Key Points
We have generated allele-specific base resolution methylomes of primary basophilic erythroblasts.
DNA demethylation during differentiation of HSPC into BasoE occurs mostly in inactive regions causing formation of PMD in 74% of methylome.
Abstract
Erythroid differentiation is associated with global DNA demethylation, but a complete methylome was lacking in the erythroid lineage. We have generated allele-specific base resolution methylomes of primary basophilic erythroblasts (BasoEs) and compared these with 8 other cell types. We found that DNA demethylation during differentiation from hematopoietic stem/progenitor cells (HSPCs) to BasoEs occurred predominantly in intergenic sequences and in inactive gene bodies causing the formation of partially methylated domains (PMDs) in 74% of the BasoE methylome. Moreover, differentially methylated regions (DMRs) between HSPCs and BasoEs occurred mostly in putative enhancer regions and were most often associated with GATA, EKLF, and AP1 binding motifs. Surprisingly, promoters silent in both HSPCs and BasoEs exhibited much more dramatic chromatin changes during differentiation than activated promoters. Unmethylated silent promoters were often associated with active chromatin states in highly methylated domains (HMDs) but with polycomb-repression in PMDs, indicating that silent promoters are generally regulated differently in HMDs and PMDs. We show that long PMDs replicate late, but that short PMDs replicate early and therefore that the partial methylation of DNA after replication during erythroid expansion occurs throughout S phase of the cell cycle. We propose that baseline maintenance methylation following replication decreases during erythroid differentiation resulting in PMD formation and that the presence of HMDs in the BasoE methylome results from transcription-associated DNA methylation of gene bodies. We detected ∼700 large allele-specific DMRs that were enriched in single-nucleotide polymorphisms, suggesting that primary DNA sequence might be a determinant of DNA methylation levels within PMDs.
Visual Abstract

Introduction
DNA methylation regulates gene expression, parental imprinting, X chromosome inactivation, and transposable elements.1,2 Most promoters and enhancers are 200- to 5000-bp regions that are generally constitutively unmethylated.3 Methylation canyons are unmethylated regions >5 kb that are mostly conserved across species and enriched in regulatory genes.4 Partially methylated domains (PMDs) are even larger megabase-sized domains that were first observed by whole genome bisulfite sequencing (WGBS) in IMR90 embryonic fibroblasts that encompass ∼40% of the genome.5 PMDs, which have been shown to contain mostly intergenic regions and silent genes, have also been observed in primary cells, tissues and tumors, and transformed cell lines,6-10 but not in H1 human embryonic stem cells.5
Several reports based on reduced representation bisulfite sequencing,11 Hpall tiny fragment enrichment by ligation-mediated assay,12 or methyl-CpG-binding domain sequencing13 have demonstrated that erythroid differentiation is associated with genome-wide demethylation that requires DNA replication to occur and that affects gene bodies, intergenic regions, and CpG shores. These previous studies were based on reduced-representation approaches and therefore could not address the presence of PMDs or analyze canyons in erythroid cells. Although widespread differentiation-associated demethylation was recently also observed in lymphoid cells, it was restricted to heterochromatin and had little functional impact on genes active in B cells,14 raising questions about the role of differentiation-associated demethylation.
Studies have shown that allelic differences in methylation were associated with PMDs in HCC1954 cells9 and that late-replicating regions were generally less methylated than early-replicating regions in primary human fibroblasts15; yet, the mechanisms of PMD formation and their functional significance remain unclear.
We have generated haplotype-resolved methylomes and transcriptomes of human primary basophilic erythroblasts (BasoEs) and analyzed them in the context of previously published WGBS, gene expression, chromatin state, and replication timing data across multiple cell lines and cell types. We show that the global demethylation during erythroid differentiation is associated with extensive PMD formation, which encompasses >74% of the cells’ genome and has only a small effect on the active part of the genome. Similarly, we found that most of the changes in promoter chromatin structure during erythroid differentiation occur either in putative enhancers or in genes that are silent in both hematopoietic stem and progenitor cells (HSPCs) and in BasoEs. We also show that silent CpG-rich unmethylated promoters have very different chromatin structure in highly methylated domains (HMDs) and PMDs, suggesting that segregating silent promoters into different genomic compartments might be one of the biological functions of PMD formation. Finally, we show that partial methylation of PMDs after replication during erythroid differentiation occurs in both early and late S phase of the cell cycle, that non–S-phase DNA methylation decreases the fraction of the genome covered by PMDs, and we observed about 700 large allele-specific differentially methylated region in the BasoE methylome which were enriched in single-nucleotide polymorphisms (SNPs). These observations support the idea that PMDs form because of a decrease in maintenance methylation in both early and late S phase and that the level of methylation within PMDs is determined in part by the primary DNA sequence.
Methods
Cell culture
Peripheral white blood cells (10-20 mL) were harvested by venipuncture from individuals FNY01_3_2 and 3_3 from family FNY01 under an approved institutional review board protocol. Mononuclear cells were isolated by density gradient centrifugation on Histopaque (Sigma-Aldrich) according to the manufacturer’s instructions. The purified cells were frozen in 2 million cell aliquots. Two million mononuclear cells were expanded and differentiated into basophilic erythroblasts in culture for 2 weeks in serum-free StemSpan media (Stem Cells Technologies, Vancouver, Canada) containing the cytokine cocktail mix described by Olivier et al16 At the end of the culture, cells were immunophenotyped by fluorescence-activated cell sorting using antibodies against CD71 (e-Bioscience 11-0719, 0.3 mg/mL) and CD235a (e-Bioscience 11-9987, 0.6 mg/mL). Cells were relatively uniform in size and >97% of the cells were double positive, demonstrating that the vast majority of cells in the culture were erythroid cells at the basophilic stage of differentiation.
P51R cells were grown as previously described in Dulbecco’s modified Eagle medium plus 10% fetal bovine serum. Confluent P51R cells were blocked in G0/G1 by replacing the medium with Dulbecco’s modified Eagle medium and 0.5% serum for 4 days. Cell-cycle analysis was performed by staining the cells with propidium iodide as described by Krishan.17
Data
Previously published data were retrieved from the data sources listed in supplemental Table 1. Most data were generated by the Roadmap Epigenomics project18; the Salk Institute,5 or the FANTOM consortium (http://fantom.gsc.riken.jp/).
Gene expression and gene annotation
We compared WGBS with RNA-sequencing (RNA-seq) data that we generated from BasoEs or that we downloaded from public databases (supplemental Table 1). Because our analysis required transcript maps that were as complete as possible, we projected the RNA-seq data on the GENCODE annotation, which contains a large number of isoforms. To identify which isoforms were expressed, we also downloaded Cap Analysis of Gene Expression (CAGE) data from the FANTOM and the ENCODE consortium databases (supplemental Table 1) and integrated both data types with custom R scripts (see the following section). All analyses were performed on autosomal genes only because the datasets used were either males or females.
RNA-seq data generation for BasoEs
Ten million BasoE cells were generated using the protocol described in the “Cell culture” section and total RNA was extracted using a Qiagen RNA extraction kit. PolyA+ libraries were then constructed using the Illumina Stranded mRNA LT Sample Prep Kit using the recommended protocols. Libraries were then sequenced in 2 to 3 different lanes on an Illumina HiSeq2500 sequencer to a depth of about 80 million 100-kb paired-end reads.
RNA-seq data processing (non–allele-specific)
The RNA-seq data that we either generated in house or downloaded was processed using the lightweight aligner Salmon19 using the default parameters to quantify expression of GENCODE (v22) transcripts lifted over to hg19 using the UCSC liftOver tool. CAGE profiles from FANTOM in the context of CAGEr analysis and high-resolution promoterome mining for integrative analyses20 was used to determine unambiguously expressed isoforms as follows.
Promoter regions were defined for each isoform as the union of all the −400 to +100 intervals around the transcription start sites (TSSs) for all of the overlapping isoforms; a CAGE score equal to the sum of the CAGE scores of the individual overlapping promoters was then assigned to these promoter regions. Each promoter region was also assigned an associated gene body equal to the longest transcript with Salmon transcript per million (TPM) score >1, and a Salmon expression score defined as the sum of TPM of all its associated isoforms. Genes were considered not expressed when the Salmon TPM of the collapsed isoforms was <1 when there was no CAGE signal associated with the collapsed promoter and when the gene was not overlapping an expressed gene. For each cell line, this expression analysis yielded ∼35 000 active or inactive promoter regions and ∼4000 ambiguous promoter regions that exhibited an RNA-seq signal and no CAGE signal. These ambiguous promoter regions were excluded from further analysis. The total number of promoter regions in each cell type differed slightly because the CAGE and RNA-seq data are cell type–specific. Similar to previous reports,21 a moderate correlation between the RNA-seq and CAGE data were observed (supplemental Figure 2A). An analogous analysis using the STAR aligner and the RefSeq annotations instead of the ENCODE/CAGE data yielded similar conclusions.
CpG-rich promoter definition
The active and inactive promoter regions were divided into CpG-rich and CpG-poor categories based on whether they overlapped with a CpG cluster as defined by the CpGCluster algorithm22 using default cutoff values (d = 50, P = 1E-5). This algorithm is based on the physical distance between neighboring CpGs on the chromosome to predict clusters of CpGs, then assigns a P value to each of these clusters; the most statistically significant ones can be predicted as CGIs. About 15 000 CpG-rich and 20 000 CpG-poor promoter regions were thus defined in each cell line.
RNA-seq data processing (allele-specific)
Fastq files from sequencing of the libraries derived from individuals FNY01_3_2 and 3_3 were aligned with the STAR aligner and SNPs were called using the GATK AseReadCounter in the Given_Allele mode taking advantage of the phased vcf that is available for these 2 individuals after having truncated the Bam files after the first SNPs to ascertain that each fragment was only counted once. SNP counts were then summed up on the RefSeq gene models using GenPlay.23 Allele-specific read counts were then assigned to overlapping RefSeq transcripts. No normalization is necessary to obtain the allele-specific ratio because the count comes from the same library and the same sequencing reactions.
