Abstract
STUDY QUESTION
Do nuclear proteins in the germinal vesicle (GV) contribute to oocyte competence acquisition during folliculogenesis?
SUMMARY ANSWER
Proteomic analysis of GVs identified candidate proteins for oocyte competence acquisition, including a key RNA processing protein–heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1).
WHAT IS KNOWN ALREADY
The domestic cat GV, which is physiologically similar to the human GV, gains the intrinsic ability to resume meiosis and support early embryo development during the pre-antral-to-antral follicle transition. However, little is known about nuclear proteins that contribute to this developmental process.
STUDY DESIGN SIZE, DURATION
GVs were enriched from pre-antral (incompetent) and antral (competent) follicles from 802 cat ovaries. Protein lysates were subjected to quantitative proteomic analysis to identify differentially expressed proteins in GVs from the two follicular categories.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Two biological replicates (from independent pools of ovaries) of pre-antral versus antral samples were labeled by tandem mass tags and then assessed by liquid chromatography–tandem mass spectrometry. Proteomic data were analyzed according to gene ontology and a protein–protein interaction network. Immunofluorescent staining and protein inhibition assays were used for validation.
MAIN RESULTS AND THE ROLE OF CHANCE
A total of 174 nuclear proteins was identified, with 54 being up-regulated and 22 down-regulated (≥1.5-fold) after antrum formation. Functional protein analysis through gene ontology over-representation tests revealed that changes in molecular network within the GVs during this transitional phase were related to chromatin reorganization, gene transcription, and maternal RNA processing and storage. Protein inhibition assays verified that hnRNPA2B1, a key nuclear protein identified, was required for oocyte meiotic maturation and subsequent blastocyst formation.
LARGE SCALE DATA
Data are available via ProteomeXchange with identifier PXD007211.
LIMITATIONS REASONS FOR CAUTION
Proteins identified by proteomic comparison may (i) be involved in processes other than competence acquisition during the pre-antral-to-antral transition or (ii) be co-expressed in other macrostructures besides the GV. Expressional and functional validations should be performed for candidate proteins before downstream application.
WIDER IMPLICATIONS OF THE FINDINGS
Collective results generated a blueprint to better understand the molecular mechanisms involved in GV competence acquisition and identified potential nuclear competence markers for human fertility preservation.
STUDY FUNDING AND COMPETING INTEREST(S)
Funded by the National Center for Research Resources (R01 RR026064), a component of the National Institutes of Health (NIH) and currently by the Office of Research Infrastructure Programs/Office of the Director (R01 OD010948). The authors declare that there is no conflict of interest.
Keywords: proteomics, oocyte, carnivore reproduction, germinal vesicle, oocyte competence
Introduction
Intraovarian oocytes at the germinal vesicle (GV) stage are arrested at first meiotic prophase. At each ovarian cycle, one or more of these oocytes become activated to grow in size and acquire the ability to mature (i.e. resume meiosis) and support future embryo development, what is often collectively referred to as ‘competency’ (Downs, 2015; Nunes et al., 2015). Most immature oocytes within each ovary never contribute to sexual reproduction, making these cells attractive targets for fertility preservation in human, livestock and endangered species (Combelles and Chateau, 2012; Comizzoli and Holt, 2014; Kikuchi et al., 2016). Compared to the whole oocyte, the GV may well be a more resilient target to minimize potential cryoinjuries and improve preservation outcomes (Comizzoli et al., 2008; Clark and Swain, 2013; Jo et al., 2012). However, there is not yet a full understanding of how the immature oocyte and its GV attain competency. Analyzing the molecular composition that renders competent oocytes will help elucidate these complex developmental processes as well as lay a foundation for advancing reproductive technologies that could exploit premature follicles, oocytes and resident GVs.
It is known that there are significant cellular and molecular changes occurring both in the GV and ooplasm during competence acquisition. Among changes observed are maternal materials production and storage for future utilization (Zuccotti et al., 2011), configurational changes in GV chromatin and termination of transcriptional activity in the fully grown oocyte (De La Fuente et al., 2004; Tan et al., 2009) and multiple epigenetic regulations (Kobayashi et al., 2012; Tang et al., 2015; Wu et al., 2015). Our laboratory studies the domestic cat (Felis catus) as a comparative model and have determined that oocyte competence is linked to transition of the GV chromatin from a filamentous to a reticular configuration (Comizzoli et al., 2011), increased histone methylation (Phillips et al., 2012), transcriptional silencing (Comizzoli et al., 2011) and protein translocation-mediated transcriptional regulation (Lee et al., 2015). Particularly noteworthy are the findings from GV transfer experiments whereby a GV from an oocyte of earlier follicular stage is transferred into the cytoplast of a fully developed oocyte. Reconstructed oocytes containing a GV from early, small or large antral follicles exhibits superior competence meiotically and developmentally compared to those from a pre-antral counterpart (Comizzoli et al., 2011). Therefore, it is clear that the GV within the reconstructed oocyte possesses intrinsic, yet undetermined, competence factors that are independent from the cytosolic composition. Moreover, these investigations have indicated that the GV achieves its full capacity during the pre-antral to antral follicular transition (Comizzoli et al., 2011). Taken together, we now can predict that GV competence factors are (i) more highly expressed in antral compared to pre-antral follicular stages and (ii) likely involved in processes that include RNA production, chromatin organization, epigenetic modifications and/or transcriptional regulation.
To date, most global scale analyses for identifying oocyte-specific competence factors have focused on mRNAs (Labrecque and Sirard, 2014). Although some transcripts have been considered as potential molecular markers in specific species (Assidi et al., 2008; Huang et al., 2013; Ashry et al., 2015; Bhardwaj et al., 2016; Kussano et al., 2016), mechanisms for how competence is established remain unclear. A key issue has been the weak correlation between mRNA and protein expression in oocytes due to accumulated stock of non-translated, maternal RNAs (Flemr et al., 2010; Schwarzer et al., 2014; Zuccotti et al., 2011). Therefore, resulting transcriptomic data may fail to reflect biological events at the time of oocyte collection. Proteomic profiles have the potential of being more precise. However, identifying protein markers indicative of oocyte quality is challenging due to the massive number of cells required for traditional techniques, such as 2D electrophoresis, that also generally yield low resolution and imprecise quantification (Virant-Klun and Krijgsveld, 2014). Although GV oocyte proteomic profiles have been generated in the mouse (Vitale et al., 2007; Wang et al., 2010; Pfeiffer et al., 2011; Cao et al., 2012; Monti et al., 2013; Schwarzer et al., 2014), only two studies so far have provided parallel information on oocyte quality based on chromatin configuration (Monti et al., 2013) or age (Schwarzer et al., 2014). Technological advancements that involve isobaric labeling strategies have circumvented some of these limitations by requiring less raw material and allowing simultaneous protein identification and relative quantification. Taking advantage of these tools, recent proteomic studies on oocyte competence have been reported. Chen et al. (2016) used iTRAQ to compare the proteome of GV oocytes, incompetent metaphase II (MII) or competent MII oocytes in the buffalo. Likewise, Powell et al. (2010) utilized ExacTag to examine the oocyte proteome and secretome from high versus low-quality pig oocytes. Although several GV transfer experiments have demonstrated that factors within both the ooplasm and GV contribute to oocyte competence (Inoue et al., 2008; Comizzoli et al., 2011), most markers identified so far have been cytosolic or membrane-derived (Monti et al., 2013; Virant-Klun and Krijgsveld, 2014). The nuclear proteome of the oocyte has been characterized only in Xenopus laevis (Wuhr et al., 2015), and as yet for no mammal.
The present GV focused study was designed to improve our understanding of nuclear factors that contribute to an oocyte’s ability to become competent. Our specific objective was to explore differentially expressed GV proteins from pre-antral (incompetent) versus antral (competent) follicles using isobaric labeling, in this case tandem mass tagging (TMT), which was combined with liquid chromatography–tandem mass spectrometry (LC-MS/MS). Our prediction was that proteins identified from the proteomic analysis would provide insights into molecular mechanisms involved in GV competence acquisition as well as markers for future GV preservation research.
