Abstract
Amyloid oligomers are considered the most neurotoxic species of amyloid aggregates. Spontaneous assembly of amyloids into aggregates is recognized as a major molecular mechanism behind Alzheimer’s disease and other neurodegenerative disorders involving protein aggregation. Characterization of such oligomers is extremely challenging, but complicated by their transient nature. Previously, we introduced a flexible nano-array (FNA) method enabling us to probe dimers assembled by the amyloid β (14–23) [Aβ (14–23)] peptide. The study presented herein modifies and enhances this approach to assemble and probe trimers of Aβ (14–23). A metal-free click chemistry approach was used, in which dibenzocyclooctyne (DBCO) groups were incorporated at selected sites within the FNA template to click Aβ (14–23) monomers at their terminal azide groups. Atomic force microscopy (AFM) force spectroscopy was employed to characterize the assemblies. The force measurement data demonstrate that the dissociation of the trimer undergoes a step-wise pattern, in which the first monomer dissociates at the rupture force ~ 48 ± 2.4 pN. The remaining dimer ruptures at the second step at a slightly larger rupture force (~ 53 ± 3.2 pN). The assembled trimer was found to be quite dynamic, and transient species of this inherently dynamic process were identified.
Graphical Abstract
INTRODUCTION
Alzheimer’s disease (AD) is associated with the formation of neurotoxic amyloid aggregates of amyloid β (Aβ) peptides in the extracellular space of the cerebral cortex of individuals affected by this disease.1–3 Amyloid plaques are mainly composed of fibrillar aggregates from the Aβ protein, primarily Aβ42 and Aβ40.4, 5 While characterization of Aβ fibrils is well established, knowledge on the structures of oligomers is quite limited. Accumulated evidences suggest that soluble oligomers, specifically dimers and trimers of Aβ, are the neurotoxic components, rather than amyloid fibrils.6–8 For example, Younkin ET AL. showed, using a Tg2576 mouse model that dimers of Aβ accumulated in the lipid raft in an age-dependent manner result in memory deficits, which strongly suggested that Aβ42 dimers are involved in the development of Alzheimer’s disease.9 Likewise, Selkoe and colleagues indicated that soluble Aβ dimers extracted from the cerebral cortex of patients with Alzheimer’s disease can interrupt the memory of a learned behavior in normal rats.10 Moreover, they also found that insoluble amyloid plaque cores from the cerebral cortex of persons with Alzheimer’s disease do not prevent long-term potentiation unless they dissociate to Aβ dimers, further suggesting Aβ dimers are neurotoxic. These early outcomes in animal models are consistent with recent findings that the human brain contains Aβ oligomers up to two decades prior to the onset of Alzheimer’s disease.11 Further, by using immuno-electron microscopy with oligomer-specific polyclonal antibodies, Aβ oligomers have been identified in amyloid precursor protein in transgenic mice as well as the brains of individuals with Alzheimer’s disease.12,13 Moreover, Aβ oligomers have also been shown to directly interact with cellular membranes, which results in the formation of pores and disrupts the proper permeability of the membranes.14 Taken together, these studies suggest that soluble amyloid oligomers play an important role in the pathogeneses of Alzheimer’s disease. Hence, characterizing and understanding the mechanism of the formation and dissociation of Aβ oligomers are essential in developing potential therapeutic drugs for Alzheimer’s disease. Given the transient nature of oligomers, IN VITRO characterization requires the development of non-traditional approaches. Fortunately, photo cross-linking of unmodified protein (PICUP) has made it possible to prepare Aβ oligomers of pre-defined sizes that remain neurotoxic regardless of crosslinking.15, 16 Time-lapse high-speed AFM imaging has also been recently applied to directly demonstrate that Aβ42 oligomers are extremely dynamic and capable of transient dissociation/assembly into dimers and trimers.17 In addition, recently we showed that the hairpin conformation plays an important role in defining amyloid aggregation pathways.18, 19 Previously, we developed AFM-based single-molecule force spectroscopy to characterize assemblies (mainly dimers) from different amyloidogenic proteins such as Aβ42, Aβ40, and α-synuclein; major findings were that those dimers are highly stable.20–22 The lifetime of the dimers was further measured using a tethered approach for probing intermolecular interactions (TAPIN), based on the use of single-molecule fluorescence.23, 24 Importantly, our group recently developed a flexible nano-array approach (FNA) in which Aβ monomers were covalently tethered to a FNA polymer at defined positions.25, 26 The FNA tether was synthesized using phosphoramidite chemistry.27 As a result, the monomers are capable of assembling into a dimer that can be probed by stretching the FNA tether.26
In this work, the FNA approach was modified and thus enabled the assembly of Aβ (14–23) monomers into trimers. A click chemistry was developed to end-tether Aβ (14–23) monomers to selected sites within the FNA template. The interaction within the trimer was probed by AFM force experiments by pulling the FNA at the ends of the polymer. These studies revealed a stepwise pattern for the dissociation of the timer. The strengths of the interactions upon the trimer stretching were also measured.
