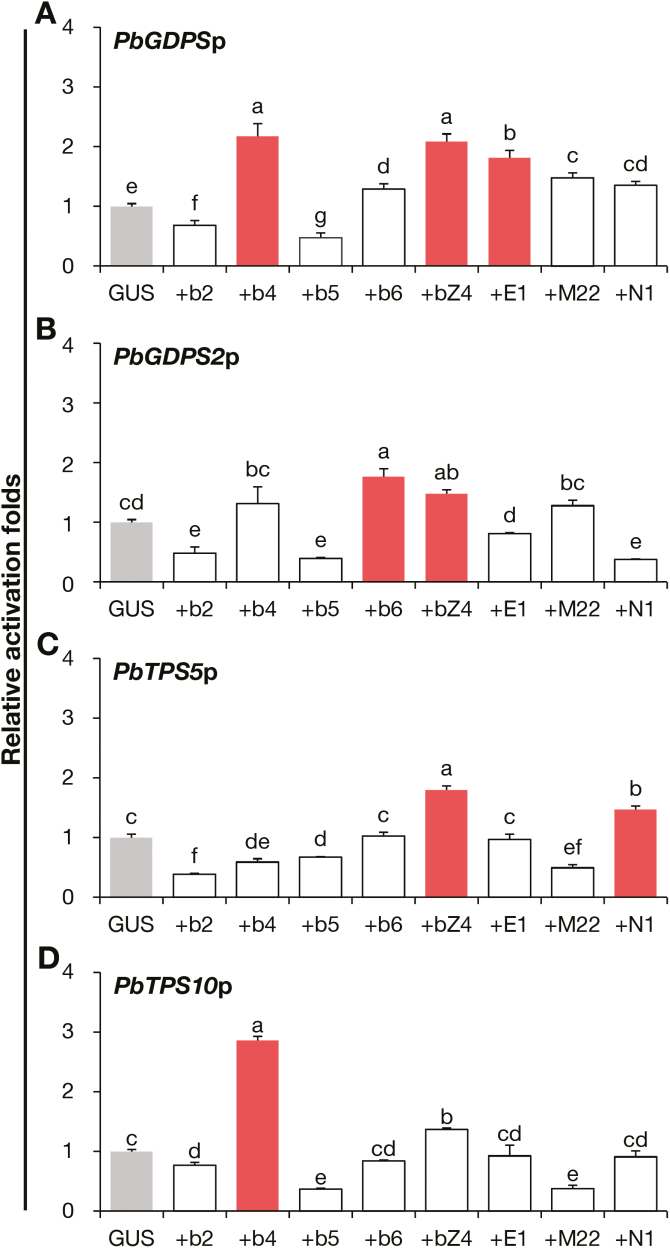
To evaluate whether PbGDPS could be transactivated by the eight TFs, we used dual luciferase assays with the coding sequences of the TFs under the control of the CaMV 35S promoter. The promoter fragments of PbGDPS2, PbTPS5, and PbTPS10 (designated PbGDPS2p, PbTPS5p, and PbTPS10p) were also analysed as they showed significant differential expression between P. bellina and P. aphrodite. Transient assays were performed in the perianths of the scentless P. aphrodite flowers, with GUS as a negative control. Among the four bHLHs, PbbHLH4 and PbbHLH6 transactivated PbGDPSp and PbTPS10p, and PbGDPS2p (Fig. 6A, B, D), but not PbTPS5p. PbbZIP4 enhanced the promoter activities of PbGDPSp, PbGDPS2p, and PbTPS5p (Fig. 6A–C), but not PbTPS10p. In contrast, PbERF1 only transactivated PbGDPSp (Fig. 6A), and PbNAC1 only transactivated PbTPS5p (Fig. 6C). PbbHLH2, PbbHLH5, and PbMYB22 did not transactivate any of the promoters tested, and in some cases even resulted in weak repression. Taken together, among the eight candidate TFs isolated by the bioinformatics analysis, five showed transactivation for monoterpene biosynthesis genes and acted as positive transactivators, namely PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNCA1.
Fig. 6.
Transactivation of the eight transcription factors (TFs) on promoter fragments of four structural genes. (A–D) Transactivation activity of the eight TFs on promoter fragments of PbGDPS (A), PbGDPS2 (B), PbTPS5 (C), and PbTPS10 (D), in P. aphrodite flowers by particle bombardment. The transient assays were performed with: GUS; +b2, PbbHLH2; +b4, PbbHLH4; +b5, PbbHLH5; +b6, PbbHLH6; +bZ4, PbbbZIP4; +E1, PbERF1; +M22, PbMYB22; and +N1 PbNAC1. The transactivation activity was evaluated according to relative fold-activity compared to the GUS control (grey). Activity with >1.5-fold change is highlighted in red. Data are means (±SE) of three biological replicates. Pairwise comparisons between groups were performed using Tukey’s honestly significant difference test, and different letters indicate significant differences at α=0.05.
