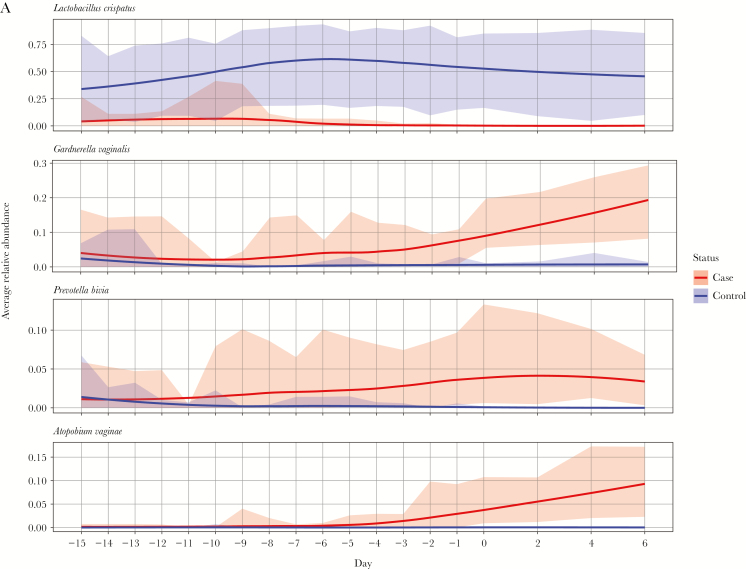
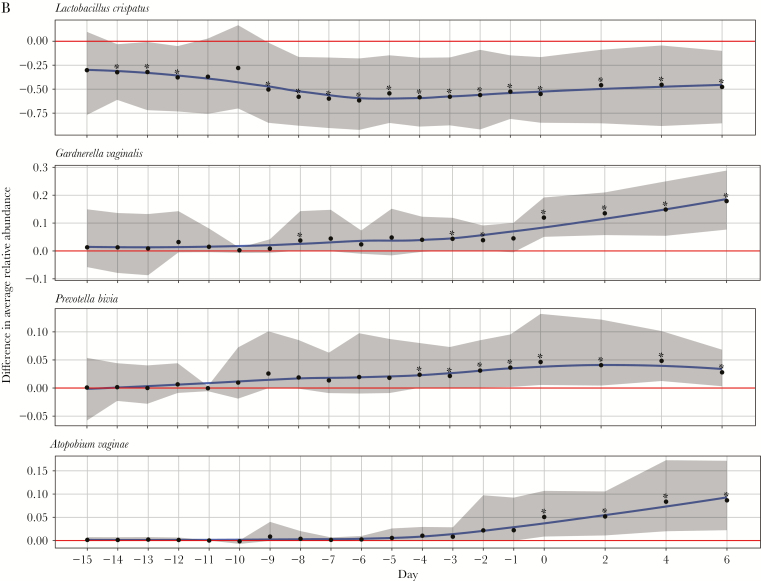
The mean relative abundance of Prevotella bivia, Gardnerella vaginalis, Atopobium vaginae, and Megasphaera type I became significantly higher among women 4 days before (P. bivia), 3 days before (G. vaginalis), and on the day of (A. vaginae and Megasphaera type I) incident bacterial vaginosis.
Keywords: Bacterial vaginosis, pathogenesis, women who have sex with women
Abstract
Background
The sequence of events preceding incident bacterial vaginosis (iBV) is unclear.
Methods
African American women who have sex with women, who had no Amsel criteria and Nugent scores of 0–3, were followed for 90 days to detect iBV (defined as a Nugent score of 7–10 on at least 2–3 consecutive days), using self-collected vaginal swab specimens. For women with iBV (cases) and women maintaining normal vaginal flora (healthy women), 16S ribosomal RNA gene sequencing targeting V4 was performed. Longitudinal vaginal microbiome data were analyzed.
Results
Of 204 women screened, 42 enrolled; of these, 45% developed iBV. Sequencing was performed on 448 specimens from 14 cases and 8 healthy women. Among healthy women, Lactobacillus crispatus dominated the vaginal microbiota in 75%. In contrast, prior to iBV, the vaginal microbiota in 79% of cases was dominated by Lactobacillus iners and/or Lactobacillus jensenii/Lactobacillus gasseri. The mean relative abundance of Prevotella bivia, Gardnerella vaginalis, Atopobium vaginae, and Megasphaera type I became significantly higher in cases 4 days before (P. bivia), 3 days before (G. vaginalis), and on the day of (A. vaginae and Megasphaera type I) iBV onset. The mean relative abundance of Sneathia sanguinegens, Finegoldia magna, BV-associated bacteria 1–3, and L. iners was not significantly different between groups before onset of iBV.
Conclusion
G. vaginalis, P. bivia, A. vaginae, and Megasphaera type I may play significant roles in iBV.
Bacterial vaginosis (BV), the most common cause of vaginal discharge [1], is associated with preterm birth, pelvic inflammatory disease, and increased risk of acquisition/transmission of sexually transmitted pathogens, including human immunodeficiency virus (HIV) [2–4]. The epidemiology of BV strongly indicates that it is acquired via sexual transmission [5]. BV is more common among women who have sex with women than among women who have sex with men [6–8].
BV is characterized by loss of lactic acid–producing lactobacilli that compose the normal vaginal flora and increases in facultative (Gardnerella vaginalis) and strict (ie, Prevotella bivia and Atopobium vaginae) anaerobes [9]. The trigger initiating these alterations is unknown [5, 10–13]. G. vaginalis has been studied most in BV pathogenesis and is present in 95%–100% of cases [14–16]. It is more virulent than other BV-associated bacteria, adhering in larger aggregates to, possessing greater cytotoxic activity for, and forming a significantly thicker biofilm on vaginal epithelial cells [17].
Our primary aim was to use molecular methods to determine the sequence of events preceding incident BV (iBV) among African American women who have sex with women. We hypothesized that G. vaginalis is the keystone pathogen whose proliferation leads to the complex changes in the vaginal flora associated with BV [13]. If G. vaginalis is the initial insult, its appearance and establishment in the vaginal microbiome will be seen in women with iBV prior to increases in other BV-associated bacteria. In addition to G. vaginalis, we also focused on several additional BV-candidate bacteria (ie, P. bivia, A. vaginae, Megasphaera type I, Sneathia sanguinegens, and BV-associated bacteria 1–3 [BVAB1–3]) [18] that may also play a role in the induction of iBV.
