Abstract
Many virulence factors have been described for opportunistic pathogens within the genus Aeromonas. Polymerase Chain Reactions (PCRs) are commonly used in population studies of aeromonads to detect virulence-associated genes in order to better understand the epidemiology and emergence of Aeromonas from the environment to host, but their performances have never been thoroughly evaluated. We aimed to determine diagnostic sensitivity and specificity of PCR assays for the detection of virulence-associated genes in a collection of Aeromonas isolates representative for the genetic diversity in the genus. Thirty-nine Aeromonas strains belonging to 27 recognized species were screened by published PCR assays for virulence-associated genes (act, aerA, aexT, alt, ascFG, ascV, ast, lafA, lip, ser, stx1, stx2A). In parallel, homologues of the 12 putative virulence genes were searched from the genomes of the 39 strains. Of the 12 published PCR assays for virulence factors, the comparison of PCR results and genome analysis estimated diagnostic sensitivities ranging from 34% to 100% and diagnostic specificities ranged from 71% to 100% depending upon the gene. To improve the detection of virulence-associated genes in aeromonads, we have designed new primer pairs for aerA/act, ser, lafA, ascFG and ascV, which showed excellent diagnostic sensitivity and specificity. Altogether, the analysis of high quality genomic data, which are more and more easy to obtain, provides significant improvements in the genetic detection of virulence factors in bacterial strains.
Introduction
Aeromonas are common inhabitants of aquatic environments and can be involved in fish and human diseases. They are frequently found in drinking water and food, including meat, fish, vegetables, and, more recently in ready-to-eat foods [1–6]. Among other illnesses, aeromonads are responsible for gastrointestinal syndromes following ingestion of contaminated food, and wound infection following water exposure in human [1–6], and for furunculosis and septicemia in fish causing major losses in the aquaculture sector [7].
The pathogenesis of Aeromonas infection is only partially elucidated [1,5,6,8] although a wide repertoire of virulence factors contributing to biofilm formation, cell adherence, invasion and cytotoxicity have been described [8]. It has been suggested that only certain subsets of strains within a species, referred as “pathotypes”, produce disease in certain individuals [1,9,10]; presumably due to differences in the content of virulence-associated genes. From this assumption, many studies leading to an abundant literature searched for virulent strains from water, fish, food or clinical samples using Polymerase Chain Reaction (PCR) for detecting virulence factors [11–14], notably for epidemiologic surveys (e. g., [15–17]). The accurate inference of virulence in such an approach depends on how accurate the PCR-based methods are for detecting genetic markers of virulence in aeromonads. But these methods have often been developed from a small number of strains, few species and for a specific purpose, and their performances have never been evaluated for the whole genus. Before raising conclusions on pathotype patterns in aeromonads, PCR performances at the genus level deserve to be questioned considering the high level of genetic polymorphism in the genus Aeromonas [18,19].
The recent availability of whole genome sequences (WGS) data from many Aeromonas isolates from various species [20,21] allows the search for virulence-associated genes by sequence comparison and the evaluation of performance of molecular methods. In the present study, we aimed to evaluate the accuracy of several widely used virulence PCRs by matching PCR results against available genomes. When necessary, accuracy was improved by the genome-driven design of new PCR tools.
Materials and methods
Bacterial strains and DNA extraction
Thirty-nine Aeromonas spp. strains were chosen to cover the whole genus. They belonged to 27 species (Table 1) among the 30 validated species in the Aeromonas genus at the time of January 2018. The current taxonomic affiliations are indicated for every strain with previously published names of strains indicated inside braces, where appropriate. Strains were grown on Trypticase Soy Agar at 35 °C for 16–24 h, and genomic DNA was extracted using the MasterPure™ DNA Purification Kit (Epicentre, USA).
Table 1. General features of the strains and genomes used in this study.
| Strain (n = 39) | Source of isolation | Geno-me size (Mbp) | No of scaffolds | Average genome coverage | N50 (nt) | G+C content (%) | No of predicted CDSs | Level of assem-bly | Genome accession number | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| A. allosacharophila CECT 4199T | Infected eel | 4.66 | 120 | 87 | 114,541 | 58.4 | 4,173 | IHQ | PRJEB7019a | [20] |
| A. australiensis CECT 8023T | Irrigation water system | 4.11 | 113 | 128 | 95,095 | 58.1 | 3,733 | IHQ | PRJEB7021a | |
| A. bestiarum CECT 4227T | Fish | 4.68 | 41 | 53 | 237,067 | 60.5 | 4,223 | IHQ | PRJEB7022a | |
| A. bivalvium CECT 7113T | Cockles | 4.28 | 69 | 30 | 149,050 | 62.3 | 3,909 | IHQ | PRJEB7023a | |
| A. caviae CECT 838T | Guinea pig | 4.47 | 111 | 95 | 101,663 | 61.6 | 4,081 | IHQ | PRJEB7024a | |
| A. dhakensis CECT 7289T {A. aquariorum}† | Aquaria of ornamental fish | 4.69 | 78 | 117 | 163,504 | 61.7 | 4,266 | IHQ | PRJEB7020a | |
| A. dhakensis CIP 107500 {A. hydrophila subsp. dhakensis}† | Human diarrheic stool | 4.71 | 73 | 84 | 165,885 | 61.8 | 4,284 | IHQ | PRJEB7048a | |