Transient ectopic expression of the five candidate TFs in the scentless orchid
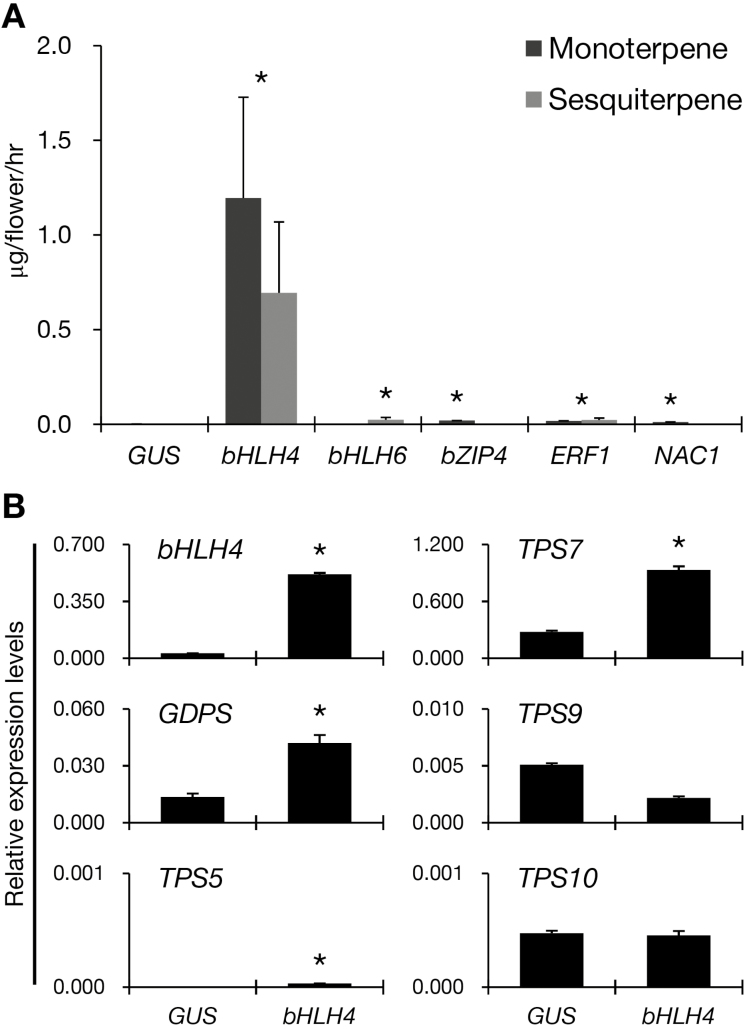
The above results demonstrated that low expression of TFs in the scentless phenotype P. aphrodite was associated with low expression of GDPS and MTPSs. We hypothesized that ectopic expression of the five transactivators in P. aphrodite might induce a scented phenotype. Efficient stable transformation systems for Phalaenopsis orchids are lacking, with those available having low transformation efficiency as well as long regeneration times. Therefore, to test our hypothesis we used an efficient and rapid transient expression assay by utilizing Agrobacterium infiltration in the perianths of P. aphrodite (Hsu et al., 2015). The coding sequences of PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNAC1 were constructed under the control of double CaMV 35S promoters and introduced into P. aphrodite flowers on the day of anthesis (D0). The floral scents emitted from these flowers were measured and identified using GC-MS analysis at 4 d post-infiltration. We detected a profound enhancement (~950-fold) in a group of monoterpenoid and sesquiterpenoid emissions in PbbHLH4-expressing P. aphrodite flowers as compared with the GUS controls (1.89 μg versus 0.002 μg h–1 per flower) (Fig. 7A). The monoterpenoids were dominated by terpineol derivatives, such as α-terpineol (the major component), β-terpineol, γ-terpineol, 1-terpineol, and terpinolene, together with a trace amounts of 1,4-cineole, limonene, fenchone, and camphor. Two sesquiterpenoids, α-cedrene and β-cedrene, were also detected in the infiltrated flowers. Transient expression of PbbHLH6, PbbZIP4, PbERF1, and PbNAC1 also induced monoterpenoid and/or sesquiterpenoid emissions, although to a much lower extent of ~10–20-fold (Fig. 7A).
Fig. 7.
Scent compounds produced by the transient ectopic expression of transcription factors (TFs) in flowers of scentless P. aphrodite. (A) Content of emitted terpenes from flowers with transient ectopic expression of GUS, PbbHLH4, PbbHLH6, PbbZIP4, PbERF1, and PbNAC1 was analysed at 5 d post-infiltration. GUS was used as a control. Data are means (±SE) from three infiltrations. (B) Expression levels of bHLH4, GDPS, and four MTPSs in the flowers of PbbHLH4-expressing P. aphrodite as determined by quantitative real-time PCR. Expression was normalized to that of Actin1. Data are means (±SE) from three replicates. Statistical analysis in comparison with the GUS control were performed using Student’s t-test at α=0.05.
To further examine this high induction of monoterpenoids in PbbHLH4-expressing P. aphrodite flowers, we examined the expression of structural genes for monoterpene biosynthesis. Both GDPS and TPS7 showed 3-fold increases in their expression, in addition to the significant increase of PbbHLH4 transcripts in the infiltrated flowers (Fig. 7B). This suggested that of the TPS genes it was specifically TPS7 that may have accounted for the great enhancement of terpineol in the PbbHLH4-expressing P. aphrodite flowers.
