ABSTRACT
In mammals, it has become increasingly clear that the gut microbiota influences not only gastrointestinal physiology but also modulates behavior. In domestic birds, ceca have the greatest gastrointestinal microbial population. Feather-pecking (FP) behavior in laying hens is one of the most important unsolved behavioral issues in modern agriculture. The aim of the present study was to assess the cecal microbial community of divergently selected high (HFP; n = 20) and low (LFP; n = 20) feather-pecking birds at 60 wk of age. The cecal samples were subjected to community profiling of 16S rRNA and in silico metagenomics using a modified bar-coded Illumina sequencing method on a MiSeq Illumina sequencer. Our results revealed that compared to HFP birds, LFP birds are characterized by an increased overall microbial diversity (beta diversity) shown by a difference in the Bray–Curtis index (R2 = 0.171, P < 0.05). Furthermore, operational taxonomic unit comparisons showed an increased presence of Clostridiae and decreased presence of Lactobaccillacae in HFP birds when compared to LFP birds (False Discovery Rate < 0.05, Mann–Whitney comparisons). Our data indicate that there may be differences in the cecal profile between these 2 lines of laying hens. More research, building on this first study using sequencing technology for profiling the chicken cecal microbiome, will be needed in order to reveal if and how there exists a functional link between the performance of FP and the cecal microbial community.
Keywords: cecal microbiota, gut microbiome, chicken, bird, feather pecking
INTRODUCTION
Feather pecking (FP) in birds kept for egg laying is a serious condition which affects up to 80% of birds in all housing systems (Gunnarsson, 1999). Genetic factors (Kjaer et al., 2001), including candidate genes, linked to the serotonergic system (Lutz et al., 2017), and nutritional factors, including dietary tryptophan levels, related to serotonin neurotransmission have also been identified to impact the risk of FP (van Hierden et al., 2004). Feather pecking is identified based on core behavioral symptoms, such as increased repetitive pecking at and pulling out of feathers from other birds (Savory, 1995), and damage to the feather cover. In addition to behavioral symptoms, FP birds display a wide range of simultaneously existing, neurological symptoms, which include fearfulness (de Haas, 2014) and hyperactive behavior (Kjaer, 2009), as well as non-neurological symptoms such as tryptophan/serotonin dysregulation (Birkl et al., 2017) and gastrointestinal abnormalities (Harlander-Matauschek et al., 2006). Gastrointestinal symptoms, including the strong desire for feathers (McKeegan and Savory, 2001; Harlander-Matauschek et al., 2009), altered intestinal motility (Harlander-Matauschek et al., 2006), and intestinal microbial metabolism most pronounced in the ceca (Meyer et al., 2013) of FP birds, suggest that gastrointestinal issues, such as altered composition of the intestinal microbiota, could contribute to the manifestation of FP. In humans, deviations from healthy microbial communities have been linked with diseases, including hyperactivity disorders (Jia et al., 2008), depressive disorders (Dash et al., 2015), and autism spectrum disorders (Mulle et al., 2013). Hence, the aim of our present study was to test the hypothesis that the cecal microbial community of FP birds show distinct differences to low-FP birds measured in cecal droppings. As avian ceca empty their contents separately and distinguishably from regular excreta by their mustard/dark brown color, sample collection does not require sacrificing birds for sampling their cecal content via dissection (Pauwels et al., 2015). Additionally, cecal samples are less variable in composition than fecal samples (Stanley et al., 2014), harbor the highest microbial cell densities, have the longest residence time of digesta (Oakley et al., 2014), and might be more representative of the gut microbial community. Hence, it was our goal to investigate the cecal microbial community, representative of the overall gut-microbiome. Microbial community structure in ceca droppings were profiled in 2 lines of laying hens selected divergently for FP (high- and low-FP birds) to determine the level and nature of similarity in cecal microbial structure between the 2 lines.
