Abstract
The importance of the rumen microbiota on nutrient cycling to the animal is well recognized; however, our understanding of the influence of the rumen microbiome composition on feed efficiency is limited. The rumen microbiomes of two large animal cohorts (125 heifers and 122 steers) were characterized to identify specific bacterial members (operational taxonomic units [OTUs]) associated with feed efficiency traits (ADFI, ADG, and G:F) in beef cattle. The heifer and steer cohorts were fed a forage-based diet and a concentrate-based diet, respectively. A rumen sample was obtained from each animal via esophageal tubing and bacterial community composition was determined through 16S rRNA gene sequencing of the V4 region. Based on a regression approach that used individual performance measures, animals were classified into divergent feed efficiency groups. Within cohort, an extreme set of 16 animals from these divergent groups was selected as a discovery population to identify differentially abundant OTUs across the rumen bacterial communities. The remaining samples from each cohort were selected to perform forward stepwise regressions using the differentially abundant OTUs as explanatory variables to distinguish predictive OTUs for the feed efficiency traits and to quantify the OTUs collective impact on feed efficiency phenotypes. OTUs belonging to the families Prevotellaceae and Victivallaceae were present across models for heifers, whereas OTUs belonging to the families Prevotellaceae and Lachnospiraceae were present across models for steers. Within the heifer cohort, models explained 19.3%, 25.3%, and 19.8% of the variation for ADFI, ADG, and G:F, respectively. Within the steer cohort, models explained 27.7%, 32.5%, and 26.9% of the variation for ADFI, ADG, and G:F, respectively. Overall, this study suggests a substantial role of the rumen microbiome on feed efficiency responses.
Keywords: beef cattle, feed efficiency, rumen microbiome
INTRODUCTION
The U.S. population is projected to increase 20% by 2050 (United Nations, 2017). Given a per capita beef consumption of 25 kg (ERS and USDA, 2017), an additional production of 1.7 billion kg of beef will be required to meet the future demand. However, compared to the production of pork, chicken, eggs, or milk, production of beef has the most land (27 to 49 m2/kg) and energy (34 to 52 MJ) use and higher global warming potential (14 to 32 kg CO2 equivalents) (de Vries and de Boer, 2010). Thus, beef producers are presented with the challenge of increasing beef supply while maintaining an economically and environmentally sustainable enterprise. As such, to increase beef production in the presence of limited resources, continuous improvement in the animals’ feed efficiency (ability to convert feed to gain) needs to be achieved (Capper, 2011).
To date, most feed efficiency studies in beef cattle have concentrated on the host genomics (Snelling et al., 2011; Lu et al., 2013) and have reported that estimates of heritability for feed efficiency measures are moderate, ranging from 0.06 to 0.62 (Berry and Crowley, 2013). However, further improvements in feed efficiency are needed. With the described role of the rumen microbiome on the nutritional status of the ruminant host (Storm et al., 1983; Bergman, 1990), one area that is poorly explored is the manipulation of the rumen microbiome to improve feed efficiency and animal production. To date, only a few studies have systematically evaluated the influence of the rumen microbiome on feed efficiency (McCann et al., 2014; Myer et al., 2015). These studies have observed shifts in certain phyla, families, and genera across cattle with different feed efficiency phenotypes, yet failed to demonstrate the collective influence of these potentially important ruminal population groups on feed efficiency. Partly, this is due to the small size of the animal populations used.
The main objective of the study was to identify predominant rumen bacterial groups that explained the variation of feed efficiency traits (ADFI, ADG, and G:F) in a large population of beef cattle using linear regression models.
MATERIALS AND METHODS
All procedures used in this study were approved by the U.S. Meat Animal Research Center (USMARC) Animal Care and Use Committee. Data were collected from a cohort of heifers (n = 125) during 2009 and a cohort of steers (n = 122) during 2014. These animals were part of the USMARC Germplasm Evaluation project (GPE) (Schiermiester et al., 2015) and included composite animals with varying percentages of: Angus, Beefmaster, Brahman, Brangus, Braunvieh, Charolais, Chiangus, Gelbvieh, Hereford, Limousin, Maine Anjou, MARC II (composite of ¼ Simmental, ¼ Gelbvieh, ¼ Hereford, and ¼ Angus), MARC III (composite of ¼ Pinzgauer, ¼ Red Poll, ¼ Hereford, and ¼ Angus), Red Angus, Red Angus × Simmental, Romosinuano, Salers, Santa Gertrudis, Shorthorn, and Simmental.
Heifers were fed a growing diet for 84 d comprised of 70% corn silage and 30% alfalfa hay (DM basis) and steers were fed a finishing diet for 78 d comprised of 57.6% dry-rolled corn, 30% wet distillers grains with solubles, 8% alfalfa hay, and 4.4% vitamin and mineral supplement (DM basis). For each animal, individual intake was measured daily using an Insentec Feeding System (Marknesse, The Netherlands). Radio frequency identification tags were placed in the right ear of each animal prior to the experiment. Each pen contained eight electronic feeding stations allowing for the measurement of individual DMI. BW was measured prior to feed delivery on two consecutive days at the beginning and end of the experiment and on 1 d every 3 wk during the experiment. In addition, rumen samples were collected via esophageal tubing approximately 14 d prior to breeding (14 mo of age) for heifers and approximately 30 d prior to shipment to the commercial abattoir for harvest for steers. Collection of rumen samples was spread over 3 d and done from 0730 to 0930 h. Following collection, rumen samples were snap-frozen in liquid nitrogen and stored at −80 °C until used for DNA extraction. A study by Paz et al. (2016) reported the microbial community composition of samples collected via esophageal tubing with addition of particles retained in the strainer to be similar to samples collected via rumen fistula. Hence, the samples collected herein adequately represented the microbial community within each animal.
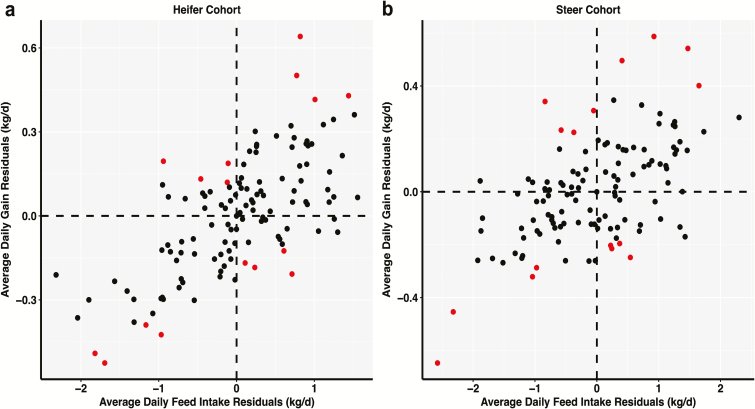
At the end of the feeding period, ADFI and ADG were calculated for each animal. ADFI was calculated by summing the total DMI for each animal over the entire period and dividing by days on the study and ADG was calculated by regressing BW gain on days on feed. Gain-to-feed was calculated as ADG divided by ADFI. Breed composition of all animals was estimated via a multi-generational pedigree. Within cohort, a linear model with breed fractions fitted as covariates was employed for both ADFI and ADG to obtain the residuals that were used as the corrected phenotypes for further analysis. This was performed to account for the inherent breed differences in ADFI and ADG (Schenkel et al., 2011). Preliminary evaluation showed overall bacterial community composition differed between heifer and steer cohorts (permutational multivariate analysis of variance [PERMANOVA], P < 0.001; Figure 1 and Supplementary Figure S1). Within cohort, boxplots were created using R v.3.3.1 (R Core Team, 2017) to screen outliers (1.5 times the interquartile range above the third quartile or below the first quartile) from the residuals of ADFI and ADG (Supplementary Figure S2). Two residual observations of ADG (one from each cohort) were classified as outliers and excluded from further analyses. Classification of animals into divergent feed efficiency groups was performed as described by Myer et al. (2015) with the exception of using residuals instead of observations that had not been corrected for fixed effects. Residuals of ADG were regressed on residuals of ADFI and quadrants were created by subdividing the axes where both ADG and ADFI reached zero (Figure 2). This approach resulted in four feed efficiency groups (represented by each Cartesian quadrant) from the combination of the two levels of ADG (high and low) and ADFI (high and low). The four feed efficiency quadrants were high ADG and high ADFI (ADGH–ADFIH), high ADG and low ADFI (ADGH–ADFIL), low ADG and high ADFI (ADGL–ADFIH), and low ADG and low ADFI (ADGL–ADFIL). The four most extreme animals from each quadrant (n = 16 animals/cohort) were selected (Figure 2) and used as the discovery population to detect differentially abundant features of the microbiome that influence feed efficiency traits. The selection of four extreme animals from each quadrant for a total extreme population of 16 animals was similar to the strategy employed by Myer et al. (2015). The remaining samples (n = 109 for heifers and n = 106 for steers) were used to develop and test linear regression models to predict ADG, ADFI, and G:F.
