Bat and mouse beige adipocytes induced from white adipocytes provide insights into therapy against obesity-related disease.
Abstract
Beige adipocytes can be induced from white adipocytes and precursors upon stimulation by cold temperatures and act like brown adipocytes to increase energy expenditure. Most in vivo studies examining the mechanisms for the induction of beige adipocytes have focused on subcutaneous white adipose tissue (sWAT; benign fat) in the mouse. How intra-abdominal WAT (aWAT; malignant fat) develops into beige adipocytes remains obscure, largely because there is a lack of a good animal model for the induction of beige adipocytes from aWAT. To better understand the development of beige adipocytes from mammalian WATs, especially aWAT, we induced beige adipocytes from bat aWAT and mouse sWAT by exposure to cold temperatures and analyzed their molecular signatures. RNA sequencing followed by whole genome–wide expression analysis shows that beige adipocytes induced from bat aWAT, rather than sWAT, have molecular signatures resembling those of mouse sWAT-induced beige adipocytes and exhibit dynamic profiles similar to those of classical brown adipocytes. In addition, we identified molecular markers that were highly enriched in beige adipocytes and conserved between bat aWAT and mouse sWAT, a set that included the genes Uqcrc1 and Letm1. Furthermore, knockdown of Uqcrc1 and Letm1 expression shows that they are required not only for beige adipocyte differentiation but also for preadipocyte maturation. This study presents a new model for research into the induction of beige adipocytes from aWAT in vivo, which, when combined with models where beige adipocytes are induced from sWAT, provides insight into therapeutic approaches for combating obesity-related diseases in humans.
INTRODUCTION
Brown and white adipose tissue (BAT and WAT, respectively) have different physiological roles in mammals and can be distinguished by their appearance and metabolic features (1). Recent studies reported that brown-like (beige) adipocytes could be a distinct type of thermogenic fat cell and are induced from WAT by exposure to cold and other stimuli (2). The inducible nature of beige adipocytes has gained significant attention owing to its potential application in the treatment of obesity and obesity-related diseases (3). Ectopic abdominal WAT (aWAT) is associated with metabolic dysfunction, such as diabetes and cardiovascular disease, while the presence of increased levels of subcutaneous WAT (sWAT) is associated with only a low risk of metabolic disease (4). Therefore, discovering mechanisms to induce beige fat from aWAT in vivo is of great clinical importance.
Most in vivo studies examining the molecular signatures and regulatory pathways of beige fat development have focused on sWAT in mice, since the major sWAT depot, inguinal WAT, is highly susceptible to browning. However, intra-abdominal epididymal WAT of male mice is quite resistant to browning (5, 6). Further studies reported that the mechanism for beige fat cell induction in epididymal WAT is distinct from that seen in inguinal WAT (7). In adult humans, abdominal sWAT and intra-abdominal omental WAT were reported to have the capacity to activate a brown fat–like differentiation program by differentiating cells in vitro (8, 9). Our recent work in bat (great roundleaf bat) indicated that aWAT (WAT depots between the anterior abdominal wall and the visceral mass) predominantly transdifferentiated into beige fat rather than sWAT (subcutaneous fat at the anterior abdominal wall) when exposed to a cold environment (10). To understand the precise biological mechanism of aWAT browning in vivo, we collected bat aWAT, as well as sWAT and iBAT (interscapular BAT), and compared their molecular signatures to different mouse adipose depots. To provide insights into the therapeutic approach of combating obesity in human, we focused on molecular signatures that are shared between bat aWAT and mouse sWAT, which are more likely to be conserved in the human browning process.
BAT and beige fat expend chemical energy to produce heat and function as a natural defense system against hypothermia, particularly in infants, small mammals, and hibernating animals (11). Besides BAT, which is strikingly observed in seasonal mammals to maintain body temperature during periods of hibernation, beige fat is also found to be essential for the survival of small mammals in cold environments. The great roundleaf bat (Hipposideros armiger) (12), as a small and hibernating mammal, evolved with strong cold-adaptive mechanisms. Our previous study demonstrated that aWAT rather than sWAT in the great roundleaf bat displayed a greater number of beige fat characteristics both in morphology and in expression profile of BAT marker genes (10). All these observations make the great roundleaf bat a good potential model for investigating the mechanisms for the induction of beige fat from aWAT in vivo.
Here, we examine in greater detail the morphological ultrastructure and the expression profiles of all genes in distinct types of adipose tissue from traditional mouse and our novel bat models. We apply mRNA sequencing (mRNA-seq) technology to adipose tissues from both bat and mouse after exposure to cold temperatures and thermoneutral temperatures. Transcriptome profiles revealed that bat aWAT and mouse sWAT show changes in gene expression that are similar with those of classical BAT after cold exposure. Highly expressed marker genes of BAT and beige fat were uncovered and mapped to metabolic and regulatory pathways to identify induction programs shared by beige adipocytes from both aWAT and sWAT. Finally, we assayed the function of two novel beige marker genes, Uqcrc1 and Letm1, which were found to promote adipogenesis and induce beige-like adipocytes. Furthermore, inhibition of adipogenesis could be partially recovered by increasing insulin in Letm1 knockdown cell lines. Our studies suggest that common regulatory mechanisms of the induction of beige fat might exist among mammalian species and thus potentially could be targets for therapy of adipose tissue–mediated diseases in humans.
