Abstract
Apurinic/apyrimidinic (AP) endonucleases play critical roles in the repair of abasic sites and strand breaks in DNA. Complete genome sequences of Helicobacter pylori reveal that this bacterial specie has a single AP endonuclease. An H. pylori homolog of Xth (HpXth) is a member of exonuclease III family, which is represented by Escherichia coli Xth. Currently, it remains unknown whether this single AP endonuclease has DNA repair activities similar to those of its counterpart in E. coli and other bacteria. We report that HpXth possesses efficient AP site cleavage, 3’-repair phosphodiesterase, and 3’-phosphatase activities but not the nucleotide incision repair function. Optimal reaction conditions for HpXth’s AP endonuclease activity are low ionic strength, high Mg2+ concentration, pH in the range 7–8, and temperature 30 °C. The kinetic parameters measured under steady-state conditions showed that HpXth removes the AP site, 3’-blocking sugar-phosphate, and 3’-terminal phosphate in DNA strand breaks with good efficiency (kcat/KM = 1240, 44, and 5,4 μM–1·min–1, respectively), similar to that of E. coli Xth. As expected, the presence of HpXth protein in AP endonuclease—deficient E. coli xth nfo strain significantly reduced the sensitivity to an alkylating agent and H2O2. Mutation of active site residue D144 in HpXth predicted to be essential for catalysis resulted in a complete loss of enzyme activities. Several important structural features of HpXth were uncovered by homology modeling and phylogenetic analysis. Our data show the DNA substrate specificity of H. pylori AP endonuclease and suggest that HpXth counteracts the genotoxic effects of DNA damage generated by endogenous and host-imposed factors.
Introduction
Cellular DNA is continuously and inevitably assaulted by a variety of endogenous and environmental genotoxic agents. Reactive oxygen species (ROS) produced by aerobic respiration generate oxidized bases, sugars, and DNA strand breaks [1–2]. In addition, hydrated DNA is prone to spontaneous decomposition such as base loss, resulting in apurinic/apyrimidinic (AP) sites, and hydrolytic deamination, yielding mostly uracil and hypoxanthine [3]. The majority of endogenous DNA base lesions and single-strand breaks with 3’-blocking groups is removed through two overlapping pathways: base excision repair (BER) and nucleotide incision repair (NIR) [4–6]. The common BER pathway consists of two sequential excision or incision steps. First, one of many DNA glycosylases hydrolyzes the N-glycosidic bond of the damaged base and produces either an AP site or a single-stranded DNA break with a single-nucleotide gap flanked by a 5’-phosphate and 3’-blocking group, which may be a phosphate or an unsaturated aldehyde remnant of deoxyribose [7–9]. In the second step of BER, an AP endonuclease cleaves 5′ to the AP site or the 3’-blocking groups, leaving a free 3’-hydroxyl and either a 5’-terminal deoxyribose phosphate moiety, or a single-nucleotide gap with a proper 5’-phosphate [10]. Although BER can handle the majority of oxidized DNA bases, some lesions, such as α-anomers of deoxynucleosides (αdN) generated by free radicals under anoxic conditions, are not repaired by DNA glycosylases but rather by AP endonucleases via the NIR pathway [11–13]. In this glycosylase-independent pathway, an AP endonuclease directly cleaves DNA 5’ to the lesion and generates a single-strand break with a 5’-dangling damaged nucleotide [5, 14]. AP endonucleases also use their 3’-repair phosphodiesterase function to clean single-strand DNA breaks with “dirty ends” generated either by ROS or by bifunctional DNA glycosylase/AP lyases, producing 3’-hydroxyl termini that can be used by DNA polymerases to initiate the repair synthesis [15–16]. Noteworthy, human major AP endonuclease 1, APE1, in addition to NIR and BER functions contains abasic RNA cleavage activity, including the ability to process abasic ribonucleotides embedded in DNA [17].
Despite this multitude of functions, both the primary activity (AP site cleavage) and less efficient but also important 3’-phosphodiesterase, 3’→5’ exonuclease, 3’-phosphatase, and NIR activities of AP endonucleases are catalyzed by a single active site [15, 18–21]. All known AP endonucleases are grouped in two distinct superfamilies based on their structural and sequence similarity to either Escherichia coli exonuclease III (Xth; Endonuclease—Exonuclease—Phosphatase superfamily, or EEP) or endonuclease IV (Nfo) [10]. AP endonucleases from the EEP superfamily, including E. coli Xth and human APE1, require Mg2+ or other divalent metal ions for catalysis. Both Xth and Nfo homologs are present in prokaryotes and eukaryotes, and have been biochemically characterized in E. coli, Saccharomyces cerevisiae, Caenorhabditis elegans, and some other species [16, 22]. Not all AP endonucleases have all repair functions; for example, human APE1 contains very weak 3′-phosphatase activity, whereas the NIR activity in bacteria is generally present in homologs of Nfo, but not of Xth. Importantly, bacterial AP endonucleases as key genome protection enzymes are required for successful infection in view of host defense systems, thus representing a promising target for antimicrobial drugs [23–26].
Helicobacter pylori is a gram-negative bacterium that colonizes human gastric mucosa. The presence of H. pylori is sometimes pathogenic causing peptic ulcers and gastroesophageal reflux disease but may also have beneficial effects such as alleviation of childhood asthma and other allergies [27]. The genome of H. pylori can undergo dramatic changes that may help this bacterium to escape from host immune surveillance. Surprisingly, as compared to E. coli and non-symbiotic bacteria, H. pylori has a limited number of genes, with little redundancy in the BER pathway [28]. Whole-genome sequencing has found a single AP endonuclease, XthA (hereafter referred to as HpXth), homologous to E. coli Xth, encoded by ORF HP1526 [29–30]. Cell-free extracts from the mutant H. pylori strain with disrupted ORF HP1526 no longer have Mg2+-dependent AP site cleavage activity [31]. The H. pylori single xthA and double xthA mutY mutant strains show a four- and 37-fold increase in the spontaneous mutation rate in comparison with the wild-type control [32]. Taken together, these data strongly indicate that HpXth is involved in the repair of spontaneously occurring AP sites and other mutagenic DNA lesions in vivo. Nevertheless, at present, detailed biochemical characterization of this enzyme is lacking.
Since H. pylori seems to lack Nfo homologs, it was interesting to define the spectrum of DNA repair activities in its only AP endonuclease, HpXth. Here, we demonstrate that HpXth possesses AP site cleavage and 3’-repair phosphodiesterase functions. The kinetic parameters of HpXth-catalyzed repair activities were measured and compared to those of homologous AP endonucleases. The ability of HpXth to rescue an AP endonuclease—deficient E. coli strain after exposure to an alkylation agent and H2O2 was studied. The evolution, mechanisms of DNA damage recognition, and biological roles of HpXth in the defence against spontaneous and host-induced damage to bacterial DNA are discussed.
Materials and methods
Bacterial strains, plasmids, and reagents
The plasmids pET28c(+) (Novagen, UK) and pBluescript II SK+ (further referred as pBSK) were used to construct the expression vector. The plasmid pBW21 encoding the E. coli Nfo proteins was generously provided by Dr. B. Weiss (Emory University School of Medicine, Atlanta, GA). Restriction enzymes, T4 DNA ligase, and Phusion High-Fidelity DNA Polymerase (Thermo Scientific, USA) were employed for the amplification and cloning of the target gene. H2O2 and Methyl methanesulfonate (MMS) were purchased from Sigma-Aldrich Chimie S.a.r.l. (Lyon, France). E. coli strain BH110 (nfo::kanR [Δ(xthA-pncA)90X::Ƭn10]) comes from the laboratory stock, and E. coli expression strain Rosetta 2 (DE3) was acquired from (Novagen, Merck4Biosciences, France). E. coli BH110 was lysogenized with a helper phage (λDE3) harboring a copy of the T7 RNA polymerase gene, using the DE3 lysogenization kit (Novagen, Merck4Biosciences, France). The lysogenized E. coli BH110 (DE3) cells were transformed with pBSK, pBW21-Nfo, pBSK-HpXth and pBSK-HpXth-D144N vectors.
Cloning and expression of H. pylori AP endonuclease in E. coli and purification of the HpXth protein
Genomic DNA of H. pylori was provided by the Republican Collection of Microorganisms (Astana, Kazakhstan). Reference gene sequence of xthA of H. pylori was retrieved from the published genome sequences: CP011330.1 (range: 1144587–1145339). The following PCR primers (shown in the 5’→3’ direction) were applied to amplify the target HpXth cDNA: HpXth-NdeI, d(GGGAATTCCATATGAAACTGATTTCATGGAATGTGAAC) and HpXth-BamHI, d(CGCGGATCCGTTAAACTAATTCCAACCCTACCGG). The ORF of XthA from H. pylori was cloned into the pET28c(+) vector at the NdeI/BamHI sites, resulting in expression plasmid pET28c/HpXth. The encoded HpXth protein carries an N-terminal 6xHis-tag sequence. The DNA sequence of plasmid insert was verified against the sequence from the GenBank database in the Vector NTI software Advance™ v11.0 (Invitrogen, USA).
Site-directed mutation D144N in the coding sequence of HpXth in plasmid vector pET28c/HpXth and pBlueScript II SK+/HpXth were generated with the QuickChange site-directed mutagenesis kit (Quickchange® XL, Stratagene). The following oligonucleotide primers (shown in the 5’→3’ direction) were used to generate HpXth-D144N mutant protein: HpXth-F-D144N, d(GTCATTGTGTGTGGGAACTTGAATGTGGCCC); HpXth-R-D144N, d(GGGCCACATTCAAGTTCCCACACACAATGAC).
To express the recombinant proteins, E. coli Rosetta 2 (DE3) cells were transformed with plasmids, and the isolated KnR transformants were grown in the Luria-Bertani (LB) broth with 50 μg/mL kanamycin. At optical density at 600 nm (OD600) of 0.6, the cells were induced by 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubation was continued at room temperature with shaking (100 rpm) overnight. The induced cells were collected by centrifugation at 6000 × g for 7 min at 4 °C. The bacterial pellet containing HpXth was resuspended in a buffer consisting of 20 mM NaCl and 20 mM Tris-HCl (pH 8.0) and supplemented with the Complete Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland). The cells were disrupted by a French press at 18,000 psi (1250 bar), and then sonicated on ice at 40% amplitude. The lysate was cleared by centrifugation at 40,000 × g for 60 min at 4 °C. The supernatant was adjusted to 500 mM NaCl and 20 mM imidazole and loaded onto a 1 mL HiTrap Chelating HP column (GE Healthcare) charged with Ni2+. The bound proteins were eluted in a 20–500 mM imidazole gradient and the fractions containing a recombinant HpXth protein were pooled and loaded onto a 1 mL HiTrap Heparin column (GE Healthcare). The bound proteins were eluted with a 50–1000 mM NaCl gradient. Fractions containing the recombinant protein were stored at -20 °C in 50% glycerol.
Oligonucleotides
All the oligodeoxyribonucleotides with modifications and their regular complementary counterparts were purchased from Eurogentec (Seraing, Belgium) and are presented in the 5’→3’ direction. They included the 30mer X-RT d(TGACTGCATAXGCATGTAGACGATGTGCAT) where X is tetrahydrofuran (THF, a synthetic analog of an abasic site), α-2’-deoxyadenosine (αdA), 5-hydroxycytosine (5ohC), or 5,6-dihydrouracil (DHU), and complementary 30mer oligonucleotides containing dA, dG, dC, or T opposite the lesion. DNA substrates were constructed as described previously [26, 33]. Briefly, the complementary oligonucleotides were annealed; the resulting duplex oligonucleotides are referred to as X•C (G, A, T), respectively, where X is a modified residue. The DNA sequence contexts have previously been used to study the DNA substrate specificity of bacterial, yeast, and plant AP endonucleases [26, 34–35]. Chemical structures of DNA adducts used in this work are shown in S1 Fig.