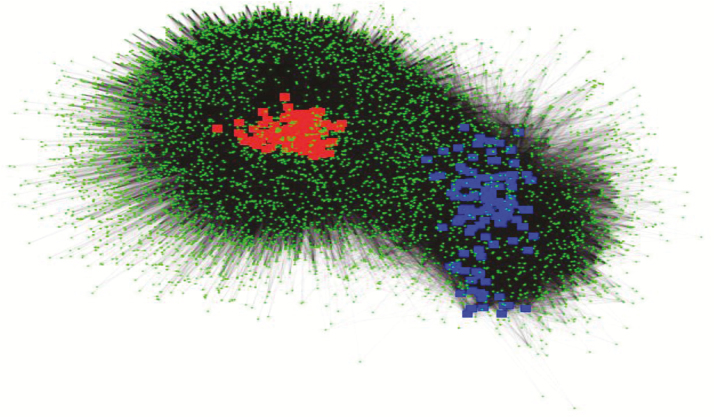
Figure 1.
Bipartite network showing significant (PERMANOVA, P < 0.001) difference in bacterial community composition between heifer (red squares) and steer (blue squares) cohorts. Green circles represent the distribution of OTUs.
Figure 2.
Discovery population sampling method. Within heifer (n = 125) and steer (n = 122) cohorts, linear models with breed fractions fitted as covariates were performed for ADFI and ADG and residuals were extracted. Residuals of ADG were regressed on residuals of ADFI. Each Cartesian quadrant represented a feed efficiency group from the combination of the two levels of ADG (high and low) and ADFI (high and low). A subsample of four animals (red circles) from each quadrant was selected for a total of 16 animals for both the (a) heifer and (b) steer cohorts.
Phenotyping the Rumen Bacterial Community
DNA extraction, library preparation, and sequencing.
Total DNA was extracted from the rumen samples (0.25 g) using the PowerMag Soil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s protocol with the modifications described below. During cell lysis, two bead-beating steps were performed in a TissueLyser (Qiagen Inc., Valencia, CA, USA) for 3 min at 30 Hz and samples were incubated in a 95 °C water bath for 5 min between the two bead-beading steps to ensure cell lysis. Following the removal of PCR inhibitors, nucleic acids were precipitated similar to the procedure describe by Yu and Morrison (2004). Briefly, 850 µL of sample supernatant and 260 μL of sodium acetate (10 mM) were mixed in 1.5 mL Eppendorf tubes, vortexed, and incubated on ice for 5 min followed by a centrifugation at 16,000 × g for 15 min at 4 °C. One volume (650 µL) of supernatant was mixed with one volume of isopropanol and incubated on ice for 30 min followed by a centrifugation at 16,000 × g for 15 min at 4 °C. The supernatant was discarded and the nucleic acid pellet was wash with ice-cold ethanol (70%). The wash was discarded and the nucleic acid pellet was dried under vacuum for 3 min and then dissolved in 450 µL of Tris (10 mM, pH 8).
Amplicon libraries of the 16S rRNA gene (V4 region) were prepared as described by Kozich et al. (2013). Briefly, each 20 μL PCR amplification reaction contained 0.5 μL Terra PCR Direct Polymerase Mix (0.625 Units), 7.5 μL nuclease-free, sterile water, 10 μL 2× Terra PCR Direct Buffer, 1 μL indexed fusion primers (10 μM), and 1 μL DNA (20 to 70 ng DNA). The cycling conditions included an initial denaturation of 98 °C for 3 min, followed by 25 cycles of 98 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s; and a final extension of 68 °C for 4 min. Following amplification, PCR products from each sample were normalized (1 to 2 ng/µL) using the SequalPrep Normalization Plate Kit (Invitrogen, Carlsbad, CA, USA) as described by the manufacturer. The normalized libraries were pooled (10 µL/sample) and purified using the MinElute PCR Purification Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s protocol. The resulting concentrated samples were size selected using the Pippin Prep (Sage Science, Inc., Beverly, MA, USA) automated size selection instrument using 1.5% agarose gel cassettes. The resulting libraries were quality controlled using the Agilent BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and quantified using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The resulting libraries were sequenced using the Illumina Miseq System (Illumina, San Diego, CA, USA) using the V2 500 cycles kit according to the manufacturer’s protocol. Raw sequences have been deposited at the NCBI Sequence Read Archive (SRA) under the accession no. SRP100776.
Data processing.
Detailed information about the bioinformatics pipeline to reproduce the analyses described in this study is available at https://github.com/FernandoLab/2017_RumenMicrobiome_Beef. Assembly of contigs and subsequent quality filtering including removal of sequences with ambiguous bases, incorrect length, or improperly assembled were performed using MOTHUR v.1.38.1 (Schloss et al., 2009). Quality-filtered sequences were clustered into operational taxonomic units (OTUs) using the UPARSE pipeline (USEARCH v7.0.1090) (Edgar, 2013). Clustering steps included dereplication, sorting by cluster size (descending and not retaining singletons), mapping sequences to OTUs at a 97% identity, and filtering of chimeric sequences using UCHIME (Edgar et al., 2011) with ChimeraSlayer gold.fa as the reference database. Representative OTU sequences were aligned against the SILVA reference alignment database v123 to identify OTUs that mapped to the V4 region. Sequences that did not align correctly were discarded to ensure all sequences overlapped the V4 region. The resulting alignment was used to construct a phylogenetic tree using Clearcut (Sheneman et al., 2006). Representative sequences were assigned taxonomy using QIIME v.1.9.1 (Caporaso et al., 2010) with assignments done as described in MOTHUR (Schloss et al., 2009) using a Naive Bayes classifier similar to the RDP Classifier (Wang et al., 2007), using the Greengenes database (gg_13_8_otus) (McDonald et al., 2012) reference sequences. OTUs classified as Archaea and Cyanobacteria were removed from the data set. The primers used to amplify the bacterial community are not designed to amplify all Archaea from the rumen and thus the generated data may be misleading on Archaea distribution. Cyanobacteria were present in very low abundance across samples (averaged 0.006% of total quality-filtered sequences) and these sequences were removed as they likely represented plant chloroplast contamination (Giovannoni et al., 1988) and were assessed to have no impact on the feed efficiency traits investigated in this study. However, it is noteworthy that members of the orders YS2, SM1D11, and mle1-12 from the phyla Cyanobacteria have been proposed to be a new class (Soo et al., 2014) or a completely new phylum termed Melainabacteria (Di Rienzi et al., 2013) and have been reported to be present in the gut of mammals, plants, and soil (Di Rienzi et al., 2013; McGorum et al., 2015). These taxa were not found within the cyanobacterial reads. Alpha metrics were used to describe bacterial richness (observed OTUs), diversity (Shannon–Weiner index [logarithm base 2]) (Shannon and Weaver, 1949), and dominance (1-Simpson index). Observed OTUs were also used to construct feed efficiency quadrant-based rarefaction curves. To reduce the intrinsic effect of animal-to-animal variation in rumen bacterial community composition, a core measurable microbiome (CMM) was defined as OTUs that were present in all four selected animals within each feed efficiency quadrant in the discovery population.