MATERIAL AND METHODS
Animals and Housing
This study was approved by the animal care committee at the University of Guelph (animal user protocol No. 3206). Birds that performed severe FP were required for this experiment. They were sampled from a total flock of 160 birds consisting of 40 high feather-pecking (HFP), 30 low feather-pecking (LFP) and 90 unselected control birds. For this experiment, we chose to compare the extremes of the 2 selected lines (LFP and HFP) for which the phenotype (expression of FP behavior) matched the genotype, as described by Meyer et al. (2013). These non-beak-trimmed White Leghorn hens were derived from a selection experiment in which a strain of laying hens was divergently selected for their propensity to feather-peck (Kjaer et al., 2001). The hatching and rearing environment was identical for all 3 lines and apart from a standard vaccination protocol, birds were not exposed to any medical treatment (antibiotics), nor did they receive probiotics at any time. Birds were kept in 10 groups of 16 birds (4 HFP, 3 LFP, 9 unselected) each and kept in identical enriched floor pens (183 L × 244 W × 290 H cm) as described in Kozak et al. (2016) under commercial management conditions at the Arkell Research Station in Guelph, Ontario, Canada. Birds were fed an Arkell Research Station layer mash diet with the following nutritional specifications; crude protein (min): 18%, crude fat (min): 5.5%, crude fiber (max): 2%, calcium (actual): 4.24%, phosphorus (actual): 0.68%, sodium (actual): 0.18%, vitamin A (min) 16,500 IU/kg, vitamin D (min) 4,130 IU/kg, vitamin E (min): 60 IU/kg.
Behavioral Recordings of FP
At 54 wk of age, each home pen was video recorded for 10 min (1× in the morning between 10 and 11 am, 1× in the afternoon between 2 and 3 pm) twice per week for 6 wk prior to fecal sampling. The number of FP bouts per individual was recorded on an all-occurrence basis (Altmann, 1974), with 1 bout being defined as a sequence of pecks at the same bird that is not interrupted for more than 4 s (Zeltner et al., 2000). Individuals were identified using continuously numbered silicone backpacks (8 × 6 × 0.5 cm), fastened onto the hens around the wings via 2 elastic straps secured to the backpacks with metal eyelets (Harlander-Matauschek et al., 2009).
Cecal Dropping Sampling
After observing pecking activity for 6 wk, 20 HFP with the highest and 20 LFP with the lowest FP activity were chosen and transferred to individual cages (45.7 L × 45.7 W × 40.5 H cm). The feed trough was placed at the front and a nipple drinker at the back of the cage. The hens had visual contact with, but no physical access to, their neighbors. Clean white PVC sheets were placed underneath each cage to collect feces from each bird over a period of 2 d. Approximately 1 g of cecal feces material, as identified by its characteristic homogeneous, smooth, and creamy texture and dark color, was sampled using a disposable pharmacist’s spatula for each bird. Samples were taken from the middle of cecal discharge to avoid contamination with non-cecal excreta. The cecal samples were transferred into 1.5 mL Eppendorf tubes and stored at –80°C until further processing of the sample.
16s rRNA Analysis of Bacterial Cecal Composition
DNA extraction was carried out as previously described (Bharwani et al., 2017). Bacterial community profiling of the 16S rRNA gene was carried out using a modified bar coded Illumina sequencing method (Bartram et al., 2011). Paired end reads of the V3 region were performed using the 341F and 518R primers (Muyzer et al., 1993). The 250 nucleotide paired-end sequencing was carried out on a MiSeq Illumina sequencer as per manufacturer's instructions at the McMaster Genome Center (McMaster University, Canada). The MiSeq data were processed by an in-house bioinformatics pipeline (Whelan et al., 2014). Sequencing results produced a minimum of 14,813 and a maximum of 1,34,898 reads per sample. Using QIIME (Caporaso et al., 2010), singletons were excluded and operational taxonomic unit (OTU) tables underwent 10 rarefactions at multiple sequencing depths to enable equal reads across samples. For alpha diversity analysis, Chao1 and Phylogenetic Diversity metrics was recruited using the alpha diversity workflow scripts, using the same number of sequences as the most indigent sample. For beta-diversity analyses, Jackknife resampling at a sequencing depth equal to 80% of the most indigent sample was used to generate Bray–Curtis distance matrices. Differences between the group microbiota profiles was assessed using the Monte Carlo Permutation Procedure (999 permutations) and similarities using the Analysis of similarities test. To analyze differential abundance of OTUs in group, the Mann–Whitney U-test followed by the Benjamini-Hochberg correction for multiple comparisons (False Discovery Rate < 0.05) was implemented using data first rarefied to even sequencing depth and then filtered to eliminate OTUs observed in fewer than 25% of the samples.