RESULTS
Experimental design and FNA assembly
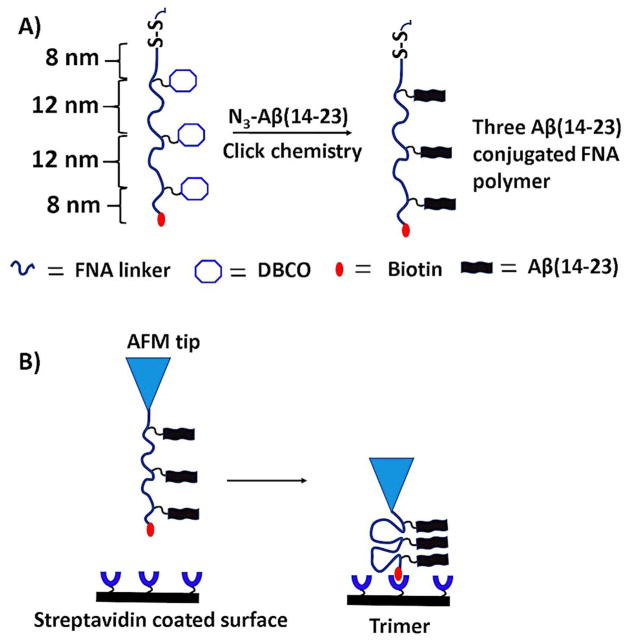
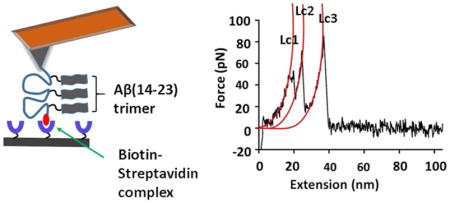
Figure 1A schematically shows the strategy for immobilization of peptides with the FNA tether. The polymer was synthesized with the Mermade 12 DNA synthesizer (Bioautomation, Irving, TX) using non-nucleotide phosphoramidite chemistry. The FNA was designed in such a way that it contained terminal thiol and biotin groups, along with three internal dibenzocyclooctyne (DBCO) units for covalent linkage of Aβ peptides. Three DBCO groups were placed at a contour length distance of ~12 nm; whereas distances of terminal groups (thiol and biotin) to closest DBCO groups was ~8 nm each. Using metal-free click chemistry, three molecules of azide-Aβ (14–23) were attached to the FNA tether (Figure 1A). The product was purified and characterized by analytical HPLC and mass spectroscopy (Supporting information, Figure S1). The flexibility of the tether allows for three Aβ (14–23) monomers to form a trimer. The FNA was stretched in the AFM force spectroscopy experiment to probe the interactions within the trimer.
Figure 1.
Schematic of the FNA-based approach for probing oligomers. (A) The scheme showing the peptide-conjugated FNA construct. The polymer had a thiol group and biotin (red blob) at the terminals; also, three DBCO groups were located at specified distances. Three Aβ (14–23) molecules were attached using a metal-free click reaction. The distances are shown according to the experimental design. (B) Force spectroscopy set up. The AFM tip was functionalized with an FNA construct and the surface with streptavidin. The Aβ(14–23) trimer is formed by association of three monomers. Note, schemes are not to scale.
Force spectroscopy experiments
As shown in Figure 1B, the FNA construct was immobilized at the maleimide-functionalized AFM tip by a covalent bond between the maleimide moiety and the FNA terminal thiol group. In the AFM force-probing experiment, the AFM probe terminated with the FNA construct was approached to a mica surface functionalized with streptavidin. To ensure single-molecule probing without contribution from adjacent streptavidin molecules conditions for the sparse coating of mica with streptavidin was developed. The density of the surface coating by streptavidin was controlled with AFM imaging prior to force measurements. The AFM image in Figure S2B shows a low density of streptavidin proteins on the surface. Additionally, the volume measurement (Figure S2C) show that majority of streptavidin are in tetrameric form (volume ~93 nm3), which further ensures that the suitability of the functionalized substrate for force measurements.
During the approach step, a strong intermolecular bond is formed between the biotin moiety on the FNA end and streptavidin on the surface.25 As a result, the FNA is anchored between the tip and the surface; therefore, the interaction between the monomers of the trimer is probed during the retraction step. Biotin-streptavidin is a robust and stable complex that requires a large force for its rupture and is used in numerous force spectroscopy experiments that utilize AFM force spectroscopy,28, 29 optical30, 31 and magnetic tweezers,29 and single-molecule fluorescence spectroscopy.32, 33 Notably, the unbinding peak for this complex plays a role of the internal reference used for characterizing other peaks that correspond to peptide-peptide interactions. To assign peaks for biotin-streptavidin complexes in force-distance (FD) curves, control experiments with the FNA containing no peptides were performed (Supporting information, Figure S3). A nice overlap of FD curves (Figure S3B) indicates a high reproducibility of rupture of biotin-streptavidin complexes, with an average force of 86 ± 2.1 pN and contour length value of 41 ± 0.3 nm. A similar force was reported in other studies for biotin-streptavidin complexes.34, 35
Rupture of Aβ (14–23) trimer
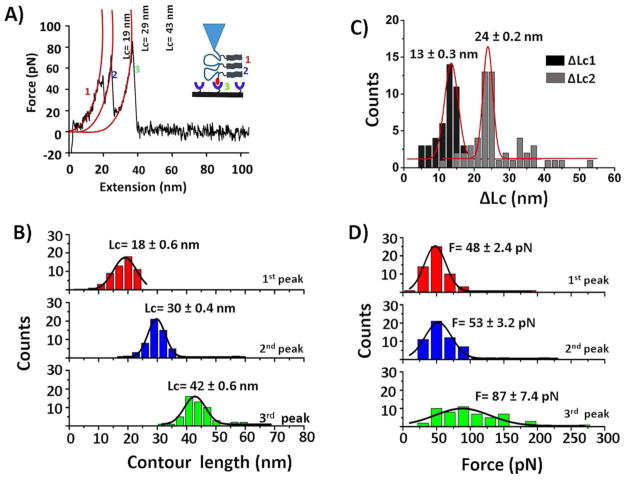
Figure 2A shows a typical FD curve that contains three peaks. Similarly, such a fingerprint may arise if monomers are assembled into a trimer, and dissociation of this trimer follows a step-wise mechanism. To characterize those peaks, individual peaks in force-distance curves were fitted using the worm-like chain model36, which resulted in force values of 48 pN, 51 pN, and 86 pN, and contour lengths 19 nm, 29 nm, and 43 nm for first, second, and third peaks, respectively (Figure 2A). Based on our design, the third peak is expected to be the biotin-streptavidin dissociation. To confirm, the force and contour length values for the third peaks were compared with a control experiments (Figure S3) where no peptide was attached. The force and contour length (Lc) values were similar to control experiments (87 ± 7.4 pN vs. 86 ± 2.1 pN, and 42 ± 0.6 nm vs. 41 ± 0.3 nm). Therefore, the third peak represents the biotin-streptavidin bond rupture. Notably, force values for first and second peak (~50 pN) were similar to the Aβ (14–23) dimer rupture force value found in references.25, 26, 37
Figure 2.