Statistical analyses.
OTU tables were rarefied based on sequencing depth, where the lowest depth of 9,081 reads for the heifer cohort and of 12,430 reads for the steer cohort was used. Rarefaction was performed using QIIME v.1.9.1 (Caporaso et al., 2010) implementing the Mersenne Twister pseudo-random number generator. Within cohort, diversity indices and statistical comparisons of the CMM across feed efficiency quadrants were conducted on the discovery population. Alpha diversity metrics were compared using a nonparametric (Monte Carlo permutations to calculate P-value) two-sample t-test with multiple comparisons corrected for false discovery rate (Benjamini and Hochberg, 1995). The Good’s coverage (Good, 1953) was calculated to evaluate adequate sampling depth. Overall CMM differences across feed efficiency quadrants were evaluated in R (R Core Team, 2017) (adonis function vegan package [Oksanen et al., 2017]) using the weighted UniFrac distance matrix as an input for PERMANOVA using the feed efficiency quadrant as the main effect. Pairwise comparisons of CMM across feed efficiency quadrants were tested with the linear discriminatory analysis (LDA) effect size (LefSe) (Segata et al., 2011) to identify differentially abundant OTUs/bacterial features among the feed efficiency quadrants. LefSe was executed using default parameters with an alpha value of 0.05 for the factorial Kruskal–Wallis test among classes and a threshold of LDA score of 2.0 for discriminative features. For each cohort, the top 10 (highest LDA scores) significant differentially abundant OTUs in each comparison were identified for downstream analysis.
Regression models.
Within cohort, differentially abundant OTUs identified by LefSe were assessed as potential microbial features predictive of ADFI, ADG, and G:F using the test population. Data were transformed using an arcsine square root function and feature selection was performed using forward stepwise regressions to identify subsets of predictive OTUs for each trait. Akaike’s information criteria (AIC) were used to select the final models and significance of predictive OTUs was declared at P ≤ 0.10. For each model, colinearity of the independent variables (variance inflation factor) was evaluated. Additionally, assumptions of linearity (observed vs. predicted values plot) (Piñeiro et al., 2008), normality (quantile–quantile plot), and homoscedasticity (residuals vs. fitted values plot) were evaluated. To evaluate model accuracy, heifer data were used to assess the steer model and in turn the heifer model was assessed using the steer data. In addition to OTU-based models, taxa-based models at the family level were assessed to predict ADG, ADFI, and G:F. Within cohort, the CMM across feed efficiency quadrants was summarized at the family level and pairwise comparisons and regression models were performed as described for OTU-based models.
Predicting functional profile from model selected bacterial features.
The online phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) (Langille et al., 2013) method (v1.1.1) available at http://galaxy.morganlangille.com/ was used to predict function based on 16S rRNA gene data. OTUs selected across feed efficiency models from both the heifer and steer cohorts were filtered from the representative OTUs sequences file generated from the UPARSE pipeline. Then the biom-formatted OTU table was generated by close reference picking against the Greengenes database (default gg_13_5) followed by normalization by copy number and metagenome predictions using the KEGG Orthologs option.
RESULTS
A total of 9,281,130 quality-filtered sequences were generated across the two cohorts. Before rarefying samples (Supplementary Figure S3) within cohort to similar sequence depth, the heifer discovery set (16 animals) included 541,804 quality-filtered sequences and the steer discovery (16 animals) set included 828,950 quality-filtered sequences. To determine if sampling effort adequately represented the rumen bacterial communities across feed efficiency quadrants, Good’s coverages were calculated and demonstrated that the sampling depth obtained for the heifer population represented 93.9% to 94.5% of the total bacterial community. For steers, the Good’s coverages predicted that 98.6% to 98.9% of the bacterial populations were represented suggesting that adequate sampling depth was obtained to evaluate the bacterial community composition. Additionally, rumen samples were collected via esophageal tubing (Paz et al. 2016) after more than 100 d of diet adaptation where the microbial community was expected to be adapted and stable at sampling time based on previous reports (Anderson et al., 2016).
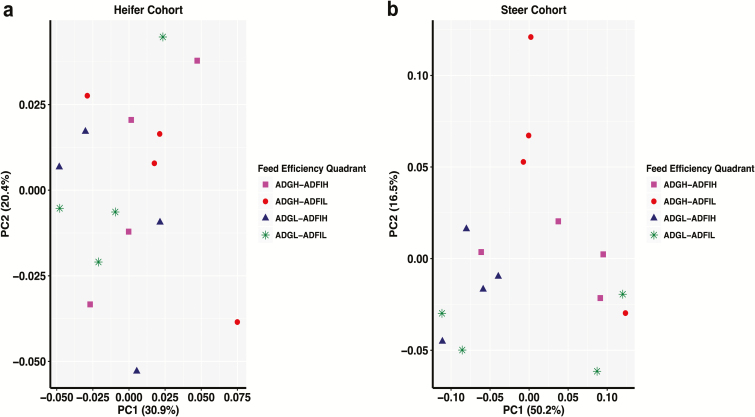
Alpha metrics for richness (P ≥ 0.83), diversity (P ≥ 0.40), and dominance (P ≥ 0.35) were similar across feed efficiency quadrants for both heifers and steers (Supplementary Figure S4). Bacteriodetes, Firmicutes, and Proteobacteria were the most abundant phyla and combined accounted for 85.9% and 94.8% of the total reads for heifers (Supplementary Figure S5) and steers (Supplementary Figure S6), respectively. Additional phyla (relative abundance > 1%) included Fibrobacteres, Tenericutes, and Verrucomicrobia for heifers and phylum Spirochaetes for both heifers and steers. Phyla composition was more variable across feed efficiency quadrants from the steer cohort compared to the heifer cohort. Substantial inter-animal variation in the rumen microbiome composition has been reported (Hernandez-Sanabria et al., 2010; Jami and Mizrahi, 2012). To reduce animal-to-animal variation, a CMM was defined for each feed efficiency quadrant. For heifers, the CMM was composed of 503, 433, 445, and 444 OTUs for the ADGH–ADFIH, ADGH–ADFIL, ADGL–ADFIH, ADGL–ADFIL feed efficiency quadrant, respectively. The overall CMM for the heifer cohort, resulting from the combined and unique OTUs across the feed efficiency quadrants, was composed of 777 OTUs (23.3% of total OTUs), which represented 88.4% of the rarefied quality-filtered reads. For steers, the CMM was composed of 147, 124, 143, and 77 OTUs for the ADGH–ADFIH, ADGH–ADFIL, ADGL–ADFIH, ADGL–ADFIL feed efficiency quadrant, respectively. The overall CMM for the steer cohort was composed of 240 OTUs (15.2% of total OTUs) which represented 82.1% of the rarefied quality-filtered reads. Bacterial communities did not cluster by feed efficiency quadrant in the principal coordinates analysis (PCoA) plots for both heifer and steer cohorts (Figure 3). PERMANOVA supported no overall bacterial community composition differences across feed efficiency quadrants within heifer (P = 0.64) and steer (P = 0.16) cohorts.