RESULTS
Gene expression profiles of in vivo brown and beige fat tissues after exposure to cold
To characterize the morphological differences between aWAT and sWAT adipocytes after cold stimulation, we observed the ultrastructure of these tissues in bats and mice under thermoneutral and cold conditions. We also observed BAT adipocytes for comparison. Similar to sWAT in mice after exposure to cold temperatures for 1 week, bat aWAT adipocytes showed relatively large, round, and condensed mitochondria with numerous transverse cristae surrounding smaller lipid droplets (Fig. 1, E and K). Mouse sWAT and bat aWAT changed to a deeper color after cold stimulation, which is indicative of the browning of white adipocytes (Fig. 1, E and K). After cold stimulation, both bat aWAT and mouse sWAT changed to generate an ultrastructure that was more similar to that seen in thermoneutral iBAT (Fig. 1, C, E, I, and K) than to that in bat sWAT and mouse aWAT at thermoneutral temperatures (Fig. 1, D and J).
Fig. 1. Photographs of the anatomical and morphological ultrastructures of different types of fat depots in bat and mouse.
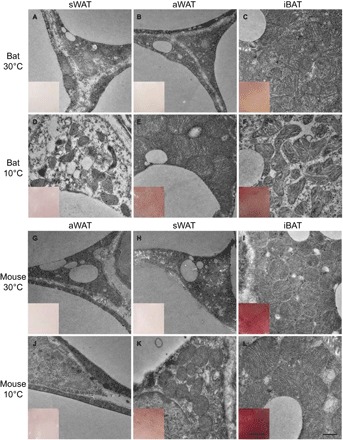
(A to F) Bat sWAT, aWAT, and iBAT under 30°C (A to C) and 10°C (D to F). (G to L) Mouse aWAT, sWAT, and iBAT under 30°C (G to I) and 10°C (J to L). Scale bars, 0.5 μm (for ultrastructure) and 1 mm (for the dissected tissues in the inset windows).
To investigate changes in gene expression patterns upon exposure to cold, we performed RNA-seq for aWAT, sWAT, and iBAT from bats and mice exposed to 30° or 10°C for 1 week. A total of 11,166 genes were expressed in all 20 samples, and 14,871 genes were expressed in at least one of the samples. The Ensembl gene IDs of mapped genes and raw read counts in the 20 samples are listed in table S1. Raw mRNA-seq data files and FPKM (expected fragments per kilobase of transcript per million fragments sequenced) values were submitted to the Gene Expression Omnibus (GEO) database with accession no. GSE72603.
Similar BAT-like gene expression profiles of bat aWAT and mouse sWAT after cold stimulation
Genome-wide hierarchical clustering revealed that adipocytes could be divided into two main clusters that represent the species (Fig. 2A). sWAT from mice at 10°C clustered with classical brown adipocytes (iBAT), with the closest clustering with BAT in a cold-induced state (10°C iBAT). In contrast, in the bat, 10°C aWAT, rather than sWAT, showed a profile similar to that of iBAT and was distinct from sWAT (Fig. 2A). As an independent approach, a multidimensional scaling (MDS) analysis of differentially expressed genes (DEGs) suggested that the gene expression profiles of the bat 10°C aWAT and mouse 10°C sWAT were more similar to those of iBAT than those from tissue at 30°C (Fig. 2B). To demonstrate concordant changes in mouse sWAT and bat aWAT after cold stimulation, we examined the correlation of gene expression between these tissues before and after cold exposure. The Pearson’s correlation coefficients increased from 0.61 at thermoneutral condition to 0.89 after cold exposure, which suggests similar dynamics for the molecular signatures between mouse sWAT and bat aWAT after cold exposure. These results suggest that cold-stimulated bat aWAT and mouse sWAT exhibit similar dynamic changes in gene expression profiles as they were induced into BAT-like beige adipocytes.
Fig. 2. Analysis of gene expression and markers for bat aWAT and mouse sWAT.
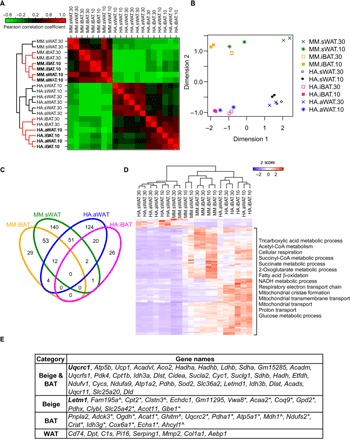
(A) Genome-wide gene expression profile and hierarchical clustering. (B) MDS of the DEGs from bat and mouse adipocytes. (C) Venn diagram of the DEGs of 10°C versus 30°C groups in bat and mouse iBATs and beige adipocytes. (D) Gene ontology (GO) analysis of the overlapping marker genes in bat and mouse. NADH, reduced form of nicotinamide adenine dinucleotide. (E) Top common markers of beige adipocytes, BAT, and WAT in bat and mouse. *Mouse beige adipocytes and BAT markers. ^Bat beige adipocytes and BAT markers. HA, H. armiger; MM, Mus musculus.
