Abstract
Bos indicus typically perform better than Bos taurus when consuming a low-quality diet; however, the response to supplementation is generally greater in B. taurus. The underlying mechanisms supporting these responses have not been fully elucidated. Characterization of differences in rumen prokaryotic populations and their functional role in the two subspecies may provide additional insight. Ten cannulated steers (5 Angus and 5 Brahman) were used in concurrent 5 × 5 Latin squares. Animals were offered ad libitum access to rice straw (4.7% CP). Treatments consisted of an unsupplemented control diet and two levels (50 or 120 mg N/kg BW) of isonitrogenous supplements (30% CP), that were either high (H; 74%) or low (L; 26%) in undegradable intake protein. Rumen samples were collected at 0 and 4 h postfeeding and separated into liquid and solid fractions. Rumen bacterial taxa were sequenced utilizing a Roche 454 platform based on the 16s rRNA gene. At 97% sequence similarity, 97,826 operational taxonomic units were identified, which included 24 phyla, 108 families, and 255 genera. Analysis included SAS PROC mixed model, QIIME, and PICRUSt. Across all samples, Bacteroidetes and Firmicutes accounted for 65% and 28% of total bacterial abundance, respectively. The families Prevotellaceae (P = 0.05) and Ruminococcaceae (P = 0.004) and the genera Prevotellaceae (family; P = 0.003) within the phyla Bacteroidetes differed significantly in relative abundance with added protein when compared to the control. Consistent differences in the relative abundance of family and genus taxa between B. indicus and B. taurus suggest roles the symbiotic rumen microbiome may have in the capacity of B. indicus to utilize low-quality forage over a range of supplement types and levels including (Prevotella, Ruminococcus [family], Sphingobacteriaceae [family], Bacteroidales [order], Pontibacter, Bacteroides, Succiclasticum, Barnesiella, and Xylanibacter). Overall bacterial community diversity differences across parameters were limited. Rice straw is recalcitrant to bacterial digestion because of high levels of silica in the epidermis making this straw more resistant to bacterial attachment. Thus, this analysis represents the bacterial diversity and function of the rumen under conditions depleted CP, recalcitrant fiber matrix and restricted digestibility which appear to limit the microbial population to those capable of attaching and digesting complexed structural carbohydrates, resulting in reduced plasticity, and more evenness in diversity across parameters.
Keywords: 16s rRNA, Angus, Bos, indicus, Bos, taurus, Brahman, low-quality forage, microbiome, protein supplementation
INTRODUCTION
Comparative studies of digestion and metabolism indicate Bos taurus are more responsive to protein supplementation while Bos indicus maintain greater levels of performance when consuming a low-quality diet (Habib et al., 2011). Dietary protein in ruminant nutrition is classified based on its availability to the microbes and the host. Degradable intake protein (DIP) is used directly by microbes and is the most direct means of supplying nitrogen to the ruminal ecosystem. Undegradable intake protein (UIP) escapes the rumen to enter the small intestine, thereby increasing protein flow to the animal, resulting in improved forage utilization (Bandyk et al., 2001; Wickersham et al., 2009) indirectly, UIP can be made available to ruminal microbes by being metabolized to urea in the liver and subsequently be recycled to the rumen (Wickersham et al., 2004).
The underlying causes of Bos sub-species differences in response to protein supplementation and consumption of low-quality diets remain unclear, but has been variously attributed to differences in energy requirements (Frisch and Vercoe, 1984), ruminal retention time, urea recycling capacity, and fermentation rate (Hunter and Siebert, 1985). Additionally, we hypothesize that differences in composition and function of the rumen microbiome may contribute to these sub-species differences. To test these hypotheses, we conducted a study with B. taurus and indicus fed a very low-quality rice straw hay with increasing levels of protein supplementation either DIP or UIP to identify supplementation driven shifts in the rumen microbial community using the 16s rRNA gene and 454 pyrosequencing technology. Developing a better understanding how the rumen microbiome adapts to the effects of source (UIP vs. DIP) and level (low vs. high) protein supplementation when fed low-quality forage could enable improved diet formulation, facilitate new probiotics discovery, and enhance our basic understanding of nitrogen metabolism.
MATERIALS AND METHODS
The experimental protocol was approved by the Institutional Animal Care and Use Committee at Texas A&M University.
Experimental Design
Five Angus steers (initial BW = 303 ± 10 kg), subspecies B. taurus, and 5 Brahman steers (initial BW = 323 ± 28 kg), subspecies B. indicus, were fitted with ruminal and proximal duodenal cannulas. Steers were housed in an enclosed, climate controlled barn with continuous lighting. Steers were then used in concurrent 5 × 5 Latin square experiments. Each steer received a subcutaneous vitamin injection (3 mL/animal; Vitamin AD Injection, Sparhawk Laboratories, Inc., Lenexa, KS) at the onset of the trial to prevent deficiencies. They were provided ad libitum access to fresh water and a trace mineral-salt block (≥ 96.0% NaCl, 1.00% S, 0.15% Fe, 0.25% Zn, 0.30% Mn, 0.009% I, 0.015% Cu, 0.0025% Co, and 0.001% Se; United Salt Corporation, Houston, TX)). Rice straw (4.7% CP, 73% NDF) was processed through a tub grinder (76 mm screen) to facilitate feed delivery daily at 0730 h and was provided at 130% of the average intake of the previous 4 d.
Treatments consisted of a rice straw only control (0 mg of N/kg BW daily) and isonitrogenous supplements: two levels of high UIP and two levels of low UIP (50 and 150 mg of N/kg BW daily). High UIP was 100% distillers’ grains (27% CP, 74% UIP, and 88% TDN), while low UIP consisted of soybean meal, corn oil, urea, and soyhulls (27% CP, 74% DIP, and 88% TDN; Table 1). Treatments were given for 15 d periods, with a 9 d adaptation period in between treatment changes. Intake, digestion, and N balance were measured from day 11 through day 14. Samples of rice straw (400 g) were collected from day 10 through day 13 and composited within each period to correspond with urine and feces collected from day 11 through day 14. Orts were collected just before the daily feeding, and orts from day 10 through day 13 were composited for each steer. Feces collected over each 24-h period were thoroughly mixed and 3% of fecal material was collected and composited within animal for each period.
Table 1.
Chemical composition of forage and supplements
| Item | Rice straw2 | L-DIP1,3 | H-DIP1,4 |
|---|---|---|---|
| % DM | |||
| OM | 84.9 | 94.5 | 93.6 |
| CP | 4.7 | 26.7 | 26.9 |
| TDN | NM | 88.0 | 88.0 |
| DIP | NM | 28.0 | 72.0 |
| NDF | 72.8 | 41.8 | 35.0 |
| ADF | 52.3 | 19.0 | 12.4 |
| Acid detergent insoluble ash | 8.8 | 0.3 | 0.4 |
1Treatments: cattle were fed 0, 50 or 150 mg N/kg BW (CON, 50 L-DIP, 150 L-DIP, 50 H-DIP, or 150 H-DIP).
2Ad libitum access.
3L-DIP = low degradable intake protein supplement (100% dried distillers’ grains).
4H-DIP = high degradable intake protein supplement (69.5% wheat middling, 30% soybean meal, and 0.5% urea).