Figure 3.
Principal coordinates analysis (PCoA) using the weighted UniFrac distance matrix displaying no structuring of bacterial communities by feed efficiency quadrant for (a) heifer and (b) steer cohorts.
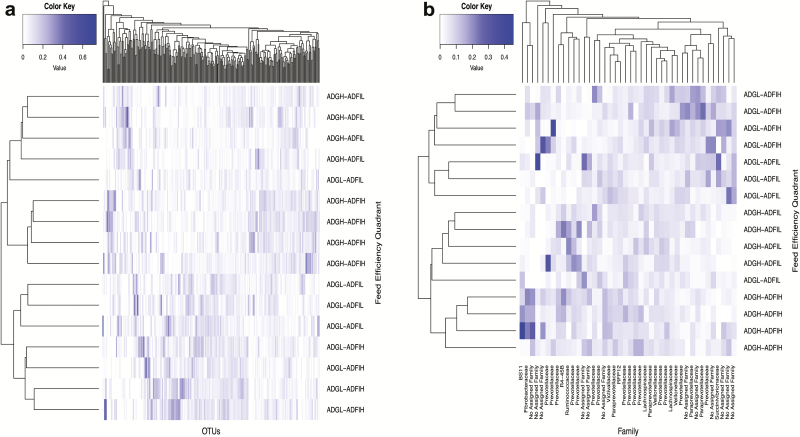
To further investigate potential rumen bacterial community differences across feed efficiency quadrants, differentially abundant OTUs across the CMM were identified using the LefSe algorithm. A total of 259 and 98 significant differentially abundant OTUs with LDA scores ≥ 2 were identified across pairwise comparisons of the feed efficiency quadrants for the heifer (Figure 4a) and steer (Figure 5a) cohorts, respectively. Overall, differentially abundant OTUs were distinctive between cohorts (Supplementary Figure S7) with only six differentially abundant OTUs in common among some samples from both cohorts. Three of the shared differentially abundant OTUs belonged to the family Lachnospiraceae and the remaining belonged to the families Paraprevotellaceae, Prevotellaceae, and Veillonellaceae. Subsets of 42 and 47 uniquely differentially abundant OTUs among heifers (Figure 4b) and steers (Figure 5b) were identified, respectively, for subsequent analysis to identify features of the microbiome that influence ADFI, ADG, and G:F using a forward stepwise regression approach.
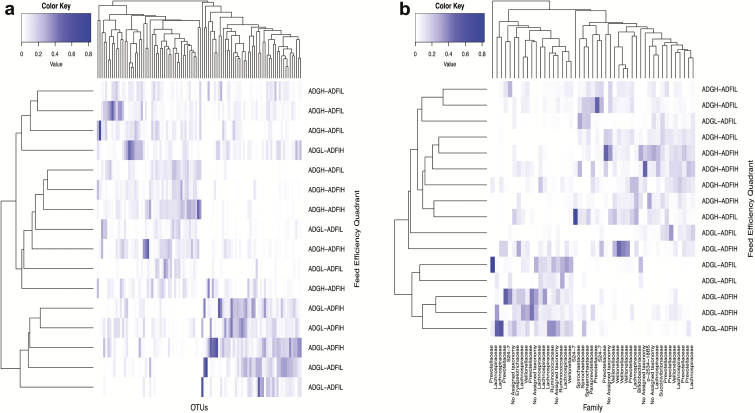
Figure 4.
Average linkage hierarchical clustering based on Bray–Curtis dissimilarity of differentially abundant OTUs identified through pairwise comparisons of the CMM across all feed efficiency quadrants within heifer cohort. (a) All differentially abundant OTUs across comparisons and (b) selected differentially abundant OTUs used in forward stepwise regression analysis to identify predictive OTUs for feed efficiency traits.
Figure 5.
Average linkage hierarchical clustering based on Bray–Curtis dissimilarity of differentially abundant OTUs identified through pairwise comparisons of the CMM across all feed efficiency quadrants within steer cohort. (a) All differentially abundant OTUs across comparisons and (b) selected differentially abundant OTUs used in forward stepwise regression analysis to identify predictive OTUs for feed efficiency traits.
Rumen Bacterial Features Affecting ADFI, ADG, and G:F
Final models for predicting ADFI, ADG, and G:F in heifer and steer cohorts are presented in Tables 1 and 2, respectively. Diagnostic plots did not expose patterns or deviations from normality in the distribution of the residuals.
Table 1.
Final linear models constructed using forward stepwise regression for predicting ADFI, ADG, and G:F for the heifer cohort
| Trait | Predictor | Coefficient | SEa | t-statistic | P-value | AICb | R 2c | Taxonomyd |
|---|---|---|---|---|---|---|---|---|
| ADFI | Intercept | −1.5397 | 0.6206 | −2.481 | 0.014813 | −53.73 | 0.1933 | |
| OTU233 | −13.4688 | 4.9111 | −2.743 | 0.007249 | Order Bacteroidales | |||
| OTU6532 | 32.1372 | 8.6851 | 3.700 | 0.000356 | Order Bacteroidales | |||
| OTU257 | 9.0497 | 8.7212 | 1.038 | 0.301976 | Paraprevotellaceae | |||
| OTU2045 | −23.0393 | 9.5493 | −2.413 | 0.017696 | Prevotellaceae | |||
| OTU125 | 13.8999 | 6.4527 | 2.154 | 0.033682 | Victivallaceae | |||
| OTU517 | 14.6939 | 7.6863 | 1.912 | 0.058836 | Ruminococcaceae | |||
| OTU5323 | 6.6179 | 4.6455 | 1.425 | 0.157453 | Prevotellaceae | |||
| OTU139 | 4.0825 | 2.5744 | 1.586 | 0.116011 | BS11 | |||
| OTU216 | 10.3417 | 6.8966 | 1.500 | 0.136951 | Prevotellaceae | |||
| OTU5133 | 13.4651 | 9.9425 | 1.354 | 0.178757 | Order Clostridiales | |||
| ADG | Intercept | −0.07717 | 0.08848 | −0.872 | 0.385160 | −402.03 | 0.2526 | |
| OTU233 | −4.33233 | 0.95934 | −4.516 | 0.000017 | Order Bacteroidales | |||
| OTU139 | 1.50292 | 0.51951 | 2.893 | 0.004666 | BS11 | |||
| OTU6532 | 6.09776 | 1.73445 | 3.516 | 0.000656 | Order Bacteroidales | |||
| OTU125 | 3.78944 | 1.30628 | 2.901 | 0.004558 | Victivallaceae | |||
| OTU2045 | −5.17327 | 1.89001 | −2.737 | 0.007313 | Prevotellaceae | |||
| OTU89 | 2.14906 | 1.20975 | 1.776 | 0.078640 | Prevotellaceae | |||
| G:F | Intercept | 0.004628 | 0.008792 | 0.526 | 0.599771 | −874.92 | 0.1979 | |
| OTU233 | −0.435453 | 0.109636 | −3.972 | 0.000133 | Order Bacteroidales | |||
| OTU139 | 0.145833 | 0.058925 | 2.475 | 0.014976 | BS11 | |||
| OTU125 | 0.503240 | 0.173214 | 2.905 | 0.004500 | Victivallaceae | |||
| OTU6532 | 0.497373 | 0.198486 | 2.506 | 0.013798 | Order Bacteroidales | |||
| OTU2045 | −0.495401 | 0.217485 | −2.278 | 0.024821 | Prevotellaceae | |||
| OTU4675 | −0.381987 | 0.215910 | −1.769 | 0.079850 | Fibrobacteraceae |
aStandard error.
bAkaike information criteria.
cAdjusted R-squared.
dFamily level classification, unless otherwise specified.