| A. dhakensis BVH28b | Human wound | 4.89 | 68 | 130 | 150,860 | 61.7 | 4,466 | IHQ | PRJEB9016a | [22] |
| A. diversa CECT 4254T | Human wound | 4.06 | 37 | 116 | 203,531 | 61.5 | 3,711 | IHQ | PRJEB7026a | [20] |
| A. encheleia CECT 4342T | Fish | 4.47 | 35 | 112 | 380,984 | 61.9 | 4,076 | IHQ | PRJEB7027a | |
| A. enteropelogenes CECT 4255T {A. trota}† | Human stool | 4.34 | 27 | 66 | 640,249 | 60.0 | 3,917 | IHQ | PRJEB7043a | |
| A. eucrenophila CECT 4224T | Fresh water fish | 4.54 | 22 | 50 | 441,212 | 61.1 | 4,113 | IHQ | PRJEB7029a | |
| A. fluvialis LMG 24681T | River water | 3.90 | 76 | 48 | 108,949 | 58.2 | 3,609 | IHQ | PRJEB7030a | |
| A. hydrophila subsp. hydrophila CECT 839T | Tin of milk with fishy odor | 4.74 | 1 | - | 4,744,448 | 61.5 | 4,119 | C | CP000462b | [23] |
| A. hydrophila BVH25a | Human respiratory tract | 5.10 | 130 | 44 | 84,371 | 60.9 | 4,598 | IHQ | PRJEB9013a | [22] |
| A. jandaei CECT 4228T | Human stool | 4.50 | 58 | 55 | 161,393 | 58.7 | 4,065 | IHQ | PRJEB7031a | [20] |
| A. media CECT 4232T | River water | 4.48 | 233 | 60 | 37,608 | 60.9 | 4,075 | IHQ | PRJEB7032a | |
| A. media LMG 13464 {A. caviae}† | Infected fish | 4.45 | 99 | 87 | 103,746 | 61.3 | 4,014 | IHQ | PRJEB12347a | [21] |
| A. media CECT 7111 | Oyster | 4.41 | 92 | 70 | 108,504 | 61.6 | 3,998 | IHQ | PRJEB12345a | |
| A. molluscorum CIP 108876T | Wedge-shells | 4.23 | 309 | 9 | 21,565 | 59.2 | 3,946 | IHQ | AQGQ01b | [24] |
| Aeromonas sp. genomospecies paramedia CECT 8838 | Human diarrheic stool | 4.46 | 128 | 99 | 78,349 | 62.2 | 4,086 | IHQ | PRJEB12349a | [21] |
| A. piscicola LMG 24783T | Salmon | 5.18 | 91 | 99 | 150,424 | 59.0 | 4,713 | IHQ | PRJEB7033a | [20] |
| A. popoffii CIP 105493T | Drinking water production plant | 4.76 | 105 | 67 | 113,495 | 58.4 | 4,331 | IHQ | PRJEB7034a | |
| A. rivipollensis LMG 13459 {A. caviae}† | Infected fish | 4.49 | 111 | 76 | 107,760 | 61.7 | 4,091 | IHQ | PRJEB12346a | [21] |
| A. rivipollensis 76c {A. media}† | Human diarrheic stool | 4.69 | 137 | 79 | 93,768 | 61.3 | 4,255 | IHQ | PRJEB8966a | [22] |
| A. rivipollensis BVH40 {A. media} | Human stool | 4.70 | 123 | 79 | 105,841 | 61.4 | 4,204 | IHQ | PRJEB9017a | |
| A. rivuli DSM 22539T | Freshwater | 4.53 | 102 | 99 | 155,151 | 60.0 | 4,149 | IHQ | PRJEB7035a | [20] |
| A. salmonicida subsp. salmonicida CIP 103209T | Salmon | 4.74 | 128 | 117 | 89, 543 | 58.5 | 4,442 | IHQ | PRJEB7036a | |
| A. sanarellii LMG 24682T | Human wound | 4.19 | 98 | 121 | 82,664 | 63.1 | 3,828 | IHQ | PRJEB7037a | |
| A. schubertii CECT 4240T | Human wound | 4.13 | 111 | 260 | 108,810 | 61.7 | 3,808 | IHQ | PRJEB7038a | |
| A. simiae CIP 107798T | Healthy monkey | 3.99 | 100 | 86 | 73,112 | 61.1 | 3,654 | IHQ | PRJEB7039a | |
| A. sobria CECT 4245T | Fish | 4.68 | 52 | 34 | 188,072 | 58.6 | 4,160 | IHQ | PRJEB7040a | |
| A. taiwanensis LMG 24683T | Human wound | 4.24 | 106 | 66 | 85,294 | 62.8 | 3,884 | IHQ | PRJEB7041a | |
| A. tecta CECT 7082T | Human diarrheic stool | 4.76 | 51 | 89 | 238,229 | 60.1 | 4,278 | IHQ | PRJEB7042a | |
| A. veronii bv. veronii CECT 4257T | Human respiratory tract | 4.52 | 52 | 59 | 181,171 | 58.8 | 4,070 | IHQ | PRJEB7044a | |
| A. veronii bv. sobria LMG 13067 | Environment | 4.74 | 72 | 46 | 147,470 | 58.3 | 4,171 | IHQ | PRJEB7051a | |
| A. veronii BVH25b | Human respiratory tract | 4.66 | 35 | 63 | 241,725 | 58.7 | 4,185 | IHQ | PRJEB9014a | [22] |
| A. veronii BVH26b | Human wound | 4.58 | 48 | 73 | 180,501 | 58.7 | 4,107 | IHQ | PRJEB9015a | |
| A. veronii 77c | Human diarrheic stool | 4.61 | 42 | 78 | 230,104 | 58.6 | 4,124 | IHQ | PRJEB9012a |
a: Performed at the Microbial Analysis, Resources and Services (MARS) facility at the University of Connecticut (Storrs, USA)
b: Obtained from GenBank, National Center for Biotechnology Information
T: Type strain
† Previously published names are indicated inside braces.
Abbreviations: IHQ, Improved high quality draft genome; C, complete genome.
Genome sequences
Thirty-seven draft genomes of Aeromonas spp., sequenced during previous works [20–22], were provided at the Microbial Analysis, Resources and Services (MARS) facility at the University of Connecticut (Storrs, USA) (Table 1) with a high quality level of draft genome on the basis of the quality of the sequencing and assembling (e.g., number of average genomes coverage and number of scaffolds) and of the verification of the automated annotation (16 housekeeping genes and 47 ribosomal protein-coding genes checked in every genome). Two other genomes were downloaded from GenBank (Table 1). To check the taxonomical assignation of genomes employed, we performed a ML phylogenetic tree of all the genomes (n = 39) calculated by the program kSNP3.1 [25], a kmer based alignment free method (see S1 Fig).
Genome analysis
Genomic contigs and circular genomes were annotated by using the RAST annotation server to identify RNAs and protein-coding genes [26]. The genomes were queried for genes encoding for a set of virulence factors described in Aeromonas spp. by using reference protein sequences (Table 2), either using translated sequences of the validated subset of UniProtKB-SwissProt or annotated genes in UniprotKB-TrEMBL database. Sequence comparisons with reference protein sequences were performed with SEEDviewer that uses bidirectional protein-protein BLAST (BlastP) sequence comparison of translated open reading frames. Proteins with amino acid sequence similarities ≥65% and E-value ≤10−10 were considered to be homologous [27].
Table 2. Virulence-associated genes detected by genome analysis.