Organic matter digestibility (OMD) of the control rice straw diet was 53.4% to 54.4% for B. indicus and B. taurus steers, respectively. OMD of B. taurus steers did not change with protein supplementation (53.7%), whereas B. indicus OMD (57%) increased (P < 0.02) with protein supplementation (Weldon, 2013).
Control diet forage organic matter intake (FOMI) of B. taurus steers was greater (P < 0.05) (16.5 g/kg BW) than for B. indicus steers (13.5 g/kg BW). B. taurus and B. indicus FOMI increased (P < 0.05) (1.4 g/kg BW and 2 g/kg BW, respectively) with protein supplementation. Total organic matter intake (TOMI) similarly responded to protein supplementation in both sub-species (2.7g/kg BW B. taurus and 2.9 g/kg BW B. indicus) (Weldon, 2013).
Microbiome Sampling
On day 15 of each treatment period, rumen contents were collected at hours 0 and 4 after feeding for 16s rRNA gene analysis. Rumen contents were strained through four layers of cheesecloth to separate liquid and solid fractions. Samples were transferred into a 15-mL polypropylene centrifuge tube and snap frozen in liquid nitrogen. The samples were then transported to the laboratory and archived at −80 °C. The DNA was extracted using the QIAamp stool DNA mini kit (Qiagen, Valencia, CA). Amplification of the V4-V6 segment of the 16S rRNA gene was conducted with barcoded primer tags and the universal eubacterial primers 530Fand 1100R, as previously described (Dowd et al., 2008). Pyrosequencing was performed with a Genome Sequencer FLX System (Roche, Branford, CT) using Titanium chemistry at the MR DNA Molecular Research Lab (Shallowater, TX).
Sequence Analysis
Sequencing data was processed using the QIIME 1.9.0 pipeline (Caporaso et al., 2010) under default parameters. Sequences were depleted of barcodes and primers, then sequences with less than 150 bp, ambiguous base calls, homopolymer runs exceeding 6 bp were removed. Sequences were then denoised and truncated using Denoise Wrapper and AmpliconNoise, with chimera removal at the average base pair quality score less than 25 (Caporaso et al., 2010; Quince et al., 2011). Operational taxonomic units (OTUs) were clustered using UCLUST at 97% similarity (Edgar, 2010). The Greengenes database (5.13) with PyNAST was used to align cluster representative sequences (DeSantis et al., 2006; McDonald et al., 2012). Singleton OTUs were removed. Taxonomic classification was performed using RDP Classifier (Wang et al., 2007). Samples were standardized to the lowest sequencing depth level (2,400 sequences per sample), then alpha, beta, and OTU richness was calculated. UniFrac-based Principle Coordinate Analysis (PCoA) was conducted to visualize the grouping of similar microbiome environments.
Resulting OTUs were then inputted into PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), http://picrust.github.com/picrust) (Langille et al., 2013). Functional predictions were made following the microbiome workflow described by the developers.
Statistical Analysis
Relative abundances of prokaryotic taxa were analyzed with PROC MIXED on SAS 9.4. Terms included in the model were treatment, sub-species × treatment, fraction × treatment, time × sub-species, time, time × treatment, sub-species × fraction, sub-species, and treatment. Period and animal were random effects. Treatment and breed means were calculated using LSMEANS. Student’s T-test, ADONIS, and Bonferroni were used to analyze differences between core microbiome/diversity indexes, PCoA, and functional predictions respectively.
RESULTS
The 16s rRNA gene analysis of rumen contents generated a total of 2,229,152 reads from 200 samples (11,145 reads/sample), which were then used for downstream analysis, including a total of 97,826 OTUs at 97% similarity. These OTUs taxonomically separated into 26 phyla, 108 families, 255 genera, and 394 species. Some OTUs could not be assigned to the highest taxonomic resolution, in this case the lower resolution assignment is reported along with the taxonomic rank in parentheses. Average sequence length for all sequences passing the quality filter was 499 bp. Diversity analyses included chao1 and Shannon indices, which did not differ (Table 2) across treatment, fraction, sub-species, or hour (Student’s t-test: P > 0.05).
Table 2.
Effect of experimental parameters on operational taxonomic unit richness and diversity at 97% similarity after rarefaction
| Treatment1 | Shannon2 | Shannon Error | chao13 | chao1 Error |
|---|---|---|---|---|
| CON | 6.256 | 0.155 | 579.4 | 119.8 |
| 50 H UIP | 6.335 | 0.073 | 604.3 | 111.3 |
| 150 H UIP | 6.298 | 0.13 | 630.7 | 142.1 |
| 50 L UIP | 6.288 | 0.12 | 603.7 | 125.9 |
| 150 L UIP | 6.344 | 0.108 | 640.2 | 127.1 |
| Fraction | ||||
| L | 6.323 | 0.118 | 641.6 | 135.0 |
| S | 6.285 | 0.128 | 581.6 | 111.8 |
| Hour | ||||
| 0.0 | 5.490 | 0.102 | 646.0 | 126.7 |
| 4.0 | 5.466 | 0.095 | 577.3 | 118.9 |
| Sub-species | ||||
| Bos taurus | 5.488 | 0.101 | 615.8 | 125.6 |
| Bos indicus | 5.469 | 0.096 | 607.4 | 129.3 |
1CON = supplemented, 50 H DIP = 50 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5% urea, 150 H DIP = 150 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5%, 50 L DIP = 50 mg N/kg BW 100% distillers’ grains, 150 L DIP = 150 mg N/kg BW 100% distillers’ grains.
2Shannon = diversity indices as calculated by Shannon and Weaver, 1949.
3chao1 = species richness as calculated by Chao et al., 2013.
Core Microbiome
Prokaryotic taxa ubiquitous across all samples were considered members of the core microbiome. There were 32 ubiquitous OTUs shared by all experimental treatments. Bacteroidales, particularly Prevotella, dominated this core microbiome. There were 361 unique OTUs that were shared across all experimental conditions, but were not found ubiquitously across all samples.
Thirty-six unique, but rare species were found in the solid fraction, but not the liquid fraction (Table 3). Collectively, these 36 species accounted for 0.001% of relative abundance, and 9.1% of the total species diversity. Taxonomically, under the conditions of this experiment cattle subspecies B. taurus and B. indicus did not differ in their core microbiome (Student’s t-test: P > 0.05). Across all OTUs, B. taurus had two unique OTUs, while B. indicus had seven (Table 3).
Table 3.
Core and unique OTUs across treatments in Bos taurus and Bos indicus steers (n = 5) fed a low-quality forage
| Measurement | Parameter | Unique, but not ubiquitous |
|---|---|---|
| Fraction | Liquid | 0 |
| Solid | 36 | |
| Treatment | Con | 10 |
| 60 H UIP | 8 | |
| 60 L UIP | 7 | |
| 120 H UIP | 10 | |
| 120 L UIP | 15 | |
| Sub-species | Bos taurus | 2 |
| Bos indicus | 7 | |
| Time | 0 h | 32 |
| 4 h | 0 |
Several unique species were associated with protein type and supplementation level (Table 3). However, similar to fraction, these taxa are found at very low abundance and not ubiquitously across each treatment. In samples collected at hour 0 there were 32 rare, nonubiquitous species compared to hour 4 (Table 3).