DNA end labeling were performed as described previously [26]. Briefly, oligonucleotides were either 5’-end labeled by T4 polynucleotide kinase (or by phosphatase minus mutant PNK) (New England Biolabs, OZYME, France) in the presence of γ/32P]ATP (3000 Ci·mmol−1) (PerkinElmer SAS, France) or 3’-end labeled by terminal transferase (New England Biolabs) in the presence of α[32P]3’-dATP (cordycepin 5’-triphosphate, 5000 Ci·mmol–1) (PerkinElmer) as recommended by the manufacturers. The labeled oligonucleotides were annealed to their appropriate complementary oligonucleotides in a buffer consisting of 50 mM KCl and 20 mM HEPES-KOH (pH 7.5) at 65 °C for 3 min and cooled down to room temperature for 2 h. The resulting duplex oligonucleotides are referred to as X•C (G, A, T), respectively, where X is a modified residue.
The following oligonucleotides were used to measure 3’→5’ exonuclease and 3’-repair phosphodiesterase activities: Exo20, d(GTGGCGCGGAGACTTAGAGA); Exo20THF, d(GTGGCGCGGAGACTTAGAGAX), where X is 3’-terminal THF; Exo20P, d(GTGGCGCGGAGACTTAGAGAp), where p is 3’-terminal phosphate; 5P-Exo19, d(pATTTGGCGCGGGGAATTCC), where p is 5’-terminal phosphate; and complementary Rex-T, d(GGAATTCCCCGCGCCAAATTTCTCTAAGTCTCCGCGCCAC) containing T opposite of THF. The recessed duplex Exo20•RexTRec was composed of Rex-T and Exo20, whereas nicked duplexes Exo20THF•RexTNick and Exo20P•RexTNick were composed of 5P-Exo19 and Rex-T, and Exo20THF or Exo20P, respectively. Structures of the nicked and recessed duplex oligonucleotides used in the present work are shown in S1 Fig. To measure the 3’-phosphatase activity of the AP endonucleases, we prepared a 20mer (Exo20) oligonucleotide containing a 3’-terminal radioactive phosphate residue: 32P, as described [26]. Previously, we have shown that human tyrosyl-DNA phosphodiesterase 1 (Tdp1) can remove a 3’-terminal cordycepin nucleoside, thereby producing an oligonucleotide fragment with a phosphate residue at the 3’ end [36]. For this purpose, Exo20 was 3’-end labeled with cordycepin [α-32P]3’-dATP to obtain a 21mer Exo20-[32P]3’-dAMP fragment, which was then treated with recombinant Tdp1 (generously provided by Prof. Olga Lavrik, Novosibirsk, Russia). The resulting 20mer Exo20-3’-[32P] fragment and 5P-Exo19 oligonucleotide were annealed to the complementary Rex-T oligonucleotide, and the resultant 3’-[32P]labeled Exo20p•RexTNick nicked duplex oligonucleotide served as the DNA substrate to measure the 3’-phosphatase activity of AP endonucleases under steady-state reaction conditions.
DNA repair assays
The standard reaction buffer (20 μL) for measuring HpXth’s DNA repair activities consisted of 10 nM [32P]labeled duplex oligonucleotide carrying a modified residue, 5 mM MgCl2, 25 mM KCl, 50 mM Tris-HCl (pH 8.0), 0.01% Nonidet P-40, 0.05 mM DTT, 0.1 mg·mL−1 BSA, and 0.2 nM enzyme, with incubation at 30 °C for 5 min, unless specified otherwise. To measure the AP site cleavage activity of human APE1, we used the buffer BER (20 μL) which consisted of 10 nM [32P]labeled THF•T duplex oligonucleotide, 100 mM KCl, 5 mM MgCl2, 10 mM Tris-HCl (pH 8.0), 0.01% Nonidet P-40, 0.05 mM DTT, 0.1 mg·mL−1 BSA, and 50 nM enzyme, with incubation at 37 °C for 1 h, unless specified otherwise. To measure nucleotide incision activities toward DNA base modifications, we applied the buffer NIR (20 μL), optimal for nucleotide incision activity of human APE1, which consisted of 10 nM [32P]labeled αdA•T duplex oligonucleotide, 25 mM KCl, 0.1 mM MgCl2, 50 mM HEPES-KOH (pH 6.8), 0.01% Nonidet P-40, 0.05 mM DTT, 0.1 mg·mL–1 BSA, and a limited amount of an enzyme, with incubation at 37 °C for 1 h, unless specified otherwise.
The kinetic parameters of repair reactions were measured as described [37]. Briefly, 10–6000 nM duplex oligonucleotide substrate was incubated under the appropriate reaction conditions. The reaction products were quantified on a PhosphorImager after separation by denaturing polyacrylamide gel electrophoresis (PAGE). The kinetic constants were measured at the initial velocity, and KM and kcat values were determined by fitting the data to the one-site binding model in Prism 4 (GraphPad Software, USA). The kinetic parameters for AP endonuclease-catalyzed 3’→5’ exonuclease reaction were determined by quantifying the reaction products expressed as a percentage of the total substrate. When an exonuclease generated multiple DNA products, the value obtained for each degradation fragment was multiplied by the number of catalytic events required for its formation, and the total exonuclease activity was calculated as the sum of these products.
The products of the reactions were analyzed as described [26]. Briefly, all reactions were stopped by adding 10 μL of a solution consisting of 0.5% SDS and 20 mM EDTA and then desalted by passing through homemade spin-down columns filled with Sephadex G25 (GE Healthcare) equilibrated in 7.5 M urea. The desalted reaction products were separated by electrophoresis in a denaturing 20% (w/v) polyacrylamide gel (7.5 M urea, 0.5× TBE). A Fuji Phosphor Screen was exposed to the gels and then scanned with Fuji FLA-9500 and analyzed in the Image Gauge v.4.0 software. At least three independent experiments were conducted for all quantitative and kinetic measurements and also gel images.
Preparation of anti-HpXth antibodies and Western blotting
To produce polyclonal antibodies, a six-month-old rabbit was immunized with the recombinant purified HpXth protein. On the first day of immunization, 0.3 mg of the HpXth protein diluted in phosphate buffer was injected subcutaneously in the back of the rabbit with complete Freund’s adjuvant. After 7 days, a rabbit was immunized with 0.15 mg of the HpXth protein in the same buffer. The third, fourth and fifth immunizations with 0.15 mg of the antigen were carried out every 7 days. Two days later, the sixth immunization was carried out, and the next day the blood serum was collected and the production of anti-HpXth antibodies was checked by immunoblotting. Animal maintenance and experimental procedures were in accordance with the Kazakhstan national regulations based on the provisions of the Central Ethics Commission of Ministry of Health of the Republic of Kazakhstan (Astana, Kazakhstan) and the Legislation of the European Convention for the Protection of Vertebrate Animals Used for Experimental and other Scientific Purposes (ETS 123, Strasbourg, 1986). The protocol used in the present study was approved on January 5, 2016 by the Local Ethical Commission of the RSE "National Center for Biotechnology", Astana, Kazakhstan.
Western blot was performed according to the standard protocol [38]. Briefly, four μg of the cell-free extracts from E. coli BH110 (DE3) strains harboring plasmids expressing the E. coli Nfo and H. pylori HpXth and HpXth-D144N proteins were separated in a 12% (w/v) SDS-PAGE and electro-transferred to the PVDF membrane. To verify protein transfer the membrane was stained with Ponceau S dye [0.1% (w/v) Ponceau S in 5% (v/v) acetic acid]. After membrane blocking with 5% (w/v) dry milk in Tween 20/Tris-buffered saline (TBST, 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% [w/v] Tween 20), the HpXth protein was detected with rabbit anti-HpXth polyclonal antibodies (1:5000) as primary antibodies and goat anti-rabbit horseradish peroxidase-conjugated antibody (Sigma-Aldrich Chimie S.a.r.l., Lyon, France) (1:10,000) as secondary antibodies. The bands were detected by chemiluminescent substrate (Applichem GmbH, Darmstadt, Germany) and exposed to an X-ray film (AgfaPhoto GmbH, Germany).
Alkylation and oxidative DNA damage sensitivity
Drug treatment was performed as previously described [26]. In brief, overnight bacterial cultures were diluted 100-fold in LB broth containing 150 μg/mL ampicillin and 0.05 mM IPTG and incubated at 28 °C until the mid-exponential phase (OD600 = 0.6). The cells were collected by centrifugation, washed once, and resuspended in phosphate-buffered saline (PBS). Ten-fold serial dilutions were prepared in PBS as well. Alkylating agent MMS was added to 2.5 mL of molten soft agar (0.6% agar in LB) at 46 °C containing 1 mM IPTG, followed immediately by 0.25 mL of each cell dilution, and the mixture was poured onto the surface of a 1.5% solid LB agar (25 mL) plate. For an oxidizing-agent sensitivity test, each cell dilution was exposed to 10 mM H2O2 for different periods, and then the cells were mixed with molten soft agar and poured onto LB plates. Colonies were scored after 1–2 days of incubation at 37 °C. Data used in survival curve construction and analysis are representative of at least three independent experiments.
Phylogenetic analysis and model building
The sequences of the conserved domains of E. coli Xth, human APE1 and APE2, Neisseria meningitidis Nape, and Methanothermobacter thermautotrophicus Mth212 were used to search the nonredundant NCBI database of protein sequences by means of BLAST [39]. One representative sequence from each family per taxonomic class was selected for analysis, unless the same species had two paralogs from the same family, in which case, both were selected. Structural model of HpXth was built by multiple iterative threading with searching of the full PDB database using I-TASSER [40]. All sequence alignments and phylogenetic trees were built in Clustal Omega [41]. The conservation analysis was carried out with the Taylor set of amino acid properties [42] and the hierarchical analysis approach outlined elsewhere [43], except that the conservation at any position was expressed as the mean of the number of crossed borders in the Venn diagrams covering all sequence pairs in the compared alignments and normalized to the number of pairs. Images were prepared in the PyMol software (Schrödinger, New York, NY). Phylogenetic trees were visualized using iTOL [44].
Liquid chromatography with tandem mass spectrometry (LC-MS/MS)
Electrophoretic separation of proteins was performed by SDS-PAGE in a 4–14% Bis-Tris gradient gel. The protein bands were excised from the gel and processed as described previously [45–46]. Next, 15 μL of 190 mM ammonium bicarbonate was added to 100 μL of 13.33 ng/μL trypsin (Promega, V5280) for activation of the enzyme, and the samples were incubated at 37°C for 16 h. The peptide mixture was next purified in ZipTipC18 pipette tips (Millipore, 0.6 μL bed, ZTC18S096). The peptide-containing supernatants were dried at 35 °C in a SpeedVac for 30 min, then resuspended in 10 μL of 0.1% trifluoroacetic acid for mass spectrometry experiments.
We used a trapping column setup (Acclaim PepMap100 C18 precolumn, 5 mm × 300 μm; 5 μm particles; Thermo Scientific) and a Dionex high-performance liquid chromatography pump (Ultimate 3000 RSLCnano System, Thermo Scientific). For this experiment, peptides were separated on a Acclaim Pep-Map RSLC column (15 cm × 75 μm, 2 μm particles; Thermo Scientific) with a 75 min multistep acetonitrile gradient (Buffer A: 0.1% formic acid; buffer B 90% acetonitrile/10% H2O in 0.1% formic acid) at a flow rate of 0.3 μL/min, according to the following time table: 0 min, 2% B; 10 min, 2% B; 58 min, 50% B; 59 min, 99% B; 69 min, 99% B; 70 min, 2.0% B; 75 min, 2.0% B. The unmodified CaptiveSpray ion source (Capillary 1300 V, dry gas 3.0 L/min, dry temperature 150 °C) was employed to interface the LC system with an Impact II (Bruker). For quantification, full-scan MS spectra were acquired at a spectral rate of 2.0 Hz followed by acquisition of one MS/MS spectrum. For data acquisition from the sample, the two most abundant precursor ions were selected for fragmentation, resulting in a total cycle time of 3 s. The mass range of the MS scan was set to extend from m/z 150 to 2200 in positive ion polarity mode.