| Strain (n = 39) | aerA/act | ser | ast | alt/pla | stx1a, stx2a | aexT | aexU | ascFG‡ | ascV | lafA |
|---|---|---|---|---|---|---|---|---|---|---|
| P09167S (AerA) Q44063E (Act) | A4SNU7E (Ahe2) P31339S (AspA) Q9RG23E (Ahe2) | Q8VRN3E | Q44061E (Alt) O87651E (Pla) | E2DQN2E (Stx1a) E2DQN6E (Stx2a) | Q93Q17S | D5LUP3E | Q6WG33E (AscF) Q6WG32E (AscG) | A4SUH2E | Q93TL9E | |
| A. allosacharophila CECT 4199T | - | + | - | + | - | - | + | - | - | - |
| A. australiensis CECT 8023T | + | + | - | + | - | - | - | - | - | - |
| A. bestiarum CECT 4227T | + | + | + | + | - | - | - | - | - | - |
| A. bivalvium CECT 7113T | - | + | - | + | - | - | - | - | - | + |
| A. caviae CECT 838T | - | - | - | + | - | - | - | - | - | + |
| A. dhakensis CECT 7289T {A. aquariorum}† | + | + | - | + | - | - | - | - | - | - |
| A. dhakensis CIP 107500 {A. hydrophila subsp. dhakensis}† | + | + | - | + | - | - | + | + | + | + |
| A. dhakensis BVH28b | + | + | - | + | - | - | + | + | + | + |
| A. diversa CECT 4254T | + | - | - | + | - | - | - | + | + | + |
| A. encheleia CECT 4342T | - | + | - | + | - | - | + | + | + | + |
| A. enteropelogenes CECT 4255T {A. trota} | + | + | + | + | - | - | - | - | - | + |
| A. eucrenophila CECT 4224T | + | + | - | + | - | - | - | - | - | + |
| A. fluvialis LMG 24681T | - | - | - | - | - | - | - | - | - | - |
| A. hydrophila subsp. hydrophila CECT 839T | + | + | + | + | - | - | - | - | - | - |
| A. hydrophila BVH25a | - | + | + | + | - | - | + | + | + | + |
| A. jandaei CECT 4228T | + | + | - | + | - | - | - | + | + | + |
| A. media CECT 4232T | - | - | - | + | - | - | - | - | - | - |
| A. media LMG 13464 {A. caviae}† | - | - | - | + | - | - | - | - | - | - |
| A. media CECT 7111 | - | - | - | + | - | - | - | - | - | - |
| A. molluscorum CIP 108876T | + | - | - | + | - | - | - | - | - | - |
| Aeromonas sp. genomospecies paramedia CECT 8838 | - | + | - | + | - | - | - | - | - | - |
| A. piscicola LMG 24783T | + | + | + | + | - | - | + | + | + | + |
| A. popoffii CIP 105493T | + | + | - | + | - | - | - | - | - | - |
| A. rivipollensis LMG 13459 {A. caviae}† | - | + | - | + | - | - | - | - | - | + |
| A. rivipollensis 76c {A. media}† | - | + | - | + | - | - | - | - | - | - |
| A. rivipollensis BVH40 {A. media}† | - | + | - | + | - | - | - | - | - | + |
| A. rivuli DSM 22539T | - | - | - | + | - | - | - | - | - | + |
| A. salmonicida subsp. salmonicida CIP 103209T | + | + | - | + | - | + | - | - | - | + |
| A. sanarellii LMG 24682T | - | - | - | + | - | - | - | - | - | - |
| A. schubertii CECT 4240T | + | - | - | + | - | - | - | + | + | + |
| A. simiae CIP 107798T | - | - | - | + | - | - | - | - | - | + |
| A. sobria CECT 4245T | + | + | + | + | - | - | - | - | - | - |
| A. taiwanensis LMG 24683T | - | - | - | + | - | - | - | - | - | + |
| A. tecta CECT 7082T | + | + | - | + | - | - | - | + | + | + |
| A. veronii bv. veronii CECT 4257T | + | + | - | + | - | - | - | - | - | - |
| A. veronii bv. sobria LMG 13067 | + | + | - | + | - | - | - | - | - | - |
| A. veronii BVH25b | + | + | - | + | - | - | - | - | - | - |
| A. veronii BVH26b | + | + | - | + | - | - | + | + | + | + |
| A. veronii 77c | + | + | - | + | - | + | + | + | + | + |
S/E: Accession numbers correspond to protein sequences in Swiss-prot (S) or TrEMBL databases (E).
T: Type strain
† Previously published names are indicated inside braces.
‡ ascF and ascG are flanking genes.
Polymerase Chain Reaction (PCR)
Both selected and new primers were used to amplify DNA from aerolysin/enterotoxin cytotoxic (aerA/act), heat-stable cytotonic enterotoxin (ast), heat-labile cytotonic enterotoxin (alt), lipase (lip), serine protease (ser), ADP-ribosylating toxins (aexT and aexU), shiga toxins 1 (stx1) and 2 (stx2a), T3SS needle proteins (ascFG), T3SS inner membrane channel protein (ascV) and lateral flagellin A (lafA) genes. PCR assays were carried out in a 25 μL reaction mixture containing 0.2 μM of each primer (Sigma-Aldrich, Saint-Louis, US), 0.2 mM of each deoxynucleoside triphosphate (dNTP) (Euromedex, Souffelweyersheim, France), 2.5 mM MgCl2 (Promega, Madison, US), and 1.25 U of GoTaq DNA polymerase (Promega, Madison, WI) in the appropriate reaction buffer containing 1.5 mM MgCl2 and 25 ng of genomic DNA as the template. The amplification conditions performed on a GeneAmp PCR System 9700 (Applied Biosystems, Foster city, US) were as follows: initial denaturation for 3 min at 95°C, followed by (1) main amplification program of the reference source specified in the Table 3 for already published PCR, with length of extension adapted to the efficacy of the DNA polymerase used (1 min/kb) when necessary, and a final extension step at 72°C for 10 min or (2) 32 amplification cycles as indicated in Table 4 for newly developed PCR. The PCR products and the 50 bp DNA ladder (New England BioLabs, Ipswich, US) were separated in 1.5% agarose gels in 0.5X TBE buffer-ethidium bromide 500 μg/mL and revealed under UV. Positive and negative controls were added in each PCR to assess the validity and specificity of amplification reaction. The positive controls of amplification corresponded to genomic DNA of one of the strains included in the study and for which the presence of gene of interest was checked on the basis of genomic analysis as follows: A. veronii 77C (aer/act, aexT and ser), A. dhakensis CIP 107500T (alt, ascFG, ascV and lafA), A. hydrophila BVH 25a (ast). Specificity of the assays was confirmed using PCR-grade water and DNA from A. fluvialis LMG 24681T which harbors in its genome none of the genes of interest. All strains were tested for all primer sets. For all the PCR assays and for each strain, the results of absence/presence of amplification was determined by two different readers and in two independent experiments on the basis of the expected size of amplified products and of the positive control.
Table 3. Performance of several PCRs used for the detection of virulence-associated genes in the genus Aeromonas.