DEGs and functional assessment of marker genes
To find common key factors in the browning process of aWAT and sWAT and compare them with iBAT markers, we identified genes that were in the overlapping sets of enriched genes in bat and mouse iBATs and beige adipocytes (Fig. 2C). We first analyzed the overlapping gene sets between bat and mouse beige adipocyte–enriched genes. We found a total of 65 genes to be enriched in both bat and mouse beige adipocytes (Fig. 2C). Among these genes, 14 are also differentially expressed in classical brown adipocytes in mice (12 genes) or classical brown adipocytes in bats (2 genes) upon cold stimulation. These genes include the previously defined beige markers Ucp1, Cidea, and Cox7a1 (6), as well as new markers such as Uqcrc1 and Letm1. In addition, some of the markers were also identified in human brown adipocytes, whose profile is similar to mouse beige adipocytes, such as Acadvl, Pdk4, and Dld (13). We identified a total of 140 and 124 species-specific beige marker genes in mice and bats, respectively. BAT and WAT marker genes were also identified. Top markers for beige adipocytes, BAT, and WAT are listed in Fig. 2E, with all potential marker genes listed in table S2. Only seven WAT markers were identified in both bat and mouse, but a substantial number of WAT markers were identified either in bat or in mouse. These species-specific genes may contribute to the biological difference between sWAT and aWAT.
To examine the importance and function of the marker genes, we clustered the expression profiles of the marker genes in common between mouse and bat across 20 samples. Mouse sWAT-induced beige depots and bat aWAT-induced beige depots were grouped into the cluster of iBAT depots, all other WAT samples were clustered together (Fig. 2D). A similar cluster tree was obtained for all the marker genes including species-specific markers (fig. S1). Subsequent GO analysis showed that the common marker genes were enriched in metabolic pathways involved in beige and BAT thermogenesis, such as tricarboxylic acid metabolism, respiratory electron transport chain, mitochondrial transmembrane transport, cellular respiration, fatty acid β-oxidation, mitochondrial cristae formation, and acetyl CoA (coenzyme A) metabolism (Fig. 2D), which indicate a thermogenic role for these marker genes. In addition, genes involved in macrophage activation and cell-cell adhesion were significantly enriched in the white adipocytes from bat, and genes related to the immune response were enriched in white adipocytes from mice, which is in accord with previous studies (14–17).
Functional analysis of new marker genes in beige adipocytes
We validated the expression of six beige marker genes enriched in beige adipocytes in both mouse and bat using real-time quantitative polymerase chain reaction (RT-qPCR; fig. S2). The results were in agreement with those from mRNA-seq in both species. We then selected two novel beige marker genes, Uqcrc1 and Letm1, for functional analysis. Uqcrc1 (ubiquinol-cytochrome c reductase core protein 1) encodes a subunit of mitochondrial respiratory complex III, which is a key component of the respiratory chain (18). Letm1 (leucine zipper-EF-hand containing transmembrane protein 1) is a conserved mitochondrial inner membrane protein. RNA-seq results revealed that the expression of both genes was significantly up-regulated in bat aWAT and mouse sWAT after exposure to cold compared to thermoneutral conditions (table S1), but their functions during the browning process were unknown.
To study the biological function of Uqcrc1 and Letm1, we experimentally differentiated preadipocyte 3T3-L1 cells into white adipocytes and beige-like adipocytes (Fig. 3, A and B). We found that the mRNA expression of Uqcrc1 and Letm1 was significantly higher in the induced beige-like adipocytes than in the white adipocytes and preadipocytes (Fig. 3, C and D), suggesting potential functions in beige adipocytes. Preadipocyte 3T3-L1 cells were transfected with small interfering RNAs (siRNAs) targeting Uqcrc1 and Letm1, followed by induction into white adipocytes and beige-like adipocytes. RNA and protein expression of both Uqcrc1 and Letm1, estimated by RT-qPCR and Western blot, respectively, was decreased substantially compared to control siRNA-transfected cells in preadipocytes, white adipocytes, and beige-like adipocytes (Fig. 3, C to E, and fig. S3). Induced beige-like adipocytes depleted of Uqcrc1 (LV-Uqcrc1-sh) or Letm1 (LV-Letm1-sh) showed significantly (P < 0.001) lower mRNA and protein expressions of Ucp1 and Pgc-1α [encoding peroxisome proliferator–activated receptor γ (PPARγ) coactivator 1α], compared to control and LV-Scr-sh–transfected cells (Fig. 3, F and G, and fig. S3). Expression of brown adipocyte– and beige adipocyte–selective molecular markers, such as C/EBPβ, Prdm16, Cd137, Tmem26, Cited1, and Tbx1, was significantly lower in cells transfected with the siRNAs targeting Uqcrc1 and Letm1 than in control siRNA-transfected cells (Fig. 3G). These results indicate that Uqcrc1 and Letm1 affect other beige marker genes and are essential for the induction of beige adipocytes.