We used Mascot software to perform searches against the SwissProt 2014_08 database (546,238 sequences; 194,363,168 residues). Search parameters were set as follows: variable modifications, oxidation (M), fragment ion mass tolerance, 20 ppm; parent ion tolerance, 20 ppm.
Results
cDNA cloning and purification of the HpXth protein
The cDNA coding for AP endonuclease HpXth was prepared by PCR from the DNA of H. pylori, and oligonucleotide primers as described in Materials and Methods. The gel-purified PCR 753 bp fragment was inserted into pET28c(+) to create pET28c/HpXth expression plasmid. Competent E. coli DH5α cells were transformed with the ligation mixture, and plasmids containing inserts were isolated from the transformant colonies. The primary structures of inserts in the plasmids were verified by sequencing. A comparison of the cloned cDNA with the respective reference sequences present in GenBank revealed polymorphism in HpXth, manifested in the form of amino acid substitution: G181D. The calculated molecular mass of His-tagged HpXth is 31.6 kDa.
To characterize the DNA repair activities of the recombinant HpXth protein, we affinity-purified the bacterial AP endonuclease from the E. coli Rosetta 2 (DE3) strain expressing the N-terminal His-tagged form of HpXth as described in Materials and Methods. After that, the homogeneity of protein preparations was assessed by SDS-PAGE in a 12% gel, revealing >95% purity of the HpXth preparations (S2 Fig). To confirm the identity of the purified protein, the bands were excised from the gel and in-gel digested with trypsin. The resulting peptide mixtures were subjected to matrix-assisted laser desorption ionization-time of flight MS and electrospray ionization tandem mass spectrometry, respectively, and individual peptide sequences were identified based on their molecular weights. Most of the identified peptides matched the expected amino acid sequences of the HpXth protein, with ~80% sequence coverage of the protein. These results indicated that the purified protein is indeed the AP endonuclease from H. pylori.
Characterization of AP site cleavage activity of the H. pylori AP endonuclease in the presence of metal cofactors
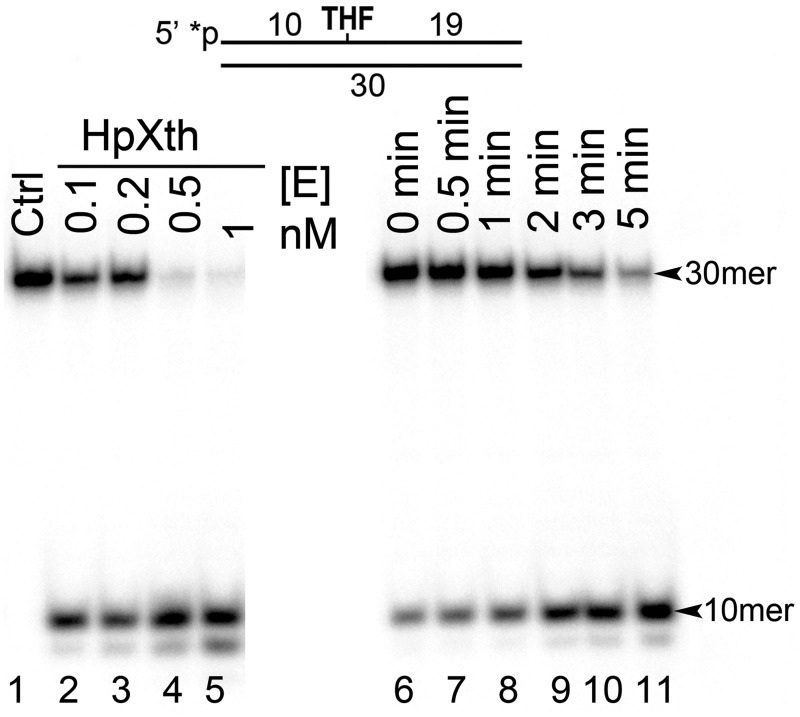
Previous study has shown Mg2+-dependent AP site cleavage activity in a cell-free extract of H. pylori dependent on the function of gene XthA coding for a homolog of E. coli Xth [31]. Nonetheless, it remained unclear whether the HpXth protein, the product of XthA, has efficient AP endonuclease and other DNA repair activities. To clarify this issue, we measured incision activities of the purified HpXth protein on the 5’-[32P]labeled 30mer oligonucleotide duplex THF•T, containing THF, an abasic site analog, at position 11, in the presence of 5 mM MgCl2. As shown in Fig 1, incubation of THF•T with HpXth generated a fast-migrating 10mer cleavage product (lanes 2–11) indicating the presence of AP endonuclease activity. HpXth exhibited a very efficient AP site cleavage activity because 0.5 nM enzyme was able to cleave >95% of 10 nM oligonucleotide substrate after only 5 min incubation under our experimental conditions (lanes 4 and 11). Incubation of THF•T with 1 nM HpXth generated the second minor cleavage product migrating similarly to the 9mer fragment (lane 5) suggesting degradation of the initial 10mer cleavage product by the 3’→5’ exonuclease activity of the enzyme.
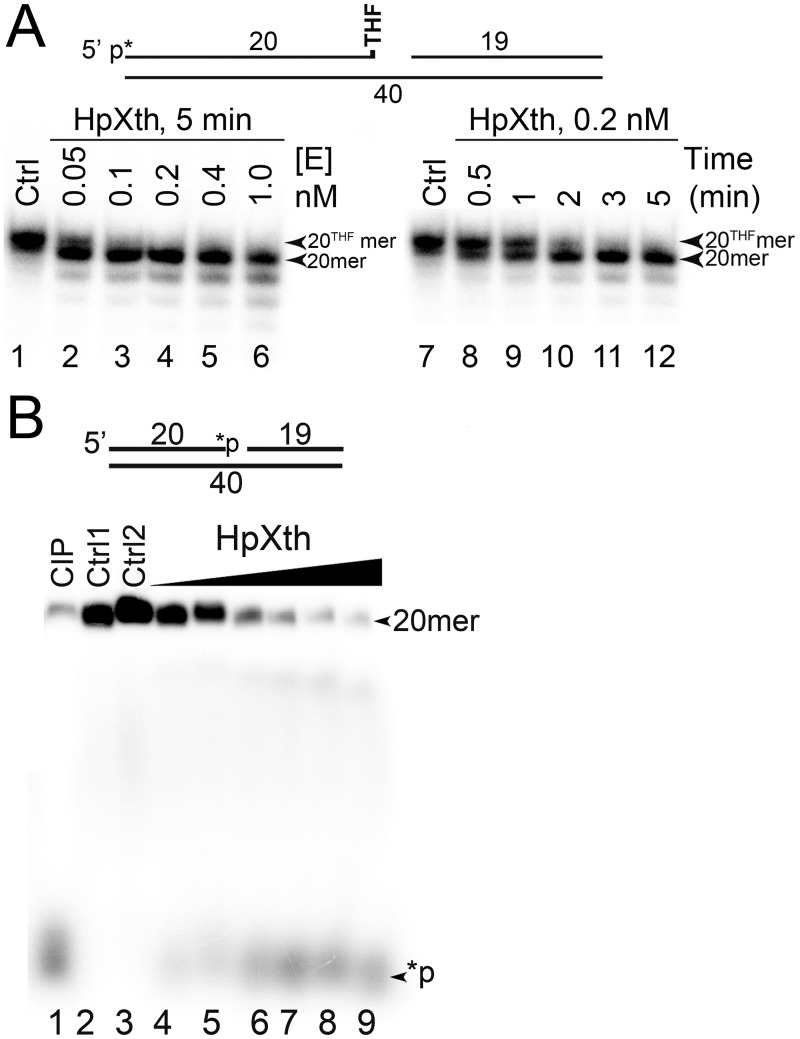
Fig 1. Purified H. pylori AP endonuclease, HpXth, has an AP site cleavage activity.
In brief, 10 nM 5’-[32P]labeled 30mer THF•T duplex was incubated with various concentrations of HpXth for 0–5 min at 30 °C in the standard reaction buffer. Denaturing PAGE analysis of products of the reaction. Lane 1, control THF•T, no enzyme; lanes 2–5, as in lane 1 but 0.1, 0.2, 0.5, and 1 nM HpXth with incubation for 5 min, respectively; lanes 6–11, as in lane 1 but incubated with 0.5 nM HpXth for 0, 0.5, 1, 2, 3, or 5 min, respectively. For details, see Materials and methods.
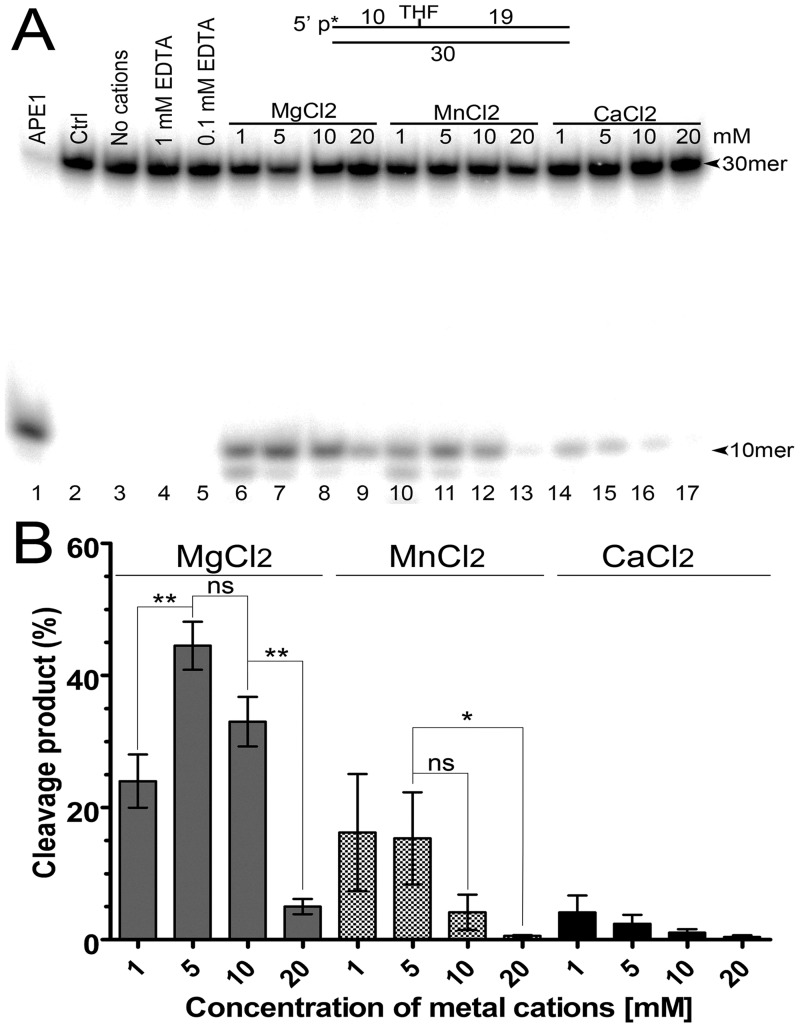
To characterize the AP site cleavage activity of H. pylori AP endonuclease in more detail, we measured the HpXth protein—mediated cleavage of THF•T in a buffer supplemented with various concentrations of divalent metal chlorides and sulphate, including MgCl2, MnCl2, CaCl2, CoCl2, NiCl2, CuSO4, ZnCl2, or FeCl2, (Fig 2 and S3 Fig). As shown in Fig 2A, in the presence of 1 or 5 mM Mg2+ or Mn2+, the purified HpXth protein exerted an efficient AP site cleavage activity by generating fast-migrating 10mer cleavage fragments (lanes 6 and 7 and 10 and 11) which migrated similarly to the 10mer product generated by the control enzyme, human APE1 (lane 1). Nonetheless, higher concentrations of Mg2+ or Mn2+ (10–20 mM) resulted in a strong decrease in the HpXth-catalyzed AP site cleavage (Fig 2A, lanes 9 and 13, and Fig 2B). The presence of 1 or 5 mM Ca2+ caused much less efficient stimulation of HpXth’s AP site cleavage activity as compared to Mg2+ or Mn2+ (Fig 2A, lanes 14 and 15). Again, high concentrations (10–20 mM) of Ca2+ resulted in strong inhibition of the AP endonuclease activity (lanes 16 and 17). It is noteworthy that no cleavage of AP site by HpXth was observed in the absence of any metal cations or in the presence of EDTA (lanes 3–5), indicating the absolute necessity of divalent metal ions for enzymatic catalysis. Taken together, these findings mean that HpXth has a nonlinear dependence on the presence of various concentrations of Mg2+ and Mn2+: its AP site cleavage activity increased in the presence of 1–5 mM and then decreased in the presence of 10–20 mM MgCl2 and MnCl2 (Fig 2).