PCoA Analysis
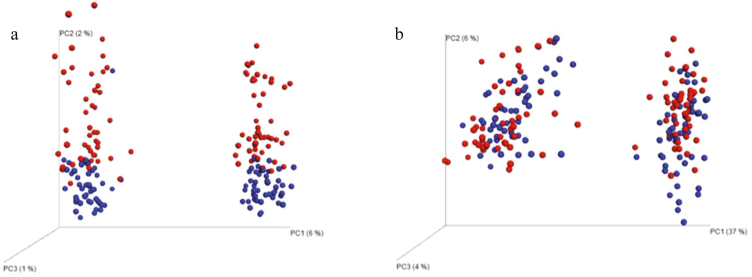
Weighted and unweighted UniFrac PCoA exhibited separation of the prokaryotic community across the liquid and solid fraction (Fig. 1). The solid fraction was more spatially heterogeneous based upon the unweighted and weighted PCoA (Fig. 1). Separation was discernible with the solid and liquid fraction patterns (Fig. 1). The rumen bacterial community of B. taurus displayed greater spatial heterogeneity compared to B. indicus independent of weighting (Fig. 1). Unweighted and weighted UniFrac distances indicated there was no difference in the rumen bacterial communities among diets (control, high or low UIP and DIP), and time (0 and 4 h) (ADONIS: Treatment P = 0.052, Time P = 0.132).
Figure 1.
Rumen bacterial community beta diversity differentiation based of sub-species (Bos taurus-red and Bos indicus-blue, right cluster-liquid and left cluster-solid) using PCoA of unweighted (a) and weighted (b) Unifrac distance (ADONIS: Subspecies unweighted P = 0.001, R2 = 1.25; weighted P = 0.003, R2 = 2.07, Fraction unweighted P = 0.001, R2 = 5.18; weighted P = 0.001, R2 = 29.85).
Effect on Prokaryote Taxa
Across all samples, Bacteroidetes and Firmicutes accounted for 65% and 28% of total bacterial abundance, respectively. An additional 24 phyla were identified. The top 10 in descending order of mean abundance for all samples were: Bacteroidetes, Firmicutes, Spirochaetes, Fibrobacteres, Proteobacteria, TM7, Tenericutes, Chloroflexi, Actinobacteria, and Synergistetes (Table 4). Additional phyla cumulatively accounted for less than 5% of observed sequences. The most abundant 20 bacterial families are listed in Table 5 and the top 20 genera, representing 85% of the total microbiome, are listed in Table 6.
Table 4.
Effect of protein supplementation (treatment), fraction, sub-species (Bos taurus and Bos indicus), and time (0 h and 4 h postfeeding) on the relative abundance of the most abundant phyla
| Phylum1 | Relative abundance | Fraction P-value | Fraction | |
|---|---|---|---|---|
| Liquid | Solid | |||
| Bacteroidetes5 | 65.44 | <0.0001 | 70.00 | 60.80 |
| Firmicutes2,5 | 28.13 | <0.0001 | 23.40 | 32.90 |
| Spirochaetes3,5 | 2.22 | <0.0001 | 1.90 | 2.50 |
| Fibrobacteres3 | 1.14 | >0.05 | ||
| Proteobacteria5 | 0.94 | <0.0001 | 1.40 | 0.50 |
| Tm7 | 0.55 | 0.01 | 0.51 | 0.59 |
| Tenericutes | 0.48 | <0.0001 | 0.79 | 0.17 |
| Chloroflexi5 | 0.2 | <0.0001 | 0.09 | 0.31 |
| Actinobacteria | 0.19 | <0.0001 | 0.11 | 0.27 |
| Synergistetes | 0.18 | <0.0001 | 0.11 | 0.24 |
| Verrucomicrobia4 | 0.17 | >0.05 | ||
1No significant sub-species × treatment or fraction × treatment differences.
2Treatment (P = 0.025) Control = 27.2%, 50 L UIP = 29.1%, 150 L UIP = 27.8%, H UIP = 29.5%, and 150 H UIP = 27.1%, CON= supplemented, 50 H DIP = 50 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5% urea, 150 H DIP = 150 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5%, 50 L DIP = 50 mg N/kg BW 100% distillers’ grains, 150 L DIP = 150 mg N/kg BW 100% distillers’ grains.
3Fraction × sub-species (P < 0.05), Spirochaetes: Angus × liquid 1.67%, Brahman × liquid 2.15%, Angus × solid 2.63%, Brahman × solid 2.44% and Fibrobacteres: Angus × liquid 0.89%, Brahman × liquid 1.25%, Angus × solid 1.29%, Brahman × solid 1.16%.
4Sub-species (P = 0.01), Angus 0.19% vs. Brahman 0.13%.
5Time (P < 0.006) Bacteroidetes 0 h 66.7% vs. 4 h 64.1%, Firmicutes 0 h 27.4% vs. 4 h 29.0%, Spirochaetes 0 h 2.0% vs. 4 h 2.4%, Proterobacteria 0 h 0.9% vs. 4 h 1.0%, Chloroflexi 0 h 0.15% vs. 4 h 0.24%.
Table 5.
Effect of protein supplementation (treatment), fraction, sub-species (Bos taurus and Bos indicus), and time (0 h and 4 h postfeeding) on the relative abundance of the most abundant families
| Sub-species | Fraction | Time | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Family | Sub-species P-value | Bos taurus | Bos indicus | Fraction P-value | Liquid | Solid | Time P-value | 0 h | 4 h |
| Prevotellaceae1,2,3 | 0.04 | 43.82 | 42.28 | 0.01 | 44.01 | 42.19 | 0.001 | 44.05 | 42.15 |
| Ruminococcaceae1 | 0.03 | 13.56 | 14.27 | <0.0001 | 10.90 | 16.99 | 0.01 | 13.56 | 14.33 |
| Sphingobacteriaceae | 0.04 | 7.77 | 8.91 | <0.0001 | 10.03 | 6.59 | 0.01 | 8.73 | 7.87 |
| Lachnospiraceae | 0.64 | 7.23 | 7.05 | <0.0001 | 4.09 | 10.25 | 0.04 | 6.85 | 7.49 |
| Bacteroidales (order)2 | <0.0001 | 2.9 | 4.3 | <0.0001 | 5.72 | 1.44 | 0.14 | 3.42 | 3.73 |
| Cytophagaceae | <0.0001 | 4.18 | 2.57 | 0.27 | 3.25 | 3.45 | 0.55 | 3.39 | 3.31 |
| Bacteroidaceae | 0.003 | 2.4 | 2.73 | <0.0001 | 2.36 | 2.75 | 0.77 | 2.58 | 2.54 |
| Porphyromonadaceae2 | 0.02 | 2.35 | 2.13 | 0.01 | 2.36 | 2.11 | 0.20 | 2.19 | 2.28 |
| Spirochaetaceae3 | 0.19 | 2.14 | 2.28 | <0.0001 | 1.90 | 2.53 | 0.00 | 2.01 | 2.42 |
| Erysipelotrichaceae | 0.85 | 1.89 | 1.86 | <0.0001 | 3.19 | 0.54 | 0.35 | 1.81 | 1.93 |
| Veillonellaceae3 | 0.01 | 1.71 | 1.41 | 0.001 | 1.71 | 1.41 | 0.09 | 1.62 | 1.50 |
| Clostridiaceae | 0.9 | 1.17 | 1.16 | <0.0001 | 0.86 | 1.48 | 0.28 | 1.14 | 1.20 |
| Fibrobacteraceae3 | 0.34 | 1.09 | 1.19 | 0.19 | 1.07 | 1.23 | 0.08 | 1.05 | 1.24 |
| Marinilabiaceae3 | 0.12 | 0.9 | 0.99 | 0.39 | 0.93 | 0.97 | 0.05 | 0.90 | 1.00 |
| Leuconostocaceae | 0.28 | 0.7 | 0.75 | <0.0001 | 0.84 | 0.60 | 0.44 | 0.71 | 0.73 |
| Cryomorphaceae2 | 0.22 | 0.53 | 0.6 | 0.001 | 0.67 | 0.46 | 0.01 | 0.62 | 0.50 |
| Rikenellaceae | 0.46 | 0.6 | 0.53 | <0.0001 | 0.43 | 0.71 | 0.04 | 0.62 | 0.52 |
1Treatment (P ≤ 0.05), Prevotellaceae CON 44.66%, 50 L UIP 41.99% 150 L UIP 43.18%, Ruminococcaceae CON 13.49% 50 L UIP 14.45% 150 L UIP 13.81% 50 H UIP 14.94% 150 H UIP 13.00, CON= supplemented, 50 L UIP = 50 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5% urea, 150 L UIP = 150 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5%, 50 H UIP = 50 mg N/kg BW 100% distillers’ grains, 150 H UIP = 150 mg N/kg BW 100% distillers’ grains.
