A new rice gene, OsMFT1, was identified that both regulates rice heading date and panicle architecture and is different from its homologs in other species.
Keywords: Branch meristem, Ghd7, heading date, OsLFL1, OsMFT1, panicle architecture, spikelet meristem
Abstract
Heading date and panicle architecture are important agronomic traits in rice. Here, we identified a gene MOTHER OF FT AND TFL1 (OsMFT1) that regulates rice heading and panicle architecture. Overexpressing OsMFT1 delayed heading date by over 7 d and greatly increased spikelets per panicle and the number of branches. In contrast, OsMFT1 knockout mutants had an advanced heading date and reduced spikelets per panicle. Overexpression of OsMFT1 significantly suppressed Ehd1 expression, and Ghd7 up-regulated OsMFT1 expression. Double mutants showed that OsMFT1 acted downstream of Ghd7. In addition, transcription factor OsLFL1 was verified to directly bind to the promoter of OsMFT1 via an RY motif and activate the expression of OsMFT1 in vivo and in vitro. RNA-seq and RNA in situ hybridization analysis confirmed that OsMFT1 repressed expression of FZP and five SEPALLATA-like genes, indicating that the transition from branch meristem to spikelet meristem was delayed and thus more panicle branches were produced. Therefore, OsMFT1 is a suppressor of flowering acting downstream of Ghd7 and upstream of Ehd1, and a positive regulator of panicle architecture.
Introduction
Heading date (flowering time) in rice (Oryza sativa) is crucial for plants to adapt to the growing environment and for the improvement of yield potential. It is determined by the interaction of endogenous signals and environmental factors. Florigen is a key endogenous signal that is synthesized in the leaves and moves to the shoot apex to induce flowering. Many environmental factors could induce or suppress florigen expression. Photoperiod (day length) is the most important environmental factor affecting flowering time (Song et al., 2015). Rice is a typical short-day (SD) plant, one whose flowering is promoted by short daylength. Many genes have been identified as involved in the photoperiod-mediated flowering pathway. Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1), which are homologous to Arabidopsis thaliana FLOWERING LOCUS T (FT), are florigen genes of rice (Kojima et al., 2002; Tamaki et al., 2007; Komiya et al., 2008; Komiya et al., 2009). In rice, there are two pathways determining floral induction. One is the Heading date 1 (Hd1)–Hd3a pathway, which is conserved with the Arabidopsis CONSTANS (CO)–FT pathway. The other is a unique rice pathway, the Ghd7–Ehd1–Hd3a/RFT1 pathway (Song et al., 2015). Hd1, homologous to Arabidopsis CO, promotes Hd3a expression under SD conditions and suppresses Hd3a under long-day (LD) conditions (Yano et al., 2000). Early heading date 1 (Ehd1) activates Hd3a and RFT1 expression independent of Hd1 in both LD and SD conditions (Doi et al., 2004). Grain number, plant height, and heading date 7 (Ghd7) suppresses Ehd1 expression. Ghd7 expression is sensitive to photoperiod. Under LD conditions, Ghd7 expression is induced and thus Ehd1 and Hd3a expression is suppressed. Under SD conditions, Ghd7 has low expression and the suppression of Ehd1 is relieved, which allows Ehd1 to induce Hd3a expression (Xue et al., 2008). In rice, a number of flowering genes are found to function by directly or indirectly regulating Ehd1, and thus Ehd1 acts as a floral integrator (Song et al., 2015). A recent discovery showed that Ghd7 interacts with Hd1 to delay heading, which indicates these two pathways are not independent in regulating heading date (Nemoto et al., 2016; Zhang et al., 2017).
The rice inflorescence has a branch structure and is usually referred to as a panicle. A rice panicle consists of a main stem, often referred to as the rachis, several primary branches, one or more secondary branches on the primary branch, and occasionally tertiary branches on the secondary branch (Xing and Zhang, 2010). After the transition from the vegetative phase to the reproductive phase, the shoot apical meristem is converted into the inflorescence meristem, and then the inflorescence meristem produces branch meristems. Each branch meristem can continue to produce new branch meristems or transform into a spikelet meristem, and then the spikelet meristem is converted to a floral meristem. In rice, each spikelet meristem generates one flower. Therefore, spikelet meristem identity determines the termination of branch meristem activity. Currently, many genes have been identified as being responsible for the initiation, maintenance, and activity of these meristems (Tanaka et al., 2013; Zhang and Yuan, 2014). A major yield quantitative trait locus (QTL), GRAIN NUMBER 1a (GN1a), encodes a cytokinin oxidase, OsCKX2. High levels of cytokinin caused by loss-of-function of OsCKX2 increase branch meristem activity, which is responsible for the increased branch and spikelet number (Ashikari et al., 2005). Rice FRIZZY PANICLE (FZP) regulates the transition of branch meristems to spikelet meristems and has a crucial role in establishing the spikelet meristem identity. fzp mutants produce branches but fail to form normal spikelets, and spikelets are replaced by branches in mutants carrying severe fzp alleles (Chujo et al., 2003; Komatsu et al., 2003; Bai et al., 2016). Another rice yield gene, TAWAWA1 (TAW1), also regulates spikelet number through suppression of the transition from branch meristems to spikelet meristems. TAW1 promotes branch meristem activity and suppresses the phase change to spikelet meristem identity through positively regulating the SVP family MADS-box genes, leading to prolonged branch formation (Yoshida et al., 2013). Thus, promotion of inflorescence meristem or branch meristem activity and appropriate delay of spikelet meristem identity formation could help to increase spikelet number.