METHODS
In Birmingham, Alabama, we recruited potential study participants through radio advertisements, flyers at college campuses and throughout Birmingham, local events, and from walk-ins at the Jefferson County Department of Health sexually transmitted diseases clinic. Screening inclusion criteria were African American race, female sex, age of 18–45 years, history of sex with a woman during the past 12 months, and having a current female sex partner. Screening exclusion criteria were use of antibiotics within the past 14 days, HIV infection, pregnancy, and menses.
Women meeting inclusion criteria provided a detailed sexual history and underwent a urine pregnancy test. They self-collected 1 vaginal swab specimen for determination of Amsel criteria [19] and vaginal Gram stain for Nugent scoring [20]. A Nugent score of 0–3 was designated as normal vaginal flora; 4–6, as intermediate flora; and 7–10, as BV flora [20]. Enrollment inclusion criteria were absence of all Amsel criteria and a Nugent score of 0–3 with no G. vaginalis morphotypes on Gram stain (interpreted by a research clinician and confirmed by a second reader in our laboratory; if there was disagreement, a third reader was used). Enrollment exclusion criteria were pregnancy, detection of trichomoniasis on a wet mount slide, and symptomatic vaginal yeast infection.
Enrolled participants completed an interviewer-administered study questionnaire on sociodemographic characteristics, sexually transmitted infection history, sexual history, douching, vaginal lubricant use, and contraception use. A pelvic examination was performed, with a cervical swab specimen obtained for Trichomonas vaginalis, Chlamydia trachomatis, and Neisseria gonorrhoeae nucleic acid amplification testing, using the BD ProbeTec Qx CTQ/GCQ/TVQ assays [21, 22]. Women practiced completion of a daily diary (comprising a yes/no checklist about behaviors and events such as sexual activity and menses) by making a day-1 entry, and they self-collected 2 vaginal swab specimens for Gram stain (for enrollment Nugent score determination by a second reader) and future vaginal microbiota sequencing. Instructions for vaginal self-swabbing and smearing of swabs onto slides were provided. Research staff observed participants smearing the vaginal swab on a slide and provided feedback. Women testing positive for T. vaginalis, C. trachomatis, or N. gonorrhoeae by nucleic acid amplification testing were removed from the study because these sexually transmitted pathogens may alter the vaginal microbiota [23–26]. The remaining women were followed for 90 days for iBV, defined as a Nugent score of 7–10 on at least 2–3 consecutive days.
Daily vaginal swab specimens for microbiome analysis were collected in individual tubes and stored in a freezer at the participant’s home. In addition to daily diaries and vaginal Gram stains, specimen tubes were transferred on ice to the study site weekly until iBV and for 1–2 weeks thereafter (owing to the lag between specimen drop-off and Gram stain interpretation) or for 90 days. Gram stains of daily vaginal specimens were evaluated to determine the development of iBV, based on Nugent score. Vaginal specimens were stored at −80oC until sequencing. Daily diary entries were scanned into a database, using Teleform (OpenText, Waterloo, Canada). Similar methods have been performed by our group [27] and others [28, 29].
For participants who developed iBV (hereafter, “cases”) and age-comparable women who maintained a normal vaginal flora for the majority of the study (ie, ≥95% of days; hereafter, “healthy women”), DNA extraction and 16S ribosomal RNA (rRNA) gene sequencing were performed on stored vaginal specimens collected from cases during the 21 days leading up to iBV and every other day for 1 week thereafter. For healthy women, samples for sequencing were chosen on the basis of menstrual cycle day, which was aligned with cases.
DNA extraction and sequencing were performed by the Louisiana State University School of Medicine Microbial Genomics Resource Group (available at: http://metagenomics.lsuhsc.edu/mgrg/). The genomic DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Germantown, MD), modified to include bead beating. Two steps of amplification were performed to prepare the sequencing library, using the AccuPrime Taq high-fidelity DNA polymerase system (Thermo-Fisher/Invitrogen/Life Technologies, Carlsbad, CA) [1]. First, the 16S ribosomal DNA hypervariable region V4 was amplified using 20 ng of genomic DNA and the following gene-specific primers with Illumina adaptors: 5ʹ-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGT GCCAGCMGCCGCGGTAA-3ʹ (forward) and 5ʹ-GTCTCGT GGGCTCGGAGATGTGTATAAGAGACAGGGACT ACHVGGGTWTCTAAT-3ʹ (reverse) [2]. Second, purified amplicon DNA from the last step of 25 cycles of the polymerase chain reaction was amplified for 8 cycles, using the following primers with different molecular barcodes: 5ʹ-AATGATACG GCGA CCACCGAGATCTACAC [i5] TCGTCGGCAGCGTC- 3ʹ (forward) and 5ʹ- CAAGCAGAAGACGGCATACGAGAT [i7] GTCTCGTGGGCTCGG-3ʹ (reverse). The normalized and pooled libraries were then run with paired-end sequencing on an Illumina MiSeq machine (Illumina, San Diego, CA), using the 2 × 250bp V2 sequencing kit. All sequence reads have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA417968.
Sequences were filtered and trimmed using DADA2 (v1.4.0) [30]. The DADA2 filterAndTrim function was run with truncLen set to 230 bp and TrimLeft set to 10 bp for both forward and reverse reads. Error rates were learned over a subset of >1 × 106 reads and used by the dada algorithm to infer sequence variants. Sequence variants were merged, and bimeras were removed. Taxonomic assignment was performed using PECAN [31] and the vaginal_319_806_rc_MCo7p2 database (available at: http://ravel-lab.org/pecan). To achieve full taxonomy assignments, the Ribosomal Database Project (RDP) [32] and GreenGenes_13_8 [33] were merged with PECAN assignments, using a custom, in-house python script (available at: https://github.com/gblanchard4/pecan_greengenes_hybridizer). The PECAN classifier reports only genus- and species-level classifications and omits higher levels of taxonomy. Hence, the RDP classification was merged with the PECAN classification by adding higher levels (kingdom–family) to the genus and species levels reported by PECAN. Whenever the RDP gave a more specific classification than PECAN, the RDP classification was used. PECAN classifications were used whenever genus/species-level discrepancies between PECAN and RDP arose, since PECAN is specifically curated for the vaginal microbiota.
