Abstract
Spray-dried porcine plasma (SDPP) has been considered as an alternative for in-feed antibiotics to improve pig growth performance; however, the effect of SDPP on gut microbiota is unknown. The objective of this study was to evaluate effects of feeding SDPP on fecal microbial communities of nursery pigs. Ninety-six weaned pigs were assigned to 16 pens, which were allotted to two dietary treatments, including the control or the control + SDPP (5% and 2.5% SDPP inclusion in phase 1 and 2, respectively) diet. Fecal samples were collected at d 0, 7, 14, 21, and 28. Multiplex sequencing of V3 region of the 16S rRNA gene was used to characterize the bacterial community structure of fecal samples. Pearson’s correlation tests were performed in Calypso to identify bacterial taxa that were either positively or negatively associated with overall growth performance. Feeding SDPP altered microbial structure at family, genus, and operational taxonomic unit (OTU) classifications; however, fecal microbes shifted with time. At the family level, Clostridiaceae increased (P < 0.001) on d 14, but decreased (P < 0.05) on d 28 in SDPP-fed pigs compared with control pigs. Decreased Veillonellaceae (P < 0.05; d 14) and Lachnospiraceae (P = 0.001; overall) were observed in SDPP-fed pigs compared with control pigs. Feeding SDPP increased lactic acid–producing bacteria (Lactobacillus delbrueckii, d 7) and cellulolytic bacteria (Ruminococcus albus, d 7; Clostridium thermocellum, d 7 and 14; and Clostridium saccharoperbutylacetonicum/beijerinckii, d 14; and Megasphaera elsdenii, d 21). On d 28, feeding SDPP decreased (P < 0.05) Clostridium difficile compared with control pigs. In conclusion, feeding SDPP altered fecal microbial communities in nursery pigs. The results of this study may provide information to help explain the positive effects associated with feeding SDPP on nutrient digestibility and gut health of nursery pigs.
Keywords: fecal microbiota, pigs, spray-dried porcine plasma
In-feed antibiotics are used as growth promoters for pigs and other farm animals. The mode of action of antibiotics may be mediated by modulation of the gut microbiota, as suggested by a lack of an effect in germ-free animals (Coates et al., 1963; Dibner and Richards, 2005). For example, antibiotics can reduce the total number of bacteria and change the bacterial composition in the gastrointestinal tract (GIT) of the host, particularly increasing Lactobacillus (Collier et al., 2003; Kim et al, 2012). Given that it has been reported that energy expenditure of the portal drained viscera (intestines, pancreas, spleen, and stomach) may account for 20–35% of whole-body energy expenditure (Yang et al., 2016), it has been hypothesized that a reduction in bacterial load in the GIT may substantially increase energetic efficiency of animals (Allen et al., 2013). In addition, changes in microbial communities attributed to antibiotics may be beneficial with respect to energy production and conversion leading to improved growth performance in pigs fed antibiotics (Looft et al., 2012).
There is strong evidence to suggest that spray-dried porcine plasma (SDPP) can improve growth performance (Grinstead et al., 2000; Pierce et al., 2005; Tran et al., 2014), enhance gut barrier function (Perez-Bosque et al., 2006; Peace et al., 2011; Tran et al., 2014), and modulate the immune system (Nofrarias et al., 2006) of pigs and other species. Therefore, SDPP has been considered an alternative to in-feed antibiotics. However, the effects of SDPP on gut microbiota are mostly unknown. Pigs infected with Escherichia coli and fed diet containing SDPP and Bacillus subtilis exhibited an increased microbial richness and diversity compared with negative-control pigs; however, feeding SDPP resulted in a decreased response in bacterial richness and diversity compared with feeding antibiotics (Bhandari et al., 2008). In another study, supplementation of plasma powder to weaned pigs infected with F18+ E. coli decreased the proliferation of this pathogen (Nollet et al., 1999). It was hypothesized that the glycan fraction of glycoprotein components in plasma powder blocks the receptors on enterocytes leading to the reduction of E. coli adhesion to the gut mucosa (Sanchez et al., 1993). In addition, results from Hermes et al. (2012) indicated that glycoproteins in plasma may be responsible for increased lactobacilli as demonstrated in the case of casein-derived glycoproteins and Torrallardona et al. (2003), using culture-dependent techniques, concluded that inclusion of spray-dried animal plasma in weaned pig diets favored the growth of lactobacilli in the ileum and cecum.
Results from this experiment documenting the positive effects of SDPP on growth performance and gut health in nursery pigs have been previously published (Tran et al, 2014). In light of the fact that, to our knowledge, there is little to no available data to characterize the diversity and composition of gut microbes in pigs fed SDPP at the community-wide level, we hypothesized that SDPP dietary inclusion may drive changes in microbial communities, resulting in positive outcomes in growth performance. Therefore, the objective of this study was to evaluate the shift of 16S rRNA of fecal microbes in response to dietary SDPP using a next generation sequencing approach, particularly Ion Torrent Platform (Life Technologies, South San Francisco, CA).
MATERIALS AND METHODS
The experimental protocol was reviewed and approved by the Institutional Animal Care and Use Committee of the University of Nebraska, Lincoln.
Animal and Experimental Design
A total of 96 weaned pigs (NE female × Danbred sire; age, 20 ± 1.2 d; initial BW, 6.06 ± 0.02 kg) were sorted by initial BW and sex and randomly assigned to 16 pens (n = 8 pens/treatment). Pens were randomly allocated to one of two dietary treatments: 1) control (no SDPP); and 2) control + SDPP. Diets were formulated to meet or exceed the nutrient requirement of swine according to National Research Council (1998). Antibiotics were excluded from all diets. The ingredient composition and calculated analysis of experimental diets are presented in Table 1. The feeding experiment consisted of two phases: phase 1 (wk 1 and 2; with 5% SDPP) and phase 2 (wk 3 and 4; with 2.5% SDPP). Pigs were housed in a temperature-controlled room (27 to 29 °C) and provided ad libitum access to feed and water throughout the study.
Table 1.
Ingredient and chemical composition of the experimental diets (%, as-fed basis)
| Phase 1 (wk 1 to 2) | Phase 2 (wk 3 to 4) | |||
|---|---|---|---|---|
| SDPP | Control | SDPP | Control | |
| Ingredient, % | ||||
| Corn | 34.10 | 34.10 | 47.96 | 47.96 |
| Soybean meal, 46.5% CP | 10.00 | 10.00 | 20.50 | 20.50 |
| SDPP | 5.00 | 0.00 | 2.50 | 0.00 |
| Select menhaden fish meal | 6.00 | 6.00 | 8.00 | 8.00 |
| Spray-dried whey | 20.00 | 20.00 | 7.50 | 7.50 |
| DairyLac 801 | 7.00 | 7.00 | 5.75 | 5.75 |
| Extruded soy protein concentrate | 8.00 | 8.00 | 0.00 | 0.00 |
| Corn starch | 5.00 | 8.64 | 2.50 | 4.29 |
| Dicalcium phosphate, 18.5% P | 0.28 | 0.66 | 0.50 | 0.68 |
| Limestone | 0.40 | 0.20 | 0.18 | 0.10 |
| Salt | 0.30 | 0.30 | 0.30 | 0.30 |
| Zinc oxide | 0.30 | 0.30 | 0.30 | 0.30 |
| Vitamin premix2 | 0.25 | 0.25 | 0.25 | 0.25 |
| Trace mineral premix3 | 0.15 | 0.15 | 0.15 | 0.15 |
| l-Lysine·HCl | 0.10 | 0.51 | 0.31 | 0.51 |
| dl-Methionine | 0.13 | 0.28 | 0.16 | 0.23 |
| l-Threonine | 0.00 | 0.23 | 0.13 | 0.25 |
| l-Tryptophan | 0.00 | 0.07 | 0.03 | 0.07 |
| Corn oil | 3.00 | 3.00 | 3.00 | 3.00 |
| l-Isoleucine | 0.00 | 0.09 | 0.00 | 0.06 |
| l-Valine | 0.00 | 0.23 | 0.00 | 0.11 |
| Total | 100.00 | 100.00 | 100.00 | 100.00 |
| Analyzed composition, % | ||||
| Lys | 1.512 | 1.52 | 1.47 | 1.48 |
| Met + Cys | 0.79 | 0.76 | 0.79 | 0.73 |
| Thr | 0.98 | 0.97 | 0.96 | 0.95 |
| Trp | 0.29 | 0.28 | 0.28 | 0.26 |
| CP | 23.12 | 20.38 | 22.80 | 20.91 |
| Calculated composition, % | ||||
| Total lysine | 1.60 | 1.58 | 1.58 | 1.57 |
| Standardized ileal digestible lysine | 1.47 | 1.47 | 1.45 | 1.45 |
| CP | 23.17 | 20.21 | 22.19 | 20.72 |
| ME, kcal/kg | 3,500 | 3,503 | 3,491 | 3,492 |
| Ca | 0.85 | 0.85 | 0.83 | 0.83 |
| P | 0.72 | 0.72 | 0.74 | 0.73 |
| Available P | 0.51 | 0.51 | 0.49 | 0.49 |
| Lactose | 20.00 | 20.00 | 10.00 | 10.00 |
1Dairylac 80 is a sweet and dried whey soluble product (International Ingredient Corporation, St. Louis, MO) containing 3.2% CP, and 0.06% Lys (analyzed composition) and 80% lactose.
2Supplied per kg of diet: vitamin A (as retinyl acetate), 5,500 IU; vitamin D (as cholecalciferol), 550 IU; vitamin E (as α-tocopheryl acetate), 30 IU; vitamin K (as menadione dimethylpyrimidinol bisulfate), 4.4 mg; riboflavin, 11.0 mg; d-pantothenic acid, 22.05 mg; niacin, 33.0 mg; vitamin B12 (as cyanocobalamin), 33.0 mg.
3Supplied per kg of diet: copper (as CuSO4·5H2O), 10 mg; iodine (as Ca(IO3)·H2O), 0.25 mg; iron (FeSO4·2H2O), 125 mg; manganese (MnO), 15 mg; selenium (Na2SeO3), 0.3 mg; and zinc (ZnSO4·H2O), 125 mg.
