Abstract
Dolutegravir (DTG) is a preferred drug for initial treatment of human immunodeficiency virus type 1 infection. We present next-generation sequencing analysis of integrase genotypes during a period of virologic failure in a treatment-naive man who initiated tenofovir disoproxil fumarate/emtricitabine plus DTG.
Keywords: antiretroviral therapy, drug resistance, integrase strand transfer inhibitors
Integrase (IN) strand transfer inhibitors (INSTIs) have quickly become a mainstay of treatment for human immunodeficiency virus type 1 (HIV-1) infection. Dolutegravir (DTG) is particularly attractive because of it high potency, tolerability, and high barrier to resistance. Overall prevalence of INSTI drug resistance is low, with the most common mutations being Q148H/R/K, G140A/S/C, E138A/K/T, N155H, and/or Y143C/R in the IN gene, which confer resistance primarily to raltegravir and elvitegravir, but little cross-resistance to DTG. DTG failure has been observed primarily through acquisition of multiple mutations [1].
To date, clinically significant DTG resistance has been described primarily in treatment-experienced patients [2], including no significant resistance noted among virologic failures in treatment-naive individuals in clinical trials [3–5]. A recent retrospective cohort study found 2 cases of virologic failure in treatment-naive individuals among total 392 DTG-treated participants in the first year of therapy [6]. One patient, treated with abacavir-lamivudine plus DTG, developed a T66I IN mutation in the setting of poor medication adherence (50–79% INSTI adherence). In the other patient, treated with tenofovir disoproxil fumarate–emtricitabine plus DTG, treatment failure occurred with only M184V detected at standard population genotype testing and self-reported medication adherence >95% [6]. Here we report what we believe to be the first case of early virologic failure during a DTG-containing initial regimen with evidence of rapid emergence of INSTI-resistance mutations during treatment.
CASE REPORT AND RESULTS
A 46-year-old man with no significant prior medical history was admitted to the hospital with progressive dyspnea, fatigue, and weight loss. At the time of admission, he was hypoxic and Pneumocystis jirovecii direct fluorescent antibody was positive from a bronchoalveolar lavage sample. He was treated for Pneumocystis pneumonia with trimethoprim-sulfamethoxazole and prednisone. HIV-1 screening antibody and confirmatory Western blot results were positive, though gp41, p31, and p24 bands were consistently indeterminate at repeated testing during this period. The patient’s plasma HIV RNA level was 1970000 copies/mL, and the absolute CD4+ T-lymphocyte count was 78/μL (12% of the T-cell subset). Standard population HIV-1 genotype (reverse-transcriptase and protease genes) revealed wild-type virus (Quest Diagnostics). He started antiretroviral therapy (ART) with tenofovir disoproxil fumarate–emtricitabine plus DTG before discharge from the hospital (Figure 1A), and his prednisone dosage was tapered according to standard guidelines.
