The epigenetic mechanism of DNA methylation optimizes gene expression in endosymbiosis between cnidarians and their microalgae.
Abstract
The symbiotic relationship between cnidarians and dinoflagellates is the cornerstone of coral reef ecosystems. Although research has focused on the molecular mechanisms underlying this symbiosis, the role of epigenetic mechanisms, that is, the study of heritable changes that do not involve changes in the DNA sequence, is unknown. To assess the role of DNA methylation in the cnidarian-dinoflagellate symbiosis, we analyzed genome-wide CpG methylation, histone associations, and transcriptomic states of symbiotic and aposymbiotic anemones in the model system Aiptasia. We found that methylated genes are marked by histone 3 lysine 36 trimethylation (H3K36me3) and show significant reduction of spurious transcription and transcriptional noise, revealing a role of DNA methylation in the maintenance of transcriptional homeostasis. Changes in DNA methylation and expression show enrichment for symbiosis-related processes, such as immunity, apoptosis, phagocytosis recognition, and phagosome formation, and reveal intricate interactions between the underlying pathways. Our results demonstrate that DNA methylation provides an epigenetic mechanism of transcriptional homeostasis that responds to symbiosis.
INTRODUCTION
Coral reefs are ecologically important marine ecosystems, which cover less than 0.2% of our oceans but sustain an estimated ~25% of the world’s marine species and 32 of 33 animal phyla (1). Coral reefs are also economically important because they provide food and livelihood opportunities to at least 500 million people; worldwide, they have a net present value of almost USD 800 billion, and they generate USD 30 billion in net economic benefits annually (1). The trophic and structural foundation of these ecosystems relies on the symbiotic relationship between reef-building corals and their dinoflagellate endosymbionts in the genus Symbiodinium. Through this metabolic symbiosis, the coral host recycles waste nitrogenous products in exchange for photosynthetically fixed carbon from the symbiont, thereby allowing corals to thrive in the nutrient-poor environment of tropical oceans. Unfortunately, coral reef ecosystems are under severe threat from anthropogenic stressors. Increasing sea surface temperature, pollution, and other stressors can cause coral bleaching, that is, the expulsion of the intracellular symbionts from the coral host tissue, leading to increased mortality and global coral reef decline. Despite increasing efforts to study the mechanisms underlying the regulation and environmental stress-related breakdown of this symbiotic association (2), we still lack knowledge of the basic molecular processes involved. Specifically, the role of epigenetic mechanisms in the regulation of this symbiosis and their potential to contribute to increased resilience in response to environmental stress, as reported in other organisms (3), remains unstudied.
DNA methylation is an epigenetic mechanism by which a methyl group (CH3) is added to the DNA. This chemical modification, which is mostly added to cytosine residues, does not constitute a base change in the DNA but rather modifies how the DNA interacts with regulatory proteins involved in transcriptional regulation and DNA packaging. DNA methylation plays an important role in many biological processes of plants and animals (4). It has been proposed as a mechanism for organisms to adjust their phenotype in response to their environment to optimize the organismal response to changing environmental conditions (5, 6). For instance, recent findings in mice show an important function for gene body methylation in optimizing gene expression by inhibiting the initiation of transcription from cryptic sequences within the gene body. These spurious transcription events can produce partial (that is, “spurious”) transcripts that, if translated, result in the expression of truncated proteins that might interfere with native protein function (7). It was also shown that gene body methylation in mice is induced by active gene expression, allowing the organism to optimize transcription of actively expressed genes in response to transcriptional changes. Similar functions for DNA methylation have also been proposed in plants, suggesting a general role of DNA methylation in the maintenance of transcriptional homeostasis (8).
Several studies on DNA methylation in cnidarians have been published recently (9, 10); however, its role and function in cnidarians is, at present, largely unknown (11). The sea anemone Aiptasia is an emerging model to study the cnidarian-dinoflagellate symbiosis. Like corals, Aiptasia establishes a stable but temperature-sensitive symbiosis with dinoflagellates of the genus Symbiodinium but can be easily maintained in an aposymbiotic state. These features, combined with the ease of Aiptasia culture, provides a tractable system to study the molecular mechanisms underlying symbiosis without the impeding stress responses associated with coral bleaching stress (12, 13).
Using the model system Aiptasia (strain CC7, sensu Exaiptasia pallida), we obtained whole-genome CpG DNA methylation and chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) data from aposymbiotic and symbiotic individuals to study the function of DNA methylation in transcriptional regulation and its role in the cnidarian-dinoflagellate symbiosis.
RESULTS
Aiptasia DNA methylation patterns change with symbiotic state
To assess changes in DNA methylation in response to symbiosis, we performed whole-genome bisulfite sequencing (WGBS) with an average coverage of 53× per individual on 12 anemones, providing six biological replicates per symbiotic state (symbiotic versus aposymbiotic). Methylation calling using the combined data set identified 710,768 methylated CpGs (6.37% of all CpGs in the Aiptasia genome), that is, methylated sites in the Aiptasia genome. Notably, the percentage of methylated CpGs is much lower than that of mammals (60 to 90%) (14) but comparable to that of the coral Stylophora pistillata (7%) (15). We identified 10,822 genes (37% of all 29,269 gene models identified in the Aiptasia genome) with at least five methylated CpG sites that were subsequently defined as methylated genes. On average, these genes had 18.4% CpGs methylated, 3-fold higher than the average methylation density across the entire genome (chi-square test, P < 2.2 × 10−16) and 167-fold higher than the methylation levels in noncoding regions. These findings indicate that the distribution of CpG methylation is nonrandom and mainly located in gene bodies, similar to corals (15, 16) and other invertebrate species (17, 18).
To analyze the relationship between methylation density (percentage of CpGs) and gene density [the number of genes per 10,000 base pairs (bp)], we ran a sliding window (window size, 40 kb; step, 30 kb) and visualized the results in a Circos plot (fig. S1) (19). The correlation of CpG content and distribution of methylation showed a negative correlation (Pearson correlation coefficient, r = −0.31; P < 2.2 × 10−16), suggesting that methylation tends to preferentially occur in CpG-poor regions (fig. S2). Gene density had a positive correlation with methylation density (r = 0.21; P < 2.2 × 10−16), consistent with the finding that methylation is predominantly located in gene bodies (Fig. 1). We also observed that within gene bodies, introns showed significantly higher methylation densities than exons (Fig. 1B).
Fig. 1. DNA methylation landscape.
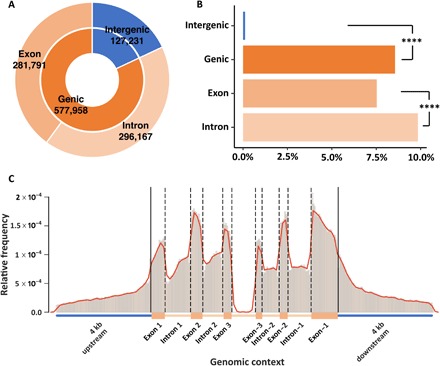
(A) Relative distribution of methylated CpGs across intergenic (18%), genic (82%), intronic (42%), and exonic (40%) regions in the Aiptasia genome. (B) Normalized frequencies of methylated CpGs in different regions. Chi-square test shows significant differences between intergenic and genic regions and between exons and introns (****P < 0.0001). (C) Relative frequencies of methylated positions across a normalized gene model. All frequencies were normalized to the lengths of the corresponding regions.