Community state type (CST; a cluster of community states that are similar in terms of the kinds and relative abundances of observed phylotypes) [28, 29] assignments were performed as follows and visualized using multidimensional scaling. Bray-Curtis distances between samples were calculated, and the most significant principal coordinate analysis eigenvectors were extracted. The R algorithm pam (partitioning around medoids) was applied to the principal coordinate analysis distances and the number of clusters (k = 5) were determined from the gap statistic [34], which is a goodness of cluster measurement [35]. The Bray-Curtis distance was chosen as a nonphylogenetically aware distance measure to better separate vaginal communities dominated by closely related but functionally distinct Lactobacillus species.
Mean ages for cases versus healthy women were compared using a t test. Proportions for sociodemographic characteristics, sexually transmitted infection history, sexual behavior history, contraception use at the time of enrollment, and other variables among cases and healthy women were compared using the Fisher exact tests in SAS 9.4 (Cary, NC). P values <.05 were considered statistically significant.
Statistical analysis of sequencing data was performed using R 3.4.0. Longitudinal microbiome data for BV-candidate bacteria (G. vaginalis, P. bivia, A. vaginae, Megasphaera type I, S. sanguinegens, and BVAB1–3) and several lactobacilli of interest (L. crispatus, L. iners, L. jensenii, and L. gasseri) were analyzed using the phyloseq v1.20.0 library [36]. Given the distributional nature of the data and the small sample size, statistical models that rely on normality assumptions were deemed inappropriate. Instead, we used a bootstrapping approach; 10000 samples were collected with replacement from cases and healthy women to create 95% confidence intervals (CIs) comparing the mean relative abundance difference between groups. Our bootstrapping approach selected the complete set of longitudinal values for each individual to maintain and account for the correlation structure among the repeated observations. Mean relative abundance differences between cases and healthy women were calculated for each day within each bootstrap. Bootstrap CIs were estimated using a Bonferroni correction that adjusted the 0.05 type I error rate for the 19 days under investigation (0.05/19 = 0.0026) and took the 0.26th and 99.74th percentile from the 10000 bootstraps to define the empirical CI for each mean difference. If the bootstrap 95% empirical CI did not include 0, it was concluded that the groups significantly differed.
This study was approved by the University of Alabama–Birmingham Institutional Review Board (protocol F131127001), the Jefferson County Department of Health Research Review Committee, and the Louisiana State University Institutional Review Board (protocol 8738). Written informed consent was obtained and included permission to use stored vaginal specimens for microbiota sequencing.
RESULTS
Between September 2014 and November 2017, 204 African American women who have sex with women were screened; 42 (21%) met enrollment inclusion criteria, and 162 (79%) met exclusion criteria (Supplementary Table 1). Of 42 enrolled, 2 were withdrawn from the study because of a Nugent score of >3 at second read, 1 was withdrawn because G. vaginalis morphotypes were detected on her enrollment Gram stain at second read, 3 were withdrawn because of positive results of nucleic acid amplification testing for C. trachomatis (n = 2) and T. vaginalis (n = 1), and 5 were lost to follow-up. Thus, 31 completed the study, of whom 14 (45%) developed iBV. Specimens from 8 healthy women with an age range (24–38 years) similar to that of cases (22–41 years) were available for sequencing. Sequencing was performed on specimens (n = 448) from 14 cases and 8 healthy women.
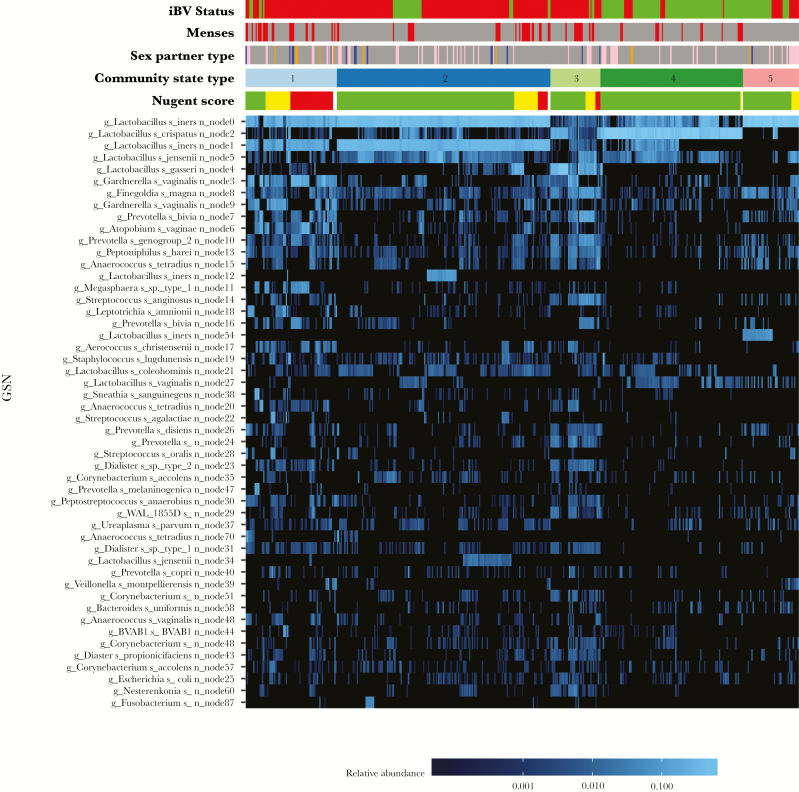
Table 1 displays sociodemographic and clinical characteristics for 14 cases and 8 healthy women. Supplementary Table 2 displays the metadata. Of 448 vaginal specimens sequenced, 5 did not have a sufficient number of reads to classify the vaginal bacterial communities. Thus, a data set of 6675726 high-quality classifiable 16S rRNA gene sequences were obtained from 443 specimens, with a mean (±SD) of 15069 ± 11317 sequences/sample. The heat map in Figure 1 displays the top 50 most highly abundant microorganisms across all specimens, which composed >95% of all reads. Sequence variants identified by the DADA2 algorithm are given as node numbers and included as unique identifiers for individual inferred sequence variants found in the sequence reads [30]. The top 10 most highly abundant organisms, in descending order, were L. iners node 0, L. crispatus, L. iners node 1, L. jensenii, L. gasseri, G. vaginalis node 3, F. magna, G. vaginalis node 9, P. bivia, and A. vaginae.
Table 1.