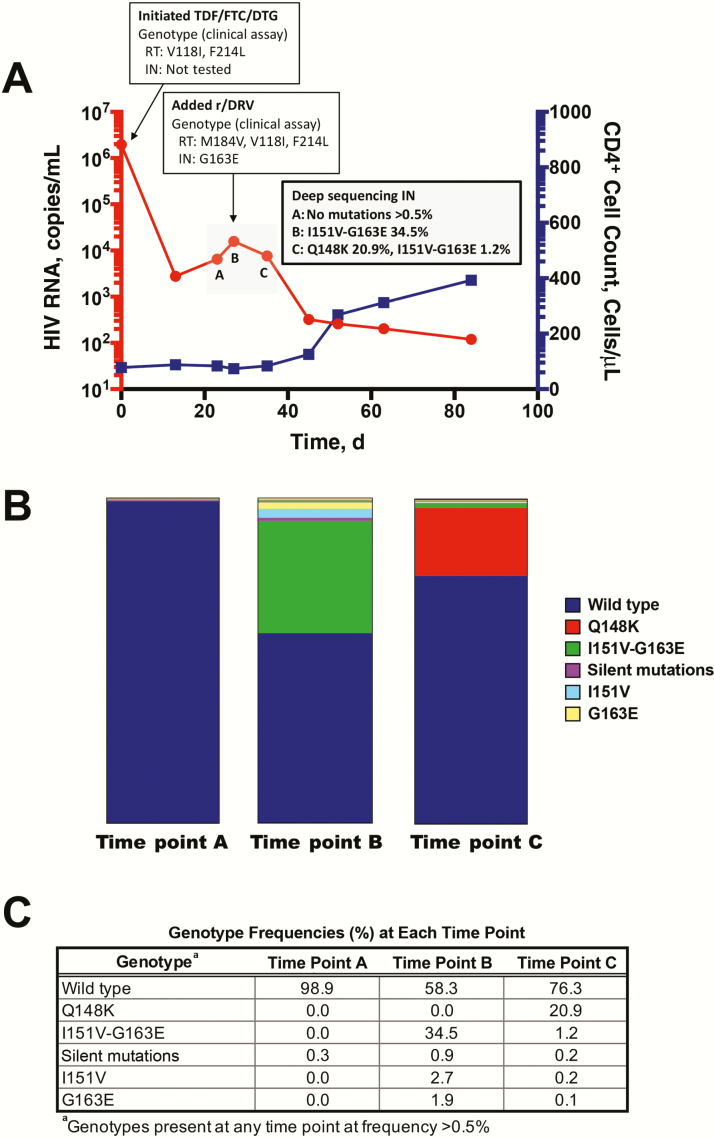
Figure 1.
Rapid development of integrase (IN) strand transfer inhibitor resistance mutations. A, Time course of human immunodeficiency virus type (HIV-1) viremia and CD4+ T lymphocytes. Plasma HIV RNA and absolute CD4+ T-lymphocyte counts from clinical laboratory measurements are plotted from the initiation of antiretroviral therapy (ART). Time points designated A, B, and C indicate those used for paired-end deep sequencing analysis (panel B), and correspond to days 27, 30, and 25 after initiation of ART, respectively. B, Graph of genotypes from IN gene amino acid region 142–165. Deep sequencing analysis was performed on preamplified region of the IN gene from 3 time points during the period of virologic inflection (panel A). Graph shows distribution of genotypes as portion of total reads. C, Complete list of all genotypes from IN gene amino acids 142–165 detected at >0.5% of total reads. Time points refer to those depicted in panel A. Abbreviations: DTG, dolutegravir; FTC, emtricitabine; r/DRV, ritonavir-boosted darunavir; RT, reverse transcriptase; TDF, tenofovir disoproxil fumarate
The patient returned to the hospital 3 days later with worsened respiratory symptoms necessitating intensive care unit admission. His plasma HIV RNA level initially decreased to 2770 copies/mL after 2 weeks of ART but then increased to 6510 and 15700 copies/mL 23 and 27 days, respectively, after ART initiation. He received no divalent cation–containing therapies and was directly observed to be taking his ART during the hospital admission. ART was intensified with ritonavir-boosted darunavir on day 30. Standard HIV-1 population genotyping from the sample on day 27 showed M184V and V118I and IN genotype showed G163E. The patient’s plasma HIV RNA level decreased to 7660 copies/mL on day 35. His clinical symptoms improved, and he was discharged. His plasma HIV RNA level decreased further to 320 copies/mL on day 45, and his CD4+ T-cell count increased to 125/μL (Figure 1A). Ritonavir-boosted darunavir was later replaced with rilpivirine because of rash on day 73, and at this writing the patient has remained virologically suppressed for >2 years during treatment with tenofovir alafenamide–emtricitabine–rilpivirine with DTG.