Methylated genes are marked by histone 3 lysine 36 trimethylation
Analysis of methylation patterns (see above) within gene bodies showed rapidly increasing methylation levels after the transcription start site that are maintained before slowly decreasing toward the transcription termination site (fig. S3A). We found that gene body methylation in Aiptasia is positively correlated with expression (Fig. 2A), suggesting that DNA methylation either increases the expression of genes or is increased as a consequence of transcription, whereby increased expression results in methylation of the respective loci. The latter interpretation would be in line with recent findings in mouse embryonic stem cells (7), which demonstrated that gene body methylation is established and maintained as a result of active transcription by RNA polymerase II and recruitment of the histone-modifying protein SetD2 that trimethylates histone H3 at lysine 36 (H3K36me3). This histone mark is specifically bound via the PWWP (Pro-Trp-Trp-Pro) domain present in the DNA methyltransferase Dnmt3b, which in turn methylates the surrounding DNA accordingly, resulting in the inhibition of transcription initiation from cryptic promoters within the gene body and thus a significant reduction of spurious transcription.
Fig. 2. DNA methylation is associated with higher expression.
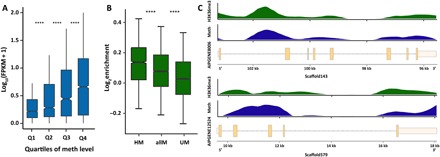
(A) Gene expression is positively correlated with median methylation levels. t test P values are 7.65 × 10−21, 3.75 × 10−14, and 1.75 × 10−13 for Q1 (first quartile) and Q2 of methylation (meth) levels, Q2 and Q3, and Q3 and Q4, respectively. FPKM, fragments per kilobase per million mapped fragments. ****P < 0.0001. (B) ChIP-seq analysis of H3K36me3 signals shows significant enrichment in methylated genes. t test P values are 2.48 × 10−20 for highly methylated genes (HM) and all methylated genes (allM) and 1.06 × 10−72 for unmethylated genes (UM) and all methylated genes. Highly methylated genes show the strongest enrichment of H3K36me3, followed by all methylated genes. In contrast, unmethylated genes show only weak enrichment of H3K36me3 over input controls. ****P < 0.0001. (C) Distribution of H3K36me3 enrichment and DNA methylation levels across two exemplary gene models. H3K36me3 and DNA methylation show coinciding distribution patterns over genes.
Analysis of the Aiptasia gene set identified a Dnmt3 gene (AIPGENE24404) that also encodes a PWWP domain as reported for the mouse homolog. To test whether the mechanism previously described in mice is conserved in Aiptasia, we performed a ChIP-seq experiment using a validated antibody against H3K36me3 (figs. S4 and S5). As predicted, our analysis confirmed a significantly higher association of H3K36me3 with methylated genes (P = 2.48 × 10−20 for highly methylated genes and all methylated genes; Fig. 2, B and C), suggesting that DNA methylation in Aiptasia might be a consequence of expression. We then analyzed whether methylated genes also exhibited significantly lower levels of intragenic initiation of transcription in Aiptasia. To assess the level of spurious transcripts in genes of different methylation levels, we calculated the expression level for each exon and computed the natural log fold change of expression relative to exon 1. Our rationale was that spurious transcripts that start somewhere within the gene body contribute more to observed increases in expression of more distal exons but less so to more proximal exons. Consequently, higher levels of spurious transcription would result in higher ratios of expression for more distal exons.
Analysis of transcriptional profiles of methylated and unmethylated genes showed significantly lower levels of spurious transcription along the gene body of methylated genes (P < 2 × 10−6; Fig. 3A). Regression analysis of median methylation levels and the coefficient of transcriptional variation of genes showed that, given the same expression level, methylated genes always exhibited lower levels of transcriptional variation (Fig. 3B).
Fig. 3. DNA methylation regulates transcriptional homeostasis.
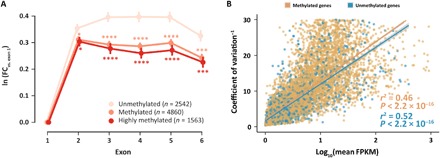
(A) Spurious transcription in gene bodies is significantly lower in methylated and highly methylated genes. The y axis shows the natural logarithm of the coverage fold change (FC) of exons 1 to 6 versus exon 1, with lower values meaning less spurious transcription. *P < 0.05, ***P < 0.001, ****P < 0.0001. (B) There is a linear relationship between the inverse of transcriptional noise and log expression level [log10(mean FPKM)]. Given the same expression level, methylated genes always show lower levels of transcriptional noise. Methylated genes, n = 8561, r2 = 0.46, P < 2.2 × 10−16; unmethylated genes, n = 2491, r2 = 0.52, P < 2.2 × 10−16.
DNA methylation regulates transcriptional homeostasis during symbiosis
On the basis of our above analyses, we investigated whether DNA methylation might also be involved in the regulation of symbiosis by identifying differentially methylated genes (DMGs) between symbiotic and aposymbiotic Aiptasia. Comparison of DNA methylation patterns using principal components analysis (PCA) separated symbiotic and aposymbiotic individuals by the first principal component, which accounted for ~18% of the variance (Fig. 4 and fig. S6). This analysis echoed the findings from a PCA on gene expression where symbiosis state was separated by the first principal component accounting for ~25% of the variance (Fig. 4B) (20) and highlighted that specific changes in DNA methylation patterns occurred in response to symbiosis. Analysis of DNA methylation and expression profiles using correlation analyses further confirmed this finding, providing additional evidence that the observed changes of both gene expression and DNA methylation were associated with symbiotic state (fig. S6).
Fig. 4. PCA and KEGG pathway enrichment analysis.
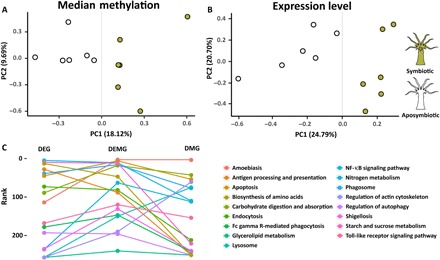
(A and B) PCA of gene expression and median methylation levels of Aiptasia genes. Both gene expression and DNA methylation separate samples by symbiosis state. PC1, first principal component; PC2, second principal component. (C) KEGG pathway enrichment analysis of previously identified symbiosis-related pathways. The y axis shows the ranks of these pathways in the pathway enrichment analyses of DEGs, DEMGs, and DMGs, respectively. The combined set of DEMG provides significant lower P values (front ranks) for symbiosis-related pathways. NF-κB, nuclear factor κB.