Table 2.
Final linear models constructed using forward stepwise regression for predicting ADFI, ADG, and G:F for the steer cohort
| Trait | Predictor | Coefficient | SEa | t-statistic | P-value | AICb | R 2c | Familyd |
|---|---|---|---|---|---|---|---|---|
| ADFI | Intercept | −0.009771 | 0.330669 | −0.030 | 0.97649 | −28.42 | 0.2766 | |
| OTU3879 | 11.920275 | 5.015395 | 2.377 | 0.01949 | Veillonellaceae | |||
| OTU103 | −6.724807 | 2.719246 | −2.473 | 0.01519 | S24-7 | |||
| OTU88 | −7.112484 | 2.603974 | −2.731 | 0.00753 | Lachnospiraceae | |||
| OTU50 | −8.319334 | 2.474744 | −3.362 | 0.00112 | Order Bacteroidales | |||
| OTU25 | 3.368074 | 1.240464 | 2.715 | 0.00788 | Paraprevotellaceae | |||
| OTU252 | 12.699986 | 4.035271 | 3.147 | 0.00221 | Bifidobacteriaceae | |||
| OTU301 | −22.733747 | 7.482761 | −3.038 | 0.00308 | Lachnospiraceae | |||
| OTU1874 | 9.573695 | 3.494031 | 2.740 | 0.00735 | Lachnospiraceae | |||
| OTU41 | 3.467929 | 1.793902 | 1.933 | 0.05622 | Prevotellaceae | |||
| OTU3670 | −22.559447 | 13.208508 | −1.708 | 0.09095 | Veillonellaceae | |||
| OTU2441 | 13.008597 | 8.933797 | 1.456 | 0.14869 | Veillonellaceae | |||
| ADG | Intercept | −0.15129 | 0.07448 | −2.031 | 0.045166 | −421.2 | 0.3253 | |
| OTU3081 | −1.24675 | 0.36013 | −3.462 | 0.000822 | Prevotellaceae | |||
| OTU17 | 1.41205 | 0.46712 | 3.023 | 0.003261 | Lachnospiraceae | |||
| OTU14 | 1.55317 | 0.37485 | 4.143 | 0.0000771 | Ruminococcaceae | |||
| OTU301 | −5.60318 | 1.45543 | −3.850 | 0.000221 | Lachnospiraceae | |||
| OTU2441 | 0.81738 | 1.37135 | 0.596 | 0.552646 | Veillonellaceae | |||
| OTU60 | 1.21132 | 0.62366 | 1.942 | 0.055231 | Prevotellaceae | |||
| OTU65 | 0.94873 | 0.34944 | 2.715 | 0.007945 | S24-7 | |||
| OTU87 | −0.76844 | 0.39937 | −1.924 | 0.057496 | Prevotellaceae | |||
| OTU9 | 0.23146 | 0.14044 | 1.648 | 0.102815 | S24-7 | |||
| OTU218 | −1.41796 | 0.65253 | −2.173 | 0.032405 | Unclassified | |||
| OTU50 | −0.73642 | 0.36059 | −2.042 | 0.044055 | Order Bacteroidales | |||
| OTU227 | 1.55665 | 0.84442 | 1.843 | 0.068555 | Lachnospiraceae | |||
| OTU738 | −5.74874 | 2.56477 | −2.241 | 0.027457 | Erysipelotrichaceae | |||
| OTU3879 | 0.69638 | 0.37874 | 1.839 | 0.069264 | Veillonellaceae | |||
| OTU36 | 0.81395 | 0.54440 | 1.495 | 0.138377 | Ruminococcaceae | |||
| G:F | Intercept | 0.026852 | 0.006685 | 4.017 | 0.000119 | −860.39 | 0.2691 | |
| OTU41 | −0.145112 | 0.039828 | −3.644 | 0.000440 | Prevotellaceae | |||
| OTU60 | 0.168961 | 0.078669 | 2.148 | 0.034305 | Prevotellaceae | |||
| OTU12 | −0.035520 | 0.021140 | −1.680 | 0.096228 | Prevotellaceae | |||
| OTU4409 | −0.454518 | 0.202494 | −2.245 | 0.027140 | Spirochaetaceae | |||
| OTU103 | 0.159020 | 0.055477 | 2.866 | 0.005123 | S24-7 | |||
| OTU25 | −0.042017 | 0.022642 | −1.856 | 0.066632 | Paraprevotellaceae | |||
| OTU3879 | −0.054002 | 0.028532 | −1.893 | 0.061482 | Veillonellaceae | |||
| OTU218 | −0.134513 | 0.080154 | −1.678 | 0.096633 | Unclassified | |||
| OTU3081 | −0.093784 | 0.044315 | −2.116 | 0.036961 | Prevotellaceae | |||
| OTU48 | −0.139982 | 0.067582 | −2.071 | 0.041072 | Lachnospiraceae | |||
| OTU168 | 0.147043 | 0.101650 | 1.447 | 0.151346 | Order Bacteroidales |
aStandard error.
bAkaike information criteria.
cAdjusted R-squared.
dFamily level classification, unless otherwise specified.
ADFI Models
The model for heifers explained 19.3% of the variation in breed-corrected ADFI. OTUs belonging to the families Ruminococcaceae, Victivallaceae, and an unclassified OTU belonging to the order Bacteroidales were associated with an increase in ADFI. In contrast, an OTU belonging to the family Prevotellaceae and an unclassified OTU belonging to the order Bacteroidales were associated with a decrease in ADFI.
The model for steers explained 27.7% of the variation in breed-corrected ADFI. OTUs belonging to the families Bifidobacteriaceae, Lachnospiraceae, Paraprevotellaceae, Prevotellaceae, and Veillonellaceae were associated with an increase in ADFI, whereas OTUs belonging to the families Lachnospiraceae, S24-7, Veillonellaceae, and an unclassified OTU belonging to the order Bacteroidales were associated with a decrease in ADFI.
ADG Models
Models explained 25.3% and 32.5% of the variation in breed-corrected ADG for heifers and steers, respectively. Five out of the six OTUs in the ADG model were shared with the ADFI model in heifers. The remaining OTU was of the Prevotellaceae family, which was indicative of an increase in ADG. For steers, the ADG model shared only two OTUs with the ADFI model. Additionally, the ADG model consisted of OTUs with positive coefficients, which included Lachnospiraceae, Prevotellaceae, Ruminococcaceae, S24-7, and Veillonellaceae families and OTUs with negative coefficients, which included Erysipelotrichaceae, Lachnospiraceae, and Prevotellaceae families and an unclassified OTU from the order Bacteroidales. Furthermore, we also identified an OTU that had no taxonomic classification beyond kingdom bacteria.
G:F Models
The model of G:F for heifers accounted for 19.8% of the variation. The five OTUs shared between the ADFI and ADG models were also found in the G:F model. In addition, an OTU of the Fibrobacteraceae family with negative coefficient was identified. The model of G:F for steers accounted for 26.9% of the variation. The model shared four OTUs with the ADFI model and four OTUs with the ADG model. The model also included OTUs belonging to Lachnospiraceae, Prevotellaceae, and Spirochaetaceae families which were associated with a decrease in G:F.