Statistical Analysis
The number of bouts (count data) between lines were compared using a GLIMMIX procedure (Version 9.4 SAS Institute Inc., Cary, NC, USA) with fixed factor line (HFP, LFP) on the means per week data. As we had count data, a Poisson distribution was assumed, pen used as a random effect, subject = bird ID, week as a repeated effect and due to the repeated measures on the same group of birds at different time points (weeks), a first-order autoregressive covariance structure was fitted to the pen by week effect. Results are presented as least square means ± standard error. Mann–Whitney U-test was used to assess statistical significance of measures derived from alpha diversity metrics. Spearman's rank-order correlation was run to determine the relationship between the individual level of FP bouts and associable OTUs.
RESULTS AND DISCUSSION
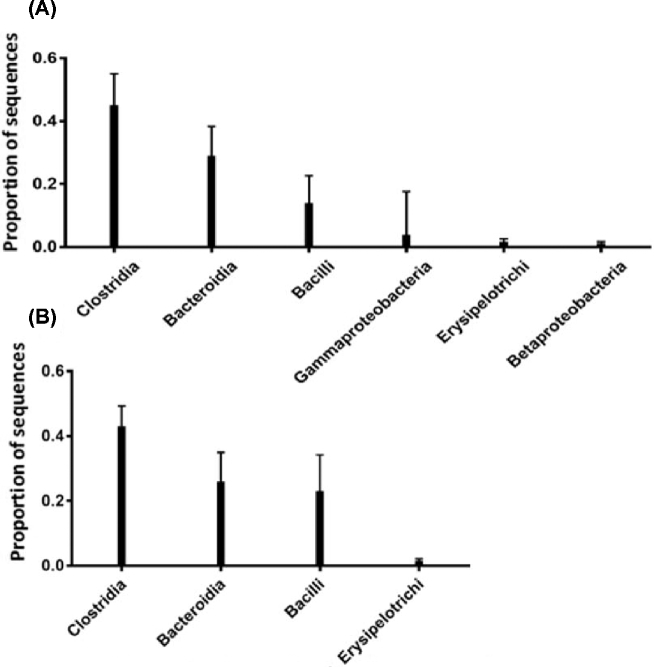
We aimed to investigate whether genetic selection for or against FP simultaneously resulted in colonization of different spectra of ceca microbiota when fed the same diet and reared under the same conditions. We compared the cecal microbial community of these 2 FP lines by using the 16S rRNA sequencing. Behavioral observations in home pens revealed that the 20 HFP birds showed a significantly higher number of FP bouts per minute than the 20 LFP birds (0.41 ± 0.311 vs 0.04 ± 0.092, F1, 19.5 = 4.89, P < 0.05), confirming that the birds’ observed phenotype matched the genotype. Table 1 presents an overview of Spearman's rank-order correlation between the individual levels of FP bouts and associable OTUs. The dominant ceca microbiota in adult laying hens belonged to the phyla Firmicutes and Bacteriodetes, which accounted for 88.7% in HFP and 92.8% in LFP birds (Figure 1). The relatively abundant genera of Firmicutes includes Clostridium, Bacillus, and Erysipelotrichus in descending order, whereas the Bacteroidetes was dominated by Bacteroides (Figure 2) in HFP and LFP birds. The relatively abundant genera of Proteobacteria, including Gammaproteobacteria and Betaproteobacteria, were higher in HFP as compared to LFP birds (Figure 2). Our results indicate Firmicutes, Bacteroidetes, and Proteobacteria as the most common phyla in the chicken ceca, with Actinobacteria accounting for the remainder and where finer scales of taxonomic resolutions show that the majority belongs to Clostridiales.
Table 1.
Spearman's Rank Order Correlation for Individual Feather-Pecking Bouts and Operational Taxonomic Units (OTUs).
| OTU ID | Test statistics | P-value (False Discovery Rate corrected) | Taxonomy |
|---|---|---|---|
| 94 | 0.48757492 | <0.05 | Root; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides |
| 147 | 0.602505853 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales |
| 153 | 0.522699938 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Bacteroides |
| 784 | 0.570215068 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillospira |
| 1042 | 0.444533386 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Veillonellaceae; g__Phascolarctobacterium |
| 1213 | 0.557725008 | <0.05 | Root; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__; g__ |
| 1435 | 0.457496421 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales; f__; g__ |
| 2392 | 0.431969949 | <0.05 | Root; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__ |
| 2566 | 0.431969949 | <0.05 | Root; p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella |
| 2607 | 0.423154236 | <0.05 | Root; p__Tenericutes; c__Mollicutes; o__RF39; f__; g__ |
Figure 1.