Materials and Methods
Ovarian follicle collection and GV enrichment
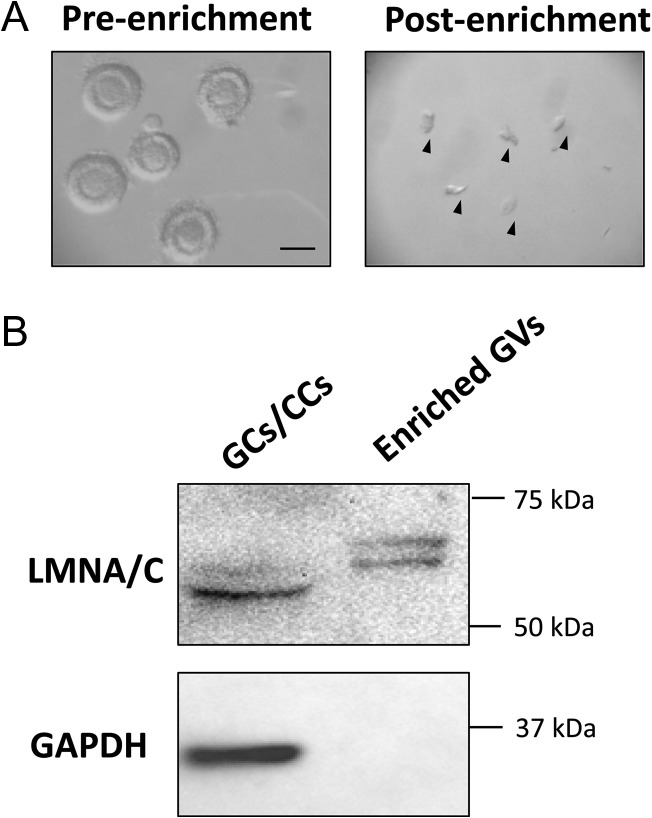
Ovaries from adult domestic cats were recovered after routine ovariohysterectomy at local veterinary clinics and transported (at 4°C) to the laboratory within 6 h of excision. Follicles and cumulus cell–oocyte complexes (COCs) were mechanically isolated into HEPES (Sigma-Aldrich, USA)-buffered minimum essential medium (MEM; Life Technologies, USA) supplemented with 2 mM l-glutamine, 1 mM pyruvate (Sigma-Aldrich, USA), 100 IU/ml penicillin (Sigma-Aldrich, USA), 100 μg/ml streptomycin (Sigma-Aldrich, USA) and 4 mg/ml bovine serum albumin (BSA; Sigma-Aldrich, USA). Follicles with diameters ranging from 100 to 400 μm were pooled into the pre-antral group, whereas counterparts containing a fluid-filled antrum (generally >400 μm) were assigned to the antral group. To enrich a large number of GVs simultaneously, pooled samples were disrupted by a sonic dismembrator (Fisher Scientific, USA) at 20% amplitude with short bursts until no or only a few follicles were visible. Pre-antral and antral groups were then applied onto 50 and 70 μm pre-separation filters (Sysmec-Partec, Germany, and Miltenyi Biotec, Germany), respectively, to remove un-disrupted follicles and oocytes. Filters were selected based on the known diameter of oocytes (from pre-antral follicles, 63–74 μm versus antral, 94–116 μm) and GVs (from pre-antral follicles, 17–25 μm versus antral, 31–41 μm) (Comizzoli et al., 2011). After being washed with phosphate-buffered saline (PBS), filtrates were pelleted at 300 g and the supernatant decanted. Isolated materials were examined immediately under a stereomicroscope (Nikon SMZ1000) to ensure isolation of GVs without retaining any intact oocytes in the final pellet (Fig. 1A). Pellets were snap-frozen in liquid nitrogen and stored at −80°C until protein extraction. Granulosa and cumulus cells (GCs/CCs) from the remaining tissue were also snap-frozen to serve as control samples for western blot analysis.
Figure 1.
(A) Efficacy of germinal vesicle (GV) enrichment observed by light microscopy. Representative pre-enriched pre-antral follicles (left panel) and post-enriched GVs (right panel; arrowheads) were shown. Bar = 200 μm. (B) Verification of nuclei isolation by western blots using LMNA/C as the nuclear marker and GAPDH as the cytosolic marker. Granulosa and cumulus cells (GCs/CCs) served as controls.
Western blotting
To examine the efficacy of nuclear enrichment, western blots were performed using both nuclear (Lamin A + C; LMNA/C) and cytosolic (glyceraldehyde 3-phosphate dehydrogenase; GAPDH) markers. Somatic cell (GCs/CCs) extract was used as a positive control. A small portion of the collected GVs and somatic cells was lysed with Laemmli sample buffer (Bio-Rad, USA) and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred onto polyvinylidene difluoride membranes (Bio-Rad, USA), blocked with 3% BSA in PBS and then incubated at 4°C overnight with LMNA/C antibody (1:200 dilution; Abcam, USA). Anti-mouse IgG antibody conjugated with horseradish peroxidase (HRP; Invitrogen, USA) was used as the secondary antibody. Immunoreactivity was detected with Clarity Western ECL Substrate (Bio-Rad, USA) and imaged with ChemiDoc XRS imaging system (Bio-Rad, USA). The membrane was then stripped and re-blotted with primary antibody against GAPDH (1:500 dilution; Novus, USA) and anti-rabbit IgG-HRP secondary antibody (Santa Cruz Biotechnologies, USA).
Protein extraction
GVs collected from the same cohort of ovaries were considered as one biological replicate, each consisting of one pre-antral and one antral sample. Two replicates were collected for proteomic assessment, resulting in a total of four samples (two pre-antral and two antral groups). Each GV sample was lysed with 2D protein extraction buffer-V (GE Healthcare, USA) in the presence of proteinase inhibitor mix (GE Healthcare, USA). GV lysate then was purified with a 2D clean-up kit (GE Healthcare, USA) following manufacturer’s instructions. In brief, proteins were precipitated by a trichloroacetic acid-containing precipitant, washed with an acetone-based buffer, and resuspended in an appropriate volume of extraction buffer.
Proteomics
The following protein identification procedures were performed by ITSIBiosciences, USA.
Protein sample preparation and TMT labeling
Total protein was precipitated from the four samples using a ToPrep kit, the pellets reconstituted in ToPI buffer and protein concentration determined with a ToPA Bradford protein assay kit (both kits and buffer, ITSIBiosciences, USA). To determine if residual BSA from MEM medium interfered with protein identification, a test run was conducted using a single sample containing the highest protein concentration. Briefly, the test aliquot was split into two equal parts, with only one undergoing an additional BSA removal step with an ASKc albumin segregation kit (ITSIBiosciences, USA). After LC-MS/MS analysis and database search, this testing revealed that BSA removal was not required. Therefore, subsequent analyses were carried out without BSA removal. Equal amounts (25 μg) of the four samples were reduced, alkylated, and trypsin-digested overnight and then individually labeled with four unique TMT reagents (Thermo Scientific, USA) as follows: pre-antral group, replicate 1, TMT-126; antral group, replicate 1, TMT-127; pre-antral group, replicate 2, TMT-128; antral group, replicate 2, TMT-130.
Multidimensional protein identification technology
All labeled samples were combined prior to multidimensional protein identification technology (MudPIT) analysis (Lohrig and Wolters, 2009), with the pooled sample separated by strong cation exchange chromatography into three fractions eluted at 50, 250 and 450 mM ammonium acetate, followed by ZipTip cleanup to remove salts. Each sample was loaded onto a PicoFrit C18 nanospray column (New Objective, USA) using a Surveyor Autosampler (Thermo Scientific, USA) operated in the no-waste injection mode. A linear acetonitrile gradient from 5 to 45% acetonitrile separated the tryptic peptides based on hydrophobicity over 230 min. Peptides then were subjected to an LTQ XL mass spectrometer (Thermo Scientific, USA) for sequencing via a nanospray source with the spray voltage set to 1.8 kV and the ion transfer capillary at 180°C. A data-dependent Top 5 method was used in which a full MS scan from m/z 300 to 1600 was followed by MS/MS scans on the five most abundant ions.
Protein identification and quantitation
Raw data files were searched against the database for Felis catus from Uniprot, using the Proteome Discoverer 1.4 Sequest HT search algorithm (Thermo Scientific, USA) and the MudPIT database search protocol. Search parameters included a maximum of two missed trypsin cleavages per peptide, cysteine carbamidomethylation, N-terminal TMT 6-plex and lysine TMT 6-plex as static modifications, and methionine oxidation and threonine TMT 6-plex as variable modifications. Proteins were considered as identified with high confidence peptides when one or more unique peptides had a Xcorr score >1.5, >2.0 and >2.5 for respective charge states of +1, +2 and +3, respectively. For statistical assessment of the confidence of peptide assignments, the Target Decoy Peptide-Spectrum Match (PSM) Validator algorithm was applied in database searches. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD007211. Quantitative changes in protein expression were detected between pre-antral and antral protein samples in each replicate. Proteins with a TMT ratio (TMT-127/TMT-126 and TMT-130/TMT-128) ≥1.50 and ≤0.66 were considered up- and down-regulated, respectively, during the pre-antral-to-antral transition. Keratin and albumin proteins were excluded from the data for further analysis as these proteins likely resulted from contaminations during surgery or sample handling.
Protein functional analysis
The Protein ANalysis THrough Evolutionary Relationships classification system (PANTHER, http://pantherdb.org/) and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, http://string-db.org/) were online bioinformatics tools used for functional interpretation of the proteomic data. Gene ontology (GO) analysis was performed with PANTHER v10.0 against the Felis catus database. Firstly, the GO_cellular component term ‘nucleus’ (GO:0 005 634) was used as an additional filter to identify nuclear proteins. Differentially expressed nuclear proteins then were subjected to statistical over-representation tests of both the GO_biological process and GO_molecular function terms using default parameters. The Bonferroni correction was used for multiple testing. Proteins associated with ontology terms were considered enriched when P < 0.05. The functional association network of proteins was constructed with STRING v10 against the Felis catus database with the minimum required interaction score set as 0.4.
Immunofluorescent staining and imaging
To validate expression of identified proteins in the GV, immunofluorescent staining was performed as previously described (Lee et al., 2015) using antibodies against heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) at 1:150 (Sigma-Aldrich, USA) or high-mobility group nucleosome binding domain 1 (HMGN1) at 1:500 (Cell Signaling, USA). Oocytes from antral follicles were stripped of cumulus cells and processed together with pre-antral counterparts for immunostaining. Oocytes fixed in 4% paraformaldehyde (PFA) in PBS (USB Corporation, USA) were rinsed with wash solution (PBS with 2% fetal calf serum [FCS; Irvine Scientific, USA] and 0.5% Triton X-100 [Sigma-Aldrich, USA]) and blocked with 20% FCS and 0.5% Triton X-100 in PBS for 30 min at 38°C. Oocytes were then incubated with primary antibodies in blocking solution overnight at 4°C. A negative control in which the primary antibodies were omitted was included in each trial. Non-immune IgG from the mouse and rabbit were used at the same concentration as their counterparts to serve as additional negative controls. After washing off primary antibodies, oocytes were incubated with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (anti-mouse IgG-FITC at 1:200 [Sigma-Aldrich, USA] or anti-rabbit IgG-FITC at 1:200 [Sigma-Aldrich, USA]) for 1 h at 38°C. At the end of the procedure, oocytes were washed and mounted with Vectashield mounting medium containing DAPI (Vector Laboratories, USA) to visualize DNA. Resulting images were obtained using a Zeiss LSM 710 confocal microscope (Imaging Core Facility, University of Maryland, USA) with Zen 2009 software (Zeiss, Germany). The excitation and detection wavelengths were 488 nm and 492–544 nm for FITC, respectively, and 405 nm and 415–487 nm for DAPI. Z-stack images were taken from the top to the bottom of the nucleus at a constant interval of 0.9 μm. Oocytes from both pre-antral, and antral follicles from the same batch of ovaries were processed together and images were taken under the same laser power configuration to allow stage comparisons. Total fluorescent intensity (arbitrary unit) within a GV was quantified using ImageJ software (National Institutes of Health, USA) with the Bio-Formats plugin (The Open Microscopy Environment). Background fluorescent signal from the cytoplasm was subtracted before calculating total intensity. Fluorescent intensity of candidate proteins was normalized with DAPI intensity in each GV.