Subsequently, we analyzed changes in DNA methylation and gene expression between symbiotic and aposymbiotic Aiptasia to assess their correlation with potential biological functions in symbiosis. We determined DMGs using a generalized linear model from Foret et al. (21) that was modified to allow for replicate-aware analysis. This approach identified 2133 DMGs [false discovery rate (FDR) ≤ 0.05; table S1] that specifically changed their methylation status in response to symbiosis. To verify these results, we sequenced a subset of 14 DMGs using bisulfite polymerase chain reactions (PCRs). The results show a strong correlation (aposymbiotic, r2 = 0.815 and P = 1 × 10−5; symbiotic, r2 = 0.922 and P = 5.2 × 10−8) to our WGBS and confirm the observed methylation changes within these loci (fig. S7).
Analysis of gene expression changes in the same 12 samples (that is, 6 symbiotic and 6 aposymbiotic anemones) identified 1278 differentially expressed genes (DEGs; FDR ≤ 0.05; table S2) between symbiotic and aposymbiotic Aiptasia, of which 14 genes were subsequently confirmed via quantitative PCR (qPCR; fig. S8). However, analysis of the overlap between DMGs and DEGs showed only 103 genes that were shared, suggesting that DEGs are not necessarily the same cohort of genes that are differentially methylated. This was further reflected in the lack of correlation between DNA methylation and gene expression changes; that is, changes in DNA methylation did not correlate with respective changes in gene expression (fig. S9). Functional enrichment analyses based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of all DMGs and DEGs identified several symbiosis-relevant functions and pathways in both groups (tables S3 to S10).
On the basis of the finding that gene body DNA methylation is likely a consequence of active transcription, we hypothesized that changes in DNA methylation patterns might also provide a record of transcriptional activity over longer periods of time. We therefore tested whether differential methylation and acute transcriptional changes, obtained from our RNA-seq analysis, provide a complementary view of the processes underlying symbiosis. For this, we compared enrichment of symbiosis-specific pathways across the sets of 2133 DMGs, 1278 DEGs, and the combined set of both DMGs and DEGs (3308 DEMGs). We observed that the combined data set (DEMGs) provided significantly lower P values for previously identified symbiosis-related pathways, including apoptosis, phagosome formation, nitrogen metabolism, and arginine biosynthesis, among others (paired t test: DEMG versus DEG, P = 0.015; DEMG versus DMG, P = 0.009; Fig. 4C and table S11). This result suggested that changes in methylation and transcription provide complementary information with regard to transcriptional adjustments in response to symbiosis.
DMGs and DEGs are involved in all stages of symbiosis
Analysis of the combined DMG and DEG gene set showed significant enrichment of genes involved in the distinct phases of symbiosis, which are symbiosis establishment, maintenance, and breakdown (2). Using an integrated pathway analysis based on known molecular interactions between proteins, we found that these processes are linked through several DMGs and/or DEGs (Figs. 5 and 6, tables S11 and S12, and the Supplementary Materials).
Fig. 5. Schematic diagram of symbiosis establishment– and breakdown-associated genes.
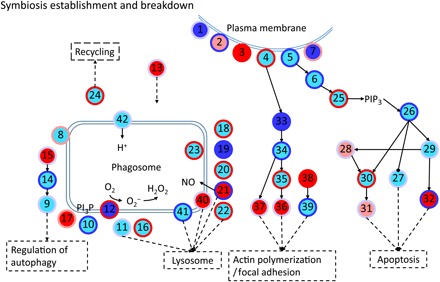
Every node represents a category of genes and generally has multiple corresponding genes. The inside colors of nodes represent the expression changes of corresponding genes, including non-DEGs (cyan), up-regulated (red), down-regulated (blue), and up- and down-regulated DEGs (light red). The colors of node edges represent the methylation level changes of corresponding genes, including non-DMGs (light blue), hypermethylated (red), hypomethylated (blue), and hypermethylated and hypomethylated DMGs (light red). Numbers in circles denote genes/proteins: 1, complement receptor; 2, scavenger receptor; 3, C-type lectin; 4, integrin; 5, Toll-like receptor; 6, Ras-related C3 botulinum toxin substrate 1 [RAC1; Ras homolog (Rho) family]; 7, collagen; 8, vesicle-associated membrane protein; 9, autophagy-related protein 16 (ATG16); 10, Ras-related protein 5 (Rab5); 11, Rab7; 12, reduced form of nicotinamide adenine dinucleotide oxidase; 13, syntaxin 12; 14, ATG5; 15, ATG10; 16, programmed cell death 6–interacting protein; 17, sorting nexin (SNX); 18, cytoplasmic dynein; 19, tubulin alpha chain; 20, tubulin beta chain; 21, nitric oxide synthase (NOS); 22, lysosome-associated membrane glycoprotein/cluster of differentiation; 23, cathepsin L; 24, kinesin; 25, phosphatidylinositol 4,5-bisphosphate 3-kinase; 26, RAC serine/threonine-protein kinase; 27, B cell lymphoma 2 (Bcl-2) antagonist of cell death; 28, tumor necrosis factor receptor–associated factor; 29, NF-κB; 30, caspase-8 (CASP8); 31, CASP7; 32, apoptosis regulator/Bcl-2 (BCL2); 33, Rho; 34, Rho-associated protein kinase; 35, phosphatidylinositol 4-phosphate 5-kinase/phosphatidylinositol 5-phosphate 4-kinase/phosphatidylinositol 3-phosphate 5-kinase; 36, vinculin; 37, radixin; 38, profilin; 39, actin; 40, CD63; 41, lysosomal-associated transmembrane protein; 42, V-type proton adenosine triphosphatase (ATPase); PI3P, phosphatidylinositol-3-phosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate.
Fig. 6. Schematic diagram of symbiosis maintenance–associated genes.
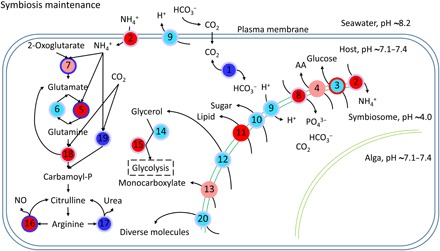
Every node represents a category of genes and generally has multiple corresponding genes. The inside colors of nodes represent the expression changes of corresponding genes, including non-DEGs (cyan), up-regulated (red), down-regulated (blue), and up-regulated and down-regulated DEGs (light red). The colors of node edges represent the methylation level changes of corresponding genes, including non-DMGs (light blue), hypermethylated (red), hypomethylated (blue), and hypermethylated and hypomethylated DMGs (light red). Numbers in circles denote genes/proteins: 1, carbonic anhydrase (CA); 2, ammonium transporter; 3, glucose transporter; 4, amino acid transporter; 5, glutamine synthetase; 6, glutamate synthase; 7, glutamate dehydrogenase; 8, phosphate transporter; 9, V-type proton ATPase; 10, sugar transporter; 11, lipid transfer protein; 12, aquaporin 3 (glycerol transporter); 13, monocarboxylate transporter; 14, alcohol dehydrogenase; 15, aldehyde dehydrogenase; 16, NOS; 17, arginase; 18, carbamoyl-phosphate synthase/aspartate carbamoyltransferase/dihydroorotase; 19, carbamoyl-phosphate synthase (ammonia); 20, ABC transporter.