BasoE DNA methylation profiles
Genomic DNA was extracted from ∼50 million basophilic erythroblasts, and libraries were produced using a protocol developed at the Einstein epigenomic facility. Extracted genomic DNA was fragmented (400-500 bp) with Covaris, end-repaired, dA tailed, and premethylated adapters (Illumina TruSeq adapters) were ligated at the ends of the fragmented DNA. The adapter ligated DNA samples were purified with AMPure XP beads (1:1 dilution) to eliminate adapter dimers and products with short insert, and then treated with sodium bisulfite using EZ DNA Methylation lightning kit (Zymo Research). The bisulfite-treated products were used as a template for polymerase chain reaction (PCR) amplification using the following condition: 25 µL of 2× KAPA HiFi HotStart Uracil+ ReadyMix, 1.5 µL of 10 µM of primer P5, and 1.5 µL of 10 µM of primer P7 and bisulfite treated library in a final volume of 50 µL; 98°C for 2 minutes, then 10 cycles of 98°C for 30 seconds, 60°C for 30 seconds and 72°C for 4 minutes followed by 10 minutes at 72°C for final extension. Amplified libraries were purified with MinElute PCR purification kit, and a size selection was performed with the MinElute Gel Extraction kit (the insert size was 250-850 bp). For each of the 2 individuals analyzed, paired-end 2× 100-bp reads were generated on 6 lanes of an Illumina HiSeq 2500.
The reads were then aligned with BSMAP24 and high- resolution unphased methylomes were generated by calculating the methylation fraction for every CpG in the genome. Because in these cells <0.1% of the methylated cytosines were in a non-CpG context, non-CpG-methylation was not studied any further.
SNPs were called using Bis-SNP25 and phased methylomes were generated using the custom software MethylPhase developed for this purpose. This software was designed to be easily incorporated in a Methyl-Seq analysis pipeline. MethylPhase requires aligned bisulfite-treated reads and a phased vcf file containing all the SNPs present in the samples. When provided with these inputs, MethylPhase can phase align bisulfite-treated fragments using the phased vcf as a reference. The phasing is done in 2 steps. First, each read spanning a methylation site is independently phased. Second, the methylation profile of each site is summarized.
Step 1: BS reads phasing.
For each read spanning a methylation site, MethylPhase inspects all of the SNPs on the read and its mate-pair. Each SNP is compared with the genotype from a VCF file. Only SNPs corresponding to heterozygous genotypes are processed. A SNP can be marked as either: (1) error state, if it does not correspond to 1 of the alleles previously genotyped; (2) allele 1, if the base from the BS-treated read corresponds to the first allele; (3) allele 2, if it corresponds to the second allele; or (4) ambiguous, if the allele cannot be determined. The allele of origin of a SNP cannot always be determined because DNA bisulfite treatment followed by PCR amplification leads to the conversion of unmethylated Cs to Ts. This implies that on the forward and reverse Crick strands (“++” and “+−”), a thymine base on the BS-treated read can actually correspond to a thymine or a converted cytosine from the sample DNA, and an adenine on the Watson strands (“−+” and “−”) can correspond to an adenine or a guanine. In these cases, if both alleles are compatible, the SNP is marked as ambiguous.
Once all the SNPs are processed, the read is marked as (1) “error,” if 1 or more of its SNPs are in error states or if different SNPs on the same read come from different alleles; (2) “ambiguous,” if all the SNPs on the read are marked as ambiguous, or (3) “allele 1” or “allele 2” if the read is not already marked as error and if at least 1 SNP can be used to determine the allele of origin.
Step 2: Methylation sites summarization.
For each methylation site, MethylPhase determines the following: (1) the number of methylated reads spanning the site on the maternal allele; (2) the number of unmethylated reads spanning the site on the maternal allele; (3) the number of methylated reads spanning the site on the paternal allele; (4) the number of unmethylated reads spanning the site on the paternal allele; (5) the number of error reads; and (6) the number of ambiguous reads. A bedgraph is then generated. The program, its source code, and its documentation are available at https://github.com/JulienLajugie/MethylPhase.git.
About 16 million CpG sites out of ∼54 million CpG present in the genome (30%) could be phased with this approach. Haplotype-resolved methylomes were then generated by calculating the methyl fraction for the paternal and the maternal chromosomes of each individual.
Reduced representation Methyl-Seq
One million P51R cells were encapsulated in agarose plugs, DNA was extracted, restricted with PacI, and separated on a CHEF pulse field electrophoresis apparatus as described by Zang et al.26 Size markers were prepared by ligating λ DNA digested with NheI, or Kas1. About 50 to 100 ng of size-purified genomic DNA was obtained. Libraries were prepared as follows: Extracted genomic DNA was fragmented (400-500 bp) with Covaris, end-repaired, and dA tailed; premethylated adapters (Illumina TruSeq adapters) were then ligated at the ends of the fragmented DNA. The adapter ligated DNA samples were purified with AMPure XP beads (1:1 dilution) to eliminate adapter dimers and products with short insert and then treated with sodium bisulphite using EZ DNA Methylation lightning kit (Zymo Research). The bisulphite-treated products were used as a template for 10 cycles of PCR amplification. Amplified libraries were purified with MinElute PCR purification kit; size selection was then performed with MinElute Gel Extraction kit (insert size, 250-850 bp). The libraries were sequenced using Illumina HiSeq2500 (100 bp paired-end reads). A total of 45 237 815 (G0/G1) and 65 658 904 (cycling cells) pairs were obtained, of which 81% could be aligned using Bismark.27 After alignment, the data were deduplicated and counts from both strands were combined for each CpGs. The data were then filtered to retain only the PacI fragments that contain an average read depth >5 for both the G0/G1 and the cycling cells. About 70% of all called CpGs were retained by this filter. The 2 632 238 and 2 576 685 CpGs corresponding to coverage of 13.9 and 10.9 for the G0/G1 and for the cycling cells were obtained. The informative PacI fragments covered ∼255 × 106 bases or ∼8.5% of the genome.
Methyl-Seq data processing
WGBS data generated in house or retrieved from the sources listed in supplemental Table 1 was processed as follows: Counts of strand-specific cytosine methylation in CpG dinucleotides were pooled using custom R scripts and data analysis was restricted to CpG sites with at least 10× coverage (96.3% of all CpG were assessed in the case of the BasoEs). Methyl-fraction was calculated as meC/(meC + C) for each CpG dinucleotide.
Meta-gene profiles
CpG dinucleotides overlapping the designated regions relative to each gene (eg, 10 kb upstream of TSS to 10 kb downstream of transcription end site [TES]) in each indicated category were ranged with respect to their position relative to the TSS and TES of each gene in a strand-specific manner using custom scripts in R/Bioconductor. Subsequently, the position and methylation of all CpGs upstream or downstream of the gene was determined for all genes. For CpG dinucleotides within genes, their position in 10 gene-specific bins of equal length between TSS and TES was determined relative to the TSS, and the average methylation of each of these bins was calculated. Composite profiles from upstream, gene-body, and downstream regions were plotted after smoothing the extragenic regions over 100-bp windows.
Segmentation of the methylomes
We used MethylSeekR28 and additional custom R scripts to segment the genome into unmethylated regions (UMRs), low methylated regions (LMRs), HMDs, and PMDs as follows: DNA methylation was summarized by CpG, CpGs with coverage <10 were discarded, and methylation β values (fraction of methylated over total read counts) were called.
MethylSeekR was then used to call UMRs and LMRs using the segmentUMRsLMRs function with meth.cutoff of 0.3, nCpG.cutoff = 5. With these parameters, MethylSeekR first smoothes methylation levels over 3 consecutive CpGs, and hypomethylated regions are identified as regions containing at least 5 CpGs (meth.cutoff) with a smoothed fraction of methylation below the 30% methylation (nCpG.cutoff). Regions thus identified that contain at least 30 consecutive CpGs are called as UMRs. Regions containing between 5 and 30 consecutive CpG are called as LMRs.
To call HMDs, we removed all UMRs and LMRs and large gaps and calculated a running mean of the methyl fraction across 101 CpGs and defined HMDs as the regions with runmean >0.78. Consecutive HMDs separated by <10 CpGs were then merged. The regions not called as HMDs were then called as PMDs. PMDs <10 kb (that disrupted HMDs) were assigned as HMDs; PMDs >250 kb were called l-PMDs. Those between 10 and 250 kb were called short-PMDs.
Finally, and specifically for K562 cells, LMRs and UMRs >10 kb that were within PMDs in K562 cells were designated as PMDs. This correction was necessary to take into account the large regions of very low methylation in K562 cells that do not appear to be bona fide UMRs or LMRs.
Association with ChromHMM states
We retrieved ChromHMM data according to the 15 coreMarks model from the Epigenome Roadmap project. For BasoE cells, we used the same parameters and chromatin immunoprecipitation-seq marks as used in the 15 core marks model using processed data from GSE12646 (supplemental Table 1) to generate ChromHMM regions using ChromHMM (v. 1.10).29 The files indicated in supplemental Table 1 were used to generate a ChromHMM segmentation of hg18; liftOver was used to lift the coordinates to hg19 because all other analyses were performed in hg19.
To generate heat maps, we determined the fraction of each element (promoter, gene body, or methylation domain) covered by each of the 15 ChromHMM categories and averaged the fraction over the indicated groups. To generate Circos plots, each promoter, gene body, or methylation domain was assigned to the ChromHMM category that encompassed the largest amount of bases of this element.
DNase hypersensitivity and TFBS data
DNase hypersensitive sites were retrieved from ENCODE in narrowPeak format for K562 (accession: ENCFF941ITD), H1 (ENCFF001UVM), HepG2 (ENCFF873IZM) and IMR90 cells (ENCFF001UWF). Transcription factor binding site (TFBS) data were downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wgEncodeRegTfbsClusteredWithCellsV3.bed.gz.
