Abstract
Improvement in growth and fatness traits are the main objectives in pig all breeding programs. Tenth rib backfat thickness (10RIBBFT) and days to 100 kg (D100), which are good predictors of carcass lean content and growth rate, respectively, are economically important traits and also main breeding target traits in pigs. To investigate the genetic mechanisms of 10RIBBFT and D100 of pigs, we sampled 1,137 and 888 pigs from 2 Yorkshire populations of American and British origin, respectively, and conducted genome-wide association study (GWAS) through combined analysis and meta-analysis, to identify SNPs associated with 10RIBBFT and D100. A total of 11 and 7 significant SNPs were identified by combined analysis for 10RIBBFT and D100, respectively. And in meta-analysis, 8 and 7 significant SNPs were identified for 10RIBBFT and D100, respectively. Among them, 6 and 5 common significant SNPs in two analysis results were, respectively, identified associated with 10RIBBFT and D100, and correspondingly explained 2.09% and 0.52% of the additive genetic variance of 10RIBBFT and D100. Further bioinformatics analysis revealed 10 genes harboring or close to these common significant SNPs, 5 for 10RIBBFT and 5 for D100. In particular, Gene Ontology analysis highlighted 6 genes, PCK1, ANGPTL3, EEF1A2, TNFAIP8L3, PITX2, and PLA2G12, as promising candidate genes relevant with backfat thickness and growth. PCK1, ANGPTL3, EEF1A2, and TNFAIP8L3 could influence backfat thickness through phospholipid transport, regulation of lipid metabolic process through the glycerophospholipid biosynthesis and metabolism pathway, the metabolism of lipids and lipoproteins pathway. PITX2 has a crucial role in skeletal muscle tissue development and animal organ morphogenesis, and PLA2G12A plays a role in the lipid catabolic and phospholipid catabolic processes, which both are involved in the body weight pathway. All these candidate genes could directly or indirectly influence fat production and growth in Yorkshire pigs. Our findings provide novel insights into the genetic basis of growth and fatness traits in pigs. The candidate genes for D100 and 10RIBBFT are worthy of further investigation.
Keywords: combined analysis, genome-wide association study, growth traits, meta-analysis, population stratification
INTRODUCTION
Improving pig performance has always been considered an important issue for the pork industry. In a pig breeding program, economically important traits, such as tenth rib backfat thickness (10RIBBFT) and days to 100 kg (D100), are frequently measured to indicate the pig carcass lean content and growth rate, respectively. Even though many difficulties exist in traditional artificial selection to improve the 10RIBBFT and D100 together, given the weak positive genetic relationship between 10RIBBFT and D100 (Bereskin et al., 1987; Bidanel et al., 1994), marker-assisted selection and even genomic selection could provide an efficient strategy for the improvement of these traits.
In recent years, the high-density SNP array was very rapidly developed. This new technique is able to simultaneously genotype hundreds of thousands of SNP markers that cover the whole target genome. Additionally, the low cost for each individual genotyped makes it practical for large animal and human populations. One of its attractive applications is the genome-wide association study (GWAS), which allows researchers to detect the nucleotide variation associated with traits of interest by performing genome-wide mapping with higher resolution. Currently, GWASs have been successfully implemented in a growing list of areas, not only on disease diagnosis or pharmaceutical research (Lutz et al., 2015; Qayyum et al., 2015) but also for agricultural practice (Goddard and Hayes, 2009). For swine, particularly, GWAS has made great progress in exploring various traits of economic importance such as growth and fatness traits (Fernández et al., 2012; Do et al., 2014; Guo et al., 2015).
The objectives of this study were to (i) conduct a GWAS in 2 Yorkshire populations with different genetic backgrounds to detect the candidate genes or genomic regions associated with D100 and 10RIBBFT, and (ii) compare combined analysis and meta-analysis with a single-population GWAS analysis.
MATERIALS AND METHODS
Ethics Statement
The whole procedure for collecting ear tissue samples was carried out in strict accordance with the protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the China Agricultural University. The IACUC of the China Agricultural University specifically approved this study (permit number DK996).
Animals and Phenotype
A total of 2,025 Yorkshire pigs used in this study were sampled from two breeding farms, including 1,137 progeny of American Yorkshires and 888 progeny of British Yorkshires. The progeny of American Yorkshires were born in 2011–2015 and came from 106 sire families (10 to 70 offspring in each family with an average of 13), and the progeny of British Yorkshires were born in 2007–2013 and came from 129 sire families (10 to 71 offspring in each family with an average of 7). There was no genetic connectedness between the 2 populations according to the pedigree information. Performance testing was carried out at these 2 farms. Phenotypic records included D100 and 10RIBBFT. Tenth rib backfat thickness was measured between the 10th and 11th ribs of pigs at a weight of approximately 100 kg by B ultrasound (HS1500; Honda, Japan). The descriptive statistics of the phenotypic values of 10RIBBFT and D100 are presented in Table 1. According to the Shapiro–Wilk normal distribution test, both traits followed a normal distribution in the 2 populations. The official conventional EBV based on a 2-trait animal model, which was separately implemented in each population, were obtained from the National Swine Genetic Improvement Center of China (http://www.cnsge.org.cn/); afterwards, corrected phenotypic values were calculated as EBV plus the estimated residual for each individual in each population.