Expression of bHLH4 is concomitant with monoterpene biosynthesis in Phalaenopsis orchids
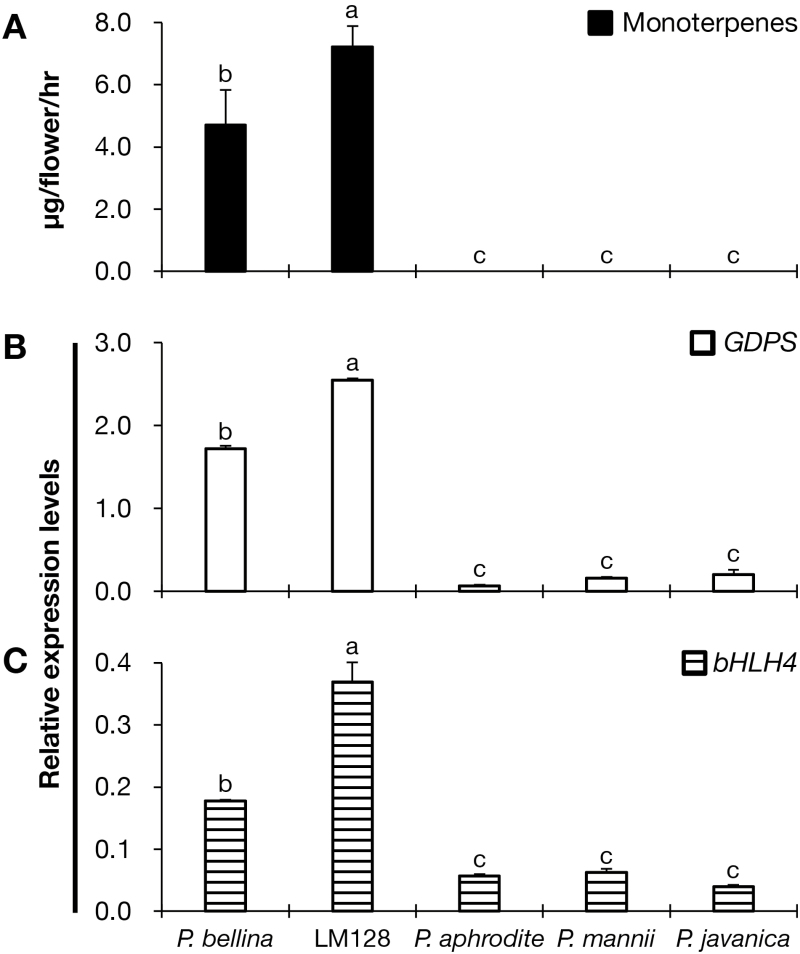
As transient expression of PbbHLH4 resulted in monoterpene biosynthesis in P. aphrodite, we were interested to examine its potential role in producing a scented phenotype in other Phalaenopsis orchids. To this end, the expression levels of both GDPS and bHLH4 were examined in other scented and scentless orchids (Fig. 8). Both genes were highly up-regulated in the commercial scented cultivar P. Meidarland Bellina Age ‘LM128’, an offspring of P. bellina that emitted high levels of monoterpenoids. In contrast, the expression levels of both genes were down-regulated in two native scentless orchids, P. javanica and P. mannii. The differential expression patterns of GDPS and bHLH4 were concomitant with monoterpene production, which implied their strong association with the scented orchid phenotype.
Fig. 8.
Expression patterns of GDPS and bHLH4 in orchid flowers. (A) Levels of emitted monoterpenes in scented P. bellina (from Fig. 1B) and P. Meidarland Bellina Age ‘LM128’ (labeled as LM128), and scentless P. aphrodite (from Fig. 1B), P. mannii and P. javanica at the D+5 floral stage (5 d after anthesis). (B, C) Expression of GDPS (B) and bHLH4 (C) in D+5 flowers. Expression was normalized to that of Actin1. Data are means (±SE) from three replicates. Pairwise comparisons between groups were performed using Tukey’s honestly significant difference test, and different letters indicate significant differences at α=0.05.
Discussion
The multigene families of the MEP pathway in P. bellina
In this study, putative MEP pathway genes were isolated by analysing the annotation results of floral transcriptomic data, and this revealed the participation of multigene families, including DXS, MCT, MDS, and HDS, in several steps in the pathway. We noticed that in contrast to only one MDS gene in scentless P. aphrodite, seven MDS genes were identified in the scented P. bellina transcriptome (Fig. 2B). Analysis showed that these seven PbMDSs shared extremely high similarity in coding sequence regions, with differences of only two or three residues (Fig. S4A at Dryad). In addition, three PbHDS genes displayed high similarity in their sequences (Fig. S4B at Dryad). We used Trinity for de novo transcriptome assembly in the analysis of P. bellina floral transcriptomic data, which defines alternative isoforms and duplicated genes (Grabherr et al., 2011). The seven detected MDS genes were deemed to be paralogous, but the three detected HDS genes were from alternative splicing. To confirm this finding, we searched for these gene sequences in the whole-genome sequence of P. equestris (Cai et al., 2015). Six MDS genes distributed in six individual scaffolds were identified, but only one HDS gene was discovered in P. equestris (data not shown). Therefore, we considered MDS to be present in Phalaenopsis orchids at a high gene-copy number, but the three HDS genes in P. bellina were likely to have resulted from alternative splicing. Because the seven PbMDS genes had various expression patterns (Fig. 2B), we concluded that they may have distinct roles in scented orchids for differential terpenoid biosynthesis.
The whole-genome sequence of P. aphrodite has been released (Chao et al., 2018), and we identified only one MDS gene within it, consistent with the result of the transcriptome analysis. This indicated that the copy number of MDS genes varies among Phalaenopsis orchids.