Probing of the Aβ (14–23) trimer. (A) An illustration of trimer formation is schematically shown in the inset; a typical force-distance trace (retract) showing stepwise dissociation of three events is numbered as 1, 2, and 3. The contour values of three individual peaks were estimated by fitting the peaks with the worm-like chain model36 (red curves). (B) Contour length histograms for three individual events. Red and blue histograms correspond to peptide-peptide interactions (events 1 and 2), whereas green represents biotin-streptavidin complexes (event 3). (C) Representative histograms for ΔLc1 (black) and ΔLc2 (grey). (D) Force histograms for three events. Histograms were fitted with Gaussian functions. Values are shown as mean ± S.E.M. The number of data points is 56.
The force-distance curves containing three peaks were found in approximately one-fourth of all successful rupture events, and these force-distance curves were analyzed similarly to that discussed above. An overlay of such force-distance curves is shown in Figure S4. The contour length values for each peak were assembled into histograms, and the most probable values were calculated by fitting the histograms with Gaussian functions (Figure 2B). These histograms separated well into three peaks, with rupture distances of 18 ± 0.6 nm, 30 ± 0.4 nm, and 42 ± 0.6 nm for peaks 1, 2 and 3, respectively. To assign the three events to the trimer dissociation pathway, the distances between the peaks Lc were measured. Lc1 was calculated by subtracting the Lc of the second peak from Lc of third peak or Lc of first peak from Lc of second peak. Similarly, Lc2 was determined by subtracting Lc of first peak from Lc of third peak. According to this design (Figure 1A), Lc1 and Lc2 are expected to be ~12 nm (one loop distance) and 24 nm (two-loop distance), respectively. Experimentally obtained Lc1 and Lc2 histograms (Figure 2C) show these values to be 13 ± 0.3 nm and 24 ± 0.2 nm, which resemble the expected values.
Figure 2D shows the force distributions for each type of rupture event. In these histograms, F= 48 ± 2.4 pN corresponds to the dissociation of the monomer within the trimer (red, peak 1); F= 53 ± 3.2 pN corresponds to the dissociation of another monomer from the dimer (blue, peak 2), and F= 87 ± 7.4 pN denotes biotin-streptavidin (green, peak 3). Reproducibility of the data was examined by performing two individual experiments; results are listed in Table S1. In the second experiments, mean force values of 44 ± 1.2 pN, 50 ± 1.0 pN, and 84 ± 3.6 pN were obtained for first, second, and third type of events. These values resemble those for the first experiment shown in the top row.
Characterization of Aβ(14–23) dimers
In the pulling experiments, force-distance curves were observed that had only two peaks (found in approximately half of all successful force-rupture events). There were two types of such two-peak events in force-distance data, described below.
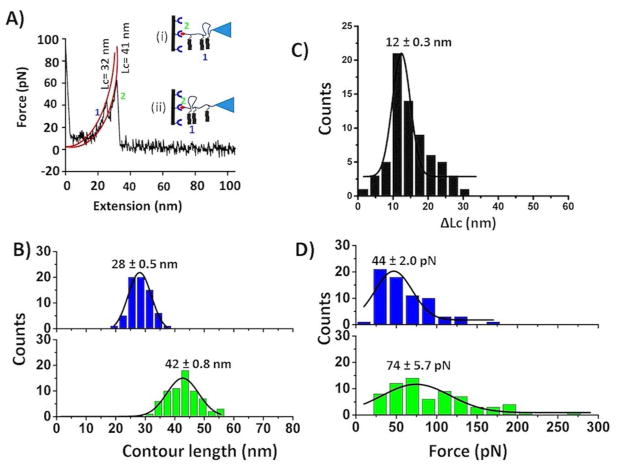
Figure 3A shows a typical force curve for the pattern in which two events are close to each other, and they correspond to the schematic in insets (i) and (ii). The contour lengths measurements were used to prove these events. Indeed, the contour length for the last peak corresponded to the biotin-streptavidin rupture (Lc= 41 nm), and the first peak ruptured at the contour length 32 nm, which corresponds to the dissociation of the peptide-peptide interaction [Figure 3A, insets (i) and (ii)]. Figure S5 shows an overlay of such force-distance curves. All force-distance curves that fall under this category were fitted with the worm-like chain (WLC) model, and data were assembled into force and contour histograms. The contour length analysis (Figure 3B) demonstrates that such types of dimers dissociate at a distance of 28 ± 0.5 nm (blue), and subsequent biotin-streptavidin unbinding events occur at 42 ± 0.8 nm (green). For the two peaks obtained in the related force-distance curves, Lc has been calculated to obtain the distance between the rupture events. Figure 3C shows that the Lc is 12 ± 0.3 nm, which is the same as expected value. Force histograms, as shown in Figure 3D, suggest the dimer rupture force is ~44 ± 2.0 pN (blue), whereas biotin-streptavidin requires a force of 74 ± 5.7 pN (green) to dissociate.