Table 1.
Descriptive statistics and normality test of 10RIBBFT and D100 in 2 Yorkshire populations
| Trait | Source | Unit | No. | Mean | SD | W-value1 | P-value |
|---|---|---|---|---|---|---|---|
| 10RIBBFT | American line | mm | 1,137 | 12.31 | 2.145 | 0.9237 | 0.22 |
| British line | 888 | 11.69 | 1.42 | 0.9048 | 0.13 | ||
| D100 | American line | d | 1,137 | 167 | 11.11 | 0.9453 | 0.57 |
| British line | 888 | 150 | 5.13 | 0.9506 | 0.44 |
1 W-value = Shapiro–Wilk test value.
Genotyping and Quality Control
Genomic DNA was extracted from blood samples using a TIANamp Blood DNA Kit (catalog number DP348; Tiangen, Beijing). Genotyping was performed using a PorcineSNP80 BeadChip (Illumina, San Diego, CA), which includes 68,528 SNPs across the entire pig genome. Genotype quality control was carried out using PLINK 1.9 software (Chang et al., 2015) separately for each population. First, individuals with call rates (CR) less than 90% were removed and then SNP with CR less than 90%, minor allele frequencies (MAF) <3%, or significant deviation from the Hardy–Weinberg equilibrium (HWE; P < 10 × 10−6) were removed. After genotype quality control, 2,009 individuals and 53,233 SNPs remained for further analysis.
Population structure.
Because the genetic background of 2 Yorkshire populations in this study is different, a principal component analysis (PCA) was carried out to detect the population stratification using GCTA software (Yang et al., 2011). In order to keep the independence of SNPs, the adjacent SNPs with r2 > 0.2 were further pruned after genotype quality control, and in total 29,229 SNPs were used in PCA. The linkage disequilibrium within each population was calculated using PLINK software as well. Meanwhile, a quantile–quantile (Q–Q) plot was generated to assess the influence of population stratification on the GWAS.
Statistical Analysis
Single-population GWAS using a linear mixed model was carried out in each pig population separately. Based on the single-population analysis, the meta-analysis was conducted. Meanwhile, the combined analysis, which combined the two pig populations in the same linear mixed model used in the single-population analysis, was also implemented.
Combined Analysis and Single-Population Analysis
Both the single-population analysis and combined analysis used the same linear mixed model. The main difference between them is that the latter utilizes the information of two populations simultaneously to contruct the genomic additive relationship (G) matrix.
Linear mixed model.
A linear mixed model was implemented to detect the association of SNP with growth and fatness traits. The model in this study is a single SNP regression model. The model includes a random polygenic effect to account for shared genetic effects of related individuals and to control for population stratification. The statistical model is described below:
where yc is the vector of phenotypes (corrected phenotypic values); 1 is a vector of ones; μ is the overall mean; b is the average effect of the gene substitution of a particular SNP; x is a vector of the SNP genotype (coded as 0,1, or 2); g is a vector of random polygenic effects with a normal distribution g ~ N(0, Gσa2), in which σa2 is the polygenic variance and G is the genomic additive relationship matrix and was constructed using all markers following VanRaden (2008); Z is an incidence matrix relating phenotypes to the corresponding random polygenic effects; and e is a vector of residual effects with a normal distribution N(0, Iσe2), in which σe2 is the residual variance. The software GCTA (Yang et al., 2011) was used to fit the model, and 10,000 permutations were performed for multiple test correction to identify significant SNP. For each trait, the phenotypic values of all individuals were shuffled in each replicate, and the maximum statistic value in each permutation was gathered to establish the empirical distribution of test statistic for GWAS. The genome-wide critical value at the significance level of 0.05 was obtained at the 5th percentile in the ordered vector of maximum. Similarly, the critical value for each chromosome (chromosome-wide) could be calculated based on the maximum statistic on each chromosome.
Meta-analysis.
Based on the results of GWAS separately in the American and British Yorkshire populations through single-population analysis, a meta-analysis based on Fisher’s method was carried out to combine P-value probabilities from each test into one test statistic (X2) using the formula
Where Pi is the raw P-value of tth study for t = 1, …, T, in which T is the number of independent studies. When all the null hypotheses are true, this combined test statistic follows a χ2 distribution with 2T of degree of freedom. Therefore, the new P-value from the meta-analysis was calculated using
where χ22T is a χ2 variable with 2T of degree of freedom. In our study, we used the common SNP in the American line and the British line by Fisher’s method to calculate a meta-analysis P-value. Afterwards, Bonferroni correction at a significance level of 0.05 was used to identify significant SNP. There were 47,498 common SNPs in the American and British populations, the threshold P-value for each SNP at a significance level of 0.05 was 1.05 × 10−6 (0.05/47498).
Identification of candidate genes.