2Fraction × sub-species (P < 0.05).
3Fraction × treatment (P < 0.001).
Table 6.
Effect of protein supplementation (treatment), fraction, sub-species (Bos taurus and Bos indicus), and time (0 h and 4 h postfeeding) on the relative abundance of the most abundant genera
| Sub-species | Fraction | Time | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Family | Sub-species P-value | Bos taurus | Bos indicus | Fraction P-value | Liquid | Solid | Time P-value | 0 h | 4 h |
| Prevotella 1,2 | 0.01 | 28.85 | 31.00 | <0.0001 | 32.81 | 28.17 | <.0001 | 30.89 | 29.09 |
| Prevotellaceae (family)2,3 | 0.13 | 11.67 | 10.96 | 0.002 | 10.70 | 11.85 | 0.97 | 11.31 | 11.32 |
| Ruminococcaceae (family)1 | 0.004 | 8.70 | 8.21 | 0.21 | 8.36 | 8.56 | <.0001 | 8.10 | 8.82 |
| Sphingobacteriaceae (family) | 0.03 | 7.55 | 6.48 | <0.0001 | 8.46 | 5.54 | 0.02 | 7.35 | 6.64 |
| Saccharofermentans | 0.36 | 3.82 | 3.64 | <0.0001 | 1.39 | 6.10 | 0.97 | 3.74 | 3.75 |
| Bacteroidales (order) | <0.0001 | 4.30 | 2.90 | <0.0001 | 5.72 | 1.43 | 0.14 | 3.42 | 3.74 |
| Pontibacter | <0.0001 | 2.69 | 4.36 | 0.59 | 3.45 | 3.55 | 0.5 | 3.55 | 3.46 |
| Bacteroides | 0.01 | 2.86 | 2.54 | <0.0001 | 2.43 | 2.94 | 0.69 | 2.71 | 2.66 |
| Treponema | 0.76 | 1.73 | 1.71 | <0.0001 | 1.10 | 2.38 | 0.001 | 1.54 | 1.91 |
| Erysipelotrichaceae (family) | 0.61 | 1.69 | 1.76 | <0.0001 | 2.98 | 0.45 | 0.47 | 1.67 | 1.76 |
| Succiniclasticum | 0.01 | 1.36 | 1.62 | 0.001 | 1.63 | 1.34 | 0.06 | 1.56 | 1.42 |
| Blautia | 0.2 | 1.41 | 1.49 | <0.0001 | 0.88 | 2.03 | 0.57 | 1.47 | 1.44 |
| Clostridium | 0.98 | 1.29 | 1.30 | <0.0001 | 0.93 | 1.66 | 0.22 | 1.26 | 1.33 |
| Fibrobacter 4 | 0.34 | 1.19 | 1.09 | 0.19 | 1.07 | 1.23 | 0.08 | 1.05 | 1.24 |
| Ruminococcus | 0.29 | 1.03 | 0.96 | <0.0001 | 0.53 | 1.48 | 0.72 | 1.00 | 1.01 |
1Sub-species × treatment (P < 0.05).
2Fraction × treatment (P < 0.05)
3Treatment (P = 0.0029) CON 11.70% 50 H UIP 11.47% 150 H UIP 12.97% 50 L UIP 10.04% 150 L UIP 10.40%, CON= supplemented, 50 H DIP = 50 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5% urea, 150 H DIP = 150 mg N/kg BW 69.5% wheat middling, 30% soybean meal, and 0.5%, 50 L DIP = 50 mg N/kg BW 100% distillers’ grains, 150 L DIP = 150 mg N/kg BW 100% distillers’ grains.
4Fraction × sub-species (P < 0.05).
Fraction
Of the experimental parameters studied, the most consistent differences in the rumen bacterial community were seen when comparing the liquid and solid fractions. Four out of the five most abundant phyla differed significantly between fractions (P < 0.05; Table 4). Bacteroidetes and Tenericutes were more prevalent in the liquid fraction. Firmicutes, Spirochaetes, Proteobacteria, Chloroflexi, Actinobacteria, and Synergistetes were all more abundant in the solid fraction (P <0.001). Fibrobacteres and Tm7 did not differ between fractions (P > 0.05). No significant differences were observed in any Archaea populations (P > 0.05).
When families were examined, Prevotellaceae, Ruminococcaceae, Sphingobacteriaceae, Lachnospiraceae, Bacteroidales (Order), Bacteroidaceae, Porphyromonadaceae, Spirochaetaceae, Erysipelotrichaceae, Veillonellaceae, Clostridiaceae, Leuconostocaceae, Cryomorphaceae, and Rikenellaceae all differed in their abundance between the solid and liquid fractions (P ≤ 0.01; Table 5). The relative abundance differed significantly between solid and liquid fraction for 15 out of the 20 top genera (Prevotella, Prevotellaceae [family], Sphingobacteriaceae [family], Saccharofermentans, Bacteroidales [order], Bacteroides, Treponema, Erysipelotrichaceae, Succiniclasticum, Blautia, Clostridium, Fibrobacter, Ruminococcus, Butyrivibrio, Barnesiella, and Xylanibacter) (P ≤ 0.002; Table 6).