Characteristics of African American Women Who Have Sex With Women, by Emergence (Cases) or Nonemergence (Healthy Women) of Incident Bacterial Vaginosis (BV) During a BV Pathogenesis Study in Birmingham, Alabama
| Characteristic | Cases (n = 14) |
Healthy Women (n = 8) |
P a |
|---|---|---|---|
| Age, y, mean ± SD | 29.1 ± 8.2 | 30.5 ± 6.2 | .67 |
| Education level | .79 | ||
| Less than high school | 1 (7) | 0 (0) | |
| High school | 5 (36) | 4 (50) | |
| Some college | 8 (57) | 4 (50) | |
| Tobacco use in past 30 d | .18 | ||
| Yes | 9 (64) | 2 (25) | |
| No | 5 (36) | 6 (75) | |
| Douching in past 30 d | - | ||
| No | 14 (100) | 8 (100) | |
| Vaginal lubricant use in past 3 mo | .19 | ||
| Yes | 6 (43) | 1 (13) | |
| No | 8 (57) | 7 (87) | |
| STI historyb | 1.00 | ||
| Yes | 13 (93) | 8 (100) | |
| No | 1 (7) | 0 (0) | |
| Sex partner history | .20 | ||
| Women only | 9 (69) | 3 (38) | |
| Men and women | 4 (31) | 5 (62) | |
| Contraception use | 1.00 | ||
| Current | 1 (7) | 1 (13) | |
| Past | 6 (43) | 4 (50) | |
| Never | 7 (50) | 3 (37) |
Data are no. (%) of study participants, unless otherwise indicated. Numbers in subgroups may not equal sums in columns because of missing data on selected variables.
Abbreviation: STI, sexually transmitted infection.
aA t test was used to compare groups for age and Fisher’s exact test was used to compare groups for the other variables.
bDefined as Trichomonas vaginalis infection, Neisseria gonorrhoeae infection, Chlamydia trachomatis infection, genital herpes, and syphilis.
Figure 1.
Heat map displaying the top 50 most highly abundant microorganisms (in descending order) composing >95% of all reads in 443 vaginal swab specimens. Nodes, identified by the DADA2 algorithm, are unique identifiers for individual inferred sequence variants found in the sequence reads [30]. The node level classification is included in this heat map and others because there could be patterns to different sequence variants of a given species that may have biological relevance or importance. Metadata at the top of the heat map include sexual activity, by partner sex (female, pink; male, blue; and unknown, orange); incident bacterial vaginosis (iBV) status (present, red; and absent, green); menstruation status (menses, red); Nugent score (0–3, green; 4–6, yellow; and 7–10, red); and community state type (CST1, light blue; CST2, dark blue; CST3, light green; CST4, dark green; and CST5, dark pink). The scale bar (at the bottom of the heat map) provides the fractional relative abundance of a given species, on a log scale. As the blue color bar gets brighter, the relative abundance of each microorganism increases.
The vaginal bacterial communities were classified into 1 of 5 CSTs, based on differences in species composition and relative abundance (Supplementary Figure 1A–E). CST 1, the CST most associated with iBV, was characterized by high proportions of L. iners nodes 0 and 1, G. vaginalis node 3, A. vaginae, Megasphaera type I, and P. bivia. CST 2 was dominated by L. iners nodes 0 and 1. CST 3 had high proportions of L. gasseri and L. jensenii and moderate proportions of G. vaginalis node 3, F. magna, and P. bivia, but it was not associated with iBV. CST4 was dominated by L. crispatus. CST5 was dominated by L. iners node 0, with moderate proportions of L. iners node 54 and F. magna. High Nugent scores were associated with CST 1; low scores were associated with CSTs 2–5. All but 1 case (K028) entered CST1. CST3 primarily belonged to 2 cases (K018 and K031). All healthy women remained in CSTs 2, 4, and 5, with an occasional venture into CSTs 1 and 3. CST5 was not detected during menses (Supplementary Figures 2–21).
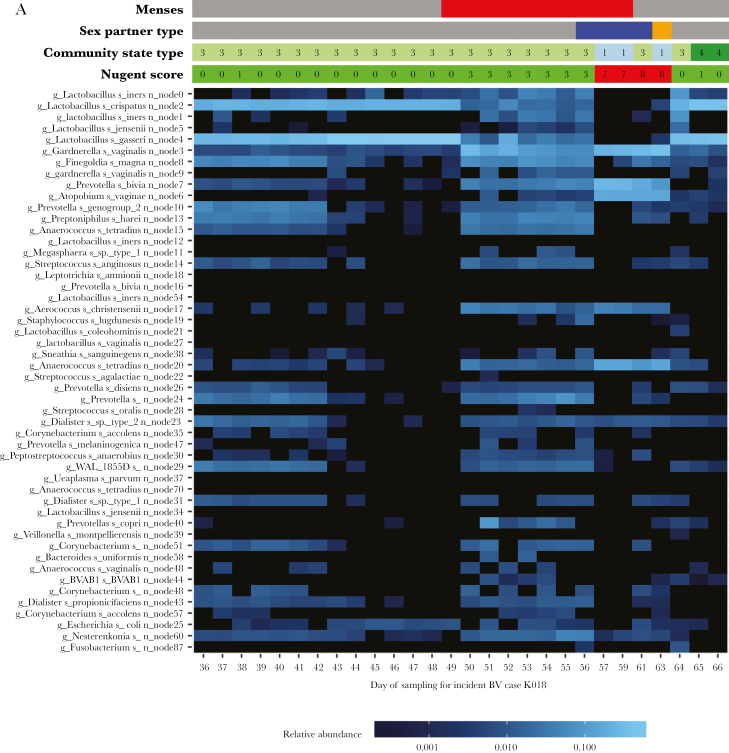
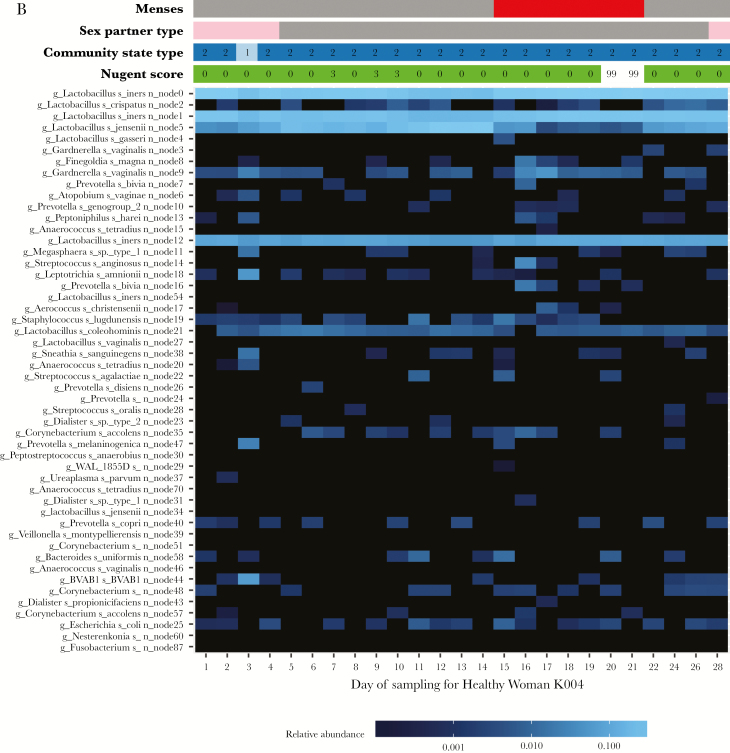