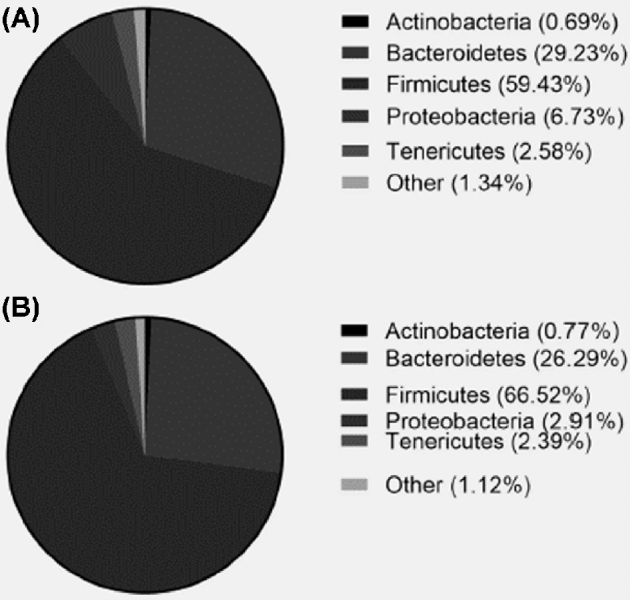
Relative abundance of bacterial diversity at phylum level in cecal samples of laying hens at 60 wk of age, comparing HFP birds (A) and LFP birds (B).
Figure 2.
Relative abundance of bacterial diversity at family level in cecal samples of laying hens at 60 wk of age, comparing HFP birds (A) and LFP birds (B).
Alpha-diversity analysis of cecal samples revealed no significant differences within the microbial community composition of each group. There were no significant differences in either the overall diversity (Faith's phylogenetic diversity, Figure 3A, Mann–Whitney U = 127, P < 0.05) or richness of the microbial community (Chao1, Figure 3B Mann–Whitney U = 129, P < 0.05). Based on these results, all samples from the 2 lines had a similar number and distribution of taxa with no strongly dominant taxa. These results could in part be explained by the birds being exposed to the same environmental sources of microorganisms, such as litter (Lu et al., 2003; Lovanh et al., 2007), and feed in particular (Muegge et al., 2011), which is known to affect overall diversity and richness of the microbiome in poultry (Oakley et al., 2014). In contrast to the lack of alpha diversity, beta diversity, measured by within-group distances, in the LFP group was significantly different from between-group distances between LFP and HFP (Bonferroni-correct non-parametric P < 0.05). Analysis of similarities revealed statistically significant differences (R = 0.171, P < 0.05) between LFP and HFP birds, which revealed that the phylogenetic composition of the ceca community in the HFP line was significantly different from the LFP line (Figure 4). Operational taxonomic units that discriminated between the HFP and LFP lines, (Benjamini-Hochberg False Discovery Rate correction, Mann–Whitney U, P < 0.05, Figure 4) included in HFP birds, OTU 22 (U = 330), 147 (U = 312), 504 (U = 294), and 72 (U = 299) with increased abundance of bacteria within the order of Clostridiales, whereas OTU 196 pointed towards increased abundance of the genus Anaerobiospirillum, order Aeromonadales. The OTU 6 (U = 303), however, indicated decreased abundance of bacteria of the genus Lactobacillus, order Lactobacillales in HFP birds. Although the majority of sequences belong to the various members of Clostridiales in a healthy laying hen ceca (Wei et al., 2013), an increased abundance of Clostridiales in cecal feces of HFP birds could be an important link in understanding how gut health may play a key role in the development of FP. This is further supported by Meyer et al. (2012) who found increased abundance of Clostridia in laying hens that ingested feathers. In humans, functional links between increased abundance of Clostridiales species and the development of behavioral disorders have been identified (Williams et al., 2011; Luna et al., 2017). A potential role for Clostridia in the development of abnormal behavior could involve microbial mediated production of abnormal metabolites (Clayton, 2012), such as p-cresol, which is known to directly affect the monoaminergic system via inhibiting dopamine β-hydroxylase (Goodhart et al., 1987). This consideration remains to be determined with further studies on abnormal metabolites in laying hens. Furthermore, our analysis revealed that the cecal microbiota of HFP birds is characterized by lower amounts of Lactobacilli. Interestingly, Lactobacillus can metabolize tryptophan into indole-derivates (Xu et al., 2002), which play a role in alteration of immune-related signaling pathways in the gut regulating inflammation (Keszthelyi et al., 2009). Additionally, Lactobacillus has been shown to have a direct, potentially positive effect on neurotransmission in the central nervous system, by impacting the signaling of visceral sensory inputs towards the hippocampus via vagal afferents (Bravo et al., 2011), potentially reducing sensitivity to stress-related behavior (Zagon, 2001), such as fearfulness, which has previously been linked to the development of FP (de Haas, 2014). However, whether the above activities of Lactobacillus play a central role in the development of FP in laying hens needs to be determined. Future experiments may be able to demonstrate not only an associative role for the microbiome in the development of FP, but also elucidate the relevant underlying physiological mechanisms and metabolic pathways. Furthermore, it remains to be determined whether the development of FP is fostered by changes in the gut microbiome or whether FP occurrence precedes microbial changes in the gastrointestinal tract. The results of the present study, however, provide an important first step in identifying the potential role of certain gut microbes in the development of FP.