To identify functionally plausible candidate genes near the significant SNP, the genes located in or overlapping the region between the 0.5 Mb upstream and 0.5 Mb downstream of the significant SNP were obtained using Ensemble (http://www.ensembl.org/Sus_scrofa/Info/Index; Sscrofa 10.2 genome version). Gene Ontology analysis was carried out using the DAVID bioinformatics resource (https://david.ncifcrf.gov/). Pathway analysis was conducted using the online KEGG (http://www.kegg.jp/kegg/pathway.html) and GeneCards (http://www.genecards.org/) tools.
RESULTS
Population Structure
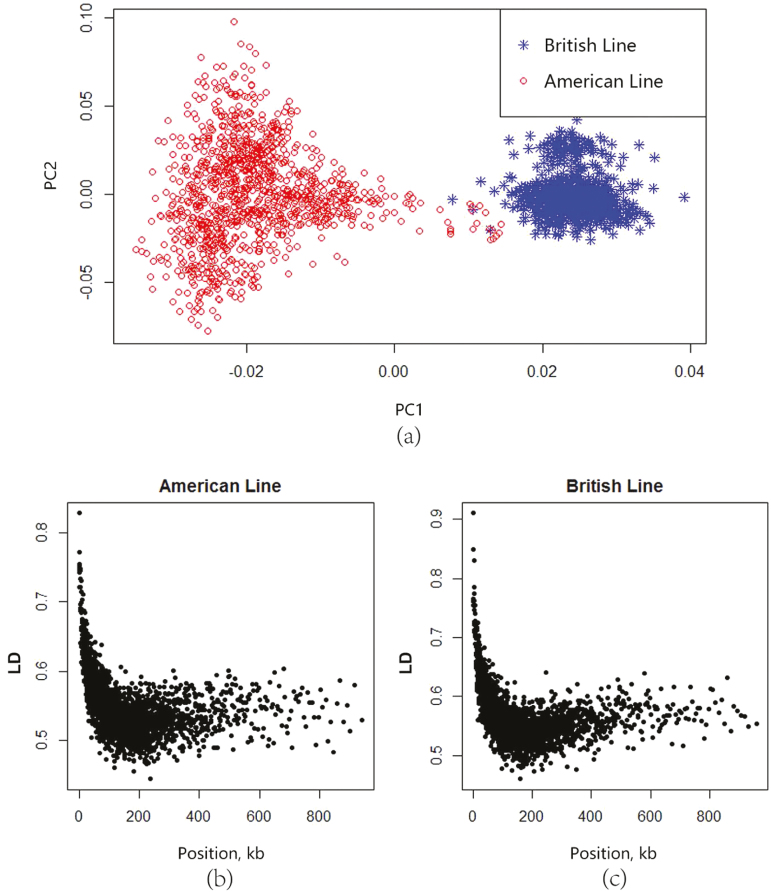
As shown in Figure 1a, the 2 Yorkshire populations can be clearly identified through principal component analysis. Within each population, all individuals were classified nearly into one cluster, implying no significant or slight genetic differentiation among them. However, although the genetic background of the 2 Yorkshire populations was different, they shared a similar linkage disequilibrium (LD) pattern. Figure 1b and c illustrates that LD similarly decayed in both populations. The average linkage disequilibrium between adjacent SNP measured with r2 was 0.563 and 0.571 in American and British Yorkshires, respectively, and the correlation of r2 between the 2 populations was 0.47. Supplemental Figure S1 indicates the impact of population stratification on GWAS. The x-axis and y-axis represent the expected and observed −log10(P-value) of all filter SNPs. The inflation factor λ, the regression of observed values on expected ones, can assess the population stratification. As λ approaches 1.0, population stratification is reduced. Without any population stratification control (Supplemental Figure S1a and S1b), the average genomic inflation factor (λ) for 10RIBBFT and D100 was 2.03 and 2.74, respectively. When the additive genomic relationship (G) matrix was used in the linear mixed model, the average genomic inflation factor (λ) for 10RIBBFT and D100 decreased to 1.21 and 1.15, respectively, indicating that population stratification was properly controlled (Supplemental Figure S1c and S1d).
Figure 1.
Population structure and distribution of LD for 2 Yorkshire populations. (a) Principal components analysis, (b) distribution of LD in the American Yorkshire line, and (c) distribution of LD in the British Yorkshire line; PC1 = first principal component; PC2 = second principal component.