Figure 2A is a heat map for a case (K018), and Figure 2B is a heat map for a healthy woman (K004). The vaginal microbiota of case K018 was initially dominated by L. gasseri and L. crispatus on days 36–49, and her Nugent score was normal. Menses occurred on day 49. By day 50, the relative abundance of L. gasseri and L. crispatus decreased, while the relative abundance of multiple anaerobes increased, and her Nugent score was 3. This continued through day 56, when she reported sex with a man. On day 57, the relative abundance of G. vaginalis node 3, P. bivia, and A. vaginae increased dramatically (from low levels), and the patient’s Nugent score was 7. iBV lasted through day 63 (during which time she continued to have sex with a male). By day 64 and in the absence of BV treatment, her Nugent score dropped to 0; the relative abundance of G. vaginalis node 3, P. bivia, and A. vaginae decreased; and the relative abundance of L. gasseri and L. crispatus increased. In contrast, healthy woman K004 maintained normal vaginal flora for the majority of the study. Her vaginal microbiota was dominated by L. iners nodes 0, 1, and 12, in addition to L. jensenii, and was not significantly altered by sex with a woman or menses.
Figure 2.
A, Heat map of a study participant with incident bacterial vaginosis (case K018). Time runs from left to right on the bottom of the heat map and is indicated by the days of study enrollment. Metadata at the top of the heat map includes sexual activity, by partner sex (female, pink; male, blue; and orange, unknown); menstruation status (menses = red); Nugent score (0–3, green; 4–6, yellow; 7–10, red; and 99, missing data); and community state type (CST1, light blue; CST2, dark blue; CST3, light green; CST4, dark green; and CST5, dark pink). The scale bar (at the bottom of the heat map) provides the fractional relative abundance of a given species, on a log scale. As the blue color bar gets brighter, the relative abundance of each microorganism increases. B, Heat map of a study participant with normal vaginal flora for the majority of the study (healthy woman K004). Time runs from left to right on the bottom of the heat map. Metadata at the top of the heat map include sexual activity, by partner sex (female, pink; male, blue; unknown, orange); menstruation status (menses, red); Nugent score (0–3, green; 4–6, yellow; and 7–10, red; missing data, 99); and community state type (CST1, light blue; CST2, dark blue; CST3, light green; CST4, dark green; and CST5, dark pink). The scale bar (at the bottom of the heat map) provides the fractional relative abundance of a given species, on a log scale. As the blue color bar gets brighter, the relative abundance of each microorganism increases.
Regarding heat maps for all cases and healthy women (Figure 2A and 2B and Supplementary Figures 2–21), the majority of healthy women (6 of 8 [75%]) had a vaginal microbiota dominated by L. crispatus. In contrast, prior to iBV, the majority of cases (11 of 14 [79%]) had a vaginal microbiota dominated by L. iners nodes 0 and 1 and/or L. jensenii and L. gasseri. Menses destabilized the vaginal microbiota, preceding iBV by 1–3 days in 5 of 14 cases (36%). In 3 cases (K018, K026, and K028), iBV resolved on its own after menses ceased. Sex immediately preceded iBV by 1–3 days in 8 of 14 cases (57%). The majority of cases (13 of 14 [93%]) had detectable G. vaginalis and/or other BV-associated bacteria (ie, A. vaginae, P. bivia, and BVAB1), at low levels, prior to iBV.
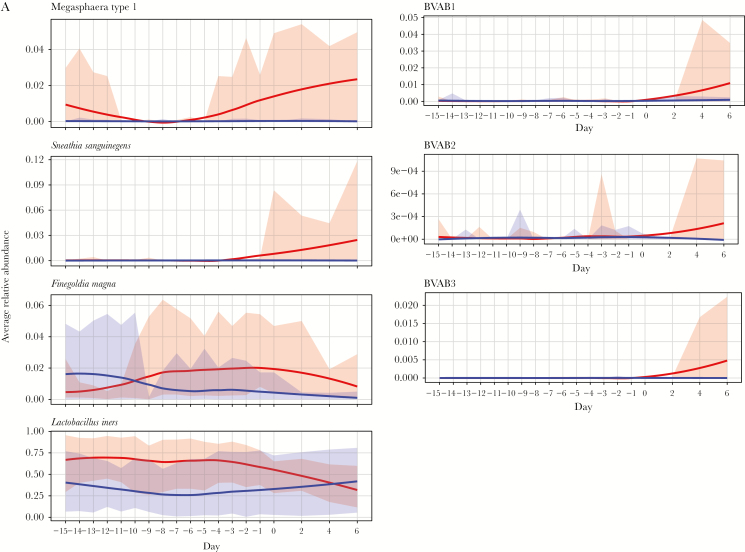
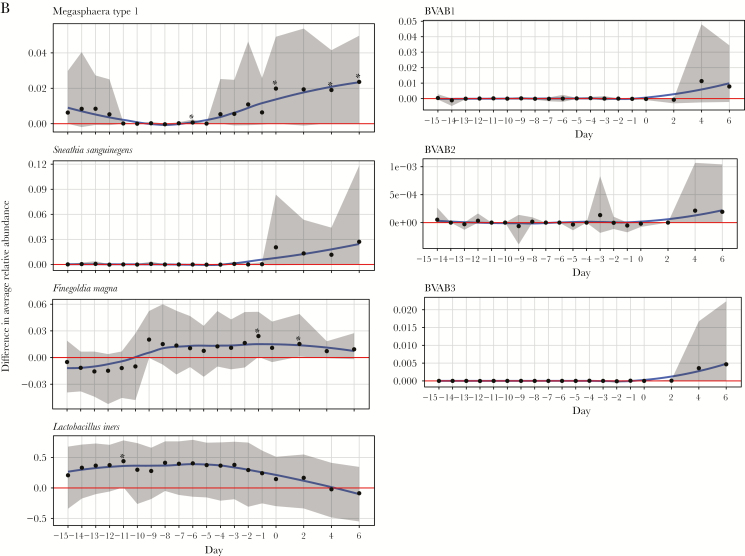
Figure 3A and 3B and Figure 4A and 4B display the difference in mean relative abundance over time (day 0 is first day of iBV) of G. vaginalis, other BV-candidate bacteria, F. magna (because it was among the top 10 most highly abundant bacteria), and L. crispatus, L. iners, L. jensenii, and L. gasseri between cases and healthy women. The mean relative abundance of L. crispatus became significantly lower in cases starting 14 days before iBV and continuing through day 6 after. The mean relative abundance of P. bivia and G. vaginalis became significantly higher in cases starting 4 days and 3 days before iBV, respectively. The mean relative abundances of A. vaginae and Megasphaera type I became significantly higher in cases on the day of iBV. There was no significant difference in the mean relative abundance of S. sanguinegens, BVAB1–3, L. iners, L. jensenii (data not shown), and L. gasseri (data not shown) between cases and healthy women leading up to iBV or thereafter. The mean relative abundance of F. magna became significantly higher in cases 1 day before iBV and 2 days after iBV. However, these were isolated findings.