| Gene | Virulence factor | Prevalence estimated from WGS analysis (n = 39) | PCRRef. | Primers (nt.) | No of sequences in align-ment | No of primer mis-matches† (min-max) | DiagnosticSe % (CI95%) |
Comments on primers / diagnostic sensitivity relationship | DiagnosticSp (%) (CI95%) |
Comments on primers / specificity relationship |
|---|---|---|---|---|---|---|---|---|---|---|
| aerA/ act | Aerolysin AerA/ Cytotoxic enterotoxin Act |
56% (n = 22) |
[15] | AHCF1 (22) AHCR1 (22) |
32 32 |
0–6 0–6 |
64 (41;83) | High variability in hybridization sequences of forward and reverse primers | 100 (80;100) | - |
| [30] | aer-f (20) aer-r (20) |
32 32 |
0–4 0–2 |
91 (71;99) | - | 82 (57;96) | aer-r: high GC content and self-end dimers | |||
| This study | aer-1FX (20) aer-2R (19) |
32 32 |
0–1 0–2 |
100 (85;100) | aer-1FX: degenerate nucleotides in the forward primer aer-f | 94 (71;100) | Degenerate bases did not decrease specificity aer-2R: identical to aer-r but one 5’ end base deleted | |||
| ser | Serine protease |
69% (n = 27) |
[13] | Serine-f (20) Serine-r (20) |
39 39 |
0–7 0–11 |
59 (39;78) | High variability in hybridization sequences of forward and reverse primers One nucleotide deleted in the reference sequences used to design the reverse primer (ENA X67043 and ENA AAF22245) |
92 (62;100) | - |
| This study | ser-1FX (21) ser-2RX (20) |
39 39 |
0–2 0–0 |
96 (81;100) | Selection of less variable sequences Degenerate bases |
92 (62;100) | Degenerate bases did not decrease specificity | |||
| ast | Cytotonic heat-stable enterotoxin | 15% (n = 6) |
[31] | ast-F (20) ast-R (20) |
7 7 |
0–4 0–3 |
100 (54;100) | - | 91 (76;98) | - |
| alt | cytotonic heat-labile enterotoxin | 97% (n = 38) |
[31] | alt-F (20) alt-R (19) |
39 39 |
0–10 0–5 |
34 (20;51) | High variability in hybridization sequences of forward and reverse primers | - | Unevaluable specificity: insufficient number of strains for which alt was not detected in WGS (1/39) |
| [13] | lip-f (18) lip-r (20) |
39 39 |
0–1 0–2 |
68 (51;83) | - | - | ||||
| stx1a | Shiga toxin 1 subunit A | 0% (n = 0) |
[32] | Stx1-a (20) Stx1-b (20) |
- | - | - | Not evaluable sensitivity: no strain for which stx1a or stx2a was detected in WGS | 100 (82;100) | - |
| stx2a | Shiga toxin 2 subunit A | 0% (n = 0) |
[33] | S2Aup (19) S2Alp (19 |
- | - | - | 100 (82;100) | - | |
| aexT | ADP- ribosylating toxin AexT | 5% (n = 2) |
[34] | RASEXOS-L (18) RASEXOS-R (18) |
9 9 |
0–1 0–1 |
2/2 | - | 100 (91;100) | - |
| aexT and/or aexU | ADP-ribosylating toxins | 23% (n = 9) |
This study | aexTU-1FX (19) aexTU-2RX (20) |
21 21 |
0–2 0–2 |
100 (66;100) | - | 100 (88;100) | - |
| asc-FG | T3SS needle proteins | 28% (n = 11) |
[16] | ascF-G-fwd (20) ascF-G-rev (20) |
38 23 |
0–5 na |
45 (17;77) | High variability in hybridization sequences of forward and reverse primers | 74 (54;89) | ascF-G-fwd: high GC content, self-end dimers and 3’-end unstability |
| This study | ascFG-1F (21) ascFG-2RX (20) |
38 23 |
0–4 0–1 |
91 (59;100) | Selection of less variable sequences Degenerate bases |
100 (88;100) | Degenerate bases did not decrease specificity | |||
| asc-V | T3SS inner membrane channel protein | 28% (n = 11) |
[16] | ascV-fwd (20) ascV-rev (20) |
21 21 |
1–2 4–7 |
55 (23;83) | High variability in hybridization sequences of forward and reverse primers | 71 (51;87) | ascV-fwd: high GC content |
| This study | ascV-1F (19) ascV-2R (20) |
21 21 |
0–1 0–2 |
100 (72;100) | Selection of less variable sequences | 100 (88;100) | - | |||
| lafA | Lateral flagellin A | 54% (n = 21) |
[35] | Laf1 (18) Laf2 (17) |
58 58 |
4–6 0–3 |
55 (34;80) | High variability in hybridization sequences of forward and reverse primers Wrong supplementary nucleotide (cytidine) in the 3’ region of laf1‡ |
100 (82;100) | - |
| This study | lafA-1FX (19) lafA-2RX (18) |
58 58 |
0–5 0–3 |
86 (64;97) | Selection of less variable sequences Degenerate bases Sensitivity <90%: high nucleotide variability throughout lafA sequence 55.8% of polymorphic positions‡ |
100 (81;100) | Degenerate bases did not decrease specificity |
†Evaluated from the virulence-associated-gene/primers multiple alignments
‡ Evaluated from lafA multiple alignments
Abbreviations: CI95%, confidence interval 95%; na, not available; PCR, Polymerase Chain Reaction; Ref., References; WGS, Whole Genome Sequences.
Table 4. Primers designed in this study and amplification conditions of PCRs.
| Virulence-associated gene | Primer sequences (5’-3’) | Amplified sequence length (bp) | Amplification conditions† | ||
|---|---|---|---|---|---|
| °C | Time (sec) | Number of cycles | |||
|
aerA (syn: act) |
aer-1FX: 5’-CCTAYRGYCTSAGCGAGAAG-3’ aer-2R: 5’-CAGTTCCAGTCCCACCACT-3’ |
430 | 94 | 60 | 32 |
| 56 | 60 | ||||
| 72 | 60 | ||||
| ser |
ser-1FX: 5’-GACAAYCGVGTSTTCAAAGAG-3’ ser-2RX: 5’-ACCACCARGTTCCAGAAGTT-3’ |
262 | 94 | 60 | 32 |
| 59 | 60 | ||||
| 72 | 30 | ||||
| aexT/aexU |
aexTU-1FX: 5’-TGGCVMTSAAAGAGTGGAT-3’ aexTU-2RX: 5’-GCARDGSRCCRTTGCCRGTC-3’ |
225 | 94 | 60 | 32 |
| 63 | 60 | ||||
| 72 | 45 | ||||
| asc-FG |
ascFG-1F: 5’-CAAGATCAACAAATGGTCGGT-3’ ascFG-2RX: 5-TTCAYCARRGADGASAGGCG-3’ |
262 | 94 | 60 | 32 |
| 60 | 60 | ||||
| 72 | 30 | ||||
| asc-V |
ascV-1F: 5’-CGCAAGGACATCATGCTGG-3’ ascV-2R: 5’-ATGATGATGAGGCCCGCGAT-3’ |
578 | 94 | 60 | 32 |
| 65 | 60 | ||||
| 72 | 45 | ||||
| lafA |
lafA-1FX: 5’-GATGYTGRGYACYGCCATG-3’ lafA-2RX: 5’-CATRTTGGARAGGTTRTTGAC-3’ |
619 | 94 | 60 | 32 |
| 64 | 60 | ||||
| 72 | 60 | ||||
Degenerate bases are underlined.
†The amplification conditions of PCR are preceded by an initial denaturation step at 95°C during 3 minutes and followed by a final elongation step at 72°C during 10 minutes.
Primer design strategy
Nucleic sequences were aligned using the Clustal ω2 program within Seaview 4 package [28] (see S1 Files). Primers displaying a length of 18–21 nucleotides were designed from the conserved regions, and AmplifX 1.7.0 (CNRS, Aix-Marseille Université, France, http://crn2m.univ-mrs.fr/pub/amplifx-dist) was used to assess their intrinsic properties such as complexity (polyX and triplet repetitions), 3’ stability and self-dimer formation. The specificity of the new primers was checked with NCBI nucleotide-nucleotide BLAST.