For instance, we found numerous symbiosis-related receptors that respond to symbiosis on a transcriptional and/or methylation level (Fig. 5), including C-type lectins (Fig. 5, 3), Toll-like receptors (Fig. 5, 5), and the scavenger receptor SRB1 (Fig. 5, 2) that has been previously implicated in symbiont recognition in the sea anemone Anthopleura elegantissima (22). Following symbiont recognition, we also found several known engulfment and sorting-related genes to change in methylation and/or expression such as Rab5 (Fig. 5, 10), SNX (Fig. 5, 17), RAC1 (Fig. 5, 6), the lysosomal-associated membrane protein 1/2 (Fig. 5, 22), and many genes related to the cytoskeleton and movement (Fig. 5, 33 to 39).
As expected in a metabolic symbiosis (2), we also identified a large number of genes known to be involved in nutrient exchange to be differentially methylated and/or expressed in our study. These included genes involved in the provision of inorganic carbon in the form of CO2 or bicarbonate (HCO3−) to fuel symbiont-driven photosynthesis (Fig. 6, 1) (23) and genes involved in the exchange of fixed carbon in the form of lipids (Fig. 6, 11), sugars (Fig. 6, 10), and amino acids (Fig. 6, 4) (24). Concordantly, we also found that genes involved in nitrogen acquisition, such as ammonium transporter genes (Fig. 6, 2), and genes involved in glutamate metabolism (Fig. 6, 5 to 7) differentially methylated and/or differentially expressed in symbiotic anemones.
Finally, our analyses also highlighted genes putatively involved in the expulsion or degradation of symbionts in response to environmental stress or as a means to control symbiont densities. Autophagy is of interest in this regard because it links to other membrane trafficking pathways and to apoptosis, and evidence suggests that autophagy also plays a role in removal of symbionts during bleaching (25). Intracellular degradation of the symbiont is a result of reengagement of the phagosomal maturation process or autophagic digestion of the symbiont by the host cell (2), and we find both apoptosis- and autophagy-related genes to significantly change in their methylation and/or expression level. These include the apoptosis genes RAC serine/threonine-protein kinase (Fig. 5, 25), CASP7 (Fig. 5, 31), CASP8 (Fig. 5, 30), nitric oxide synthase (Fig. 5, 21), and Bcl-2 (Fig. 5, 27), as well as the autophagy proteins 5 and 10 (Fig. 5, 14 and 15), among others.
DISCUSSION
To assess the role of CpG methylation in the cnidarian-dinoflagellate symbiosis, we undertook a global analysis of changes in the DNA methylomes and transcriptomes of aposymbiotic and symbiotic Aiptasia. In contrast to their vertebrate counterparts, only 6.37% of all CpGs in the Aiptasia genome are methylated, but their distribution is significantly nonrandom (t test; P < 3 × 10−300), and the methylation ratio is highest in gene bodies (18.4% of CpGs are methylated). Analysis of the distribution of the histone modification H3K36me3 further showed significant enrichment of this epigenetic marker in methylated genes, echoing findings in mammals and invertebrates (26). We found that methylated genes show significant reduction of spurious transcription and transcriptional noise (Fig. 2B), suggesting that both the underlying mechanism of epigenetic cross-talk and the biological function of DNA methylation are evolutionarily conserved throughout metazoans. These results highlight a tight interaction of transcription and epigenetic mechanisms in optimizing gene expression in response to changing transcriptional needs (7). Further support for such a role is provided by the analysis of DMGs and DEGs, which, when combined, showed significant increase in enrichment of symbiosis-relevant processes. This suggests that DNA methylation and transcriptome analyses provide complementary views of cellular responses to symbiosis whereby methylation changes provide a transcriptional record of longer-term transcriptional adjustments.
While our analysis identified several genes, processes, and pathways previously reported to be involved in symbiosis, it further highlights their intricate molecular interactions. Symbiosis recognition, sorting, and breakdown are interconnected processes, which is reflected in the observed changes in methylation and expression. The molecular machinery involved in phagosome maturation is tightly linked to autophagy and apoptosis, enabling the host to respond to potential pathogen invasion and to degrade and remove dead or unsuitable symbionts. This is strongly supported by immunofluorescence examinations of Aiptasia gastrodermal cell macerates, showing that Rab5 appears around healthy, newly ingested, and already established Symbiodinium but is replaced by Rab7 in heat-killed or dichlorophenyl dimethylurea–treated newly ingested Symbiodinium. Conversely, Rab7 is absent from untreated newly infected or already-established Symbiodinium (27). However, Rab5 has also been reported to be involved in other functions during symbiosis. Specifically, it was shown that Rab5-positive phagosomes are acidified in Caenorhabditis elegans, indicating that Rab5 is necessary to recruit V-type ATPases to acidify the phagosome by pumping in protons (28). Here, V-type H+-ATPase functions in concert with CA by pumping protons into the surrounding seawater, producing a locally low pH environment that increases the conversion to CO2 and facilitates carbon flux to the symbiont (29). Previous studies in corals have shown that this process appears to be conserved where the symbiosome is also acidified to very low pH (pH ~4.0) to promote photosynthesis (29).
Rab5 is also required for the exosomal release of CD63 (30), which mediates the endocytotic sorting process and transport to lysosomes (31). This process is further regulated by Rac1 (32) in conjunction with SNX (one DEG and five DMGs) and the guanosine triphosphatase Rho (one DEG and DMG), all of which were also identified in our analyses. The sorting of phagocytosed Symbiodinium is critical to symbiosis establishment as Symbiodinium cells are phagocytosed at the apical end and transported to the base of the cell, where they are protected from digestion. In contrast, Symbiodinium staying at the apical end of the cell are degraded (33). Previous studies have also suggested a role for apoptosis in postphagocytic selection of compatible symbionts (34) and the regulation of host and symbiont cell growth and proliferation (2). Note that the response of apoptotic genes observed here and in other studies (35) is quite complex with changes in DNA methylation and expression observed for both apoptosis activators, such as CASP7 and CASP8, and apoptosis inhibitors, such as Bcl-2. The high variability of apoptosis gene expression observed across symbiosis studies might reflect the different roles this process fulfills in symbiosis establishment, maintenance, and breakdown. It is conceivable that these processes require different transcriptional responses to either induce or inhibit apoptosis. A significant increase in caspase activity, for instance, has been reported for larvae from the coral Fungia scutaria when challenged with incompatible symbionts (36). Conversely, the apoptosis inhibitor Bcl-2 has frequently been found to be up-regulated in corals under thermal stress, suggesting a role in the prevention of symbiosis breakdown and survival (37).
Similar to the processes of symbiosis initiation and breakdown, we also found significant enrichment of genes involved in nutrient exchange, including transporters for amino acids, sugars, lipids, and others. Many of these transporters have previously been implicated in symbiosis maintenance (2, 38). Notably, this also includes genes involved in the transport and assimilation of ammonium. Nitrogen is a main limiting nutrient on coral reefs (39), and the coral-Symbiodinium symbiosis has been proposed to increase the efficiency of nitrogen utilization by both partners (40). While the underlying nature of this mechanism is currently debated (41), the changes in methylation and gene expression of central genes involved in ammonium assimilation, such as glutamate dehydrogenase and glutamate synthetase, are consistent with previous findings in Aiptasia and corals (27).