Cattle Sub-species
Cattle sub-species influenced the relative abundance of 7 out of the top 20 predominant families and 9 out of the top 20 predominant genera (Prevotella, Ruminococcus [family], Sphingobacteriaceae [family], Bacteroidales [order], Pontibacter, Bacteroides, Succiclasticum, Barnesiella, and Xylanibacter) (P ≤ 0.05; Table 6). The average percent difference between the cattle sub-species was 12% at the genus and family level. The greatest differences were observed in Sphingobacteriaceae (family) 15.3%, Bacteroidales (order) 38.9%, Pontibacter 47.4%, and Succiniclasticum 17.4%.
Treatment
Firmicutes responded to most protein supplementation treatments with a maximum of 8.1% difference observed between the control and high UIP (27.1% vs. 29.5%; P = 0.02), however at higher resolution taxa no differences were observed. The families Prevotellaceae (P = 0.05) and Ruminococcaceae (P = 0.004) and the genera Prevotellaceae (family) (P = 0.003), a decrease in relative abundance was observed with protein supplementation over the control, however these differences in treatment did not conform to linear or quadratic response contrast models (P > 0.09). No differences in diversity or relative abundance of methanogenic and pathogenic taxa were attributable to type or level of protein supplementation protein supplementation (P > 0.05).
Time
Four of the five most abundance phyla differed across time (P < 0.006; Table 4). Relative abundance of Bacteroidetes decreased from hour 0 to hour 4 postfeeding, while Firmicutes increased at hour 4 postfeeding (P < 0.006; Table 4). Proteobacteria and Chloroflexi increased after feeding (P < 0.006; Table 4). The relative abundance of Fibrobacteres was not influenced by time (P > 0.05). Of the 20 most abundant families, 8 families exhibited significant differences between pre- and postfeeding (P ≤ 0.01; Table 5). At the genera level, Prevotella and Sphingobacteriaceae (genus) decreased, while Ruminococcaceae (genus) and Treponema increased at hour 4 postfeeding (P ≤ 0.001; Table 6). These changes accounted for a range of 6.0% to 21.5% difference in relative abundance and averaged 6.6% (Table 6).
16s rRNA Gene Inferred Functional Pathways
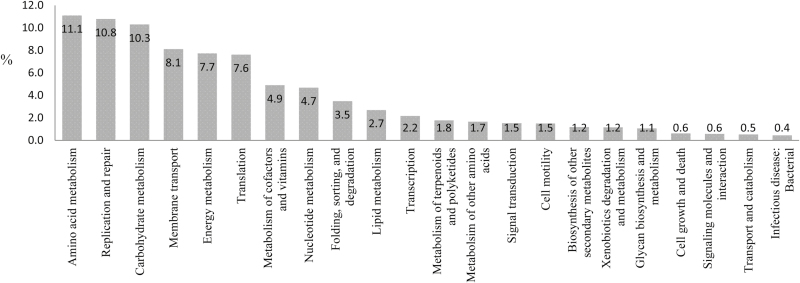
PICRUSt was used to conduct a functional potential analysis of the samples based on the 16s rRNA gene by proxy. The predominant level 2 KEGG pathways were amino acid metabolism (11.0%), replication and repair (10.8%), and carbohydrate metabolism (10.3%; Fig. 2).
Figure 2.
Functional analysis (KEGG PICRUSt) of pathways in the rumen of Bos taurus and Bos indicus.
The solid fraction showed greater relative abundance of level 1 KEGG pathways for cellular processes, environmental information processing, and metabolism (Bonferroni, P < 0.05). Similarly, all level 2-pathway assignments were greater in the solid fraction except for the transport and catabolism category (Table 7). When KEGG assignments were compared across sub-species, no differences were observed, as was the case for supplementation treatment (Bonferroni, P > 0.05). All level 2-pathways were more abundant at hour 4 postfeeding compared to prefeeding (hour 0) (change from hour 0 to hour 4 >8%, Bonferroni P ≤ 0.05).
Table 7.
Functional analysis via PICRUSt of the most abundant pathways in the solid and liquid fraction of the rumen in Bos taurus and Bos indicus steers (n = 5) fed a low-quality forage
| Level 2 KEGG gene pathway | % Solid | % Liquid | % Difference |
|---|---|---|---|
| Replication and repair* | 10.89 | 9.64 | 12.2 |
| Amino acid metabolism* | 10.26 | 9.94 | 3.2 |
| Carbohydrate metabolism* | 10.38 | 9.21 | 11.9 |
| Membrane transport* | 8.61 | 6.83 | 23.1 |
| Translation* | 7.67 | 6.84 | 11.4 |
| Metabolism of cofactors and vitamins* | 4.93 | 4.4 | 11.4 |
| Glycan biosynthesis and metabolism | 3.38 | 3.27 | 3.3 |
| Folding, sorting, and degradation* | 3.48 | 3.14 | 10.3 |
| Lipid metabolism* | 2.73 | 2.41 | 12.5 |
| Transcription* | 2.29 | 1.83 | 22.3 |
| Metabolism of terpenoids and polyketides* | 1.76 | 1.61 | 8.9 |
| Metabolism of other amino acids* | 1.64 | 1.51 | 8.3 |
| Signal transduction* | 1.6 | 1.3 | 20.7 |
| Cell motility* | 1.78 | 1.06 | 50.7 |
| Biosynthesis of other secondary metabolites* | 1.17 | 1.07 | 8.9 |
| Xenobiotics degradation and metabolism* | 1.16 | 1.04 | 10.9 |
| Signaling molecules and interaction | 0.55 | 0.54 | 1.8 |
| Transport and catabolism | 0.49 | 0.5 | 2.0 |
*Indicates P value < 0.05 based on Kruskal–Wallis analysis of variance with Bonferroni corrections.
DISCUSSION
In this study, 16s rRNA gene sequences were used to characterize phylogenetic diversity and potential functional capability of the rumen microbiota of B. taurus and indicus. To the best of our knowledge, no previous study has reported rice straw diet and protein supplementation-dependent changes of the rumen microbes comparing these two sub-species of cattle. Rice straw is a unique low-quality forage in that it contains much more silica (12% to 16%) compared to other straws, which typically have 3% to 5% silica (Jackson, 1977). Soluble silica in the rumen is known to inhibit cellulolytic digestion which leads to a depression in forage digestion; this is theorized to occur via a direct enzymatic depression by the silica and a reduction in the availability of minerals to the rumen microbes (Shimojo and Goto, 1989). Therefore, this study provides a better understanding of the rumen ecosystem comparing these two sub-species under a diet with low fermentation potential, which was previously lacking in the rumen microbial ecology literature.
The rumen core microbiome is the population of microbes that remain stable regardless of conditions. Deviations in and the size of the core microbiome may be indicators of changes in response to diet, time, host effect, or other experimental treatments (Li et al., 2012; Petri et al., 2013; McCann et al., 2014; Omoniyi et al., 2014). For example, there were 32 OTUs across experimental parameters within the core microbiome of steers fed a low-quality rice straw diet in the current study. While, steers fed a higher-quality Bermuda grass diet were found to have 22 core OTUs (McCann et al., 2014). Dairy cows fed a high-quality silage-based diet were found to have 26 unique OTUs (Li et al., 2012). Similarly, Petri et al. (2013) found 11 core OTUs in a high-grain diet and 38 in a high-forage diet. Likewise, Omoniyi et al. (2014) observed six core OTUs with goat fed a higher quality tree-based diet and 11 OTUs present in animals fed low-quality grass. The camel core microbiome consists of approximately 746 OTUs, this study utilized very low-quality natural forages and woody shrubs (Gharechahi et al., 2015).