Taxa-Based Models at the Family Level
Compared to OTU-based models, models at the family level explained less of the variation in ADFI, ADG, and G:F in both heifers and steers (Supplementary Tables S1 and S2). For heifers, models explained 7.79%, 12.0%, and 14.2% of the variation in ADFI, ADG, and G:F, respectively. For steers, models explained 11.8%, 6.43%, and 8.80% of the variation in ADFI, ADG, and G:F, respectively.
Predicting the Functional Role of Bacterial Features in the Models
To gain insight of the functions from the bacterial OTUs identified in the feed efficiency models and how they potentially influence feed efficiency, PICRUSt was used to predict functional features. Within heifers, bacterial OTUs (89, 125, 139, 233, and 6532) identified across feed efficiency models were predicted to have functional categories related to glycolysis and gluconeogenesis, glycan degradation, protein degradation (peptidases), nitrogen metabolism, and biosynthesis of essential AA such as lysine, valine, leucine, or isoleucine. Similarly, within steers, bacterial OTUs (14, 17, 60, 65, 87, 227, 301, 738) identified across feed efficiency models were predicted to have functional categories related to glycolysis and gluconeogenesis, glycan degradation, protein degradation (peptidases), and starch and sucrose metabolism. Interestingly, a majority of the OTUs identified through the regression models were predicted to have higher number of transporters. Further investigation of the distribution of transporters revealed a numerically higher association with positive coefficient OTUs compared to negative coefficient OTUs (Supplementary Figure S8).
DISCUSSION
Feed efficiency is an economically important trait for sustainable beef production. Multiple factors such as nutrition and management practices (de Ondarza and Tricarico, 2017), genetics, and physiological mechanisms (Herd and Arthur, 2009) influence feed efficiency responses. Moreover, the rumen microbial community mediates energy available to the animal through pregastric fermentation, which suggests a role in feed efficiency. Since bacteria are the prevalent microorganism in the rumen (1011 viable cells/g rumen content) (Mackie et al., 2001), we evaluated the rumen bacterial community composition to investigate its influence on feed efficiency.
Microbial Community Composition in the Steer and Heifer Cohorts
Across feed efficiency quadrants for both cohorts (steer and heifer), no differences in rumen bacterial richness and diversity were observed. Similar observations in beef cattle have been previously reported when evaluating different variable regions (V1–V3 [Myer et al., 2015] and V4–V6 [McCann et al., 2014]). The overall bacterial community composition was significantly different between heifer and steer cohorts (PERMANOVA, P < 0.001; Figure 1). This difference in rumen bacterial community composition is confounded by diet, gender, and time. Therefore, in our subsequent analyses and interpretations, we analyzed and described the cohorts independently. The main phyla identified in both cohorts included Bacteriodetes, Firmicutes, and Proteobacteria. These phyla have been observed to be predominant in beef cattle fed either high-forage or high-concentrate diets (Petri et al., 2013; McCann et al., 2014; Myer et al., 2015). Comparable to other studies (McCann et al., 2014; Myer et al., 2015), the overall rumen bacterial community composition was similar across feed efficiency quadrants within cohorts (PERMANOVA, P ≥ 0.16; Figure 3). A greater number of significant OTUs were observed in heifers fed a forage-based diet compared to steers fed a grain-based diet. This was not surprising as dietary increase of highly fermentable substrates has been observed to decrease rumen microbial diversity as microbes that more efficiently utilize these substrates dominate the microbial community structure (Fernando et al., 2010).
To investigate the role of the rumen microbiome on feed efficiency, we evaluated differences in the rumen bacterial community using defined feed efficiency phenotypes based on ADFI and ADG. To this end, a discovery population within each cohort was used to define a CMM for each feed efficiency quadrant. Then, differentially abundant OTUs that potentially described each feed efficiency phenotype were identified (see Materials and Methods). In heifers, 10 OTUs were included in the ADFI model. Among the 10 OTUs identified, five OTUs were significantly associated with ADFI and mainly (3/5) belonged to the order Bacteroidales (Table 1). Out of the three OTUs belonging to Bacteroidales, only one was classified beyond order and belonged to the Prevotellaceae family. The remaining two significant OTUs belonged to the Ruminococcaceae and Victivallaceae families. Ruminococcaceae members are well known to be present in the rumen (Russell et al., 2009) and to possess cellulolytic activity (White et al., 1993). Additionally, the family Victivallaceae has been isolated from human feces and has been shown to ferment cellobiose (Zoetendal et al., 2003). The observation of bacterial members related to fiber degradation influencing ADFI in the heifer cohort is not surprising given that the diet fed was composed exclusively of corn silage and alfalfa hay. In steers, half (5/10) of the OTUs with a significant effect on ADFI belonged to the order Clostridiales and included microbes belonging to families Lachnospiraceae and Veillonellaceae (Table 2). Myer et al. (2015) found an OTU of the Veillonellaceae family and an OTU of the Clostridiales order to be associated with ADFI. Four of the remaining significant OTUs belonged to the order Bacteroidales and included members of the families S24-7, Paraprevotellaceae, and Prevotellaceae. In addition, Bifidobacteriaceae was identified to be associated with ADFI. Interestingly, although not the same OTU, Prevotellaceae was significantly associated with ADFI in both heifers and steers suggesting that members of this predominant family may be associated with ADFI independent of diet.
The ADG model for the heifer cohort included six significant OTUs (Table 1). Five OTUs belonged to the order Bacteroidales (BS11, Prevotellaceae, and unclassified families) and one OTU belonged to the order Victivallales (Victivallaceae family). For steers, the ADG model included 13 significant OTUs from families belonging to the orders Bacteroidales (six OTUs), Clostridiales (five OTUs), and Erysipelotrichales (one OTU). Taxonomic analysis revealed that in both heifer and steer cohorts, members of the Prevotellaceae family were classified as Prevotella at the genus level. Within the rumen microbiome, Prevotella is a dominant bacterial genus (Stevenson and Weimer, 2007) with roles in the digestion of polysaccharides (Matsui et al., 2000) and protein (Wallace, 1996). Prevotella represented ~28.5% of the rarefied quality-filtered reads in both heifer and steer cohorts. In steers, OTUs of the Lachnospiraceae were classified as Butyrivibrio at the genus level. Butyrivibrio species have hemicellulolytic, proteolytic, and uricolytic activities (Cotta and Hespell, 1986; Kelly et al., 2010). The ability to break the aforementioned compounds paralleled the dietary supply, as the diet contained a high concentration of wet distillers grains with solubles, a feed composed of mainly protein, fiber, and fat (Klopfenstein et al., 2007). Associations between ADG and members of the families Lachnospiraceae, Prevotellaceae, Veillonellaceae, and Victivallaceae have previously been observed in beef cattle (Myer et al., 2015). For steers, the taxa-based model (Supplementary Table S1) for ADG included the Lachnospiraceae family supporting an important role of this family on ADG when feeding high-concentrate diets.
The G:F model included six significant OTUs for the heifer cohort (Table 1). Families belonged to the orders Bacteroidales (four OTUs), Fibrobacterales (one OTU), and Victivallales (one OTU). All the OTUs in the G:F model that were shared with the ADFI and ADG models kept the direction of their effect. For instance, if an OTU had a positive coefficient in either ADFI or ADG, it also had a positive coefficient on G:F. For the steer cohort, the G:F model included 10 significant OTUs mainly (four OTUs) of the Prevotellaceae family. None of the four OTUs shared between the G:F and ADFI models had similar direction of their effects, whereas three out the four OTUs shared between the G:F and ADG models had the same direction of their effects and belonged to Prevotellaceae and unclassified families. McCann et al. (2014) identified an OTU of the order Bacteroidales to be associated with more efficient steers (negative residual feed intake) in grazing conditions. However, Shabat et al. (2016) found members of the order Bacteroidales to be more abundant in inefficient (positive residual feed intake) dairy cows. Interestingly, in the previously mentioned study, only 2 out 18 differentially abundant species were associated with efficient dairy cows and complemented with lower richness and higher dominance values in efficient compared to inefficient cows suggested a less diverse microbiome in efficient cows. In the current study, we identified members of the order Bacteroidales with positive or negative coefficients on G:F in both heifer and steer cohorts. Consistent with our results, Prevotella spp. have been observed to have both positive (Hernandez-Sanabria et al., 2012) and negative (Carberry et al., 2012; Hernandez-Sanabria et al., 2012; McCann et al., 2014) associations with feed efficiency. In dairy cows, members of the Prevotellaceae family have been associated with inefficient cows (Shabat et al., 2016). Based on microbial transcriptome profiles, Li et al. (2016) reported Lachnospiraceae and Veillonellaceae to be associated with less efficient steers and is similar to the results observed in the G:F model of steers in this study.