CONCLUSIONS
This study provides the first analysis of the function and role of DNA methylation in the cnidarian-dinoflagellate symbiosis. Our results show that the epigenetic cross-talk between the histone mark H3K36me3 and gene body methylation is conserved in cnidarians and reveal a role of gene body methylation in the reduction of spurious transcription and transcriptional noise. Furthermore, we show that changes in DNA methylation patterns are associated with symbiotic states and imply a function in the establishment, maintenance, and breakdown of the important cnidarian-dinoflagellate symbiotic association. Our findings therefore provide evidence for a role of DNA methylation as an epigenetic mechanism involved in the maintenance of transcriptional homeostasis in the cnidarian-dinoflagellate symbiosis. The premise that epigenetic mechanisms play a role in organismal acclimation warrants future experiments targeted to investigate whether DNA methylation could also contribute to resilience through the epigenetic adjustment of transcription in response to environmental stress in Aiptasia and corals.
MATERIALS AND METHODS
Aiptasia culture and DNA/RNA extraction
Aiptasia (E. pallida) of the clonal strain CC7 (42) was used for this study. Anemones were maintained in polycarbonate tubs with autoclaved seawater at ~25°C on a 12-hour light/12-hour dark cycle at 20 to 40 μmol photons m−2 s−1 light intensity and fed freshly hatched Artemia nauplii (brine-shrimp) approximately twice per week. To generate aposymbiotic anemones and reduce potential symbiosis-related DNA methylation patterns, animals were subjected to multiple cycles of cold-shock treatment and the photosynthesis inhibitor diuron (Sigma-Aldrich), as described in Baumgarten et al. (12). We note that while this approach provided 12 months for the aposymbiotic anemones to change their methylomes to the new symbiosis state, the possibility cannot be excluded that residual methylation signals of symbiosis persisted for longer time periods due to long-term memory. In the same way, it should be noted that the bleaching process involving diuron and cold stress treatment could also induce long-term changes in DNA methylation patterns (43). Aposymbiotic anemones were kept individually in 15 ml of autoclaved seawater in six-well plates and inspected by fluorescence stereomicroscopy to confirm complete absence of dinoflagellates. To exclude potential batch effects as a source of DNA methylation changes, we first generated four independent batches of aposymbiotic anemones. The four batches of aposymbiotic anemones were then maintained in parallel for a period of 1 year before beginning the experiment described below to minimize any potential effects on the DNA methylation pattern caused by the bleaching treatment.
To generate symbiotic anemones, we then separately infected aposymbiotic CC7 individuals from each of the four aposymbiotic cultures described above using the compatible Clade B Symbiodinium strain SSB01, originally isolated from Aiptasia strain H2 (12, 44). The four batches of symbiotic anemones were maintained for a further 12 months under regular culture conditions, as described above. The corresponding four aposymbiotic cultures were maintained in darkness until 3 months before collection. For the last 3 months, individuals from these aposymbiotic cultures were subjected to the same culture conditions as the symbiotic cultures to monitor for unwanted spontaneous reestablishment of symbiosis under light.
After the 12-month experimental period, we collected six biological replicates from each of the four aposymbiotic and symbiotic cultures (one additional replicate was taken from batches 1 and 2 of each symbiotic state) for subsequent DNA and RNA extraction as described below. For each symbiotic state, six biological replicates, weighing 20 to 28 mg (wet weight), were extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen). The manufacturer’s protocol was followed with the omission of the optional step 4 (temporal storage at 4°C if not performing DNA purification immediately). Both DNA and RNA for subsequent experiments (RNA-seq, bisulfite sequencing, and validations) were generated from the same pools of extracted nucleic acids. DNA concentrations were determined using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). RNA concentrations and integrity were determined using the Bioanalyzer Nano RNA Kit (Agilent Technologies).
RNA-seq and bisulfite sequencing
Directional mRNA libraries were produced using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s protocol. Bisulfite DNA libraries were prepared following a modified version of the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB). Methylated TruSeq Illumina adapters (Illumina) were used during the adapter ligation step, followed by bisulfite conversion with the EpiTect Bisulfite Kit (Qiagen), with the following cycling conditions: 95°C for 5 min, 60°C for 25 min, 95°C for 5 min, 60°C for 85 min, 95°C for 5 min, and 60°C for 175 min, and three cycles of 95°C for 5 min, 60°C for 180 min, and hold at 20°C for ≤5 hours.
The final libraries were enriched with the KAPA HiFi HotStart Uracil+ ReadyMix (2X) (Kapa Biosystems) following the standard protocol for bisulfite-converted next generation sequencing library amplification. Final libraries were quality-checked using the Bioanalyzer DNA 1K Chip (Agilent Technologies), quantified using Qubit 2.0 (Thermo Fisher Scientific), pooled in equimolar ratios, and sequenced on the HiSeq 2000.
Identification of methylated CpGs
Sequencing of the 12 libraries (2 conditions, 6 biological replicates each) resulted in 819 million read pairs from 8 lanes of the Illumina HiSeq 2000 platform. Adapters were trimmed from the raw sequences using Cutadapt v1.8 (45). Subsequently, trimmed reads were mapped to the Aiptasia genome (12) using Bowtie2 v2.2.3 (46), and methylation calls were performed using Bismark v0.13 (47).
Three filters were used to reduce false positives. First, for each position with k methylated reads mapping to it, the probability of it occurring through sequencing error (that is, unmethylated position appearing as methylated) was modeled using a binomial distribution B(n, p), where n is the coverage (methylated + unmethylated reads) and p is the probability of sequencing error (set to 0.01). We kept positions with k methylated reads if P(X ≥ k) < 0.05 (post-FDR correction). Second, retained methylated positions had to have ≥1 methylated read in all six biological replicates of at least one growth condition. Finally, median coverage of retained positions across all 12 samples had to be ≥10.
Assignment of genomic context to methylated cytosines
On the basis of the gene annotation of the Aiptasia genome (GFF3 file) (12) and the positional coordinates of the methylated cytosines produced by Bismark, we annotated every methylated cytosine based on the genomic context, including whether the methylated position resides in a genic or intergenic region, and the distances to the 5′ and 3′ ends of each genomic feature (gene/intergenic region/exon/intron).
CpG bias
Methylated cytosines are frequently spontaneously deaminated to uracil, which can be subsequently converted to thymine after DNA repair. As a result of this process, methylated CpGs are expected to decrease in abundance over evolutionary time, and the ratio of observed to expected CpGs (CpG O/E) has been previously used to predict putatively methylated and unmethylated genes (48). CpG O/E ratios of Aiptasia protein-coding genes were calculated according to Zeng and Yi (49).
Identification of DMGs
Using the methylation level of aposymbiotic genes as a control, generalized linear models were implemented in R to identify genes that were differentially methylated in the symbiotic Aiptasia. The general formula used was
glm(methylated, non_methylated ~ symbiotic state × position, family = “binomial”)
where “methylated, non_methylated” was a two-column response variable denoting the number of methylated and nonmethylated reads at a particular position. For predictor variables, “position” denoted relative position of the methylated site in the gene, while “symbiotic state” denoted symbiotic or aposymbiotic conditions. Data from individual replicates were entered separately to assign equal weightage to each replicate, as pooling results in a disproportionate skew toward the replicate with the highest coverage. Genes with <5 methylated positions were filtered out to reduce type I errors, and genes with FDR ≤ 0.05 were considered as DMGs.