Fig. 3. Functional analysis of Uqcrc1 and Letm1 in beige adipocyte differentiation.
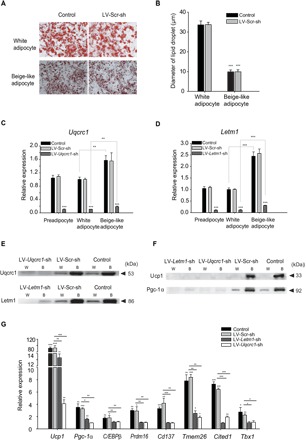
(A) Oil Red O staining of differentiated white adipocyte cultures and beige-like adipocyte cultures. Scale bar, 50 μm. (B) Diameter of lipid droplets was calculated using ImageJ software. (C and D) Expression of Uqcrc1 and Letm1 in 3T3-L1 preadipocytes and experimentally differentiated adipocytes. (E and F) Protein expression of Uqcrc1, Letm1, Ucp1, and Pgc-1α. W and B represent white and beige adipocytes, respectively. (G) Gene expression of Ucp1, Pgc-1α, C/EBPβ, Prdm16, Cd137, Tmem26, Cited1, and Tbx1. n = 3 for all groups. Data are means ± SD for all bar graphs. *P < 0.05; *P < 0.01; ***P < 0.001. Significant analysis was performed by one-sided Student’s t test.
Function of Letm1 in adipogenesis and sensitivity to insulin
During the induction of siRNA-affected beige adipocytes, we identified additional roles for Uqcrc1 or Letm1 in adipogenesis. Expression of the adipogenic marker Fabp4 (fatty acid–binding protein 4) was significantly higher in differentiated white adipocytes than in preadipocytes but substantially decreased in white adipocytes depleted of Uqcrc1 or Letm1 (Fig. 4A). As assessed by Oil Red O staining, preadipocyte 3T3-L1 cells transfected with siRNAs targeting Uqcrc1 (LV-Uqcrc1-sh) or Letm1 (LV-Letm1-sh) had lower adipogenic potential when induced into white adipocytes and displayed a significantly decreased lipid droplet content and size compared to control and LV-Scr-sh–transfected cells (Fig. 4B). These results suggest that silencing of Uqcrc1 and Letm1 suppresses preadipocyte differentiation and further inhibits adipocyte browning. When trying to induce beige adipocytes using insulin, we found that increasing insulin concentrations generated increased numbers of lipid droplets and that expression of Fabp4 increased in parallel with insulin in white adipocytes (Fig. 4, C and D). In contrast, we observed no significant changes in Uqcrc1 knockdown adipocytes even at the highest insulin concentration. These results suggest that the inhibition of adipogenesis by depleted Letm1 could be partially recovered by increasing insulin dose, which suggests that Letm1 has a role equivalent with insulin. Previous studies reported that Letm1 is associated with the development of insulin resistance through the insulin/PI3K (phosphoinositide 3-kinase)/PKB (protein kinase B) signaling pathway (19) and that insulin, through PI3K, regulates adipogenesis in both brown and white fat (20, 21). On the basis of these findings, it is possible that Letm1 controls adipogenesis and adipocyte browning by regulating insulin through PI3K (Fig. 4E).
Fig. 4. Letm1 function in adipogenesis and sensitivity to insulin concentration.
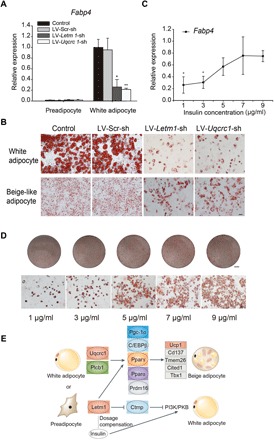
(A) Fabp4 expression in 3T3-L1 preadipocytes and differentiated white adipocytes. (B) Oil Red O staining of differentiated white adipocyte cultures and beige-like adipocyte cultures. (C) Fabp4 expression in differentiated white adipocytes from Letm1-depleted preadipocytes treated with increasing insulin concentrations. (D) Oil Red O staining of differentiated white adipocytes treated with increasing insulin concentrations. Scale bars, 5 mm (for the round dishes) and 50 μm (for microscopic Oil Red O staining). (E) Proposed regulatory pathways of Uqcrc1 and Letm1 in the development of beige adipocytes and Letm1 function in adipogenesis. n = 3 for all groups. Data are means ± SD for all bar graphs. *P < 0.05; **P < 0.01. Significant analysis was performed by one-sided Student’s t test.