Rice MOTHER OF FT AND TFL1 (MFT) belongs to the family of phosphatidylethanolamine-binding proteins (PEBPs). The PEBP family is a family of evolutionarily conserved genes widely present in eukaryotes (Karlgren et al., 2011). In higher plants, the PEBP gene family consists of three main homologous subfamilies, FT-like, TERMINAL FLOWER1 (TFL1)-like and MFT-like genes (Chardon and Damerval, 2005). As the name suggests, the MFT-like subfamily is a homolog of FT and TFL1 and is thought of as the evolutionary ancestor to them (Hedman et al., 2009). In Arabidopsis, there are six PEBP family genes: two FT-like genes (FT and TSF), three TFL-like genes (TFL1, BFT, and ATC) and one MFT-like gene (MFT) (Danilevskaya et al., 2008). Both FT and TFL1 are key regulators of floral transition but have antagonistic roles. FT has been shown to be florigen and induces flowering while TFL1 has been identified as a flowering suppressor (Alvarez et al., 1992; Kardailsky et al., 1999; Hanzawa et al., 2005). In addition to repressing flowering, TFL1 plays a crucial role in determining inflorescence architecture. In Arabidopsis, a main shoot apical meristem produces either indeterminate flowers or indeterminate lateral axes after floral transition. TFL1 prevents the meristems from assuming the floral identity and accounts for indeterminate growth of the inflorescence shoot. Thus, 35S::TFL1 transgenic plants exhibited an extended vegetative phase and branched inflorescence while loss-of-function of TFL1 produced terminal flowers at the shoot apex (Alvarez et al., 1992; Benlloch et al., 2007; Liu et al., 2013). ARABIDOPSIS THALIANA CENTRORADIALIS (ATC) and BROTHER OF FT AND TFL1 (BFT) inhibit flowering similarly to TFL1 (Huang et al., 2012; Yoo et al., 2010), and TSF promotes flowering similarly to FT (Yamaguchi et al., 2005). As a gene homologous to both FT and TFL1, MFT seems to have no major effect on flowering. Overexpression of MFT led to slightly early flowering while loss of MFT function did not exhibit an obvious phenotype in flowering (Yoo et al., 2004). Later studies showed MFT is involved in the regulation of seed germination via ABA and GA signaling pathways. Loss-of-function of MFT led to hypersensitivity to ABA in seed germination. MFT is directly bound by ABA-INSENSITIVE 3 (ABI3) and ABI5 on the promoter. MFT is suppressed and promoted by ABI3 and ABI5, respectively. In addition, DELLA proteins, the major repressors of GA signaling, could directly bind to the MFT promoter and promote its expression. On the other hand, MFT exerts a negative feedback regulation of ABA signaling by directly repressing ABI5 (Xi et al., 2010). Besides Arabidopsis, there are several MFT homologs reported to regulate seed germination in other species. In wheat, TaMFT is a repressor of seed germination and co-localizes with a seed dormancy QTL (Nakamura et al., 2011). Through ectopic overexpression in Arabidopsis, a Soyben homolog of MFT (GmMFT) negatively regulates seed germination, and strawberry homolog of MFT (FvMFT) regulates germination via participating in GA and ABA signaling (Li et al., 2014; Hu et al., 2016).
In rice, 19 PEBP genes were identified based on genome wide analysis, of which there were 13 FT-like genes, four TFL-like genes, and two MFT-like genes (Chardon and Damerval, 2005; Danilevskaya et al., 2008). Among them, the most well-studied homolog of FT is Hd3a. Ha3a is a mobile flowering signal that moves from leaf to shoot apical meristem where it interacts with 14-3-3 protein and OsFD1 to form a florigen activation complex, which is essential for the activation of the inflorescence meristem identity gene (Tamaki et al., 2007; Taoka et al., 2011). The four TFL1 homologs in rice are named RCN1, RCN2, RCN3, and RCN4. Overexpression of RCN1, RCN2, and RCN3 exhibited branched dense panicle architecture and delayed heading date, while knocking down of all RCNs produced reduced branches and small panicles (Nakagawa et al., 2002; Zhang et al., 2005; Liu et al., 2013). So far, two MFT homologs in rice, OsMFT1 and OsMFT2, have not been identified yet. A previous study proposed that OsMFT1 was positively regulated by Ghd7 in the flowering pathway through an expression QTL (eQTL)-guided function-related co-expression analysis (Wang et al., 2014). It is very likely that OsMFT1 regulates heading date and panicle architecture. Here, we confirmed that OsMFT1 acts downstream of Ghd7 and elucidated its mechanism in controlling heading and panicle architecture by identification of its upstream and downstream genes.
Materials and methods
Plant material and growth condition
The japonica rice variety Zhonghua 11 (ZH11) was used as the wild type and recipient for genetic transformation. Ghd7-related material, including NIL(mh7), NIL(zs7), OX-Ghd7ZH11, and Ami-Ghd7, were from previous studies (Xue et al., 2008; Weng et al., 2014); the ghd7 mutant had an SNP mutation resulting in a premature stop codon in the ZH11 background. For measurement of the agronomic traits, rice plants grown at Wuhan were under natural LD conditions, whereas plants grown at Hainan were under SD conditions. Germinated seeds were sown in the seed beds and 1-month-old seedlings were transplanted to the fields with 10 plants in a row. The heading date was the day when the first panicle of the plant emerged. Plants in the middle of each row were harvested individually and used to score the traits of spikelets per panicle, number of primary branches, and number of secondary branches.
Vector construction and genetic transformation
To generate the overexpression vector, coding sequences of OsMFT1 were isolated from ZH11 leaf cDNA and cloned into T-vector (Promega), then digested with KpnI and XbaI and cloned into the KpnI–XbaI sites of pCAMBIA1301S. For generating CRISPR mutants, a specific single guide RNA (sgRNA) targeting OsMFT1 was designed and assembled into the vector pCXUN-CAS9 (sgRNA was driven by the U3 promoter). The constructs were introduced into ZH11 callus by Agrobacterium-mediated transformation.
RNA extraction and qRT-PCR analysis
Samples of leaves, young panicles, and other tissues were frozen in liquid nitrogen immediately after being collected from the plants. Total RNA was extracted using Trizol reagent (TransGen Biotech, Beijing). Then 3 μg of total RNA was digested by DNase I and reverse transcribed by Superscript III reverse transcriptase (Invitrogen, USA) to obtain the first-strand cDNA according to the manufacturer’s protocol. Real-time PCR was performed in a 96-well plate in an ABI Prism 7500 real-time PCR system (Applied Biosystems, USA) using SYBR Premix ExTaq reagent (TaKaRa, Dalian). The relative expression levels were calculated according to the method proposed previously (Livak and Schmittgen, 2001), with the rice ubiquitin gene serving as an internal control. Primers used for real-time PCR are listed in Supplementary Table S1 at JXB online.
Subcellular localization
To confirm the subcellular localization of OsMFT1, the coding sequence of OsMFT1 was amplified and inserted into the pM999 vector driven by the CaMV 35S promoter. The fusion construct 35S::OsMFT1::YFP was co-transformed into rice protoplasts with 35S::GHD7::CFP, which was used as a nuclear marker. The construct 35S::YFP was used as a control. Rice protoplasts transformation was conducted as previously described (Xie and Yang, 2013). After transformation into rice protoplasts and incubation in the dark for 12–16 h, the fluorescence was observed by confocal microscopy (Leica Microsystems).
Yeast one-hybrid assay
AD-OsLFL1 was constructed by inserting the coding sequence of OsLFL1 into the vector pB42AD (Clontech, USA). The promoter fragments of OsMFT1 were cloned into the vector pLacZi2μ to construct pro::LacZ. The yeast one-hybrid assay was performed as previously described (Tang et al., 2012). Briefly, the AD-OsLFL1 and pro::LacZ were co-transformed into yeast strain EGY48 and spread on the selective medium SD/−Trp/−Ura (Clontech). The grown transformants were transferred to SD/−Trp/−Ura medium containing raffinose, galactose, and X-gal (Sigma-Aldrich, USA) for developing the blue color.