To better understand the resistance dynamics during the period of apparent virologic failure, a portion of the IN gene was amplified for deep sequencing from serial plasma samples collected over 8 days after peak viremia (time points A, B, and C in Figure 1A). Viral RNA was isolated, and an IN gene fragment from amino acids 142–165 was amplified using reverse-transcription polymerase chain reaction (see Supplementary Methods). This region was chosen to include as many potential reported mutations within the fragment size optimal for sequencing with an Illumina HiSeq 2000 sequencing platform. Owing to limited plasma samples from this clinical period, we were able to analyze only this portion of the IN gene. Sequencing of these samples generated a mean of 2483155 reads covering the region of interest. Paired-end reads were used for error correction and referenced to NL4-3 molecular clone sequence (see Supplementary Methods). The relative frequency of each mutation was calculated for each sample as raw read count to the total sequencing depth (Figure 1B and 1C).
Sequencing analysis revealed primarily wild-type virus at time point A (Figure 1B). At time point B, 3 days later, the primary mutation genotype was I151V-G163E. Population genotype performed for clinical care on this same day also confirmed presence of the G163E mutation, in addition to M184V mutation in reverse-transcriptase (Figure 1A). Time point C, 5 days after time point B, showed Q148K emergence and continued presence of I151V-G163E (Figure 1B). A full list of all mutations present at >0.5% is shown in Figure 1C.
DISCUSSION
Development of DTG resistance in first-line treatment is a rare and not fully understood event [6, 7]. This report presents a case of rapid emergence of multiple potential resistance mutations in the IN gene during a time of increasing plasma HIV RNA level after initial rapid decay during on first-line DTG-containing 3-drug ART. We believe this is the first such report and as such could have significant clinical implications for the planned scale-up of DTG-containing regimens for first-line and second-line therapy globally. Only 1 previous cohort has identified cases of treatment-naive virologic failure during DTG treatment [6]. Risk of treatment failure in this retrospective study was associated with viremia >100000 HIV-1 RNA copies/mL and CD4+ T-cell counts <200/μL [6], both of which were also characteristic of our patient.
We demonstrated a case of rapid development of Q148K mutation (20.9%), along with the presence of numerous minority mutations (Figure 1). The role of minority variants, which includes viral populations below the 20%–30% detection limit of standard clinical genotype assays, can affect response to nonnucleoside reverse-transcriptase inhibitors and protease inhibitors [8, 9], but the role of minority variants in INSTI treatment failure is not clear. Studies have similarly demonstrated evolution of INSTI resistance during raltegravir therapy, but this has not been consistently shown to alter outcomes in larger studies [8]. In our case, it is unlikely that Q148K alone conferred clinical DTG resistance, but more likely this in addition to other mutations collectively resulted in the observed clinical outcome.
The role of the existing M184V mutation in the observed clinically obtained population genotype is unclear, but intriguing. There have been reports of reverse-transcriptase mutations, specifically M184V, influencing IN resistance. A post hoc analysis of a phase 3 trial of a regimen including elvitegravir, cobicistat, emtricitabine, and tenofovir disoproxil fumarate showed that primary INSTI mutations often occurred in the setting of preexisting M184V [10], with similar findings in other studies of treatment-experienced patients [11]. Although we observed both M184V and Q148K, we do not know whether these are on the same virion or separate quasispecies. The lack of additional clinical specimens for phenotypic testing limits our ability to determine the relative influence of each observed mutation on the virologic failure. One possibility is that M184V emergence led to the initial loss of virologic control, which then predisposed to the rapid selection of DTG resistance.
Our analysis is limited by the IN gene region available for sequencing (amino acids 142–165), because this does not allow for assessment of other potentially relevant mutations in other regions, including T66, E138, G140, R263, or other de novo mutations. Similarly, the absence of IN gene sequencing before ART initiation limits our ability to evaluate for transmitted IN gene resistance, although such transmitted resistance has been a rare event to date [12, 13]. Unfortunately, neither phenotypic testing nor DTG levels were available owing to limited specimens. Medication adherence was unlikely to have contributed to virologic failure in this case, because the patient was hospitalized most of the time and therefore had directly observed therapy. Medication administration records were reviewed in detail, and there was no concurrent administration of divalent cations or other medications that could similarly compromise absorption of DTG.