DISCUSSION
Here, we identified a new model for examining the developmental programs of beige fat in aWAT. Most studies in adult humans were performed on BAT depots, which were reported to be analogous to beige fat in rodents (2, 22). However, increased aWAT is a major risk factor for metabolic disease in humans. The precise induction program for beige adipocytes in human WAT depots remains obscure. Full inducibility of beige fat under relatively mild temperatures and easy access to great roundleaf bats make them a potential model system for studying beige fat formation from aWAT in humans. Although human-specific signatures might exist, this study focused on conserved signatures for the browning of aWAT and sWAT across mammals to provide new opportunities for obesity therapy.
Uqcrc1 is reported to interact with Plcb1 (23, 24), which affects insulin secretion by targeting PPARγ in pancreatic cells (25). Here, depletion of Uqcrc1 led to down-regulated expression of Ucp1, Prdm16, C/EBPβ, and Pgc-1α. Pgc-1α is a coactivator of PPARγ (26). These results support the possibility that Uqcrc1 interacts with Plcb1 to regulate the expression of Prdm16, C/EBPβ, and Pgc-1α by targeting PPARγ and affecting insulin secretion to control the development of beige adipocytes, therefore up-regulating the thermogenic marker gene Ucp1 (Fig. 4E).
This study discovered beige-selective markers in aWAT induced by exposure to cold temperatures in vivo. Our genome-wide RNA-seq data analyses demonstrate that beige fat induced from bat aWAT displays molecular signatures that resemble that of mouse sWAT, which indicates that WAT from different developmental origins (27) or species have common factors for beige formation. To gain further insights into therapeutic potential for human obesity, we compared the identified markers to the brown fat– and beige fat–selective genes previously reported to be enriched in the human supraclavicular region and mouse classical brown and subcutaneous beige fats (27). Around 30% of these 99 genes are found in our brown or beige marker groups, suggesting pivotal roles for the browning process or thermogenic function since they are conserved across different species and tissues.
Although this study focused on markers that overlap between mouse and bat, a large number of species-specific or tissue-specific markers were also identified (table S2). For the marker genes enriched specifically in bat beige adipocytes, 24.3% of them are observed to have induced expression with more than twofold changes but did not reach the stringent criteria for identifying the marker genes, such as Grpel1, Mrpl12, and Mcee. About 13.9% genes show moderate increased expression upon cold stimulation in mouse sWAT, such as Lace1, Rbp4, and Tubb4b. Only 7.3% genes show undetectable expression in mouse. Other genes are detected either without significant changes (19.1%) or at low level (35.3%) in mouse. These specific beige marker genes need further experimental validation and should be valuable for probing species- or tissue-specific markers in the browning process.
About 40% of the identified markers are supported by data from recent reports on beige adipocytes in mouse or human; however, many additional novel markers, including Uqcrc1 and Letm1, were identified. Together, this research presents potential targets for therapeutic intervention in obesity and related disease and constitutes an important foundation for future studies on beige adipocytes, WAT, and BAT.
MATERIALS AND METHODS
Adipose tissue sampling and mRNA-seq library design
Experimental procedures were conducted according to our previous methods including those for the collection of bat samples, housing of bats and mice, temperature acclimation, sacrifice, and tissue sample collection (10). All animal experiments were conducted under the Guidelines and Regulations for the Administration of Laboratory Animals (Decree No. 2, the State Science and Technology Commission of the People’s Republic of China, 14 November 1998) and were approved by the Animal Ethics Committee of East China Normal University (ID no. AR2012/03001). In brief, bats and mice were housed in individual cages at temperatures of 30° and 10°C for 7 days, with sufficient food and water supply. The 30°C group was used as the control, as this is the normal summer environmental temperature for these bats. Three types of adipose depots were collected (iBAT, sWAT, and aWAT) for each species. iBAT depots were collected from the largest interscapular region. sWAT was collected from adipocytes beneath the skin and above the abdominal wall in bats and from the inguen in the mice. aWAT was collected from the fat tissue between the abdominal wall and the viscera. Two biological replicates (a male and a female) were used for mRNA-seq, and three other biological replicates were used for RT-qPCR. Total RNA from adipose tissues was extracted using the RNeasy Lipid Tissue Mini Kit (Qiagen). mRNA libraries were sequenced by the HiSeq 2000 sequencing system (Illumina) according to standard protocols.
Sequencing analysis
Clean data were obtained after trimming adapter sequences from the raw sequence reads and removing reads with unknown “N” bases. Filtered reads from bats were mapped to the known reference H. armiger genome (12). Gene symbols for bat transcripts were assigned on the basis of their best alignment by BLAST to mouse genes (Ensembl). For the mouse data, we conducted a similar analysis based on the mouse genome (mm74) and mouse gene annotation from Ensembl. Raw read counts were extracted from the bam file using Bedtools. The abundances of the mRNA-seq reads were reported in FPKM using the TopHat-Cufflinks pipeline to evaluate the expression level of transcripts (28). DEGs between the 10° and 30°C samples were identified using edgeR package and were those showing a twofold or more changes after log2-transformed and a false discovery ratio (FDR) <0.05 with induced beige expression greater than 10 counts per million reads. BAT- and WAT-selective markers were identified by comparing the expression profile between BAT and WAT with log2 fold change ≥1.5 and FDR <0.05 and basal expression of markers greater than 95% for all expressed genes in each adipose depot.