When steer cohort data were used to evaluate the heifer models and vice versa, adjusted R2 values for ADFI, ADG, and G:F were substantially decreased (Table 3). This is likely attributable to many factors including diet, gender, and age being different between the two cohorts. Figure 4 clearly depicts different bacterial community clustering between the cohorts. Yet, the G:F model developed for steers was able to predict 11% of the variation when using the heifer data, even with OTUs not being similar across the two cohorts. It is important to remember that various management or environmental conditions (diets, breeds, gender, etc.) affect microbial community. As such the models proposed herewith may not be robust across other management and environmental parameters and further testing will be required to determine robustness in different populations on similar diets. In the taxa-based models, when steer cohort data were used to evaluate the heifer models and vice versa (Supplementary Table S3), the models were incompatible. Significant models were observed for ADG and G:F within the steer cohort and accounted for 4% and 5% of the variation, respectively. This suggests that the abundance of certain bacterial species might affect feed efficiency in cattle rather than overall changes in the microbial community taxa. The current study built models focused on the associations between bacteria and feed efficiency traits; however, inclusion of other rumen microorganisms could potentially lead to improved models through a more holistic approach.
Table 3.
Evaluation of model accuracy to predict ADFI, ADG, and G:F for the heifer and steer cohorts
| Traita | P-value | R 2b |
|---|---|---|
| ADFI | ||
| Heifer | 0.50 | <0.01 |
| Steer | 0.40 | <0.01 |
| ADG | ||
| Heifer | 0.08 | 0.05 |
| Steer | 0.16 | 0.05 |
| G:F | ||
| Heifer | 0.85 | <0.01 |
| Steer | 0.01 | 0.11 |
aHeifer data were used to assess the steer model and steer data were used to assess the heifer model.
bAdjusted R-squared.
As expected, functional prediction of bacterial OTUs in the feed efficiency models identified metabolic pathways involved in starch and carbohydrate metabolism and protein metabolism. Feed efficiency is greatly influenced by the ability of the microbes to extract energy from the diet and the capacity of the microbes to produce microbial cell protein as a protein source for the host. However, the prediction of increased number of transporters in the OTUs identified in the models was surprising. Previous studies have reported the increased abundance of transporters in the rumen and their role in mediation of nutrient uptake (Popova et al., 2017). It is possible that in addition to increased metabolism, efficient and broader uptake of nutrients by the microbes can influence animal performance and efficiency. As such, further investigating how nutrient transport and the abundance of nutrient transporters such as ABC transporters affect animal efficiency might be interesting. Future studies utilizing shotgun metagenome sequencing of the rumen microbiome in different feed efficiency phenotypes may provide more insight into the role of nutrient transporters and the type of transporters that may influence feed efficiency in the ruminant animal.
CONCLUSIONS
The critical role of the rumen microbiome in feed digestion within the ruminant animal suggests microbial features influence feed efficiency. This study identified a subset of bacterial OTUs that impact feed efficiency in heifers and steers in growing and finishing diets, respectively. Additionally, this study showed that approximately 20% of the variation in feed efficiency traits (ADFI, ADG, G:F) can be explained using the rumen microbiome in beef cattle. The rumen microbiome is an important factor that influences feed efficiency and research that includes the rumen microbiome functional capacity could provide novel opportunities to improve our understanding of genes and mechanisms that influence feed efficiency towards increasing the productivity of animal operations.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
Conflict of interest statement.
USDA is an equal opportunity provider and employer. S.C.F., author of this publication, has disclosed a significant financial interest in NuGUT LLC. In accordance with its Conflict of Interest policy, the University of Nebraska-Lincoln’s Conflict of Interest in Research Committee has determined that this must be disclosed. The rest of the authors have nothing to disclose.
ACKNOWLEDGMENTS
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
LITERATURE CITED
- Anderson C.L., Schneider C.J., Erickson G.E., MacDonald J.C., and Fernando S.C.. 2016. Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs. J. Appl. Microbiol. 120:588–599. doi:10.1111/jam.13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289–300. [Google Scholar]
- Bergman E.N. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590. [DOI] [PubMed] [Google Scholar]
- Berry D.P., and Crowley J.J.. 2013. Cell Biology Symposium: genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 91:1594–1613. doi:10.2527/jas.2012-5862 [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I.,. et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper J.L. 2011. The environmental impact of beef production in the United States: 1977 compared with 2007. J. Anim. Sci. 89:4249–4261. doi:10.2527/jas.2010-3784 [DOI] [PubMed] [Google Scholar]
- Carberry C.A., Kenny D.A., Han S., McCabe M.S., and Waters S.M.. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78:4949–4958. doi:10.1128/AEM.07759-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta M.A., and Hespell R.B.. 1986. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 52:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ondarza M.B., and Tricarico J.M.. 2017. Review: advantages and limitations of dairy efficiency measures and the effects of nutrition and feeding management interventions. Prof. Anim. Sci. 33:393–400. doi:10.15232/pas.2017-01624 [Google Scholar]
- de Vries M., and de Boer I.J.M.. 2010. Comparing environmental impacts for livestock products: a review of life cycle assessments. Livest. Sci. 128:1–11. doi:10.1016/j.livsci.2009.11.007 [Google Scholar]
- Di Rienzi S.C., Sharon I., Wrighton K.C., Koren O., Hug L.A., Thomas B.C., Goodrich J.K., Bell J.T., Spector T.D., Banfield J.F.,. et al. 2013. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2:e01102. doi:10.7554/eLife.01102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economic Research Service (ERS) and U.S. Department of Agriculture (USDA).. 2017. Food availability (per capita) data system. [Google Scholar]
- Edgar R.C. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996–998. doi:10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando S.C., Purvis H.T., Najar F.Z., Sukharnikov L.O., Krehbiel C.R., Nagaraja T.G., Roe B.A., and Desilva U.. 2010. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76:7482–7490. doi:10.1128/AEM.00388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S.J., Turner S., Olsen G.J., Barns S., Lane D.J., and Pace N.R.. 1988. Evolutionary relationships among cyanobacteria and green chloroplasts. J. Bacteriol. 170:3584–3592. doi:10.1128/JB.170.8.3584-3592.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good I.J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. doi:10.1093/biomet/40.3-4.237 [Google Scholar]
- Herd R.M., and Arthur P.F.. 2009. Physiological basis for residual feed intake. J. Anim. Sci. 87:E64–E71. doi:10.2527/jas.2008-1345 [DOI] [PubMed] [Google Scholar]