Fig 2. Divalent-metal ion dependence of HpXth’s AP site cleavage activity.
In brief, 10 nM 5’-[32P]labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min at 37 °C in the standard reaction buffer containing various concentrations of metal cations. (A) Denaturing PAGE analysis of products of the reaction. Lane 1, THF•T and 0.1 nM APE1 in buffer BER; lane 2, control THF•T, no enzyme; lane 3, as in lane 2 but HpXth in the standard reaction buffer but without cations; lanes 4 and 5, as in lane 3 but 1 and 0.1 mM EDTA, respectively; lanes 6–9, as in lane 3 but 1, 5, 10, and 20 mM MgCl2, respectively; lanes 10–13, as in lanes 6–9 but 1–20 mM MnCl2; lanes 14–17, as in lanes 6–9 but 1–20 mM CaCl2. The arrows denote the position of the 30mer substrate and 10mer cleavage product. (B) Graphical representation of data from panel A. The statistical significance of the differences among the mean values were evaluated using two-tailed Student’s test (***P < 0.001; **P < 0.01; *P < 0.05 and nsP > 0.05). For details, see Materials and methods.
Further characterization of HpXth regarding the divalent-metal requirement showed that at a very low (0.01 mM) concentration, Zn2+, Ni2+, Fe2+, or Cu2+ stimulated the AP endonuclease activity, while the same metal cations at 0.1–1.0 mM strongly inhibited AP site cleavage activity of the bacterial enzyme (S3 Fig). To further substantiate inhibitory effects of the above metals, we measured HpXth activity toward THF•T in a buffer supplemented with various concentrations of both metals MgCl2 and ZnCl2. The results revealed that even low concentrations of ZnCl2 (0.1–1 mM) in the reaction mixture already containing 1–5 mM MgCl2, strongly inhibited AP site cleavage activity of HpXth (S4 Fig), suggesting that Zn2+ and heavier metal cations can efficiently compete with Mg2+ for the enzyme’s metal-binding site. Finally, all these results suggest that among metal cations heavier than Mg, only Mn2+ and Ca2+ can stimulate HpXth AP endonuclease activity in the concentration range 1–5 mM, whereas all other cations, except Co2+, stimulate this enzymatic activity only at very low concentrations.
Effects of pH, temperature, ionic strength, and substitution of a conserved amino acid on H. pylori AP endonuclease activity
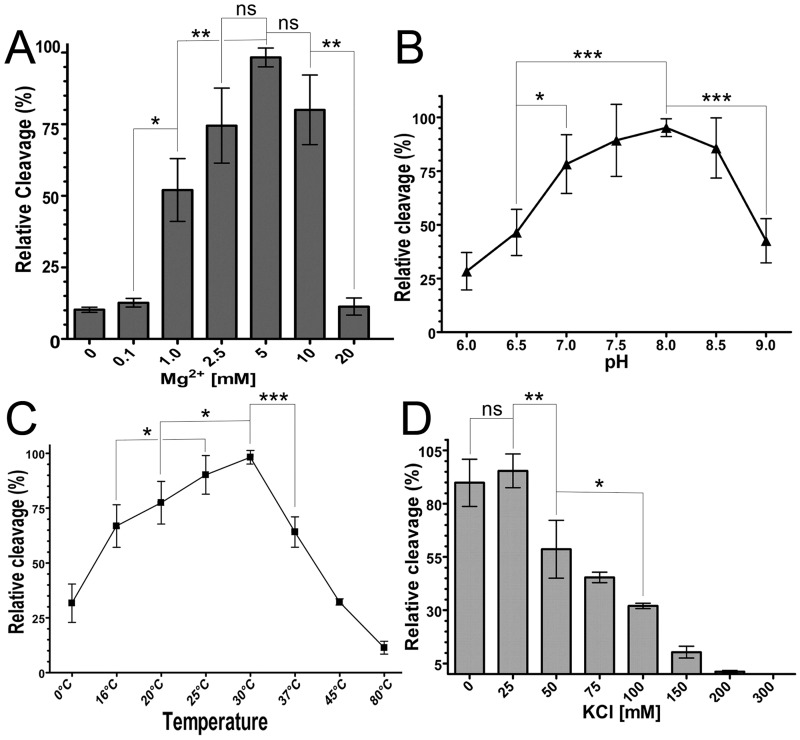
To confirm the biochemical properties of the purified HpXth protein, we measured cleavage of the 30mer THF•T duplex at various MgCl2 concentrations, ionic-strength levels, pH levels, and temperatures. As shown in Fig 3, MgCl2 (panel A), pH (panel B), and temperature (panel C) dependence of the HpXth-catalyzed AP site cleavage was clearly bell-shaped. HpXth manifested the highest AP endonuclease activity at 5 mM MgCl2, pH 8.0, and 30 °C. Notably, the H. pylori AP endonuclease was sensitive to elevated temperature and ionic strength and was inhibited at 37 °C and 50 mM KCl; furthermore 10-fold inhibition was observed at 150 mM KCl (Fig 3D). These results suggest that the requirements for reaction conditions of HpXth are different from those of E. coli Xth and human APE1 AP endonucleases.
Fig 3. Dependence of HpXth’s AP site cleavage activity on reaction conditions.
Briefly, 10 nM 5’-[32P]labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min in the standard reaction buffer but with varying temperature, pH, concentrations of Mg2+ and salt. Products of the reaction were resolved by denaturing PAGE, then visualized by phosphorimaging and quantified in ImageQuant. (A) Graphical representation of Mg2+ concentration dependence. (B) Graphical representation of pH dependence. (C) Graphical representation of temperature dependence. (D) Graphical representation of ionic-strength dependence. The statistical significance of the differences among the mean values were evaluated using two-tailed Student’s test (***P < 0.001; **P < 0.01; *P < 0.05 and nsP > 0.05). For details, see Materials and methods.
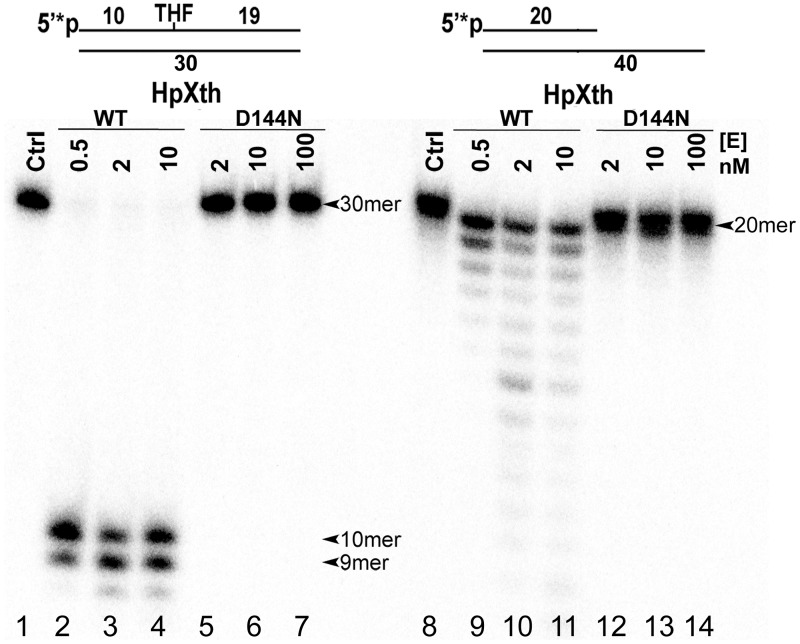
To ensure that the observed AP site cleavage activity is not due to trace contamination via expression of host endonucleases, we constructed mutant of HpXth by site-directed mutagenesis, then purified it by the same scheme as that for the wild-type enzyme. We introduced the mutation D144N into HpXth because in the Xth family enzymes, the residue homologous to D144 of HpXth forms a coordination bond with the catalytic metal ion. In the human APE1 protein, the corresponding mutation D210N reduces the enzymatic activity ~10,000-fold [47]. The purified HpXth-D144N mutant protein was incubated with the 5’-[32P]labeled THF•T and recessed Exo20•RexTRec duplexes to measure AP site cleavage and 3’→5’ exonuclease activities, respectively. Note that the recessed Exo20•RexTRec duplex oligonucleotide was composed of a long regular 40mer RexT template fragment and a short complementary regular 5’-[32P]labeled 20mer Exo20 fragment. As shown in Fig 4, mutant H. pylori AP endonuclease, even when present in excess, did not show any detectable AP site cleavage activity (lanes 5–7) as compared to wild-type HpXth (lanes 2–4). Of note, HpXth D144N mutant concomitantly lost its nonspecific 3’→5’ exonuclease activity toward the Exo20•RexTRec duplex, whereas HpXth-WT exerted a DNA-degrading activity under the same reaction conditions (Fig 4, lanes 12–14 versus lanes 9–11). Altogether, these results indicated that D144 of HpXth is essential for DNA repair activities of the H. pylori protein and that the preparations of recombinant enzymes used in this study are not contaminated with host AP endonucleases.
Fig 4. Comparative characterization of AP site cleavage and 3’→5’ exonuclease activities of wild-type HpXth and mutant HpXth-D144N.
In brief, 10 nM 5’-[32P]labeled THF•T and Exo20•RexTRec were incubated at 30 °C with increasing amounts of the HpXth-WT and HpXth-D144N proteins for 5 min at 30 °C in the standard reaction buffer. Lanes 1–7, THF•T; lane 1, control, no enzyme; lanes 2–4, 0.5, 2, and 10 nM HpXth-WT, respectively; lanes 5–7, 2, 10, and 100 nM HpXth-D144N, respectively; lanes 8–14, same as in lanes 1–7 but with Exo20•RexTRec. Products of the reaction were analyzed by denaturing PAGE. The arrows denote the position of the 30mer and 20mer substrates and 10mer and 9mer cleavage products. For details, see Materials and methods.
Characterization of 3’-repair and NIR activities of pure H. pylori AP endonuclease
Hydrolytic AP endonucleases, in addition to their classic AP site cleavage activity, catalyze the removal of 3’-blocking sugar-phosphates and 3’-terminal phosphate remnants from DNA strand breaks generated by ROS or DNA glycosylases/AP lyases. To test whether HpXth has the 3’-repair phosphodiesterase activities, we prepared nicked Exo20THF•RexTNick and Exo20P•RexTNick duplexes composed of a long 40mer RexT template fragment and two short complementary DNA fragments: the regular 19mer and 20mer Exo20 fragment, containing a 3’-terminal THF and 3’-phosphate residue, respectively. Of note, Exo20THF•RexTNick and Exo20•RexTRec duplexes with 5’-[32P]labeled Exo20 were used to measure the 3’-repair phosphodiesterase and 3’→5’ exonuclease activities, respectively. The Exo20P•RexTNick duplex with 3’-[32P]labeled Exo20 was employed to measure the 3’-phosphatase activity.