Replication timing data
Timing data for IMR90, K562 cells were retrieved from http://www.replicationdomain.com/data.php. The data for the non-allele–specific, the allele-specific, and the core asynchronously replicated domains (c-ARDs) in BasoEs were generated in-house using the TimEX methods and are available as described elsewhere.30-32 TimEX scores in 5-kb windows were calculated as the ratio of the normalized number of reads in S-phase cells/the number of normalized reads in G1 cells. Quantile normalization of a matrix containing the timing values for each of these 4 cell lines was performed on the autosomes using the normalize.quantiles function from the Bioconductor preprocessCore library33 to compensate for the fact that the data were obtained in different laboratories using different approaches.
Identification of DMRs between HSPC and BasoE
Differentially methylated regions (DMRs) between HSPCs and BasoEs were called with the Bioconductor DSS package,34 using all CpGs with at least 10× coverage. The core of DSS is a procedure based on Bayesian hierarchical model to estimate and shrink CpG site–specific dispersions, then conduct Wald tests for detecting differential methylation. Briefly, differentially methylated loci were first identified using δ = 0.1, and p.threshold = 0.001, after smoothing CpGs <500 bp. Then, DMRs were called with the parameters p.threshold = 0.001, δ = 0.2, minlen = 100, minCG = 10, dis.merge = 20, and pct.sig = 0.6. DMRs called because of the presence of PMDs in BasoEs were excluded. To generate Circos plots, association with ChromHMM categories was assessed by assigning each DMR the ChromHMM category with the largest overlap. Similar results were observed when focusing on DMRs fully covered by only 1 ChromHMM category (not shown). To generate heat maps, we averaged the fraction of each DMR covered by each ChromHMM category. Analysis of DNA sequence motifs was performed with Homer (http://homer.ucsd.edu/homer/motif/) using default settings and motif lengths of 6, 8, 10, and 12.
Identification of allele-specific DMRs (a-DMRs)
DMRs between alleles were called using the R/Bioconductor package DSS, using phased genome-wide paired CpG methylation data (paired CpG = combined number of methyl or unmethyl reads on both strands) after removal of paired CpGs within UMRs and LMRs and those with low coverage (<5×). Smoothing (smoothing.span = 50 000) was applied before calling differentially methylated loci (p.threshold = 0.05) and DMRs (p.threshold = 0.05, δ = 0, minlen = 100, minCG=10, dis.merge = 10 000, pct.sig = 0.5) with DSS. Resulting DMRs with an absolute methylation difference >0.05 were retained.
The smoothed data were exported as paternal and maternal bedgraph files. The complete data tracks can be visualized by downloading the associated GenPlay project (see the following section).
To calculate the correlation between allele-specific methylation in a-DMRs and timing, we further filtered the a-DMRs to maintain a minimum read count of 300 for total methylation reads of maternal or paternal allele and a minimum number of timing SNPs of ≥140 in G1 phase per DMR.
Mutation spectrum analysis on a-DMRs
We used SnpEff (http://snpeff.sourceforge.net/) to determine SNP frequencies, mutation spectrum, and Ti/Tv ratio (supplemental Figure 6) within a-DMR and non–a-DMR regions with sufficient coverage to allow to call DMRs. Specifically, we tiled the genome into 10-kb tiles, determined read coverage, and retained all 10-kb windows with ≥100 reads/10 kb and used that as the space to potentially be able to call a-DMRs. The minimum average read count in a-DMRs was 113/10 kb.
Results
BasoEs were produced from a 2-week culture30 of peripheral blood progenitor cells collected from 2 sisters in family FNY01, a quartet of healthy individuals that had their genomes completely sequenced and phased.35 Haplotype-resolved methylomes were then generated by WGBS yielding unphased methylomes for most CpGs in the genome and phased methylomes for about 30% of all CpGs.
To compare BasoEs to other cells, we reanalyzed published WGBS data of HSPCs, which are the BasoE precursors, 2 additional types of SPCs, 3 untransformed differentiated cells, and 2 transformed cell lines (Table 1; supplemental Table 1).
Table 1.
Panel of cells
| Name | Source | Category |
|---|---|---|
| H1 | Cultured embryonic stem cells | Stem and progenitor |
| HSPC | Mobilized peripheral blood | Stem and progenitor |
| Ganglionic Neurospheres | Differentiated from human embryonic stem cells | Stem and progenitor |
| BasoE | Cultured primary basophilic erythroblasts | Somatic |
| IMR90 | Cultured primary fetal lung fibroblasts | Somatic |
| Liver | Primary tissue | Somatic |
| Pancreas | Primary tissue | Somatic |
| K562 | Chronic myelogenous leukemia | Transformed |
| HepG2 | Hepatocyte carcinoma | Transformed |
Long PMDs are gene-poor but short PMDs are as rich as, or richer, in genes than HMDs
As expected, visual inspection revealed that all methylomes contained short unmethylated regions, corresponding to regulatory elements,3 embedded either into HMDs or PMDs (Figure 1A). To segment the methylome, we used MethylSeekR28 to call LMRs and UMRs. Subsequently, we masked these and separated the genome into PMDs and HMDs using an experimentally determined cutoff of 78% methylation over sliding windows of 101 CpGs with at least 10× coverage. Adjacent HMDs separated by PMDs <10 kb in size were merged. PMDs were separated into short PMDs (s-PMDs; <250 kb) and long PMD (l-PMDs; ≥250 kb) (see “Methods” for details). Calling PMDs first and then LMRs and UMRs as previously published28 yielded similar results in most cell types but caused problems in transformed cells.
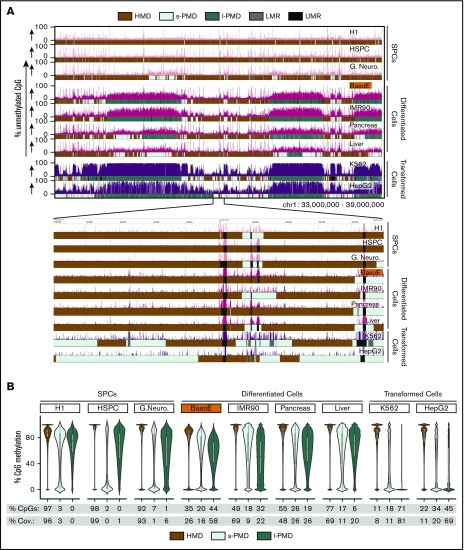
Figure 1.
Diversity of global methylation patterns in HSPC, differentiated, and transformed cells. (A) The percentage of unmethylated CpG dinucleotides is plotted for a typical region on chr1. Genplay screenshot of a 6-megabase region on chromosome 1 that illustrates the Methyl-Seq data and the segmentation of the methylome. The inset below the main panel represents the indicated 200-kb region at the center of the 6-megabase region to illustrate the fine structure of the UMRs and LMRs. The colored boxes below each track illustrate the cell-specific segmentation of the genome into HMDs, s-PMDs, l-PMDs, LMRs, and UMRs. The methylome of SPCs is composed mostly of HMDs and of small unmethylated regions (UMRs and LMRs) that correspond to regulatory elements. Methylomes of differentiated and transformed cells also contain HMDs, UMRs, and LMRs, but PMDs represent a large fraction of the genome of these cells. PMDs are ∼30% to 70% methylated in differentiated cells but are almost completely unmethylated in transformed cells. (B) Violin plots illustrating DNA methylation density in HMDs and PMDs. The percentages of CpGs and coverage (% Cov.) of the genome that are in HMDs, l-PMDs, and s-PMDs are indicated below the plots. SPCs are composed mostly of HMDs; differentiated cells contain variable amounts of HMDs and PMDs while transformed cells contain small amounts of HMDs.
This revealed that BasoEs had the highest PMD content of all nontransformed cells, covering 74% of their genome. BasoEs were most similar to other differentiated cells that harbored PMDs covering between 31% and 52% of their genome. As previously reported,36 methylation within IMR90 PMDs was particularly variable, as illustrated by the very flat violin plot in PMDs (Figure 1B), but this pattern was unique to these cells and was not observed in BasoEs.
By contrast, PMDs covered only 1% to 6% of the genome of SPCs and as much as 85% to 90% of transformed cells, respectively (Figure 1B). PMDs in transformed cells differed from those of BasoEs because most of them were almost completely unmethylated. They were nevertheless classified as PMDs rather than as LMRs or UMRs because of their very large size. We considered creating a special category for these regions because they were different from other PMDs but ultimately decided not to because PMDs in differentiated and transformed cells often overlapped significantly.
Analysis of TSS in BasoEs and other cells suggested an inverse relationship between the length of PMDs and TSS density. Splitting PMDs into short (<250 kb) and long (>250 kb) fractions revealed that, as previously reported for IMR90 cells, l-PMDs were poorer in TSSs than HMDs36 (supplemental Figure 1A-B). However, s-PMDs were as rich as, or richer, in TSSs than HMDs in all cells tested (supplemental Figure 1B).
Active genes are located almost exclusively in HMDs but silent genes are split between HMDs and PMDs
To study gene expression in HMDs and PMDs, we generated RNA-seq data for BasoEs and downloaded public RNA-seq data for the other cell types (supplemental Table 1). Given that many transcripts identified by RNA-seq are not part of RefSeq gene models, we quantified expression using the GENCODE annotation, which is more complete, but contains many intragenic promotors that are difficult to classify as active or inactive. To improve this transcript model, we downloaded CAGE data and combined these with RNA-seq data to identify all active GENCODE promoters that were subsequently divided into CpG-rich and CpG-poor categories using CpGcluster.22 CAGE and RNA-seq data were generally in good agreement (supplemental Figure 2A). In all cells, >60% of the CpG-rich but only 2% to 4% of CpG-poor CAGE and RNA-seq–defined promoter regions were active (supplemental Figure 2B), suggesting that many of the annotated CpG-poor promoters might not be functional.