Identification of DEGs
RNA-seq generated 889 million raw read pairs from six lanes on the Illumina HiSeq 2000 platform. Adapters, primers, and low-quality bases were removed from the ends of raw reads using Trimmomatic v0.33 (ILLUMINACLIP:TruSeq2-PE.fa:4:25:9 LEADING:28 TRAILING:28 SLIDINGWINDOW:4:30 MINLEN:50). The resulting trimmed reads were mapped to the Aiptasia genome using HISAT v2.0.1 (50), and transcripts were assembled on the basis of the Aiptasia gene models (GFF3 file) using StringTie v1.2.2 (51). Trinity (align_and_estimate_abundance.pl – Bowtie2 v2.2.7/RSEM v1.2.22/edgeR v3.10.5) (52) was run against the transcripts using trimmed reads for expression abundance estimation, and then DEGs between symbiotic and aposymbiotic anemones were identified with FDR ≤ 0.05.
PCA and correlation matrices
Median methylation levels of methylated genes and log2FPKM (base 2) of expressed genes were shifted to be zero-centered and analyzed by PCA using the prcomp function in R. Using the same data, we calculated correlation matrices (Pearson correlation coefficient) and clustered samples with hclust implemented in R using complete linkage and Euclidean distance.
Spurious transcription analysis
Trimmed reads were mapped to the Aiptasia genome using HISAT2 v2.1.0, and mapping coverage per position was extracted using BEDTools v2.17.0. To assess spurious transcription levels, we determined the coverage per exon normalized across all six replicates (assuming every replicate had a coverage of 1 million sequences in total) and then calculated the average coverage ratios of exons 2 to 6 versus exon 1 for every gene.
GO enrichment of DMGs and DEGs
GO annotation was based on the previously published Aiptasia genome (12). Functional enrichment of DMGs and DEGs was carried out with topGO (53) using default settings. GO terms with P ≤ 0.05 were considered significant, and the occurrence of at least five times in the background set was additionally required for DMGs. Multiple testing correction was not applied to the resulting P values as the tests are considered to be nonindependent (53).
KEGG enrichment of DMGs and DEGs
KEGG orthology annotation was carried out by combining the KEGG annotations provided in the original Aiptasia genome publications and a separate set of annotations based on the KEGG Automatic Annotation Server (www.genome.jp/tools/kaas/; parameters: GHOSTZ, Eukaryotes, Bidirectional Best Hit) (54). A KEGG pathway enrichment analysis of both DMGs and DEGs was carried out using Fisher’s exact test, and pathways with P ≤ 0.05 were considered significant.
Validation of gene expression changes from RNA-seq by qPCR
Three randomly picked RNA libraries per symbiotic state were used for qPCR validation of RNA-seq results. Complementary DNA was synthesized using the Invitrogen SuperScript III First-Strand Synthesis SuperMix kit. A total of 14 genes were validated for differential expression using qPCR (tables S13 to S15). RPS7, RPL11, and NDH5 were used as internal reference standards (35). qPCR was carried out using the Invitrogen Platinum SYBR Green qPCR SuperMix-UDG kit on an Applied Biosystems 7900HT Fast Real-Time PCR System according to the manufacturer’s instructions.
Validation of methylation changes using bisulfite PCR
Three DNA samples per symbiotic state were randomly picked for the validation of observed methylation changes and bisulfite converted using the EZ-96 DNA Methylation-Gold Kit (Zymo Research). We selected 18 DMGs to design primers, of which 14 were amplified using the Promega PCR Master Mix (Promega; table S16). The resulting PCR fragments were indexed using the Illumina Nextera XT Index Kit, purified via AmpureXP bead clean up (Beckman Coulter), and sequenced on the Illumina MiSeq platform. Finally, an average of 1870 sequence reads per replicate was obtained to identify methylated CpGs using Bismark, as described above. The correlation between whole-genome bisulfite conversion and bisulfite PCR was calculated using generalized linear model.
Chromatin immunoprecipitation
We used the Zymo-Spin ChIP Kit (Zymo Research) to conduct histone bound chromatin extraction, with minor adjustments to the manufacturer’s protocol. Briefly, three biological replicates, each consisting of two symbiotic anemones, were used for this experiment. Each anemone was first washed with phosphate-buffered saline (PBS) with 0.1% Triton X-100. Anemones were then fixed in 1× PBS with 1% formaldehyde for 15 min. To stop cross-linking reactions, glycine was added to the solution and left to rest for 10 more minutes. Following the manufacturer’s protocol, we centrifuged and washed whole anemones. We prepared the Nuclei Prep Buffer according to the protocol and crushed the two anemones of each replicate together using a douncer for homogenization. Samples were then sonicated for 15 cycles on ice (15 s on and 30 s cooling) to ensure fragmentation to 200 to 500 bp. Thereafter, the protocol was followed without further modifications.
Immunoprecipitation was achieved using a target-specific antibody to H3K36me3 (ab9050, Abcam), which has been validated in many eukaryotic species, including mouse (55), Arabidopsis thaliana (56), yeast (57), and zebrafish (58, 59). Comparison of Aiptasia histone H3 to the respective homologs from several species for which this antibody has been previously validated showed high conservation of the N-terminal tail containing position H3K36 (fig. S4), whereby 100% conservation to the zebrafish homolog was observed.
Corresponding input controls for each of the three replicates were generated as suggested by the manufacturer. DNA fragment quality and quantity were confirmed using High Sensitivity DNA Reagents (Agilent Technologies) on a bioanalyzer, after which ChIP libraries were constructed using the NEBNext ChIP-seq Library Prep Master Mix Set (NEB).
Sequencing resulted in 10 million read pairs per replicate. These read pairs were trimmed using Trimmomatic v0.33 and mapped to the Aiptasia genome using HISAT2, as described above. H3K36me3 enrichments were calculated as log2(average signal/average input control) for all genes, unmethylated genes, and highly methylated genes (methylation level > 70; methylation density > 40). P values were calculated using a t test.
Antibody affinity validation through Western blotting
Total protein was extracted from a snap-frozen anemone crushed in 10% trichloroacetic acid. The homogenized sample was left to incubate overnight at −20°C to allow proteins to precipitate. The solution was centrifuged at 20,000g at 4°C for 20 min to collect suspended proteins. The pellet was then washed three times in 80% acetone and then spun down again as previously described. The final pellet was then air-dried for 10 to 15 min to remove residual acetone. The final protein was suspended in urea lysis buffer [7 M urea and 2 M thiourea (pH 7.5)] by vortexing for 2 hours.