Protein inhibition assay
A protein inhibition assay was utilized to evaluate the role of hnRNPA2B1 and HMGN1 in oocyte competence acquisition. An antibody to phosphodiesterase 3A (PDE3A; Millipore, USA), a well-documented oocyte protein essential for meiotic maturation (Richard et al., 2001; Jensen et al., 2005; Li et al., 2012), was used as a positive control to determine technical proficiency of the assay. Chariot (Active Motif, USA), a peptide nanoparticle-mediated transfection reagent, has been proven effective for introducing antibodies into the mouse oocyte through the zona pellucida without side-effects (Li et al., 2016). COCs were collected from adult cat ovaries. To prepare the Chariot–antibody complex, 2 μl of Chariot in 50 μl of H2O was mixed with 1.5 μg of antibody in 50 μl of PBS (or without antibody for mock transfection) at room temperature for 30 min. The complexes then were added to the COCs in equal amount of culture medium (MEM [Sigma-Aldrich, USA] supplemented with 1 mM pyruvate, 2 mM l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin and 4 mg/ml BSA). COCs were incubated at 38°C with 5% CO2 for 3 h before adding another 200 μl of culture medium and further incubation for a total of 24 h. To prevent meiosis resumption, 250 nM of milrinone (Sigma-Aldrich, USA) was added to the medium (except for the fresh control group) during the course of transfection (Grupen et al., 2006).
IVM, IVF and embryo culture
After 24 h of transfection, excess antibodies were rinsed off with SAGE blastocyst medium (SAGE, Denmark) before incubating COCs in 50 μl microdrops (5–15 COCs per drop) of IVM medium consisting of 1 μg/ml equine FSH (Bioniche Animal Health, USA) and 1 μg/ml ovine LH (National Hormone and Pituitary Program, USA) in SAGE blastocyst medium. After a 24 h maturation culture (38°C in 5% CO2), COCs were inseminated with 1 × 106/ml fresh or frozen–thawed domestic cat epididymal spermatozoa. At 24 h post-insemination, oocytes were denuded and cleaned by gentle pipetting. Presumptive zygotes were cultured (38°C in 5% CO2) in SAGE blastocyst medium in 50 μl microdrops (5–15 embryos per drop) for up to 7 days before fixation with 4% PFA in PBS and staining with DAPI. Oocyte stages were determined by assessing chromosomal configuration and alignment as well as the presence of polar bodies. Embryo stages were determined by the number of blastomeric nuclei. An embryo with 25–63 blastomeres without a blastocoele was classified as a morula, whereas one with a visible blastocoele and at least 64 blastomeres was considered a blastocyst (Comizzoli et al., 2011).
cAMP ELISA
PDE3A induces meiotic resumption by catalyzing hydrolysis of cAMP (Conti et al., 2002; Thomas et al., 2002). Therefore, cAMP levels were measured to validate effective antibody inhibition of PDE3A. A direct cAMP ELISA kit (ENZO Life Sciences, USA) was used following manufacturer’s instructions for acetylated assay. COCs were lysed in 0.1 M HCl for 10 min followed by centrifugation (15 000 g, 15 min, 4°C) and supernatant frozen (−80°C) until thawed for assay. All standards, controls and samples were analyzed in technical duplicates. Optical density at 405 nm was measured with a BioTek ELx808 microplate reader (BioTek Instruments, USA).
Experimental design and statistical analysis
Experiment 1: Proteomic analysis of nuclear proteins differentially expressed in GVs from pre-antral versus antral follicles
Two biological replicates of GV samples, each prepared from independent pools of cat ovaries, were evaluated to provide a confident selection of identified proteins. The first replicate contained GVs isolated from 3059 pre-antral and 2402 antral follicles (525 ovaries), with the other including 1012 and 1091 GVs from pre-antral and antral follicles (277 ovaries), respectively. Nuclei enrichment was verified by western blot analysis with non-detectable expression of GAPDH in the protein extract from the purified GV sample (Fig. 1B). The lamin A/C antibody used (recognizing both lamin A and C isoforms) provided additional evidence to confirm purity of the GV suspension. Specifically, two isoforms were detected both in the control (somatic cells) and the enriched GVs samples (Fig. 1B); however, the molecular weights of the double-bands differed ~5–10 kDa between the two groups (likely due to different post-translational modifications). This finding suggested that the control and the enriched samples consisted of different cell populations. The absence of the detected lower-weight isoforms in the control indicated that there was no detectable somatic cell contamination in the enriched GVs. As described above, protein extract of each sample was TMT-labeled and subjected to MudPIT simultaneously to allow protein identification and relative quantification. To evaluate the quantification reproducibility, we performed correlation analysis of proteins in two replicates. Among all resulting nuclear proteins, ones that were up-regulated (i.e. antral > pre-antral) or down-regulated (i.e. antral < pre-antral) over 1.5-fold in both sets were further analyzed by (i) over-representative tests of association with GO terms in biological process and molecular function databases (using PANTHER) and (ii) the functional association network (via STRING).
Experiment 2: Functional validation of selected candidate proteins identified in the proteomic analysis
Based on the proteomic analysis and a literature review of differentially expressed nuclear proteins, hnRNPA2B1 and HMGN1 were selected for expressional and functional validation. Specificity of antibodies against these target proteins in the cat ovary was confirmed by western blot. Using these antibodies, the up-regulation of each candidate protein was confirmed by immunofluorescent staining as described above. Spatiotemporal expression was examined (n = 118 oocytes in 2 replicates) from both pre-antral and antral follicles collected from the same batch of ovaries. Z-stack images were captured and total fluorescent intensity within the GV was measured and normalized for each oocyte as described above.
Prior to functional validation, the effectiveness of protein inhibition via Chariot transfection in the cat oocytes was investigated using PDE3A antibody as an example. COCs were mock transfected, transfected with PDE3A antibody or mock transfected in the presence of the PDE3 inhibitor milrinone (Thomas et al., 2002) in groups of 10 (n = 30 COCs per treatment in three replicates) and then examined for cAMP levels 24 h later. Once the method was verified, protein inhibition assays were used to establish the association of the selected proteins with oocyte competence acquisition. A total of 245 COCs (in 3–8 replicates) was subjected to one of the following treatments: (i) no treatment (fresh control); (ii) no transfection (overnight culture with neither Chariot nor antibodies); (iii) mock transfection (Chariot added without antibodies); (iv) transfection of PDE3A antibody (positive control); (v) transfection of hnRNPA2B1 antibody; or (vi) transfection of HMGN1 antibody. Then using IVM, IVF and/or 7 days long embryo culture, oocytes in each group were assessed and compared for maturation success (reaching MII stage) and developmental competence (embryo cleavage, morulae and blastocyst formation).
Statistical analyses
Correlation of the quantitative proteomic results from the two replicates was determined by Pearson correlation coefficient. Distribution of relative fluorescent quantification data was examined by a Shapiro–Wilk normality test. Because results revealed that data were not normally distributed, pairwise comparison between stages was evaluated using a Mann–Whitney test. ELISA data were analyzed by ANOVA followed by Tukey’s multiple test. Proportional data from protein inhibition assays were analyzed by Chi-square evaluations. Blastomere numbers in formed blastocysts per group were analyzed using a Kruskal–Wallis test. The PDE3A antibody-treated group was excluded from analyses because only one blastocyst formed. For all comparisons, differences were considered significant at P < 0.05 (Prism v6.05; GraphPad Software, USA).