We identified another gene encoding geranyl diphosphate synthase, PbGDPS2. Its expression during the different floral developmental stages was similar to that of PbGDPS, with both being initiated on the day of anthesis (D0) and peaking on D+5 (Fig. 5A, B). Interestingly, PbGDPS2 was also expressed in scentless P. aphrodite (Fig. 5O). In scented P. bellina, PbGDPS shows flower-specific expression (Hsiao et al., 2008), while PbGDPS2 is also expressed in leaves and roots (data from Orchidstra 2.0, see Table S3 at Dryad; Chao et al., 2017). In addition to the biosynthesis of monoterpenoids, GDPS is required for biosynthesis of gibberellins in tomato, and might be involved in the biosynthesis of other terpenoids, including di-, tri-, tetra-, and/or polyterpenes (van Schie et al., 2007). Thus, it is plausible that PbGDPS2 may play a different role from PbGDPS in Phalaenopsis orchids.
Conserved bHLH factors for terpene regulation
The subgroup IVa of the bHLH protein family has previously been shown to be a conserved module for plant terpenoid biosynthesis regulated by jasmonate (Mertens et al., 2016). Phylogenetic analysis showed that PbbHLH4 falls into the subgroup IIIb, close to the subgroup IIIe in which AtMYC2 resides, the only regulator controlling floral sesquiterpenes in the Arabidopsis inflorescence (Hong et al., 2012) (Fig. S5 at Dryad). Intriguingly, AtMYC2 is also grouped with other terpene-regulation bHLHs, including AabHLH1 (Ji et al., 2014), CrMYC2 (Zhang et al., 2011), and SlMYC1 (Spyropoulou et al., 2014b), indicating a well-conserved regulation of terpenoids by bHLHs in the plant kingdom.
In this study, we also isolated PbbHLH9 as being classified in the subgroup IIIe (Fig. S5 at Dryad). However, we applied two criteria to characterize the upstream factor for regulating PbGDPS, and PbbHLH9 was excluded because its expression pattern belonged to the STEM profile ‘5’ (Fig. S3 at Dryad) and its expression showed little differences between P. bellina and P. aphrodite (Fig. 4). Nevertheless, because most genes in the MEP pathway showed similar expression between P. bellina and P. aphrodite (Fig. 2B), the possibility of the involvement of PbbHLH9 in the regulation of floral terpenes in orchids needs to be further examined. In addition, we do not preclude the possibility that additional bHLH genes might play similar roles to PbbHLH4 in the regulation of terpene biosynthesis in Phalaenopsis orchids.
Developmental regulation for floral monoterpene biosynthesis
For the correct the timing of a plant event, a set of internal mechanisms is often co-ordinated to occur in coincidence with, or prior to, the event proceeding. The emission of monoterpenes in P. bellina began on D0 (the start of blossoming), while their release strikingly increased on D+1 (by ~30-fold, Fig. 1C) as the flowers became fully opened. The crucial enzyme PbGDPS exhibited induction on D0 (Fig. 5A) prior to emission of volatiles, and its upstream TFs showed either much earlier expression at the big-bud stage (D–1) (PbbHLH4 and PbbZIP4, Fig. 5H, K), or expression coinciding with volatile emission (PbbERF1, Fig. 5L). On the other hand, PbMTPSs, the final step of the pathway, were expressed coincidentally with the formation of volatiles (Fig. 5C, D, E, F). This well-designed regulation of the metabolic pathway contributed to monoterpene emission in the early floral stages of P. bellina.
The emission of monoterpenes in P. bellina flowers gradually increased over the first 5 d after anthesis, remained relatively steady up to D+9, and decreased thereafter (Fig. 1C). We speculate that this developmental modulation of floral scents might be designed to facilitate pollination. A study of P. ‘Tianxiang’ showed that the receptivity of the stigma increases after anthesis, but becomes weaker around half the life span of the flower (Yu and Li, 2017). The decreasing emission of scent with ageing might also be related to energy conservation (Dudareva et al., 1999).
The phylogenetic analysis showed that PbbHLH4 was grouped with AtbHLH33/ICE2/SCRM2 and AtbHLH116/ICE1/SCRM (Fig. S5 at Dryad), which play dual roles in cold acclimation and stomatal differentiation (Kanaoka et al., 2008; Kim et al., 2015). Jasmonate is a critical upstream signal for an ICE-mediated cold-stress response pathway (Hu et al., 2013). Moreover, jasmonate, or jasmonic acid, is a general inducer of terpenoid biosynthesis for defense responses (Martin et al., 2002; Fäldt et al., 2003; Miller et al., 2005; Erbilgin et al., 2006; Lundborg et al., 2016; Yoshitomi et al., 2016). Several reports have described the involvement of jasmonic acid in stress responses in Phalaenopsis orchids, such as enhanced cold, heat, or drought tolerance in seedlings (Huageng et al., 2011; Zou et al., 2011) and reduced flower-bud abortion during shipping (Chen, 2005).
Overall, PbbHLH4 may regulate the biosynthesis of floral monoterpenes in P. bellina in order to attract pollinators and also for stress responses, but additional studies are needed.
Possible negative regulators for the biosynthesis of floral monoterpenes
Using transactivation assays, we found that the three TFs, namely PbbHLH2, PbbbHLH5, and PbMYB22, showed negative effects on the promoter activities of structural genes for monoterpene biosynthesis (Fig. 6), suggesting that they might repress the expression of these structural genes to some extent. Two studies in spearmint have reported repressors for monoterpene biosynthesis, MsYABBY5 and MsMYB, which were found to be highly expressed in peltate glandular trichomes where monoterpenes are accumulated (Wang et al., 2016; Reddy et al., 2017). It would be worthwhile studying the roles of these factors and how they interact with other activators in regulating the expression of the structural genes involved in monoterpenes biosynthesis.