Samples were then prepared for Western blot by adding 4× sampling buffer [0.38 M tris base, 8% SDS, 4 mM EDTA, 40% glycerol, and bromophenol blue (4 mg/ml)] to a final concentration of 1×. After a 2-min incubation at 90°C, samples were ready to be run on a gel at 10 to 12 mA. The gel was transferred to a polyvinylidene difluoride nitrocellulose membrane, rinsed with tris-buffered saline (TBS) buffer [150 mM NaCl, 25 mM tris (pH 7.4), and 0.1% Triton X-100], and blocked for 30 min at room temperature (RT) in TBS containing 5% fat-free powder milk. The primary antibody was diluted in TBS/milk and incubated on an undulating orbital shaker overnight at 4°C. After three washes in TBS for 10 min each, the membrane was again blocked in TBS/milk for 20 min at RT before proceeding with secondary antibody staining. The horseradish peroxidase (HRP)–linked antibody [anti-rabbit immunoglobulin G (IgG) HRP conjugate W4011 and anti-mouse IgG HRP conjugate W4031, Promega] was diluted in TBS/milk (1:10,000) and incubated for 2 hours at RT. After final triplicate 10-min washes in TBS, membranes were developed.
Supplementary Material
Acknowledgments
We thank J. Pringle for the provision of the initial Aiptasia CC7 and Symbiodinium SSB01 cultures. We thank S. Baumgarten for his help with establishing the Aiptasia cultures and critical reading of the manuscript. We also thank the four anonymous reviewers for their effort and valuable suggestions, which greatly improved this study. Funding: Research reported in this publication was supported by baseline funding from the King Abdullah University of Science and Technology to M.A. Author contributions: M.A. conceived and coordinated the project. Y.L., G.C., M.J.C., and N.Z. performed experiments. M.A., C.R.V., and Y.J.L. provided tools and/or data. C.T.M. constructed libraries for WGBS, ChIP-seq, and RNA-seq. Y.L., Y.J.L., and M.A. analyzed expression, methylation, and ChIP-seq data. M.A. and Y.L. wrote the manuscript with input from Y.J.L. and C.R.V. All authors read and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Sequencing data of bisulfite sequencing, RNA-seq, and ChIP-seq were deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject code PRJNA415358. Scripts are available on https://github.com/YongLiedu/Aiptasia_DNA_Meth. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat2142/DC1
Fig. S1. Circos visualization of different data at the genome-wide level.
Fig. S2. Methylated genes in Aiptasia have lower CpG O/E.
Fig. S3. Methylation patterns.
Fig. S4. Sequence conservation of histone H3 homologs.
Fig. S5. Western blot.
Fig. S6. Correlation matrices of replicates.
Fig. S7. Validation of methylation level.
Fig. S8. qPCR validation of gene expression levels.
Fig. S9. Correlation between DNA methylation changes and gene expression changes.
Table S1. Aiptasia DMGs.
Table S2. Aiptasia DEGs.
Table S3. GO enrichment analysis of DMGs—biological process.
Table S4. GO enrichment analysis of DMGs—molecular function.
Table S5. GO enrichment analysis of DMGs—cellular component.
Table S6. GO enrichment analysis of DEGs—biological process.
Table S7. GO enrichment analysis of DEGs—molecular function.
Table S8. GO enrichment analysis of DEGs—cellular component.
Table S9. KEGG pathway enrichment analysis of DEGs.
Table S10. KEGG pathway enrichment analysis of DMGs.
Table S11. KEGG pathway enrichment analysis of DEMGs and statistics of symbiosis related pathways’ ranks.
Table S12. Cnidarian-dinoflagellate symbiosis–related Aiptasia genes.
Table S13. qPCR primers for gene expression validation.
Table S14. qPCR raw Ct (cycle threshold) values for gene expression validation.
Table S15. qPCR: ΔΔCt approach for fold-change calculations.
Table S16. Primers for bisulfite PCR-based methylation validation.
REFERENCES AND NOTES
- 1.Sylvain J. C., How to protect a coral reef: The public trust doctrine and the law of the sea. Sustain. Dev. Law Policy 7, 32–35 (2006). [Google Scholar]
- 2.Davy S. K., Allemand D., Weis V. M., Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lämke J., Bäurle I., Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones P. A., Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Rando O. J., Verstrepen K. J., Timescales of genetic and epigenetic inheritance. Cell 128, 655–668 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Richards E. J., Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 7, 395–401 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S., Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Zilberman D., An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 18, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Putnam H. M., Davidson J. M., Gates R. D., Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon G. B., Bay L. K., Matz M. V., Evolutionary consequences of DNA methylation in a basal metazoan. Mol. Biol. Evol. 33, 2285–2293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torda G., Donelson J. M., Aranda M., Barshis D. J., Bay L., Berumen M. L., Bourne D. G., Cantin N., Foret S., Matz M., Miller D. J., Moya A., Putnam H. M., Ravasi T., van Oppen M. J. H., Vega Thurber R., Vidal-Dupiol J., Voolstra C. R., Watson S.-A., Whitelaw E., Willis B. L., Munday P. L., Rapid adaptive responses to climate change in corals. Nature Clim. Chang. 7, 627–636 (2017). [Google Scholar]
- 12.Baumgarten S., Simakov O., Esherick L. Y., Jin Liew Y., Lehnert E. M., Michell C. T., Li Y., Hambleton E. A., Guse A., Oates M. E., Gough J., Weis V. M., Aranda M., Pringle J. R., Voolstra C. R., The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 11893–11898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voolstra C. R., A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol. Ecol. 22, 4366–4368 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Tucker K. L., Methylated cytosine and the brain: A new base for neuroscience. Neuron 30, 649–652 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Liew Y. J., Zoccola D., Li Y., Tambutté E., Venn A. A., Michell C. T., Cui G., Deutekom E. S., Kaandorp J. A., Voolstra C. R., Forêt S., Allemand D., Tambutté S., Aranda M., Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci. Adv. 4, eaar8028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.G. B. Dixon, L. K. Bay, M. V. Matz, Patterns of gene body methylation predict coral fitness in new environments. bioRxiv:184457 (2017).