Analysis of expression revealed dramatic differences between the DNA methylation domains of BasoE because both s- and l-PMDs were almost devoid of active genes, with 98% of the genes in l-PMDs inactive (Figure 2A). Expressed genes were therefore almost exclusively located in HMDs, which also contained ∼20% silent genes. A similar dichotomy was found for all other cells, although PMDs of transformed cells contained a slightly higher proportion of active genes.
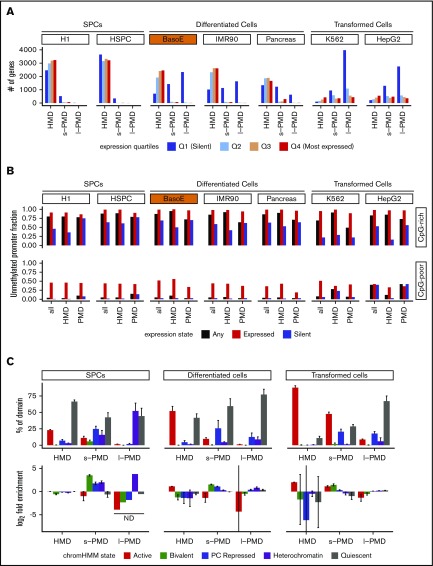
Figure 2.
PMDs contain mostly silent genes, whereas HMDs contain a mixture of expressed and silent genes. (A) GENCODE gene models were divided into 4 expression quartiles based on RNA-seq and CAGE data, and the number of genes within HMDs, s-PMDs, and l-PMDs were plotted for each cell type. Genes overlapping both HMDs and PMDs were excluded from the analysis. Q1 represents unexpressed genes, whereas Q2 through Q4 represent tertiles of increasing expression. Almost all active genes are in the HMD compartment, except for transformed cell lines where most of the genome is in PMDs. (B) Bar graph illustrating the fraction of all active and silent promoters that are unmethylated, separated by CpG density of their promoters into CpG-rich (top) and CpG-poor (bottom). y-axis represents the fraction of promoter that are expressed or silent. Unmethylated promoters were defined as promoters overlapping either an UMR or LMR. Silent promoters were defined as promoters associated with genes exhibiting no RNA-seq (TPM < 1) and no CAGE signal; expressed promoters as promoters associated with genes exhibiting both an RNA-seq (TPM > 1) and a CAGE signal (see “Methods”). In both HMDs and PMDs, almost all active and a significant fraction of silent promoters are unmethylated. (C) ChromHMM analysis based on the 15-state core model from the Roadmap Epigenome Project. Top: bar graphs illustrating the average fractional chromatin composition within HMDs, s-PMDs, and l-PMDs. For each cell line, the percentage of the genome in each of the 3 methylation domains that is covered by the 5 chromatin states was calculated. Results were then averaged per each cell category (SPC, differentiated or transformed). Error bars indicate the standard error of the mean for the respective groups of cell types analyzed. For clarity, the 15 ChromHMM classes were summarized into 5 larger categories: active (red: TssA, TssAFlnk, TxFlnk, Tx, TxWk, EnhG, Enh, ZNF.Rpts), bivalent (green: TssBiv, BivFlnk, EnhBiv), repressed (blue: RepPC, ReprPCWk), heterochromatin (purple), and quiescent (gray). The y-axis represents the percentage of HMDs or PMDs that overlap with a chromatin states as defined previously. HMDs of differentiated cells contain most of the active chromatin, whereas PMDs are composed mostly of repressed and quiescent chromatin. Bottom, graphs are organized as previously, but the y-axis represents the enrichment, calculated as (# of bases in HMDs, s-PMDs, or l-PMDs covered with a particular ChromHMM state/# of bases in HMDs, s-PMDs or l-PMDs)/(# of bases in the genome covered with a particular ChromHMM state/# number of bases in the genome). The bar represents the average log2 of the enrichment (± standard deviation) for each cell category and each ChromHMM category. Some of the error bars are very large because the fraction of the genome covered by some of the cell categories was very low and therefore highly variable in different cell types.
To categorize the methylation state of promoter regions, we determined whether they overlapped with a UMR or LMR (considered as unmethylated) or not (methylated). In BasoE and in all nontransformed cells in our panel, >98% of the expressed and ∼40% to 70% of the silent CpG-rich promoter regions were unmethylated (Figure 2B). Silent CpG-rich promoter regions were generally more often unmethylated in PMDs than in HMDs (Figure 2B).
Expressed CpG-poor promoters were variably methylated (Figure 2B); however, many of these contained no or very few CpG dinucleotides, making the relevance of DNA methylation for the regulation of this group of promoters questionable. Therefore, we focused most analyses on the CpG-rich promoters.
To determine if HMDs and PMDs were associated with different chromatin structure, we used ChromHMM analysis from the Roadmap Epigenomics project29 in which 5 histone marks were combined to segment the genome into 15 chromatin states. In differentiated cell types, an average (± standard error of the mean) of 46.3 ± 8.7% of the genome was located in HMDs, and 52.1 ± 7.1% of the chromatin of the HMDs were in 1 of the active chromatin states with most of the remainder in the quiescent state (Figure 2C; supplemental Figure 2C). By contrast, an average of 20.0 ± 2.2% and 33.7 ± 9.9% of the genome were located in s- and l-PMDs, respectively, and only 9.6 ± 1.9% and 1.2 ± 0.3% of the s- and l-PMDs were associated with active chromatin states with most of the remainder in either bivalent repressed or heterochromatic states (Figure 2C) .
Enrichment analysis (Figure 2C, bottom) revealed that in differentiated cells, HMDs were enriched in active marks, whereas s- and l-PMDs were depleted (paired Student t test P value respectively = 0.021 and 0.015).
Gene body methylation and transcriptional activity are directly correlated
Consistent with other studies, bodies of active genes were highly methylated in BasoEs and all cells in our panel (Figure 3A; supplemental Figure 3). This was true for most genes except for highly expressed small genes such as the globin genes, which are completely embedded within large UMRs.37 One major difference between BasoEs and their precursors, the HSPCs, was that the bodies of inactive genes were partly methylated in the former cell type but highly methylated in the latter. Gene body demethylation during erythroid differentiation was therefore specific to inactive genes. This observation was not restricted to BasoEs and HSPCs but reflected differences between stem and progenitor cells (which have a very heavily methylated genome) and differentiated cells which contain PMDs.
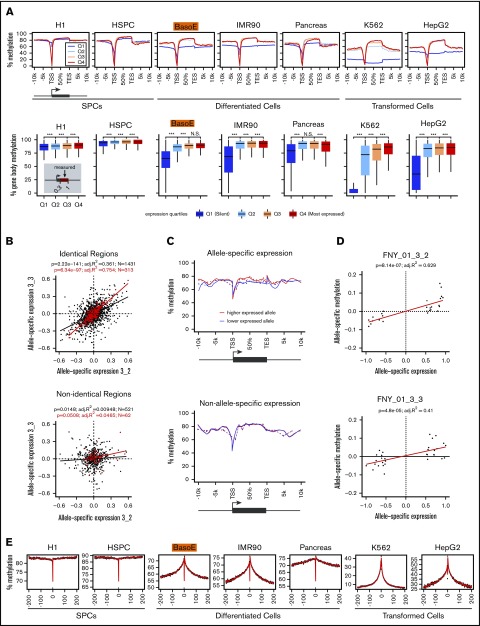
Figure 3.
Methylation analysis of gene bodies and surrounding regions. (A, top) Meta-gene analysis illustrating gene body methylation as a function of expression. The average percentage of methylated CpG dinucleotides of aggregated signal from all genes with indicated coordinates (in kb) relative to the TSS or TES is shown for unexpressed genes (Q1) and for expressed genes in tertiles of expression (Q2-Q4). Gene bodies of expressed genes were highly methylated in all cells. Gene bodies of silent genes were highly methylated in cells composed mostly of HMDs (SPCs) but were partially methylated in cells rich in PMDs (differentiated and transformed cells). Level of gene-body methylation of silent genes was cell type–specific and similar to the average level of methylation of the PMDs. Methylation levels dramatically dropped near the promoters reflecting the presence of UMRs and LMRs in these regions. (A, bottom) Box plots depicting average gene body methylation (starting at 30% of the gene length after the TSS [to eliminate the effect of the presence of UMRs/LMRs] and ending at the TES) of the genes from the 4 expression quartiles. Statistical significance assessed by Student t test is indicated (***P < .001). (B) Scatter plots illustrating Pearson r2 correlation between the difference of maternal and paternal gene expression in individuals FNY01_3_2 and 3_3 in the haplo-identical but not in the nonidentical fraction of their genomes. The red dots represent the highly expressed genes (>500 RNA-seq reads); black dots, genes with >20 RNA-seq reads. Only autosomal protein-coding genes were considered. x-axis and y-axis represent allele-specific expression defined as (βmaternal − βpaternal) with βmaternal = (# of maternal reads)/(# of total reads) and βparternal = (# of paternal reads)/(# of total reads). The high correlation in the haplo-identical genes (r2 = 0.754 for the genes >500 reads and r2 = 0.361 for the genes >20 reads) suggests that most of the allele-specific variation in gene expression detected at the cell population level is genetic in origin. (C) Allele-specific meta-gene body methylation analysis illustrating that the most highly expressed allele is more highly methylated than the least expressed allele. The top and bottom graph respectively represent gene body methylation for the genes expressed in an allele-specific manner, or in a non–allele-specific manner in FNY01_3_2 and FNY01_3_3. The difference between the 2 curves in the top graph was statistically significant (paired Student t test between the average gene body methylation of the most expressed allele vs least expressed allele P < .0006 for both individuals separately, and P < 1.2×10E-09 combined). Autosomal protein-coding genes exhibiting at least a twofold difference in allele expression were analyzed. Genes with <20 RNA-seq reads and <100 methylation counts were filtered out yielding a list of 26 and 34 genes for FNY01_3_2 and FNY01_3_3, respectively. The dotted line represents the raw data (the averaged methylation per windows joined by lines); the continuous lines the loess smooth of the same data. The meta-gene analysis was performed as described in panel A. (D) Scatter-plot illustrating the relationship between allele-specific expression and methylation in FNY01_3_2 and FNY01_3_3. Data processing and definition of allele-specific expression are as in panels B and C. Allele-specific methylation is defined as (βmaternal − βpaternal) with β = (# of methylated reads)/(# of total reads). (E) Meta-plots illustrating the progressive decay of DNA methylation in the sequences flanking active gene bodies (at position 0). The methylation fraction for 1-kb windows of 5′ and 3′ flanking sequences was averaged to generate the plots (see “Methods”). The x-axis represents the distance in kilobytes of each window to either the TSS (negative numbers) or to the TES (positive number). The black dots represent the raw data; the red curve represents the kernel regression smooth of the same data. The inverted spikes in the middle are caused by the UMRs that are present near the promoters of each gene. For both the 3′ and 5′ flanking sequences, the methylation fraction was maximal in the gene body and progressively decreased in the flanking sequences until it reached the average methylation level that is typical of the PMDs for each cell type.