Possible reasons for low expression of bHLH4 in P. aphrodite
We conclude that the lack of activators, especially bHLH4, led to the scentless phenotype in P. aphrodite. To examine why bHLH4 was expressed at a low level in P. aphrodite, we first compared the coding sequence (CDS) of PabHLH4 with PbbHLH4. Interestingly, two single-nucleotide insertions were found in the CDS of PabHLH4, leading to a frame-shift and a premature stop codon in the coding region (Fig. S6A at Dryad). This could be the main reason for the low expression of PabHLH4 in P. aphrodite. In addition, a large insertion and numerous nucleotide substitutions were also found in the promoter fragment of PaHLH4 (PaHLH4p) as compared to that of PbHLH4p (Fig. S6B at Dryad).
Potential interactions of bHLH4 with other activators
We examined the expression profiles of the TFs and their transactivation activity on the various structural genes in order to determine whether there were any interactions or additive activation effects between the TFs. Previous studies have shown interactions between different classes of TFs in regulating terpene biosynthesis. For instance, CrWRKY1 is an upstream regulator for CrORCA3, CrMYC2, and CrZCTs in C. roseus (Pan et al., 2016), and SlMYC1 together with SlEOT1 have synergistic effects that induce SlTPS5 promoters in tomato (Spyropoulou et al., 2014b).
To examine this in Phalaenopsis orchids, we performed co-infiltration experiments in P. aphrodite flowers by equimolar mixing with Agobacterium cells harboring a single TF. However, these infiltrated flowers wilted at 3–4 d post-infiltration (DPI), indicating that the floral tissue could not stand the stress of co-infiltrated. We tried diluting the bacterial solution, but there were no effects in the infiltrated flowers.
Instead, we used a Phalaenopsis cultivar, P. I-Hsin Venus, which emits a small amount of linalool (Chuang et al., 2017). Although many of the infiltrated flowers also wilted at 3–4 DPI, a 2-fold induction of monoterpenes was detected in PbbHLH4-expressing flowers as compared to the GUS control (Fig. S7A at Dryad). The increased monoterpenoids included a large proportion of linalool and trace amounts of ocimene and eucalyptol. With a combination of PbbHLH4 and PbbZIP4, the monoterpene level showed a 3-fold increase as compared to the GUS control. Gene expression analysis showed that this combination induced increases in the transcript levels of GDPS, TPS5, TPS7, and TPS9 (Fig. S7B at Dryad). PbZIP4 alone did not have any activation effects (data not shown), suggesting that there was a synergistic effect between PbbHLH4 and PbbZIP4. In contrast, the combination of PbbHLH4 and PbNAC1 did not further enhance the levels of monoterpenes as compared to PbbHLH4 alone (Fig. S7A at Dryad). This suggested that the interactive effects between these two TFs were not as high as PbbHLH4 and PbbZIP4. Furthermore, the combination of all three TFs decreased the emission of monoterpenes as well as the expression levels of individual TFs, as compared to plants infiltrated with one or two TFs. This could have been due to dilution effects or competition between the 35S promoters for all three TFs.
Overall, the results indicate that PbbHLH4 may interact with PbbZIP4 and/or PbNAC1 in regulating monoterpene biosynthesis in Phalaenopsis orchids to varying degrees.
Manipulation of floral volatile terpenoids by TFs in orchids
PbbHLH4 was able to transactivate the upstream promoters of PbGDPS and PbTPS10 in transient assays (Fig. 6), which indicated that they are downstream target genes of PbbHLH4. PbGDPS is a dual-function prenyltransferase as recombinant PbGDPS yields GDP and FDP with IDP/DMADP and IDP/GDP as substrates for monoterpene and sesquiterpene production, respectively (Hsiao et al., 2008). The transactivating ability of PbbHLH4 on PbGDPS might account for it facilitating both monoterpene and sesquiterpene biosynthesis in the transient-expression flowers. Ectopic expression of a spearmint R2R3MYB, MsMYB, whose target gene is GDPS affects both monoterpenes and sesquiterpenes in sweet basil (Reddy et al., 2017). Moreover, ectopic transient expression of PbbHLH4 in the scentless commercial cultivar P. Sogo Yukidian ‘V3’ increased the internal pools of sesquiterpenes (Fig. S8 at Dryad), so PbbHLH4 might control the metabolic flux in different branches of terpene biosynthesis.
It was interesting that the transient expression of PbbHLH4 in the perianths of scentless P. aphrodite induced the production of terpineol (Fig. 7A), which has an odor similar to lilac and a sweet smell (Dionísio et al., 2012). Terpineol derivatives were found at very low levels in the floral scent profile of P. bellina (data not shown). We speculate that the terpineol derivatives might be generated from the downstream MTPS in the scentless orchid. By analysing the expression of structural genes for monoterpene biosynthesis in the PbbHLH4-expressing P. aphrodite flowers, we found that expression of both GDPS and TPS7 showed a 3-fold increase (Fig. 7B), indicating that PbbHLH4 was able to regulate GDPS and terpene synthase. In contrast, the expression of PaTPS10 was not activated by PbbHLH4, which might be a result of differences in their promoter sequences (Fig. S9B at Dryad). We do not exclude the possibility that TFs other than PbbHLH4 may have played more important roles in regulating PbTPS5 and PbTPS10; however, given the lack of higher GDPS induction, the production of linalool and geraniol was naturally at a lower level in the infiltrated flowers.