As shown in Fig 5A, HpXth cleaves the nicked 5’-[32P]labeled Exo20THF•RexTNick duplex generating a 20mer DNA fragment that migrates faster than the 20mer Exo20THF substrate, indicating that the enzyme removes the 3’-terminal THF residue, thus leaving a Exo20 fragment with 3’-OH (lanes 2–6 and 10–12). Even at a low (0.05 nM) enzyme concentration, HpXth removed the 3’-THF residue in more than 90% of Exo20THF•RexTNick duplex molecules in 5 min (lane 2), whereas 0.2 nM HpXth repaired most of the DNA substrate after only 2 min of incubation (lane 10), suggesting that H. pylori AP endonuclease has a highly efficient 3’-phosphodiesterase activity. When acting on the 3’-[32P]labeled Exo20P•RexTNick duplex, HpXth generates a free radioactive phosphate [32P] residue, which migrates at the bottom of the gel (Fig 5B, lanes 6–9), similar to the product generated by calf intestinal alkaline phosphatase (CIP; lane 1), indicating that the bacterial AP endonuclease removes the 3’-terminal [32P] residue leaving an unlabeled Exo20 fragment. Of note, a higher protein concentration of HpXth (5 nM) was required to remove more than 50% of phosphate residues from the Exo20P•RexTNick duplex (lane 7) as compared to the Exo20THF substrate.
Fig 5. 3’-Repair phosphodiesterase and 3’-phosphatase activities of the HpXth protein.
(A) Enzyme concentration and time-dependent cleavage of the 5’-[32P]labeled Exo20THF•RexTNick oligonucleotide duplex by HpXth. In brief, 10 nM Exo20THF•RexTNick was incubated with increasing amounts of HpXth in the standard reaction buffer at 30 °C for various periods. Lanes 1–6, time-dependent cleavage; lanes 7–12, enzyme concentration—dependent cleavage. The arrows denote the position of substrate “20THFmer” and cleavage product “20mer.” (B) Enzyme concentration—dependent cleavage of the 3’-[32P]labeled Exo20P•RexTNick oligonucleotide duplex by HpXth. In brief, 10 nM Exo20P•RexTNick was incubated with various concentrations of HpXth in the standard reaction buffer for 5 min at 30 °C. Lane 1, Exo20P•RexTNick incubated with 1 U of calf Intestinal alkaline phosphatase, CIP; lanes 2–3, control, no enzyme; lanes 4–9, as in lane 2 but 0.5, 1, 2, 5, 10 and 20 nM HpXth. The arrows denote the position of the 20mer substrate (20mer) and 3’-terminal phosphate product (*p). Products of the reaction were analyzed by denaturing PAGE. For details see Materials and methods.
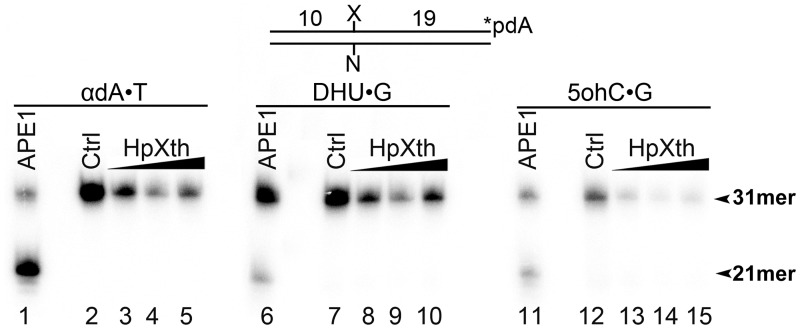
We further examined the DNA substrate specificity of H. pylori enzyme by means of 3’-[32P]labeled 30mer duplex oligonucleotides containing a damaged nucleotide: αdA, DHU, or 5ohC. In control experiments, APE1 cleaved αdA•T, DHU•G, and 5ohC•G duplexes (Fig 6, lanes 1, 6, and 11). No cleavage 5’ to the lesion site of DNA duplexes was observed in the presence of HpXth (lanes 3–5, 8–10, and 13–15), suggesting that H. pylori AP endonuclease does not possess a NIR activity under the experimental conditions tested.
Fig 6. An assay of the nucleotide incision activity of H. pylori HpXth toward a 30mer oligonucleotide duplex containing a modified base.
In brief, 10 nM 3’-[α-32P]-dAMP-labeled 31mer oligonucleotide duplexes containing various DNA base lesions were incubated with increasing amounts of the purified HpXth protein in the buffer NIR for 10 min at 30 °C. Products of the reaction were analyzed by denaturing PAGE. Lanes 1–5, αdA•T; lane 1, 2 nM APE1; lane 2, control no enzyme; lanes 3–5, 5, 20, and 100 nM HpXth, respectively; lanes 6–10, same as in lanes 1–5 but DHU•G; lanes 11–15, same as in lanes 1–5 but 5ohC•G. The arrows denote the position of the 31mer substrate and 21mer cleavage product. The statistical significance of the differences among the mean values were evaluated using two-tailed Student’s test (***P < 0.001; **P < 0.01; *P < 0.05 and nsP > 0.05). For details see Materials and methods.
Steady-state kinetic parameters of the DNA repair reactions catalyzed by H. pylori AP endonuclease
To further substantiate the DNA substrate specificity of the purified HpXth protein, we determined steady-state kinetic parameters of the repair activities and calculated KM, kcat, and kcat/KM values for the cleavage of various DNA substrates. The 30mer THF•T and 40mer Exo20THF•RexTNick, Exo20P•RexTNick, and Exo20•RexTRec duplex oligonucleotides with an AP site residue and a 3’-THF, 3’-P, and 3’-OH group were applied to measure AP endonuclease, 3’-phosphodiesterase, 3’-phosphatase, and 3’→5’ exonuclease activities, respectively. For comparison of the kinetic parameters, E. coli Xth and human APE1 data are presented. As shown in Table 1, steady-state kinetic parameters of HpXth-catalyzed repair reactions revealed that the H. pylori enzyme has a strong substrate preference for AP sites in duplex DNA. The kcat/KM value of HpXth for AP site cleavage is 28- and 9-fold higher than that of its 3’-phosphodiesterase and 3’→5’ exonuclease activities. It is noteworthy that the kcat/KM values for AP site cleavage and 3’-phosphodiesterase activities of HpXth toward the THF•T and Exo20THF•RexTNick duplexes were quite similar to those of E. coli Xth, but nearly 10-fold lower than those of human AP endonuclease 1 (APE1). Similar to human APE1, HpXth has a weak 3’-phosphatase activity with ~220-fold lower kcat/KM as compared to that of its AP endonuclease function. Overall, these kinetic parameters suggested that H. pylori AP endonuclease is similar to E. coli Xth and can be considered a major cellular enzyme that processes AP sites and DNA strand breaks containing 3’-blocking groups in vivo.
Table 1. A comparison of kinetic parameters of AP endonucleases from E. coli, H. pylori, and humans.
| Protein | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HpXth | E. coli Xthb | Human APE1c | |||||||||
| DNA substratea | KM, nM | kcat, min–1 | kcat/KM, min–1·μM–1 | KM, nM | kcat, min–1 | kcat/KM, min–1·μM–1 | kcat/KM ratio, HpXth/EcXth | KM, nM | kcat, min–1 | kcat/KM, min–1·μM–1 | kcat/KM ratio, HpXth/APE1 |
| THF•T | 15.0 ± 2.2 | 18.5 | 1240 | 31 | 40 | 1300 | 1 | 7.7 | 97 | 12,700 | 0.10 |
| Exo20THF•RexTNick | 202 ±100 | 8.8 | 43.6 | 731 | 32.2 | 44 | 1 | 6.0 | 20 | 3300 | 0.13 |
| Exo20P•RexTNick | 40 ±14 | 0.22 | 5.4 | - | - | - | - | 20 | 3.6 | 180 | 0.03 |
| Exo20•RexTRec | 6800 ± 1000 | 920 ± 140 | 135 | - | - | - | - | 2.4 | 0.86 | 360 | 0.38 |
aEach substrate was used to measure a specific DNA repair function under the optimal reaction conditions: the THF•T duplex for the AP endonuclease activity; the Exo20THF•RexTNick nicked DNA duplex for the 3’-repair phosphodiesterase activity; the Exo20P•RexTNick nicked DNA duplex for the 3’-phosphatase activity; and the Exo20•RexTRec recessed DNA duplex for the 3’→5’ exonuclease activity (see Materials and methods).
bKinetic parameters of the E. coli Xth-catalyzed AP site cleavage and 3’-phosphodiesterase reaction were taken from other studies [26, 59].
cKinetic parameters of human APE1-catalyzed DNA repair reactions were taken from ref. [37].
Homology modeling and phylogenetic analysis of HpXth
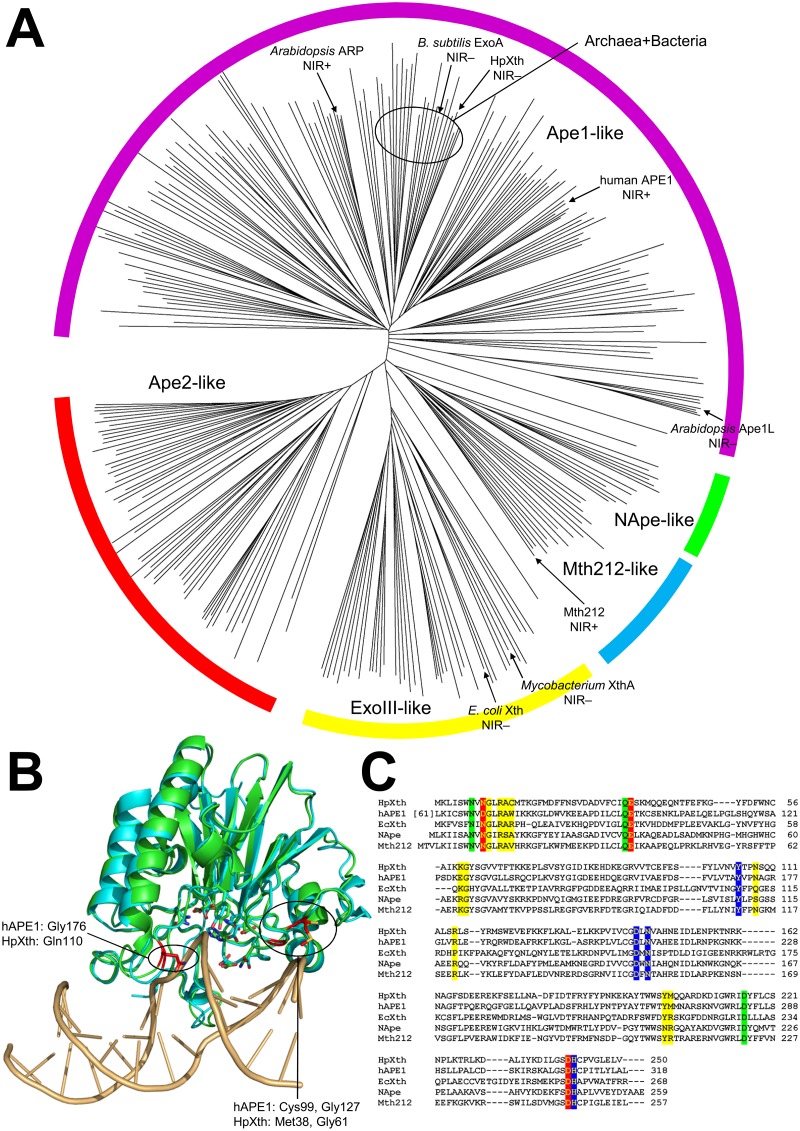
AP endonucleases belonging to the Endonuclease—Exonuclease—Phosphatase (EEP) superfamily are classified into five families based on their sequence homology: ExoIII-like (with E. coli Xth being the prototypical member), NApe-like (similar to Neisseria NApe endonuclease), Mth212-like (similar to Methanothermobacter Mth212 endonuclease), Ape1-like and Ape2-like (similar to human APE1 and APE2, respectively). Unlike its E. coli homolog, HpXth is classified as an Ape1-like endonuclease by the NCBI Conserved Domain Database [48]. We re-examined the phylogenetic relations among the EEP superfamily AP endonucleases using a set of 284 sequences evenly spread over all domains of life (Fig 7A and S1 File). HpXth, together with several other bacterial and archaeal sequences, formed a separate clade among the Ape1-like family members (labeled as Archaea+Bacteria in Fig 7A). Bacillus subtilis ExoA protein, an efficient AP endonuclease with a low NIR activity [49], also fell into this clade. A closer look at Epsilonproteobacteria, the class to which H. pylori belongs, revealed that basal Epsilonproteobacteria possess AP endonucleases from ExoIII-like, Mth212-like, and Ape1-like families, but some of those were lost during the evolution: in Helicobacteraceae, only Ape1-like sequences remain, while its sister group, Campylobacteraceae, retained only Mth212-like AP endonucleases.