Figure 3.
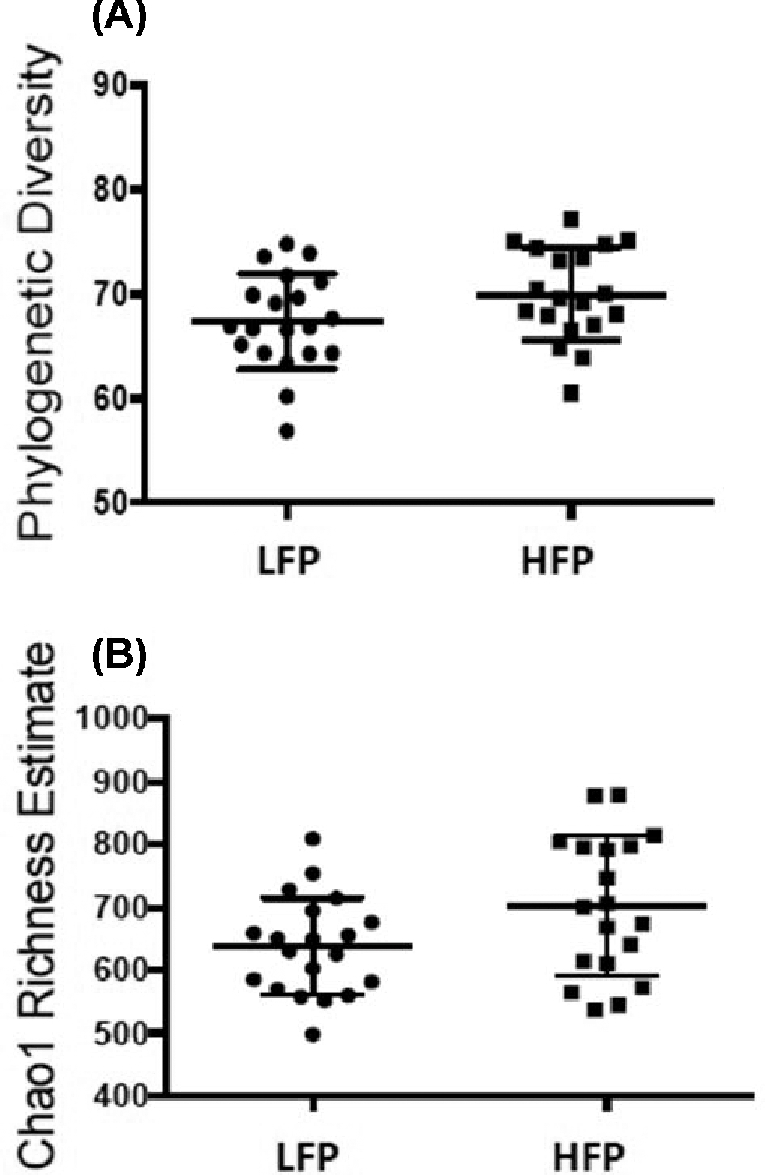
Alpha diversity: phylogenetic diversity (A) and Chao1 richness estimate (B) for LFP and HFP birds.
Figure 4.