Figures 3.
Variation in trends of mean relative abundance of Lactobacillus crispatus, Gardnerella vaginalis, Prevotella bivia, and Atopobium vaginae over time between study participants with incident bacterial vaginosis (iBV; cases) and study participants who maintained normal vaginal flora (healthy women). 16S ribosomal RNA gene sequencing was performed on stored vaginal specimens obtained from cases for 21 days leading up to iBV (and aligned menstrual cycle days for healthy women) and every other day for 1 week thereafter. To perform this analysis, the sequencing data were organized so that the first day of iBV was labeled as day 0 and aligned between cases. The x-axis on each figure represents time, while the y-axis represents the average difference in relative abundance of each microorganism between cases and healthy women. A, Mean relative abundances of L. crispatus, G. vaginalis, P. bivia, and A. vaginae over time among cases (red line, with 95% confidence intervals [CIs] shaded in red) and healthy women (purple line with 95% CIs shaded in purple) are displayed as smooth curves, using the loess smoothing technique. B, Difference in mean relative abundances of these microorganisms between cases and healthy women over time is represented by a blue smooth curve, again using the loess smoothing technique (with 95% CIs shaded in gray). The horizontal red line indicates no difference in the mean relative abundance of the microorganism between cases and healthy women. Days with a statistically significant difference in the mean relative abundance of the microorganism between cases and healthy women are denoted by asterisks. Scales may differ among y-axes. Standardizing these scales would make a subset of the figures difficult to read.
Figures 4.
Variation in trends of mean relative abundance of Megasphaera type I, Sneathia sanguinegens, Finegoldia magna, bacterial vaginosis (BV)–associated bacteria 1–3 (BVAB1–3), and Lactobacillus iners over time between study participants with incident bacterial vaginosis (iBV; cases) and study participants who maintained normal vaginal flora (healthy women). 16S ribosomal RNA gene sequencing was performed on stored vaginal specimens obtained from cases for 21 days leading up to iBV (and aligned menstrual cycle days for healthy women) and every other day for 1 week thereafter. To perform this analysis, the sequencing data were organized so that the first day of iBV was labeled as day 0 and aligned between cases. The x-axis on each figure represents time, while the y-axis represents the difference in mean relative abundance of each microorganism between cases and healthy women. A, Mean relative abundances of Megasphaera type I, S. sanguinegens, F. magna, BVAB1–3, and L. iners over time in cases (red line with 95% confidence intervals [CIs] shaded in red) and healthy women (purple line with 95% CIs shaded in purple) are displayed as smooth curves, using the loess smoothing technique. B, Differences in mean relative abundances of these microorganisms between cases and healthy women over time are represented by a blue smooth curve, again using the loess smoothing technique (with 95% CIs shaded in gray). The horizontal red line indicates no difference in the mean relative abundance of the microorganism between cases and healthy women. Days with a statistically significant difference in the mean relative abundance of the microorganism between cases and healthy women are denoted by asterisks. Scales may differ among y-axes. Standardizing these scales would make a subset of the figures difficult to read.
DISCUSSION
To our knowledge, this is the first prospective study with daily vaginal specimen collection to investigate the induction of iBV. Our results indicate that the initial decrease in the mean relative abundance of L. crispatus, a marker of a healthy vaginal microbiota [29, 37], and the subsequent sequential increase in mean relative abundances of P. bivia, G. vaginalis, A. vaginae, and Megasphaera type I (anaerobes commonly found in BV [15]) play important roles. Although other microorganisms have been suggested as potential BV pathogens, there was no significant differences between cases and healthy women regarding the mean relative abundance of S. sanguinegens, F. magna, BVAB1–3, or L. iners leading up to iBV.
In vitro, G. vaginalis is more virulent than other BV-associated bacteria [17]. It is a facultative anaerobe that may tolerate the high oxidation-reduction (redox) potential of a healthy vaginal microbiota, unlike strict anaerobes [13]. Similar to facultative anaerobes involved in the initiation of oral disease [38], it is possible that pathogenic strains of G. vaginalis [39], perhaps at a certain threshold, create a lower redox potential in the vaginal microbiota that is suitable for increases in the abundance of other BV-associated bacteria [13]. Interestingly, in this study, the mean relative abundance of P. bivia became significantly higher among cases 1 day before that of G. vaginalis became significantly higher prior to iBV. Evidence of a commensal, symbiotic relationship between G. vaginalis and P. bivia has been noted [40], which could further stimulate their growth. Ammonia produced by P. bivia stimulates G. vaginalis growth, and amino acids produced by G. vaginalis enhance P. bivia growth [40]. Because of our small sample size and only once daily vaginal swab specimen collection, we cannot pinpoint the exact time that the mean relative abundances of P. bivia and G. vaginalis increased within this 1-day time frame to state with certainty that the abundance of 1 microorganism increased before the other. Nevertheless, our results suggest that synergy between P. bivia and G. vaginalis may be an important event prior to iBV.