In this study, we selected P. aphrodite as the scentless orchid with which to make comparisons because it has overtaken many other varieties in terms of its commercialization. However, P. aphrodite has a small genome size (2.80 pg/2C) as compared with P. bellina (15.03 pg/2C) (Lin et al., 2001) and there is usually cross-incompatibility between P. bellina and P. aphrodite, or its offspring. In some cases, despite successful crossing, the offspring are infertile. This is one of the obstacles faced by orchid breeders in trying to induce the scented trait into commercial cultivars with a P. aphrodite background. Therefore, because transient ectopic expression of PbbHLH4 strongly induced the scent volatiles in P. aphrodite, it has great potential for molecular breeding of scented cultivars.
Data Deposition
The following figures, tables and datasets are available at the Dryad Data Repository: https://doi.org/10.5061/dryad.kt056q7.
Fig. S1. The putative protein sequences of GDPS and four TPSs from both P. bellina and P. aphrodite.
Fig. S2. Phylogenetic tree of the putative PbMTPS.
Fig. S3. Gene cluster analysis of transcription factors as determined using the STEM software.
Fig. S4. Multiple alignment of the MEP pathway gene families in the P. bellina
transcriptome.
Fig. S5. Phylogenetic tree inferred from the amino sequences of PbbHLH4 with terpene-related bHLHs and the IIIb and IIIe subgroups of Arabidopsis bHLHs.
Fig. S6. Multiple alignment of the coding region and promoter fragments of PabHLH4 and PbbHLH4.
Fig. S7. Scent compounds produced by the transient ectopic expression of PbbHLH4 with PbbZIP4 and/or PbNAC1 in flowers of P. I-Hsin Venus.
Fig. S8. Scent compounds produced by the transient ectopic expression of PbbHLH4 in flowers of P. Sogo Yukidian ‘V3’.
Fig. S9. Multiple alignment of the promoter fragments of TPS5 and TPS10 isolated from P. aphrodite and P. bellina.
Table S1. TFs involved in the regulation of terpene biosynthesis.
Table S2. Sequences of all primers used in this study.
Table S3. Expression levels of GDPS and GDPS2 in vegetative organs in P. bellina.
Dataset 1. Expression levels of reference, structural, and TF genes in the two transcriptomes examined.
Dataset 2. STEM clustering results for associating expression patterns of the transcription factors examined.
Acknowledgments
We thank Dr Tuan-Hua David Ho and Dr Shu-Hsing Wu (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan), and Dr Yu-Yun Hsiao (Orchid Research and Development Center, NCKU, Taiwan) for helpful discussions. We thank Dr Chi-Kuang Wen (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China) and Dr Michel Delseny (Laboratoire Génome et Développement des Plantes, France) for careful reading of the manuscript. We thank Dr Chuan-Ming Yeh (Division of Strategic Research and Development, Graduate School of Science and Engineering, Saitama University, Japan) and Ya-Lan Chang (Department of Life Sciences, NCKU, Taiwan) for their assistance in the statistical analysis. This work was supported by the Ministry of Science and Technology, Taiwan (no. MOST-102-2313-B-006-001-MY3).
References
- Blight MM, Le Métayer M, Delègue M-HP, Pickett JA, Marion-Poll F, Wadhams LJ. 1997. Identification of floral volatiles involved in recognition of oilseed rape flowers, Brassica napus by honeybees, Apis mellifera. Journal of Chemical Ecology 23, 1715–1727. [Google Scholar]
- Byers KJ, Bradshaw HD Jr, Riffell JA. 2014. Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). The Journal of Experimental Biology 217, 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Liu X, Vanneste K, et al. 2015. The genome sequence of the orchid Phalaenopsis equestris. Nature Genetics 47, 65–72. [DOI] [PubMed] [Google Scholar]
- Caputi L, Aprea E. 2011. Use of terpenoids as natural flavouring compounds in food industry. Recent Patents on Food, Nutrition & Agriculture 3, 9–16. [DOI] [PubMed] [Google Scholar]
- Chao YT, Chen WC, Chen CY, et al. 2018. Chromosome‐level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnology Journal, in press. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YT, Yen SH, Yeh JH, Chen WC, Shih MC. 2017. Orchidstra 2.0—A transcriptomics resource for the orchid family. Plant & Cell Physiology 58, e9. [DOI] [PubMed] [Google Scholar]
- Chen M-Y. 2005. Effects of environmental factor, orchid mycorrhizal fungi and plant growth substances on the growth and flower quality of Phalaenopsis spp. with flower stalk after transport. Masters Thesis, National Taiwan University. [Google Scholar]
- Chen YH, Tsai YJ, Huang JZ, Chen FC. 2005. Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Research 15, 639–657. [DOI] [PubMed] [Google Scholar]
- Chuang Y-C, Hung Y-C, Tsai W-C, Chen W-H, Chen H-H. 2018. Data from: PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. Dryad Digital Repository. doi: 10.5061/dryad.kt056q7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y-C, Lee MC, Chang YL, Chen WH, Chen HH. 2017. Diurnal regulation of the floral scent emission by light and circadian rhythm in the Phalaenopsis orchids. Botanical Studies 58, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionísio AP, Molina G, de Carvalho DS, dos Santos R, Bicas J, Pastore G. 2012. Natural flavourings from biotechnology for foods and beverages. In: Baines D, Seal R, eds. Natural food additives, ingredients and flavourings. Cambridge, UK: Woodhead Publishing Limited, 231–259. [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J. 2004. Biochemistry of plant volatiles. Plant Physiology 135, 1893–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Piechulla B, Pichersky E. 1999. Biogenesis of floral scent. Horticultural Reviews 24, 31–54. [Google Scholar]