Fecal Sample Collection and DNA Extraction
Fecal samples (1 pig/pen) were collected at the beginning (d 0) and weekly thereafter (d 7, 14, 21, and 28). Each treatment had the same number of gilts (n = 4) and barrows (n = 4) and the same pigs were sampled throughout the study. Clean and disinfected plastic loops were inserted into the rectum of pigs for fecal sampling. The collected samples were put in 2-mL autoclaved tubes and stored at −20 °C for subsequent analysis.
The genomic DNA was extracted from 0.3 g of fecal sample following the procedure described by Martinez et al. (2009). The DNA pellet was resuspended in Tris-HCl buffer (10 mM, pH 8.0). The DNA concentration and purity was measured using Nanodrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). The extracted DNA samples were checked on 1% agarose gel for integrity. The DNA samples were subsequently aliquoted and stored at −20 °C.
PCR Amplicon Preparation and Sequencing of 16S rRNA Ion Tags Using Ion Torrent (PGM) Platform
The preparation of amplicon library and sequencing process were conducted in the Fernando lab (Department of Animal Science, University of Nebraska). Briefly, V3 region of the 16S rRNA gene was amplified by PCR using barcoded primers. The PCR mixture (25 µL) consisted of 0.25 µL of Terra PCR Direct Polymerase Mix (1.25 U/µL), 12.5 µL of 2 × PCR Direct reaction buffer, 0.5 µL of 341 forward primers (25 µmol), 1 µL of barcoded 518 reverse primers (10 µmol), 0.25 µL of BSA, and 2 µL of genomic DNA (20–50 ng). Amplification condition was 98 °C for 3 min; 30 cycles of 98 °C for 30 s, 53 °C for 30 s, and 68 °C for 40 s; and a single final extension step at 68 °C for 4 min. All amplicons from the individual PCR mixture were pooled to equal concentrations based on the preferred band intensity on a 2.0% agarose gel using GeneTools 1D (Syngene, Frederick, MD) gel analysis. The pooled amplicons were purified using MinElute PCR Purification Kit (Cat. No. 28004; Qiagen, Valencia, CA). The DNA fragment corresponding to V3 region in the pooled amplicon mixture was picked on an E-gel SizeSelect 2% agarose (Cat. No. G6610-02; Invitrogen, Life Technologies, South San Francisco, CA). The quality and concentration of selected DNA fragment were measured using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). The DNA samples were diluted to 15 pM for templating to Ion Sphere particles and subsequent emulsion PCR using the Ion OneTouch 2 instrument (Life Technologies, South San Francisco, CA). Templated-Ion Sphere particles were sequenced on a 316-chip kit with the Ion Torrent Personal Genome Machine (Life Technologies, South San Francisco, CA) according to manufacturer’s protocols.
Quality Control of the Sequence Tags and Sequence Analysis to Characterize Microbial Communities
The obtained sequences were filtered using PGM software (Torrent Suite version 3.4.2) to remove low quality and polyclonal reads. The BaseCaller parameters were set with quality cutoff of 15, quality window of 30, and adaptor cutoff of 16. The sequences were de-multiplexed to remove barcodes and primers. After trimming, only sequences containing 80 to 176 base pairs (length of the amplified region) were selected for subsequent analyses. After quality control, n = 8 samples for each treatment at a timepoint, except n = 6 for control at d 0; n = 7 for control at d 7; n = 3 for SDPP at d 7; n =7 for SDPP at d 14; and n = 7 samples for control at d 21.
To characterize the microbial communities from the obtained sequences, a combination of phylogenetic and operational taxonomic unit (OTU)-based approaches was used. The analyses were conducted based on the total and core sequences using Mothur (Schloss et al., 2009; http://www.mothur.org/wiki/454_SOP), QIIME (Caporaso et al., 2010), and Ribosomal Database Project (RDP) pipelines (http://pyro.cme.msu.edu). First, the trimmed unique sequences were submitted to RDP pipelines for alignment using Aligner tool. Subsequently, in Mothur environment, chimeras were checked and removed from the aligned sequences using uchime v4.2.0 (Edgar et al., 2011) and a reference database for 16S rRNA genes (http://drive5.com/uchime/gold.fa). The cleaned sequences were clustered into OTU with a sequence identity threshold of 97%. In QIIME, an OTU table was constructed from the OTU abundances generated in Mothur.
Total sequence analysis.
Representative sequences chosen for each OTU in Mothur environment were assigned using the RDP Bayesian Classifier trained on Greengenes taxonomy (DeSantis et al., 2006). Cyanobacteria were filtered out before the OTU table was used to determine the diversity of samples. The number of sequences was not the same among samples; therefore, total sequences of each sample were subsampled to the lowest sequences (4,618 sequences) using QIIME before statistical comparisons between treatments were done. Chao1 and Shannon indices were used as measurements of alpha-diversity of the microbial communities. Chao1 index measures the microbial richness by taking the number of observed species divided by the ratio of singletons (species captured once) over doubletons (species captured twice). Shannon index takes into account a total number of sequences and measures the microbial species richness (amount) and evenness.
Core sequence analysis.
In QIIME, core sequences were selected to be present in more than 63% of a treatment at a given timepoint after removal of singletons from the total sequences. In order to statistically compare abundance of core OTUs among treatments, the number of sequences per each OTU was normalized to the number of total core sequences of each sample. The MIXED procedure of SAS (SAS Inst. Inc, Cary, NC) was used to identify OTUs that had a significant treatment × time interaction. These OTUs were sorted by the abundance and aligned to the most closely related species in NCBI using a Basic Local Alignment Search Tool (BLASTn). A phylogenetic tree was constructed from the representative sequence per OTU using CLEARCUT command in Mothur. The generated tree was used for subsequent beta-diversity analysis in QIIME. Unweighted Unifrac and UPGMA dendrograms were generated to visually compare beta-diversity between treatments. In some cases, OTUs that were initially identified distinct had high sequence similarities (≥97%; ClustalW2; http://www.ebi.ac.uk/Tools/services/web/toolform.ebi?tool=clustalw2); thus, they were combined and considered as single OTUs.
Shared and unique core OTUs between treatments.
In QIIME, core taxa were pooled by treatment × time interaction (control and SDPP at d 0, 7, 14, 21, and 28) and a pooled OTU table was created. To obtain core sequences and OTUs that were shared between control and SDPP treatments at each timepoint, Venn diagrams were generated using VENN command in Mothur. The unique OTUs among groups were determined by using FILTER command in QIIME after excluding the list of shared OTUs. In order to statistically compare between control and SDPP treatment at each timepoint, the number of shared or unique sequences was normalized to total core sequences of each group at a timepoint. The top 10 most abundantly shared OTUs in each treatment at a timepoint were selected and subjected to BLASTn for alignment against the closest species. Because the unique OTUs accounted for less abundant OTUs, only the OTUs having greater than 0.02% core sequences were selected and subjected to BLASTn for alignment. To visually display the distribution of shared OTUs among groups, a heatmap and phylogenetic tree were generated by Java TreeView (http://jtreeview.sourceforge.net/) and Mega5 (Tamura et al. 2011; www.megasoftware.net.), respectively.
Correlation of bacterial taxa and growth performance.
Calypso (Zakrzewski et al., 2016) was used for Pearson’s analysis to determine the correlation of the top 100 core OTUs and overall ADG and BW on d 7, 14, 21, and 28. The sequence data were normalized using total-sum normalization, which divides sequence reads by the total number of reads in each sample. Rare taxa with less than 0.01% were also removed before the analysis. Analyses based on taxonomy assignments used the websites Calypso at http://bioinfo.qimr.edu.au/calypso. The differential abundance OTUs were subjected to BLASTn for alignment.
Statistical Analysis
Data were analyzed as a randomized complete design using MIXED procedure of SAS (SAS Inst. Inc, Cary, NC) with pig as the experimental unit. The statistical model included treatment, day, and their interaction as fixed effects. The pdiff option was used for pair-wise comparisons. Means were presented as least-squares means ± SEM. Statistical comparison of the differences in beta diversity was conducted on the unweighted unifrac distance matrices using ANOSIM function of Qiime. A P-value of ≤0.05 was considered significant and a P value of >0.05 but ≤0.10 was considered as a trend.
RESULTS
Alteration of Fecal Microbiota by Consumption of SDPP: Phylogenetic-Based Analysis
Multiplex sequencing of 16S rRNA amplicons of 70 fecal DNA samples produced an average of 38,367 sequences (total of 2,685,690 sequences) and 717 OTUs (total of 50,205 OTUs) per sample after quality control and chimeric checking. For core sequence analysis, average of 36,629 core sequences (total 2,564,076 sequences) and 14 OTUs per sample were identified (total of 1,042 OTUs). Core sequences covered 95% of total sequences of 70 samples in this study, indicating that a core set of bacteria were shared and present in most pigs.
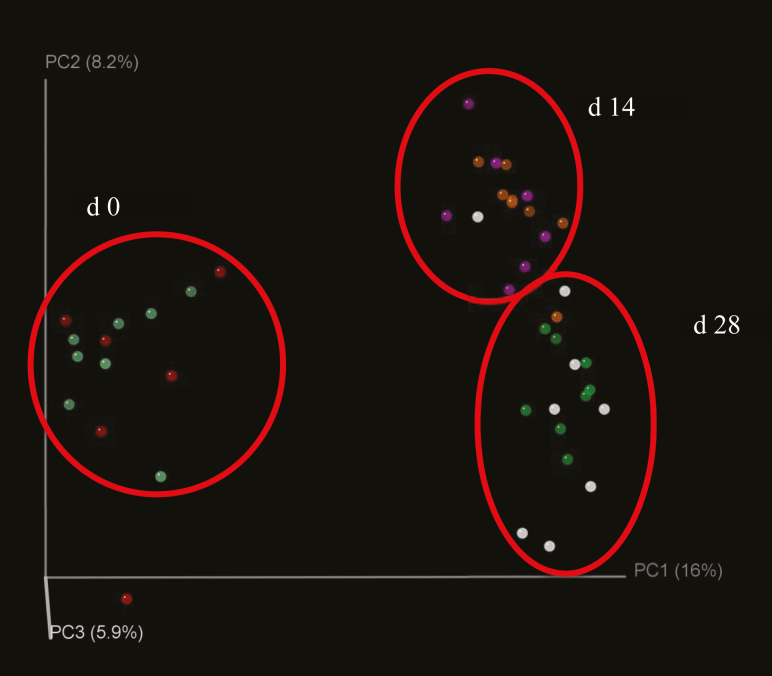
Feeding SDPP did not affect alpha-diversity measurements estimated by Shannon and Chao1 indices (Table 2); except for a reduction (P = 0.001) of Chao1 index in pigs fed SDPP compared with control pigs on d 14. Similar, unweighted unifrac displaying beta diversity (Figure 1) at d 0, 14, and 28 of core bacterial taxa indicates a clear difference (day; P < 0.01) between bacterial community structure at weaning (d 0, before solid feed consumption) and after weaning (d 14 and 28, when pigs were fed solid feed). Alpha-diversity was affected by age (time) of pigs, indicated by greater Chao1 and Shannon indices on d 0, 7, and 14 compared with those of d 21 and 28 postweaning.