- 17.Feng S., Cokus S. J., Zhang X., Chen P.-Y., Bostick M., Goll M. G., Hetzel J., Jain J., Strauss S. H., Halpern M. E., Ukomadu C., Sadler K. C., Pradhan S., Pellegrini M., Jacobsen S. E., Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S A. 107, 8689–8694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Romero R., Suárez-Ulloa V., Rodríguez-Casariego J., García-Souto D., Diaz G., Smith A. J., Pasantes J. J., Rand G. M., Eirín-López J. M., Effects of Florida Red Tides on histone variant expression and DNA methylation in the Eastern oyster Crassostrea virginica. Aquat. Toxicol. 186, 196–204 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A., Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.W. N. Venables, B. D. Ripley, in Modern Applied Statistics with S. (Springer-Verlag New York, 2002), chap. 11. [Google Scholar]
- 21.Foret S., Kucharski R., Pellegrini M., Feng S., Jacobsen S. E., Robinson G. E., Maleszka R., DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl. Acad. Sci. U.S.A. 109, 4968–4973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubauer E. F., Poole A. Z., Detournay O., Weis V. M., Davy S. K., The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ 4, e2692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rädecker N., Pogoreutz C., Wild C., Voolstra C. R., Stimulated respiration and net photosynthesis in Cassiopeia sp. during glucose enrichment suggests in hospite CO2 limitation of algal endosymbionts. Front. Mar. Sci. 4, 1–4 (2017). [Google Scholar]
- 24.Oakley C. A., Ameismeier M. F., Peng L., Weis V. M., Grossman A. R., Davy S. K., Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell. Microbiol. 18, 1009–1023 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Downs C. A., Kramarsky-winter E., Martinez J., Kushmaro A., Woodley C. M., Loya Y., Ostrander G. K., Symbiophagy as a cellular mechanism for coral bleaching. Autophagy 5, 211–216 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Nanty L., Carbajosa G., Heap G. A., Ratnieks F., van Heel D. A., Down T. A., Rakyan V. K., Comparative methylomics reveals gene-body H3K36me3 in Drosophila predicts DNA methylation and CpG landscapes in other invertebrates. Genome Res. 21, 1841–1850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M.-C., Cheng Y.-M., Sung P.-J., Kuo C.-E., Fang L.-S., Molecular identification of Rab7 (ApRab7) in Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae. Biochem. Biophys. Res. Commun. 308, 586–595 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S., A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10, 556–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barott K. L., Venn A. A., Perez S. O., Tambutté S., Tresguerres M., Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 607–612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., Zimmermann P., David G., Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Latysheva N., Muratov G., Rajesh S., Padgett M., Hotchin N. A., Overduin M., Berditchevski F., Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 26, 7707–7718 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anitei M., Stange C., Parshina I., Baust T., Schenck A., Raposo G., Kirchhausen T., Hoflack B., Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin–AP-1-coated carrier biogenesis at the TGN. Nat. Cell Biol. 12, 330–340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAuley P. J., Smith D. C., The green hydra symbiosis. V. Stages in the intracellular recognition of algal symbionts by digestive cells. Proc. R. Soc. B Biol. Sci. 216, 7–23 (1982). [Google Scholar]
- 34.Dunn S. R., Weis V. M., Apoptosis as a post-phagocytic winnowing mechanism in a coral-dinoflagellate mutualism. Environ. Microbiol. 11, 268–276 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Lehnert E. M., Mouchka M. E., Burriesci M. S., Gallo N. D., Schwarz J. A., Pringle J. R., Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 4, 277–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis V. M., Reynolds W. S., deBoer M. D., Krupp D. A., Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbiodinium spp. and planula larvae of the scleractinian coral Fungia scutaria. Coral Reefs 20, 301–308 (2001). [Google Scholar]
- 37.Pernice M., Dunn S. R., Miard T., Dufour S., Dove S., Hoegh-Guldberg O., Regulation of apoptotic mediators reveals dynamic responses to thermal stress in the reef building coral Acropora millepora. PLOS ONE 6, e16095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin S., Cheng S., Song B., Zhong X., Lin X., Li W., Li L., Zhang Y., Zhang H., Ji Z., Cai M., Zhuang Y., Shi X., Lin L., Wang L., Wang Z., Liu X., Yu S., Zeng P., Hao H., Zou Q., Chen C., Li Y., Wang Y., Xu C., Meng S., Xu X., Wang J., Yang H., Campbell D. A., Sturm N. R., Dagenais-Bellefeuille S., Morse D., The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Radecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C., Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 23, 490–497 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Douglas A. E., Nitrogen recycling or nitrogen conservation in an alga-invertebrate symbiosis? J. Exp. Biol. 201, 2445–2453 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Aranda M., Li Y., Liew Y. J., Baumgarten S., Simakov O., Wilson M. C., Piel J., Ashoor H., Bougouffa S., Bajic V. B., Ryu T., Ravasi T., Bayer T., Micklem G., Kim H., Bhak J., LaJeunesse T. C., Voolstra C. R., Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., Weis V. M., Medina M., Schwarz J. A., Generation and analysis of transcriptomic resources for a model system on the rise: The sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10, 258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown B. E., Dunne R. P., Edwards A. J., Sweet M. J., Phongsuwan N., Decadal environmental ‘memory’ in a reef coral? Mar. Biol. 162, 479–483 (2015). [Google Scholar]
- 44.Xiang T., Hambleton E. A., DeNofrio J. C., Pringle J. R., Grossman A. R., Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity1. J. Phycol. 49, 447–458 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics Action 17, 10–12 (2011). [Google Scholar]
- 46.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki M. M., Kerr A. R. W., De Sousa D., Bird A., CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 17, 625–631 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng J., Yi S. V., DNA methylation and genome evolution in honeybee: Gene length, expression, functional enrichment covary with the evolutionary signature of DNA methylation. Genome Biol. Evol. 2, 770–780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Brian Couger M., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C. N., Henschel R., LeDuc R. D., Friedman N., Regev A., De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. Alexa, J. Rahnenfuhrer, topGO: Enrichment Analysis for Gene Ontology. R package version 2.24.0 (2016).
- 54.Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M., KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soboleva T. A., Parker B. J., Nekrasov M., Hart-Smith G., Jin Tay Y., Tng W.-Q., Wilkins M., Ryan D., Tremethick D. J., A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B. PLOS Genet. 13, e1006633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wollmann H., Stroud H., Yelagandula R., Tarutani Y., Jiang D., Jing L., Jamge B., Takeuchi H., Holec S., Nie X., Kakutani T., Jacobsen S. E., Berger F., The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janke R., Iavarone A. T., Rine J., Oncometabolite D-2-hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. eLife 6, e22451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vastenhouw N. L., Zhang Y., Woods I. G., Imam F., Regev A., Shirley Liu X., Rinn J., Schier A. F., Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu S.-F., Zhang H., Cairns B. R., Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat2142/DC1
Fig. S1. Circos visualization of different data at the genome-wide level.
Fig. S2. Methylated genes in Aiptasia have lower CpG O/E.
Fig. S3. Methylation patterns.
Fig. S4. Sequence conservation of histone H3 homologs.
Fig. S5. Western blot.
Fig. S6. Correlation matrices of replicates.
Fig. S7. Validation of methylation level.
Fig. S8. qPCR validation of gene expression levels.
Fig. S9. Correlation between DNA methylation changes and gene expression changes.
Table S1. Aiptasia DMGs.
Table S2. Aiptasia DEGs.
Table S3. GO enrichment analysis of DMGs—biological process.
Table S4. GO enrichment analysis of DMGs—molecular function.
Table S5. GO enrichment analysis of DMGs—cellular component.
Table S6. GO enrichment analysis of DEGs—biological process.
Table S7. GO enrichment analysis of DEGs—molecular function.
Table S8. GO enrichment analysis of DEGs—cellular component.
Table S9. KEGG pathway enrichment analysis of DEGs.
Table S10. KEGG pathway enrichment analysis of DMGs.
Table S11. KEGG pathway enrichment analysis of DEMGs and statistics of symbiosis related pathways’ ranks.
Table S12. Cnidarian-dinoflagellate symbiosis–related Aiptasia genes.
Table S13. qPCR primers for gene expression validation.
Table S14. qPCR raw Ct (cycle threshold) values for gene expression validation.
Table S15. qPCR: ΔΔCt approach for fold-change calculations.
Table S16. Primers for bisulfite PCR-based methylation validation.