- Erbilgin N, Krokene P, Christiansen E, Zeneli G, Gershenzon J. 2006. Exogenous application of methyl jasmonate elicits defenses in Norway spruce (Picea abies) and reduces host colonization by the bark beetle Ips typographus. Oecologia 148, 426–436. [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt J, Martin D, Miller B, Rawat S, Bohlmann J. 2003. Traumatic resin defense in Norway spruce (Picea abies): methyl jasmonate-induced terpene synthase gene expression, and cDNA cloning and functional characterization of (+)-3-carene synthase. Plant Molecular Biology 51, 119–133. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature Biotechnology 29, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. 2003. The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. 2012. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. The Plant Cell 24, 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YY, Jeng MF, Tsai WC, Chuang YC, Li CY, Wu TS, Kuoh CS, Chen WH, Chen HH. 2008. A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2–4D motif. The Plant Journal 55, 719–733. [DOI] [PubMed] [Google Scholar]
- Hsiao YY, Tsai WC, Chen WH, Chen HH. 2011. Biosynthetic regulation of floral scent in Phalaenopsis. In: Chen WH, Chen HH, eds. Orchid biotechnology II. Singapore: World Scientific, 145–180. [Google Scholar]
- Hsiao YY, Tsai WC, Kuoh CS, Huang TH, Wang HC, Wu TS, Leu YL, Chen WH, Chen HH. 2006. Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biology 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Pan ZJ, Lai PH, et al. 2013. Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. Journal of Experimental Botany 64, 3869–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH. 2015. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiology 168, 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Wu PS, Chen TC, Yu CW, Tsai WC, Wu K, Wu WL, Chen WH, Chen HH. 2014. Histone acetylation accompanied with promoter sequences displaying differential expression profiles of B-class MADS-box genes for Phalaenopsis floral morphogenesis. PloS ONE 9, e106033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D. 2013. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. The Plant Cell 25, 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huageng Y, Suliang Y, Huijuan C, Chongfa Y, Fusun Y, Zifan L. 2011. Effect of exogenous methyl jasmonate, calcium and salicylic acid on the heat tolerance in Phalaenopsis seedlings under high temperature stress. Chinese Agricultural Science Bulletin 28, 029. [Google Scholar]
- Huang M, Sanchez-Moreiras AM, Abel C, Sohrabi R, Lee S, Gershenzon J, Tholl D. 2012. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytologist 193, 997–1008. [DOI] [PubMed] [Google Scholar]
- Ji Y, Xiao J, Shen Y, et al. 2014. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant & Cell Physiology 55, 1592–1604. [DOI] [PubMed] [Google Scholar]
- Junker RR, Gershenzon J, Unsicker SB. 2011. Floral odor bouquet loses its ant repellent properties after inhibition of terpene biosynthesis. Journal of Chemical Ecology 37, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. 2008. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. The Plant Cell 20, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Lee M, Lee JH, Lee HJ, Park CM. 2015. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Molecular Biology 89, 187–201. [DOI] [PubMed] [Google Scholar]
- Li CY, Leopold AL, Sander GW, Shanks JV, Zhao L, Gibson SI. 2013. The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biology 13, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Leopold AL, Sander GW, Shanks JV, Zhao L, Gibson SI. 2015a. CrBPF1 overexpression alters transcript levels of terpenoid indole alkaloid biosynthetic and regulatory genes. Frontiers in Plant Science 6, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang H, Li F, et al. 2015b. The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. The Plant Journal 84, 296–308. [DOI] [PubMed] [Google Scholar]
- Li X, Xu Y, Shen S, Yin X, Klee H, Zhang B, Chen K, Hancock R. 2017. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. Journal of Experimental Botany 68, 4929–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee H-C, Chen W-H, Chen C-C, Kao Y-Y, Fu Y-M, Chen Y-H, Lin T-Y. 2001. Nuclear DNA contents of Phalaenopsis sp. and Doritis pulcherrima. Journal of the American Society for Horticultural Science 126, 195–199. [Google Scholar]
- Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang DC, Yeh HH. 2007. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiology 143, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, Lv Z, Wang G, Tang K. 2013. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytologist 198, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Lundborg L, Nordlander G, Björklund N, Nordenhem H, Borg-Karlson AK. 2016. Methyl jasmonate-induced monoterpenes in scots pine and norway spruce tissues affect pine weevil orientation. Journal of Chemical Ecology 42, 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z, Wang S, Zhang F, et al. 2016. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant & Cell Physiology 57, 1961–1971. [DOI] [PubMed] [Google Scholar]
- Ma D, Pu G, Lei C, et al. 2009. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant & Cell Physiology 50, 2146–2161. [DOI] [PubMed] [Google Scholar]
- Martin D, Tholl D, Gershenzon J, Bohlmann J. 2002. Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiology 129, 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Van Moerkercke A, Vanden Bossche R, Pollier J, Goossens A. 2016. Clade IVa basic helix-loop-helix transcription factors form part of a conserved jasmonate signaling circuit for the regulation of bioactive plant terpenoid biosynthesis. Plant & Cell Physiology 57, 2564–2575. [DOI] [PubMed] [Google Scholar]