Correspondingly, A. vaginae, which is highly specific for BV, rarely occurs in the absence of G. vaginalis, suggesting synergism between these microorganisms [41]. Women with G. vaginalis and A. vaginae have higher rates of recurrent BV than women with G. vaginalis alone [41]. A. vaginae stimulates an innate immune response from vaginal epithelial cells in greater magnitude than G. vaginalis, leading to localized cytokine and β-defensin production [42], suggesting that it is a potent component of the host response to BV. This may contribute to adverse outcomes associated with BV (ie, preterm birth and pelvic inflammatory disease), as increased vaginal inflammatory cytokines and neutrophils are predictive of both [43, 44]. Activation of the nuclear factor кB signaling pathway by A. vaginae may also have a role in increased HIV expression, as this pathway is critical in the transcriptional induction of the HIV-1 promotor [42].
Data on Megasphaera type I are limited. It is highly associated with BV [45], rare or absent in sexually naive women [18], and associated with female-female sex and lifetime number of sex partners [18]. Further in vitro studies on the interactions of P. bivia, G. vaginalis, A. vaginae, and Megasphaera type I are needed.
Of healthy women, 75% had a vaginal microbiota dominated by L. crispatus, while 79% of cases had a vaginal microbiota dominated by L. iners and/or L. jensenii and L. gasseri prior to iBV. This is similar to a study of pregnant women in which L. crispatus promoted the stability of normal vaginal flora whereas L. iners and/or L. gasseri predisposed women to abnormal vaginal flora [37]. Similar to other prospective quantitative polymerase chain reaction–based or vaginal microbiome studies [29, 46], menses and sexual activity in this study led to fluctuations in bacterial community composition over time, at times rapidly. This is in contrast to a study that found that penile-vaginal sex did not alter the consistency of vaginal bacterial communities [39].
Our understanding of shifts in the vaginal microbiota and their influence on women’s health continues to evolve [47], underscoring the need to better understand the pathogenesis of iBV. In a prospective study of HIV-negative women [48], those with diverse vaginal bacterial communities dominated by anaerobes had a >4-fold higher risk of acquiring HIV. Specific bacterial taxa (including Prevotella) were identified that were linked with inflammation and HIV. Additionally, in a tenofovir microbicide study [49], HIV incidence was reduced by 61% in Lactobacillus-dominant women receiving tenofovir, compared with 18% in women with non–Lactobacillus-dominant vaginal bacteria (including G. vaginalis).
Although our data represent a novel way to study the pathogenesis of iBV, there are limitations. The sample size was small, primarily because of a large prevalence of abnormal Nugent scores at baseline, which limited the pool of potential enrollees among African American women who have sex with women. Future studies should include populations with a lower BV prevalence, to increase the proportion of eligible women. Second, survey questionnaire and daily diary data were based on self-report and subject to social-desirability and recall bias. Third, despite using experienced Nugent score readers, Nugent score sensitivity can be as low as 65% [50], and some cases of iBV may have been missed. Fourth, sexually experienced women may not be the ideal population to study iBV pathogenesis, as their baseline vaginal microbiota (even if in a normal Nugent score range) may be colonized by G. vaginalis and other BV-associated bacteria, owing to prior episodes of BV (as seen in our study). Virginal women who become sexually active and develop iBV may be a more ideal study population. Finally, our results are solely based on 16S rRNA sequencing data, which only provide relative abundance estimates. This method may not have detected bacterial species present at very low levels in the vaginal microbiota that could also play a role in iBV. Additional in vitro studies, including animal model research, are needed to better understand the interactions between key vaginal bacteria in the pathogenesis of iBV.
Despite these limitations, our results suggest that G. vaginalis, P. bivia, A. vaginae, and Megasphaera type I may play significant roles in the induction of iBV, although the roles of other organisms cannot be ruled out. Additional prospective studies with larger sample sizes are needed to continue to investigate iBV pathogenesis.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Shaunte Fisher, Laurie Hillsman, Sanquetta McClendon, and Joy Brown Lewis, for assisting with participant recruitment; Hanne Harbison MHS, MSN, CRNP, Saralyn Richter, RN, Rhonda Whidden, RN, Meghan Whitfield, CRNP, Cynthia Poore, CRNP, and Christen Press, CRNP, for assisting with participant screening and enrollment; Charles Rivers, PhD, MSPH, Cheri Aycock, and Keonte Graves, for assisting with laboratory analyses; and the laboratory of Barbara Van Der Pol, PhD, MPH, for performing the BD ProbeTec Qx CTQ/GCQ/TVQ assays.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant K23AI106957 to C. A. M.), the National Institute of General Medical Sciences (grant P20GM103424 to the Louisiana Biomedical Research Network), and the National Center for Advancing Translational Sciences (grant UL1TR001417 to the University of Alabama-Birmingham Center for Clinical and Translational Science).
Potential conflicts of interest. D. H. M. is on the advisory boards of BioFire Diagnostics and GlaxoSmithKline. J. R. S. has been a consultant for and received research support from Hologic/GenProbe, BD Diagnostics, Cepheid, Quidel, Symbiomix, StarPharma, and Toltec. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 5th Annual Louisiana Conference on Computational Biology and Bioinformatics, New Orleans, Louisiana, 7–8 April 2017.