Challenging PCR accuracy characteristics
The accuracy of the PCR assays were evaluated by comparing the results of the PCR with those obtained by genome analysis, with WGS results considered as gold standard. Diagnostic sensitivity was defined as the percentage of ‘positive PCR results among all the strains for which the gene was detected in WGS’, while specificity was defined as the percentage of ‘negative PCR results among all the strains for which the gene was not detected in WGS’. Diagnostic sensitivity and specificity values below 90% were considered as insufficient. Exact confidence interval of a frequency, i.e., binomial confidence interval, was determined for each percentage, as described elsewhere [29] by using the web server http://statpages.info/confint.html.
Results
Virulence-associated gene selection
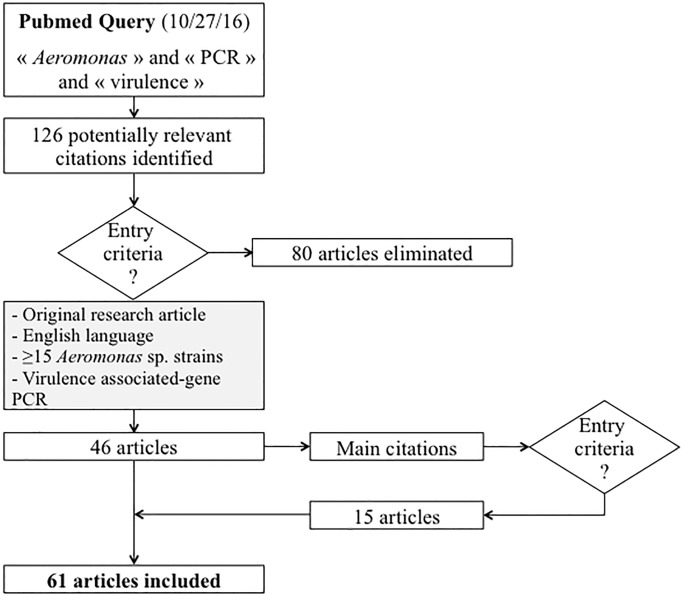
The literature analysis was used to inventory virulence genes commonly searched in Aeromonas and to identify the reference of the PCRs used in the different studies, as summarized in Fig 1. We selected 61 original research studies that performed virulence-associated gene PCRs for populations of at least 15 Aeromonas strains (Fig 1). From these 61 population studies, the origin of strains and distribution of Aeromonas species, the virulence-associated genes and the references of the PCRs method used are presented in the S1 Table. Among these 61 articles (see S1 Table for references), the most studied genes were aerA (n = 51), alt (n = 35), act (n = 30), ast (n = 30), lip (n = 19), hlyA (n = 17), fla (n = 15), aexT (n = 11), aexU (n = 2), ascV (n = 11), ser (n = 10), gcat (n = 9), lipH3 (n = 8), pla (n = 8), lafA (n = 8), ascFG (n = 6), exu (n = 6), ahyB (n = 5), stx1 (n = 5), stx2 (n = 5) and alp-1 (n = 5). These genes corresponded to different categories of virulence factors that are included in the pathotypes definition: hemolysins and related genes (aerA, act, hlyA), cytotonic enterotoxins genes (alt, ast), lipases genes (lip, gcat, lipH3, pla, alp-1), flagellar appendages genes (fla, lafA), type III secretion system effectors AexT and AexU genes (aexT, aexU), type III secretion system components genes (ascV, ascFG), serine protease gene (ser), nuclease gene (exu), elastase gene (ahyB) and shiga-toxins genes (stx1, stx2). We selected one or several genes from these virulence categories: aerA (aerolysin), act (cytolytic enterotoxin), ast (heat-stable cytotonic enterotoxin), alt (heat-labile cytotonic enterotoxin), pla (phospholipase A), ser (serine protease), aexT and aexU (ADP-ribosylating toxins), stx1 and stx2 (shiga-toxins), ascFG (type III secretion system needle proteins), ascV (type III secretion system inner membrane channel protein) and lafA (lateral flagellin A).
Fig 1. Flowchart for the literature analysis.
The automatic Pubmed query using « Aeromonas », « PCR » and « virulence » as keywords generated 126 articles (October 27, 2016), among which 46 corresponded to the entry criteria. The reading of the relevant references quoted in these 46 selected articles led to the manual inclusion of 15 additional articles.
Genome content in virulence-associated genes
We detected homologues for all the studied genes except for stx1 and stx2 (Table 2). The reference sequences of aerolysin gene aerA from A. bestiarum Ah65 {considered in the original source as A. hydrophila Ah65} (SwissProt P09167) and Cytolytic enterotoxin gene act from A. dhakensis SSU {considered in the original source as A. hydrophila SSU} (UniprotKB-TrEMBL Q44063) shared a very high level of identity (95.1%) and similarity (97.2%) in protein sequence (493 amino-acids both). Similarly, the phospholipase A gene pla from A. piscicola AH-3 {considered in the original source as A. hydrophila AH-3} (UniprotKB-TrEMBL O87651) and the heat-labile cytotonic enterotoxin gene alt from A. dhakensis SSU (UniprotKB-TrEMBL Q44061) shared a very high level of identity (96.9%) and similarity (98.5%) in protein sequence, from an alignment of 89% query-cover of alt coding-sequence (368/385 amino-acids), pla coding-sequence spanning 805 amino-acids. These homology data and the presence of common flanking genes (data not shown), led to consider aerA/act and alt/pla as two single virulence-associated genes for all the strains included in the study. The ser gene that is putatively involved in the aerolysin activation by proteolytic cleavage was detected in 14 out of 17 aerA/act positive strains (Table 2). The genes coding for structural components of type III secretion system (T3SS) were detected simultaneously in the 11 T3SS gene positive strains (Table 2). The 248 residues from the N-terminal domain of the two ADP-ribosyltransferase toxins, AexT from A. salmonicida subsp. salmonicida ATCC 33658T (SwissProt Q93Q17; 475 amino-acids) and AexU from A. veronii bv. sobria AeG1 (UniprotKB-TrEMBL D5LUP3, 512 amino-acids) presented a high level of identity (89.5%) and similarity (94.8%) in protein sequence, but they differed significantly in their C-terminal domain (< 20% similarity). All the strains that harbor the T3SS effector genes aexT/aexU, except A. salmonicida subsp. salmonicida CIP 103209T, also possessed the T3SS structural components. For the lateral flagellin gene, from 1 to 4 copies were detected in the genomes of the 21-lafA positive strains.