Electrophoretic mobility shift assay
To get the OsLFL1 protein, the coding sequence of OsLFL1 was amplified and cloned into the expression vector pSPUTK (Smaczniak et al., 2012). Proteins were synthesized using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega). The oligonucleotides were synthesized and labeled with 5′-biotin by the Shanghai Sangon Company. Double-stranded oligonucleotides were generated by mixing equal amounts of the complementary single-stranded oligonucleotides and heating for 2 min at 95 °C, then cooling down to 25 °C. Biotin-labeled probes were incubated with the OsLFL1 protein in the binding buffer [10 mM Tris (pH 7.5), 50 mM KCl, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, 50 ng μl−1 Poly (dI–dC), 2.5% glycerol and 0.05% NP-40] for 20 min at room temperature. For the competition reaction, 10-, 20-, 50-, and 100-fold non-labeled probes were mixed with the labeled probes. The reaction mixture was loaded onto a 6% native polyacrylamide gel and run at 4 °C. The DNA shift was detected by developing the biotin signal using the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Dual luciferase transcriptional activity assay in rice protoplasts
To test the transcriptional activity of the OsLFL1 protein, the coding sequence of OsLFL1 was fused in-frame with the GAL4 DNA-binding domain GAL4BD as the effector vector, with CaMV35S-Gal4-LUC as the reporter. To test the transcriptional activation activity of OsLFL1 on OsMFT1, the coding sequence of OsLFL1 was driven by CaMV35S as an effector with the luciferase driven by the promoter of OsMFT1 as a reporter. The effectors and corresponding reporters were co-transformed into rice protoplasts with the internal control vector CaMV35S-LUC as previously described (Xie and Yang, 2013). The Dual-Luciferase Reporter Assay System (Promega) was used to measure the luciferase activity. Briefly, the rice protoplasts were lysed with Passive Lysis buffer after incubation overnight. The supernatant of the lysate was incubated with luciferase assay substrate and the firefly luciferase (fLUC) activity was measured with the TECAN Infinite M200 System. After the measurement of fLUC, Stop & Glo substrate buffer was added to the reaction and then the Renilla luciferase (rLUC) activity was measured. Three independent transformations for each combination were performed, and the relative luciferase activity was calculated by the ratio fLUC/rLUC.
RNA in situ hybridization
The probes for hybridization were amplified from the OsMFT1 coding sequence using specific primers and inserted into the pGEM-T vector (Promega) for RNA transcription in vitro. The respective sense and antisense probes were produced using SP6 and/or T7 transcriptase labelled with the Digoxigenin RNA labeling kit (Roche). Young panicle tissues were collected and fixed in FAA solution (50% ethanol, 5% acetic acid and 3.7% formaldehyde) at 4 °C overnight. RNA in situ hybridization and immunological detection were performed as previously described (Zhao et al., 2009).
Seed germination test
ZH11 and the transgenic homozygous OX-OsMFT1 lines were grown during normal growing seasons in Wuhan. We marked the panicles when they appeared from the leaf sheath, then harvested the panicles 40 d after their heading. After being dried under sunlight for 3 d, grains were threshed and used for a germination test. Fully filled grains were spread on plates with wet filter paper and immediately moved into an incubator in the dark at 28 °C. Each plate was filled with 50 seeds, and three plates were used for each genotype. Germination was defined as the emergence of the radical, and the number of germinated seeds was counted every half-day after imbibition.
Results
OsMFT1 overexpression and knockout plants showed altered heading date and panicle architecture in rice
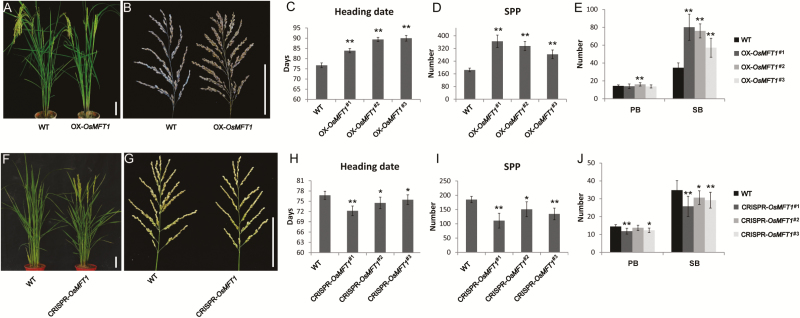
To identify the function of OsMFT1, we overexpressed OsMFT1 using the CaMV35S promoter in a japonica variety Zhonghua 11 (ZH11) and obtained 50 T0 transgenic plants, of which 18 were positive (see Supplementary Fig. S1). The positive plants showed delayed heading date and increased spikelet number per panicle compared with wild type (WT) (Fig. 1A, B). Overexpression of OsMFT1 (OX-OsMFT1) in the T1 generation significantly delayed heading date by over 7 d and almost doubled spikelet number per panicle (Fig. 1C, D). Overexpression lines had greatly increased number of branches, especially secondary branches (Fig. 1E), resulting in dense panicles. Three lines of OsMFT1 knockout mutants each having a 1 bp deletion, and 1 bp and 2 bp insertion in the first exon were generated using a CRISPR–Cas9 strategy (Supplementary Fig. S2). A slight but significant promotion in heading date and decrease of spikelets per panicle were observed in all OsMFT1 knockout mutants compared with WT (Fig. 1F, I). The number of primary branches and secondary branches was significantly reduced (Fig. 1J). Taken together, OsMFT1 is a suppressor of heading and positive regulator of spikelets per panicle in rice.
Fig. 1.
Phenotypes of OsMFT1 overexpression and knockout transgenic plants grown in the field in summer (long-day condition) in Wuhan. (A, B) Phenotypes of OsMFT1 overexpression lines (right) and wild type (left) whole plant (A) and panicle (B). Scale bars, 10 cm. (C–E) Comparison of three T1 overexpression lines with the wild type for heading date (C), spikelet number per panicle (SPP) (D), primary branches (PB) and secondary branches (SB) (E). (F, G) Phenotypes of OsMFT1 CRISPR plants (right) and wild type (left) whole plant (F) and panicle (G). Scale bars, 10 cm. (H–J) Comparison of three T1 CRISPR lines with the wild type for heading date (H), SPP (I), PB, and SB (E). Error bars indicate the standard deviation (SD), n≥15 each. *P<0.05, **P<0.01, t-test.