Figure 3.
Identification of Aβ (14–23) dimers within trimer. (A) A typical force-distance curve showing two peaks close to each other; possible interaction patterns are shown in the inset. Individual peaks were fitted with the worm-like chain model36 (red line), and corresponding Lc values are shown. (B) Contour length histograms for two separate events. (C) ΔLc histogram related to two events. (D) Force histograms corresponding to peptide-peptide (upper, blue) and biotin-streptavidin interactions (below, green). Histograms were fitted with Gaussian functions, and values are shown as mean ± S.E.M. The number of data points is 68.
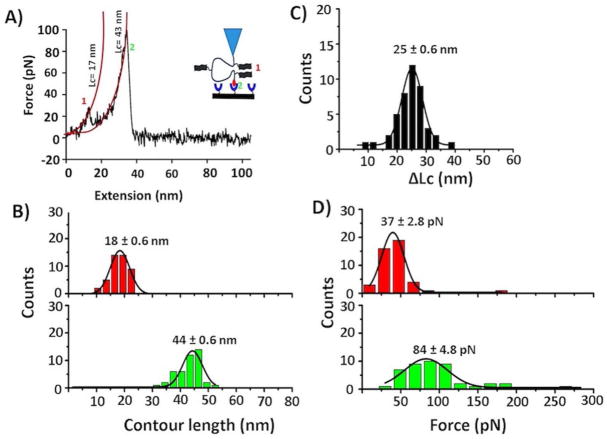
Figure 4A schematically shows another type of two-peak containing force-distance curves for which two events are far. To characterize those peaks, again WLC approximation was done. The contour length measurements produce Lc = 17 nm for the first peak, which corresponds to the dimer dissociation. The second peak (Lc= 43 nm) corresponds to biotin-streptavidin unbinding events. Figure S6A shows the overlay of such force-distance curves. Analysis over a set of such events was performed; the data were assembled as histograms, as shown in Figures 4B–D.
Figure 4.
Characterization of Aβ (14–23) dimers. (A) A typical force-distance curve showing two peaks far from each other; Lc values for each individual peaks were estimated by the worm-like chain model (red lines). (B) Contour length histograms for two different events; red = Aβ (14–23) dimer and green = Biotin-streptavidin interaction. (C) ΔLc distribution corresponding to a long distance between two peaks. (D) Force histograms corresponding to peptide-peptide (upper, red) and biotin-streptavidin interactions (below, green). Histograms are fitted with Gaussian function, and values are presented as mean ± S.E.M. The number of data points is 44.
Contour length distributions for those two events show that Aβ (14–23) dimers dissociate at an average distance of Lc = 18 ± 0.6 nm (Figure 4B, upper, red), suggesting that the dimer is formed by peptides 1 and 3 (lengths 8 nm + 8 nm; see Figure 1A). This assignment is supported by measuring the distances between the two peaks, Lc. The data are shown in Figure 4C, and Gaussian fitting produces a value of Lc =25 ± 0.6 nm, which is approximately the expected value of 24 nm according to the design (Figure 1A). For the dimer dissociation, the force distributions, as shown in Figure 4D (top, red), resulted in a mean value of 37 ± 2.8 pN. Again, rupture events for biotin-streptavidin occur at an Lc value of ~44 ± 0.6 nm (Fig. 4B, bottom, green), and the rupture force is ~ 84 ± 4.8 pN (Figure 4D, bottom, green), which aligns with control experiments (Figure S3). The reproducibility of the data was confirmed by performing another set of experiments under similar experimental conditions (Table S2). The results from two independent experiments showed very similar values in terms of force and contour lengths.
Force curves with no peptide-peptide interactions
In the data set for this study, ~24% of force-distance curves were found to have only one peak, as shown in Figure S7A. These rupture curves were fitted with the WLC model; force and contour length values were assembled into histograms. Figures S7B and S7C show that the average force = 84 ± 4.7 pN and Lc = 41 ± 0.4 nm, which is similar to control experiments (Figure S3), indicating that this peak corresponds to the dissociation of the biotin-streptavidin complex. Note that these events correspond to situations in which the dimers and trimers dissociated prior to probing, suggesting that the complexes are quite dynamic.