SNPs Significantly Associated with 10RIBBBFT
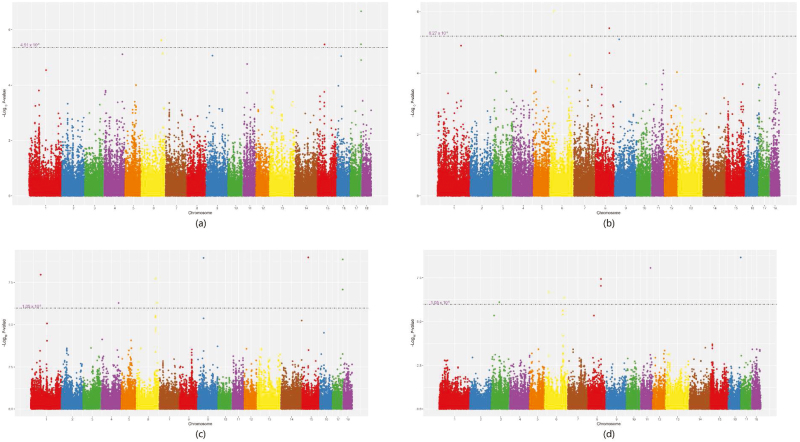
The GWAS results of all significant SNPs associated with 10RIBBFT in combined analysis and meta-analysis are illustrated in Table 2 and Figure 2a and c. For combined analysis, the empirical P-value of a permutation test at the genome-wide significance level of 0.05 was 4.51 × 10−6. Similarly, the empirical P-values at the chromosome-wide significance level of 0.05 were also obtained for each chromosome. For SSC1, SSC4, SSC6, SSC9, SSC11, SSC16, and SSC17, where the chromosome-wide significant SNPs were identified, the empirical P-values of a permutation test at the chromosome-wide significance level were 3.11 × 10−5, 1.23 × 10−5, 2.75 × 10−5, 2.22 × 10−5, 1.87 × 10−5, 1.72 × 10−5, and 2.75 × 10−5, respectively. A total of 11 SNPs were identified to be significantly associated with 10RIBBFT in combined analysis, explaining 2.77% additive genetic variance of 10RIBBFT. Among them, 4 SNPs reached the genome-wide significance level and 7 reached the chromosome-wide significance level (Figure 2a). The 4 genome-wide significant SNPs were located on SSC6, SSC15, and SSC17, and the 7 chromosome-wide significant SNPs were located on SSC1, SSC4, SSC6 and SSC9, SSC11, SSC16, and SSC17.
Table 2.
Significant SNP and associated genes for 10RIBBFT in the combined analysis and meta-analysis
| SNP name | SSC | Location, bp | P (combined)-value1 | P (meta-analysis)-value2 | Associated gene3 |
Distance,4 bp | Gene function | Effect6(%) |
|---|---|---|---|---|---|---|---|---|
| MARC0023432 | 1 | 63,194,355 | 1.29 × 10−3 | 1.09 × 10−8 | CNR1 | −8478 | Signal transduction | 0.042788 |
| ASGA0004384 | 1 | 134,037,038 | 2.89 × 10−5 | 8.30 × 10−6 | TNFAIP8L3 | −82,677 | Phospholipid transport; regulation of lipid metabolic process | 0.63985 |
| MARC0101639 | 4 | 129,079,594 | 7.81 × 10−6 | 5.23 × 10 −7 | VCAM1 | +25,002 | Cell-matrix adhesion | 0.060221 |
| WU_10.2_6_138496555 | 6 | 138,496,555 | 2.45 × 10−6 | 1.87 × 10−8 | ANGPTL3 | +385,265 | Glycerol, fatty acid, and phospholipid metabolic process | 0.537356 |
| H3GA0019109 | 6 | 143,021,550 | 7.37 × 10−6 | 4.99 × 10 −7 | DAB1 | Introns | Neuron migration | 0.151069 |
| WU_10.2_9_29774722 | 9 | 29,774,722 | 8.73 × 10−6 | 1.12 × 10 −9 | EEF1A2 | +132,165 | Positive regulation of lipid kinase activity | 0.386885 |
| WU_10.2_11_22253325 | 11 | 22,253,325 | 1.74 × 10−5 | 2.19 × 10−3 | ENSSSCG000000 23738 |
Introns | Intracellular protein transport | 0.043826 |
| MARC0109867 | 15 | 38,679,115 | 1.79 × 10−4 | 1.06 × 10 −9 | ENSSSCG000000 29846 |
+407,341 | Striated muscle contraction | 0.047907 |
| ASGA0091668 | 15 | 41,967,209 | 3.46 × 10 −6 | 3.19 × 10−4 | ENSSSCG000000 23144 |
+354,274 | NA5 | 0.170797 |
| WU_10.2_16_22135509 | 16 | 22,135,509 | 9.11 × 10−6 | 2.94 × 10−5 | SPEF2 | Introns | NA | 0.30447 |
| WU_10.2_17_64641249 | 17 | 64,641,249 | 3.41 × 10 −6 | 5.41 × 10−6 | BMP7 | −103,697 | Organ morphogenesis and development | 0.099771 |
| WU_10.2_17_65044153 | 17 | 65,044,153 | 1.26 × 10−5 | 8.15 × 10 −8 | PCK1 | −50,630 | Gluconeogenesis; lipid and glucose metabolic process | 0.059291 |
| ALGA0096314 | 17 | 65,125,689 | 2.21 × 10−7 | 1.37 × 10 −9 | PCK1 | +24,938 | Gluconeogenesis; lipid and glucose metabolic process | 0.320835 |
1 P (combined)-value = P-value from the combined analysis. The bold data in this column represent the significant SNP at genome-wide significant level; otherwise at the chromosome-wide significant level.
2 P (meta-analysis)-value = P-value from the meta-analysis. The bold data in this column represent the significant SNP at genome-wide significant level.