Clustering analysis, MDS plot, and GO analysis
Hierarchical clustering was performed using gplots package in R based on the FPKM data for all genes after z score transformation according to a previous study (29, 30). The MDS plot was performed by edgeR using 164 significant DEGs (log2 fold change ≥1.5 and FDR <0.05) upon cold stimulation in both the bat aWAT and mouse sWAT. Heat maps and hierarchical clustering of overlapped beige adipocyte, BAT, and WAT marker genes in bat and mouse were constructed using gplots (30). GO analysis was performed using PANTHER (9) with significant P values (P < 0.05) by filtering out the top hierarchical GO terms.
Real-time quantitative polymerase chain reaction
RNA (1 μg) was reversely transcribed using the ABI High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-qPCR was performed using SYBR Premix Ex Taq (TaKaRa) on an ABI/Prism 7300 instrument (Applied Biosystems) with three biological replicates. Relative gene expression level was evaluated by the ΔΔCt method with endogenous control gene Tbp (31). Primer sequences used in this study are listed in table S3.
Transmission electron microscopy
Samples were fixed in 2.5% glutaraldehyde in 100 nM phosphate buffer (pH 7.4) at 4°C overnight, followed by 1% osmium tetroxide, and then embedded in Epon 812. After being stained with uranyl acetate and lead citrate, ultrathin sections of 70 nm were obtained and observed using a JEM-2100 transmission electron microscope (JEOL Ltd.), with the images recorded by a Gatan CCD 832 digital detector.
Cell culture and differentiation
Cell lines, including 3T3-L1 preadipocytes and 293T, were cultured in normal growth media consisting of Dulbecco’s modified Eagle’s medium (Hyclone, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Hyclone, Thermo Fisher Scientific). All cells were grown at 37°C in a humidified atmosphere of 5% CO2. To induce white adipocytes, 3T3-L1 preadipocytes were treated with differentiation medium I (DMI) from days 2 to 5 (postconfluence designated as day 0). DMI is composed of normal growth media, insulin (1 μg/ml; Sigma), dexamethasone (1 μM; Sigma), and 3-isobutyl-1-methyl-xanthine (0.5 mM; IBMX, Sigma). Cells were then switched to DMII, which includes normal growth medium and insulin (1 μg/ml), from days 5 to 9, after which they were placed back into normal growth medium and refed every 2 days (32). To induce beige adipocytes, 0.5 μM rosiglitazone (Rosi, Sigma) and 0.5 mM IBMX were added into DMII, and then, the cells were exposed to 10 μM isoproterenol (Iso, Sigma) for 4 hours before harvesting (33, 34).
Oil Red O staining
Differentiated 3T3-L1 cells were rinsed with phosphate-buffered saline twice and fixed with 4% paraformaldehyde for 60 min at room temperature. After being washed with 60% isopropanol, cells were stained for 10 min in a diluted Oil Red O solution. Stained cells were washed four times with double-distilled H2O, visualized using light microscopy (Olympus) and photographed (35).
Lentivirus production and viral infection
To produce a plasmid that expresses short hairpin RNA, the oligonucleotides were annealed and ligated into the pLKO.1 vector (Institute Pasteur of Shanghai Chinese Academy of Sciences). The oligo sequences are as follows: Letm1, 5′-CCGGTGATAATAAGGACGGCAATATCTCGAGATATTGCCGTCCTTATTATCATTTTTG-3′ (forward) and 5′-AATTCAAAAATGATAATAAGGACGGCAATATCTCGAGLTATTGCCGTCCTTATTATCA-3′ (reverse); Uqcrc1, 5′-CCGGGAATGCTTTGGTGTCTCATTTCTCGAGAAATGAGACACCAAAGCATTCTTTTTG-3′ (forward) and 5′-AATTCAAAAAGAATGCTTTGGTGTCTCATTTCTCGAGAAATGAGACACCAAAGCATTC-3′ (reverse). A random sequence for LV-Scr-sh is as follows: 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′ (forward) and 5′-AATTCAAAAACAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG-3′ (reverse). For lentivirus production, a recombinant vector plasmid and packaging plasmids (9.2ΔR and VSV-G) were cotransfected into 293T cells using polyethylenimine (Polysciences). Media were refreshed, and the supernatant was collected after 48 and 72 hours and filtered through 0.45-μm filters. 3T3-L1 preadipocytes were transduced with lentivirus in the presence of polybrene (8 μg/ml; Sigma). After 5 hours, the media were changed. For selection of successfully transduced cells, infected cells were incubated in fresh medium supplemented with puromycin (2 μg/ml; Amresco) the next day and then cultured in this medium for 72 hours. After harvesting the cells, the knockdown efficiency in the 3T3-L1 preadipocytes was analyzed by RT-qPCR.