Results
Experiment 1.1: Nuclear proteins differentially expressed in GVs from pre-antral versus antral follicles
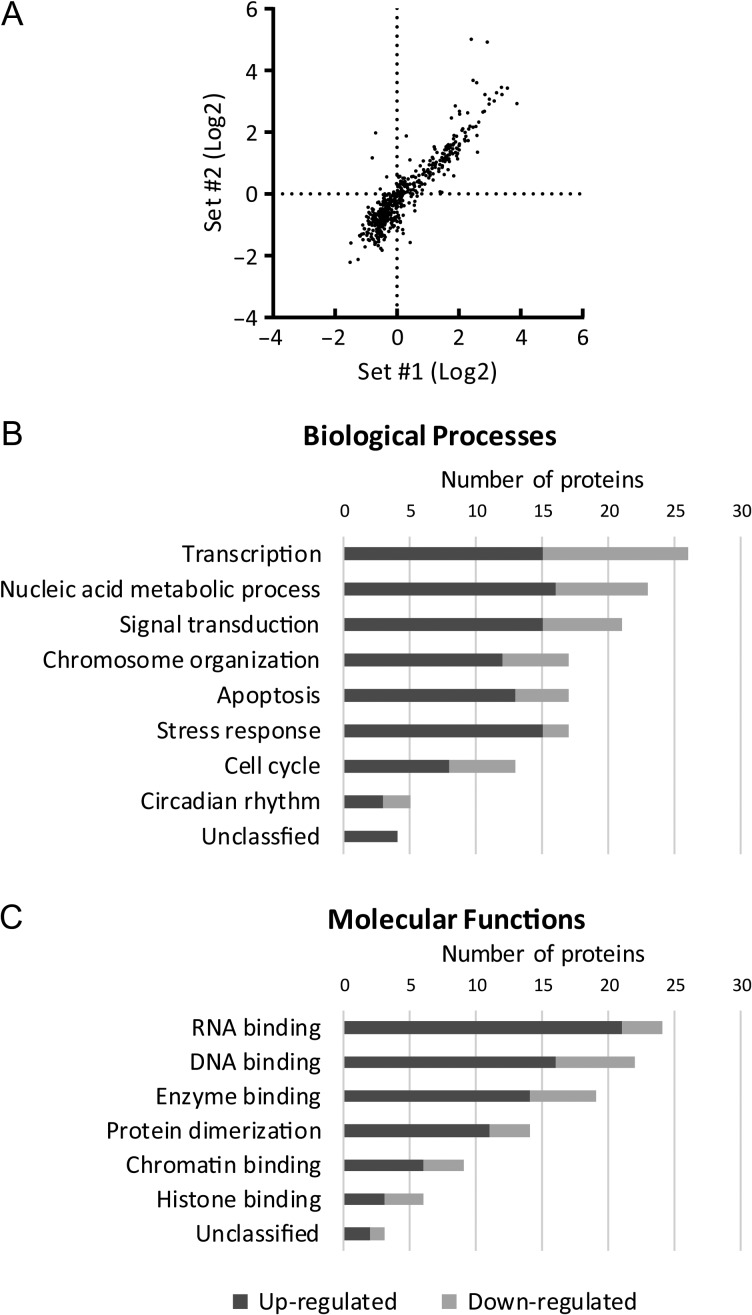
A total of 913 domestic cat proteins was unequivocally identified with high confidence peptides; of these, 501 had TMT tags in all four samples that permitted quantitative comparison. There was a high correlation (r = 0.92, R2 = 0.84) (Fig. 2A) between the quantification results from the two replicates of pre-antral versus antral samples. GO analysis assigned cellular component terms to 489 of the feline proteins. Specifically, the term ‘nucleus’ was associated with 174 proteins (36%). Additionally, 21, 13 and 12% of the proteins were assigned to GO terms ‘extracellular exosome’, ‘cytoskeleton’ and ‘mitochondrion’, respectively, whereas 16% were unclassified. To focus solely on nuclear proteins, we applied the GO term ‘nucleus’ as an exclusive bioinformatics filter. Among the 174 nuclear proteins identified, 76 were differentially expressed (≥1.5-fold) between pre-antral and antral groups in both replicates; 54 proteins were up-regulated and 22 down-regulated at the antral compared to pre-antral stage (Tables I and II). Up-regulated proteins were expressed 1.5- to 32.1-fold more in the antral groups. Among the most predominate were HMGN1, bromodomain adjacent to zinc finger domain 2B (BAZ2B), calreticulin (CALR), protein disulfide-isomerase A3 (PDIA3), epithelial cell transforming 2 (ECT2) and cyclin-dependent kinase inhibitor 2B (CDKN2B) (Table I). Down-regulated proteins produced more modest fold changes between the two follicular stages, from 1.5- to 2.5-fold, including polyhomeotic homolog 1B (PHC1), IKAROS family zinc finger 2 (IKZF2), optic atrophy 1 (OPA1) and breast cancer 1 early onset (BRCA1) (Table II).
Figure 2.
(A) Correlation of quantification data (ratio of antral over pre-antral groups) from the two replicates. r = 0.92, R2 = 0.84. Over-represented Gene Ontology categories from the (B) ‘biological process’ and (C) ‘molecular function’ databases using PANTHER against the Felis catus database.
Table I.
Proteins up-regulated in antral versus pre-antral GVs grouped by main function (n = 54 total proteins expressing at least a 1.5-fold difference in both sets of proteomic quantification). Selected candidate proteins for further validation are marked in bold.
| Gene symbol | Gene name | Fold difference | |
|---|---|---|---|
| Set #1 | Set #2 | ||
| Chromosome organization | |||
| HMGN1 | High-mobility group nucleosome binding domain 1 | 9.4 | 9.7 |
| HIST1H2BD | Histone cluster 1, H2bd | 3.9 | 2.8 |
| NPM1 | Nucleophosmin 1 | 3.8 | 2.4 |
| BAZ2B | Bromodomain adjacent to zinc finger domain, 2B | 3.7 | 7.2 |
| HIST1H2BB | Histone cluster 1, H2bb | 3.5 | 2.5 |
| SMC3 | Structural maintenance of chromosomes 3 | 3.1 | 2.8 |
| Histone H2A | 3.1 | 2.3 | |
| Histone H3 | 2.9 | 2.2 | |
| BANF1 | Barrier to autointegration factor 1 | 2.9 | 1.5 |
| H2AFV | H2A histone family, member V | 2.4 | 1.8 |
| Transcription/signal transduction | |||
| CALR | Calreticulin | 8.8 | 8.1 |
| PDIA3 | Protein disulfide-isomerase A3 | 6.9 | 6.3 |
| ECT2 | Epithelial cell transforming 2 | 4.1 | 6.4 |
| MED1 | Mediator complex subunit 1 | 3.9 | 2.5 |
| TOPORS | Topoisomerase I binding, arginine/serine-rich | 3.6 | 3.1 |
| C1QBP | Complement component 1 Q subcomponent-binding protein | 3.5 | 3.5 |
| PHB | Prohibitin | 3.1 | 2.2 |
| TCF7L2 | Transcription factor 7-like 2 | 1.9 | 2.1 |
| PPP1CA | Protein phosphatase 1 catalytic subunit, alpha isoform | 1.7 | 1.9 |
| RNA processing | |||
| RBMX | Similar to RNA binding motif protein, X-linked | 6.0 | 3.7 |
| SFPQ | Splicing factor proline/glutamine-rich | 4.5 | 3.4 |
| BCAS2 | Breast carcinoma amplified sequence 2 | 3.5 | 2.8 |
| RALYL | RALY RNA binding protein-like | 3.4 | 2.6 |
| hnRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 3.0 | 2.3 |
| SART3 | Squamous cell carcinoma antigen recognized by T cells 3 | 2.9 | 2.7 |
| hnRNPF | Heterogeneous nuclear ribonucleoprotein F | 2.6 | 1.9 |
| SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 | 2.5 | 1.7 |
| hnRNPU | Heterogeneous nuclear ribonucleoprotein U | 2.4 | 2.0 |
| IARS | Isoleucine-tRNA ligase | 2.3 | 1.6 |
| hnRNPD | Heterogeneous nuclear ribonucleoprotein D | 2.0 | 1.7 |
| Metabolism/stress response | |||
| HSPA5 | Heat shock 70 kDa protein 5 | 5.5 | 4.5 |
| HSP90B1 | Endoplasmin | 4.6 | 4.0 |
| ATP5A1 | ATP synthase subunit alpha | 4.6 | 3.3 |
| HADH | Hydroxyacyl-CoA dehydrogenase | 3.3 | 2.6 |
| SCP2 | Sterol Carrier Protein 2 | 3.2 | 3.0 |
| HSPB1 | Heat shock 27 kDa protein 1 | 3.2 | 2.2 |
| PARP9 | Poly (ADP-ribose) polymerase family, member 9 | 2.7 | 2.3 |
| DLST | Dihydrolipoamide S-succinyltransferase | 2.5 | 2.5 |
| HMOX1 | Heme oxygenase 1 | 2.3 | 1.6 |
| PRDX1 | Peroxiredoxin 1 | 2.2 | 2.3 |
| PKM | Pyruvate kinase | 2.2 | 2.2 |
| MDH2 | Malate dehydrogenase 2 | 2.2 | 1.8 |
| Others | |||
| RPS23 | Ribosomal protein S23 | 10.5 | 9.3 |
| CTSB | Cathepsin B | 7.8 | 7.5 |
| ASPSCR1 | Alveolar soft part sarcoma chromosome region, candidate 1 | 6.0 | 12.1 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B | 5.3 | 32.1 |
| SLC25A6 | Solute carrier family 25 member 6 | 4.0 | 3.0 |
| RPS19 | Ribosomal protein S19 | 4.0 | 2.7 |
| ASRGL1 | Asparaginase like 1 | 3.8 | 3.7 |
| LRRC41 | Leucine rich repeat containing 41 | 3.8 | 2.8 |
| LMNB2 | Lamin B2 | 3.7 | 2.5 |
| EEF1A1 | Elongation factor 1-alpha 1 | 3.3 | 2.9 |
| LMNA | Lamin A | 2.6 | 1.8 |
| TMEM201 | Transmembrane protein 201 | 2.3 | 1.9 |
Table II.