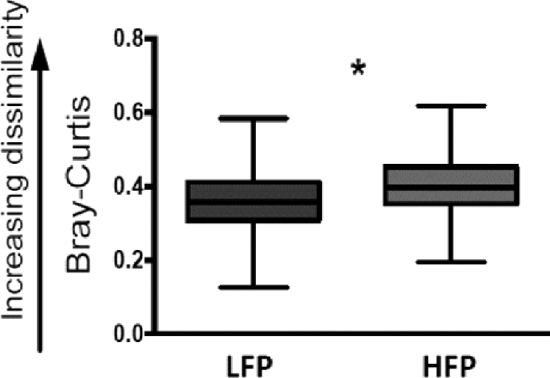
Beta diversity: analysis of Bray-Curtis distance between LFP and HFP birds.
ACKNOWLEDGMENTS
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC DG Grant No. 0531194), the Egg Farmers of Canada (EFC), and the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA).
REFERENCES
- Altmann J. 1974. Observational study of behavior: sampling methods. Behaviour 49:227–266. [DOI] [PubMed] [Google Scholar]
- Bartram A. K., Lynch M. D., Stearns J. C., Moreno-Hagelsieb G., Neufeld J. D.. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 77:3846–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharwani A., Mian M. F., Surette M. G., Bienenstock J., Forsythe P.. 2017. Oral treatment with Lactobacillus rhamnosus attenuates behavioural deficits and immune changes in chronic social stress. BMC Med. 15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkl P., Franke L., Rodenburg T. B., Ellen E., Harlander-Matauschek A.. 2017. A role for plasma aromatic amino acids in injurious pecking behavior in laying hens. Physiol. Behav. 175:88–96. [DOI] [PubMed] [Google Scholar]
- Bravo J. A., Forsythe P., Chew M. V., Escaravage E., Savignac H. M., Dinan T. G., Bienenstock J., Cryan J. F.. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. 108:16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A.. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton T. A. 2012. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS Lett. 586:956–961. [DOI] [PubMed] [Google Scholar]
- Dash S., Clarke G., Berk M., Jacka F. N.. 2015. The gut microbiome and diet in psychiatry. Curr. Opin. Psychiatry 28:1–6. [DOI] [PubMed] [Google Scholar]
- de Haas E. N. 2014. The fearful feather pecker: applying the principles to practice to prevent feather pecking in laying hens. PhD Diss.Wageningen Univ., Wageningen, The Netherlands.
- Goodhart P. J., DeWolf W. E. Jr., Kruse L. I.. 1987. Mechanism-based inactivation of dopamine .beta.-hydroxylase by p-cresol and related alkylphenols. Biochemistry 26:2576–2583. [DOI] [PubMed] [Google Scholar]
- Gunnarsson S. 1999. Effect of rearing factors on the prevalence of floor eggs, cloacal cannibalism and feather pecking in commercial flocks of loose housed laying hens. Br. Poult. Sci. 40:12–18. [DOI] [PubMed] [Google Scholar]
- Harlander-Matauschek A., Beck P., Piepho H. P.. 2009. Taste aversion learning to eliminate feather pecking in laying hens, Gallus gallus domesticus. Anim. Behav. 78:485–490. [Google Scholar]
- Harlander-Matauschek A., Piepho H. P., Bessei W.. 2006. The effect of feather eating on feed passage in laying hens. Poult. Sci. 85:21–25. [DOI] [PubMed] [Google Scholar]
- Jia W., Li H., Zhao L., Nicholson J. K.. 2008. Gut microbiota: a potential new territory for drug targeting. Nat. Rev. Drug. Discov. 7:123–129. [DOI] [PubMed] [Google Scholar]
- Keszthelyi D., Troost F. J., Masclee A. A. M.. 2009. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. J. Neurogastroenterol. Motil. 21:1239–1249. [DOI] [PubMed] [Google Scholar]
- Kjaer J. B. 2009. Feather pecking in domestic fowl is genetically related to locomotor activity levels: implications for a hyperactivity disorder model of feather pecking. Behav. Genet. 39:564–570. [DOI] [PubMed] [Google Scholar]
- Kjaer J. B., Sørensen P., Su G.. 2001. Divergent selection on feather pecking behaviour in laying hens (Gallus gallus domesticus). Appl. Anim. Behav. Sci. 71:229–239. [DOI] [PubMed] [Google Scholar]
- Kozak M., Tobalske B., Martins C., Bowley S., Wuerbel H., Harlander-Matauschek A.. 2016. Use of space by domestic chicks housed in complex aviaries. Appl. Anim. Behav. Sci. 181: 115–121. [Google Scholar]
- Lovanh N., Cook K. L., Rothrock M. J., Miles D. M., Sistani K.. 2007. Spatial shifts in microbial population structure within poultry litter associated with physicochemical properties. Poult. Sci. 86:1840–1849. [DOI] [PubMed] [Google Scholar]