PCR performance assessment
Results of the evaluation of PCR accuracy are presented in Table 3. Ability to amplify gene of interest was assessed for all genes but stx1a or stx2a because there were no strains for which the genes were detected in WGS (Table 2). The ability of the PCRs to amplify the gene of interest (i.e., diagnostic sensitivity, hereafter sensitivity) ranged from 34 to 100%. The PCR screening aerA/act that uses primers “aer”, and the PCRs screening the ast and aexT genes displayed high sensitivities with the tested set of strains, with values of 91%, 100% and 100%, respectively. However, the corresponding confidence intervals for ast and aexT PCR were large (range >40%) because the number of positive genomes was low, 6 and 2 out of the 39 genomes studied, respectively. On the opposite, with a sensitivity value of 64%, the PCR usually employed for screening aerA/act genes using primers “AHC” was disappointing and was associated with frequent false negative results. Similarly, low values of sensitivity were observed for the following genes: ser (59%), alt/pla (primers “alt”, 34%; primers “lip”, 68%), ascFG (45%), ascV (55%) and lafA (55%). Depending on the gene, the lack of sensitivity resulted from either an excessive variability in annealing of forward and/or reverse primers, or even errors in primer sequence transcript (e.g., Serine-r and Laf1; Table 3).
The specificities of the PCRs that could be evaluated ranged from 71% to 100% depending on the gene considered (Table 3). Specificity was not determined for alt because only one out of the 39 WGS lacked alt gene (A. fluvialis LMG 24681T). PCRs used for screening the following genes were highly specific: aerA/act-using primers “AHC” (100%), ser (92%), ast (91%), aexT (100%) and lafA (100%). The test method was insufficiently specific for aerA/act genes using primers “aer” (82%), ascF-G (74%) and ascV (71%) genes. Therefore, an overestimation of the prevalence of these virulence-associated factors by generation of false positive results was expected. Depending on the gene, the default of specificity likely resulted from PCR primers properties such as high GC content, self-end dimer formation and/or 3’-end unstability (Table 3).
Genome driven design and evaluation of new PCR primers
Considering the low performance of some PCRs, we designed new primer pairs on the basis of multiple alignments (Table 4) with partial and full-length sequences retrieved from Genbank or EMBL databases (S2 Table), and from the WGS provided in this study (Table 1). To design the new primers, we selected low variable areas of sequence alignments and used degenerate bases in case of polymorphism. Newly designed primers were associated with an increased sensitivity of the virulence gene detection (Table 3). Meanwhile, the use of degenerate primers did not impair the specificity (Table 3). For screening of ADP-ribosylating toxins, we have designed primers matching in the homologous part shared by aexT and aexU genes that were associated with a sensitivity of 100%.
Discussion
Many virulence factors are described in the genus Aeromonas, but to date, the pathogenicity of specific strains still cannot be predicted from the genome content. The genus Aeromonas is characterized by intraspecific and interspecific genetic diversities of both ribosomal and housekeeping genes that impair the definition of species and lead to the population structure of the genus in several complexes of species [19,36]. However, using the whole genome sequences and the average nucleotide identity index to compare them, all the species could be clearly delineated [20,21]. The huge variability observed in some housekeeping genes also occurs in virulence-associated genes, as already shown for the “S-layer protein” gene (vapA) that harbored variability up to 15% at the amino-acid level among the strains of different subspecies of A. salmonicida [18].
This study focuses on the performances of molecular techniques used for the detection of virulence-associated genes in Aeromonas strains. Some previous work aiming to detect virulence factors in the genus Aeromonas reported discrepancies between studies due to molecular methods used (e.g., [37]). The choice of strains used for the primer design was particularly questioned. The present study does not question the validity of previously published PCRs, especially when the original study aimed to screen the presence of virulence-associated genes in particular groups or species (e.g., in A. caviae, [38]), but aims to evaluate PCR performances at the genus level. The comparison of PCR results and high-quality WGS with automatic annotation and careful manual checking allows for the first time the proper evaluation of the PCR performances for detecting virulence-associated genes in aeromonads. Most new primers and PCR conditions proposed herein improve PCR performances although the design of degenerate primers was required for some virulence genes, i.e. when sequence polymorphisms are scattered all along the gene. Neither diagnostic sensitivity nor specificity was hampered in these cases.
Similar to most other population studies, our study suffers from some limitations. Studying whether strains are virulent may be challenging for opportunistic pathogens but at first, tools need to be accurate in establishing whether a gene is present or absent from genomes. Our study may have some potential bias in the strain sampling. But the collection of strains included in the study aimed to be as representative of the diversity found in the genus as possible, covering the interspecies diversity in the whole genus, given the studied genes are sought in a wide range of species [39], and the intra-species diversity of species that are the most frequently isolated in human clinical contexts [1,40]. Illustrating the unavoidable sampling bias, the shiga-toxin coding genes stx were not detected in this study by PCR nor by genome analysis. However, the stx1 gene seems to be quite rare in the genus [41]. Another explanation would be stx instability due to the mobility of the stx phage that can lead to the gene loss after subculture [41] or may occur during storage before genome sequencing and PCR. The prevalence of some virulence markers was either too low (e.g., stx) or too high (e.g., lip, alt) so that PCR assay diagnostic sensitivity or specificity could not be evaluated, respectively. Despite the limitation in strain and sequence collections, this study provides a meaningful highlight in accuracy of widely used tools together with virulence gene content in a large collection of strains.
Besides performance assessment and method improvements, this study provides some interesting data on virulence in aeromonads, like the possible matching between several virulence markers identified in different Aeromonas species that could be established or confirmed. For instance, the homology between aerolysin gene aerA from A. hydrophila and cytotoxic enterotoxin gene act from A. dhakensis has previously been considered [42]. In fact, both toxins exhibit hemolytic, cytotoxic and enterotoxic activities and both cause lethality in mice [43–47]. Moreover, a similar mechanism of action, i.e., oligomerization followed by pore-formation, has been reported for these 2 toxins [46,48]. These two toxins are closely related but it is still possible that the aerolysin/cytotonic enterotoxin from A. dhakensis SSU possesses some structural and functional originality [46], and further characterization of the purified homologous proteins is required to clarify this point. The aerA/act PCR based on newly designed primers improved the diagnostic sensitivity for the panel of aeromonads covering the whole genus from 64–91% to 100%, and dramatically improved sensitivity for the species A. dhakensis (from 0–67% to 100%). Given the clinical importance of this species [49], an optimized performance for detecting the well-studied aerA virulence gene is critical, and should provide advances in the patho-epidemiological knowledge of the species. The putative correspondence between phospholipase A gene (pla) from A. piscicola AH-3 and heat-labile cytotonic enterotoxin gene (alt) from A. dhakensis SSU, was also observed by Balsalobre et al. [50]. In this case, the nomenclature of the virulence factor depends on the study [51,52]. The biological features are possibly divergent as mentioned by Merino et al. [52] and could depend on the allele of the gene alt/pla but this requires further studies.