The effect of the low-quality, slowly fermentable rice straw diet was more pronounced in the solid fraction than the liquid fraction. As such, there was a total of 36 very rare, unique, nonubiquitous species in the solid fraction that were absent in the liquid fraction similar to the results of McCann et al. (2014). This is best illustrated in the nonweighted PCoA, which exhibited greater spatial heterogeneity in solid fraction grouping as compared to the liquid. The solid fraction of rice straw diets represent a more biochemically diverse, nutrient dense, and physiochemically stratified environment (Bae et al., 1997; Van Soest, 2006; Chen et al., 2008) than the liquid fraction; which results in an increased number of ecological niches available that can support greater taxonomic diversity and abundance. Correspondingly, there was an associated increase in metabolic pathway abundance, as reflected in the KEGG analysis (Table 7). The increase in OTUs in the solid fraction and across samples in low-quality diets compared to high-quality diets may be a natural consequence of fermentable substrate diversity in a high forage or low-quality diet or could be a response to another feature of the diet such as an increase in secondary plant compounds or silica, which may support greater bacterial diversity (Bae et al., 1997; Van Soest, 2006; Chen et al., 2008).
While numerous significant differences across experimental parameters were found, the absolute change in the relative abundance of most bacterial taxa was modest, particularly in the context of the differences in protein type and levels among diets and fractions. We had hypothesized that treatment, sub-species, and time would result in noticeable microbial community changes, however beta diversity metrics did not differ across these parameters (Table 2). Other studies examining the rumen microbiome with differing dietary conditions or protein supplementation have consistently found changes in relative abundance (>20%) and significant differences in diversity metrics across most taxa (Callaway et al., 2010; Fernando et al., 2010; Pitta et al., 2010). In retrospect, this is likely the result of the basal rice straw diet, which was very recalcitrant to digestion compared to previously reported straw and hay basal diets, with low carbohydrate diversity and CP, with associated elevated levels of indigestible silica, cutins and fiber, creating a matrix, which is resistant to initial microbial attachment and digestion (Jackson, 1977; Bae et al., 1997; Van Soest, 2006). Consequentially, total tract digestion (%) was low (53.4% Bi vs. 54.4% Bt, P =0.01) and no differences were observed in treatment (National Research Council, 2000; Weldon et al., 2013). These substrate attributes apparently limit attachment and biofilm consortia establishment to a comparatively narrow range of taxa. Thus, resulting in a more even bacterial community observed across all samples, in particularly the solid fraction, and effectively reducing the observed plasticity of the rumen microbial environment. As such, this diet likely selected for certain microbial community structures and functions, such that neither an increase in protein level or type, nor sub-species affectively altered the bacterial community. This is in line with previous studies also showing basal diet to be the major factor in influencing the microbiome (Carberry et al., 2012; Hernandez-Sanabria et al., 2012). Likewise, McCann et al. (2014) who similarly fed a low-quality oat straw diet and supplemented protein in the form of postextraction algal residue did not distinguish PCoA separation with treatment.
A few significant differences were observed with dietary treatment. A high level of DIP supported the greatest number of rare species (15 unique, rare species in high DIP compared to 10 in control). This could reflect DIP supplementation providing fermentable substrate and improving rumen function supporting greater diversity compared to no protein supplement or UIP, which largely escaped the rumen. Likewise, protein supplementation also appeared to increase the prevalence of mobility and chemotaxis pathways, which again indicates a response to the protein supplementation by rumen microbes. But as previously mentioned overall limited dietary effects on the relative abundance and diversity of bacteria taxa were attributed to the low digestion potential of rice straw (Nutrient requirements of beef cattle, 2000).
Two of the greatest concerns to beef cattle producers and consumers are the safety of their product and the environmental repercussions (Galyean et al., 2011). This study detected no differences in potentially pathogenic taxa including Salmonella, Escherichia, and Campylobacter with the inclusion of distillers’ grains, which had previously shown to increase with the inclusion of certain distillers’ grains (Jacob et al., 2007). This is in line with the work of Callaway et al. (2010) whom found no E. coli O157:H7 with increasing inclusion of dried distillers’ grains. No differences were observed in the relative abundance or diversity in any methanogenic taxa. This was confirmed via measurement of methanogen concentrations using quantitative polymerase chain reaction which showed no subspecies, treatment, or time differences observed (Bell, 2015).
Despite differing digestive phenotypes, B. taurus and indicus did not differ in the diversity of their core microbiome under our experimental conditions. Although B. taurus has two unique OTUs and B. indicus has seven, these OTUs do not represent novel taxa. This does contribute to the greater spatial heterogeneity in B. taurus over B. indicus reflected in the unweighted PCoA analysis, and why the weighed PCoA exhibited no separation between the two biological types. There were no true diversity differences between the microbiomes of the two sub-species and dissimilarities are only seen in the relative abundance of some taxa on low-quality rice straw diets.
Although no significant differences in diversity were observed across sub-species, there were taxa that differed in their relative abundance between B. taurus and indicus, which may have been modulated by certain ruminal host traits. For example, B. indicus have greater rumen ammonia concentrations due to a faster digestion rate (Hunter and Siebert, 1985), and which was observed in this experiment as well (Weldon, 2013). It was been hypothesized that this may be partially due to their rumen bacteria being more efficient in utilizing dietary protein and recycled ammonia (Howes et al., 1963). Or physiological differences between sub-species such as ammonia concentration may select for changes in the abundance of bacterial taxa. In other words, it is unclear if the rumen microbiome is the cause or a consequence of differing rumen environments. However, due to the lack of data on the metabolic potential of most of the rumen microbiome, it is difficult to associate individual taxa or functional groups with the physiological capacity of the animal. For example, Pontibacter, within the phylum Bacteroidetes, family Cytophageae, displayed large and significant sub-species differences (Table 6) with nearly double the relative abundance in B. taurus as compared to B. indicus. However, nothing is known about its metabolic potential or role in the rumen, so we cannot posit on why or how it is more abundant in B. taurus. Likewise, Succiniclasticum was significantly higher in B. taurus (Table 6). This genus has members that exclusively derive energy from the conversion of succinate to propionate (Van Gylswyk, 1995). However, this was not reflected in the VFA profile as the molar percentage of propionate was not different across sub-species or treatments (Weldon, 2013). Xylanibacter was also significantly higher in B. taurus. Although its function in the rumen is unknown, it is likely to be involved in the breakdown of plant polysaccharides (Li and Zhao, 2015). It has previously been established to be more abundant in the solid fraction (Petri et al., 2013). The genus Barnesiella was significantly higher in B. indicus. Dietary starch is known to positively increase the relative abundance of Barnesiella in nonlactating Holsteins (Zened et al., 2013). It is unclear if the rumen of B. indicus would create an environment more hospitable for this genus or other taxa via starch availability or other physiological differences across sub-species under current experimental conditions.