DISCUSSION
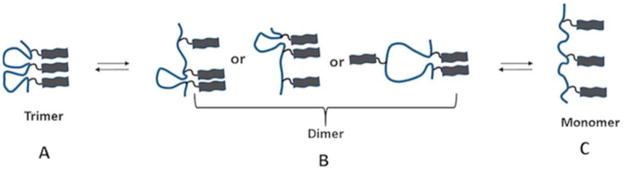
The results for this study demonstrate that Aβ monomers being tethered to FNA can assemble into trimers. Force-distance curves contain three peaks, suggesting that dissociation of the trimer occurs in a step-wise pattern. The analysis of force spectroscopy data shows that the trimer dissociates initially into a dimer and monomer followed by the dissociation of the remaining dimers into monomers. Given the ability of the force spectroscopy experiments to measure the contour lengths, each peak of the force curves was identified. Various combinations of oligomerization possibilities and the results are summarized schematically in Figure 5, in which all possible dimers are shown.
Figure 5.
Scheme showing the dissociation pathway for the Aβ (14–23) trimer. The trimer (A) first dissociates into dimers and monomers (B), and subsequently, the dimer dissociates into monomers (C). Schemes are not to scale.
Rupture forces for the initial trimer dissociation steps and dimer dissociation were measured (Table S1). Although the values for the first and second steps were similar (48± 2.4 pN and 53 ± 3.2 pN), the rupture force for the first step, the monomer dissociation within the trimer was slightly smaller compared with the second step. A similar trend was confirmed by an independent experiment (44 ± 1.2 pN and 50 ± 1.0 pN for peaks 1 and 2, respectively; Table S1). The force values for dissociation of the dimers were very similar to our previous experiments in which the Aβ (14–23) dimer was tethered to FNA with the use of a different coupling method (48 ± 6 pN).25 Importantly, a very similar value (49 ± 5.8 pN) was obtained during AFM force spectroscopy experiments performed by probing Aβ (14–23) monomers immobilized onto the AFM tip and surface.37 This suggests that FNA immobilization does not change the dimers structure and stability.
The assembled trimer is very dynamic and capable of spontaneous dissociation into dimer plus monomer assemblies (Figure 5B), followed by the full dissociation of the monomers (Figure 5C). These states correspond to the force spectroscopy experiments with three-peak and two-peak events, respectively. The yield of the two-peak events (50%) is two-fold greater than the three-peak events (25%), which is consistent with lower rupture forces for the first peak compared with the second peak (see Table S1 and the paragraph above). The increased stability of dimers compared with trimers is consistent with findings for the highest yield of dimers among oligomers extracted from the brain.38
Monomers in oligomers can be arranged in parallel or antiparallel orientations. We have shown previously that these arrangements can be distinguished by rupture forces. For example, Aβ (14–23) monomers arranged in an anti-parallel fashion rupture at forces of 48 ± 8 pN, whereas the rupture force for the parallel dimer is considerably lower, 30 ± 4 pN.37, 39 Using this criterion and rupture force values of 48 ± 2.4 pN and 53 ± 3.2 pN for the first and second peaks, respectively, it can be concluded that the monomers in the FNA-tethered trimers are arranged in an antiparallel fashion.25, 26 A similar type of anti-parallel monomer arrangement has been observed within Aβ dimers and trimers in a number of computer simulation studies.40–42
CONCLUSIONS
In conclusion, this study shows that the FNA platform can be extended to the assembly Aβ trimers. Though evidence suggests that small oligomers such as dimers or trimers are neurotoxic, preparing defined sizes of oligomers is difficult task because they spontaneously assemble into a large population of different sizes of oligomers. Previously, we hypothesized that the use of the FNA platform can allow for the assembly of oligomers larger than dimers; trimers were assembled for the current paper. By applying AFM force spectroscopy, we found that assembly of three monomers arranged in an anti-parallel fashion form a trimer, and during rupture, the trimer follows a step-wise dissociation pathway. Moreover, our results suggest this trimer is highly dynamic when dissociating into dimers and monomers. Given that the FNA platform is created using DNA synthesis chemistry, much longer FNA polymers capable of assembly into large oligomers can be synthesized using a regular DNA synthesizer. In addition to structural and dynamic studies of the FNA-tethered oligomers, these assemblies have the potential for use in numerous translational studies, such as the use them as antigens for producing oligomer-specific antibodies for Alzheimer’s disease.
MATERIALS AND METHODS
For synthesis of FNA tether following reagents were purchased from Glen Research (Sterling, VA): biotin-linked CPG (3′-PROTECTED Biotin Serinol CPG; 20-2993), Spacer 18 phosphoramidite (18-O Dimethoxytritylhexaethyleneglycol, 1-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite, 10–1918), DBCO-dT-CE phosphoramidite (5′-Dimethoxytrityl-5-[(6-oxo-6-(dibenzo[b,f]azacyclooct-4-yn-1-yl)-capramido-N-hex-6-yl)-3-acrylimido]-2′-deoxy-Uridine, 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite, 10-1539), Thiol-Modifier C6 S-S, (1-O-Dimethoxytrityl-hexyl-disulfide,1′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite, 10-1936).
The azide-labeled Aβ (14–23) peptide [K(N3)HQKLVFFVAED] was synthesized and purified by Peptide 2.0 Inc. (Chantilly, VA). Tris (2-carboxyethyl) phosphine (TCEP) hydrochloride was from Hampton Research (Aliso Viejo, CA). N-(γ-Maleimidobutyryloxysuccinimide ester) GMBS was from Pierce Biotechnology (Grand Island, NY). Streptavidin protein was purchased from Sigma-Aldrich (St. Louis, MO). 1-(3-aminopropyl) silatrane (APS) was synthesized as previously described.43 All other reagents or solvents were purchased from Sigma Aldrich (St. Louis, MO).