- Hernandez-Sanabria E., Goonewardene L.A., Wang Z., Durunna O.N., Moore S.S., and Guan L.L.. 2012. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78:1203–1214. doi:10.1128/AEM.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanabria E., Guan L.L., Goonewardene L.A., Li M., Mujibi D.F., Stothard P., Moore S.S., and Leon-Quintero M.C.. 2010. Correlation of particular bacterial PCR-denaturing gradient gel electrophoresis patterns with bovine ruminal fermentation parameters and feed efficiency traits. Appl. Environ. Microbiol. 76:6338–6350. doi:10.1128/AEM.01052-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E., and Mizrahi I.. 2012. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:e33306. doi:10.1371/journal.pone.0033306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W.J., Leahy S.C., Altermann E., Yeoman C.J., Dunne J.C., Kong Z., Pacheco D.M., Li D., Noel S.J., Moon C.D.,. et al. 2010. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS One 5:e11942. doi:10.1371/journal.pone.0011942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein T.J., Erickson G.E., and Bremer V.R.. 2007. Board-invited review: use of distillers by-products in the beef cattle feeding industry. J. Anim. Sci. 86:1223–1231. doi:10.2527/jas.2007-0550 [DOI] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., and Schloss P.D.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. doi:10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R.,. et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotech. 31:814–821. doi:10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhou M., Ominski K., and Guan L.L.. 2016. Does the rumen microbiome play a role in feed efficiency of beef cattle?J. Anim. Sci. 94:44. doi:10.2527/jas.2016-0524 [Google Scholar]
- Lu D., Miller S., Sargolzaei M., Kelly M., Vander Voort G., Caldwell T., Wang Z., Plastow G., and Moore S.. 2013. Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J. Anim. Sci. 91:3612–3633. doi:10.2527/jas.2012-5716 [DOI] [PubMed] [Google Scholar]
- Mackie R.I., McSweeney C.S., and Aminov R.I.. 2001. Rumen. In: Encyclopedia of life sciences. Chichester (UK): John Wiley & Sons, Ltd. [Google Scholar]
- Matsui H., Ogata K., Tajima K., Nakamura M., Nagamine T., Aminov R.I., and Benno Y.. 2000. Phenotypic characterization of polysaccharidases produced by four prevotella type strains. Curr. Microbiol. 41:45–49. doi:10.1007/s002840010089 [DOI] [PubMed] [Google Scholar]
- McCann J.C., Wiley L.M., Forbes T.D., Rouquette F.M., and Tedeschi L.O.. 2014. Relationship between the rumen microbiome and residual feed intake-efficiency of Brahman bulls stocked on bermudagrass pastures. PLoS One 9:e91864. doi:10.1371/journal.pone.0091864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., and Hugenholtz P.. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. doi:10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGorum B.C., Pirie R., Glendinning L., McLachlan G., Metcalf J.S., Banack S.A., Cox P.A., and Codd G.A.. 2015. Grazing livestock are exposed to terrestrial cyanobacteria. Vet. Res. 46:16. doi:10.1186/s13567-015-0143-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P.R., Smith T.P.L., Wells J.E., Kuehn L.A., and Freetly H.C.. 2015. Rumen microbiome from steers differing in feed efficiency. PLoS One 10:e0129174. doi:10.1371/journal.pone.0129174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., and Wagner H.. 2017. vegan: Community Ecology Package. R package version 2.4-5. https://CRAN.R-project.org/package=vegan [Google Scholar]
- Paz H.A., Anderson C.L., Muller M.J., Kononoff P.J., and Fernando S.C.. 2016. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front. Microbiol. 7:1206. doi:10.3389/fmicb.2016.01206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J., and McAllister T.A.. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS One 8:e83424. doi:10.1371/journal.pone.0083424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro G., Perelman S., Guerschman J.P., and Paruelo J.M.. 2008. How to evaluate models: observed vs. predicted or predicted vs. observed?Ecol. Modell. 216:316–322. doi:10.1016/j.ecolmodel.2008.05.006 [Google Scholar]
- Popova M., McGovern E., McCabe M.S., Martin C., Doreau M., Arbre M., Meale S.J., Morgavi D.P., and Waters S.M.. 2017. The structural and functional capacity of ruminal and cecal microbiota in growing cattle was unaffected by dietary supplementation of linseed oil and nitrate. Front. Microbiol. 8:937. doi:10.3389/fmicb.2017.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team.. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- Russell J.B., Muck R.E., and Weimer P.J.. 2009. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 67:183–197. doi:10.1111/j.1574-6941.2008.00633.x [DOI] [PubMed] [Google Scholar]
- Schenkel F.S., Miller S.P., and Wilton J.W.. 2011. Genetic parameters and breed differences for feed efficiency, growth, and body composition traits of young beef bulls. Can. J. Anim. Sci. 84:177–185. doi:10.4141/A03-085 [Google Scholar]
- Schiermiester L.N., Thallman R.M., Kuehn L.A., Kachman S.D., and Spangler M.L.. 2015. Estimation of breed-specific heterosis effects for birth, weaning, and yearling weight in cattle. J. Anim. Sci. 93:46–52. doi:10.2527/jas.2014-8493 [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J.,. et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. doi:10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi:10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabat S.K. Ben G. Sasson A. Doron-Faigenboim T. Durman S. Yaacoby M.E. Berg Miller B.A. White N. Shterzer, and Mizrahi I.. 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10:2958–2972. doi:10.1038/ismej.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E., and Weaver W.. 1949. The mathematical theory of communication. Urbana (IL): University of Illinois Press. [Google Scholar]
- Sheneman L., Evans J., and Foster J.A.. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823–2824. doi:10.1093/bioinformatics/btl478 [DOI] [PubMed] [Google Scholar]
- Snelling W.M., Allan M.F., Keele J.W., Kuehn L.A., Thallman R.M., Bennett G.L., Ferrell C.L., Jenkins T.G., Freetly H.C., Nielsen M.K.,. et al. 2011. Partial-genome evaluation of postweaning feed intake and efficiency of crossbred beef cattle. J. Anim. Sci. 89:1731–1741. doi:10.2527/jas.2010-3526 [DOI] [PubMed] [Google Scholar]
- Soo R.M., Skennerton C.T., Sekiguchi Y., Imelfort M., Paech S.J., Dennis P.G., Steen J.A., Parks D.H., Tyson G.W., and Hugenholtz P.. 2014. An expanded genomic representation of the phylum cyanobacteria. Genome Biol. Evol. 6:1031–1045. doi:10.1093/gbe/evu073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson D.M., and Weimer P.J.. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165–174. doi:10.1007/s00253-006-0802-y [DOI] [PubMed] [Google Scholar]
- Storm E., Orskov E.R., and Smart R.. 1983. The nutritive value of rumen micro-organisms in ruminants. 2. The apparent digestibility and net utilization of microbial N for growing lambs. Br. J. Nutr. 50:471–478. [DOI] [PubMed] [Google Scholar]
- United Nations.. 2017. Department of Economic and Social Affairs, Population Division. World Population Prospects: the 2017 Revision. [Google Scholar]
- Wallace R. 1996. The proteolytic systems of ruminal microorganisms. Ann. Zootech. 45:301–308. doi:10.1051/animres:19960653 [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. doi:10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B.A., Mackie R.I., and Doerner K.C.. 1993. Enzymatic hydrolysis of forage cell walls. In: Lung, H. G., D. R. Buxton, R. D. Hatfield, and J. Ralph, editors. Forage cell wall structure and digestibility. Madison (WI): American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; p. 455–484. [Google Scholar]
- Yu Z., and Morrison M.. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812. [DOI] [PubMed] [Google Scholar]
- Zoetendal E.G., Plugge C.M., Akkermans A.D.L., and de Vos W.M.. 2003. Victivallis vadensis gen. nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int. J. Syst. Evol. Microbiol. 53:211–215. doi:10.1099/ijs.0.02362-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.