Biological interpretation of the presence of virulence genes needs to be cautious because the presence of a virulence gene does not imply its expression in the host. In addition, we have to consider the pathogenic and non-pathogenic interactions depending on the context. For example, the aeromonads T3SS system is important for both pathogenicity and mutualism as demonstrated inside the leech microbiota [53]. In parallel, post-translational modifications for activation or effector translocation should be examined. The joint presence of the ser gene and aer/act should be considered because aerolysin is activated by a serine protease [54]. The ser gene was absent from the genomic data of three aerA/act positive strains included in this study: A. diversa CECT 4254T, A. molluscorum CIP 108876T and A. schubertii CECT 4240T. Similarly, the presence of T3SS effectors AexT/U should be considered with respect to the presence of T3SS components, e.g., ascFG and ascV. Their presence is required for the delivery of AexT/U [55,56]. The presence of both genes aexT and aexU that we observed with the strains A. veronii 77C and BVH26b has already been mentioned for other A. veronii isolates and appears to be a frequent situation for strains carrying T3SS in the A. veronii group [37]. Therefore, for screening aexT and aexU genes in aeromonads, we propose an aexT/U PCR, followed by a molecular search of the aexT gene alone when the screening assay is positive.
In epidemiologic and pathogenesis studies of Aeromonas, large collections of strains were screened for their content in virulence-associated genes. Given the polymorphisms in virulence-associated genes highlighted in this study and because PCR assays to detect virulence associated genes were widely used without prior evaluation of their performance (diagnostic sensitivity and specificity) at the genus level (S1 Table), the published prevalence of virulence genes should be interpreted with caution. When the PCR performance is poor, the estimated prevalence is blurred with assay errors and does not reflect the true prevalence of the virulence factors. Importantly, assumptions on the pathogenic behavior of aeromonads have been made from virulence patterns obtained from defective/inaccurate tool results, and this may have led to wrong conclusions. This may contribute to the current complexity of virulence patterns in aeromonads, to the poor understanding of aeromonad virulence and to the frequent lack of links between the virulence factor pattern and the pathogenic behavior. Optimization of PCR conditions and primer is crucial to avoid biased data in applied and clinical microbiology, and in epidemiology.
The bacterial genomes, today widespread and easy to obtain, have a very high potential in numerous research areas, including the design of consolidated microbiological diagnostic tools, although they are maybe currently underused for this purpose. In our study, genomes allowed an accuracy evaluation of virulence-associated gene PCRs commonly used in the genus Aeromonas and their improvement by designing several improved oligonucleotide primers. WGS could obviously be used as primary data for searching virulence associated genes but WGS are still expensive when a large panel of strains is studied. Therefore, we propose here an accurate approach based on these genomic data to improve PCR methods aimed to detect virulence-associated genes in environmental bacteria.
Further advances in the knowledge of the genetic diversity and of the evolution of virulence-associated genes should improve the understanding of aeromonad adaptation to pathogenic behavior or patho-adaptation, and thereby the revision of pathotype definition.
Supporting information
(DOCX)
(ZIP)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Association des Biologistes de l’Ouest (ABO) and the Association pour la recherche et le développement en microbiologie et pharmacie (ADEREMPHA) to ETR. Part of this research was supported by USDA and ARS agreement 58-1930-4-002 to JG.
References
- 1.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23: 35–73. 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khajanchi BK, Fadl AA, Borchardt MA, Berg RL, Horneman AJ, Stemper ME, et al. Distribution of Virulence Factors and Molecular Fingerprinting of Aeromonas Species Isolates from Water and Clinical Samples: Suggestive Evidence of Water-to-Human Transmission. Appl Environ Microbiol. 2010;76: 2313–2325. 10.1128/AEM.02535-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker JL, Shaw JG. Aeromonas spp. clinical microbiology and disease. J Infect. 2011;62: 109–118. 10.1016/j.jinf.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 4.Nyenje ME, Odjadjare CE, Tanih NF, Green E, Ndip RN. Foodborne pathogens recovered from ready-to-eat foods from roadside cafeterias and retail outlets in Alice, Eastern Cape Province, South Africa: public health implications. Int J Environ Res Public Health. 2012;9: 2608–2619. 10.3390/ijerph9082608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueras MJ, Beaz-Hidalgo R. Aeromonas infections in humans Aeromonas. Caister Academic Press, Norfolk, UK, ed Graf J; 2015. pp. 65–68. [Google Scholar]
- 6.Teunis P, Figueras MJ. Reassessment of the Enteropathogenicity of Mesophilic Aeromonas Species. Front Microbiol. 2016;7: 1395 10.3389/fmicb.2016.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hossain MJ, Sun D, McGarey DJ, Wrenn S, Alexander LM, Martino ME, et al. An Asian Origin of Virulent Aeromonas hydrophila Responsible for Disease Epidemics in United States-Farmed Catfish. mBio. 2014;5: e00848–14. 10.1128/mBio.00848-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomás JM. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012: 256261 10.5402/2012/256261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janda JM, Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding Panorama of species, disease presentations, and unanswered questions. Clin Infect Dis Off Publ Infect Dis Soc Am. 1998;27: 332–344. [DOI] [PubMed] [Google Scholar]
- 10.Joseph S.W., Carnahan A. M. Update on the genus Aeromonas. ASM News; 2000; 218–223. [Google Scholar]
- 11.Albert MJ, Ansaruzzaman M, Talukder KA, Chopra AK, Kuhn I, Rahman M, et al. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J Clin Microbiol. 2000;38: 3785–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins LM, Marquez RF, Yano T. Incidence of toxic Aeromonas isolated from food and human infection. FEMS Immunol Med Microbiol. 2002;32: 237–242. [DOI] [PubMed] [Google Scholar]
- 13.Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003;84: 269–278. [DOI] [PubMed] [Google Scholar]
- 14.Senderovich Y, Ken-Dror S, Vainblat I, Blau D, Izhaki I, Halpern M. A molecular study on the prevalence and virulence potential of Aeromonas spp. recovered from patients suffering from diarrhea in Israel. PloS One. 2012;7: e30070 10.1371/journal.pone.0030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingombe CI, Huys G, Tonolla M, Albert MJ, Swings J, Peduzzi R, et al. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl Environ Microbiol. 1999;65: 5293–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chacón MR, Soler L, Groisman EA, Guarro J, Figueras MJ. Type III secretion system genes in clinical Aeromonas isolates. J Clin Microbiol. 2004;42: 1285–1287. 10.1128/JCM.42.3.1285-1287.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nawaz M, Khan SA, Khan AA, Sung K, Tran Q, Kerdahi K, et al. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010;27: 327–331. 10.1016/j.fm.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 18.Lund V, Mikkelsen H. Genetic diversity among A-proteins of atypical strains of Aeromonas salmonicida. Dis Aquat Organ. 2004;61: 257–262. 10.3354/dao061257 [DOI] [PubMed] [Google Scholar]