Table 2.
Alpha-diversity indices of total sequences in pigs fed SDPP or control diet1
| Day | Treatment2 | Total sequences | Subsampled to 4,618 sequences | ||
|---|---|---|---|---|---|
| Chao13 | Shannon4 | Chao13 | Shannon4 | ||
| 0 | CTL | 4,232 | 4.82 | 1,178 | 4.68 |
| SDPP | 9,197 | 5.73 | 1,625 | 5.52 | |
| 7 | CTL | 3,289 | 5.02 | 1,410 | 4.90 |
| SDPP | 5,113 | 5.78 | 1,673 | 5.67 | |
| 14 | CTL | 6,108 | 5.26 | 1,522 | 5.10 |
| SDPP | 4,451 | 5.01 | 1,069* | 4.90 | |
| 21 | CTL | 4,233 | 4.29 | 1,202 | 4.16 |
| SDPP | 8,175 | 3.77 | 929 | 3.64 | |
| 28 | CTL | 4,794 | 3.74 | 963 | 3.65 |
| SDPP | 4,233 | 3.28 | 790 | 3.17 | |
| SEM | – | – | 118.1 | 0.35 | |
| Statistics | P, treatment | – | – | 0.60 | 0.70 |
| P, day | – | – | <0.001 | <0.001 | |
| P, treatment × day | – | – | 0.001 | 0.106 | |
1Total number of processed sequences was 2,685,690. The average number of sequences per sample was 38,367. Number of sequences of each sample was standardized to 4,618, which is the lowest number of sequences of a sample. The normalized values were used for statistical analysis.
2Treatment included control (CTL) and SDPP.
3Chao1 measures the microbial richness. It is estimated by the number of observed species divided by the ratio of singletons and doubletons.
4Shannon measures the microbial richness and evenness.
*P < 0.01: comparison between SDPP and CTL group at the corresponding time.
Figure 1.
Unweighted Unifrac displaying beta-diversity at d 0, 14, and 28 of core bacterial taxa in pigs fed SDPP or control diets.
Table 3 summarizes the most abundant core bacterial taxa in fecal samples of pigs fed control or SDPP diets. There were 11 phyla identified in this study. When averaged among all timepoints, the most dominant phyla were Firmicutes (80.71%), Bacteroidetes (9.94%), and unclassified bacteria (6.52%). At lower abundance were Proteobacteria (1.43%), Actinobacteria (1.14%), Tenericutes (0.13%), Synergistetes (0.09%), and Chlamydiae (0.02%) phyla. Feeding SDPP to pigs had no effect on fecal microbes at the phylum level; however, a numerical increase was observed for Bacteroidetes on d 7 (12.08% vs. 5.03%) and 14 (11.41% vs. 5.34%) in SDPP pigs. In addition, microbial communities were shifted (P < 0.05; Table 3) by time at the phylum level except for Tenericutes phylum.
Table 3.
Abundance of core bacterial taxa in fecal samples of pigs fed control or SDPP diets
| Taxonomy | Day 0 | Day 7 | Day 14 | Day 21 | Day 28 | SEM | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTL1 | SDPP2 | CTL | SDPP | CTL | SDPP | CTL | SDPP | CTL | SDPP | trt3 | Day | trt×day | ||
| Phylum | ||||||||||||||
| Firmicutes | 59.49 | 65.73 | 79.57 | 72.84 | 82.30 | 80.81 | 90.32 | 92.05 | 93.02 | 90.98 | 4.23 | 0.860 | <0.001 | 0.65 |
| Bacteroidetes | 27.93 | 23.56 | 5.03 | 12.08 | 5.34 | 11.41 | 5.20 | 2.53 | 1.63 | 4.74 | 3.50 | 0.410 | <0.001 | 0.37 |
| Unclassified | 4.66 | 3.44 | 12.20 | 12.53 | 8.60 | 6.16 | 4.36 | 4.17 | 5.08 | 4.03 | 1.70 | 0.380 | <0.001 | 0.94 |
| Proteobacteria | 7.42 | 6.54 | 0.03 | 0.00 | 0.03 | 0.06 | 0.00 | 0.23 | 0.00 | 0.00 | 1.00 | 0.880 | <0.001 | 0.99 |
| Actinobacteria | 0.05 | 0.07 | 3.10 | 2.27 | 3.38 | 1.30 | 0.09 | 0.90 | 0.08 | 0.18 | 1.10 | 0.560 | 0.040 | 0.66 |
| Tenericutes | 0.03 | 0.18 | 0.05 | 0.26 | 0.36 | 0.08 | 0.02 | 0.11 | 0.12 | 0.05 | 0.10 | 0.740 | 0.400 | 0.07 |
| Synergistetes | 0.43 | 0.47 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.920 | <0.001 | 0.99 |
| Family | ||||||||||||||
| Streptococcaceae | 0.54 | 1.61 | 1.08 | 5.23 | 0.21 | 5.48 | 41.16 | 47.85 | 52.04 | 58.19 | 6.84 | 0.270 | <0.001 | 0.99 |
| Lactobacillaceae | 1.50 | 13.10 | 20.43 | 20.42 | 39.05 | 29.74 | 17.15 | 21.54 | 4.87 | 10.85 | 7.12 | 0.560 | <0.001 | 0.58 |
| Lachnospiraceae4 | 10.20 | 9.00 | 33.74 | 18.51 | 18.22 | 10.93 | 9.35 | 8.04 | 9.81 | 6.86 | 2.67 | 0.001 | <0.001 | 0.10 |
| Ruminococcaceae | 4.75 | 10.99**a | 7.63 | 10.14a | 6.75 | 7.64a | 4.86 | 3.26b | 4.09 | 3.32b | 1.49 | 0.110 | 0.001 | 0.05 |
| Clostridiaceae | 3.88 | 4.99a | 1.27 | 3.68a | 5.36 | 15.92***b | 4.48 | 1.63a | 10.74 | 5.40*a | 2.30 | 0.400 | 0.001 | 0.001 |
| S24_7 | 6.20 | 4.41 | 3.64 | 7.76 | 2.73 | 10.34 | 2.60 | 0.92 | 0.46 | 4.01 | 2.74 | 0.160 | 0.210 | 0.28 |
| Erysipelotrichaceae | 17.54 | 4.56 | 1.35 | 1.01 | 0.45 | 0.45 | 0.22 | 0.24 | 0.21 | 0.19 | 2.79 | 0.120 | <0.001 | 0.06 |
| Bacteroidaceae | 16.01 | 4.46 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.71 | 0.170 | 0.040 | 0.10 |
| Clostridiales spp. | 7.83 | 8.69 | 5.59 | 5.98 | 2.47 | 4.53 | 3.29 | 1.80 | 5.37 | 3.26 | 1.49 | 0.950 | <0.001 | 0.51 |
| Prevotellaceae | 2.05 | 6.10 | 0.90 | 2.57 | 1.86 | 0.77 | 1.49 | 1.53 | 1.01 | 0.50 | 1.27 | 0.290 | 0.060 | 0.20 |
| unclassified bacteria | 4.66 | 3.44 | 12.20 | 12.53 | 8.60 | 6.16 | 4.36 | 4.17 | 5.08 | 4.03 | 1.70 | 0.380 | <0.001 | 0.94 |
| Erysipelotrichaceae | 17.54 | 4.56 | 1.35 | 1.01 | 0.45 | 0.45 | 0.22 | 0.24 | 0.21 | 0.19 | 2.79 | 0.120 | <0.001 | 0.06 |
| Enterobacteriaceae | 6.96 | 5.97 | 0.03 | 0.00 | 0.02 | 0.06 | 0.00 | 0.23 | 0.00 | 0.00 | 1.41 | 0.860 | <0.001 | 0.99 |
| Coprobacillaceae | 0.02 | 0.02 | 1.05 | 0.60 | 2.44 | 0.31 | 4.77 | 1.99 | 1.96 | 0.06 | 1.58 | 0.140 | 0.210 | 0.87 |
| Bifidobacteriaceae | 0.01 | 0.00 | 3.10 | 2.27 | 3.36 | 1.30 | 0.09 | 0.90 | 0.08 | 0.18 | 1.10 | 0.560 | 0.040 | 0.66 |
| Veillonellaceae | 1.52 | 0.30a | 0.45 | 1.14ab | 2.06 | 0.51*a | 1.65 | 3.15b | 0.12 | 0.19a | 0.62 | 0.780 | <0.001 | 0.05 |
| Unclassified Firmicutes | 2.36 | 1.90 | 0.91 | 1.89 | 0.83 | 0.81 | 0.79 | 0.25 | 0.62 | 0.50 | 0.39 | 0.900 | <0.001 | 0.43 |
| Unclassified Bacteroidales | 2.25 | 4.69 | 0.15 | 1.43 | 0.57 | 0.19 | 0.70 | 0.02 | 0.04 | 0.09 | 0.47 | 0.250 | <0.001 | 0.17 |
| Genus | ||||||||||||||
| Streptococcus | 0.54 | 1.59 | 1.07 | 5.21 | 0.21 | 5.47 | 41.11 | 47.79 | 51.98 | 58.11 | 6.84 | 0.270 | <0.001 | 0.993 |
| Lactobacillus | 1.50 | 13.08 | 20.43 | 20.41 | 39.04 | 29.72 | 17.14 | 21.54 | 4.86 | 10.85 | 7.12 | 0.564 | <0.001 | 0.580 |
| Unclassified Lachnospiraceae5 | 10.12 | 8.96 | 31.56 | 17.64 | 16.59 | 9.61 | 8.34 | 7.28 | 8.72 | 5.96 | 2.54 | 0.002 | <0.001 | 0.113 |
| unclassified Clostridiaceae | 1.72 | 3.24a | 0.89 | 3.41a | 5.18 | 15.37***b | 3.98 | 1.47a | 10.10 | 5.02*ab | 2.14 | 0.314 | 0.001 | 0.002 |
| Unclassified Clostridiales | 7.83 | 8.69 | 5.59 | 5.98 | 2.47 | 4.53 | 3.29 | 1.80 | 5.37 | 3.26 | 1.49 | 0.950 | 0.001 | 0.512 |