Expression characterization of OsMFT1 and subcellular localization
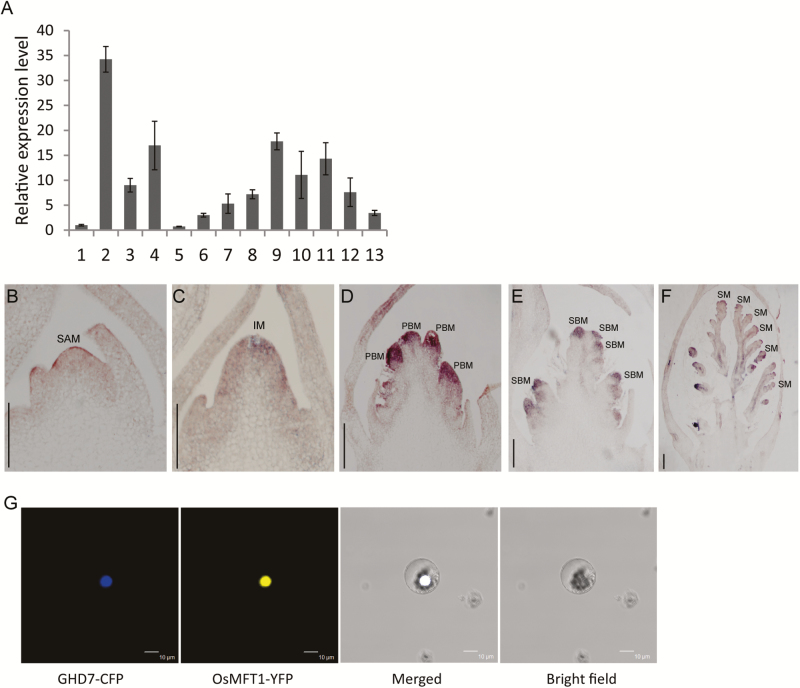
To characterize the spatial–temporal expression pattern of OsMFT1, the RNA transcript level of OsMFT1 was examined in roots, stems, leaves, sheaths, and developing young panicles using quantitative real-time PCR. OsMFT1 was preferably expressed in stem, leaves, sheath, and developing young panicles (Fig. 2A). RNA in situ hybridization revealed that OsMFT1 was slightly expressed in the shoot apical meristem and inflorescence meristem (Fig. 2B, C), and strongly expressed in the primary branch meristem (Fig. 2D), secondary branch meristem (Fig. 2E) and spikelet meristem (Fig. 2F). To determine the subcellular localization of OsMFT1, the full length coding sequence (CDS) of OsMFT1 was fused to the yellow fluorescent protein (YFP) reporter gene driven by the CaMV 35S promoter. Then, the OsMFT1-YFP and GHD7-CFP plasmids were co-transformed into protoplasts. The OsMFT1-YFP fusion protein was luminescent in the nucleus and the YFP fluorescence overlapped with cyan fluorescent protein (CFP) fluorescence, which indicated that OsMFT1 co-localized with GHD7, a nuclear protein (Fig. 2B). Thus, OsMFT1 is a nuclear protein.
Fig. 2.
Expression pattern and subcellular localization of OsMFT1. (A) RNA expression level of OsMFT1 in Zhonghua11 in 13 different tissues, including (1) root, (2) stem, (3) sheath, (4) leaf of 30-day plant, (5) 1–2 mm young panicles, (6) 2–5 mm young panicles, (7) 0.5–1 cm young panicles, (8) 1–3 cm young panicles, (9) 3–5 cm young panicles, (10) leaf at 1–2 mm panicle stage, (11) sheath at 1–2 mm panicle stage, (12) leaf at 5 cm panicle stage, and (13) sheath at 5 cm panicle stage. Error bars indicate SD based on three biological replicates. (B–F) RNA in situ hybridization analysis of OsMFT1 expression in the shoot apical meristem (B), inflorescence meristem (C), young panicle at primary branch initiation stage (D), secondary branch initiation stage (E), and spikelet meristem differentiation stage (F). Scale bars, 100 µm. IM, inflorescence meristem; PBM, primary branch meristem; SAM, shoot apex meristem; SBM, secondary branch meristem; SM, spikelet meristem. (G) OsMFT1 colocalized with the transcription factor GHD7 in the nucleus of rice protoplasts.
The role of OsMFT1 in regulating heading date
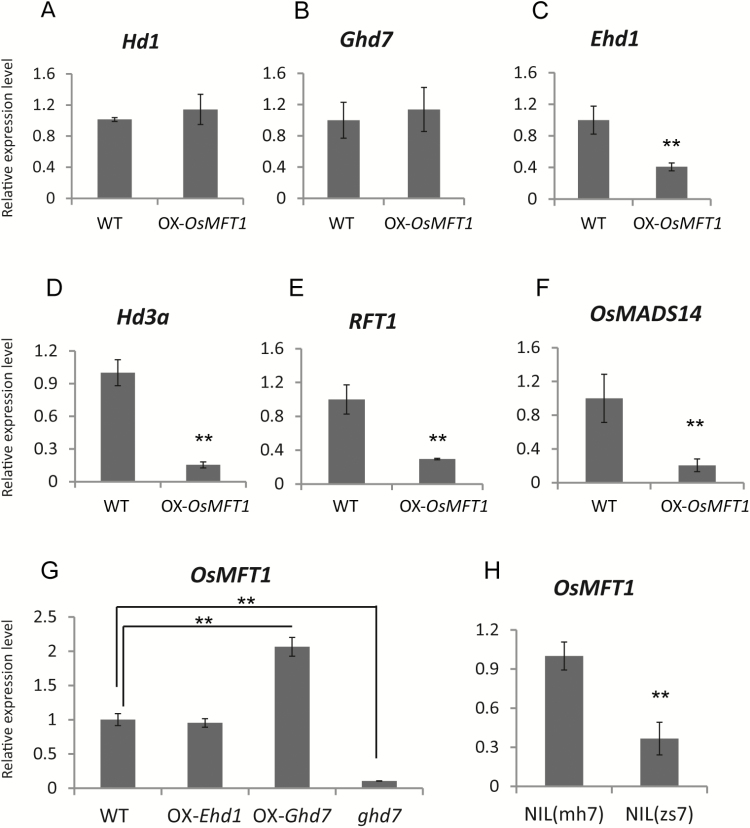
OsMFT1 overexpression lines and wild type were used to examine the transcriptional level of key genes involved in the photoperiodic flowering pathway. There were no significant differences in the expression of Ghd7 and Hd1 between OsMFT1 overexpression lines and wild type, while the expression levels of Ehd1, Hd3a, RFT1, and MADS14 were greatly suppressed in overexpression lines (Fig. 3A–G), indicating that OsMFT1 acted upstream of Ehd1. To further elucidate the regulatory relationship between OsMFT1 and these genes, the RNA expression level of OsMFT1 in an Ehd1 overexpression line (OX-Ehd1), a Ghd7 overexpression line (OX-Ghd7), a Ghd7 mutant line (ghd7), and a pair of Ghd7 near isogenic lines was examined. Compared with the wild type ZH11, the expression level of OsMFT1 did not vary in the OX-Ehd1 line, indicating that Ehd1 did not regulate OsMFT1 in turn (Fig. 3G). The expression level of OsMFT1 was approximately 4-fold more than that in the OX-Ghd7 line and was reduced by 3-fold in the Ghd7 mutant compared with wild type (Fig. 3G). Similarly, the expression level of OsMFT1 in NIL(mh7) was 3-fold of that in NIL(zs7), which has completely lost Ghd7 (Fig. 3H). In general, OsMFT1 expression is regulated by Ghd7 but not by Ehd1 and, in contrast, OsMFT1 suppressed Ehd1 expression. The expression of other flowering genes was also examined in OsMFT1 overexpression lines and wild type, but no significant differences were detected (see Supplementary Fig. S3). Thus, it is suggested that OsMFT1 acts downstream of Ghd7 and upstream of Ehd1 in the photoperiod flowering pathway.