Synthesis of Aβ (14–23) conjugated FNA polymer
The polymer was synthesized in a DNA synthesizer (MerMade 12, Bioautomation; Irving, TX), using phosphoramidite chemistry as described in our previous papers.26, 27, 44 Briefly, synthesis was started with biotin-linked controlled porous glass (CPG), and then following units were added to the sequence: 6 units of spacer S18, 1 unit DBCO, 6 units spacer S18, 1 unit DBCO, 6 units spacer S18, 1 unit DBCO, 4 units spacer S18, and then 1 unit 5′ thiol modifier. To conjugate Aβ (14–23) peptides with FNA, CPG was dispersed in azide terminated Aβ (14–23) solution in 10 mM sodium phosphate buffer (pH 7.4) at a molar ratio of peptide:FNA of 6:1 and stirred for 4 hours in the dark. The unreacted peptide was filtered. The construct was then cleaved from CPG, purified by reverse phase-high performance liquid chromatography (RV-HPLC) (Figure S1), and characterized by mass spectroscopy. Details of the synthesis are described in the supporting information.
Functionalization of AFM tip
The construct was covalently attached to the AFM tip using similar protocol as discussed in references 26, 45. Briefly, AFM tips (MSNL10, Bruker Corporation, Camarillo, CA) were cleaned with ethanol and water, and dried in gentle flow of argon gas. They were further cleaned and oxidized by treatment with UV light (λ =366 nm) for 45 minutes. Tips were functionalized with amine by treatment with 10 YM APS solution for 30 minutes and followed by multiple rinses with DI water. Tip surfaces were then converted to maleimide functionalities by treatment with 100 YM GMBS solution in DMSO for 1 hour, followed by washing with DMSO and DI water. A solution containing 10 nM of FNA construct was prepared in sodium phosphate buffer (10 mM, pH 7.4) containing 10 μM TCEP 30 minutes before applying to the surface to ensure that the S-S bond in FNA was cleaved. A droplet of the FNA solution was placed on a piece of parafilm; the tip was covered with the FNA droplet and kept at 4°C overnight. The parafilm was placed on a wet tissue paper in a closed petri dish to minimize evaporation of solution. After the reaction, the tips were washed with DI water, and unreacted maleimide groups were quenched with 10 mM of β-mercaptoethanol. Finally, the tips were washed with DI water and stored in 10 mM sodium phosphate buffer (pH 7.4) until use.
Mica surface modification with streptavidin
For force measurements, streptavidin was covalently attached to the mica surface. A small piece of mica (1.5 cm x 1.5 cm approx.) was glued to a glass slide using epoxy glue EPO-TEK-301 (Epoxy Technology; Billerica, MA), and upper surface of mica was cleaved with scotch tape. The surface was covered with 167 YM APS solution and incubated for 30 minutes in humidified chamber, followed by washing with DI water. The surface was then treated with 0.05% of aqueous glutaraldehyde for 30 minutes and rinsed with DI water. Then the solution of streptavidin (0.001 μg/mL) was applied to the surface for 2 hours, and finally, the surface was washed with DI water.
To check the surface coverage of streptavidin proteins on the surface, similar functionalization steps were performed on a small piece of mica. After functionalization, the surface was dried with argon gas. The surface was imaged with Nanoscope III (Bruker Corporation, Camarillo, CA ) in a tapping mode using AFM probes (TESPA, nominal spring constant 42\N/m; Bruker Corporation, USA). The images were processed with Femtoscan Online AFM software (Advanced Technologies Center; Lomonosov Moscow State University, Moscow) (see details in supporting information and Figure S2).
Force measurement and data analysis
Force measurements were performed in 10 mM sodium phosphate buffer at pH 7.4 containing 0.1 mM ethylene-diamine-tetraacitic acid (EDTA) at room temperature with the MFP-3D AFM instrument (MFP-3D, Asylum Research; Santa Barbara, CA). Silicon nitride (Si3N4) AFM tips (MSNL10, Bruker Corporation, USA) with a nominal spring constant of 0.02–0.03 N/m were used for force measurements. The actual spring constants of the AFM tips were calculated by using a thermal method (Igor Pro 6.37). To maximize probability of oligomer formation, a 100 pN trigger force and 1s dwell time were set. The probe was retracted at a speed of 500 nm/s each time. Several thousand force-extension curves were acquired to obtain a dataset with several hundred rupture events. Force curves were estimated with the worm-like-chain36 method using the following equation: F(x)= kBT/Lp[1/4(1−x/Lc)−2−1/4+x/Lc], where F(x) is the force at the distance of x, kB is the Boltzmann constant, T is absolute temperature, and Lp and Lc are the persistence length and contour length, respectively. The data were assembled into histograms and fitted with the Gaussian function to estimate the most probable force and contour length for the specific rupture events. The mean values (maxima in the Gaussians) ± SEM were calculated from the datasets. The reproducibility of the data was demonstrated by performing two independent experiments (see Table S1 and S2).
Supplementary Material
Acknowledgments
We thank Lyubchenko lab members for their suggestions to improve this project. We specially thank to Dr. A. Gall and Dr. E. Viazovkina (Cepheid INC, Bothell, USA) for their help with synthesis of the FNA construct. We thank Melody A. Montgomery for the professional editing of this manuscript. The work was supported by grants to Y.L.L. from the National Institutes of Health (NIH: GM096039 and GM118006).