Proteins down-regulated in antral versus pre-antral GVs grouped by main function (n = 22 total proteins expressing at least a 1.5-fold difference in both sets of proteomic quantification).
| Gene symbol | Gene name | Fold difference | |
|---|---|---|---|
| Set #1 | Set #2 | ||
| Chromosome organization | |||
| PHC1 | Polyhomeotic homolog 1B | 2.0 | 2.5 |
| HDAC1 | Histone deacetylase 1 | 1.8 | 1.6 |
| CHD8 | Chromodomain helicase DNA binding protein 8 | 1.6 | 1.6 |
| Transcription/signal transduction | |||
| PTK6 | Protein tyrosine kinase 6 | 1.8 | 1.6 |
| IKZF2 | IKAROS family zinc finger 2 | 1.7 | 2.0 |
| PTPN6 | Tyrosine-protein phosphatase non-receptor type | 1.7 | 1.9 |
| BRD2 | Bromodomain containing 2 | 1.6 | 1.9 |
| USP2 | Ubiquitin carboxyl-terminal hydrolase | 1.6 | 1.8 |
| UHRF2 | Ubiquitin-like with PHD and ring finger domains 2 | 1.5 | 2.2 |
| BHLHE40 | Basic helix-loop-helix family, member e40 | 1.5 | 1.6 |
| RNA processing | |||
| ELAC2 | ElaC homolog 2 | 2.1 | 2.2 |
| KIAA1429 | KIAA1429 | 1.5 | 1.8 |
| Metabolism/stress response | |||
| OPA1 | Optic atrophy 1 | 2.3 | 2.5 |
| BRCA1 | Breast cancer 1, early onset | 1.8 | 2.6 |
| HSPH1 | Heat shock protein family H member 1 | 1.6 | 1.6 |
| Others | |||
| SACS | Sacsin | 2.1 | 2.4 |
| KATNA1 | Katanin p60 ATPase-containing subunit A1 | 1.9 | 2.0 |
| CEP290 | Centrosomal protein 290 kDa | 1.8 | 1.9 |
| CERS3 | Ceramide synthase 3 | 1.8 | 1.6 |
| NAMPT | Nicotinamide phosphoribosyltransferase | 1.7 | 1.8 |
| DPY19L4 | Dpy-19-like protein 4 | 1.7 | 1.6 |
| CCNE1 | Cyclin E1 | 1.5 | 2.0 |
Experiment 1.2: Gene ontology analysis of proteins differentially expressed in GVs from pre-antral versus antral follicles
Of the 76 differentially expressed proteins, 72 were associated with at least one GO term in the biological process database. Overall, 36% of proteins were related to transcription (15 up-regulated, 11 down-regulated), 32% to nucleic acid metabolic process (16 up-regulated, 7 down-regulated) and 29% to signal transduction (15 up-regulated, 6 down-regulated). Other significant categories included chromosome organization, apoptosis and stress response, each consisting of 17 proteins (24%) (Fig. 2B).
Overall, 73 proteins were assigned at least one GO annotation in the molecular function database. The most prevalent were 33% related to RNA binding (21 up-regulated, 3 down-regulated), 30% to DNA binding (16 up-regulated, 6 down-regulated) and 26% to enzyme binding (14 up-regulated, 5 down-regulated). Proteins associated with protein dimerization (19%; 11 up-regulated, 3 down-regulated), chromatin binding (12%; 6 up-regulated, 3 down-regulated) and histone binding (8%; 3 up-regulated, 3 down-regulated) also were prominently enriched (Fig. 2C).
Experiment 1.3: Protein interaction network of differentially expressed proteins
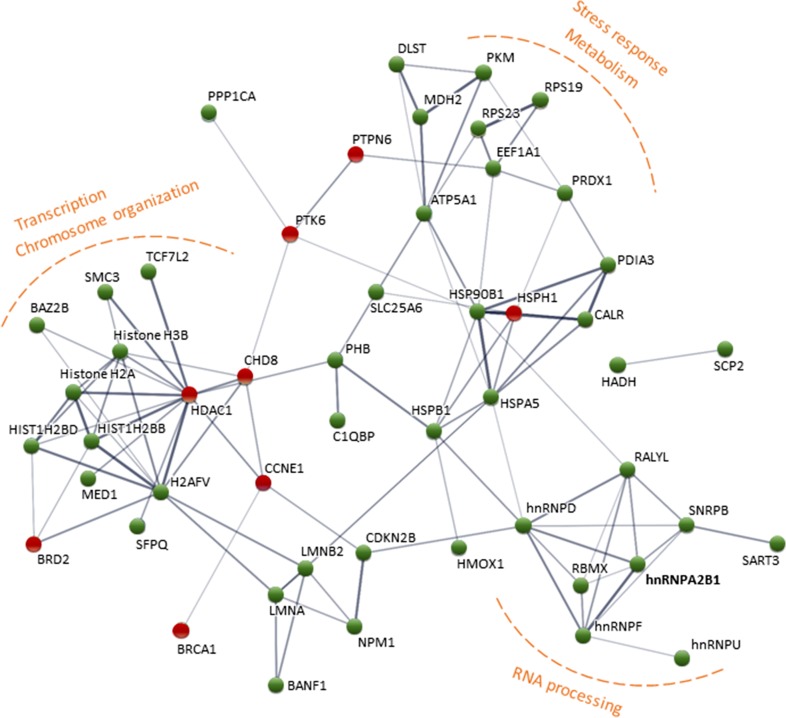
The interaction network constructed by STRING demonstrated significant (P = 8.57E-11) interactions among identified proteins. Three main clusters were evident (Fig. 3), the first around chromosome modifiers and transcription regulators, including BAZ2B, histone deacetylase 1 (HDAC1), chromodomain helicase DNA binding protein 8 (CHD8), mediator complex subunit 1 (MED1), transcription factor 7-like 2 (TCF7L2) and several histones and variants (Histone H2A, H2AFV, HIST1H2BB, HIST1H2BD and H3F3B). The second cohort consisted of proteins associated with RNA splicing and processing, including several heterogeneous nuclear ribonucleoproteins (hnRNPs: hnRNPA2B1, hnRNPF, hnRNPU and hnRNPD), small nuclear ribonucleoprotein polypeptides B and B1 (SNRPB), RNA binding motif protein X-linked (RBMX) and RALY RNA binding protein-like (RALYL). The third group included proteins involved in stress response and metabolism, including the heat shock proteins (HSPs), ATP synthase subunit alpha (ATP5A1), elongation factor 1-alpha 1 (EEF1A1), ribosomal protein S23 (RPS23), malate dehydrogenase 2 (MDH2) and pyruvate kinase (PKM) (Fig. 3).
Figure 3.
Protein–protein interaction network constructed with STRING (disconnected nodes hidden for simplification). Up-regulated proteins are depicted in green and down-regulated in red. Line thickness indicates the strength of data supporting the interaction.
Experiment 2: Expressional and functional validation of hnRNPA2B1 and HMGN1
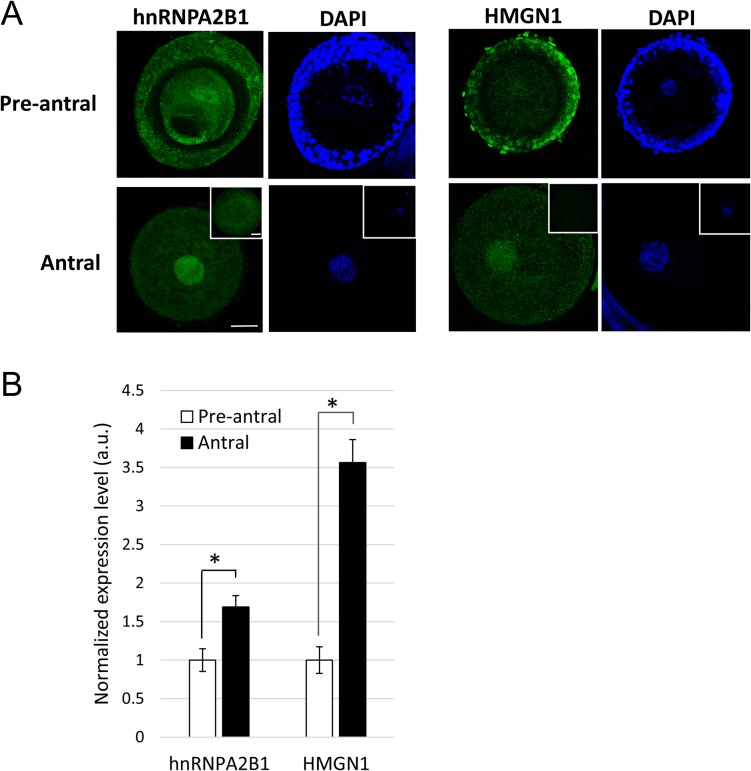
Two candidate proteins, hnRNPA2B1 and HMGN1, were selected for additional validation. The antibodies used here recognized protein products corresponding to predicted sizes of hnRNPA2B1 (37 kDa) and HMGN1 (18 kDa) in cat ovary (Supplemental Fig. S1). Immunofluorescent staining confirmed that both were localized in the GV (Fig. 4A). Proteomic screening revealed an average 2.6-fold increase in protein expression in the antral compared to the pre-antral group for hnRNPA2B1 and a 9.6-fold increase for the same comparison for HMGN1. Paired assessments of hnRNPA2B1 expression based on fluorescence quantification confirmed up-regulation (1.7-fold; P = 0.0107) (Fig. 4B). HMGN1 was marginally detectable in GVs from pre-antral follicles, with expression increasing 3.6-fold (P = 1.593E-10) in the antral counterparts (Fig. 4B).
Figure 4.
Presence of hnRNPA2B1 and HMGN1 in GVs from pre-antral and antral oocytes. (A) Total fluorescence within a GV was imaged and (B) quantified after immunostaining. Non-immune IgG from mouse and rabbit (insets) were included as negative controls. Bars = 30 μm. Average fluorescent values from the pre-antral groups were set as 1. Values are means ± SEM. *P < 0.05.
PDE3A antibody transfection via Chariot led to increased (P = 0.0116) cAMP level similar to that achieved by incubating with the well-characterized PDE3 inhibitor milrinone (Supplemental Fig. S2). This observation confirmed the efficacy of the Chariot transfection system in the cat model.
While fresh (untreated) controls tended to have better meiotic and developmental competence, neither the no transfection treatment (with milrinone) nor mock transfection influenced (P ≥ 0.05) the proportion of oocytes achieving MII, cleaving post-insemination, or forming a morula or blastocyst (Table III). Numbers of blastomeres per blastocyst after a 7-day culture were similar (P ≥ 0.05) among the control groups (fresh, 135.4 ± 29.2 cells [mean ± SD]; no transfection, 126.5 ± 30.9; mock transfection, 101.2 ± 24.8). Transfecting PDE3A antibody maintained GV arrest with only a few oocytes achieving MII or fertilization. Only a single oocyte from this group developed into a blastocyst (75 blastomeres). Again, overall findings from this treatment confirmed efficacy of the protein inhibition approach. When hnRNPA2B1 was inhibited, the proportion of oocytes achieving MII markedly decreased to <20% compared to >60% in fresh (P = 2.43E−06), no transfection (P = 2.29E−07) or mock transfection (P = 6.62E−06) controls. Moreover, none of these antibody-treated, matured oocytes formed morulae (Table III). By contrast, inhibition of HMGN1 failed to influence the ability of oocytes to achieve MII. Under these conditions, proportions of embryos developing into morulae or blastocysts as well as number of blastomeres per blastocyst (110.7 ± 27.7) were comparable (P ≥ 0.05) to controls (Table III).