Most of the most abundant taxa differed significantly between solid and liquid fractions, however these differences were relatively minor in terms of percent change except for Saccharofermentans, family Clostridiaceae, phylum Firmicutes, which were notably more abundant in the solid than liquid fraction. Petri et al. (2013) found that these taxa were highly associated with a forage-based ration.
Prevotella was the most dominant genus across all communities and variables. Prevotella/Prevotellaceae were particularly sensitive to experimental conditions- changing significantly with sub-species, sub-species × treatment, fraction, fraction × treatment, fraction × sub-species, time × treatment, and time. Prevotella deceased in response to protein supplementation, was found in greater abundance in B. taurus than B. indicus, more abundant in the liquid fraction, and decreased between hour 0 and hour 4. Henderson et al. (2013) and Pitta et al. (2010) also found Prevotella to be more abundant in the liquid fraction. Prevotella/Prevotellaceae was found at a lower relative abundance in this dataset (30% on average) compared to other rumen microbiome analyses which reported upwards of 50% of the total bacterial abundance (Koike et al., 2003; Stevenson and Weimer, 2007; Bekele et al., 2010; Thoetkiattikul et al., 2013). Again, this is likely the result of the rice straw diet and resultant limited rumen available carbohydrate and protein.
Prevotella is a very diverse genus, as such; it is difficult to assign its metabolic strategies/roles (Edwards et al., 2004; Bekele et al., 2010). It is worth noting that Prevotella bryantii and Prevotella ruminicola, which dominate culture based research, only accounted for 0.6% and 3.8% of observed relative abundance respectively. Similarly, P. bryantii and P. ruminicola accounted for 0.0009% and 0.115% of total Prevotella, with unknown species accounting for over 80% of all observed diversity. Previous studies showed Prevotella decreased with grain feeding and is more abundant in forage based diets, which may be related to sensitivity to pH (Mao et al., 2015). de Menezes et al. (2011) found Prevotella to be highest in a forage-based ration. However, if these were the case, the relative abundance would be predicted to be greater in the solid portion. However, other studies have found the opposite trend, with Prevotella increasing with protein supplementation and a higher concentrate diet (Callaway et al., 2010; Fernando et al., 2010; Pitta et al., 2010; Thoetkiattikul et al., 2013; Pitta et al., 2014). Some members of Prevotella are highly associated with protein degradation, such as Prevotella ruminicola, which is one of the few groups that can degrade oligopeptides into amino acids. However, we observed a decrease in Prevotella and Prevotellaceae with both DIP and UIP protein supplementation. If Prevotella ruminicola were associated with a higher concentrate diet, it would explain why the relative abundance was lower in our dataset compared to others. It is clear that the functional roles of Prevotella in the rumen require additional studies as existing literature suggests that Prevotella spp. are both cellulolytic and proteolytic (Edwards et al., 2004; Bekele et al., 2010).
Pre- and postfeeding had a significant impact on most of the most abundant phyla, but this trend was not evident across most families and genera, or the community at large. Rather, only a few genera were observed to respond to time. This trend is illustrated at the community level, i.e., unweighted and weighted PCoA, where there is no spatial segregation between hours 0 and 4 postfeeding, further indicating only a few taxa responded to time after feeding. There was a decrease in some diversity metrics (chao1 and observed species) from time 0 to 4 h postfeeding, though this trend was not statistically significant. This is supported by the observation that more rare species were observed in the prefeeding samples. The increase in abundance of KEGG pathways from hour 0 to hour 4 at first appears to conflict with the idea that feeding drops diversity. However, this may be an artifact of presenting data in terms of relative abundance. If there is a reduction in diversity, then the relative abundance of the remaining pathways will be increased. A decrease in diversity of all other metrics from hour 0 from hour 4 could be due to the underlying lack of readily fermentable substrates at hour 0.
Overall, it is apparent inhibition of microbial function and response is due to feeding rice straw, which was previously established as very difficult to digest. As such, it was difficult to ascertain what, if any, effects the inclusion of increasing UIP or DIP at increasing levels of supplementation had on the rumen environment. This is supported by cattle weight loss over the course of the study (Weldon, 2013; unpublished data). It is reasonable to state in this study, a basal diet of rice straw was by far the dominant determining factor for the rumen microbiome composition and predicted function superseding the effects of host, protein supplementation, and time. While notable differences were observed between solid and liquid fraction prokaryotic diversity and prediction function, there were notable sub-species by treatment effects. The difference in relative abundance of taxa observed across cattle sub-species may be a reflection of differing host rumen environments between subspecies B. indicus and B. taurus such as their ability to utilize low-quality forage and protein supplementation. Therefore, differences in breeds are likely a combination of the physiology and the bacterial populations, however more data are needed to connect taxa with host traits.
LITERATURE CITED
- Bae H. D., McAllister T. A., Kokko E. G., Leggett F. L., Yanke L. J., Jakober K. D., Ha J. K., Shin H. T., and Cheng K. -J.. 1997. Effect of silica on the colonization of rice straw by ruminal bacteria. Anim. Feed Sci. Technol. 65:165–181. [Google Scholar]
- Bandyk C. A., Cochran R. C., Wickersham T. A., Titgemeyer E. C., Farmer C. G., and Higgins J. J.. 2001. Effect of ruminal vs postruminal administration of degradable protein on utilization of low-quality forage by beef steers. J. Anim. Sci. 79:225–231. [DOI] [PubMed] [Google Scholar]
- Bekele A. Z., Koike S., and Kobayashi Y.. 2010. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305:49–57. [DOI] [PubMed] [Google Scholar]
- Bell N. L. 2015. Supplementation strategies to improve efficiency of forage utilization and mitigate enteric methane production in Bos indicus and Bos taurus cattle [Thesis] http://oaktrust.library.tamu.edu/handle/1969.1/155117.
- Callaway T. R., Dowd S. E., Edrington T. S., Anderson R. C., Krueger N., Bauer N., Kononoff P. J., and Nisbet D. J.. 2010. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88:3977–3983. [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I.,. et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry C. A., Kenny D. A., Han S., McCabe M. S., and Waters S. M.. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78:4949–4958. doi:10.1128/AEM.07759-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. L., Wang J. K., Wu Y. M., and Liu J. X.. 2008. Effects of chemical treatments of rice straw on rumen fermentation characteristics, fibrolytic enzyme activities and populations of liquid- and solid-associated ruminal microbes in vitro. Anim. Feed Sci. Technol. 141:1–14. [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E., Callaway T. R., Wolcott R. D., Sun Y., McKeehan T., Hagevoort R. G., and Edrington T. S.. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Edwards J. E., McEwan N. R., Travis A. J., and Wallace R. J.. 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86:263–281. [DOI] [PubMed] [Google Scholar]
- Fernando S. C., Purvis H. T., Najar F. Z., Sukharnikov L. O., Krehbiel C. R., Nagaraja T. G., Roe B. A., and DeSilva U.. 2010. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76:7482–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch J. E., and Vercoe J. E.. 1984. Analysis of growth of different cattle genotypes reared in different environments. J. Agric. Sci. http://agris.fao.org/agris-search/search.do?recordID=US201302564386.