3 PCK1 = phosphoenolpyruvate carboxykinase 1; ANGPTL3 = angiopoietin-like 3; BMP7 = bone morphogenetic protein 7; DAB1 = disabled homolog 1; VCAM1 = vascular cell adhesion molecule 1; EEF1A2 = eukaryotic translation elongation factor 1 alpha 2; SPEF2 = sperm flagellar 2; TNFAIP8L3 = TNF alpha induced protein 8-like 3; CNR1 = cannabinoid receptor 1. The associated genes in bold in this column represent these genes were associated with traits based on annotation.
4+/− = the location of SNP in downstream/upstream of the nearest gene.
5NA = not available.
6Effect = the proportion of additive genetic variance explained by the identified SNP.
Figure 2.
Manhattan plot of different analyses for 10RIBBFT and D100. The x-axis represents the chromosomes and the y-axis represents the −log10(P-value). The red line indicates the significance threshold for the (a) combined analysis in the 2 populations for 10RIBBFT, (b) combined analysis in the 2 populations for D100, (c) meta-analysis in the 2 populations for 10RIBBFT, and (d) meta-analysis in the 2 populations for D100.
As shown in Table 2, a total of 8 SNPs were identified by meta-analysis to be significantly associated with 10RIBBFT, which were located in SSC1, SSC4, SSC6, SSC9, SSC15, and SSC17 (Figure 2c). In addition, there were 6 common significant SNPs identified by both combined analysis and meta-analysis (Supplemental Table S1), explaining 2.09% additive genetic variance of 10RIBBFT.
SNPs Significantly Associated with D100
The GWAS results of all significant SNPs associated with D100 in combined analysis and meta-analysis are illustrated in Table 3 and Figure 2b and d. For combined analysis, a total of 7 SNPs were identified to be significantly associated with D100, explaining 0.67% additive genetic variance of D100. Among them, 3 genome-wide and 4 chromosome-wide significant SNPs were identified (Figure 2b). The empirical P-value at the genome-wide significance level of 0.05 was 6.27 × 10−6. The empirical P-values of a permutation test at the chromosome-wide significance level of 0.05 for SSC1, SSC6, SSC8, and SSC9, where the chromosome-wide significant SNPs were identified, were 3.11 × 10−5, 2.75 × 10−5, 2.35 × 10−5, and 2 × 10−5, respectively. Meanwhile, 7 significant SNPs associated with D100 were detected by meta-analysis, they were located on SSC3, SSC6, SSC8, SSC11, and SSC16 (Figure 2d), and 5 significant SNPs, which located in SSC3, SSC6, and SSC8, were overlapped with those from combined analysis, and 0.52% additive genetic variance of D100 was explained by them.
Table 3.
Significant SNP and associated genes for D100 in the combined analysis and meta-analysis
| SNP name | SSC | Location, bp | P (combined)–value1 | P (meta-analysis)–value2 | Associated gene3 | Distance,4 bp | Gene function | Effect6(%) |
|---|---|---|---|---|---|---|---|---|
| MARC0055965 | 1 | 240,256,448 | 1.28 × 10−5 | NA | 7SK | −340,605 | NA | 0.070736 |
| ALGA0018931 | 3 | 52,464,041 | 6.03 × 10 −6 | 7.91 × 10 −7 | ENSSSCG000000 29325 |
+371,904 | Translation | 0.166871 |
| MARC0101536 | 6 | 139,068,649 | 2.64 × 10−5 | 4.55 × 10 −7 | ENSSSCG000000 23243 |
+217,561 | DNA-templated and replication | 0.052004 |
| H3GA0053564 | 6 | 17,434,093 | 9.77 × 10 −7 | 2.00 × 10 −7 | CCDC102A | Exons | NA | 0.027472 |
| WU_10.2_8_120377608 | 8 | 120,377,608 | 3.45 × 10 −6 | 9.10 × 10 −8 | PITX2 | +451,497 | Skeletal muscle tissue development; animal organ morphogenesis | 0.144018 |
| ASGA0096290 | 8 | 120,840,701 | 2.24 × 10−5 | 3.73 × 10 −8 | PLA2G12A | +244,286 | Lipid and phospholipid catabolic process | 0.094187 |
| ALGA0051735 | 9 | 20,355,090 | 8.01 × 10−6 | 1.90 × 10−2 | NA | NA | NA | 0.113077 |
| H3GA0032360 | 11 | 74,978,970 | 2.33 × 10−4 | 8.75 × 10 −9 | DOCK9 | −18,814 | Small GTPase-mediated signal transduction | 0.024394 |
| WU_10.2_16_83745324 | 16 | 83,745,324 | 2.96 × 10−4 | 2.24 × 10 −9 | ENSSSCG000000 17110 |
+233,640 | Protein phosphorylation | 0.022255 |
1 P (combined)–value = P-value from the combined analysis. The bold data in this column represent the significant SNP at genome-wide significant level; otherwise at the chromosome-wide significant level.
2 P (meta-analysis)–value = P-value from the meta-analysis. The bold data in this column represent the significant SNP at genome-wide significant level.
3 CCDC102A = coiled-coil domain containing 102A; PITX2 = paired-like homeodomain 2; PLA2G12A = phospholipase A2 group XIIA; DOCK9 = dedicator of cytokinesis 9. The associated genes in bold in this column represent these genes were associated with traits based on annotation.