| Unclassified Clostridia | 6.17 | 6.32 | 4.63 | 3.14 | 2.97 | 2.73 | 1.76 | 1.44 | 2.01 | 1.23 | 0.95 | 0.357 | <0.001 | 0.939 |
| Oscillospira | 2.56 | 7.32***a | 3.05 | 2.31b | 1.70 | 2.47b | 1.21 | 0.90b | 1.05 | 1.17b | 1.01 | 0.140 | 0.001 | 0.046 |
| p_75_a5 | 17.42 | 4.31** | 0.84 | 0.23 | 0.05 | 0.03 | 0.00 | 0.00 | 0.03 | 0.03 | 2.76 | 0.110 | 0.001 | 0.050 |
| Faecalibacterium | 0.01 | 0.02 | 1.91 | 2.78 | 2.27 | 0.60 | 1.81 | 1.48 | 0.59 | 0.40 | 0.05 | 0.390 | <0.001 | 0.170 |
| Unclassified Ruminococcaceae | 1.28 | 2.98** | 1.26 | 1.22 | 0.75 | 1.28 | 0.69 | 0.27 | 1.09 | 0.67 | 0.45 | 0.330 | 0.004 | 0.074 |
| Sharpea | 0.02 | 0.02 | 0.10 | 0.60 | 1.78 | 0.07 | 4.68 | 1.69 | 1.93 | 0.05 | 1.54 | 0.200 | 0.210 | 0.780 |
| Unclassified Firmicutes | 2.36 | 1.90 | 0.91 | 1.89 | 0.83 | 0.81 | 0.79 | 0.25 | 0.62 | 0.50 | 0.39 | 0.900 | <0.001 | 0.430 |
| Unclassified S24-7 | 6.20 | 4.41 | 3.64 | 7.76 | 2.73 | 10.34 | 2.60 | 0.92 | 0.46 | 4.01 | 2.74 | 0.163 | 0.218 | 0.279 |
| Bacteroides | 15.65 | 4.29*** | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 3.79 | 0.171 | <0.001 | 0.100 |
| Unclassified Bacteroidales | 2.25 | 4.69 | 0.15 | 1.43 | 0.57 | 0.19 | 0.70 | 0.02 | 0.04 | 0.09 | 0.76 | 0.250 | <0.001 | 0.171 |
| Escherichia | 6.76 | 5.77 | 0.03 | 0.00 | 0.02 | 0.05 | 0.00 | 0.23 | 0.00 | 0.00 | 1.37 | 0.860 | <0.001 | 0.990 |
| Bifidobacterium | 0.01 | 0.00 | 3.10 | 2.27 | 3.36 | 1.30 | 0.09 | 0.90 | 0.08 | 0.18 | 1.10 | 0.560 | 0.038 | 0.663 |
| Unclassified bacteria | 4.66 | 3.44 | 12.20 | 12.53 | 8.60 | 6.16 | 4.36 | 4.17 | 5.08 | 4.03 | 1.70 | 0.38 | <0.001 | 0.940 |
| Species (OTUs) | ||||||||||||||
| C. saccharoperbutylacetonicum/beijerinckii (3) | 1.53 | 3.05ac | 0.46 | 3.01ac | 4.96 | 15.07***b | 3.82 | 1.36ac | 9.82 | 4.83c | 2.13 | 0.304 | <0.001 | 0.003 |
| C. difficile (13) | 1.05 | 2.09c | 0.01 | 0.39ab | 0.26 | 1.83*c | 1.00 | 0.47a | 3.06 | 1.68*bc | 0.50 | 0.482 | <0.001 | 0.01 |
| Ruminiclostridium thermocellum (30) | 0.39 | 0.26a | 0.63 | 2.31**b | 0.28 | 1.81***b | 0.25 | 0.06a | 0.36 | 0.25a | 0.29 | 0.002 | <0.001 | 0.001 |
| E. rhusiopathiae (10) | 16.08 | 3.93***a | 0.78 | 0.22a | 0.05 | 0.03a | 0.00 | 0.00a | 0.03 | 0.02a | 2.57 | 0.110 | <0.001 | 0.051 |
| L. reuteri (14) | 0.11 | 1.14ab | 0.47 | 4.01*b | 5.76 | 3.28*b | 1.03 | 0.14a | 0.01 | 0.00a | 0.95 | 0.681 | <0.001 | 0.045 |
| M. elsdenii (25) | 0.80 | 0.03ab | 0.27 | 0.76ac | 0.41 | 0.08a | 0.49 | 1.70**c | 0.03 | 0.11ab | 0.34 | 0.514 | 0.015 | 0.024 |
| O. valericigenes (29) | 0.83 | 2.88***a | 0.24 | 0.18b | 0.13 | 0.13b | 0.19 | 0.11b | 0.05 | 0.11b | 0.44 | 0.147 | <0.001 | 0.052 |
| Ruminococcaceae spp. (47) | 0.20 | 0.02a | 0.15 | 0.46*b | 0.69 | 0.42*b | 0.22 | 0.19a | 0.33 | 0.16a | 0.09 | 0.244 | <0.001 | 0.047 |
| B. helcogenes (115) | 0.00 | 0.00 | 0.03 | 0.08 | 0.05 | 0.01 | 0.32 | 0.02** | 0.07 | 0.10 | 0.07 | 0.230 | 0.108 | 0.054 |
| Mitsuokella jalaludinii (122) | 0.04 | 0.01a | 0.01 | 0.00a | 0.30 | 0.01***a | 0.06 | 0.13b | 0.00 | 0.00a | 0.05 | 0.103 | 0.008 | 0.003 |
| L. delbrueckii subsp. bulgaricus (130) | 0.04 | 0.00a | 0.10 | 0.47***b | 0.10 | 0.03a | 0.04 | 0.02a | 0.05 | 0.05a | 0.04 | 0.024 | <0.001 | <0.001 |
| P. denticola (137 | 0.02 | 0.00a | 0.00 | 1.55***b | 0.01 | 0.03a | 0.01 | 0.01a | 0.00 | 0.00 | 0.16 | 0.003 | 0.000 | <0.001 |
| E. harbinense (162) | 0.02 | 0.01a | 0.06 | 0.23***c | 0.06 | 0.08b | 0.02 | 0.02a | 0.02 | 0.02a | 0.01 | <0.001 | <0.001 | <0.001 |
| S. suis (160) | 0.00 | 0.00a | 0.02 | 0.18***c | 0.04 | 0.02a | 0.05 | 0.03ab | 0.06 | 0.03b | 0.02 | 0.035 | <0.001 | <0.001 |
| R. albus (179) | 0.00 | 0.00a | 0.02 | 0.14***b | 0.03 | 0.02a | 0.04 | 0.02a | 0.06 | 0.04a | 0.02 | 0.254 | 0.006 | 0.01 |
| N. timonensis (84) | 0.03 | 0.02a | 0.03 | 0.37***b | 0.34 | 0.39b | 0.08 | 0.04a | 0.11 | 0.03a | 0.04 | 0.017 | <0.001 | <0.001 |
a-cPair-wise comparisons were made for SDPP treatment over time. Means without similar superscripts are different (P < 0.05).
1CTL: control. Control diet had no SDPP throughout the study.
2Pigs fed diet containing SDPP at 5% (d 0 to 14) and 2.5% (d 14 to 28).
3Treatment (trt) included CTL or SDPP.
4Treatment effect (P = 0.001; CTL = 16.3%; SDPP = 10.7%) when averaged across all timepoints.
5Treatment effect (P = 0.002; CTL = 15.1%; SDPP = 9.9%) when averaged across all timepoints.
*P < 0.05: SDPP compared with control at the corresponding timepoint.
**P < 0.01: SDPP compared with control at the corresponding timepoint.
***P < 0.001: SDPP compared with control at the corresponding timepoint.
At the family level, 46 families were identified, among which 18 families having greater than 1% of core sequences (Table 3) and belonging to 4 phyla (Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria). In Firmicutes phylum, Streptococcaceae (21.34%), Lactobacillaceae (17.87%), Lachnospiraceae (13.47%), Ruminococcaceae (6.34%), Clostridiaceae (5.74%), unclassified Clostridiales (4.88%), unclassified Clostridia (3.24%), Erysipelotrichaceae (2.62%), Coprobacillaceae (1.32%), and Veillonellaceae (1.11%), and unclassified Firmicutes (1.09%) were the most dominant families. In Bacteroidetes phylum, S24_7 (4.31%), Bacteroidaceae (2.05%), Prevotellaceae (1.88%), and unclassified Bacteroidales (1.01%) were the most dominant families. At lower abundance, Enterobacteriaceae (1.33%) and Bifidobacteriaceae (1.13%) were the two most dominant families in Proteobacteria and Actinobacteria phyla, respectively. There were 6.52% of sequences that were not classified. Feeding SDPP had limited effects on fecal microbes at the family level; however, there was a shift (P < 0.05; Table 3) in fecal microbial communities in majority of families when pigs aged. On d 0 (weaning), Ruminococcaceae was greater (10.99% vs. 4.75%; P < 0.01) in the SDPP group, but there were no differences between the two groups at other timepoints. Clostridiaceae, Veillonellaceae, and Lachnospiraceae were families affected by feeding SDPP. Specifically, Clostridiaceae had increased in pigs fed SDPP (15.92% vs. 5.36%; P < 0.001) on d 14, but decreased at the end of the experiment (5.40% vs. 10.74%; P < 0.05). Veillonellaceae had decreased (0.51% vs. 2.06%; P = 0.05) in SDPP pigs on d 14. When averaged among all timepoints, there was a treatment effect on Lachnospiraceae where SDPP pigs had a decreased (10.7% vs. 16.3%; P < 0.01) proportion of this family compared with control pigs.