Fig. 3.
Transcriptional regulation between OsMFT1 and the key flowering genes. (A–F) mRNA expression comparison of OsMFT1, Ghd7, Ehd1, Hd3a, RFT1, and MADS14 between wild type and OsMFT1 overexpression lines. (G) Expression of OsMFT1 in Zhonghua 11 (ZH11), Ehd1 overexpression lines (OX-Ehd1), Ghd7 overexpression lines (OX-Ghd7) and Ghd7 knockout mutant (ghd7). (H) Expression of OsMFT1 in Ghd7 near isogenic lines (NILs). NIL(mh7) had a strongly functional allele of Ghd7 while NIL(zs7) had a non-functional Ghd7. Plants were grown in a growth chamber (14 h light/10 h dark cycle for LD conditions) and the top–second leaves of 45-day plants were sampled for RNA extraction at 2 h after lights on when most flowering genes reached their peaks of RNA expression level. Error bars indicate SD based on three biological replicates. **P<0.01, t-test.
OsMFT1 overexpression rescued the phenotype of Ami-Ghd7
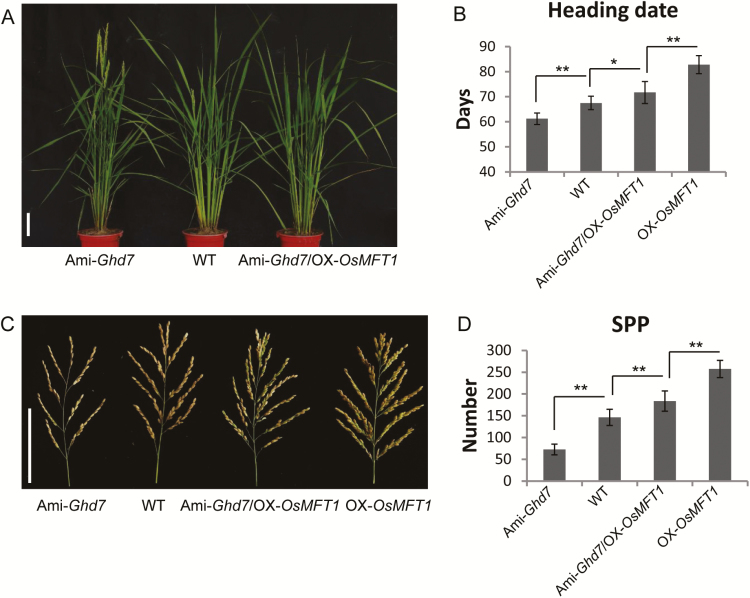
To understand the effect of OsMFT1 on Ghd7-mediated flowering and panicle architecture, a hybrid F1 was generated by crossing OX-OsMFT1 with Ami-Ghd7 (Ghd7 artificial microRNA), in which Ghd7 expression was largely suppressed. Four genotypes showing higher OsMFT1 and lower Ghd7 (Ami-Ghd7/OX-OsMFT1), higher OsMFT1 but normal Ghd7 (OX-OsMFT1), lower Ghd7 but normal OsMFT1 (Ami-Ghd7) and wild type were identified from an F2 population (see Supplementary Fig. S4). The heading date and spikelets per panicle of four genotypes in long-day conditions are displayed in Fig. 4. Compared with the wild type plants, Ami-Ghd7 showed significantly advanced heading date and smaller panicle size while the OsMFT1 overexpression line (OX-OsMFT1) showed significantly delayed heading date and denser panicle architecture. The double mutant, Ami-Ghd7/OX-OsMFT1, simultaneously overexpressing OsMFT1 and suppressing Ghd7, exhibited a 4-day delay in heading date and an approximately 25% increase in spikelets per panicle than the wild type. Similar differences in phenotypes among four genotypes were observed in SD conditions, but the genotypes had smaller phenotype values than their corresponding phenotype values in the LD conditions (Supplementary Fig. S5). These results indicated that OsMFT1 overexpression rescued the phenotype of Ami-Ghd7 in heading date and spikelets per panicle, and OsMFT1 was indeed acting downstream of Ghd7.
Fig. 4.
Comparison of heading date and panicle architecture among wild type, single mutants, and double mutants in LD condition. (A, B) Heading date of Ami-Ghd7 (artificial microRNA-mediated Ghd7 silencing in ZH11), OX-OsMFT1 (OsMFT1 overexpressing lines), their hybrid Ami-Ghd7/OX-OsMFT1 and WT. (C, D) Panicle architecture of Ami-Ghd7, OX-OsMFT1, Ami-Ghd7/OX-OsMFT1, and WT. SPP, spikelets per panicle. Error bars indicate SD; n≥10 for each.