Although DTG resistance and failure remain exceedingly rare in treatment-naive individuals, our case and detailed genotypic analysis present intriguing data showing rapid evolution of IN mutations in the presence of DTG, concomitant with increasing plasma viremia. Despite the limitations noted, our report is a reminder that, despite a high barrier to resistance, no agent as initial therapy for HIV-1 is impervious to resistance.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are most grateful to the patient described in this report for their generous participation. We also thank Grace Aldrovandi, MD, CM and Judith Currier, MD, MSc for insightful discussions.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was support by a UCLA Clinical and Translational Science Institute career development award (NIH grant KL2TR001882 to J. A. F), the UCLA Center for HIV Identification, Prevention, and Treatment Services (National Institute of Mental Health grant P30MH58107 to R. J. L), the UCLA Center for AIDS Research (NIH grant 5P30AI028697), and the UCLA Clinical Translational Science Institute (NIH grant UL1TR001881).
Potential conflicts of interest. R. J. L. has served as a consultant for Gilead Sciences. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shafer RW. Human immunodeficiency virus type 1 drug resistance mutations update. J Infect Dis 2017; 216:843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cahn P, Pozniak AL, Mingrone H, et al. ; extended SAILING Study Team Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 2013; 382:700–8. [DOI] [PubMed] [Google Scholar]
- 3. Walmsley SL, Antela A, Clumeck N, et al. ; SINGLE Investigators Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N Engl J Med 2013; 369:1807–18. [DOI] [PubMed] [Google Scholar]
- 4. Clotet B, Feinberg J, van Lunzen J, et al. ; ING114915 Study Team Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 2014; 383:2222–31. [DOI] [PubMed] [Google Scholar]
- 5. Raffi F, Jaeger H, Quiros-Roldan E, et al. ; extended SPRING-2 Study Group Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis 2013; 13:927–35. [DOI] [PubMed] [Google Scholar]
- 6. Lepik KJ, Harrigan PR, Yip B, et al. Emergent drug resistance with integrase strand transfer inhibitor-based regimens. AIDS 2017; 31:1425–34. [DOI] [PubMed] [Google Scholar]
- 7. Taiwo BO, Zheng L, Nyaku AN, et al. ACTG A5353: a pilot study of dolutegravir (DTG) + lamivudine (3TC) for initial treatment of HIV-1-infected participants with HIV-1 RNA <500000 copies/ml. 9th IAS Conference on HIV Science (IAS 2017), 23–26 July 2017, Paris, France 2017. [Google Scholar]
- 8. Stella-Ascariz N, Arribas JR, Paredes R, Li JZ. The role of HIV-1 drug-resistant minority variants in treatment failure. J Infect Dis 2017; 216:847–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simen BB, Simons JF, Hullsiek KH, et al. ; Terry Beirn Community Programs for Clinical Research on AIDS Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]
- 10. White K, Kulkarni R, Miller MD. Analysis of early resistance development at the first failure timepoint in elvitegravir/cobicistat/emtricitabine/tenofovir disoproxil fumarate-treated patients. J Antimicrob Chemother 2015; 70:2632–8. [DOI] [PubMed] [Google Scholar]
- 11. Winters MA, Lloyd RM Jr, Shafer RW, Kozal MJ, Miller MD, Holodniy M. Development of elvitegravir resistance and linkage of integrase inhibitor mutations with protease and reverse transcriptase resistance mutations. PLoS One 2012; 7:e40514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koullias Y, Sax PE, Fields NF, Walensky RP, Hyle EP. Should we be testing for baseline integrase resistance in patients newly diagnosed with human immunodeficiency virus?Clin Infect Dis 2017; 65:1274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doyle T, Dunn DT, Ceccherini-Silberstein F, et al. ; CORONET Study Group Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J Antimicrob Chemother 2015; 70:3080–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.