Antibodies and Western blotting
Cultured 3T3-L1, LV-Scr-sh 3T3-L1, LV-Letm1-sh 3T3-L1, and LV-Uqcrc1-sh 3T3-L1 cells were homogenized in lysis buffer [RIPA (radioimmunoprecipitation assay) Lysis Buffer I, Sangon Biotech) and were loaded onto the gel for electrophoresis. Proteins were then transferred onto the polyvinylidene difluoride (PVDF) membrane and immunoblotted with antibodies, Letm1, Uqcrc1, Ucp1 (Santa Cruz Biotechnology), and Pgc-1α (Abcam). Equal loading was controlled by Ponceau staining. The volume of each band on the PVDF membrane was normalized to that of the corresponding Ponceau-stained protein bands, which served as a linearity loading control as described previously (36, 37).
Supplementary Material
Acknowledgments
We thank X. Wang, S. Mu, and X. Qin for the assistance with data analysis. Funding: This work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (The National Key Research and Development Program, nos. 2016YFD0500300 and 2016YFC1200100), the National Natural Science Foundation of China (nos. 31672274 and 31570382), and the Department of Education of Liaoning Province (Climbing Scholar). Author contributions: S.Z., Z.W., and H.X. designed the study. H.X., Z.W., and Y.W. analyzed the data. Y.H., S.K., J.Z., Y.-H.P., and W.H. performed the experiments. H.X., Z.W., D.M.I., and S.Z. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The sequencing data set described in this paper has been submitted to the National Center for Biotechnology Information GEO database (www.ncbi.nlm.nih.gov/geo/; accession no. GSE72603). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaar5319/DC1
Fig. S1. Clustered profile of all marker genes.
Fig. S2. Expression levels of six beige marker genes assayed by RT-qPCR.
Fig. S3. Ponceau staining as loading control in Western blots.
Table S1. Mapped Ensembl genes and raw read counts from 20 samples.
Table S2. All beige adipocyte, BAT, and WAT marker genes.
Table S3. Primers used for qPCR.
REFERENCES AND NOTES
- 1.Cinti S., The adipose organ. Prostaglandins Leukot. Essent. Fatty Acids 73, 9–15 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A.-H., Khandekar M., Virtanen K. A., Nuutila P., Schaart G., Huang K., Tu H., van Marken Lichtenbelt W. D., Hoeks J., Enerbäck S., Schrauwen P., Spiegelman B. M., Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sidossis L., Kajimura S., Brown and beige fat in humans: Thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125, 478–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Gurmaches J., Hung C.-M., Sparks C. A., Tang Y., Li H., Guertin D. A., PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 16, 348–362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seale P., Conroe H. M., Estall J., Kajimura S., Frontini A., Ishibashi J., Cohen P., Cinti S., Spiegelman B. M., Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 121, 96–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W., Seale P., Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 17, 691–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y. H., Petkova A. P., Mottillo E. P., Granneman J. G., In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartesaghi S., Hallen S., Huang L., Svensson P. A., Momo R. A., Wallin S., Carlsson E. K., Forslöw A., Seale P., Peng X.-R., Thermogenic activity of UCP1 in human white fat-derived beige adipocytes. Mol. Endocrinol. 29, 130–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elabd C., Chiellini C., Carmona M., Galitzky J., Cochet O., Petersen R., Pénicaud L., Kristiansen K., Bouloumié A., Casteilla L., Dani C., Ailhaud G., Amri E.-Z., Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells 27, 2753–2760 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Zhu T., Ke S., Fang N., Irwin D. M., Lei M., Zhang J., Shi H., Zhang S., Wang Z., The great roundleaf bat (Hipposideros armiger) as a good model for cold-induced browning of intra-abdominal white adipose tissue. PLOS ONE 9, e112495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townsend K., Tseng Y.-H., Brown adipose tissue: Recent insights into development, metabolic function and therapeutic potential. Adipocyte 1, 13–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong D., Lei M., Hua P., Pan Y.-H., Mu S., Zheng G., Pang E., Lin K., Zhang S., The genomes of two bat species with long constant frequency echolocation calls. Mol. Biol. Evol. 34, 20–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinoda K., Luijten I. H. N., Hasegawa Y., Hong H., Sonne S. B., Kim M., Xue R., Chondronikola M., Cypess A. M., Tseng Y.-H., Nedergaard J., Sidossis L. S., Kajimura S., Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 21, 389–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axelsson J., Heimbürger O., Lindholm B., Stenvinkel P., Adipose tissue and its relation to inflammation: The role of adipokines. J. Ren. Nutr. 15, 131–136 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Zeyda M., Stulnig T. M., Adipose tissue macrophages. Immunol. Lett. 112, 61–67 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Tordjman J., Guerre-Millo M., Clément K., Adipose tissue inflammation and liver pathology in human obesity. Diabetes Metab. 34, 658–663 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Henegar C., Tordjman J., Achard V., Lacasa D., Cremer I., Guerre-Millo M., Poitou C., Basdevant A., Stich V., Viguerie N., Langin D., Bedossa P., Zucker J.