References
- 1. Allsworth JE, Peipert JF. Prevalence of bacterial vaginosis: 2001–2004 National Health and Nutrition Examination Survey data. Obstet Gynecol 2007;109:114–20. [DOI] [PubMed] [Google Scholar]
- 2. Cohen CR, Lingappa JR, Baeten JM et al. . Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med 2012; 9:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hillier SL, Nugent RP, Eschenbach DA et al. . Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med 1995; 333:1737–42. [DOI] [PubMed] [Google Scholar]
- 4. Eschenbach DA. Bacterial vaginosis and anaerobes in obstetric-gynecologic infection. Clin Infect Dis 1993; 16(Suppl 4):S282–7. [DOI] [PubMed] [Google Scholar]
- 5. Muzny CA, Schwebke JR. Gardnerella vaginalis: Still a prime suspect in the pathogenesis of bacterial vaginosis. Curr Infect Dis Rep 2013; 15:130–5. [DOI] [PubMed] [Google Scholar]
- 6. Koumans EH, Sternberg M, Bruce C et al. . The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis 2007; 34:864–9. [DOI] [PubMed] [Google Scholar]
- 7. Evans AL, Scally AJ, Wellard SJ, Wilson JD. Prevalence of bacterial vaginosis in lesbians and heterosexual women in a community setting. Sex Transm Infect 2007; 83:470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olson KM, Boohaker LJ, Schwebke JR et al. . Comparisons of vaginal flora patterns among sexual behaviour groups of women: implications for the pathogenesis of bacterial vaginosis. Sex Health 2018; 15:61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Hanlon DE, Moench TR, Cone RA. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect Dis 2011; 11:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ness RB, Hillier SL, Richter HE et al. . Douching in relation to bacterial vaginosis, lactobacilli, and facultative bacteria in the vagina. Obstet Gynecol 2002; 100:765. [DOI] [PubMed] [Google Scholar]
- 11. Muzny CA, Schwebke JR. Pathogenesis of bacterial vaginosis: discussion of current hypotheses. J Infect Dis 2016; 214(Suppl 1):S1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srinivasan S, Fredricks DN. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip Perspect Infect Dis 2008; 2008:750479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwebke JR, Muzny CA, Josey WE. Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: a conceptual model. J Infect Dis 2014; 210:338–43. [DOI] [PubMed] [Google Scholar]
- 14. Eschenbach DA, Hillier S, Critchlow C, Stevens C, DeRouen T, Holmes KK. Diagnosis and clinical manifestations of bacterial vaginosis. Am J Obstet Gynecol 1988; 158:819–28. [DOI] [PubMed] [Google Scholar]
- 15. Hill GB. The microbiology of bacterial vaginosis. Am J Obstet Gynecol 1993; 169:450–4. [DOI] [PubMed] [Google Scholar]
- 16. Catlin BW. Gardnerella vaginalis: characteristics, clinical considerations, and controversies. Clin Microbiol Rev 1992; 5:213–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patterson JL, Stull-Lane A, Girerd PH, Jefferson KK. Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 2010; 156:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fethers K, Twin J, Fairley CK et al. . Bacterial vaginosis (BV) candidate bacteria: associations with BV and behavioural practices in sexually-experienced and inexperienced women. PLoS One 2012; 7:e30633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med 1983; 74:14–22. [DOI] [PubMed] [Google Scholar]
- 20. Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol 1991; 29:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Der Pol B, Liesenfeld O, Williams JA et al. . Performance of the cobas CT/NG test compared to the Aptima AC2 and Viper CTQ/GCQ assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J Clin Microbiol 2012; 50:2244–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Der Pol B, Williams JA, Taylor SN et al. . Detection of Trichomonas vaginalis DNA by use of self-obtained vaginal swabs with the BD ProbeTec Qx assay on the BD Viper system. J Clin Microbiol 2014; 52:885–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Veer C, Bruisten SM, van der Helm JJ, de Vries HJ, van Houdt R. The cervicovaginal microbiota in women notified for chlamydia trachomatis infection: a case-control study at the sexually transmitted infection outpatient clinic in Amsterdam, The Netherlands. Clin Infect Dis 2017; 64:24–31. [DOI] [PubMed] [Google Scholar]
- 24. Martin DH, Zozaya M, Lillis RA, Myers L, Nsuami MJ, Ferris MJ. Unique vaginal microbiota that includes an unknown Mycoplasma-like organism is associated with Trichomonas vaginalis infection. J Infect Dis 2013; 207:1922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ketterer MR, Rice PA, Gulati S et al. . Desialylation of neisseria gonorrhoeae lipooligosaccharide by cervicovaginal microbiome sialidases: the potential for enhancing infectivity in men. J Infect Dis 2016; 214:1621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shafer WM. Does the cervicovaginal microbiome facilitate transmission of neisseria gonorrhoeae from women to men? implications for understanding transmission of gonorrhea and advancing vaccine development. J Infect Dis 2016; 214:1615–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ravel J, Brotman RM, Gajer P et al. . Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 2013; 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ravel J, Gajer P, Abdo Z et al. . Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011; 108(Suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gajer P, Brotman RM, Bai G et al. . Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012; 4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holm JB, Gajer P, Ravel J. PECAN: A fast, novel 16S rRNA gene sequence non-clustering based taxonomic assignment tool. 16th International Symposium on Microbial Ecology; 2016; Montreal, Canada. [Google Scholar]
- 32. Cole JR, Wang Q, Cardenas E et al. . The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37:D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald D, Price MN, Goodrich J et al. . An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Statist Soc B 2001; 62:411–23. [Google Scholar]
- 35. DiGiulio DB, Callahan BJ, McMurdie PJ et al. . Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA 2015; 112:11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol 2009; 9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruby J, Goldner M. Nature of symbiosis in oral disease. J Dent Res 2007; 86:8–11. [DOI] [PubMed] [Google Scholar]
- 39. Vodstrcil LA, Twin J, Garland SM et al. . The influence of sexual activity on the vaginal microbiota and Gardnerella vaginalis clade diversity in young women. PLoS One 2017; 12:e0171856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis 1997; 175:406–13. [DOI] [PubMed] [Google Scholar]
- 41. Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J Infect Dis 2006; 194:828–36. [DOI] [PubMed] [Google Scholar]
- 42. Libby EK, Pascal KE, Mordechai E, Adelson ME, Trama JP. Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes Infect 2008; 10:439–46. [DOI] [PubMed] [Google Scholar]
- 43. Ramsey PS, Lyon MD, Goepfert AR et al. . Use of vaginal polymorphonuclear to epithelial cell ratios for the prediction of preterm birth. Obstet Gynecol 2005; 105:139–44. [DOI] [PubMed] [Google Scholar]
- 44. Yudin MH, Hillier SL, Wiesenfeld HC, Krohn MA, Amortegui AA, Sweet RL. Vaginal polymorphonuclear leukocytes and bacterial vaginosis as markers for histologic endometritis among women without symptoms of pelvic inflammatory disease. Am J Obstet Gynecol 2003; 188:318–23. [DOI] [PubMed] [Google Scholar]
- 45. Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol 2007; 45:3270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Srinivasan S, Liu C, Mitchell CM et al. . Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One 2010; 5:e10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tuddenham S, Ghanem KG. A microbiome variable in the HIV-prevention equation. Science 2017; 356:907–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gosmann C, Anahtar MN, Handley SA et al. . Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 2017; 46:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klatt NR, Cheu R, Birse K et al. . Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 2017; 356:938–45. [DOI] [PubMed] [Google Scholar]
- 50. Chaijareenont K, Sirimai K, Boriboonhirunsarn D, Kiriwat O. Accuracy of Nugent’s score and each Amsel’s criteria in the diagnosis of bacterial vaginosis. J Med Assoc Thai 2004; 87:1270–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.