At the genus level, 87 genera were identified with 18 genera having more than 1% of core sequences and accounted for 88.13% core sequences (Table 3). Nine of the genera were classified (Streptococcus, Lactobacillus, Oscillospira, p_75_a5, Bacteroides, Escherichia, Faecalibacterium, Bifidobacterium, and Sharpea) and accounted for 50.52% of the core sequences. The other nine genera were not classified (Table 3) and comprised of 37.61% of the core sequences. Consistent with observations at the phylum level, feeding SDPP affected Lachnospiracaea spp. and Clostridiaceae spp.; however, these two genera were not taxonomically classified. At d 0, decreased (P < 0.001) Oscillospira and greater (P < 0.01) p_75_a5 were observed in the SDPP group. These results indicate variation among pigs at the baseline; however, feeding SDPP did not have an effect on these two genera at other timepoints.
Alteration of Fecal Microbiota by Consumption of SDPP: An OTU-Based Analysis
As shown in Table 3, a minimum of 37.61% of sequences were not classified at the genus level. Therefore, we employed the OTU-based approach to further characterize microbial composition. Because there were too many core OTUs (1,042), only the 20 most abundant OTUs that had significant treatment × time interactions and greater than 0.03% of the core sequences were presented (Table 3). Feeding SDPP during the first 2 wk postweaning had greater effects on fecal microbes compared with the later 2 wk. On d 7, pigs fed SDPP had greater Lactobacillus reuteri (4.01% vs. 0.47%, P < 0.05), Lactobacillus delbrueckii (0.47% vs. 0.1%, P < 0.001), Clostridium thermocellum (2.31% vs. 0.63%, P < 0.001), Prevotella denticola (1.56% vs. 0.00%, P < 0.001), Ruminococcaceae spp. (0.46% vs. 0.15%, P < 0.05), Ruminococcus albus (0.14% vs. 0.02%, P < 0.001), Ethanoligenens harbinense (0.23% vs. 0.06%, P < 0.001), Streptococcus suis (0.18% vs. 0.02%, P < 0.001), and unclassified bacteria (0.37% vs. 0.03%, P < 0.001) compared with control-fed pigs. On d 14, greater Clostridium saccharoperbutylacetonicum (15.07% vs. 4.96%, P < 0.001), Ruminiclostridium thermocellum (1.81% vs. 0.28%, P < 0.001), and Clostridium difficile (1.83% vs. 0.26%, P < 0.001) and lower L. reuteri (3.28% vs. 5.76%, P < 0.05), Ruminococcaceae spp. (0.42% vs. 0.69%, P < 0.05), and Selenomonas ruminantium subsp. lactilytica (0.01% vs. 0.30%, P < 0.001) were observed in pigs fed SDPP compared with control pigs. On d 21, feeding SDPP increased Megasphaera elsdenii (1.70% vs. 0.49%, P < 0.01) and decreased Bacteroides helcogenes (0.02% vs. 0.32%, P < 0.01) in pigs. Overall (d 28), feeding SDPP reduced the abundance of C. difficile (1.68% vs. 3.06%, P < 0.05) compared with feeding control diets.
Pigs Fed SDPP and Control Diets Shared Majority of Core OTUs and Sequences
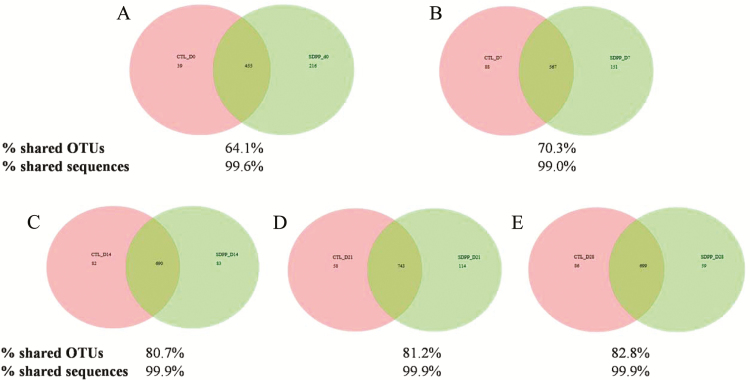
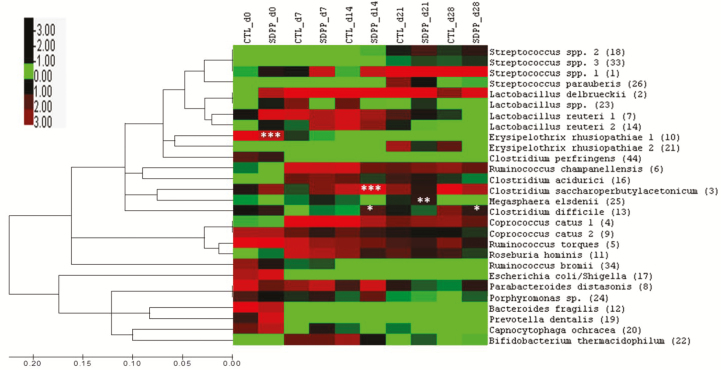
At the OTU level, pigs fed SDPP and control diets had 64.1%, 70.3%, 80.7%, 81.2%, and 82.8% shared OTUs on d 0 (at weaning), 7, 14, 21, and 28 postweaning, respectively (Figure 2). In shared OTUs, there were 99.0% to 99.9% shared sequence reads between control and SDPP-fed pigs. The most abundant shared OTUs between control and SDPP-fed pigs were presented in Table 4 and Figure 3. The OTUs that have significant treatment × d interaction (P < 0.05) were C. saccharoperbutylacetonicum, Erysipelothrix rhusiopathiae, M. elsdenii, and C. difficile. On d 14, pigs fed SDPP had greater C. saccharoperbutylacetonicum (15.07% vs. 4.96%; P < 0.001) and C. difficile (1.83% vs. 0.26%; P < 0.05). However, C. difficile had decreased (1.68% vs. 3.06%; P < 0.05) in SDPP pigs on d 28 when compared with control pigs. On d 21, feeding SDPP increased (1.70% vs. 0.49%; P < 0.01) M. elsdenii compared with feeding control diets. Lactobacillus reuteri tended (6.69% vs. 11.98%; P = 0.098) to decreased in SDPP pigs on d 14. In addition, when averaged among all sampling timepoints, Coprococcus catus 2 was reduced (3.8% vs. 7.03%; P < 0.05) in pigs fed SDPP compared with pigs fed the control diet.
Figure 2.
Distribution of core OTUs in pigs fed control and SDPP diets on d 0, 7, 14, 21, and 28 postweaning (Panels A, B, C, D, and E, respectively). Proportion of shared OTUs was calculated by number of shared OTUs divided by total OTU richness of two groups at a timepoint. Proportion of shared sequences was calculated by number of shared sequences divided by total sequences of two groups at a timepoint.
Table 4.
The most abundant shared species (OTUs) between pigs fed control or SDPP diets1
| OTU # | Species (OTUs) | E value | Day 0 | Day 7 | Day 14 | Day 21 | Day 28 | SEM | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTL | SDPP | CTL | SDPP | CTL | SDPP | CTL | SDPP | CTL | SDPP | trt5 | day | trt × day | ||||
| 2 | L. Delbrueckii | E-82 | 0.17 | 4.05 | 9.55 | 9.80 | 20.33 | 20.60 | 10.27 | 13.97 | 2.99 | 9.74 | 4.7 | 0.345 | 0.003 | 0.956 |
| 3 | Clostridium saccharoperbutylacetonicum/ beijerinckii | E-66 | 1.53 | 3.05 | 0.46 | 3.01 | 4.96 | 15.07*** | 3.82 | 1.36 | 9.82 | 4.83* | 2.0 | 0.304 | <0.001 | 0.003 |
| 5 | Ruminococcus torques | 5E-67 | 9.06 | 7.88 | 5.71 | 3.30 | 3.05 | 2.72 | 2.02 | 1.71 | 2.73 | 1.63 | 1.2 | 0.196 | <0.001 | 0.948 |
| 8 | Bacteroidetes.S247 | 2E-45 | 4.22 | 2.85 | 2.52 | 5.29 | 1.80 | 6.27 | 1.87 | 0.56 | 0.30 | 2.71 | 1.7 | 0.207 | 0.259 | 0.293 |
| 9 | Coprococcus catus | E-58 | 5.22 | 4.95 | 2.49 | 1.38 | 2.24 | 1.62 | 1.33 | 1.13 | 0.95 | 0.81 | 0.7 | 0.337 | <0.001 | 0.978 |
| 10 | Erysipelothrix rhusiopathiae | E-42 | 16.08 | 3.93*** | 0.78 | 0.22 | 0.05 | 0.03 | 0.00 | 0.00 | 0.03 | 0.02 | 2.6 | 0.110 | <0.001 | 0.051 |
| 12 | Bacteroides fragilis | 2E-76 | 15.45 | 3.99 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.6 | 0.161 | <0.001 | 0.087 |
| 17 | Escherichia coli/Shigella | E-77 | 6.66 | 5.66 | 0.03 | 0.00 | 0.02 | 0.05 | 0.00 | 0.23 | 0.00 | 0.00 | 1.3 | 0.853 | <0.001 | 0.990 |
| 19 | Prevotella dentalis | 6E-66 | 1.12 | 5.32 | 0.08 | 0.02 | 0.03 | 0.01 | 0.07 | 0.02 | 0.02 | 0.02 | 1.0 | 0.194 | 0.003 | 0.111 |
| 20 | Unclassified Bacteroidales | 5E-27 | 1.83 | 4.13 | 0.10 | 1.32 | 0.47 | 0.14 | 0.66 | 0.01 | 0.02 | 0.06 | 0.7 | 0.238 | <0.001 | 0.167 |
| 24 | Porphyromonas spp. | 4E-63 | 1.36 | 1.20 | 0.56 | 1.49 | 0.47 | 2.48 | 0.39 | 0.24 | 0.08 | 0.94 | 0.6 | 0.079 | 0.211 | 0.310 |
| 34 | Ruminococcus bromii | 6E-56 | 2.90 | 2.21 | 0.54 | 0.55 | 0.05 | 0.06 | 0.00 | 0.00 | 0.00 | 0.01 | 0.5 | 0.722 | <0.001 | 0.964 |
| 44 | Clostridium perfringens | 3E-64 | 1.72 | 1.38 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.4 | 0.788 | <0.001 | 0.988 |
| 4 | Coprococcus catus 2 | 2E-70 | 0.34 | 0.14 | 19.46 | 8.49 | 9.11 | 4.45 | 2.71 | 3.18 | 3.51 | 2.74 | 1.8 | 0.015 | <0.001 | 0.081 |
| 6 | Ruminococcus champanellensis | E-28 | 0.32 | 0.01 | 7.19 | 5.43 | 6.11 | 2.41 | 2.28 | 2.92 | 3.36 | 2.39 | 1.1 | 0.125 | 0.001 | 0.414 |
| 11 | Roseburia hominis | E-68 | 0.18 | 0.37 | 4.57 | 4.26 | 3.05 | 1.56 | 2.23 | 1.52 | 1.54 | 0.87 | 0.7 | 0.169 | <0.001 | 0.77 |
| 16 | Clostridium acidurici | 4E-38 | 0.01 | 0.02 | 1.90 | 2.76 | 2.26 | 0.59 | 1.80 | 1.47 | 0.58 | 0.40 | 0.01 | 0.390 | <0.001 | 0.165 |
| 22 | Bifidobacterium t hermacidophilum/boum/ thermophilum | 2E-45 | 0.01 | 0.00 | 3.04 | 2.21 | 3.30 | 1.27 | 0.09 | 0.88 | 0.08 | 0.18 | 0.9 | 0.554 | 0.038 | 0.657 |
| 23 | Lactobacillus spp. | 4E-63 | 0.04 | 0.61 | 1.95 | 0.12 | 1.85 | 0.12 | 0.12 | 1.34 | 0.01 | 0.05 | 0.8 | 0.518 | 0.683 | 0.267 |
| 21 | Erysipelothrix rhusiopathiae | 2E-73 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 4.66 | 1.53 | 1.92 | 0.04 | 1.4 | 0.259 | 0.099 | 0.676 |
| 25 | Megasphaera elsdenii | 6E-81 | 0.80 | 0.03 | 0.27 | 0.76 | 0.41 | 0.08 | 0.49 | 1.70** | 0.03 | 0.11 | 0.3 | 0.514 | 0.015 | 0.024 |
| 26 | Streptococcus parauberis | 3E-69 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 2.72 | 2.13 | 0.14 | 0.16 | 1.1 | 0.869 | 0.088 | 0.998 |
| 13 | Clostridium difficile | 4E-33 | 1.05 | 2.09 | 0.01 | 0.39 | 0.26 | 1.83* | 1.00 | 0.47 | 3.06 | 1.68* | 1.0 | 0.482 | <0.001 | 0.010 |
| 33 | Streptococcus spp. 3 | E-65 | 0.53 | 1.57 | 1.06 | 5.13 | 0.20 | 5.38 | 37.64 | 44.71 | 51.07 | 57.01 | 9.69 | 0.270 | <0.001 | 0.990 |
| 14 | L. reuteri 4 | 6E-76 | 1.15 | 5.91 | 5.04 | 8.08 | 11.98 | 6.69* | 3.89 | 1.81 | 0.62 | 0.11 | 2.9 | 0.99 | <0.001 | 0.098 |
1Dietary treatments included control (CTL) and SDPP. Data were presented as percent of OTUs in total sequences of a treatment.