-D., Clement K., Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 9, R14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai V. G., Lee T., Delongchamp R. R., Moland C. L., Branham W. S., Fuscoe J. C., Leakey J. E. A., Development of mitochondria-specific mouse oligonucleotide microarray and validation of data by real-time PCR. Mitochondrion 7, 322–329 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Park J., Li Y., Kim S.-H., Yang K.-J., Kong G., Shrestha R., Tran Q., Park K. Ah., Jeon J., Hur G. M., Lee C.-H., Kim D.-H., Park J., New players in high fat diet-induced obesity: LETM1 and CTMP. Metabolism 63, 318–327 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Cannon B., Nedergaard J., Brown adipose tissue: Function and physiological significance. Physiol. Rev. 84, 277–359 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Rosen E. D., MacDougald O. A., Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Sharp L. Z., Shinoda K., Ohno H., Scheel D. W., Tomoda E., Ruiz L., Hu H., Wang L., Pavlova Z., Gilsanz V., Kajimura S., Human BAT possesses molecular signatures that resemble beige/brite cells. PLOS ONE 7, e49452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piazzi M., Blalock W. L., Bavelloni A., Faenza I., D’Angelo A., Maraldi N. M., Cocco L., Phosphoinositide-specific phospholipase C β 1b (PI-PLCβ1b) interactome: Affinity purification-mass spectrometry analysis of PI-PLCβ1b with nuclear protein. Mol. Cell. Proteomics 12, 2220–2235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatr-aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N. K., O’Donnell L., Oster S., Theesfeld C., Sellam A., Stark C., Breitkreutz B.-J., Dolinski K., Tyers M., The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiume R., Ramazzotti G., Faenza I., Piazzi M., Bavelloni A., Billi A. M., Cocco L., Nuclear PLCs affect insulin secretion by targeting PPARγ in pancreatic β cells. FASEB J. 26, 203–210 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Boström P., Wu J., Jedrychowski M. P., Korde A., Ye L., Lo J. C., Rasbach K. A., Boström E. A., Choi J. H., Long J. Z., Kajimura S., Zingaretti M. C., Vind B. F., Tu H., Cinti S., Højlund K., Gygi S. P., Spiegelman B. M., A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chau Y.-Y., Bandiera R., Serrels A., Martínez-Estrada O. M., Qing W., Lee M., Slight J., Thornburn A., Berry R., McHaffie S., Stimson R. H., Walker B. R., Chapuli R. M., Schedl A., Hastie N., Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16, 367–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalek A. K., Satija R., Shuga J., Trombetta J. J., Gennert D., Lu D., Chen P., Gertner R. S., Gaublomme J. T., Yosef N., Schwartz S., Fowler B., Weaver S., Wang J., Wang X., Ding R., Raychowdhury R., Friedman N., Hacohen N., Park H., May A. P., Regev A., Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki R., Shimodaira H., Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Hua Y., Ke S., Wang Y., Irwin D. M., Zhang S., Wang Z., Prolonged treatment with 3-isobutyl-1-methylxanthine improves the efficiency of differentiating 3T3-L1 cells into adipocytes. Anal. Biochem. 507, 18–20 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Karamanlidis G., Karamitri A., Docherty K., Hazlerigg D. G., Lomax M. A., C/EBPβ reprograms white 3T3-L1 preadipocytes to a brown adipocyte pattern of gene expression. J. Biol. Chem. 282, 24660–24669 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Lone J., Choi J. H., Kim S. W., Yun J. W., Curcumin induces brown fat-like phenotype in 3T3-L1 and primary white adipocytes. J. Nutr. Biochem. 27, 193–202 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Qian S.-W., Tang Y., Li X., Liu Y., Zhang Y.-Y., Huang H.-Y., Xue R.-D., Yu H.-Y., Guo L., Gao H.-D., Liu Y., Sun X., Li Y.-M., Jia W.-P., Tang Q.-Q., BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 110, E798–E807 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero-Calvo I., Ocón B., Martinez-Moya P., Suárez M. D., Zarzuelo A., Martínez-Augustin O., de Medina F. S., Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal. Biochem. 401, 318–320 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Fortes M. A. S., Marzuca-Nassr G. N., Vitzel K. F., da Justa Pinheiro C. H., Newsholme P., Curi R., Housekeeping proteins: How useful are they in skeletal muscle diabetes studies and muscle hypertrophy models? Anal. Biochem. 504, 38–40 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H. M., Erdjument-Bromage H., Tempst P., Rudnicki M. A., Beier D. R., Spiegelman B. M., PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suenaga M., Kurosawa N., Asano H., Kanamori Y., Umemoto T., Yoshida H., Murakami M., Kawachi H., Matsui T., Funaba M., Bmp4 expressed in preadipocytes is required for the onset of adipocyte differentiation. Cytokine 64, 138–145 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaar5319/DC1
Fig. S1. Clustered profile of all marker genes.
Fig. S2. Expression levels of six beige marker genes assayed by RT-qPCR.
Fig. S3. Ponceau staining as loading control in Western blots.
Table S1. Mapped Ensembl genes and raw read counts from 20 samples.
Table S2. All beige adipocyte, BAT, and WAT marker genes.
Table S3. Primers used for qPCR.