Table III.
Impact of hnRNPA2B1 and HMGN1 inhibition in oocytes from antral follicles on meiotic and developmental competence.
| Treatment | Milrinone | n | MII | Cleavage | Morula | Blastocyst |
|---|---|---|---|---|---|---|
| Fresh control | − | 27 | 74.1%a | 63.0%a | 37.0%ab | 25.9%a |
| No transfection | + | 43 | 72.1%a | 41.9%a | 23.3%a | 9.3%ab |
| Mock transfection | + | 51 | 62.7%a | 58.8%a | 37.3%b | 11.8%ab |
| PDE3A antibody | + | 37 | 8.1%b | 5.4%b | 2.7%c | 2.7%bc |
| hnRNPA2B1 antibody | + | 42 | 16.7%b | 11.9%b | 0.0%c | 0.0%c |
| HMGN1 antibody | + | 45 | 71.1%a | 53.3%a | 28.9%ab | 15.6%ab |
Within columns, proportions with different superscripts differ (P < 0.05).
Discussion
Combining TMT-labeling, fractionation and LC-MS/MS, we identified 76 nuclear proteins differentially expressed in the GVs of oocytes recovered from pre-antral versus antral follicles. A significant proportion of these proteins was associated with biological processes known to be critical for GV competence, including modulation of chromatin configuration, regulation of gene transcription and processing of maternal RNAs. We further validated that one identified protein, hnRNPA2B1, was required for oocyte meiotic maturation and subsequent blastocyst formation. Findings provided insight into the nuclear network involved in oocyte competence acquisition, all while generating a list of candidate proteins for biomarker exploration.
In the cat, chromatin within the GV transitions from a filamentous to a reticular configuration upon antrum formation, coinciding with acquisition of nuclear competence (Comizzoli et al., 2011). Dynamic histone modifications also have been observed during oocyte development in the cat model (Phillips et al., 2012) as well as other mammalian species, including the mouse (Bao et al., 2000), sheep (Russo et al., 2013) and pig (Bui et al., 2007). Seventeen differentially expressed proteins were associated with chromatin configuration in the present study. Consistent with this function, these proteins generally were assigned to GO_molecular function categories of ‘DNA binding’, ‘chromatin binding’ and/or ‘histone binding’. Notable proteins in this category include HDAC1, nucleophosmin (NPM1) and HMGN1. HDAC1 induces chromatin compaction through histone deacetylation that leads to transcriptional repression (Seto and Yoshida, 2014). Proteomic screening revealed a 1.7-fold down-regulation of HDAC1 in the cat GV of antral compared to pre-antral follicles. This observation was consistent with findings in the mouse where expression of this protein decreased over the course of in vivo oocyte growth (Ma et al., 2012). As HDAC1 expression declines, it is likely that histone deacetylation activity in the GV of an antral follicle is compensated by HDAC2 to regulate gene transcription (Ma et al., 2012; Lee et al., 2015). In the fully grown mouse and bovine oocyte, NPM1 appears to disperse from the nucleolus into the nucleoplasm, coinciding with the termination of ribosomal transcription (Zatsepina et al., 2000; Fair et al., 2001; Lindstrom, 2011). However, evidence suggests that rather than being involved in rDNA transcription in the growing oocyte, NPM1 likely participates in ribosome biogenesis or protein shuttling (Zatsepina et al., 2000; Lindstrom, 2011; Shishova et al., 2015). Finally, HMGN1 is a non-histone chromosomal protein that reduces nucleosome compaction and promotes transcription (Bustin, 2001). Although it appears to help regulate differentiation and development of diverse cell types and organs during embryogenesis (Mohamed et al., 2001; Vigneault et al., 2004; Furusawa and Cherukuri, 2010), HMGN1’s role in oogenesis remains unknown. Previous studies have reported that this protein indirectly modulates histone modifications (H3K14 acetylation and H3S10 phosphorylation) (Lim et al., 2004, 2005) that have been linked to regulating proper meiotic division during oocyte maturation (Endo et al., 2005; Akiyama et al., 2006; Teperek-Tkacz et al., 2010). Because HMGN1 has apparent connectivity to epigenetic regulation and chromatin remodeling (both critical for GV competence), it was selected for more detailed study.
It is noteworthy that core histones and their variants have been found to be up-regulated in the oocytes of more advanced stage bovine follicles (Caixeta et al., 2009; Labrecque et al., 2015), which is consistent with our observations here in the cat. Dynamic histone expression patterns also have been observed during oogenesis and early embryogenesis in the mouse (Wu et al., 2014; Tang et al., 2015), cow (Labrecque et al., 2015) and zebrafish (Yue et al., 2013). Furthermore, a recent study suggested that alterations in histone variant expression in the oocyte is related to reproductive defects (e.g. genome reprogramming and early embryo development) observed in the diabetic mouse model (Jiang et al., 2016). Collectively, it appears that incorporating these histone variants into nucleosomes could be a critical epigenetic mechanism that is conserved among diverse species to effect appropriate gene activation in the oocyte and subsequent development of the embryo.
Oocytes remain transcriptionally active through most of folliculogenesis to produce transcripts required for development of the egg itself and the future embryo. At the end of the growth phase, global transcriptional silencing occurs in most mammalian species studied, often coinciding with configurational change of chromatin (Tan et al., 2009). Because gene transcription usually is recognized as a direct outcome of epigenetic regulation and chromatin remodeling, it was unsurprising that many of the identified epigenetic factors (e.g. HDAC1, HMGN1, NPM1 and CHD8) were also associated with transcriptions based on GO and protein network analyses. Several transcriptional factors were identified from the proteomic screening, including TCF7L2 and basic helix-loop-helix family member e40 (BHLHE40). TCF7L2 acts downstream of the Wnt signaling pathway, which is elevated in murine oocytes from antral follicular stages (Usongo et al., 2012) and has been implicated in oocyte growth, maturation and embryo cleavage (Zheng et al., 2006; Harwood et al., 2008; Spate et al., 2014). By contrast, BHLHE40 exerts transcriptional repression through recruitment of HDAC (Sun and Taneja, 2000). In our study, observed down-regulation of both BHLHE40 and HDAC1 at the antral follicular stage may have signaled de-repression of specific downstream target genes.
Proteomic screening also revealed a significant number of proteins associated with RNA processing machinery, including several hnRNPs. Most were assigned to ‘nucleic acid metabolic process’, ‘transcription’ and ‘RNA binding’ categories based on biological and molecular functions, and also were clustered in the functional network. These proteins are known to form complexes related to post-transcriptional modifications of newly synthesized RNAs and their subsequent transport (Dreyfuss et al., 2002). More specifically, hnRNPs have been implicated in localization of maternal RNA in Drosophila and Xenopus models (Palacios and St Johnston, 2001), but with no precise role yet understood in the mammalian oocyte. The observation that most of these proteins were up-regulated suggested that there may have been increased demand at the antral follicular stage, likely for both processing and shuttling of maternal RNAs before onset of transcriptional silencing. It was for this reason that hnRNPA2B1 was selected as a representative of this group for more examination for its involvement in GV competence acquisition.
Both candidates, hnRNPA2B1 and HMGN1, met the criteria for potential GV competence factors. The proteomic outcome of these proteins was validated by immunofluorescent staining, including confirming nuclear expression in the cat oocyte. Both proteins also increased in the expression in the antral versus pre-antral follicle, upholding the proteomic quantification results. Similar up-regulation of hnRNPA2B1 has been described in the mouse oocyte from microarray analysis (Liu et al., 2001). Transcript expression of HMGN1 has been compared among GV and MII oocytes as well as early embryos in the bovine model (Vigneault et al., 2004). However, our observation represents the first report of a change in HMGN1 expression during the course of oocyte growth.
Once protein expression patterns were verified, functional evaluations via antibody inhibition assays were conducted to determine if hnRNPA2B1 and HMGN1 were indeed involved in competence acquisition. This is an essential step because a complex array of molecular adjustments are occurring concurrently during the pre-antral-to-antral transition to support the (by then) rapidly growing oocyte. The peptide nanoparticle transfection system applied has been used previously in studies of the mouse oocyte and no side-effects were observed (Li et al., 2016). In the cat model, overnight incubation with the peptide nanoparticle and/or milrinone (meiosis inhibitor) alone also were safe, with no deleterious influence on oocyte meiotic or developmental competence. We also determined that transfecting PDE3A antibodies (a positive assay control) successfully prevented meiotic resumption of cat oocytes with <10% reaching MII, which was consistent with the >90% transfection success reported earlier for the mouse oocyte (Li et al., 2016). Therefore, our results demonstrated a technical proficiency in incorporating these antibodies into multiple oocytes simultaneously to cause a persistent inhibition, even after washing away excess antibodies and a protracted incubation.