4+/− = the location of SNP in downstream/upstream of the nearest gene.
5NA = not available.
6Effect = the proportion of additive genetic variance explained by the identified SNP.
Identification of Candidate Genes
Based on 11 common significant SNPs associated with 10RIBBFT and D100 identified by combined analysis and meta-analysis, 10 genes, which located within the region between the 0.5 Mb upstream and 0.5 Mb downstream of the significant SNP, were found and annotated (Tables 2 and 3). While Go analysis revealed that there were 6 annotated genes had a highlight biology function with 10RIBBFT and D100. Among these 6 annotated genes, 4 genes had function related to 10RIBBFT and 2 related to D100.
DISCUSSION
Candidate Genes
According to the results of gene annotation, a total of 6 genes were relevant to both traits. Generally, all these candidate genes could regulate or influence backfat thickness and body weight through different kinds of biological processes and pathways. For 10RIBBFT, 4 candidate genes, PCK1, ANGPTL3, EEF1A2, and TNFAIP8L3, are highlighted as promising biological candidate genes for 10RIBBFT. PCK1 is associated with gluconeogenesis and lipid and glucose metabolic processes. Pena et al. (2016) found that PCK1 takes part in the metabolic step in lipid metabolism. In Iberian pigs, PCK1 is related to enzyme kinetic and functional properties modifying fat distribution (Latorre et al., 2016). ANGPTL3 takes part in lipid digestion, mobilization, transport, and lipoprotein metabolism pathways. ANGPTL3 has been reported to have functions in the activity of lipoprotein lipase and lipoprotein metabolism in humans (Bauer et al., 2011; Adeyemo et al., 2012). It also has been identified by GWAS as a lipid-associated locus in pigs (Feng et al., 2006). As to TNFAIP8L3, it has key functions and is involved in some lipid pathway, like phospholipid biosynthesis, transport, metabolism, and lipoproteins metabolism pathway. TNFAIP8L3 has not been defined in pigs and mice, whereas in humans it was regarded as a lipid second messenger transfer protein (Cui et al., 2015). Our findings will be helpful for a better understanding of the role of TNFAIP8L3 in backfat thickness metabolism. Also, EEF1A2 has a close association with phospholipid transport and could positively regulate lipid kinase activity. In previous pig GWAS studies, the EEF1A2 gene has been reported to be associated with intramuscular fat content in longissimus muscle (Serão et al., 2011), and our study confirmed the previous investigation.
For D100, 5 significant SNPs were found to be intragenic or close to 5 genes. Among these genes, only PITX2 and PLA2G12A were relevant with D100. PITX2 has a crucial role in skeletal muscle tissue development and animal organ morphogenesis, and the transforming growth factor beta signaling pathway (Shih et al., 2007). PLA2G12A plays a role in the lipid catabolic process and the phospholipid catabolic process. Although PLA2G12A had been reported involving in lipid metabolism (Ballester et al., 2017) and relevant with intramuscular fat (Puig-Oliveras et al., 2016), it could be helpful for fat deposition in the late stage of pig production.
Population Stratification Control in Different Genetic Backgrounds
The most important problem in GWAS is the risk of false-positive results for significant SNP, which would mislead the process of gene functional verification in the next step (Tucker et al., 2014). In our study, we combined 2 Yorkshire populations with different genetic backgrounds, which can easily cause population stratification, as Figure 1a shows. Therefore, it is essential to control population stratification in GWAS. At present, there are 4 common methods to resolve the problem of population stratification, that is, genomic control (Wang et al., 2015), structured association (Fontanesi et al., 2012), PCA (Fontanesi et al., 2012), and a linear mixed model that includes polygenic effects. In the linear mixed model, a relationship matrix among individuals constructed based on pedigree or genotype data can reduce the influence of population stratification. Although pedigree information could not be used to construct a unified relationship matrix due to no genetic exchange between these 2 populations in recent years, the actual existing relationship between populations can be traced through genomic information. Some weak genetic connectedness between American Yorkshires and British Yorkshires is illustrated in Supplemental Figure S2, implying that different populations of the same breed could have some common genetic background even though gene frequencies changed and LD decayed. In our study, the genomic relationship matrix adequately controlled population stratification, as the Q–Q plot indicated (Supplemental Figure S1). This made the GWAS results more reliable.
Comparing the Single-Population Analysis with the Combined Analysis and Meta-analysis
In our study, we separately carried out GWAS in American and British Yorkshire populations using the same model as in the Materials and Methods section. However, only few significant SNPs were detected in each population, and there were no common significant SNPs among the 2 populations (data were not shown). Although many GWASs have been carried out for the same traits, unfortunately, the consistency of results from different investigations is relatively low. For example, Qiao et al. (2015) performed a GWAS using a White Duroc × Erhualian F2 intercross population and a Chinese Sutai population to reveal SNP and candidate genes related to growth and fatness traits, but no overlap of results was found in the 2 populations. Likewise, Zhu et al. (2014) carried out separate GWAS in 2 populations, one population consisted of 820 Yorkshire pigs and Large White × Landrace intercrosses, and the other consisted of 208 Berkshire × Yorkshire F2 intercrosses. Similarly, there were no overlapping significant SNPs identified in the 2 populations. Fowler (2013) performed a GWAS separately in 3 breeds, Duroc, Yorkshire, and Pietrain, to look for any association between significant SNP and fatness, but no common significant SNP or regions among the 3 breeds were identified. It might be due to the small sample size, different population structures, and the complexity of traits (Willer et al., 2010).