To determine whether transcription levels correlate with gene body methylation, we analyzed genes that were expressed in an allele-biased manner in BasoE. Focusing on these genes provides unequivocal results, similar to experimentally disrupting a regulatory element, provided that the allele-biased differences are mostly from cis-acting mutations in regulatory elements. To validate this assumption, we took advantage of the sibling relationship between FNY01 3_2 and 3_3 and compared the haplo-identical regions of their genome (where the 2 sisters inherited the same paternal and maternal chromosomes) to the haplo-non-identical regions (where they inherited opposite alleles of maternal and paternal chromosomes). Haplo-identical genes exhibited a high correlation between the maternal and paternal expression differences (r2 = 0.754 for the highly expressed genes (number of reads >500), and r2 = 0.361 for all analyzable genes (number of reads >20), whereas haplo-non-identical genes did not, demonstrating that differences in expression between alleles were indeed mostly cis-linked and genetic in origin (Figure 3B).
Respectively, 26 and 34 genes in individuals 3_2 and 3_3 exhibited twofold or higher allelic difference in expression and had sufficient coverage to assess allele-specific gene body DNA methylation. Expression and gene body methylation for this set of genes were highly positively correlated (r2 ≈ 0.45; Figure 3C-D) and the difference in gene body methylation between the highly expressed and the least expressed alleles was statistically significant (paired Student t test P = 1.2×10E-09 for the combined FNY01_3_2 and 3_3 samples), suggesting that transcription levels directly determine the levels of gene body methylation.
Methylation of the flanking sequences of active genes decays progressively
To look specifically at methylation in gene body-flanking regions, we focused on the subset of flanking sequences that were not part of a neighboring active gene (see “Methods”). This revealed that DNA methylation was maximal in the gene body and progressively decreased in the flanking sequences until it reached the average methylation level of the PMDs characteristic of each cell type (Figure 3E). This was true for both the 3′ and 5′ flanking sequences, but on the promoter side were regions with very low methylation because of the presence of UMRs characteristic of promoter-associated regulatory regions and CpG islands.
Silent promoters exhibit different chromatin structures in HMDs and PMDs
Transcriptionally active CpG-rich promoters were almost all classified as active TSS (TssA or TssFlnk) by ChromHMM (supplemental Figure 4), demonstrating a good consistency between the transcriptional and chromatin data. Because silent genes can be found in both HMDs and PMDs, we investigated whether they were regulated differently within these 2 compartments at the chromatin level.
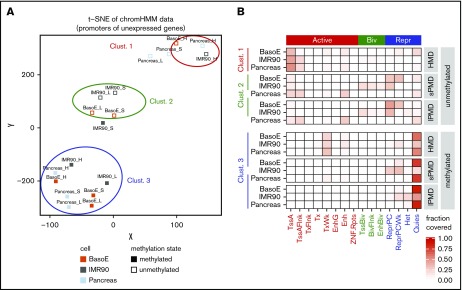
Analysis of either all or of only the CpG-rich (not shown) silent promoters of the differentiated cell types by unsupervised t-distributed stochastic neighbor embedding (t-SNE) based on their ChromHMM status revealed 3 distinct clusters (Figure 4A). Cluster 1 contained silent unmethylated promoters located in HMDs and cluster 2 contained silent unmethylated promoters located in s-PMDs and l-PMDs. Cluster 3 contained all silent methylated promoters. Promoters in cluster 1 were most often associated with active chromatin marks, but promoters in cluster 2 were most often associated with polycomb (PC) repression (Figure 4B), suggesting that silent promoters are often regulated by completely different mechanisms in HMDs and PMDs because, in HMDs, silent promoters tended to carry active marks, whereas, in PMDs, they tended to carry repressive marks. About three-quarters of the promoters in cluster 3 were classified as quiescent; the remaining as TxWk (weak transcription) or as ReprPCwk (weak repressed PC), suggesting that methylated silent promoters, by contrast to their unmethylated counterparts, tend to lose their histone marks and therefore their epigenetic identity and become similar to inactive intergenic chromatin. Methylated IMR90 promoters in s-PMDs and unmethylated pancreas promoters in PMDs did not fit neatly into these clusters, perhaps reflecting idiosyncratic features of these cells.
Figure 4.
Transcriptionally inactive promoters are regulated by different mechanisms in HMDs and PMDs. (A) Graph illustrating t-SNE analysis of the chromatin status as determined by ChromHMM of silent IMR90, pancreas, and BasoE promoters, separated by their methylation status and their location in PMDs or HMDs. The t-SNE was used with default parameters and a perplexity of 4. Inactive promoters from BasoEs, IMR90, and pancreas were divided into 6 categories: methylated (open squares) and unmethylated (closed squares) located in either HMDs (H), s-PMDs (S), or l-PMDs (L). Multiple runs of t-SNE for all inactive promoters, as well as analyses using only the inactive CpG-rich promoters resulted in very similar clusters (not shown). Inactive promoters fell into 3 clusters: cluster 1 contained unmethylated promoters located in HMDs, cluster 2 contained unmethylated promoters located in short and long PMDs, and cluster 3 contained all methylated promoters (except for the IMR90 s-PMDs). (B) Heat map illustrating the chromatin status (based on ChromHMM) of promoters in clusters 1, 2, and 3. The fractions of promoter overlapping the indicated ChromHMM state are shown. Promoters in cluster 1 generally exhibited active chromatin marks, whereas those in cluster 2 exhibited repressive or bivalent marks, demonstrating that the mechanisms of silencing of unmethylated promoters in HMDs and PMDs are different. Promoters in cluster 3 were almost all quiescent regardless of their location in HMDs or PMDs.
Promoter DNA methylation is largely invariant but chromatin structure changes dramatically, particularly in genes that are silent in both HSPCs and BasoEs
To understand how the epigenome of erythroid cells is established, we analyzed changes occurring upon differentiation of HSPCs into BasoEs. Consistent with previous reports,13 we observed a 13% decrease in the number of expressed promoters in BasoE, resulting from silencing of 2234 and activation of 813 promoters.
Almost no changes were observed in the methylation status of promoters during erythroid differentiation. Very few promoters acquired or lost any UMRs or LMRs. (Figure 5A; supplemental Figure 5A; and not shown). Erythroid differentiation was therefore associated with changes in the expression of 3047 promoters but with largely invariant methylation states between HSPCs and BasoEs with <0.5% of the CpG-rich promoters changing their methylation status during differentiation.
Figure 5.
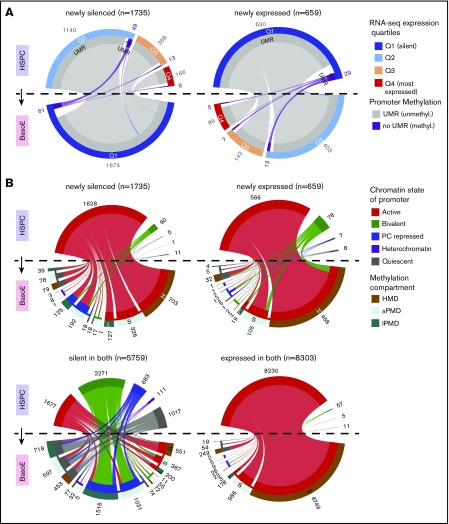
DNA methylation and chromatin changes during erythroid differentiation in CpG-rich promoters. (A) Circos plots illustrating the changes in DNA methylation status of newly silenced (left) and newly expressed (right) CpG-rich promoters during differentiation from HSPCs (top half of the circle) to BasoEs (bottom half) as a function of expression levels. The bands connecting the half-circles indicate the redistribution of genes from HSPCs into the indicated categories in BasoEs. The outer circle represents the expression levels (Q1 [silent], dark blue; Q2, blue; Q3 orange; Q4, [highly expressed], red). The inner circle segments represent the methylation status of the promoters (gray: unmethylated [UMR-containing]; purple: methylated [no-UMR]). The numbers on the outside indicate the number of promoters according to their methylation status split by expression quartile. In the plot on the left, promoters (active in HSPC [quartiles Q2-Q4] but silent in BasoE [quartile Q1]) were almost all unmethylated and remained so after differentiation. In the plot on the right, the newly expressed promoters (silent in HSPCs but active in BasoEs) were almost all already unmethylated in HSPCs. (B) Circos plots illustrating the change in chromatin status of promoters as a function of their location in HMDs or PMDs and as a function of their change in expression during differentiation from HSPCs to BasoEs (clockwise): newly silenced in BasoEs, newly expressed in BasoEs, expressed in both HSPCs and BasoEs, and not expressed in either HSPCs or BasoEs. The bands connecting the half-circles indicate the redistribution of genes from HSPCs into the indicated categories in BasoEs. The outer circles represent the location of the promoters in HMDs (H, brown), short PMDs (S, light green), or long PMDs (L, dark green). The outer circle was omitted from the HSPC side of the graph, because almost the entire genome of HSPC is located in HMDs. The inner circle represents the chromatin states. For clarity, the 15 ChromHMM classes were summarized into 5 larger categories: active (TssA, TssAFlnk, TxFlnk, Tx, TxWk, EnhG, Enh, ZNF/Rpts; red), bivalent (TssBiv, BivFlnk, EnhBiv; purple), repressed (ReprPC, ReprPCWk, green), heterochromatin (blue), and quiescent (gray). The circle segments above the dotted line represent the methylation and chromatin status of promoters in HSPCs, those below the dotted line the methylation and chromatin status of promoters in BasoEs. The numbers on the outside indicate the number of promoters in each category. The graph illustrates the observation that the promoters newly expressed in BasoEs and promoters expressed in both cell types exhibits few chromatin changes during differentiation and that the promoters newly silenced in BasoEs and promoters silent in both cell types exhibit much more dramatic chromatin structure changes.