- Galyean M. L., Ponce C., and Schutz J.. 2011. The future of beef production in North America. Anim. Front. 1:29–36. [Google Scholar]
- Gharechahi J., Zahiri H. S., Noghabi K. A., and Salekdeh G. H.. 2015. In-depth diversity analysis of the bacterial community resident in the camel rumen. Syst. Appl. Microbiol. 38:67–76. [DOI] [PubMed] [Google Scholar]
- Habib M., Pollott G., and Leaver D.. 2011. Digestibility and nitrogen balance of high- and low-quality forages supplemented with high- and low-protein concentrates fed to two breeds of cattle. J. Appl. Anim. Res. 39:303–310. [Google Scholar]
- Henderson G., Cox F., Kittelmann S., Miri V. H., Zethof M., Noel S. J., Waghorn G. C., and Janssen P. H.. 2013. Effect of DNA Extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS ONE 8:e74787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanabria E., Goonewardene L. A., Wang Z., Durunna O. N., Moore S. S., and Guan L. L.. 2012. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78:1203–1214. doi:10.1128/AEM.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes J. R., Hentges J. F., and Davis G. K.. 1963. Comparative digestive powers of Hereford and Brahman cattle 1. J. Anim. Sci. 22:22–26. [Google Scholar]
- Hunter R. A., and Siebert S. D.. 1985. Utilization of low-quality roughage by Bos taurus and Bos indicus cattle. Br. J. Nutr. 53:637–648. [DOI] [PubMed] [Google Scholar]
- Jackson M. G. 1977. Rice straw as livestock feed. World Anim. Rev. 23:34–40. [Google Scholar]
- Jacob M. E., Fox J. T., Narayanan S. K., Drouillard J. S., Renter D. G., and Nagaraja T. G.. 2007. Effects of feeding wet corn distillers grains with solubles with or without monensin and tylosin on the prevalence and antimicrobial susceptibilities of fecal foodborne pathogenic and commensal bacteria in feedlot cattle. J. Anim. Sci. 86:1182–1190. [DOI] [PubMed] [Google Scholar]
- Koike S., Yoshitani S., Kobayashi Y., and Tanaka K.. 2003. Phylogenetic analysis of fiber-associated rumen bacterial community and PCR detection of uncultured bacteria. FEMS Microbiol. Lett. 229:23–30. [DOI] [PubMed] [Google Scholar]
- Langille M. G. I., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R.,. et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W., Wu S., Baldwin R. L., Li W., and Li C.. 2012. Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PLoS ONE 7:e29392 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3257242/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., and Zhao X.. 2015. Comparative analyses of fecal microbiota in Tibetan and Chinese Han living at low or high altitude by barcoded 454 pyrosequencing. Sci. Rep. 5:14682 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4595765/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S. -Y., Huo W. -J., and Zhu W. -Y.. 2015. Microbiome–metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 18(2):525–541. [DOI] [PubMed] [Google Scholar]
- McCann J. C., Drewery M. L., Sawyer J. E., Pinchak W. E., and Wickersham T. A.. 2014. Effect of postextraction algal residue supplementation on the ruminal microbiome of steers consuming low-quality forage. J. Anim. Sci. 92:5063–5075. [DOI] [PubMed] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., and Hugenholtz P.. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menezes A. B., Lewis E., O’Donovan M., O’Neill B. F., Clipson N., and Doyle E. M.. 2011. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 78:256–265. [DOI] [PubMed] [Google Scholar]
- National Research Council.. 2000. Nutrient requirements of beef cattle. Washington (DC): National Academy Press, 1996 [i.e. 2000]. [Google Scholar]
- Omoniyi L. A., Jewell K. A., Isah O. A., Neumann A. P., Onwuka C. F. I., Onagbesan O. M., and Suen G.. 2014. An analysis of the ruminal bacterial microbiota in West African Dwarf sheep fed grass- and tree-based diets. J. Appl. Microbiol. 116:1094–1105. doi:10.1111/jam.12450 [DOI] [PubMed] [Google Scholar]
- Petri R. M., Forster R. J., Yang W., McKinnon J. J., and McAllister T. A.. 2012. Characterization of rumen bacterial diversity and fermentation parameters in concentrate fed cattle with and without forage. J. Appl. Microbiol. 112:1152–1162. [DOI] [PubMed] [Google Scholar]
- Petri R. M., Schwaiger T., Penner G. B., Beauchemin K. A., Forster R. J., McKinnon J. J., and McAllister T. A.. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 8:e83424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D. W., Parmar N., Patel A. K., Indugu N., Kumar S., Prajapathi K. B., Patel A. B., Reddy B., and Joshi C.. 2014. Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of Kankrej Cattle. PLoS ONE 9:e111710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D. W., Pinchak W. E., Dowd S. E., Osterstock J., Gontcharova V., Youn E., Dorton K., Yoon I., Min B. R., Fulford J. D.,. et al. 2010. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb. Ecol. 59:511–522. [DOI] [PubMed] [Google Scholar]
- Quince C., Lanzen A., Davenport R. J., and Turnbaugh P. J.. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M., and Goto I.. 1989. Effect of sodium silicate on forage digestion with rumen fluid of goats or cellulase using culture solutions adjusted for pH. Anim. Feed Sci. Technol. 24:173–177. [Google Scholar]
- Stevenson D. M., and Weimer P. J.. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165–174. [DOI] [PubMed] [Google Scholar]
- Thoetkiattikul H., Mhuantong W., Laothanachareon T., Tangphatsornruang S., Pattarajinda V., Eurwilaichitr L., and Champreda V.. 2013. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr. Microbiol. 67:130–137. [DOI] [PubMed] [Google Scholar]
- Van Gylswyk N. O. 1995. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 45:297–300. [DOI] [PubMed] [Google Scholar]
- Van Soest P. J. 2006. Rice straw, the role of silica and treatments to improve quality. Anim. Feed Sci. Technol. 130:137–171. [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., and Cole J. R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon K. 2013. Nitrogen metabolism in Bos indicus and Bos taurus cattle consuming low-quality forages [Thesis] http://oaktrust.library.tamu.edu/handle/1969.1/151030.
- Weldon K. K., McCann J. C., Sawyer J. E., and Wickersham T. A.. 2013. Effect of supplemental protein amount and degradability on intake and digestion in Bos indicus and Bos taurus steers fed rice straw. J. Anim. Sci. 91(Suppl. 2):26. [Google Scholar]
- Wickersham T. A., Cochran R. C., Titgemeyer E. C., Farmer C. G., Klevesahl E. A., Arroquy J. I., Johnson D. E., and Gnad D. P.. 2004. Effect of postruminal protein supply on the response to ruminal protein supplementation in beef steers fed a low-quality grass hay. Anim. Feed Sci. Technol. 115:19–36. [Google Scholar]
- Wickersham T. A., Titgemeyer E. C., Cochran R. C., and Wickersham E. E.. 2009. Effect of undegradable intake protein supplementation on urea kinetics and microbial use of recycled urea in steers consuming low-quality forage. Br. J. Nutr. 101:225–232. [DOI] [PubMed] [Google Scholar]
- Zened A., Combes S., Cauquil L., Mariette J., Klopp C., Bouchez O., Troegeler-Meynadier A., and Enjalbert F.. 2013. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 83:504–514. [DOI] [PubMed] [Google Scholar]