- Miller B, Madilao LL, Ralph S, Bohlmann J. 2005. Insect-induced conifer defense. White pine weevil and methyl jasmonate induce traumatic resinosis, de novo formed volatile emissions, and accumulation of terpenoid synthase and putative octadecanoid pathway transcripts in Sitka spruce. Plant Physiology 137, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Nishizawa Y, Minami E, Nojiri H, Yamane H, Okada K. 2015. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. Journal of Plant Physiology 173, 19–27. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Chen X, Wang MY, Matich AJ, Perez RL, Allan AC, Green SA, Atkinson RG. 2015. Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiology 167, 1243–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. 2009. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. The Journal of Biological Chemistry 284, 26510–26518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Mustafa NR, Tang K, Choi YH, Verpoorte R. 2016. Monoterpenoid indole alkaloids biosynthesis and its regulation in Catharanthus roseus: a literature review from genes to metabolites. Phytochemistry Reviews 15, 221–250. [Google Scholar]
- Pan ZJ, Chen YY, Du JS, Chen YY, Chung MC, Tsai WC, Wang CN, Chen HH. 2014. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytologist 202, 1024–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, Hu JM, Chen HH. 2011. The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant & Cell Physiology 52, 1515–1531. [DOI] [PubMed] [Google Scholar]
- Paul P, Singh SK, Patra B, Sui X, Pattanaik S, Yuan L. 2017. A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytologist 213, 1107–1123. [DOI] [PubMed] [Google Scholar]
- Pauw B, Hilliou FA, Martin VS, et al. 2004. Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. The Journal of Biological Chemistry 279, 52940–52948. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. 2010. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Research 38, D822–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VA, Wang Q, Dhar N, et al. 2017. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnology Journal 15, 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E. 2008. Biosynthesis of plant-derived flavor compounds. The Plant Journal 54, 712–732. [DOI] [PubMed] [Google Scholar]
- Shen SL, Yin XR, Zhang B, Xie XL, Jiang Q, Grierson D, Chen KS. 2016. CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. Journal of Experimental Botany 67, 4105–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou EA, Haring MA, Schuurink RC. 2014a. Expression of Terpenoids 1, a glandular trichome-specific transcription factor from tomato that activates the terpene synthase 5 promoter. Plant Molecular Biology 84, 345–357. [DOI] [PubMed] [Google Scholar]
- Spyropoulou EA, Haring MA, Schuurink RC. 2014b. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics 15, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, Hwa KT, Shih MC. 2011. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant & Cell Physiology 52, 1501–1514. [DOI] [PubMed] [Google Scholar]
- Su CL, Chao YT, Yen SH, Chen CY, Chen WC, Chang YC, Shih MC. 2013a. Orchidstra: an integrated orchid functional genomics database. Plant & Cell Physiology 54, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CL, Chen WC, Lee AY, Chen CY, Chang YC, Chao YT, Shih MC. 2013b. A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS ONE 8, e80462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. 2011. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiology 157, 2081–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. 2015. Biosynthesis and biological functions of terpenoids in plants. Advances in Biochemical Engineering/Biotechnology 148, 63–106. [DOI] [PubMed] [Google Scholar]
- Vainstein A, Lewinsohn E, Pichersky E, Weiss D. 2001. Floral fragrance. New inroads into an old commodity. Plant Physiology 127, 1383–1389. [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Steensma P, Gariboldi I, et al. 2016. The basic helix-loop-helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. The Plant Journal 88, 3–12. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Steensma P, Schweizer F, et al. 2015. The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proceedings of the National Academy of Sciences, USA 112, 8130–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CC, Ament K, Schmidt A, Lange T, Haring MA, Schuurink RC. 2007. Geranyl diphosphate synthase is required for biosynthesis of gibberellins. The Plant Journal 52, 752–762. [DOI] [PubMed] [Google Scholar]
- Wang Q, Reddy VA, Panicker D, Mao HZ, Kumar N, Rajan C, Venkatesh PN, Chua NH, Sarojam R. 2016. Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint (Mentha spicata). Plant Biotechnology Journal 14, 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Wang JW, Wang S, Wang JY, Chen XY. 2004. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiology 135, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura C, Mizutani E, Okada K, et al. 2015. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. The Plant Journal 84, 1100–1113. [DOI] [PubMed] [Google Scholar]
- Yoshitomi K, Taniguchi S, Tanaka K, Uji Y, Akimitsu K, Gomi K. 2016. Rice terpene synthase 24 (OsTPS24) encodes a jasmonate-responsive monoterpene synthase that produces an antibacterial γ-terpinene against rice pathogen. Journal of Plant Physiology 191, 120–126. [DOI] [PubMed] [Google Scholar]
- Yu L, Li J. 2017. Pollen viability and stigma receptivity of Phalaenopsis. Chinese Agricultural Science Bulletin 33, 54–58. [Google Scholar]
- Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. 2012. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Molecular Plant 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zhang F, Fu X, Lv Z, et al. 2015. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Molecular Plant 8, 163–175. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J. 2011. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. The Plant Journal 67, 61–71. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, et al. 2016. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant 9, 1667–1670. [DOI] [PubMed] [Google Scholar]
- Zou Q-C, Zhu K-Y, Liu H-C, Zhou J-H, Ma G-Y. 2011. Effect of exogenous methyl jasmonate on chlorophyll fluorescence and antioxidant characteristics in the leaves of Phalaenopsis amabilis under abiotic stress. Plant Physiology Journal 47, 913–917. [Google Scholar]