2Treatment effect (P = 0.015; CTL = 7.03%; SDPP = 3.80%) when averaged across all timepoints.
3Combined OTU1 + 18 + 33 for Streptococcus spp. (98% similarity).
4Combined OTU7 and OUT14 for Lactobacillus reuteri (98% similarity).
5Treatment.
Figure 3.
Distribution of shared OTUs in pigs fed SDPP compared with control pigs. ***P < 0.001; **P < 0.01; *P < 0.05 compared with control.
In contrast to shared OTUs, unique OTUs in each group of pigs comprised of less abundant OTUs (Table 5). On d 7 postweaning, control pigs had 0.76% E. rhusiopathiae str. Fujisawa (seven out of seven pigs), 0.07% S. suis (one out of seven pigs), 0.05% L. delbrueckii subsp. Bulgaricus (two out of seven pigs), 0.04% Acidaminococcus fermentans 1 (one out of seven pigs), and 0.03% Acidaminococcus fermentans 2 (one out of seven pigs), whereas none of these OTUs present in pigs fed SDPP. However, SDPP pigs had two P. denticola OTUs (0.17%; two out of three pigs) and one Eubacterium cellulosolvens OTU (0.03%; two out three pigs) that were not present in control pigs.
Table 5.
Unique OTUs in pigs fed SDPP and control diets on d 7 postweaning
| n | Day | #OTU ID | Species (OTUs) | E value | Number of sequences | % sequences/ CTL group | % sequences/ SDPP group | |
|---|---|---|---|---|---|---|---|---|
| CTL | SDPP | |||||||
| 1 | 0 | 143 | Desulfomonile tiedjei | 2E-21 | 11 | 0 | 0.01 | – |
| 1 | 0 | 138 | Clostridium aciduric | 8E-50 | 10 | 0 | 0.01 | – |
| 5 | 0 | 88 | Lactobacillus helveticus/amylovorus | 4E-58 | 0 | 132 | – | 0.04 |
| 6 | 0 | 325 | Lactobacillus reuteri 1 | 5E-67 | 0 | 81 | – | 0.03 |
| 5 | 0 | 295 | Lactobacillus reuteri 2 | 8E-60 | 0 | 76 | – | 0.03 |
| 4 | 0 | 289 | Lactobacillus sp. 1 | 3E-54 | 0 | 69 | – | 0.02 |
| 2 | 0 | 191 | Lactobacillus sp. 2 | 3E-69 | 0 | 47 | – | 0.02 |
| 5 | 0 | 749 | Ruminococcus sp. | 1E-32 | 0 | 50 | – | 0.02 |
| 1 | 0 | 269 | Streptococcus suis | 5E-17 | 0 | 80 | – | 0.03 |
| 7 | 7 | 60 | Catenibacterium mitsuokai | 5E-47 | 728 | 0 | 0.76 | – |
| 1 | 7 | 260 | Streptococcus suis | 2E-41 | 64 | 0 | 0.07 | – |
| 2 | 7 | 366 | L. delbrueckii subsp. Bulgaricus | 2E-56 | 52 | 0 | 0.05 | – |
| 1 | 7 | 346 | Acidaminococcus fermentans 1 | E-51 | 42 | 0 | 0.04 | – |
| 1 | 7 | 278 | Acidaminococcus fermentans 2 | E-52 | 27 | 0 | 0.03 | – |
| 2 | 7 | 611 | Prevotella denticola 1 | E-38 | 0 | 66 | – | 0.10 |
| 2 | 7 | 1006 + 1219 | Prevotella denticola 21 | 6E-66 | 0 | 29 | – | 0.04 |
| 2 | 7 | 469 | Eubacterium cellulosolvens | 2E-60 | 0 | 21 | – | 0.03 |
1OTU 1006 and OTU 1219 were combined (97% similarity).
Pearson-Based Correlation Analysis
In addition to the microbial analyses described above, the16s rRNA gene sequence dataset was used to conduct Pearson-based regression analysis to identify bacterial taxa significantly correlated with growth performance phenotypes including ADG (Table 6) and BW (Table 7). Only those bacterial taxa with a P-value greater than 0.05 are reported (25 total bacterial taxa). With respect to ADG, the bacterial taxa with the highest positive correlation was Neglecta timonensis (P < 0.02; r = 0.70), whereas the bacterial taxa with highest negative correlation was Prevotella copri (P < 0.019; r = −0.72) on d 7. With respect to BW, the bacterial taxa with the highest positive correlation was Adlercreutzia equolifaciens (P < 0.007; r = 0.79) on d 7 and the bacterial taxa with the highest negative correlation were Prevotella copri (P < 0.037; r = −0.66) on d 7 and Lactobacillus gasseri (P < 0.005; r = −0.66) on d 28.
Table 6.
Correlation of ADG1 and bacterial taxa2 at the OTU level on d 7, 14, 21, and 28 using Pearson analysis3
| Day | OTU_ID | Putative species | Family | Seq ID | Identity, % | Pearson’s R index | P 3 |
|---|---|---|---|---|---|---|---|
| 7 | 38 | Prevotella copri | Prevotellaceae | NR_113411.1 | 98% | −0.72 | 0.019 |
| 7 | 84 | Neglecta timonensis | Ruminococcaceae | NR_144736.1 | 97% | 0.70 | 0.023 |
| 7 | 14 | Lactobacillus coleohominis | Lactobacillaceae | NR_042436.1 | 98% | 0.66 | 0.037 |
| 14 | 57 | Oscillibacter valericigenes | Oscillospiraceae | NR_074793.1 | 92% | −0.68 | 0.005 |
| 14 | 1 | Streptococcus equinus | Streptococcaceae | NR_113594.1 | 98% | 0.55 | 0.036 |
| 21 | 60 | Catenibacterium mitsuokai | Erysipelotrichaceae | NR_027526.1 | 96% | 0.58 | 0.024 |
1Growth performance data from this experiment previously reported (Tran et al., 2014).
2Only taxa with P < 0.05 are shown.
3Correlation determined using Person’s R index within Calypso (Zakrzewski et al., 2016). Raw sequences were normalized to total number of sequences of each sample before the correlation was determined.
Table 7.