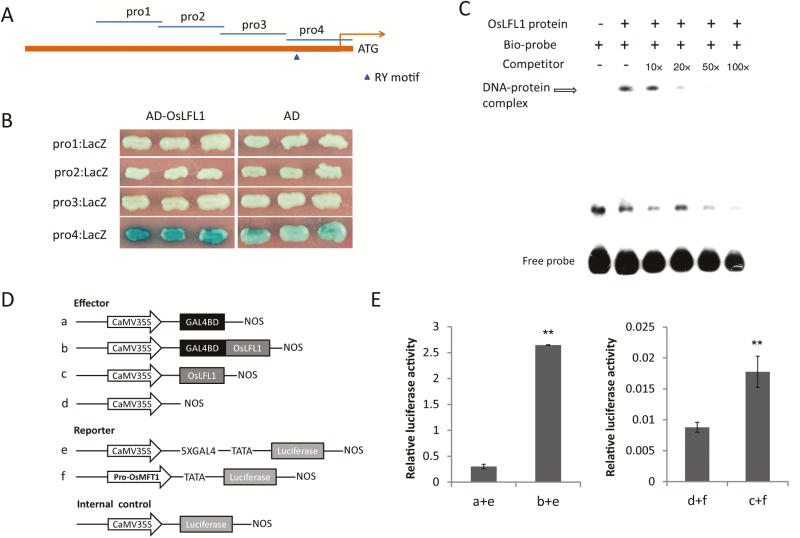
OsLFL1 activated OsMFT1 expression by directly binding to its promoter
In Arabidopsis, ABI3 directly binds to the RY motif in the promoter of AtMFT (Park et al., 2011; Mao and Sun, 2015). Among the ABI3 homologs reported in rice, OsLFL1 is a flowering repressor (Peng et al., 2008; Romanel et al., 2009). Thus, OsLFL1 is proposed to probably bind to OsMFT1 promoter directly. Then, the 1.6 kb sequence of the OsMFT1 promoter and 5′-UTR region was divided into four fragments and used to perform a yeast one-hybrid assay with OsLFL1 protein. A binding activity of OsLFL1 protein to the fourth fragment (pro4) closest to ATG was identified (Fig. 5A, B). cis-Element analysis identified an RY motif (CATGCATG) 221 bp upstream of the translation start site ATG in the OsMFT1 promoter (Fig. 5A). An electrophoretic mobility shift assay (EMSA) showed that OsLFL1 protein directly bound to the 50-bp fragment containing the RY motif in vitro (Fig. 5C). To verify how OsLFL1 regulates OsMFT1, a dual-luciferase transient assay was performed in rice protoplasts to examine the transcriptional activity of OsLFL1. As shown in Fig. 5D, E, with the firefly luciferase driven by five copies of the yeast GAL4 binding domain (GAL4BD) as a reporter, relative luciferase activity of OsLFL1 fused with GAL4BD as an effector was much higher than GAL4BD itself as an effector, indicating that OsLFL1 had significant transcriptional activation activity; with the firefly luciferase driven by the OsMFT1 promoter as a reporter, relative luciferase activity of OsLFL1 as an effector was 2-fold of empty ‘none’ as an effector indicating that OsLFL1 had activation activity on the OsMFT1 promoter. Taken together, OsLFL1 activates OsMFT1 expression by directly binding to its RY motif in the promoter.
Fig. 5.
In vivo and in vitro assay of OsLFL1 binding to the promoter of OsMFT1. (A) The promoter of OsMFT1 was divided into four fragments (pro1–4), and the RY motif was contained in pro4. (B) OsLFL1 bound to pro4 in yeast cells through a yeast one-hybrid assay on selective medium (SD/−Trp−Ura) containing X-gal for developing the blue color. (C) EMSA assay using the OsLFL1 protein and 50-bp OsMFT1 promoter containing RY motif as a probe labeled with 5′-biotin. The 10-, 20-, 50- and 100-fold non-labeled probes were used for competition. (D, E) OsLFL1 activates the expression of OsMFT1 by dual luciferase transient assay in rice protoplasts. Error bars indicate SD based on three biological replicates, **P<0.01, t-test.
Overexpression of OsMFT1 suppressed the expression of spikelet meristem identity genes
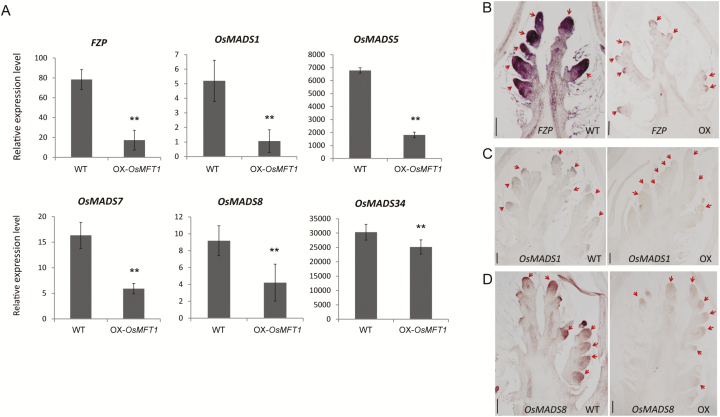
To identify downstream genes of OsMFT1, the 0.5–1 mm young panicles from OX-OsMFT1 and wild type plants were used for transcriptome sequencing to find differentially expressed genes. Compared with wild type, 84 genes were up-regulated and 86 genes were down-regulated in the OX-OsMFT1 young panicles (see Supplemenatry Table S2). Three genes regulating spikelet meristem differentiation were down-regulated, including MADS1, MADS5, and FZP. FZP is a spikelet meristem identity gene that determines the transition from panicle branching to spikelet formation. OsMADS1 and OsMADS5, together with OsMADS7, OsMADS8, and OsMADS34 are five SEPALLATA-like genes that are classified as class E genes for floral determinacy (Cui et al., 2010). A qRT-PCR assay showed that these genes were indeed down-regulated in OX-OsMFT1 (Fig. 6A). Further RNA in situ hybridization showed that the significantly weaker expression of FZP, OsMADS1, and OsMADS8 was detected in the secondary branch meristem of OX-OsMFT1 compared with wild type (Fig. 6B–D).
Fig. 6.
Differential expression of rice SEP-like genes and FZP between OsMFT1 overexpression lines and wild type. (A) RNA expression comparison of FZP, OsMADS1, OsMADS5, OsMADS7, OsMADS8, and OsMADS34 between OX-OsMFT1 and wild type using quantitative real time PCR. Error bars indicate SD based on three biological replicates. (B–D) RNA in situ hybridization of FZP (B), OsMADS1 (C), and OsMADS8 (D) in young panicles of wild type (left) and OsMFT1 overexpression lines (right). Scale bars, 100 µm. Red arrowheads show secondary branch meristems.