We, therefore, implemented combined analysis and meta-analysis to improve the detection power. Combining different populations could reveal hidden or unclear associations that may not be detected by an independent study (Willer et al., 2010). Combined analysis mainly focuses on 1 breed because they experienced a similar breeding history. Generally, the combined analysis identified more significant SNPs than single-population analysis in most situations, while it also generated larger P-values than single-population analysis in some scenarios as shown in Supplementary Tables S3 and S4. The MAF, HW, and CR of 18 significant SNPs associated with growth and fatness traits identified by combined analysis are presented in Supplementary Table S5. Zhang et al. (2015) combined 2 independent populations of Duroc from Hypor (961 samples) and Genesus (982 samples) to identify SNP and candidate genes related to meat quality, while they did not carry out single-population analysis; the advantage of combined analysis was not clear. In addition, in our study, there are 5 significant SNPs detected in the American, the British, and the combined populations (Supplementary Tables S3 and S4), and no difference was found in the allele frequency at these SNPs among the two single populations and combined population, implying that significant SNP detection was not due to issues with population stratification.
Different from combined analysis, meta-analysis decreased the P-values for all SNPs (Supplementary Tables S1 and S2). This was also confirmed in other studies, for example, Guo et al. (2015) used a meta-analysis to analyze limb bone lengths in 4 different pig populations, showing meta-analysis made the P-value smaller and more significant. Le et al. (2017) used 3 Danish pig breeds (Landrace, Yorkshire, and Duroc) and different meta-analysis methods (a within-breed meta-analysis for multiple traits and a crossbreed meta-analysis for single traits) to perform an association analysis on 4 four conformation traits. The number of significant SNPs identified in the within-breed multiple traits meta-analysis for 3 breeds was larger than in the single-trait analysis. However, meta-analysis actually recalculated new P-values of SNPs only based on those of single population analysis, and it did not take the population information into account. A meta-analysis might deduce high false-positive rate, particularly in the scenario of populations with different genetic backgrounds. Therefore, considering the balance of detection power and false-positive rate, we used the common significant SNPs obtained by combined analysis and meta-analysis for further analysis in this study.
In summary, we conducted a GWAS for traits of backfat thickness and growth in 2,025 Yorkshire pigs from 2 populations with distinct genetic backgrounds by using combined analysis and meta-analysis. A total of 11 and 7 significant SNPs were identified by combined analysis for 10RIBBFT and D100, respectively. And in meta-analysis, 8 and 7 significant SNPs were identified for 10RIBBFT and D100, respectively. Among them, 6 and 5 common significant SNPs in two analysis methods were, respectively, identified associated with 10RIBBFT and D100, and correspondingly explained 2.09% and 0.52% of the additive genetic variance of 10RIBBFT and D100. Gene Ontology analysis highlighted six genes, PCK1, ANGPTL3, EEF1A2, TNFAIP8L3, PITX2, and PLA2G12, as promising candidate genes relevant with backfat thickness and growth. Our findings provide novel insights into the genetic basis of growth and fatness in pigs.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
ACKNOWLEDGMENTS
We thank the Beijing Station of Animal Husbandry, Beijing LM Pig Breeding Technology Co., Ltd., and Beijing Shunxin Agricultural Co., Ltd. for providing blood samples.
This work was supported by grants from the earmarked fund for the China Agriculture Research System (CARS-35), the National Natural Science Foundation of China (31671327), the Beijing City Committee of Science and Technology Key Project (D151100004615004), the Program for Changjiang Scholar and Innovation Research Team in University (grant number IRT_15R621), and the Beijing Innovation Consortium of Agriculture Research System (BAIC02-2016).