Correlation of BW1 and bacterial taxa2 at the OTU level on d 7, 14, 21, and 28 using Pearson analysis3
| Day | OTU_ID | Putative species | Family | Seq. ID | Identity, % | Pearson’s R index | P |
|---|---|---|---|---|---|---|---|
| 7 | 91 | Adlercreutzia equolifaciens | Eggerthellaceae | NR_121696.1 | 91% | 0.79 | 0.007 |
| 7 | 38 | Prevotella copri | Prevotellaceae | NR_113411.1 | 98% | −0.66 | 0.037 |
| 14 | 57 | Oscillibacter valericigenes | Oscillospiraceae | NR_074793.1 | 92% | −0.65 | 0.008 |
| 14 | 117 | Ruminococcus bromii | Ruminococcaceae | NR_025930.1 | 97% | −0.6 | 0.017 |
| 14 | 42 | Butyricicoccus pullicaecorum | Clostridiaceae | NR_044490.1 | 92% | −0.56 | 0.029 |
| 14 | 99 | Romboutsia timonensis | Peptostreptococcaceae | NR_144740.1 | 98% | 0.54 | 0.037 |
| 14 | 72 | Blautia marasmi | Lachnospiraceae | NR_147395.1 | 97% | −0.52 | 0.046 |
| 14 | 122 | Mitsuokella jalaludinii | Selenomonadaceae | NR_028840.1 | 99% | −0.52 | 0.048 |
| 21 | 14 | Lactobacillus pontis | Lactobacillaceae | NR_036788.2 | 98% | −0.65 | 0.008 |
| 21 | 100 | Acetoanaerobium pronyense | Peptostreptococcaceae | NR_136796.1 | 94% | −0.64 | 0.011 |
| 21 | 20 | Imtechella halotolerans | Flavobacteriaceae | NR_117181.2 | 83% | −0.62 | 0.013 |
| 21 | 53 | Peptostreptococcus russellii | Peptostreptococcaceae | NR_115155.1 | 92% | −0.61 | 0.015 |
| 21 | 35 | Lactobacillus gasseri | Lactobacillaceae | NR_075051.1 | 98% | −0.58 | 0.023 |
| 21 | 8 | Barnesiella viscericola | Barnesiellaceae | NR_121773.1 | 88% | −0.56 | 0.030 |
| 21 | 42 | Butyricicoccus pullicaecorum | Clostridiaceae | NR_044490.1 | 92% | −0.52 | 0.048 |
| 28 | 35 | Lactobacillus gasseri | Lactobacillaceae | NR_075051.1 | 98% | −0.66 | 0.005 |
| 28 | 15 | Pseudoflavonifractor phocaeensis | Clostridiales4 | NR_147370.1 | 95% | 0.55 | 0.026 |
| 28 | 40 | Intestinimonas butyriciproducens | Clostridiales4 | NR_118554.1 | 94% | 0.55 | 0.028 |
| 28 | 153 | Eubacterium tarantellae | Clostridiaceae | NR_104741.1 | 94% | −0.51 | 0.044 |
1Growth performance data from this experiment previously reported (Tran et al., 2014).
2Only taxa with P < 0.05 are shown.
3Correlation determined using Person’s R index within Calypso (Zakrzewski et al., 2016). Raw sequences were normalized to total number of sequences of each sample before the correlation was determined.
4Unclassified clostridiales.
DISCUSSION
SDPP has been used in nursery diets to improve growth performance and health of pigs, especially when no in-feed antibiotic is included as a growth promoter (Coffey and Cromwell, 1995; Van Dijk et al., 2002; Bosi et al., 2004). However, there are a few studies evaluating the change of gut microbes in responses to diets containing plasma products (Nollet et al., 1999; Bhandari et al., 2008; Balan et al., 2011). Additionally, the techniques (e.g., Denaturing Gradient Gel Electrophoresis and Terminal Restriction Fragment Length Polymorphism) used to profile gut microbes in those studies had limitations and did not characterize the change of gut microbes at a community-wide level. We employed a next generation sequencing approach to profile the change of fecal microbial communities in pigs fed SDPP during a 4-wk feeding period. The advantage of using next generation sequencing to characterize microbial communities is that this technique allows the evaluation of global change of fecal microbes in response to dietary treatment at different taxonomic levels (from phylum to species).
In a previous publication (Tran et al., 2014), we reported that feeding SDPP in a phase-1 nursery diet increased ADG, ADFI, and G:F in pigs during the first wk postweaning; however, the improvement of ADG in pigs fed SDPP diminished in the second phase (d 14 to 28). In addition, feeding SDPP improved gut integrity of pigs, particularly in the duodenum. The mechanism for the improved gut integrity may result from the increased cell viability and proliferation of enterocytes in pigs fed SDPP. There were no treatment effects on circulating IgG, A, and total antioxidant capacity. Thus, the increased pig growth performance may be a result of the protective effect of SDPP on gut barrier function and the increased nutrient absorption in the small intestine (Tran et al. 2014).
In this study, we showed that there was no dietary effect on alpha-diversity of microbial communities; however, a lower species richness measured by Chao1 index was observed in SDPP pigs on d 14. Our results are different from previous studies which reported an increased microbial richness and diversity in pigs fed SDPP and challenged with E. coli K88 (Bhandari et al., 2008). It should be noted that the pigs used in the study of Bhandari and coworkers were challenged with E. coli K88; however, no E. coli response was detected in that study. In addition, sample size at some timepoints and differences in crude protein and synthetic amino acids between the control and SDPP diets may limit interpretation of the data in some instances. It is difficult to determine whether the reduction in microbial diversity is beneficial to gut health. Thus, there is a need to evaluate the microbial composition at different taxonomic levels.
Feeding SDPP resulted in a shift of fecal microbiota at family and OTU levels. It has been reported previously that feeding plasma products (e.g., ovine immunoglobulin) to rats increased lactic acid–producing bacteria, including Leuconostoc, Lactobacillus, and Weissella (Balan et al., 2011). In this study, we observed that feeding SDPP to pigs affected the lactic acid–producing bacteria including L. reuteri and L. delbrueckii; however, the shift of fecal bacteria greatly depended on pig age. Specifically, L. reuteri and L. delbrueckii increased on d 7, but the abundance of L. reuteri decreased on d 14. Therefore, it appears that feeding SDPP may delay the presence of L. reuteri and L. delbrueckii in the first wk postweaning, which is the most challenging time for newly-weaned pigs. The increase of these Lactobacillus species may be correlated to the improvement of feed intake, BW gain, and feed efficiency in pigs fed SDPP on d 7, which were shown in our previous study (Tran et al., 2014). On d 14, control pigs had similar feed intake and BW gain and increased L. reuteri compared with SDPP pigs. This observation could be a result of compensatory gain and a correlation between growth performance and Lactobacillus spp. (Dumonceaux et al., 2006) in control pigs.
Ruminococcus albus (d 7), Ruminiclostridium thermocellum (d 7 and 14), and C. saccharoperbutylacetonicum/beijerinckii (d 14) and M. elsdenii (d 21) were increased in pigs fed SDPP diets. C. saccharoperbutylacetonicum/ beijerinckii are acetone- and butyric acid–producing Clostridia. Ruminococcus albus and Ruminiclostridium thermocellum are cellulolytic bacteria, which are able to convert cellulosic substrates into fermentative products such as ethanol, lactate, and acetate (Shoham et al., 1999; Demain et al., 2005). Megasphaera elsdenii is a Gram-negative rumen organism, which ferments lactate to propionic acid via acrylate pathway and ferments a variable part of lactate to butyrate (Marounek et al., 1989, Marx et al., 2011, Prabhu et al., 2012). Megasphaera elsdenii can also produce VFA, which contributes to energy balance of the animals. The presence of these cellulose degraders and butyric acid–producing bacteria provides support for nutrient digestion and health of the GIT in newly-weaned pigs experience the dietary transition from sow milk to grain-based (with a greater proportion of dietary fiber) diet.
Feeding diets containing SDPP had a different impact on Clostridiaceae family and C. difficile over time. On d 14, greater Clostridiaceae family and C. difficile species were observed in SDPP vs. control pigs. However, the proportions of this family and species were reduced in pigs fed SDPP compared with control pigs at the end of the experiment. Clostridium difficile is a pathogenic bacterium, which may cause severe enteritis in infected pigs and humans (see review by Keessen et al., 2011). The reduction of this species is important to maintain pig health and to control bacteria shedding to the environment. It should be noted that immunoglobulin supplement can be used for C. difficile enteritis treatment (Tjellstrom et al., 1993). Thus, it is relevant to observe the reduction of C. difficile in pigs fed plasma product, which contains 17.9% to 22.5% IgG (Pierce et al., 2005) and significant other classes of Ig (Anderson L. and Anderson G., 2002).
Venn diagrams were created to visualize the distribution of OTUs between pigs fed SDPP and control diets (Figure 2). The majority of core sequences (99.0% to 99.9%) belonged to shared OTUs between these two groups of pigs. These OTUs accounted for 64.0% to 82.8% of core OTUs at a specific timepoint. Among shared OTUs, C. saccharoperbutylacetonicum/beijerinckii, L. reuteri, M. elsdenii, and C. difficile were affected by feeding SDPP over time.
While shared OTUs increase with age, which is consistent with previous reports indicating that bacterial phylogenetic diversity increases with age (Gaorui et al., 2016), the shared OTUs contained both abundant and less abundant OTUs and unique OTUs were comprised of only less dominant OTUs (Table 5). It is noteworthy that Catenibacterium mitsuokai was not detected in pigs fed SDPP on d 7, whereas this strain appeared in all pigs fed control diets (0.76% of core sequences). This is a significant finding because E. rhusiopathiae str. Fujisawa is a pathogenic bacterium causing swine erysipelas (Brooke and Riley, 1999).
As mentioned previously, growth performance from this experiment has already been reported (Tran et al., 2014). Here we have characterized the effects of dietary treatment on microbial populations with respect to shared and unique OTU. However, potentially of value is the correlation between phenotype (overall ADG and BW) with specific bacterial taxa. As such, overall ADG and BW were correlated with bacterial taxa on d 7, 14, 21, and 28. A negative correlation identifies bacterial taxa that are more abundant in lower performing pigs, whereas a positive correlation identifies bacterial taxa that are more abundant in higher performing pigs. Only two bacterial taxa (Prevotella copri and Oscillibacter valericigenes) were observed as having a high negative correlation across both phenotypes (ADG and BW). On d 7, where the greatest positive impact on ADG was observed (Tran et al., 2014), two bacterial taxa (Neglecta timonensis and Lactobacillus coleohominis) had high positive correlations with overall ADG and on d 28, where the greatest positive impact on BW was observed, two bacterial taxa (Pseudoflavonifractor phocaeensis and Intestinimonas butyriciproducens) had positive correlations with overall BW. Calypso allows comparisons of taxonomic data from 16s rRNA datasets (Zakrzewski et al., 2016) and has been utilized previously to identify bacterial taxa that may be associated with specific phenotypes in poultry (Stanley et al., 2016).
The significance of this study was that for the first time the microbial communities were characterized in pigs fed a SDPP diet using a next generation sequencing approach. Results from this study indicate that feeding SDPP may decrease pathogenic bacteria (C. difficile, E. rhusiopathiae, B. helcogenes), whereas feeding SDPP may increase cellulose degraders (Clostridium sp. and Ruminococcus sp.) and butyric acid–producers (Lactobacillus sp.), which may have positive impacts on nutrient digestion and gut health. In addition, irrespective of dietary treatment, further evaluation of this dataset has revealed bacterial taxa that may be correlated to important growth performance phenotypes.