Footnotes
Notes
The authors declare no competing financial interests.
The supporting Information is available free of charge on the ACS publications websites. Details of FNA synthesis, purification, and characterization, streptavidin immobilization procedure and surface imaging, and control force spectroscopy experiments are found in Figures S1–7 and Tables S1–2.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annual review of biochemistry. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Roher AE, Lowenson JD, Clarke S, Woods AS, Cotter RJ, Gowing E, Ball MJ. beta-Amyloid-(1–42) is a major component of cerebrovascular amyloid deposits: implications for the pathology of Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:10836–10840. doi: 10.1073/pnas.90.22.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mak K, Yang F, Vinters HV, Frautschy SA, Cole GM. Polyclonals to beta-amyloid(1–42) identify most plaque and vascular deposits in Alzheimer cortex, but not striatum. Brain research. 1994;667:138–142. doi: 10.1016/0006-8993(94)91725-6. [DOI] [PubMed] [Google Scholar]
- 6.Dahlgren KN, Manelli AM, Stine WB, Jr, Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. The Journal of biological chemistry. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 7.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 8.Jana MK, Cappai R, Pham CL, Ciccotosto GD. Membrane-bound tetramer and trimer Abeta oligomeric species correlate with toxicity towards cultured neurons. Journal of neurochemistry. 2016;136:594–608. doi: 10.1111/jnc.13443. [DOI] [PubMed] [Google Scholar]
- 9.Kawarabayashi T, Shoji M, Younkin LH, Wen-Lang L, Dickson DW, Murakami T, Matsubara E, Abe K, Ashe KH, Younkin SG. Dimeric amyloid beta protein rapidly accumulates in lipid rafts followed by apolipoprotein E and phosphorylated tau accumulation in the Tg2576 mouse model of Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:3801–3809. doi: 10.1523/JNEUROSCI.5543-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nature medicine. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesne SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA, Ashe KH. Brain amyloid-beta oligomers in ageing and Alzheimer’s disease. Brain : a journal of neurology. 2013;136:1383–1398. doi: 10.1093/brain/awt062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 13.Kokubo H, Kayed R, Glabe CG, Yamaguchi H. Soluble Abeta oligomers ultrastructurally localize to cell processes and might be related to synaptic dysfunction in Alzheimer’s disease brain. Brain research. 2005;1031:222–228. doi: 10.1016/j.brainres.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 14.Kayed R, Lasagna-Reeves CA. Molecular mechanisms of amyloid oligomers toxicity. Journal of Alzheimer’s disease : JAD. 2013;33(Suppl 1):S67–78. doi: 10.3233/JAD-2012-129001. [DOI] [PubMed] [Google Scholar]
- 15.Rahimi F, Maiti P, Bitan G. Photo-induced cross-linking of unmodified proteins (PICUP) applied to amyloidogenic peptides. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitan G, Teplow DB. Rapid photochemical cross-linking--a new tool for studies of metastable, amyloidogenic protein assemblies. Accounts of chemical research. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee S, Sun Z, Hayden EY, Teplow DB, Lyubchenko YL. Nanoscale Dynamics of Amyloid beta-42 Oligomers As Revealed by High- Speed Atomic Force Microscopy. ACS Nano. 2017;11:12202–12209. doi: 10.1021/acsnano.7b05434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maity S, Lyubchenko YL. Probing of Amyloid Abeta (14–23) Trimers by Single-Molecule Force Spectroscopy. Jacobs journal of molecular and translational medicine. 2015;1:004. [PMC free article] [PubMed] [Google Scholar]
- 19.Maity S, Hashemi M, Lyubchenko YL. Nano-assembly of amyloid beta peptide: role of the hairpin fold. Scientific reports. 2017;7:2344. doi: 10.1038/s41598-017-02454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv Z, Roychaudhuri R, Condron MM, Teplow DB, Lyubchenko YL. Mechanism of amyloid beta-protein dimerization determined using single-molecule AFM force spectroscopy. Scientific reports. 2013;3:2880. doi: 10.1038/srep02880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim B-H, Palermo NY, Lovas S, Zaikova T, Keana JFW, Lyubchenko YL. Single-Molecule Atomic Force Microscopy Force Spectroscopy Study of Aβ-40 Interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasnoslobodtsev AV, Volkov IL, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL. alpha-Synuclein misfolding assessed with single molecule AFM force spectroscopy: effect of pathogenic mutations. Biochemistry. 2013;52:7377–7386. doi: 10.1021/bi401037z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv Z, Krasnoslobodtsev AV, Zhang Y, Ysselstein D, Rochet JC, Blanchard SC, Lyubchenko YL. Direct Detection of alpha-Synuclein Dimerization Dynamics: Single-Molecule Fluorescence Analysis. Biophysical journal. 2015;108:2038–2047. doi: 10.1016/j.bpj.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv Z, Krasnoslobodtsev AV, Zhang Y, Ysselstein D, Rochet JC, Blanchard SC, Lyubchenko YL. Effect of acidic pH on the stability of alphasynuclein dimers. Biopolymers. 2016;105:715–724. doi: 10.1002/bip.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasnoslobodtsev AV, Zhang Y, Viazovkina E, Gall A, Bertagni C, Lyubchenko YL. A flexible nanoarray approach for the assembly and probing of molecular complexes. Biophysical journal. 2015;108:2333–2339. doi: 10.1016/j.bpj.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maity S, Viazovkina E, Gall A, Lyubchenko Y. A Metal-free Click Chemistry Approach for the Assembly and Probing of Biomolecules. Journal of nature and science. 2016;2:e187. [PMC free article] [PubMed] [Google Scholar]