Discussion
Conserved functions in regulating flowering and panicle architecture between MFT-like and TFL1-like families in rice
Most of the PEBP family genes are functionally conserved among higher plants. Previous studies indicated that FT-like genes generally induce flowering both in monocots and dicots, while TFL1-like genes delay flowering and regulate the inflorescence architecture, including TFL1 in Arabidopsis, RCNs in rice, and ZCNs in maize (Alvarez et al., 1992; Nakagawa et al., 2002; Zhang et al., 2005; Danilevskaya et al., 2010; Wickland and Hanzawa, 2015). As the evolutionary ancestor of FT and TFL1, MFT-like genes are generally related to seed germination (Xi et al., 2010; Nakamura et al., 2011; Li et al., 2014; Hu et al., 2016). As expected, OsMFT1 also regulates seed germination, which will be further studied in future (see Supplementary Fig. S6). OsMFT1 overexpression significantly delays flowering and largely increases branching, and the OsMFT1 knockout mutant promotes flowering and reduces branching. The performance of OsMFT1 overexpression plants is very similar to rice TFL1-like gene (RCN1, RCN2, and RCN3) overexpression plants (Nakagawa et al., 2002; Zhang et al., 2005). In addition, knocking down four TFL1-like genes in rice resulted in small panicles with reduced branches, similar to the OsMFT1 knockout plants (Liu et al., 2013). We examined the expression of rice TFL1-like genes (RCN1, RCN2, and RCN3) in the leaves and panicles of OsMFT1 overexpression lines and wild type, and no big differences were detected (Supplementary Fig. S7). Since OsMFT1 and rice TFL1-like genes have similar functions in regulating flowering and inflorescence architecture, functional redundancy may exist between them.
FT and TFL1 have only 39 non-conservative amino acid substitutions but have distinct functions due to a potential ligand binding residue and a divergent external loop (Hanzawa et al., 2005; Ahn et al., 2006). Lateral studies revealed that the mutation of at least four residues converts FT into a complete TFL1 mimic by affecting the protein surface charge through testing the effects of numerous mutations of FT in vivo (Ho and Weigel, 2014). We compared the crucial amino acid residues of FT-like and TFL1-like protein in both Arabidopsis and rice (see Supplementary Fig. S8); furthermore, we analyzed the potential function of OsMFT1 according to the reported mutations in FT and their corresponding phenotypes (Ho and Weigel, 2014). The results indicate that the key amino acid residues of FT-like and TFL1-like proteins are conserved between Arabidopsis and rice, and the methionine (M) at position 109, lysine (K) at 128 and alanine (A) at 138 most likely contribute to the conferred TFL1 activity of OsMFT1 (Supplementary Table S3), which could explain the functional similarity of OsMFT1 and TFL1-like genes in rice.
In Arabidopsis, MFT mainly functions in regulating seed germination instead of flowering (Yoo et al., 2004; Xi et al., 2010). In this study, OsMFT1 had effects on both seed germination and flowering (Fig. 1; Supplementary Fig. S6). It is noted that the upstream regulator Ghd7 and downstream gene Ehd1 of OsMFT1 are specific genes in rice that have no homologs identified in Arabidopsis (Doi et al., 2004; Xue et al., 2008). This is probably why MFT1 has no regulation in flowering in Arabidopsis.
Activation of OsMFT1 by both OsLFL1 and Ghd7
A previous study has proposed that Ghd7 positively regulates OsMFT1 in flowering through an eQTL-guided function-related co-expression analysis (Wang et al., 2014). Here, we have further demonstrated that Ghd7 regulates OsMFT1 expression transcriptionally and OsMFT1 plays a role in regulating flowering time and spikelets per panicle downstream of Ghd7 using double mutants. Ghd7 is a central regulator that has many downstream targets to regulate multiple traits (Weng et al., 2014), and OsMFT1 is only one of the downstream targets of Ghd7. That is probably why Ami-Ghd7/OX-MFT1 did not fully show the performance of OX-OsMFT1 and exhibited an intermediate phenotype. Both OsLFL1 and GHD7 are transcriptional factors activating OsMFT1 expression at transcriptional level. OsLFL1 directly binds to the promoter of OsMFT1 (Fig. 5), but yeast one-hybrid assay demonstrated that GHD7 didn’t bind to the OsMFT1 promoter (see Supplementary Fig. S9), indicating that GHD7 probably regulates OsMFT1 expression indirectly. Recent studies have revealed that an RY motif in FLC and EUI1 could recruit a complex for gene repression (Yuan et al., 2016; Xie et al., 2018). It is possible that an RY motif in OsMFT1 recruits a protein complex consisting of OsLFL1 and GHD7 for gene activation, or one of the Ghd7 downstream targets also directly binds to the promoter of OsMFT1.
Prolonged branch meristem differentiation caused dense panicles in OsMFT1 overexpression plants
Rice panicle is derived from the inflorescence meristem, and the inflorescence meristem develops branch meristems. The differentiation of spikelet meristems indicates that the branch meristem stops producing branch primordia and terminates to be a spikelet. Here, we found that overexpressing OsMFT1 significantly represses FZP and SEPALLATA-like genes expression. Repression or absence of FZP causes the branch meristem to produce more lateral branch primordia instead of spikelet primordia and overexpression of FZP accelerates the spikelet formation, which results in fewer branches (Komatsu et al., 2003; Bai et al., 2016). The latest reports on FZP have revealed that transcriptional silencer-mediated repression of FZP expression increases spikelet number per panicle (Bai et al., 2017). Class E genes are required for floral determinacy, including the five SEPALLATA-like genes OsMADS1, OsMADS5, OsMADS7, OsMADS8, and OsMADS34. Ectopic expression of OsMADS1 was reported to cause reduced branches and small panicles (Wang et al., 2017). Thus the delay of accumulation of RNA expression of FZP and SEPALLATA-like genes indicates the delay of acquiring spikelet meristem identity. Hence, we proposed OX-OsMFT1 has prolonged branch meristem differentiation, which leads to increased branches and spikelets. Nevertheless, the mechanism of OsMFT1 regulation of FZP and SEPALLATA-like genes is still unknown.
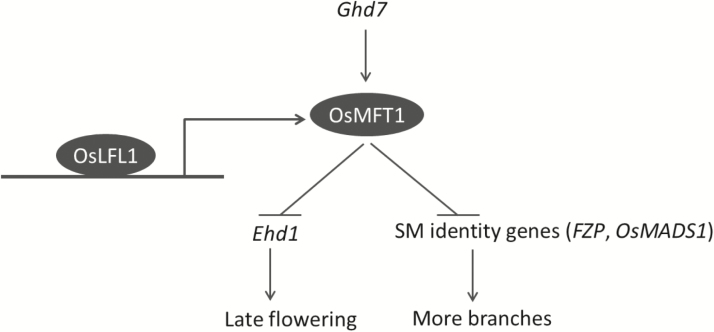
Based on these results, we propose a working model of OsMFT1 (Fig. 7). OsLFL1 protein directly binds to the OsMFT1 promoter and activates OsMFT1 expression. OsMFT1 negatively regulates flowering downstream of Ghd7 and upstream of Ehd1. Meanwhile, OsMFT1 represses the expression of genes for spikelet meristem identity and prolongs branch differentiation, which results in more panicle branches. Hence, OsMFT1 is a suppressor of heading and a positive regulator of panicle architecture in rice.