Functional evaluations revealed that hnRNPA2B1 exhibited a strong impact on the cat oocyte. When this protein was inhibited, most oocytes failed to achieve MII and none developed into a morula-stage embryo. We concluded that hnRNPA2B1 was required for meiotic resumption and subsequent embryo development and, therefore, was a potential nuclear marker for oocyte competence. Because transcriptional silencing occurs at the end of oogenesis and persists until embryonic genome activation, stored maternal RNA at this time is the predominate source for nascent protein synthesis (Sirard, 2012). Indeed it is believed that sufficient RNA accumulation is crucial for the growing oocyte to become fully competent (Eichenlaub-Ritter and Peschke, 2002). Therefore, it was logical to predict that inhibiting hnRNPA2B1 in the immature cat oocyte probably disrupted proper RNA processing and storage that, in turn, compromised subsequent developmental events. Moreover, given tight associations within the protein interaction network, it was likely that there were other RNA processing proteins (e.g. other hnRNP members) carrying out similar functions. In contrast, inhibiting HMGN1 failed to markedly alter the oocyte’s ability to mature, cleave or develop into a morula or blastocyst. Mohamed et al. (2001) previously reported delayed embryo progression when both HMGN1 and HMG17 (a.k.a. HMGN2) were inhibited by injecting antisense oligonucleotides into one-cell mouse embryos. No similar phenotype was observed when antibody inhibition was performed on the immature cat oocyte. Specifically, inhibiting HMGN1 had no subsequent influence on embryo growth rate as demonstrated by a comparable proportion of blastocysts formed and number of blastomeres per embryo compared to controls. This functional difference between the cat and the mouse may indicate species difference or that this particular protein plays some other, as yet unidentified role in oocyte growth. It also was possible that some other HMGN family members (e.g. HMG17) was compensating for the loss-of-function of HMGN1, especially as it is known that there is functional redundancy in the mouse (Furusawa et al., 2009).
As one advances to profile and quantify the GV proteome in mammals, a challenge is dealing the arduous, time-consuming task of manually isolating sufficient numbers of GVs from oocytes. In the present study, the process was simplified by using a sonic dismembrator for cell disruption followed by filtration and centrifugation to enrich GVs in bulk. Post-isolation examination by western blotting did not detect expression of the mitochondrial marker GAPDH. However, GO analysis of proteomic data revealed that heavy macrostructures and organelles (including cytoskeletons, exosomes and mitochondria) were co-isolated with GVs. The expression level of the mitochondrial proteins might be below the detection threshold of western blotting, yet detectable with the higher sensitivity of mass spectrometry. Although the isolation method enabled quick and simple mass GV enrichment from oocytes, it appeared that the higher yield was achieved at the expense of purity. This issue was circumvented through bioinformatic filtering using specific GO terms to allow focusing on desired targets (Pfeiffer et al., 2011), and eventually led to identification of potential nuclear markers. However, it should be noted that some of the identified proteins, particularly ones involved in metabolism and stress response, may have originated from co-isolated mitochondria; therefore, some caution should be exercised in interpreting those findings. For future exploration, alternative differential centrifugation may well be a favorable means to improve bulk enrichment of GVs.
In summary, we identified key nuclear players for the oocyte to achieve full capacity to mature, fertilize, and advance as a preimplantation embryo in the domestic cat model. These differentially expressed proteins revealed a framework of molecular reorganization within a GV that renders its competency. Those proteins of most significance were involved in chromatin organization, transcription and RNA processing. This network of proteins likely prepares GVs from antral follicles to respond to signals funneling through different pathways into the nucleus. Although unable to be used as predictive markers (due to the technique-induced cell damage from the protein visualization procedure), our findings lay a foundation for applying this proteomic information to reproductive technologies. For example, validated proteins may have value as nuclear markers for ensuring optimal in vitro culture conditions for immature oocytes or evaluating potential causes for infertility in human clinics. Moreover, there is great potential to utilize protein assessments for better understanding the influence and even mitigating current limitations of fertility preservation approaches. The latter includes conventional cryopreservation, as well as desiccation, an alternative novel strategy allowing GV storage at room temperature (Elliott et al., 2015). Regardless, present findings cataloging potential molecular markers will underpin future investigations for improving capacity to preserve viable maternal genomes to benefit human fertility.
Supplementary data
Supplementary data are available at Molecular Human Reproduction Online.
Authors’ roles
P.-C.L. designed and performed experiments, analyzed and interpreted data and wrote the article. D.E.W. contributed to the conception of the work and critically revised the article and article revision. P.C. contributed to study design, data interpretation and article revision.
Funding
National Center for Research Resources (R01 RR026064), a component of the National Institutes of Health (NIH) and currently by the Office of Research Infrastructure Programs/Office of the Director (R01 OD010948).
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
We thank Dr Brent Whitaker (Animal Rescue Inc.) and Dr Keiko Antoku, and their staff for providing domestic cat ovaries; Kylie White for technical assistance in GV collection; the Imaging Core at the University of Maryland (College Park) for access to the confocal microscope; and Michael Jakubasz, Smithsonian’s National Zoological Park, for use of the plate reader.
References
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci USA 2006;103:7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashry M, Lee K, Mondal M, Datta TK, Folger JK, Rajput SK, Zhang K, Hemeida NA, Smith GW. Expression of TGFbeta superfamily components and other markers of oocyte quality in oocytes selected by brilliant cresyl blue staining: relevance to early embryonic development. Mol Reprod Dev 2015;82:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assidi M, Dufort I, Ali A, Hamel M, Algriany O, Dielemann S, Sirard MA. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod 2008;79:209–222. [DOI] [PubMed] [Google Scholar]
- Bao S, Obata Y, Carroll J, Domeki I, Kono T. Epigenetic modifications necessary for normal development are established during oocyte growth in mice. Biol Reprod 2000;62:616–621. [DOI] [PubMed] [Google Scholar]
- Bhardwaj R, Ansari MM, Pandey S, Parmar MS, Chandra V, Kumar GS, Sharma GT. GREM1, EGFR, and HAS2; the oocyte competence markers for improved buffalo embryo production in vitro. Theriogenology 2016;86:2004–2011. [DOI] [PubMed] [Google Scholar]
- Bui HT, Van Thuan N, Kishigami S, Wakayama S, Hikichi T, Ohta H, Mizutani E, Yamaoka E, Wakayama T, Miyano T. Regulation of chromatin and chromosome morphology by histone H3 modifications in pig oocytes. Reproduction 2007;133:371–382. [DOI] [PubMed] [Google Scholar]
- Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci 2001;26:431–437. [DOI] [PubMed] [Google Scholar]
- Caixeta ES, Ripamonte P, Franco MM, Junior JB, Dode MA. Effect of follicle size on mRNA expression in cumulus cells and oocytes of Bos indicus: an approach to identify marker genes for developmental competence. Reprod Fertil Dev 2009;21:655–664. [DOI] [PubMed] [Google Scholar]
- Cao S, Guo X, Zhou Z, Sha J. Comparative proteomic analysis of proteins involved in oocyte meiotic maturation in mice. Mol Reprod Dev 2012;79:413–422. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhai L, Qu C, Zhang C, Li S, Wu F, Qi Y, Lu F, Xu P, Li X et al. . Comparative proteomic analysis of buffalo oocytes matured in vitro using iTRAQ technique. Sci Rep 2016;6:31795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NA, Swain JE. Oocyte cryopreservation: searching for novel improvement strategies. J Assist Reprod Genet 2013;30:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combelles CM, Chateau G. The use of immature oocytes in the fertility preservation of cancer patients: current promises and challenges. Int J Dev Biol 2012;56:919–929. [DOI] [PubMed] [Google Scholar]
- Comizzoli P, Holt WV. Recent advances and prospects in germplasm preservation of rare and endangered species. Adv Exp Med Biol 2014;753:331–356. [DOI] [PubMed] [Google Scholar]
- Comizzoli P, Pukazhenthi BS, Wildt DE. The competence of germinal vesicle oocytes is unrelated to nuclear chromatin configuration and strictly depends on cytoplasmic quantity and quality in the cat model. Hum Reprod 2011;26:2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comizzoli P, Wildt DE, Pukazhenthi BS. Impact of anisosmotic conditions on structural and functional integrity of cumulus-oocyte complexes at the germinal vesicle stage in the domestic cat. Mol Reprod Dev 2008;75:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M, Andersen CB, Richard F, Mehats C, Chun SY, Horner K, Jin C, Tsafriri A. Role of cyclic nucleotide signaling in oocyte maturation. Mol Cell Endocrinol 2002;187:153–159. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, Eppig JJ. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol 2004;275:447–458. [DOI] [PubMed] [Google Scholar]
- Downs SM. Nutrient pathways regulating the nuclear maturation of mammalian oocytes. Reprod Fertil Dev 2015;27:572–582. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 2002;3:195–205. [DOI] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Peschke M. Expression in in-vivo and in-vitro growing and maturing oocytes: focus on regulation of expression at the translational level. Hum Reprod Update 2002;8:21–41. [DOI] [PubMed] [Google Scholar]
- Elliott GD, Lee PC, Paramore E, Van Vorst M, Comizzoli P. Resilience of oocyte germinal vesicles to microwave-assisted drying in the domestic cat model. Biopreserv Biobank 2015;13:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Naito K, Aoki F, Kume S, Tojo H. Changes in histone modifications during in vitro maturation of porcine oocytes. Mol Reprod Dev 2005;71:123–128. [DOI] [PubMed] [Google Scholar]