Analysis of chromatin structure revealed that CpG-rich promoters expressed in both HSPCs and BasoEs, or newly expressed in BasoEs, exhibited the same chromatin status in both cell types in ∼95% of cases (Figure 5B; supplemental Figure 5B-D). By contrast, promoters that were not expressed in either cell type, or that were newly silenced, exhibited dramatic changes, with only 59% of the marks present in HSPCs retained in BasoEs (Figure 5B; supplemental Figure 5B-D). These changes, which do not have an obvious biological significance in the case of the 5759 genes that are silent in both lineages, were quite varied but often involved a loss of bivalent promoters which became mostly PC-repressed as well as conversion from active or PC-repressed states to a quiescent state.
The net effect of these changes was a large decrease in histones carrying posttranslational modifications. EnrichR38 analysis revealed that promoters silent in BasoEs that changed chromatin structure were highly enriched in categories related to lymphopoiesis and myeloid cells, whereas promoters expressed in BasoEs that changed chromatin structure were highly enriched in categories related to erythropoiesis (supplemental Table 2), suggesting an association with lineage restriction.
Methylation canyons often expand during erythroid differentiation
We identified 643 DNA methylation canyons (defined as UMRs >5 kb) with 457 common to BasoEs and HSPCs, 148 BasoE-specific, and 40 HSPC-specific canyons, suggesting that there might be more variability in canyon than in promoter methylation. However, DNA methylation in canyons was generally relatively stable because the differences were mostly caused by brief interruptions of canyons or by contraction of small canyons. Only 33 canyons exhibited methylation differences >10% between HSPCs and BasoEs, and most of those were in constitutive heterochromatin at the chromosome ends (not shown). Consistent with previous reports on UMR sizes,12 analyses of canyon edges revealed that the wave of demethylation associated with erythroid differentiation generally led to an increase in canyon size, although the increase was not systematic (supplemental Figure 5E).
The chromatin structure of the 455 canyons common to HSPCs and BasoEs fell into 3 categories: PC-repressed, bivalent, or active. Upon differentiation, 98% of the repressed and 83% of the active canyons maintained their state, but 96% of the bivalent canyons became PC-repressed. Differentiation was therefore associated with a massive switch from bivalent to repressed chromatin with very few cases of activation (supplemental Figure 5F).
DMRs between HSPCs and BasoEs are mostly associated with enhancers
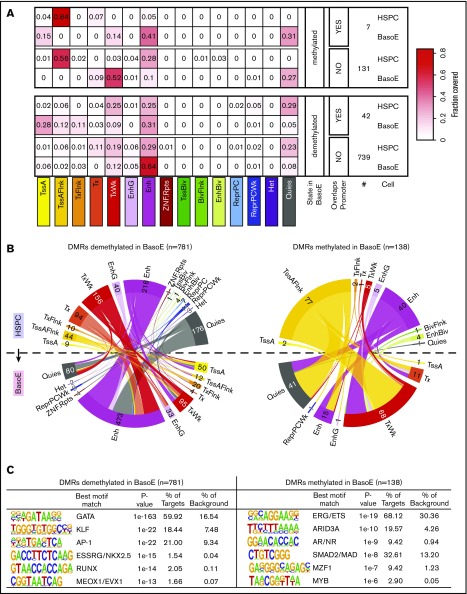
To identify DMRs between HSPCs and BasoEs, we used the DSS software package,34 limiting the analysis to regions that are HMDs in BasoEs because almost all PMDs are called as DMRs when compared with HSPCs, which contain hardly any PMDs. This revealed 138 DMRs associated with gain and 781 associated with loss of methylation in BasoE (supplemental Table 3). Eighty-two percent of these DMRs overlapped with a UMR or an LMR, but as expected from the previous analysis, only 5% overlapped with a promoter (supplemental Table 3). This suggested that many of these DMRs might be located in enhancers or other regulatory elements.
Analysis of histone marks corroborated this hypothesis and revealed that 81% of the regions covered by DMRs overlapped with regions identified as enhancers, genic enhancers, or bivalent enhancers by ChromHMM in BasoEs, HSPCs, or both (Figure 6A-B). To characterize these DMRs in more detail, we compared the chromatin state of each of these DMRs in BasoEs and HSPCs (Figure 6B). This revealed that DMRs demethylated in BasoEs generally transitioned from a quiescent or weakly transcribed ChromHMM state (which differs from quiescence by the presence of small amounts of the H3K36me3 mark) to an enhancer state. By contrast, DMRs methylated in BasoEs tended to be either in an enhancer or flanking an active transcription start site (TssAFlnk) state in HSPCs and tended to acquire a quiescent or weakly transcribed state in BasoE.
Figure 6.
DNA methylation and chromatin changes during erythroid differentiation in DMRs. (A) Heat maps illustrating the changes in chromatin state of short DMRs within HMDs between HSPCs and BasoEs. Top and bottom panels show average fractional chromatin composition by sequence length of DMRs that are more or less methylated in BasoEs than in HSPCs, respectively. They are additionally broken down into DMRs that overlap promoters and those that do not. The numbers indicate the number of DMRs in each category. The 15 ChromHMM states are indicated below the heat map. Most notable changes include disappearance of the flanking active TSS (TssAFlnk) marks and increase of enhancer marks between HSPCs and BasoEs. (B) Circos plots illustrating the changes in chromatin state of DMRs that lose (left) or gain (right) methylation during differentiation from HSPCs (top half of the circle) to BasoEs (bottom half). The bands connecting the half-circles indicate the redistribution of DMRs from HSPCs into the indicated ChromHMM categories in BasoEs. The numbers indicate the number of DMRs in each category. Again, the most notable changes include the increase in the enhancer marks and reduction of quiescence marks upon demethylation of DMRs upon differentiation (left), and, conversely, the increase in quiescence marks and weak transcription and the decrease of chromatin marks associated with enhancers and transcriptional flanks upon gain of methylation in DMRs between HSPCs and BasoEs (left). (C) Top enriched sequence motifs in DMRs identified by Homer motif analysis. Best motif matches to the identified motifs shown as positional weight matrices (PWMs) are indicated, and enrichment parameters for each motif are shown. DMRs demethylated in BasoEs were enriched in erythropoietic transcription factors (right), whereas DMRs that gained methylation in BasoEs were enriched in genes associated with stem cells and lymphoid cells, consistent with lineage restriction-associated changes in enhancer methylation.
To determine whether these DMRs were enriched in specific DNA-binding motifs, we performed motif analysis using Homer39 and found that the DMRs demethylated in BasoEs were remarkably enriched in binding sites for GATA, KLF, and AP-1 transcription factors (Figure 6C), which suggests that the changes in methylation might be associated with activation of GATA1, KLF1, and NFE-2, 3 factors known to be important for erythropoiesis. DMRs methylated in BasoEs were enriched in binding motifs for ERG/ETS, Arid3A, AR/ND, and SMAD, which are associated with self-renewal, lymphopoiesis, and myelopoiesis, suggesting that these DMRs are associated with downregulation of transcription factors normally expressed in HSPCs and downregulated in erythroid cells (Figure 6C). These results support and extend the conclusions of Yu et al,12 who stated that changes in methylation in short regulatory regions are preferentially associated with transcription factor binding, and provide a novel list of putative erythroid-specific enhancers identified by histone mark analysis and validated by changes in DNA methylation.
Change of chromatin structure during erythroid differentiation from HSPCs to BasoE
In summary, chromatin structure during erythroid differentiation was highly dynamic, but the majority of the changes affected either putative enhancers, genes not expressed in either cell type, genes that were newly silenced in BasoEs, or bivalent DNA methylation canyons. Newly activated promoters most often already carried active histone marks in HSPCs. Transcriptionally inactive promoters located in regions that remained classified as HMDs in BasoEs were more likely to retain their active marks and to lose their repressed and heterochromatic marks than promoters located in regions that switch from being HMDs in HSPCs to PMDs in BasoEs. Notably, the loss of bivalent marks occurred equally in PMDs and HMDs, whereas active marks were generally preserved in promoters located in HMDs in BasoE but erased in 50% of promoters located in PMDs in BasoEs (Figure 5B; supplemental Figure 5B). By contrast, PC-repressed and heterochromatic marks were less well preserved in HMDs than in PMDs.