Fig. 7.
A suggested working model of OsMFT1.
Supplementary data
Supplementary data are available at JXB online
Fig. S1. RNA expression level of partial T0 individuals of OX-OsMFT1.
Fig. S2. The mutation positions and mutation types of three OsMFT1 CRISPR lines.
Fig. S3. RNA expression level comparison of some rice flowering genes.
Fig. S4. Expression of Ghd7 and OsMFT1 in four genotypes from an F2 population.
Fig. S5. Comparison of heading date and panicle architecture among four genotypes.
Fig. S6. Comparison of germination speed between ZH11 and OX-OsMFT1 lines.
Fig. S7. RNA expression level comparison of rice TFL1-like genes between ZH11 and OX-OsMFT1 lines.
Fig. S8. Amino acid alignment of FT-like and TFL1-like protein segments in Arabidopsis and rice.
Fig. S9. Yeast one-hybrid assay of GHD7 and OsMFT1 promoter.
Table S1. Primers used in this study.
Table S2. Differentially expressed genes between ZH11 andOX-OsMFT1 young panicles.
Table S3. Comparison of six important amino acid residuesbetween AtFT, AtTFL1, and OsMFT1.
Acknowledgements
We thank the farm technician Mr J. B. Wang for his excellent work in the field. This work was funded by the National Natural Science Foundation of China (31571751), National Special Program for Research of Transgenic Plant of China (2011ZX08009-001-002) and National Key Research and Development Program of China (2016YFD0100301).
References
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal 25, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J, Guli CL, Yu X, Smyth DR. 1992. terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. The Plant Journal 2, 103–116. [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, et al. . 2005. Cytokinin oxidase regulates rice grain production. Science 309, 741–745. [DOI] [PubMed] [Google Scholar]
- Bai X, Huang Y, Hu Y, Liu H, Zhang B, Smaczniak C, Hu G, Han Z, Xing Y. 2017. Duplication of an upstream silencer of FZP increases grain yield in rice. Nature Plants 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bai XF, Huang Y, Mao DH, Wen M, Zhang L, Xing YZ. 2016. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Scientific Reports 6, 19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. 2007. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany 100, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Damerval C. 2005. Phylogenomic analysis of the PEBP gene family in cereals. Journal of Molecular Evolution 61, 579–590. [DOI] [PubMed] [Google Scholar]
- Chujo A, Komatsu M, Hiratsu K, Ohme-Takagi M, Kyozuka J. 2003. Function analysis of FZP, a rice floral meristem identity gene. Plant and Cell Physiology 44, S133. [Google Scholar]
- Cui R, Han J, Zhao S, et al. . 2010. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). The Plant Journal 61, 767–781. [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV. 2010. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiology 153, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR. 2008. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiology 146, 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. 2004. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes & Development 18, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Sciences, USA 102, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H, Källman T, Lagercrantz U. 2009. Early evolution of the MFT-like gene family in plants. Plant Molecular Biology 70, 359–369. [DOI] [PubMed] [Google Scholar]
- Ho WW, Weigel D. 2014. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. The Plant Cell 26, 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Gao YR, Wei W, Zhang K, Feng JY. 2016. Strawberry MOTHER OF FT AND TFL1 regulates seed germination and post-germination growth through integrating GA and ABA signaling in Arabidopsis. Plant Cell Tissue and Organ Culture 126, 343–352. [Google Scholar]
- Huang NC, Jane WN, Chen J, Yu TS. 2012. Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. The Plant Journal 72, 175–184. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U. 2011. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiology 156, 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant & Cell Physiology 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. 2003. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. 2008. Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. 2009. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. [DOI] [PubMed] [Google Scholar]
- Li Q, Fan CM, Zhang XM, Wang X, Wu FQ, Hu RB, Fu YF. 2014. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS One 9, e99642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teo ZW, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H. 2013. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Developmental Cell 24, 612–622. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mao Z, Sun W. 2015. Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. Journal of Experimental Botany 66, 4781–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. 2002. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. The Plant Journal 29, 743–750. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, et al. . 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell 23, 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Nonoue Y, Yano M, Izawa T. 2016. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. The Plant Journal 86, 221–233. [DOI] [PubMed] [Google Scholar]
- Park J, Lee N, Kim W, Lim S, Choi G. 2011. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. The Plant Cell 23, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. 2008. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. Journal of Plant Physiology 165, 876–885. [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. 2009. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4, e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muino JM, et al. . 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences, USA 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. 2015. Photoperiodic flowering: time measurement mechanisms in leaves. Annual Review of Plant Biology 66, 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. 2007. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano HY. 2013. Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant & Cell Physiology 54, 313–324. [DOI] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R. 2012. Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. The Plant Cell 24, 1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, et al. . 2011. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu H, Weng X, Xie W, Xu C, Li X, Xiao J, Zhang Q. 2014. An expression quantitative trait loci-guided co-expression analysis for constructing regulatory network using a rice recombinant inbred line population. Journal of Experimental Botany 65, 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zeng XQ, Zhuang H, et al. . 2017. Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice. Plant Growth Regulation 81, 433–442. [Google Scholar]
- Weng X, Wang L, Wang J, et al. . 2014. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiology 164, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickland DP, Hanzawa Y. 2015. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Molecular Plant 8, 983–997. [DOI] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. The Plant Cell 22, 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Yang Y. 2013. RNA-guided genome editing in plants using a CRISPR–Cas system. Molecular Plant 6, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhang Y, Han J, et al. . 2018. The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in rice. Molecular Plant 11, 720–735. [DOI] [PubMed] [Google Scholar]
- Xing Y, Zhang Q. 2010. Genetic and molecular bases of rice yield. Annual Review of Plant Biology 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. . 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. 2005. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant & cell physiology 46, 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. . 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH. 2010. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. The Plant Journal 63, 241–253. [DOI] [PubMed] [Google Scholar]
- Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH. 2004. Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Molecules and Cells 17, 95–101. [PubMed] [Google Scholar]
- Yoshida A, Sasao M, Yasuno N, et al. . 2013. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proceedings of the National Academy of Sciences, USA 110, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J, He Y. 2016. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nature Genetics 48, 1527–1534. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yuan Z. 2014. Molecular control of grass inflorescence development. Annual Review of Plant Biology 65, 553–578. [DOI] [PubMed] [Google Scholar]
- Zhang SH, Hu WJ, Wang LP, Lin CF, Cong B, Sun CR, Luo D. 2005. TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Science 168, 1393–1408. [Google Scholar]
- Zhang ZY, Hu W, Shen GJ, Liu HY, Hu Y, Zhou XC, Liu TM, Xing YZ. 2017. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Scientific Reports 7, 5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. 2009. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. The Plant Cell 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.