- Fair T, Hyttel P, Lonergan P, Boland M. Immunolocalization of nucleolar proteins during bovine oocyte growth, meiotic maturation, and fertilization. Biol Reprod 2001;64:1516–1525. [DOI] [PubMed] [Google Scholar]
- Flemr M, Ma J, Schultz RM, Svoboda P. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod 2010;82:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Cherukuri S. Developmental function of HMGN proteins. Biochim Biophys Acta 2010;1799:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Ko JH, Birger Y, Bustin M. Expression of nucleosomal protein HMGN1 in the cycling mouse hair follicle. Gene Expr Patterns 2009;9:289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupen CG, Fung M, Armstrong DT. Effects of milrinone and butyrolactone-I on porcine oocyte meiotic progression and developmental competence. Reprod Fertil Dev 2006;18:309–317. [DOI] [PubMed] [Google Scholar]
- Harwood BN, Cross SK, Radford EE, Haac BE, De Vries WN. Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev Dyn 2008;237:1099–1111. [DOI] [PubMed] [Google Scholar]
- Huang X, Hao C, Shen X, Zhang Y, Liu X. RUNX2, GPX3 and PTX3 gene expression profiling in cumulus cells are reflective oocyte/embryo competence and potentially reliable predictors of embryo developmental competence in PCOS patients. Reprod Biol Endocrinol 2013;11:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Nakajima R, Nagata M, Aoki F. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod 2008;23:1377–1384. [DOI] [PubMed] [Google Scholar]
- Jensen JT, Zelinski-Wooten MB, Schwinof KM, Vance JE, Stouffer RL. The phosphodiesterase 3 inhibitor ORG 9935 inhibits oocyte maturation during gonadotropin-stimulated ovarian cycles in rhesus macaques. Contraception 2005;71:68–73. [DOI] [PubMed] [Google Scholar]
- Jiang G, Zhang G, An T, He Z, Kang L, Yang X, Gu Y, Zhang D, Wang Y, Gao S. Effect of Type I diabetes on the proteome of mouse oocytes. Cell Physiol Biochem 2016;39:2320–2330. [DOI] [PubMed] [Google Scholar]
- Jo JW, Jee BC, Suh CS, Kim SH. The beneficial effects of antifreeze proteins in the vitrification of immature mouse oocytes. PloS One 2012;7:e37043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Kaneko H, Nakai M, Somfai T, Kashiwazaki N, Nagai T. Contribution of in vitro systems to preservation and utilization of porcine genetic resources. Theriogenology 2016;86:170–175. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y et al. . Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 2012;8:e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussano NR, Leme LO, Guimaraes AL, Franco MM, Dode MA. Molecular markers for oocyte competence in bovine cumulus cells. Theriogenology 2016;85:1167–1176. [DOI] [PubMed] [Google Scholar]
- Labrecque R, Lodde V, Dieci C, Tessaro I, Luciano AM, Sirard MA. Chromatin remodelling and histone m RNA accumulation in bovine germinal vesicle oocytes. Mol Reprod Dev 2015;82:450–462. [DOI] [PubMed] [Google Scholar]
- Labrecque R, Sirard MA. The study of mammalian oocyte competence by transcriptome analysis: progress and challenges. Mol Hum Reprod 2014;20:103–116. [DOI] [PubMed] [Google Scholar]
- Lee P-C, Wildt DE, Comizzoli P. Nucleolar translocation of histone deacetylase 2 is involved in regulation of transcriptional silencing in the cat germinal vesicle. Biol Reprod 2015;93:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu Y, Yan J, Yan LY, Zhao Y, Li R, Liu P, Hsueh AJ, Qiao J. The role of cilostazol, a phosphodiesterase 3 inhibitor, on oocyte maturation and subsequent pregnancy in mice. PLoS One 2012;7:e30649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Jin Z, Gao L, Liu P, Yang Z, Zhang D. Effective protein inhibition in intact mouse oocytes through peptide nanoparticle-mediated antibody transfection. PeerJ 2016;4:e1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J-H, West K, Rubinstein Y, Bergel M, Postinikov Y, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J 2005;24:3038–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell 2004;15:573–584. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int 2011;2011:195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-C, He Z, Rosenwaks Z. Application of complementary DNA microarray (DNA chip) technology in the study of gene expression profiles during folliculogenesis. Fertil Steril 2001;75:947–955. [DOI] [PubMed] [Google Scholar]
- Lohrig K, Wolters D. Multidimensional protein identification technology. Methods Mol Biol 2009;564:143–153. [DOI] [PubMed] [Google Scholar]
- Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci USA 2012;109:E481–E489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed OA, Bustin M, Clarke HJ. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev Biol 2001;229:237–249. [DOI] [PubMed] [Google Scholar]
- Monti M, Zanoni M, Calligaro A, Ko MS, Mauri P, Redi CA. Developmental arrest and mouse antral not-surrounded nucleolus oocytes. Biol Reprod 2013;88:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C, Silva JV, Silva V, Torgal I, Fardilha M. Signalling pathways involved in oocyte growth, acquisition of competence and activation. Hum Fertil 2015;18:149–155. [DOI] [PubMed] [Google Scholar]
- Palacios IM St, Johnston D. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol 2001;17:569–614. [DOI] [PubMed] [Google Scholar]
- Pfeiffer MJ, Siatkowski M, Paudel Y, Balbach ST, Baeumer N, Crosetto N, Drexler HC, Fuellen G, Boiani M. Proteomic analysis of mouse oocytes reveals 28 candidate factors of the ‘reprogrammome’. J Proteome Res 2011;10:2140–2153. [DOI] [PubMed] [Google Scholar]
- Phillips TC, Wildt DE, Comizzoli P. Increase in histone methylation in the cat germinal vesicle related to acquisition of meiotic and developmental competence. Reprod Domest Anim 2012;47:210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MD, Manandhar G, Spate L, Sutovsky M, Zimmerman S, Sachdev SC, Hannink M, Prather RS, Sutovsky P. Discovery of putative oocyte quality markers by comparative ExacTag proteomics. Proteomics Clin Appl 2010;4:337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard FJ, Tsafriri A, Conti M. Role of phosphodiesterase type 3A in rat oocyte maturation. Biol Reprod 2001;65:1444–1451. [DOI] [PubMed] [Google Scholar]
- Russo V, Bernabo N, Di Giacinto O, Martelli A, Mauro A, Berardinelli P, Curini V, Nardinocchi D, Mattioli M, Barboni B. H3K9 trimethylation precedes DNA methylation during sheep oogenesis: HDAC1, SUV39H1, G9a, HP1, and Dnmts are involved in these epigenetic events. J Histochem Cytochem 2013;61:75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer C, Siatkowski M, Pfeiffer MJ, Baeumer N, Drexler HC, Wang B, Fuellen G, Boiani M. Maternal age effect on mouse oocytes: new biological insight from proteomic analysis. Reproduction 2014;148:55–72. [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishova KV, Lavrentyeva EA, Dobrucki JW, Zatsepina OV. Nucleolus-like bodies of fully-grown mouse oocytes contain key nucleolar proteins but are impoverished for rRNA. Dev Biol 2015;397:267–281. [DOI] [PubMed] [Google Scholar]
- Sirard MA. Factors affecting oocyte and embryo transcriptomes. Reprod Domest Anim 2012;47:148–155. [DOI] [PubMed] [Google Scholar]
- Spate LD, Brown AN, Redel BK, Whitworth KM, Murphy CN, Prather RS. Dickkopf-related protein 1 inhibits the WNT signaling pathway and improves pig oocyte maturation. PLoS One 2014;9:e95114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci USA 2000;97:4058–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JH, Wang HL, Sun XS, Liu Y, Sui HS, Zhang J. Chromatin configurations in the germinal vesicle of mammalian oocytes. Mol Hum Reprod 2009;15:1–9. [DOI] [PubMed] [Google Scholar]
- Tang MC, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, Lim SL, Hudson DF, Kalitsis P, O’Bryan MK et al. . Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet 2015;11:e1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperek-Tkacz M, Meglicki M, Pasternak M, Kubiak JZ, Borsuk E. Phosphorylation of histone H3 serine 10 in early mouse embryos: active phosphorylation at late S phase and differential effects of ZM447439 on first two embryonic mitoses. Cell Cycle 2010;9:4674–4687. [DOI] [PubMed] [Google Scholar]
- Thomas RE, Armstrong DT, Gilchrist RB. Differential effects of specific phosphodiesterase isoenzyme inhibitors on bovine oocyte meiotic maturation. Dev Biol 2002;244:215–225. [DOI] [PubMed] [Google Scholar]
- Usongo M, Rizk A, Farookhi R. beta-Catenin/Tcf signaling in murine oocytes identifies nonovulatory follicles. Reproduction 2012;144:669–676. [DOI] [PubMed] [Google Scholar]
- Vigneault C, McGraw S, Massicotte L, Sirard MA. Transcription factor expression patterns in bovine in vitro-derived embryos prior to maternal-zygotic transition. Biol Reprod 2004;70:1701–1709. [DOI] [PubMed] [Google Scholar]
- Virant-Klun I, Krijgsveld J. Proteomes of animal oocytes: what can we learn for human oocytes in the in vitro fertilization programme? Biomed Res Int 2014;2014:856907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale AM, Calvert ME, Mallavarapu M, Yurttas P, Perlin J, Herr J, Coonrod S. Proteomic profiling of murine oocyte maturation. Mol Reprod Dev 2007;74:608–616. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016;44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 2010;107:17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BJ, Dong FL, Ma XS, Wang XG, Lin F, Liu HL. Localization and expression of histone H2A variants during mouse oogenesis and preimplantation embryo development. Genet Mol Res 2014;13:5929–5939. [DOI] [PubMed] [Google Scholar]
- Wu XF, Yuan HJ, Li H, Gong S, Lin J, Miao YL, Wang TY, Tan JH. Restraint stress on female mice diminishes the developmental potential of oocytes: roles of chromatin configuration and histone modification in germinal vesicle stage oocytes. Biol Reprod 2015;92:13. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Guttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ et al. . The nuclear proteome of a vertebrate. Curr Biol 2015;25:2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue HM, Li Z, Wu N, Liu Z, Wang Y, Gui JF. Oocyte-specific H2A variant H2af1o is required for cell synchrony before midblastula transition in early zebrafish embryos. Biol Reprod 2013;89:82. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Bouniol-Baly C, Amirand C, Debey P. Functional and molecular reorganization of the nucleolar apparatus in maturing mouse oocytes. Dev Biol 2000;223:354–370. [DOI] [PubMed] [Google Scholar]
- Zheng P, Vassena R, Latham K. Expression and downregulation of WNT signaling pathway genes in rhesus monkey oocytes and embryos. Mol Reprod Dev 2006;73:667–677. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Merico V, Cecconi S, Redi CA, Garagna S. What does it take to make a developmentally competent mammalian egg? Hum Reprod Update 2011;17:525–540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.