LITERATURE CITED
- Adeyemo A., Bentley A. R., Meilleur K. G., Doumatey A. P., Chen G., Zhou J., Shriner D., Huang H., Herbert A., Gerry N. P.,. et al. 2012. Transferability and fine mapping of genome-wide associated loci for lipids in African Americans. BMC Med. Genet. 13:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester M., Ramayo-Caldas Y., Revilla M., Corominas J., Castelló A., Estellé J., Fernández A. I., and Folch J. M.. 2017. Integration of liver gene co-expression networks and eGWAs analyses highlighted candidate regulators implicated in lipid metabolism in pigs. Sci. Rep. 7:46539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R. C., Stylianou I. M., and Rader D. J.. 2011. Functional validation of new pathways in lipoprotein metabolism identified by human genetics. Curr. Opin. Lipidol. 22:123–128. [DOI] [PubMed] [Google Scholar]
- Bereskin B. 1987. Genetic and phenotypic parameters for pig growth and body composition estimated by intraclass correlation and parent-offspring regression. J. Anim. Sci. 65:644. [DOI] [PubMed] [Google Scholar]
- Bidanel J. P., Ducos A., Guéblez R., and Labroue F.. 1994. Genetic parameters of backfat thickness, D100 at 100 kg and ultimate pH in on-farm tested French Landrace and Large White pigs. Livest. Prod. Sci. 40:291–301. [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C. A. M., Vattikuti S., Purcell S. M., and Lee J. J.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Hao C., Zhang W., Shao J., Zhang N., Zhang G., and Liu S.. 2015. Identical expression profiling of human and murine TIPE3 protein reveals links to its functions. J. Histochem. Cytochem. 63:206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do D. N., Ostersen T., Strathe A.B., Mark T., Jensen J., and Kadarmideen H.N.. 2014. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 15:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.Q., Chen X.D., Xia T., Gan L., Qiu H., Dai M.H., Zhou L., Peng Y., and Yang Z.Q.. 2006. Cloning, chromosome mapping and expression characteristics of porcine ANGPTL3 and -4. Cytogenet. Genome Res. 114:44–49. [DOI] [PubMed] [Google Scholar]
- Fernández A.I., Pérez-Montarelo D., Barragán C., Ramayo-Caldas Y., Ibáñez-Escriche N., Castelló A., Noguera J.L., Silió L., Folch J.M., and Rodríguez M.C.. 2012. Genome-wide linkage analysis of QTL for growth and body composition employing the PorcineSNP60 BeadChip. BMC Genet. 13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L., Schiavo G., Galimberti G., Calò D.G., Scotti E., Martelli P.L., Buttazzoni L., Casadio R., and Russo V.. 2012. A genome wide association study for backfat thickness in Italian Large White pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genom. 13:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler K.E., Pong-Wong R., Bauer J., Clemente E.J., Reitter C.P., Affara N.A., Waite S., Walling G.A., and Griffin D.K.. 2013. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genom. 14:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M.E., and Hayes B.J.. 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10:381–391. [DOI] [PubMed] [Google Scholar]
- Guo Y., Hou L., Zhang X., Huang M., Mao H., Chen H., Ma J., Chen C., Ai H., Ren J., and Huang L.. 2015. A meta analysis of genome-wide association studies for limb bone lengths in four pig populations. BMC Genet. 16:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre P., Burgos C., Hidalgo J., Varona L., Carrodeguas J.A., and López-Buesa P.. 2016. c.A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci. Rep. 6:19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.H., Christensen O.F., Nielsen B., and Sahana G.. 2017. Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 49:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S.M., Cho M.H., Young K., Hersh C.P., Castaldi P.J., McDonald M.-L., Regan E., Mattheisen M., DeMeo D.L., Parker M.,andet al. ECLIPSE Investigators, and COPDGene Investigators. 2015. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 16:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena R.N., Ros-Freixedes R., Tor M., and Estany J.. 2016. Genetic marker discovery in complex traits: a field example on fat content and composition in pigs. Int. J. Mol. Sci. 17:2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Oliveras A., Revilla M., Castelló A., Fernández A.I., Folch J.M., and Ballester M.. 2016. Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci. Rep. 6:31803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qayyum R., Becker L.C., Becker D.M., Faraday N., Yanek L.R., Leal S.M., Shaw C., Mathias R., Suktitipat B., and Bray P.F.. 2015. Genome-wide association study of platelet aggregation in African Americans. BMC Genet. 16:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R., Gao J., Zhang Z., Li L., Xie X., Fan Y., Cui L., Ma J., Ai H., Ren J., and Huang L.. 2015. Genome-wide association analyses reveal significant loci and strong candidate genes for growth and fatness traits in two pig populations. Genet. Sel. Evol. 47:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serão N.V., Veroneze R., Ribeiro A.M., Verardo L.L., Braccini Neto J., Gasparino E., Campos C. F., Lopes P.S., and Guimarães S.E.. 2011. Candidate gene expression and intramuscular fat content in pigs. J. Anim. Breed. Genet. 128:28–34. [DOI] [PubMed] [Google Scholar]
- Shih H.P., Gross M.K., and Kioussi C.. 2007. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc. Natl. Acad. Sci. U.S.A. 104:5907–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker G., Price A.L., and Berger B.. 2014. Improving the power of GWAS and avoiding confounding from population stratification with PC-Select. Genetics 197:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P.M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. [DOI] [PubMed] [Google Scholar]
- Wang K., Liu D., Hernandez-Sanchez J., Chen J., Liu C., Wu Z., Fang M., and Li N.. 2015. Genome wide association analysis reveals new production trait genes in a male Duroc population. PLoS One 10:e0139207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J., Li Y., and Abecasis G. R.. 2010. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., and Visscher P.M.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang Z., Bruce H., Kemp R.A., Charagu P., Miar Y., Yang T., and Plastow G.. 2015. Genome-wide association studies (GWAS) identify a QTL close to PRKAG3 affecting meat pH and colour in crossbred commercial pigs. BMC Genet. 16:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Liu X., Max R., Zhang Z., Zhao S., and Fan B.. 2014. Genome-wide association study of the backfat thickness trait in two pig populations. Front. Agric. Sci. Eng. 1:91–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.