- 27.Tong Z, Mikheikin A, Krasnoslobodtsev A, Lv Z, Lyubchenko YL. Novel polymer linkers for single molecule AFM force spectroscopy. Methods (San Diego, Calif ) 2013;60:161–168. doi: 10.1016/j.ymeth.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CY, Lou J, Wen KK, McKane M, Eskin SG, Ono S, Chien S, Rubenstein PA, Zhu C, McIntire LV. Actin depolymerization under force is governed by lysine 113:glutamic acid 195-mediated catch-slip bonds. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5022–5027. doi: 10.1073/pnas.1218407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manibog K, Yen CF, Sivasankar S. Chapter Twelve - Measuring Force-Induced Dissociation Kinetics of Protein Complexes Using Single- Molecule Atomic Force Microscopy. In: Spies M, Chemla YR, editors. Methods in Enzymology. Academic Press; 2017. pp. 297–320. [DOI] [PubMed] [Google Scholar]
- 30.Jiao J, Rebane AA, Ma L, Gao Y, Zhang Y. Kinetically coupled folding of a single HIV-1 glycoprotein 41 complex in viral membrane fusion and inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E2855–E2864. doi: 10.1073/pnas.1424995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao J, Rebane AA, Ma L, Zhang Y. Single-Molecule Protein Folding Experiments Using High-Precision Optical Tweezers. Methods in molecular biology (Clifton, NJ ) 2017;1486:357–390. doi: 10.1007/978-1-4939-6421-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W-Q, Hou X-M, Li M, Dou S-X, Xi X-G. BLM unfolds Gquadruplexes in different structural environments through different mechanisms. Nucleic Acids Research. 2015;43:4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung HS, Meng F, Kim J-Y, McHale K, Gopich IV, Louis JM. Oligomerization of the tetramerization domain of p53 probed by two- and three-color single-molecule FRET. Proceedings of the National Academy of Sciences. 2017;114:E6812. doi: 10.1073/pnas.1700357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taninaka A, Takeuchi O, Shigekawa H. Hidden variety of biotinstreptavidin/avidin local interactions revealed by site-selective dynamic force spectroscopy. Physical Chemistry Chemical Physics. 2010;12:12578–12583. doi: 10.1039/c0cp00259c. [DOI] [PubMed] [Google Scholar]
- 35.Sevim S, Shamsudhin N, Ozer S, Feng L, Fakhraee A, Ergeneman O, Pane S, Nelson BJ, Torun H. An Atomic Force Microscope with Dual Actuation Capability for Biomolecular Experiments. Scientific reports. 2016;6:27567. doi: 10.1038/srep27567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouchiat C, Wang MD, Allemand J, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophysical journal. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovas S, Zhang Y, Yu J, Lyubchenko YL. Molecular Mechanism of Misfolding and Aggregation of Aβ(13–23) The Journal of Physical Chemistry B. 2013;117:6175–6186. doi: 10.1021/jp402938p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez de la Torre A, Gay M, Vilaprinyo-Pascual S, Mazzucato R, Serra-Batiste M, Vilaseca M, Carulla N. Direct Evidence of the Presence of Cross-Linked Aβ Dimers in the Brains of Alzheimer’s Disease Patients. Analytical Chemistry. 2018;90:4552–4560. doi: 10.1021/acs.analchem.7b04936. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Lyubchenko Yuri L. The Structure of Misfolded Amyloidogenic Dimers: Computational Analysis of Force Spectroscopy Data. Biophysical journal. 2014;107:2903–2910. doi: 10.1016/j.bpj.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klimov DK, Thirumalai D. Dissecting the assembly of Abeta16–22 amyloid peptides into antiparallel beta sheets. Structure (London, England 1993) 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 41.Jang S, Shin S. Amyloid β-Peptide Oligomerization in Silico: Dimer and Trimer. The Journal of Physical Chemistry B. 2006;110:1955–1958. doi: 10.1021/jp055568e. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen PH, Li MS, Derreumaux P. Effects of all-atom force fields on amyloid oligomerization: replica exchange molecular dynamics simulations of the A[small beta]16–22 dimer and trimer. Physical Chemistry Chemical Physics. 2011;13:9778–9788. doi: 10.1039/c1cp20323a. [DOI] [PubMed] [Google Scholar]
- 43.Shlyakhtenko LS, Gall AA, Lyubchenko YL. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. Methods in molecular biology (Clifton, NJ ) 2013;931:295. doi: 10.1007/978-1-62703-056-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maity S, Viazovkina E, Gall A, Lyubchenko YL. Polymer Nanoarray Approach for the Characterization of Biomolecular Interactions. Methods in molecular biology (Clifton, NJ ) 2018;1814:63–74. doi: 10.1007/978-1-4939-8591-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maity S, Viazovkina E, Gall A, Lyubchenko YL. Single-molecule probing of amyloid nano-ensembles using the polymer nanoarray approach. Physical Chemistry Chemical Physics. 2017;19:16387–16394. doi: 10.1039/c7cp02691a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.