- 19.Roger F, Marchandin H, Jumas-Bilak E, Kodjo A, Lamy B. Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol. 2012;12: 62 10.1186/1471-2180-12-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colston SM, Fullmer MS, Beka L, Lamy B, Gogarten JP, Graf J. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio. 2014;5 10.1128/mBio.02136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talagrand-Reboul E, Roger F, Kimper J-L, Colston SM, Graf J, Latif-Eugenín F, et al. Delineation of Taxonomic Species within Complex of Species: Aeromonas media and Related Species as a Test Case. Front Microbiol. 2017;8: 621 10.3389/fmicb.2017.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosser T, Talagrand-Reboul E, Colston SM, Graf J, Figueras MJ, Jumas-Bilak E, et al. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front Microbiol. 2015;6: 1218 10.3389/fmicb.2015.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 2006;188: 8272–8282. 10.1128/JB.00621-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spataro N, Farfán M, Albarral V, Sanglas A, Lorén JG, Fusté MC, et al. Draft Genome Sequence of Aeromonas molluscorum Strain 848TT, Isolated from Bivalve Molluscs. Genome Announc. 2013;1 10.1128/genomeA.00382-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinforma Oxf Engl. 2015;31: 2877–2878. 10.1093/bioinformatics/btv271 [DOI] [PubMed] [Google Scholar]
- 26.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42: D206–214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990;87: 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27: 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 29.Lamy B, Delignette-Muller ML, Baty F, Carret G. Simple table for estimating confidence interval of discrepancy frequencies in microbiological safety evaluation. J Microbiol Methods. 2004;56: 137–139. 10.1016/j.mimet.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 30.Soler L, Figueras MJ, Chacón MR, Vila J, Marco F, Martinez-Murcia AJ, et al. Potential virulence and antimicrobial susceptibility of Aeromonas popoffii recovered from freshwater and seawater. FEMS Immunol Med Microbiol. 2002;32: 243–247. [DOI] [PubMed] [Google Scholar]
- 31.Aguilera-Arreola MG, Hernández-Rodríguez C, Zúñiga G, Figueras MJ, Castro-Escarpulli G. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol Lett. 2005;242: 231–240. 10.1016/j.femsle.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40: 3613–3619. 10.1128/JCM.40.10.3613-3619.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muniesa M, de Simon M, Prats G, Ferrer D, Pañella H, Jofre J. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect Immun. 2003;71: 4554–4562. 10.1128/IAI.71.8.4554-4562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun M, Stuber K, Schlatter Y, Wahli T, Kuhnert P, Frey J. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 2002;184: 1851–1858. 10.1128/JB.184.7.1851-1858.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merino S, Gavín R, Vilches S, Shaw JG, Tomás JM. A colonization factor (production of lateral flagella) of mesophilic Aeromonas spp. is inactive in Aeromonas salmonicida strains. Appl Environ Microbiol. 2003;69: 663–667. 10.1128/AEM.69.1.663-667.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roger F, Lamy B, Jumas-Bilak E, Kodjo A, colBVH study group, Marchandin H. Ribosomal multi-operon diversity: an original perspective on the genus Aeromonas. PloS One. 2012;7: e46268 10.1371/journal.pone.0046268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver AC, Graf J. Prevalence of genes encoding the type three secretion system and the effectors AexT and AexU in the Aeromonas veronii group. DNA Cell Biol. 2009;28: 383–388. 10.1089/dna.2009.0867 [DOI] [PubMed] [Google Scholar]
- 38.Wang G, Tyler KD, Munro CK, Johnson WM. Characterization of cytotoxic, hemolytic Aeromonas caviae clinical isolates and their identification by determining presence of a unique hemolysin gene. J Clin Microbiol. 1996;34: 3203–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Wang W, Zhu Z, Chen A, Du P, Wang R, et al. Distribution, virulence-associated genes and antimicrobial resistance of Aeromonas isolates from diarrheal patients and water, China. J Infect. 2015;70: 600–608. 10.1016/j.jinf.2014.11.004 [DOI] [PubMed] [Google Scholar]
- 40.Chen P-L, Wu C-J, Chen C-S, Tsai P-J, Tang H-J, Ko W-C. A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2013; 10.1111/1469-0691.12456 [DOI] [PubMed] [Google Scholar]
- 41.Alperi A, Figueras MJ. Human isolates of Aeromonas possess Shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clin Microbiol Infect. 2010;16: 1563–1567. 10.1111/j.1469-0691.2010.03203.x [DOI] [PubMed] [Google Scholar]
- 42.Buckley JT, Howard SP. The Cytotoxic Enterotoxin of Aeromonas hydrophila Is Aerolysin. Infect Immun. 1999;67: 466–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernheimer AW, Avigad LS. Partial characterization of aerolysin, a lytic exotoxin from Aeromonas hydrophila. Infect Immun. 1974;9: 1016–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty T, Huhle B, Bergbauer H, Goebel W. Cloning, expression, and mapping of the Aeromonas hydrophila aerolysin gene determinant in Escherichia coli K-12. J Bacteriol. 1986;167: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose JM, Houston CW, Coppenhaver DH, Dixon JD, Kurosky A. Purification and chemical characterization of a cholera toxin-cross-reactive cytolytic enterotoxin produced by a human isolate of Aeromonas hydrophila. Infect Immun. 1989;57: 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferguson MR, Xu XJ, Houston CW, Peterson JW, Coppenhaver DH, Popov VL, et al. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect Immun. 1997;65: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bücker R, Krug SM, Rosenthal R, Günzel D, Fromm A, Zeitz M, et al. Aerolysin from Aeromonas hydrophila perturbs tight junction integrity and cell lesion repair in intestinal epithelial HT-29/B6 cells. J Infect Dis. 2011;204: 1283–1292. 10.1093/infdis/jir504 [DOI] [PubMed] [Google Scholar]
- 48.Wilmsen HU, Pattus F, Buckley JT. Aerolysin, a hemolysin from Aeromonas hydrophila, forms voltage-gated channels in planar lipid bilayers. J Membr Biol. 1990;115: 71–81. [DOI] [PubMed] [Google Scholar]
- 49.Chen P-L, Lamy B, Ko W-C. Aeromonas dhakensis, an Increasingly Recognized Human Pathogen. Front Microbiol. 2016;7: 793 10.3389/fmicb.2016.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balsalobre LC, Dropa M, Matté GR, Matté MH. Molecular detection of enterotoxins in environmental strains of Aeromonas hydrophila and Aeromonas jandaei. J Water Health. 2009;7: 685–691. 10.2166/wh.2009.082 [DOI] [PubMed] [Google Scholar]
- 51.Chopra AK, Peterson JW, Xu XJ, Coppenhaver DH, Houston CW. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb Pathog. 1996;21: 357–377. 10.1006/mpat.1996.0068 [DOI] [PubMed] [Google Scholar]
- 52.Merino S, Aguilar A, Nogueras MM, Regue M, Swift S, Tomás JM. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect Immun. 1999;67: 4008–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silver AC, Kikuchi Y, Fadl AA, Sha J, Chopra AK, Graf J. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci U S A. 2007;104: 9481–9486. 10.1073/pnas.0700286104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abrami L, Fivaz M, Decroly E, Seidah NG, Jean F, Thomas G, et al. The pore-forming toxin proaerolysin is activated by furin. J Biol Chem. 1998;273: 32656–32661. [DOI] [PubMed] [Google Scholar]
- 55.Burr SE, Stuber K, Frey J. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J Bacteriol. 2003;185: 6583–6591. 10.1128/JB.185.22.6583-6591.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, et al. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb Pathog. 2007;43: 127–146. 10.1016/j.micpath.2007.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(ZIP)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.