Fig 7. Phylogenetic analysis and structural models of H. pylori AP endonuclease.
(A) An unrooted phylogenetic tree of the EEP superfamily AP endonucleases (230 sequences, one species per taxonomic class). Different families are indicated by colored arcs: yellow, ExoIII-like; green, Nape-like; blue, Mth212-like; magenta, Ape1-like; and red, Ape2-like. The clade combining HpXth, B. subtilis ExoA, and the related bacterial and archaeal sequences is labeled “Archaea+Bacteria.” The known NIR-proficient and NIR-deficient enzymes are indicated. (B) Overlay of the structure of human APE1 (1DEW, cyan) and the HpXth model (green). All active-site residues are shown but left unlabeled for clarity. The DNA groove—contacting residues possibly involved in the NIR function are indicated by red carbon atoms. (C) Alignment of HpXth and four core members of AP endonuclease families (human APE1 catalytic domain, E. coli Xth, N. meningitidis NApe, and M. thermautotrophicus Mth212). Metal-binding sites A and B are red and blue, respectively, non—metal-binding active site residues are green; other DNA-binding residues are yellow.
Multiple threading revealed the best templates for HpXth to be B. subtilis ExoA [PDB entry 5CFE, [49]], Archaeoglobus fulgidus AP endonuclease [2VOA, [50]], N. meningitidis NApe [2JC5, [51]], M. thermautotrophicus Mth212 [3FZI, [52]], and 3W2X, unpublished), and human APE1 [2O3H, [53]]. The calculations converged to a single model (S2 File) with the excellent confidence score 1.96. The model showed the best fit to the template proteins plus Leishmania major AP endonuclease LMAP [2J63, [54]], Methanosarcina mazei exonuclease III homolog Mm3148 (5J8N, unpublished), N. meningitidis AP endonuclease NExo [2JC4, [51]], and Sulfolobus islandicus exonuclease III (5EWT, unpublished). The superposition of the model and human APE1/DNA structure [1DEW, a complex with THF-containing DNA [55]] showed low r.m.s.d. (0.848 over the heavy atoms), with all catalytically important residues in the active site of APE1 conserved in HpXth and occupying similar positions (Fig 7B and S5 Fig). One exception is Asn9 of HpXth, which corresponds to Asp70 in the human protein and forms part of a catalytic Mg2+-binding site (Fig 7C). However, Ape1-like enzymes vary at this position, containing Asp, Asn, or small amino acids (Gly/Ala). Despite influencing the metal preference and/or the balance of endonuclease, exonuclease, and 3’-phosphodiesterase activities [33], both Asn and Asp are likely suitable for the biologically relevant function of EEP superfamily AP endonucleases.
Drug sensitivity of an E. coli DNA repair-deficient strain expressing the H. pylori AP endonuclease
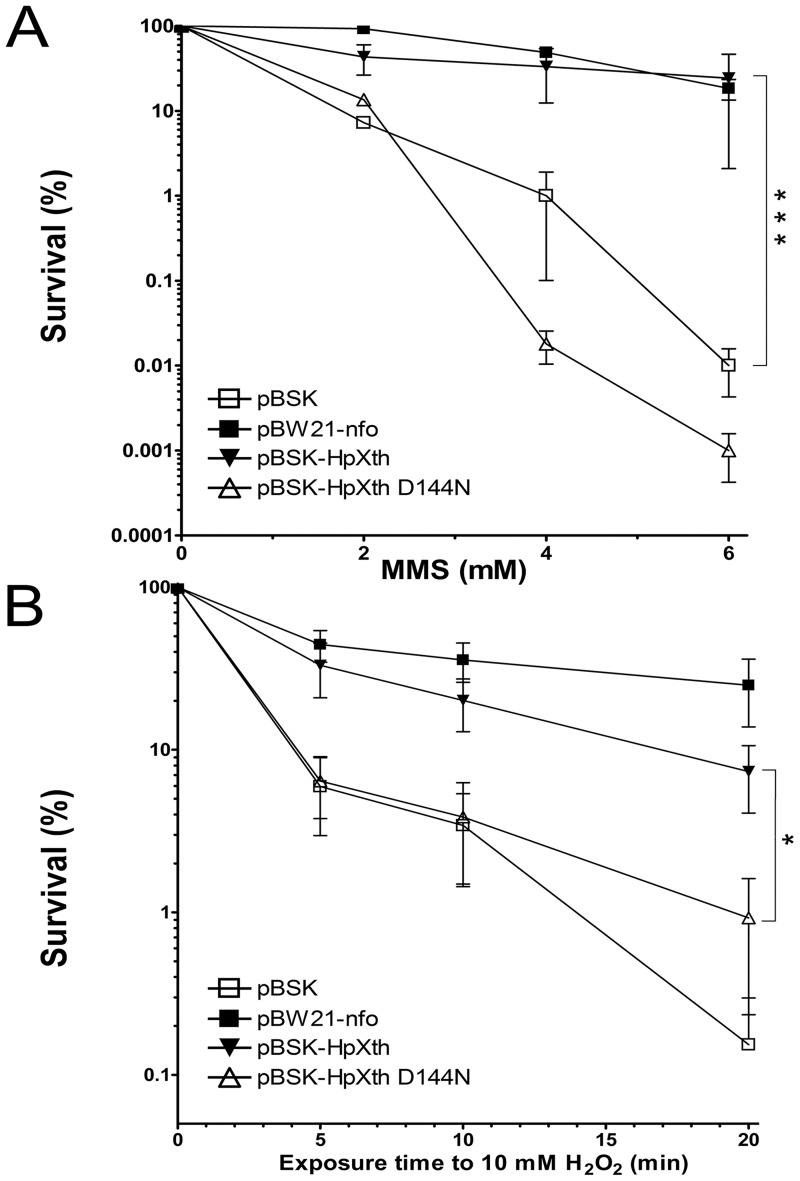
To examine the biological roles of HpXth’s activities, we employed the phenotype rescue assay of E. coli mutant. We measured the sensitivity of AP endonuclease-deficient E. coli xthA nfo BH110 (DE3) strain harboring plasmid pBW21 coding for the E. coli Nfo and plasmid pBluescript SK(+) containing inserts coding for wild-type or mutant pathogen’s AP endonuclease HpXth or HpXth-D144N, respectively, to exposure to MMS and H2O2. Western blotting using rabbit anti-HpXth polyclonal antibodies confirms the presence of the HpXth proteins in E. coli BH110 (DE3) cells harboring the plasmids that encode the AP endonuclease from H. pylori (S6 Fig). MMS methylates purines in cellular DNA, subsequently methylated purines are excised by DNA glycosylases in the BER pathway leading to the appearance of AP sites in DNA [56]. Exposure to H2O2 oxidizes DNA bases and also generates single-strand DNA breaks with blocked 3’ termini [57]. The AP endonuclease-deficient E. coli BH110 (DE3) strain lacking both Xth and Nfo is very sensitive to both agents [58]. As expected, the plasmid that encodes for E.coli Nfo rescues BH110 (DE3) cells after exposure to MMS and H2O2 (Fig 8). Consistent with our biochemical results, the pBSK-HpXth plasmid encoding HpXth restored the resistance to MMS to the level close to that of plasmid-harboring E. coli Nfo, showing a 10- to 104-fold increase in the cell survival at different concentration of MMS as compared to the control empty plasmid (Fig 8A).
Fig 8. Differential drug sensitivity of AP endonuclease—deficient E. coli strain BH110 (DE3) carrying a plasmid coding for the H. pylori AP endonuclease.
The strains are represented as follows: the BH110 (DE3) strain carrying control empty vector pBluescript II SK+ (pBSK) (□), strain BH110 (DE3) carrying pBW21-Nfo (■), or pBSK-HpXth (▼), or pBSK-HpXth-D144N (△). Each survival curve represents at least three independent experiments. (A) The survival of the MMS-treated E. coli AP endonuclease—deficient strains. (B) The survival of the E. coli AP endonuclease—deficient strains under oxidative stress. The statistical significance of the differences among the mean values were evaluated using two-tailed Student’s test (***P < 0.001; **P < 0.01; *P < 0.05 and nsP > 0.05). For details, see Materials and methods.
Plasmid pBSK-HpXth partially restored the resistance to H2O2 showing a 5- to 100-fold increase in the cell survival at different periods of exposure to H2O2 as compared to the control empty plasmid (Fig 8B). As expected, the plasmid harboring mutant HpXth-D144N did not confer resistance to MMS and H2O2, as was the case for the empty control plasmid, suggesting that this enzyme’s catalytic activities are essential for their biological functions (Fig 8). Remarkably, the E. coli BH110 (DE3) cells expressing HpXth-D144N were more sensitive to MMS as compared to cells carrying the empty control vector, suggesting that the mutant H. pylori protein may inhibit repair of AP sites by other back-up DNA repair pathways. Taken together, these results suggested that HpXth can efficiently repair AP sites and only to some extent 3’-blocking groups in E. coli.
Discussion
Unlike many bacteria, including E. coli, which possess two structurally unrelated AP endonucleases, the Helicobacter lineage has retained only a homolog of exonuclease III. Previously, it has been demonstrated that the mutants of H. pylori lacking HpXth (nucT HP1526 and xthA::cat) have undetectable Mg2+-dependent AP site cleavage activity and show a 4-fold increase in the spontaneous mutation frequency over the wild-type strain, suggesting that HpXth is a functional homolog of E. coli Xth [31–32]. Nevertheless, HpXth has not been biochemically characterized to date. In this study, we have cloned cDNA encoding the HpXth, overexpressed and purified it from E. coli, and characterized the DNA substrate specificity of the enzyme. Our results suggest that HpXth protein possesses AP site cleavage, 3’-repair phosphodiesterase, 3’-phosphatase, and 3’-exonuclease functions. Furthermore, HpXth, just as E. coli Xth, but in contrast to their human homolog APE1, has no damage-specific nucleotide incision activity.
Enzymatic activities of HpXth
In line with the well-characterized E. coli Xth, our data indicate that HpXth is a divalent metal ion—dependent enzyme requiring Mg2+ or Mn2+ for the full activity (Figs 2 and 3). Of note, HpXth was considerably inhibited in the presence of 0.1–1.0 mM ZnCl2 (S3 and S4 Figs), suggesting that Zn2+ ions could thwart AP site repair in H. pylori. The detailed characterization of the reaction conditions showed that HpXth-catalyzed AP site cleavage is optimal at a high Mg2+ concentration (5 mM), neutral pH, low ionic strength, and 30 °C (Fig 3). These reaction conditions with low salt and relatively high Mg2+ more closely resemble the optimum of E. coli Xth [59] than that of human APE1 [13]. The Mg2+ concentration, ionic strength, pH, and even temperature dependence curves of HpXth-catalyzed AP endonuclease activity are all bell-shaped. These observations may imply that the catalytically active protein conformation is sensitive to changes in the reaction conditions; perhaps in vivo, the HpXth-catalyzed reactions are tightly regulated.
The preparation of recombinant HpXth displayed nonspecific 3’→5’ exonuclease activity degrading nicked DNA after the AP site cleavage and undamaged recessed DNA duplexes (Figs 1 and 4). To confirm that the exonuclease activity is an intrinsic property of this AP endonuclease, we constructed and characterized the HpXth containing a D144N mutation that should disrupt the metal-binding shell. The mutant completely lost both the ability to cleave AP sites and the 3’→5’ exonuclease activity (Fig 4). Hence, the the 3’→5’ exonuclease function is intrinsically present in HpXth and may play an important physiological role.