ACKNOWLEDGMENTS
We sincerely thank Dr. Jens Walter for his valuable comments on this manuscript. We also extend our gratefulness to Dr. Ines Martinez for her technical support on the sequencing analysis. We thank Yanshuo Li, our graduate student at the University of Nebraska, for helping with animal care and fecal sample collection.
LITERATURE CITED
- Allen H. K., Levine U. Y., Looft T., Bandrick M. and Casey T. A.. 2013. Treatment, promotion, commotion: antibiotic alternatives in food-producing animals. Trends Microbiol. 21:114–119. doi:10.1016/j.tim.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Anderson N. L., and Anderson N. G.. 2002. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics 1:845–867. doi:10.1074/mcp.R200007-MCP200 [DOI] [PubMed] [Google Scholar]
- Balan P., Han K. S., Rutherfurd-Markwick K., Singh H., and Moughan P. J.. 2011. Ovine serum immunoglobulin has immunomodulatory effects in growing rats gavaged with Salmonella enteritidis. J. Nutr. 141:950–956. doi:10.3945/jn.110.131433 [DOI] [PubMed] [Google Scholar]
- Bhandari S. K., B Xu, C. M Nyachoti, D. W Giesting, and D. O Krause. 2008. Evaluation of alternatives to antibiotics using an Escherichia coli K88+ model of piglet diarrhea: effects on gut microbial ecology. J. Anim. Sci. 86:836–847. [DOI] [PubMed] [Google Scholar]
- Bosi, P., L. Casini, A. Finamore, C. Cremokolini, G. Merialdi, P. Trevisi, F. Nobili, and E. Mengheri. 2004. Spray-dried plasma improves growth performance and reduces inflammatory status of weaned pigs challenged with enterotoxigenic Escherichia coli k88. J. Anim. Sci. 82:1764–1772. [DOI] [PubMed] [Google Scholar]
- Brooke, C. J., and T. V. Riley. 1999. Erysipelothrix rhusiopathiae: bacteriology, epidemiology and clinical manifestations of an occupational pathogen. J. Med. Microbiol. 48(9):789–799. [DOI] [PubMed] [Google Scholar]
- Caporaso, JG, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. Gonzalez Pena, J. K. Goodrich, J. I. Gordon, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates M. E., Fuller R., Harrison G. F., Lev M., and Suffolk S. F.. 1963. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br. J. Nutr. 17:141–150. doi:10.1079/BJN19630015 [DOI] [PubMed] [Google Scholar]
- Coffey R. D., and Cromwell G. L.. 1995. The impact of environment and antimicrobial agents on the growth response of early-weaned pigs to spray-dried porcine plasma. J. Anim. Sci. 73:2532–2539. doi:10.2527/1995.7392532x [DOI] [PubMed] [Google Scholar]
- Collier C. T., Smiricky-Tjardes M. R., Albin D. M., Wubben J. E., Gabert V. M., Deplancke B., Bane D., Anderson D. B., and Gaskins H. R.. 2003. Molecular ecological analysis of porcine ileal microbiota responses to antimicrobial growth promoters. J. Anim. Sci. 81:3035–3045. [DOI] [PubMed] [Google Scholar]
- Demain A. L., Newcomb M., and Wu J. H.. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124–154. doi:10.1128/MMBR.69.1.124-154.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner J. J., and Richards J. D.. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634–643. [DOI] [PubMed] [Google Scholar]
- Dumonceaux T. J., Hill J. E., Hemmingsen S. M., and Van Kessel A. G.. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2823. doi:10.1128/AEM.72.4.2815-2823.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaorui B., Ma S., Zhu Z., Su Y., Zoetendal E. G., Mackie R., Liu J., Mu C., Huang R., Schmidt H., and Zhu W.. 2016. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ. Microbiol. 18:1566–1577. doi:10.1111/1462-2920 [DOI] [PubMed] [Google Scholar]
- Grinstead G. S., Goodband R. D., Dritz S. S., Tokach M. D., Nelssen J. L., Woodworth J. C., and Molitor M.. 2000. Effects of a whey protein product and spray-dried animal plasma on growth performance of weanling pigs. J. Anim. Sci. 78:647–657. [DOI] [PubMed] [Google Scholar]
- Hermes G.R., Molist F., Francisco Perez J., Gomez de Segura A., Ywazaki M., Davin R., Nofrarias M., Korhonen T. K., Virkola R., and Martin-Orue S. M.. 2013. Casein glycomacropeptide in the diet may reduce Escherichia coli attachment to the intestinal mucosa and increase the intestinal lactobacilli of early weaned piglets after an enterotoxigenic E. coli K88 challenge. Br. J. Nutr. 109:1001–1012. doi:10.1017/S0007114512002978 [DOI] [PubMed] [Google Scholar]
- Keessen E. C., Gaastra W., and Lipman L. J.. 2011. Clostridium difficile infection in humans and animals, differences and similarities. Vet. Microbiol. 153:205–217. doi:10.1016/j.vetmic.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Kim H. B., Borewicz K., White B. A., Singer R. S., Sreevatsan S., Tu Z. J., and Isaacson R. E.. 2012. Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. USA. 109:15485–15490. doi:10.1073/pnas.1205147109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T., Johnson T. A., Allen H. K., Bayles D. O., Alt D. P., Stedtfeld R. D., Sul W. J., Stedtfeld T. M., Chai B., Cole J. R.,. et al. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA. 109:1691–1696. doi:10.1073/pnas.1120238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marounek M., Fliegrova K., and Bartos S.. 1989. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl. Environ. Microbiol. 55:1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Wallace G., Zhang C., Legge R., Benson A. K., Carr T. P., Moriyama E. N., and Walter J.. 2009. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 75:4175–4184. doi:10.1128/AEM.00380-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H., Graf A. B., Tatto N. E., Thallinger G. G., Mattanovich D., and Sauer M.. 2011. Genome sequence of the ruminal bacterium Megasphaera elsdenii. J. Bacteriol. 193:5578–5579. doi:10.1128/JB.05861-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofrarias M., Manzanilla E. G., Pujols J., Gibert X., Majo N., Segales J., and Gasa J.. 2006. Effects of spray-dried porcine plasma and plant extracts on intestinal morphology and on leukocyte cell subsets of weaned pigs. J. Anim. Sci. 84:2735–2742. doi:10.2527/jas.2005-414 [DOI] [PubMed] [Google Scholar]
- Nollet H., Deprez P., Van Driessche E., and Muylle E.. 1999. Protection of just weaned pigs against infection with F18+ Escherichia coli by non-immune plasma powder. Vet. Microbiol. 65:37–45. [DOI] [PubMed] [Google Scholar]
- Peace R. M., Campbell J., Polo J., Crenshaw J., Russell L., and Moeser A.. 2011. Spray-dried porcine plasma influences intestinal barrier function, inflammation, and diarrhea in weaned pigs. J. Nutr. 141:1312–1317. doi:10.3945/jn.110.136796 [DOI] [PubMed] [Google Scholar]
- Perez-Bosque A., Amat C., Polo J., Campbell J. M., Crenshaw J., Russell L., and Moreto M.. 2006. Spray-dried animal plasma prevents the effects of Staphylococcus aureus enterotoxin B on intestinal barrier function in weaned rats. J. Nutr. 136:2838–2843. [DOI] [PubMed] [Google Scholar]
- Pierce J. L., Cromwell G. L., Lindemann M. D., Russell L. E., and Weaver E. M.. 2005. Effects of spray-dried animal plasma and immunoglobulins on performance of early weaned pigs. J. Anim. Sci. 83:2876–2885. [DOI] [PubMed] [Google Scholar]
- Prabhu R., Altman E., and Eiteman M. A.. 2012. Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl. Environ. Microbiol. 78:8564–8570. doi:10.1128/AEM.02443-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Kanarek L., Koninkx J., Hendriks H., Lintermans P., Bertels A., Charlier G., and Van Driessche E.. 1993. Inhibition of adhesion of enterotoxigenic Escherichia coli cells expressing F17 fimbriae to small intestinal mucus and brush-border membranes of young calves. Microb. Pathog. 15:207–219. [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J.,. et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. doi:10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham Y., Lamed R., and Bayer E. A.. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275–281. [DOI] [PubMed] [Google Scholar]
- Stanley D., Hughes R. J., Geier M. S., and Moore R. J.. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 7:187. doi:10.3389/fmicb.2016.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellstrom B., Stenhammar L., Eriksson S., and Magnusson K. E.. 1993. Oral immunoglobulin A supplement in treatment of Clostridium difficile enteritis. Lancet 341:701–702. [DOI] [PubMed] [Google Scholar]
- Torrallardona D., Conde M. R., Badiola I., Polo J. and Brufau J.. 2003. Effect of fishmeal replacement with spray-dried animal plasma and colistin on intestinal structure, intestinal microbiology, and performance of weanling pigs challenged with Escherichia coli K99. J. Anim. Sci. 81:1220–1226. doi:10.2527/2003.8151220x [DOI] [PubMed] [Google Scholar]
- Tran H., Bundy J. W., Li Y. S., Carney-Hinkle E. E., Miller P. S., and Burkey T. E.. 2014. Effects of spray-dried porcine plasma on growth performance, immune response, total antioxidant capacity, and gut morphology of nursery pigs. J. Anim. Sci. 92:4494–4504. doi:10.2527/jas.2014-7620 [DOI] [PubMed] [Google Scholar]
- Van Dijk A. J., Margry R. J., Van Der Lee A. G., Hemke G., and Beynen A. C.. 2002. Growth performance and health status in weanling piglets fed spray-dried porcine plasma under typical Northern European conditions. J. Anim. Physiol. Anim. Nutr. (Berl) 86:17–25. [DOI] [PubMed] [Google Scholar]
- Whiteley A. S., Jenkins S., Waite I., Kresoje N., Payne H., Mullan B., Allcock R., and O’Donnell A.. 2012. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) Platform. J. Microbiol. Methods 91:80–88. doi:10.1016/j.mimet.2012.07.008 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang X., Xiong X., and Yin Y.. 2016. Energy metabolism in intestinal epithelial cells during maturation along the crypt-villus axis. Sci. Rep. 6:31917. doi:10.1038/srep31917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewski M., Proietti C., Ellis J., Hasan S., Brion M. J., Berger B., and Krause L.. 2016. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 33:782–783. doi:10.1093/bioinformatics/btw725 [DOI] [PMC free article] [PubMed] [Google Scholar]