- Lu J., Idris U., Harmon B., Hofacre C., Maurer J. J., Lee M. D.. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R. A., Oezguen N., Balderas M., Venkatachalam A., Runge J. K., Versalovic J., Veenstra-VanderWeele J., Anderson G. M., Savidge T., Williams K. C.. 2017. Distinct microbiome-neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell. Mol. Gastroenterol. Hepatol. 3:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz V., Stratz P., Preuß S., Tetens J., Grashorn M. A., Bessei W., Bennewitz J.. 2017. A genome-wide association study in a large F2-cross of laying hens reveals novel genomic regions associated with feather pecking and aggressive pecking behavior. Genet. Sel. Evol. 49:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan D. E., Savory C. J.. 2001. Feather eating in individually caged hens which differ in their propensity to feather peck. Appl. Anim. Behav. Sci. 73:131–140. [DOI] [PubMed] [Google Scholar]
- Meyer B., Bessei W., Vahjen W., Zentek J., Harlander-Matauschek A.. 2012. Dietary inclusion of feathers affects intestinal microbiota and microbial metabolites in growing Leghorn-type chickens. Poult. Sci. 91:1506–1513. [DOI] [PubMed] [Google Scholar]
- Meyer B., Zentek J., Harlander-Matauschek A.. 2013. Differences in intestinal microbial metabolites in laying hens with high and low levels of repetitive feather-pecking behavior. Physiol. Behav. 110:96–101. [DOI] [PubMed] [Google Scholar]
- Muegge B. D., Kuczynski J., Knights D., Clemente J. C., González A., Fontana L., Henrissat B., Knight R., Gordon J. I.. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle J. G., Sharp W. G., Cubells J. F.. 2013. The gut microbiome: a new frontier in autism research. Curr. Psychiatry Rep. 15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., De Waal E. C., Uitterlinden A. G.. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. B., Lillehoj H. S., Kogut M. H., Kim W. K., Maurer J. J., Pedroso A., Lee M. D., Collett S. R., Johnson T. J., Cox N. A.. 2014. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 360:100–112. [DOI] [PubMed] [Google Scholar]
- Pauwels J., Taminiau B., Janssens G. P. J., De Beenhouwer M., Delhalle L., Daube G., Coopman F.. 2015. Cecal drop reflects the chickens' cecal microbiome, fecal drop does not. J. Microbiol. Methods 117:164–170. [DOI] [PubMed] [Google Scholar]
- Savory C. J. 1995. Feather pecking and cannibalism. Worlds Poult. Sci. J. 51:215–219. [Google Scholar]
- Stanley D., Hughes R. J., Moore R. J.. 2014. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 98:4301–4310. [DOI] [PubMed] [Google Scholar]
- van Hierden Y. M., de Boer S. F., Koolhaas J. M., Korte S. M.. 2004. The control of feather pecking by serotonin. Behav. Neurosci. 118:575–583. [DOI] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z.. 2013. Bacterial census of poultry intestinal microbiome. Poult. Sci. 92:671–683. [DOI] [PubMed] [Google Scholar]
- Whelan F. J., Verschoor C. P., Stearns J. C., Rossi L., Luinstra K., Loeb M., Smieja M., Johnstone J., Surette M. G., Bowdish D. M.. 2014. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann. Am. Thorac. Soc. 11:513–521. [DOI] [PubMed] [Google Scholar]
- Williams B. L., Hornig M., Buie T., Bauman M. L., Paik M. C., Wick I., Bennett A., Jabado O., Hirschberg D. L., Lipkin W. I.. 2011. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One 6:e24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. R., Hu C. H., Wang M. Q.. 2002. Effects of fructooligosaccharide on conversion of L-tryptophan to skatole and indole by mixed populations of pig fecal bacteria. J. Gen. Appl. Microbiol. 48:83–89. [DOI] [PubMed] [Google Scholar]
- Zagon A. 2001. Does the vagus nerve mediate the sixth sense? Trends Neurosci. 24:671–673. [DOI] [PubMed] [Google Scholar]
- Zeltner E., Klein T., Huber-Eicher B.. 2000. Is there social transmission of feather pecking in groups of laying hen chicks? Anim. Behav. 60:211–216. [DOI] [PubMed] [Google Scholar]