To get an idea about comparative importance of the observed enzymatic activities of HpXth, we have measured the steady-state kinetic parameters of its AP site cleavage, 3’-repair phosphodiesterase, 3’-phosphatase, and 3’→5’ exonuclease activities. The analysis of kinetic constants revealed that HpXth possesses efficient AP endonuclease and 3’-repair phosphodiesterase activities with kcat and kcat/KM values very similar to those of E. coli Xth (Table 1). For both bacterial enzymes, these constants were 2.5–10-fold lower in comparison with the human counterpart, APE1. Nevertheless, the kinetic parameters suggest that HpXth can efficiently protect H. pylori from endogenous and induced AP sites and strand breaks in genomic DNA.
Phylogenetic distribution and structural features of EEP activities
In addition to the AP site and 3′-end processing activities, certain EEP superfamily enzymes possess the NIR activity that nicks DNA at some sites containing damaged bases. Notably, the NIR activity is distributed unevenly in the phylogeny: it has been reported for human APE1 [13], Arabidopsis ARP [33], and Methanothermobacter Mth212 [60], but is lacking or negligible in ExoIII-like enzymes [13, 26], bacterial/archaeal Ape1-like enzymes [[49] and this work] and plant Ape1L [35]. Ape2-like enzymes, in view of their low AP endonuclease activity and the predominant 3’→5’-exonuclease activity [61], are also likely to be free of the NIR activity. The identification of NIR-deficient HpXth, grouped with low-NIR B. subtilis ExoA, allowed us to revisit the requirements for efficient NIR among the EEP endonucleases.
The phylogenetic analysis of the EEP superfamily AP endonucleases suggests that either the NIR function was present in an ancestral EEP endonuclease and then was lost in certain lineages (such as ExoIII-like, bacterial/archaeal Ape1-like, and plant Ape1L), or was independently acquired by APE1, ARP, and Mth212-like homologs. Because the loss of a function is more probable than independent emergence of one in several lineages, we examined the conservation of residues in different EEP groups to find which positions differentiate NIR-proficient enzymes from NIR-deficient ones, assuming the Ape1-like and Mth212-like enzymes to be NIR-proficient unless clustering with B. subtilis ExoA/HpXth or Ape1L. We identified three positions that could be related to the NIR proficiency; of note, all of them are involved in forming contacts with DNA away from the active site (Fig 7B, carbons colored red). The first two corresponded to Cys99 and Gly127 in human APE1. Gly127 was replaced by Lys/Arg in all Ape1L enzymes, whereas the homologs of Cys99 were found to have small side chains in most NIR-proficient enzymes and aliphatic bulky ones in NIR-deficient enzymes (e.g., Met in HpXth or Val in E. coli Xth). In human APE1, Cys99 and Gly127 form a part of the structure that is inserted into the DNA minor groove 5’ to the damaged nucleotide [55]. At the same side but closer to the lesion, Gly176 in APE1 corresponds to small-side chain residues in the major groove in most NIR-proficient enzymes, whereas NIR-deficient ones possess a charged or polar group at the equivalent position (Glu in E. coli Xth, Gln in HpXth). These differences could lead to different modes of DNA kinking upon binding to the enzyme, and these conditions might be crucial for the correct entry of base-containing nucleotides into the active site of AP endonucleases. A comparison of Ape1-like NIR-proficient enzymes with NIR-deficient ones reveals that the former group has Phe/Tyr in the active site pressing against the everted abasic nucleotide (Phe266 in human APE1), whereas all NIR-deficient enzymes have a bulkier Trp side chain at this position, possibly interfering with the correct eversion of the damaged-base—containing nucleotide during NIR.
Many bacterial enzymes, including AP endonucleases, are different from their eukaryotic counterparts by having no or little regulatory additions to the catalytic domains. Comparing human APE1 with bacterial Xth, it is clear that the human protein carries an additional N-terminal tail, where all regulatory Lys residues are located. Sequence alignment of human APE1 and E. coli and H. pylori Xth clearly indicate that there is no counterparts of regulatory Lys [62] and redox-active Cys residues [63] in the bacterial enzymes (S7 Fig). This observation most likely reflects the differences in the intracellular distribution of the human and bacterial enzymes and in their involvement in the transcription regulation.
Biological role of a single H. pylori AP endonuclease
Efficient AP endonuclease and 3’-end repair functions of HpXth suggest that H. pylori AP endonuclease plays an important role in the repair of AP sites and single-strand DNA breaks with blocked 3’ termini generated by alkylating agents, ROS, or DNA glycosylases. Consistent with DNA substrate specificities characterized in vitro, the expression of HpXth in E. coli AP endonuclease—deficient BH110(DE3) strain conferred resistance to MMS comparable to that provided by a plasmid encoding E. coli Nfo (Fig 8A). In addition, HpXth imparted resistance to H2O2 but to a lesser degree than E. coli Nfo did (Fig 8B); this finding may be consistent with less efficient 3’-end repair functions as compared to the AP site cleavage activity (Table 1). Also, the absence of the NIR function in HpXth found here is suggestive of possibly increased sensitivity of H. pylori to oxidizing agents such as bleomycin and organic peroxides, that induce specific lesions that are subject to NIR [58, 64]. In conclusion, our findings together with earlier reports unequivocally identify HpXth as the principal AP endonuclease and 3’-repair phosphodiesterase in H. pylori, protecting this important human pathogen from the genotoxic effects of endogenous and induced AP sites and DNA strand breaks.
Supporting information
Domains and their families were assigned according to the Conserved Domain Database [48].
(DOCX)
(PDB)
(A) Duplex oligonucleotides used to measure 3’-repair phosphodiesterase and 3’-5’ exonuclease activities. (B) Chemical structures of abasic sites and alpha-anomeric 2’deoxyadenosine.
(TIF)
Lane 1, Protein Ladder (Thermo Scientific, cat. # 26616); lane 2, 1 μg HpXth; lane 3, 1 μg HpXth-D144N. For details, see Materials and methods.
(TIF)
Briefly, 10 nM 5’-[32P]-labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min at 37 °C in the buffer containing various concentrations of metal cations. Products of the reaction were resolved by denaturing PAGE, then visualized by phosphorimaging and quantified in ImageQuant. For details, see Materials and methods.
(TIF)
In brief, 10 nM 5’-[32P]-labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min at 37 °C in the standard reaction buffer but with varying concentrations of MgCl2 and ZnCl2, unless otherwise stated. Lane 1, THF•T and 0.1 nM APE1 in the buffer BER; lane 2, control THF•T, no enzyme; lanes 3–4, as in lane 1 but supplemented with 0.1 and 1 mM ZnCl2, respectively; lane 5, THF•T and 0.2 nM HpXth in the buffer without metal cations; lane 6, as in lane 5 but 1 mM MgCl2; lane 7, as in lane 6 but 0.1 mM ZnCl2; lane 8, as in lane 6 but 1 mM ZnCl2; lane 9, as in lane 5 but 5 mM MgCl2; lane 10, as in lane 9 but 0.1 mM ZnCl2; lane 10, as in lane 9 but 1 mM ZnCl2. The arrows denote the position of the 30mer substrate and a 10mer cleavage product. For details, see Materials and methods.
(TIF)
HpXth model superimposed over human APE1 structure (1BIX). Sm3+ ion replacing catalytic Mg2+ in the crystal structure is shown as a magenta ball. Amino acids labels show numbering in HpXth first, followed by numbering in the corresponding human or E. coli protein. Cartoon traces and carbon atoms of HpXth model are colored green, those of human and E. coli structures are cyan, oxygen and nitrogen atoms are red and blue, respectively.
(TIF)
Four μg of cell-free extracts from the BH110 (DE3) strains carrying different plasmids were separated using 12% SDS-PAGE and transferred to the PVDF membrane and then analyzed by Western blotting. (A) Western blot analysis of the PVDF membrane using rabbit anti-HpXth polyclonal antiserum. Lane 1, protein molecular weight markers; lane 2, control plasmid-free E. coli BH110 (DE3) strain; lane 3, empty vector pBluescript II SK+ (pBSK); lane 4, pBSK-HpXth; lane 5, pBSK-HpXth-D144N; lane 6, 50 ng of the purified His-tagged HpXth; lane 7, as 6 but 100 ng. (B) Ponceau S-staining of the PVDF membrane. Lanes 1–7, same as in panel A. For details, see Materials and methods.
(TIF)
(TIF)
Acknowledgments
We wish to thank Dr. Olga Lavrik for the recombinant Tdp1 protein and Arman Kulyyassov, Ph.D., for advice and help with the LC-MS/MS experiments.
Abbreviations
- 3’OH
3’-terminal hydroxyl
- 3’P
3’-terminal phosphate
- 3’PA
3’-terminal phospho α,β-unsaturated aldehyde
- 3’-THF
3’-terminal THF residue
- αdA
alpha-anomeric 2’-deoxyadenosine
- αdN
alpha-anomeric 2’-deoxynucleoside
- AP
apurinic/apyrimidinic
- APE1
major human AP endonuclease 1
- BER
base excision repair
- DHU
5,6-dihydrouracil
- EEP
Endonuclease—Exonuclease—Phosphatase
- HpXth
Helicobacter pylori homolog of E. coli exonuclease III
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MMS
methylmethanesulfonate
- NIR
nucleotide incision repair
- nt
nucleotide
- ROS
reactive oxygen species
- THF
3-hydroxy-2-hydroxymethyltetrahydrofuran or tetrahydrofuran
- Xth
Escherichia coli exonuclease III
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants to S.A. from the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan AP05130820 (www.biocenter.kz), to M.S. from la Ligue Nationale Française Contre le Cancer “Equipe Labellisée LIGUE 2016” grant EL2016.LNCC/MuS and Electricité de France RB 2017 (http://www.edf.fr), to A.A.I. from ERA.Net RUS Plus (www.eranet-rus.eu) [#306] and French National Center for Scientific Research (PRC CNRS/RFBR n1074 REDOBER), and to D.O.Z. from the Russian Ministry of Science and Education (6.5773.2017/6.7) and Russian Science Foundation (17-14-01190). A.T. was supported by Ph.D. fellowships from L.N. Gumilyov Eurasian National University (www.enu.kz), Kazakhstan. The sponsors played no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
References
- 1.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531(1–2):37–80. . [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat Res. 2003;531(1–2):5–23. . [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–15. 10.1038/362709a0 . [DOI] [PubMed] [Google Scholar]
- 4.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–76. 10.1146/annurev.genet.38.072902.092448 . [DOI] [PubMed] [Google Scholar]
- 5.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415(6868):183–7. 10.1038/415183a . [DOI] [PubMed] [Google Scholar]
- 6.Yasui A. Alternative excision repair pathways. Cold Spring Harb Perspect Biol. 2013;5(6):1–8. 10.1101/cshperspect.a012617 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65(10):1544–65. 10.1007/s00018-008-7543-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace SS. Base excision repair: A critical player in many games. DNA Repair (Amst). 2014. 10.1016/j.dnarep.2014.03.030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14(12):2491–507. 10.1089/ars.2010.3466 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson DM 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485(4):283–307. . [DOI] [PubMed] [Google Scholar]
- 11.Ide H, Tedzuka K, Shimzu H, Kimura Y, Purmal AA, Wallace SS, et al. Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Biochemistry. 1994;33(25):7842–7. . [DOI] [PubMed] [Google Scholar]
- 12.Ishchenko AA, Ide H, Ramotar D, Nevinsky G, Saparbaev M. Alpha-anomeric deoxynucleotides, anoxic products of ionizing radiation, are substrates for the endonuclease IV-type AP endonucleases. Biochemistry. 2004;43(48):15210–6. 10.1021/bi049214+ . [DOI] [PubMed] [Google Scholar]
- 13.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32(1):73–81. 10.1093/nar/gkh165 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hang B, Chenna A, Fraenkel-Conrat H, Singer B. An unusual mechanism for the major human apurinic/apyrimidinic (AP) endonuclease involving 5’ cleavage of DNA containing a benzene-derived exocyclic adduct in the absence of an AP site. Proc Natl Acad Sci U S A. 1996;93(24):13737–41. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–48. 10.1146/annurev.bi.63.070194.004411 . [DOI] [PubMed] [Google Scholar]