Allele-specific DMRs are associated with the most variable DNA sequences in the genome
Attempts to analyze small allele-specific DMRs (a-DMRs) were not fruitful because the allele-specific data were too sparse. To analyze allele-specific DNA methylation at a larger scale, we used the Bioconductor DSS package to identify a-DMRs (Figure 7A). This revealed 751 and 727 a-DMRs >20 kb in FNY01_3_2 and 3_3, respectively. Eighty-seven percent of these a-DMRs were located in PMDs and 17.3% of all a-DMRs and 30.2% of the haplo-identical a-DMRs were common between FNY01_3_2 and 3_3. Further analysis based on data from the ENCODE consortium revealed that a-DMRs were not enriched in genes, in DNA repeats, or in any particular epigenetic states (not shown). However, SNP analysis revealed that, although a-DMRs had a similar mutation spectrum and Ts/Tv ratio (supplemental Figure 6), they were significantly richer in SNPs than the rest of the genome with equivalent depth of sequencing coverage and thus potential to detect SNPs (P < .001, hypergeometric permutation test; Figure 7B). This suggests that the primary DNA sequence might be a direct determinant of DNA methylation in intergenic sequences because we detected differences in DNA sequence but no striking epigenetic differences in these regions.
Figure 7.
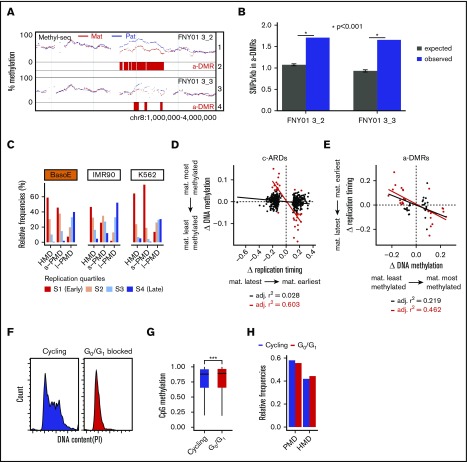
Analysis of differential methylation between alleles. (A) GenPlay genome browser view illustrating an a-DMR between parental alleles on chromosome 8. Tracks 1 and 3 illustrate smoothed average methylation levels for the paternal (blue) and maternal (red) alleles for the indicated individual; tracks 2 and 4 highlight the corresponding a-DMRs identified by DSS software as described in the “Methods” section. (B) A-DMRs are richer in heterozygous SNPs than the rest of the genome. Data were generated using SnpEff based on the previously published genome sequence of FNY01_3_2 and 3_3. The expected number of SNPs was calculated by shuffling the a-DMRs 10 000 times at random location in the genome where the coverage was similar to that of the a-DMRs. The average of ∼1 SNP/kb in the expected samples is very close to the average number of SNPs/kb genome-wide observed in these 2 individuals. (C) Bar graphs illustrating the replication timing of HMDs, short PMDs, and long PMDs in 3 cell types. Average replication timing was determined for 5-kb tiles genome-wide and separated into quartiles. The fractions of each domain in the respective quartile of replication timing are indicated. In BasoE, IMR90, and K562 cells, both HMDs and short PMDs replicated early, whereas long PMDs replicated late. (D) Scatter plots illustrating the correlation between timing of replication and DNA methylation in c-ARDs. x-axis: differential allelic replication timing ratio = maternal timing ratio − paternal timing ratio, with timing ratio being the sum of maternal or paternal TimEX read counts in S phase over the sum of maternal or paternal TimEX read counts in G1 phase, respectively, across individual regions; y-axis: differential allelic methylation = βmat − βpat with β = ratio of the sum of maternal or paternal methylated reads over the sum of maternal or paternal total read counts, respectively, across individual regions. Black dots, regression line and Pearson r2 for all c-ARDs; red dots, regression line and Pearson r2 for c-ARDs with an abs(Δ DNA methylation) >0.05. There is no correlation between timing of replication and DNA methylation in c-ARDs except for a subgroup of outlier c-ARDs (in red) that overlap with a-DMRs. (E) Scatter plots illustrating the correlation between timing of replication and DNA methylation in a-DMRs. X-axis: differential allelic methylation (as in panel D) across individual a-DMRs; y-axis: differential allelic replication timing ratio (as in panel D). All a-DMRs and c-ARDs plotted contained at least 300 informative reads for allele-specific measurement of DNA methylation or for timing of replication analysis. Black dots, regression line and Pearson r2 for a-DMRs with an abs(Δ DNA methylation) >0.05; red dots, regression line and Pearson r2 for a-DMRs with an abs(Δ DNA methylation) >0.1. There is a strong correlation between timing of replication and DNA methylation in the strong a-DMRs (red dots). (F) Histograms illustrating the DNA content as assayed by propidium iodide (PI) staining in p51R cells that were cycling or blocked in G0/G1 for 4 days by incubation in a culture medium containing 0.5% fetal bovine serum. The G0/G1 block is almost complete. (G) Box plot illustrating CpG methylation in G0/G1 and in cycling (control) cells. Cells blocked in G0/G1 are statistically significantly more methylated than the cycling cells (P < .001, Wilcoxon rank sum test). (H) Bar plots illustrating the proportion of PMDs and HMDs in G0/G1 and in cycling (control) cells. Cells blocked in G0/G1 contain fewer PMDs than cycling cells.
Timing of replication in S phase does not generally correlate with the levels of DNA methylation
Because a correlation between low methylation levels and late replication has been reported previously,15 we investigated this relationship in our data. Importantly, in BasoEs, IMR90, and K562, the 3 cell types for which data were available, ∼90% of the HMDs and the vast majority of s-PMDs replicated early, whereas almost all l-PMDs replicated late (Figure 7C), clearly suggesting that PMDs can form throughout S phase.
To assess whether the timing of replication during S phase affects the level of DNA methylation, we analyzed previously reported c-ARDs (core–asynchronously replicated regions), which are regions in which the 2 alleles do not replicate at the same time.30,31 We identified ∼500 c-ARDs in individuals FNY01_3_2 and 3_3 with enough coverage to measure methylation in an allele-specific manner. No correlation between the c-ARDs and allele-specific methylation was found except for a group of outlier c-ARDs overlapping with the regions with highest allele-specific methylation differences (Figure 7D). This suggested that, except for the outliers, the timing of replication in S phase does not determine the levels of DNA methylation. To assess the relationship between timing and methylation in a-DMRs, we calculated the allele-specific timing of replication of each allele in all a-DMRs. This revealed a strong correlation (r2 = 0.462) between the methylation and timing differentials in a-DMRs, with the least methylated allele replicating earlier than the most methylated allele (Figure 7E), suggesting that a higher level of methylation in a-DMRs causes a delay in the timing of replication.
Most maintenance methylation occurs shortly after replication, but some methylation activity has been detected outside of S phase.40-43 To assess when during the cell-cycle DNA methylation in PMDs and HMDs is maintained, we measured DNA methylation using a novel reduced representation method in human p51R mesenchymal cells (supplemental Figure 7), which are highly responsive to serum starvation-induced cell-cycle block.44 Comparison of p51R cells that were cycling or blocked for 96 hours in G0/G1 by serum starvation (Figure 7F) revealed that the genome of G0/G1-arrested cells was more methylated than that of cycling cells (P < .001, Wilcoxon rank sum test; Figure 7G) and exhibited fewer PMDs (Figure 7H), covering 55.5% of the genome compared with 58% in cycling cells. We concluded that non–S-phase DNA methylation might contribute to cycling cells having more of their genome in PMDs than stem and progenitor cells that do not cycle as often.
Discussion
We provide a genome-wide allele-specific BasoE methylome and show that BasoEs are particularly rich in PMDs. Globally, the methylome of BasoEs was more similar to other differentiated cells than to HSPCs or K562 erythroleukemia cells. These comparisons also suggested that methylomes can be divided based on their PMD content into 3 classes that do not correspond to cell lineages but are associated with varying differentiation potential, with PMDs covering, respectively, 1% to 10%, 30% to 74%, and 85% to 90% of the methylomes of stem and progenitors cells, differentiated cells, and transformed cell lines.
We have identified 919 DMRs enriched in putative enhancers, which are associated with binding sites for transcription factors known to be involved in hematopoiesis, particularly with erythropoiesis. This provides an important resource to identify novel enhancers important for erythropoiesis and supports previous studies that have shown that short regulatory regions that are not promoters were preferentially subject to lineage-specific changes in DNA methylation.45-47
However, except for these potentially functionally important changes located in regulatory regions, most of the changes in DNA methylation during erythroid differentiation have no known functions. We found that the vast majority of the global DNA demethylation during erythroid differentiation from HSPCs to BasoEs occurred in unexpressed regions, leading to PMD formation in intergenic regions and gene bodies of inactive genes. In contrast, DNA methylation status and histone marks of most promoters expressed in BasoEs did not change during erythroid differentiation, regardless of whether they were expressed or silent in HSPCs. Therefore, we conclude that, in accordance with findings from Xu J et al,48 promoter DNA methylation levels and histone marks necessary for gene expression in BasoEs are already largely preestablished in HSPCs.
We propose that, in BasoEs, the formation of PMDs is caused by a decrease in baseline maintenance methylation, as supported by the observation of Shearstone et al,49 who have provided evidence that erythroid demethylation is replication-dependent in the 5% of the genome they sequenced. We also propose that maintenance of HMDs results from gene transcription–associated methylation coupled with diffusion of the DNA methyltransferase DNMTs in the region flanking transcribed genes, because we have observed an association between gene body methylation of active genes and transcription and because methylation levels in BasoE are highest in active gene bodies and decrease gradually in the flanking sequence until reaching average methylation levels of PMDs.
These observations suggest that global methylation patterns in BasoEs are set by 3 largely independent mechanisms: UMR and LMR formation, which is driven by transcription factor binding associated with histone modification that prevents DNA methylation and causes differentiation-associated changes in DNA methylation at specific CpGs and at DMRs; transcription, which is associated with the deposition of histone marks that favor a high level of methylation3,50; and maintenance methylation after DNA replication, which sets a baseline level of methylation for the part of the genome not regulated by the other 2 mechanisms.
This raises the question of whether maintenance methylation decreases in late S phase when inactive genes tend to replicate,51 or throughout S phase. Our finding that s-PMDs replicate early, whereas l-PMDs replicate late, and that the timing of replication in asynchronously replicated regions (c-ARD) did not correlate with methylation levels show that the demethylation in BasoEs occurs independently of the timing of replication.
Analysis of p51R cells revealed a small amount of non–S-phase methylation in cells blocked in G0/G1, which led to a decrease in the number and size of PMDs. Although we had to use p51R mesenchymal cells for these experiments because BasoEs cannot be blocked in G0/G1 for a long period, this suggests that cells, such as HSPCs that spend extended periods in quiescence might have very few PMDs, in part because they slowly accumulate DNA methylation when they are not cycling. In addition, HSPCs likely also have intrinsically higher levels of maintenance methylation in S phase because we observed increased levels of DNMT1 and decreased levels of DNMT3A and DNMT3B between human HSPCs and BasoEs in our RNA-seq data (not shown). These observations, which are in accordance with the data of Shearstone et al.11 during mouse erythroid differentiation, suggest that changes in DNMT levels during differentiation contribute to PMD formation in human BasoEs because they affect the levels of methylation maintenance.
Promoters of genes silenced during erythroid differentiation and, surprisingly, promoters of genes silent in both HSPCs and BasoEs exhibited a much more dynamic chromatin structure than promoters of newly expressed genes and tended to lose chromatin marks because they transitioned from an active to a quiescent state or from a bivalent to a repressed state that both involve a loss of H3K4me3. Given that most of these genes are located in PMDs, this commonality provides another potential mechanism for the decrease in maintenance methylation responsible for PMD formation, because the factors that maintain H3K4me3 have been shown to interact directly with DNMTs.52
Except for the changes in methylation in regulatory regions that involve <0.01% of all CpGs, the broad function of DNA demethylation and chromatin changes occurring during erythroid differentiation are unclear. Whether these changes are epiphenomena associated with the presumably functionally important changes in regulatory regions or whether they suppress spurious gene expression and differentiation into other lineages once the cells are committed to an erythroid fate or serve some other purpose will have to be determined.
Importantly, we found that promoter silencing is often paradoxically associated with the retention of active marks in HMDs but with repressive polycomb marks in PMDs, demonstrating that unmethylated silent promoters are regulated by different mechanisms in HMDs and PMDs. We also found that promoters and DNA methylation canyons tend to lose active and bivalent marks in PMDs. Together, these observations suggest that PMD formation might be a mechanism to segregate a fraction of the silent promoters in a specialized genomic compartment and might indicate a biologically important function of PMDs. However, causes and consequences are unclear here.
The chromatin structure in PMDs is simpler than in HMDs because PMDs carry few histone marks and are not transcribed. This likely facilitates replication, because collisions between the replication and transcription machinery are known to stall replication.53,54 Another possible role of PMDs, which can be considered as a long succession of nondescript nucleosomes, might therefore be to facilitate the very rapid divisions that are characteristic of precursors of mature effectors such as red blood cells and mature B cells. However, a recent report that final maturation of megakaryocytes is not associated with general demethylation suggests that demethylation is lineage specific and not a general feature of terminal hematopoietic differentiation.55
We found that a-DMRs are enriched in SNPs, but failed to detect epigenetic differences between a-DMRs and the rest of the genome, suggesting that in untranscribed regions that are not regulatory sequences, the primary DNA sequence might be an important determinant of the levels of DNA methylation. We also found that the level of DNA methylation in strong a-DMRs inversely correlated with timing of replication, demonstrating that in these regions the more highly methylated allele replicates later than their less methylated allelic counterpart.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Einstein Stem Cell, flow cytometry and epigenomic facilities for essential contribution to this study.
This study was funded by grant from the National Institutes of Health, National Heart, Lung, and Blood Institute (1R01HL130764) and from NYSTEM (C030135 and C029154) (E.E.B.).
Authorship
Contribution: B.B. contributed to the bioinformatics analysis and manuscript composition; J.L. contributed to the bioinformatics analysis; Z.Y., S.Z., and R.M. performed the experiments; J.G. contributed to manuscript composition; M.S. performed experiments and contributed to bioinformatics analysis and manuscript composition; and E.E.B. designed and supervised the study and contributed to experimental design, bioinformatics analysis, and manuscript composition.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric E. Bouhassira, Department of Cell Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: eric.bouhassira@einstein.yu.edu.
References
- 1.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484-492. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204-220. [DOI] [PubMed] [Google Scholar]
- 3.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321-326. [DOI] [PubMed] [Google Scholar]
- 4.Jeong M, Sun D, Luo M, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46(1):17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells [published correction appears in Nature. 2014;514:126]. Nature. 2011;471(7336):68-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44(1):40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raddatz G, Gao Q, Bender S, Jaenisch R, Lyko F. Dnmt3a protects active chromosome domains against cancer-associated hypomethylation. PLoS Genet. 2012;8(12):e1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hon GC, Hawkins RD, Caballero OL, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hon GC, Rajagopal N, Shen Y, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45(10):1198-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334(6057):799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Mo Y, Ebenezer D, et al. High resolution methylome analysis reveals widespread functional hypomethylation during adult human erythropoiesis. J Biol Chem. 2013;288(13):8805-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogart A, Lichtenberg J, Ajay SS, Anderson S, Margulies EH, Bodine DM; NIH Intramural Sequencing Center. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22(8):1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulis M, Merkel A, Heath S, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20(4):670-680. [DOI] [PubMed] [Google Scholar]
- 16.Olivier E, Qiu C, Bouhassira EE. Novel, high-yield red blood cell production methods from CD34-positive cells derived from human embryonic stem, yolk sac, fetal liver, cord blood, and peripheral blood. Stem Cells Transl Med. 2012;1(8):604-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66(1):188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. ; The NIH Roadmap Epigenomics Mapping Consortium. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patro R, Duggal G, Kingsford C. Accurate, fast, and model-aware transcript expression quantification with Salmon. Nat Methods. 2017;14:417-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haberle V, Forrest ARR, Hayashizaki Y, Carninci P, Lenhard B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015;43(8):e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaji H, Lizio M, Itoh M, et al. ; FANTOM Consortium. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014;24(4):708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackenberg M, Previti C, Luque-Escamilla PL, Carpena P, Martínez-Aroza J, Oliver JL. CpGcluster: a distance-based algorithm for CpG-island detection. BMC Bioinformatics. 2006;7(1):446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lajugie J, Bouhassira EE. GenPlay, a multipurpose genome analyzer and browser. Bioinformatics. 2011;27(14):1889-1893. [DOI] [PubMed] [Google Scholar]
- 24.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: combined DNA methylation and SNP calling for bisulfite-seq data. Genome Biol. 2012;13(7):R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Zhang Y, Scheuring CF, Wu CC, Dong JJ, Zhang HB. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat Protoc. 2012;7(3):467-478. [DOI] [PubMed] [Google Scholar]
- 27.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burger L, Gaidatzis D, Schübeler D, Stadler MB. Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res. 2013;41(16):e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE. Allele-specific analysis of DNA replication origins in mammalian cells. Nat Commun. 2015;6(1):7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay R, Lajugie J, Fourel N, et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10(5):e1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desprat R, Thierry-Mieg D, Lailler N, et al. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 2009;19(12):2288-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185-193. [DOI] [PubMed] [Google Scholar]
- 34.Park Y, Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016;32(10):1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lajugie J, Mukhopadhyay R, Schizas M, Lailler N, Fourel N, Bouhassira EE. Complete genome phasing of family quartet by combination of genetic, physical and population-based phasing analysis. PLoS One. 2013;8(5):e64571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaidatzis D, Burger L, Murr R, et al. DNA sequence explains seemingly disordered methylation levels in partially methylated domains of Mammalian genomes. PLoS Genet. 2014;10(2):e1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lathrop MJ, Hsu M, Richardson CA, et al. Developmentally regulated extended domains of DNA hypomethylation encompass highly transcribed genes of the human β-globin locus. Exp Hematol. 2009;37(7):807-813.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiryanov GI, Kirnos MD, Demidkina NP, Alexandrushkina NI, Vanyushin BF. Methylation of DNA in L cells on replication. FEBS Lett. 1980;112(2):225-228. [DOI] [PubMed] [Google Scholar]
- 41.Spada F, Haemmer A, Kuch D, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176(5):565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hervouet E, Nadaradjane A, Gueguen M, Vallette FM, Cartron P-F. Kinetics of DNA methylation inheritance by the Dnmt1-including complexes during the cell cycle. Cell Div. 2012;7(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandiver AR, Idrizi A, Rizzardi L, Feinberg AP, Hansen KD. DNA methylation is stable during replication and cell cycle arrest. Sci Rep. 2015;5(1):17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24(8):1914-1922. [DOI] [PubMed] [Google Scholar]
- 45.Farlik M, Sheffield NC, Nuzzo A, et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Reports. 2015;10(8):1386-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda M, Glass JL, Thompson RF, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37(12):3829-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadler MB, Murr R, Burger L, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature [published correction appears in Nature. 2012;484(7395:550]. 2011;480(7378):490-495. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23(4):796-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shearstone J R, Pop R, Bock C, et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334(80):799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baubec T, Colombo DF, Wirbelauer C, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520(7546):243-247. [DOI] [PubMed] [Google Scholar]
- 51.Desprat R, Thierry-Mieg D, Lailler N, et al. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 2009;19(12):2288-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butler JS, Lee J-H, Skalnik DG. CFP1 interacts with DNMT1 independently of association with the Setd1 Histone H3K4 methyltransferase complexes. DNA Cell Biol. 2008;27(10):533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44(6):966-977. [DOI] [PubMed] [Google Scholar]
- 54.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017;170(4):774-786.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farlik M, Halbritter F, Müller F, et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell. 2016;19(6):808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.