- 16.Daley JM, Zakaria C, Ramotar D. The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat Res. 2010;705(3):217–27. 10.1016/j.mrrev.2010.07.003 . [DOI] [PubMed] [Google Scholar]
- 17.Malfatti MC, Balachander S, Antoniali G, Koh KD, Saint-Pierre C, Gasparutto D, et al. Abasic and oxidized ribonucleotides embedded in DNA are processed by human APE1 and not by RNase H2. Nucleic Acids Res. 2017;45(19):11193–212. 10.1093/nar/gkx723 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couve-Privat S, Ishchenko AA, Laval J, Saparbaev M. Nucleotide Incision Repair: An Alternative and Ubiquitous Pathway to Handle Oxidative DNA Damage In Oxidative Damage to Nucleic Acids, edited by Evans Mark D. and Cooke Marcus S. Austin: Landes Bioscience and Springer Science+Business Media; 2007. 54–66 p. [Google Scholar]
- 19.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, et al. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37(12):3946–58. 10.1093/nar/gkp275 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3’ ends justify the means. Mutat Res. 2000;460(3–4):211–29. . [DOI] [PubMed] [Google Scholar]
- 21.He H, Chen Q, Georgiadis MM. High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I. Biochemistry. 2014;53(41):6520–9. 10.1021/bi500676p . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barzilay G, Hickson ID. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays. 1995;17(8):713–9. 10.1002/bies.950170808 . [DOI] [PubMed] [Google Scholar]
- 23.Carpenter EP, Corbett A, Thomson H, Adacha J, Jensen K, Bergeron J, et al. AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. Embo J. 2007;26(5):1363–72. 10.1038/sj.emboj.7601593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson AR, Soliven KC, Castor ME, Barnes PD, Libby SJ, Fang FC. The Base Excision Repair system of Salmonella enterica serovar typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 2009;5(5):e1000451 10.1371/journal.ppat.1000451 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagorska K, Silhan J, Li Y, Pelicic V, Freemont PS, Baldwin GS, et al. A network of enzymes involved in repair of oxidative DNA damage in Neisseria meningitidis. Mol Microbiol. 2012;83(5):1064–79. 10.1111/j.1365-2958.2012.07989.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abeldenov S, Talhaoui I, Zharkov DO, Ishchenko AA, Ramanculov E, Saparbaev M, et al. Characterization of DNA substrate specificities of apurinic/apyrimidinic endonucleases from Mycobacterium tuberculosis. DNA Repair (Amst). 2015;33:1–16. 10.1016/j.dnarep.2015.05.007 . [DOI] [PubMed] [Google Scholar]
- 27.Talebi Bezmin Abadi A. Helicobacter pylori: A Beneficial Gastric Pathogen? Frontiers in Medicine. 2014;1(26). 10.3389/fmed.2014.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Veen S, Tang CM. The BER necessities: the repair of DNA damage in human-adapted bacterial pathogens. Nat Rev Microbiol. 2015;13(2):83–94. 10.1038/nrmicro3391 . [DOI] [PubMed] [Google Scholar]
- 29.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47. 10.1038/41483 . [DOI] [PubMed] [Google Scholar]
- 30.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397(6715):176–80. 10.1038/16495 . [DOI] [PubMed] [Google Scholar]
- 31.Mathieu A, O’Rourke EJ, Radicella JP. Helicobacter pylori genes involved in avoidance of mutations induced by 8-oxoguanine. J Bacteriol. 2006;188(21):7464–9. 10.1128/JB.00851-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Kang J, Blaser MJ. Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J Bacteriol. 2006;188(17):6224–34. 10.1128/JB.00477-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akishev Z, Taipakova S, Joldybayeva B, Zutterling C, Smekenov I, Ishchenko AA, et al. The major Arabidopsis thaliana apurinic/apyrimidinic endonuclease, ARP is involved in the plant nucleotide incision repair pathway. DNA Repair (Amst). 2016;48:30–42. 10.1016/j.dnarep.2016.10.009 . [DOI] [PubMed] [Google Scholar]
- 34.Ishchenko AA, Sanz G, Privezentzev CV, Maksimenko AV, Saparbaev M. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 2003;31(21):6344–53. 10.1093/nar/gkg812 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joldybayeva B, Prorok P, Grin IR, Zharkov DO, Ishenko AA, Tudek B, et al. Cloning and Characterization of a Wheat Homologue of Apurinic/Apyrimidinic Endonuclease Ape1L. PLoS One. 2014;9(3). 10.1371/journal.pone.0092963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebedeva NA, Rechkunova NI, Ishchenko AA, Saparbaev M, Lavrik OI. The mechanism of human tyrosyl-DNA phosphodiesterase 1 in the cleavage of AP site and its synthetic analogs. DNA Repair (Amst). 2013;12(12):1037–42. 10.1016/j.dnarep.2013.09.008 . [DOI] [PubMed] [Google Scholar]
- 37.Gelin A, Redrejo-Rodriguez M, Laval J, Fedorova OS, Saparbaev M, Ishchenko AA. Genetic and biochemical characterization of human AP endonuclease 1 mutants deficient in nucleotide incision repair activity. PLoS One. 2010;5(8):e12241 10.1371/journal.pone.0012241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J WE. Molecular cloning. A laboratory manual by T Maniatis, E F Fritsch and J Sambrook. pp 545. Cold Spring Harbor Laboratory, New York. 1982. $48. ISBN 0-87969-136-0. Biochemical Education. 1983;11(2):82-. 10.1016/0307-4412(83)90068-7 [DOI] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. 10.1038/nmeth.3213 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539 10.1038/msb.2011.75 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor WR. The classification of amino acid conservation. J Theor Biol. 1986;119(2):205–18. . [DOI] [PubMed] [Google Scholar]
- 43.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9(6):745–56. . [DOI] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–5. 10.1093/nar/gkw290 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulyyassov A, Shoaib M, Pichugin A, Kannouche P, Ramanculov E, Lipinski M, et al. PUB-MS: a mass spectrometry-based method to monitor protein-protein proximity in vivo. J Proteome Res. 2011;10(10):4416–27. 10.1021/pr200189p . [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. . [DOI] [PubMed] [Google Scholar]
- 47.Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, et al. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J Biol Chem. 2013;288(12):8445–55. 10.1074/jbc.M112.422774 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D3. 10.1093/nar/gkw1129 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redrejo-Rodriguez M, Vigouroux A, Mursalimov A, Grin I, Alili D, Koshenov Z, et al. Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA. Biochimie. 2016;128–129:20–33. 10.1016/j.biochi.2016.06.011 . [DOI] [PubMed] [Google Scholar]
- 50.Schmiedel R, Kuettner EB, Keim A, Strater N, Greiner-Stoffele T. Structure and function of the abasic site specificity pocket of an AP endonuclease from Archaeoglobus fulgidus. DNA Repair (Amst). 2009;8(2):219–31. 10.1016/j.dnarep.2008.10.009 . [DOI] [PubMed] [Google Scholar]
- 51.Carpenter EP, Corbett A, Thomson H, Adacha J, Jensen K, Bergeron J, et al. AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. Embo J. 2007;26(5):1363–72. 10.1038/sj.emboj.7601593 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lakomek K, Dickmanns A, Ciirdaeva E, Schomacher L, Ficner R. Crystal structure analysis of DNA uridine endonuclease Mth212 bound to DNA. J Mol Biol. 2010;399(4):604–17. 10.1016/j.jmb.2010.04.044 . [DOI] [PubMed] [Google Scholar]
- 53.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643(1–2):54–63. 10.1016/j.mrfmmm.2008.04.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidal AE, Harkiolaki M, Gallego C, Castillo-Acosta VM, Ruiz-Perez LM, Wilson K, et al. Crystal structure and DNA repair activities of the AP endonuclease from Leishmania major. J Mol Biol. 2007;373(4):827–38. 10.1016/j.jmb.2007.08.001 . [DOI] [PubMed] [Google Scholar]
- 55.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature. 2000;403(6768):451–6. 10.1038/35000249 . [DOI] [PubMed] [Google Scholar]
- 56.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21(8):668–76. . [DOI] [PubMed] [Google Scholar]
- 57.Demple B, Johnson A, Fung D. Exonuclease III and endonuclease IV remove 3’ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci U S A. 1986;83(20):7731–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168(3):1120–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi M, Lillis R, Demple B, Takeshita M. Interactions of Escherichia coli endonuclease IV and exonuclease III with abasic sites in DNA. J Biol Chem. 1994;269(34):21907–14. . [PubMed] [Google Scholar]
- 60.Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, et al. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc Natl Acad Sci U S A. 2013;110(39):E3695–703. 10.1073/pnas.1305624110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3’-5’ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34(9):2508–15. 10.1093/nar/gkl259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lirussi L, Antoniali G, Vascotto C, D’Ambrosio C, Poletto M, Romanello M, et al. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol Biol Cell. 2012;23(20):4079–96. 10.1091/mbc.E12-04-0299 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13(9):5370–6. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishchenko AA, Deprez E, Maksimenko A, Brochon JC, Tauc P, Saparbaev MK. Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc Natl Acad Sci U S A. 2006;103(8):2564–9. 10.1073/pnas.0508582103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domains and their families were assigned according to the Conserved Domain Database [48].
(DOCX)
(PDB)
(A) Duplex oligonucleotides used to measure 3’-repair phosphodiesterase and 3’-5’ exonuclease activities. (B) Chemical structures of abasic sites and alpha-anomeric 2’deoxyadenosine.
(TIF)
Lane 1, Protein Ladder (Thermo Scientific, cat. # 26616); lane 2, 1 μg HpXth; lane 3, 1 μg HpXth-D144N. For details, see Materials and methods.
(TIF)
Briefly, 10 nM 5’-[32P]-labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min at 37 °C in the buffer containing various concentrations of metal cations. Products of the reaction were resolved by denaturing PAGE, then visualized by phosphorimaging and quantified in ImageQuant. For details, see Materials and methods.
(TIF)
In brief, 10 nM 5’-[32P]-labeled 30mer THF•T duplex was incubated with 0.2 nM HpXth for 5 min at 37 °C in the standard reaction buffer but with varying concentrations of MgCl2 and ZnCl2, unless otherwise stated. Lane 1, THF•T and 0.1 nM APE1 in the buffer BER; lane 2, control THF•T, no enzyme; lanes 3–4, as in lane 1 but supplemented with 0.1 and 1 mM ZnCl2, respectively; lane 5, THF•T and 0.2 nM HpXth in the buffer without metal cations; lane 6, as in lane 5 but 1 mM MgCl2; lane 7, as in lane 6 but 0.1 mM ZnCl2; lane 8, as in lane 6 but 1 mM ZnCl2; lane 9, as in lane 5 but 5 mM MgCl2; lane 10, as in lane 9 but 0.1 mM ZnCl2; lane 10, as in lane 9 but 1 mM ZnCl2. The arrows denote the position of the 30mer substrate and a 10mer cleavage product. For details, see Materials and methods.
(TIF)
HpXth model superimposed over human APE1 structure (1BIX). Sm3+ ion replacing catalytic Mg2+ in the crystal structure is shown as a magenta ball. Amino acids labels show numbering in HpXth first, followed by numbering in the corresponding human or E. coli protein. Cartoon traces and carbon atoms of HpXth model are colored green, those of human and E. coli structures are cyan, oxygen and nitrogen atoms are red and blue, respectively.
(TIF)
Four μg of cell-free extracts from the BH110 (DE3) strains carrying different plasmids were separated using 12% SDS-PAGE and transferred to the PVDF membrane and then analyzed by Western blotting. (A) Western blot analysis of the PVDF membrane using rabbit anti-HpXth polyclonal antiserum. Lane 1, protein molecular weight markers; lane 2, control plasmid-free E. coli BH110 (DE3) strain; lane 3, empty vector pBluescript II SK+ (pBSK); lane 4, pBSK-HpXth; lane 5, pBSK-HpXth-D144N; lane 6, 50 ng of the purified His-tagged HpXth; lane 7, as 6 but 100 ng. (B) Ponceau S-staining of the PVDF membrane. Lanes 1–7, same as in panel A. For details, see Materials and methods.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.