Aspiration of oral contents can lead to pneumonia, which is a major cause of death among elderly adults susceptible to swallowing impairments. Tongue microbiota are a dominant source of oral microbial populations that are ingested with saliva. This large-scale population-based study revealed variations in the tongue microbiota among community-dwelling elderly adults. The total bacterial density was independent of the conditions of teeth surrounding the tongue, whereas the microbiota composition, especially the relative abundances of predominant commensals, showed an association with tooth conditions. Our results demonstrate that the elderly with fewer teeth, poorer dental hygiene, and more dental caries experience constantly ingest more dysbiotic microbiota, which could be harmful for their respiratory health.
KEYWORDS: community-dwelling, dental, elderly, microbiota, oral, tongue
ABSTRACT
Tongue microbiota are a dominant source of oral microbial populations that are ingested with saliva, and therefore careful attention is required for the maintenance of health of elderly adults, who are susceptible to aspiration of oral contents. This study aimed to investigate the variation in tongue microbiota among community-dwelling elderly adults. Following a dental examination, tongue coating was collected from a 15-mm-diameter circular area at the center of the tongue dorsum of 506 elderly adults aged 70 to 80 years inhabiting the town of Hisayama, Japan. The microbial composition and density were determined by a 16S rRNA gene sequencing approach using a next-generation sequencer and quantitative PCR analysis, respectively. Co-occurrence network analysis identified two cohabiting groups of predominant commensals, one of which was primarily composed of Prevotella histicola, Veillonella atypica, Streptococcus salivarius, and Streptococcus parasanguinis; these organisms have been previously associated with an increased risk of mortality due to pneumonia in the frail elderly. This bacterial group was more predominant in the elderly with fewer teeth, a higher plaque index, and more dental caries experience, whereas the total bacterial density was independent of these traits. A higher density of fungi was also observed in the elderly with these traits, as well as in individuals who wore dentures. These results suggest that elderly adults with poorer oral health swallow a more dysbiotic microbiota formed on the tongue.
IMPORTANCE Aspiration of oral contents can lead to pneumonia, which is a major cause of death among elderly adults susceptible to swallowing impairments. Tongue microbiota are a dominant source of oral microbial populations that are ingested with saliva. This large-scale population-based study revealed variations in the tongue microbiota among community-dwelling elderly adults. The total bacterial density was independent of the conditions of teeth surrounding the tongue, whereas the microbiota composition, especially the relative abundances of predominant commensals, showed an association with tooth conditions. Our results demonstrate that the elderly with fewer teeth, poorer dental hygiene, and more dental caries experience constantly ingest more dysbiotic microbiota, which could be harmful for their respiratory health.
INTRODUCTION
The human oral cavity is densely colonized by diverse microorganisms on the intraoral surfaces, which are constantly shed into saliva. Normally, the detached organisms ingested with saliva are transported via the esophagus into the stomach and are then inactivated by gastric acid and proteolytic enzymes (1). However, impairments in swallowing and cough reflux with aging allow for aspiration into the lower respiratory tract and subsequent pulmonary infection (2). Swallowing disorders are highly prevalent in elderly patients with pneumonia (3, 4), which is a major cause of death among elderly adults. Mechanical oral hygiene to decrease the oral microbial burden is recognized as an effective approach to reduce the mortality rate from aspiration pneumonia (5) and is introduced as standard nursing care to the frail elderly in the hospital and nursing home settings.
Although aspirated saliva contains microorganisms colonizing various oral sites, their bacterial composition indicates that the dominant source is the microbiota formed on the tongue (6–8). The dorsum of the tongue has a large surface area with papillary structures, which can retain numerous microorganisms, including both aerobes and anaerobes (9). The microbial community is packed less densely than dental plaque, allowing the bacterial cells ready access to sufficient nutrients via saliva (10). The loose community structure and the desquamation of epithelial cells facilitate the release of resident microorganisms into the saliva (11). These features imply that careful attention should be given to the tongue microbiota in the elderly susceptible to swallowing impairments (12). Furthermore, increased tongue coating in edentulous elderly adults, scored by visual inspections, was associated with aspiration pneumonia (13) and febrile status (14).
Using a 16S rRNA gene next-generation sequencing approach, we recently reported that the dysbiotic composition of tongue microbiota was also associated with a higher risk of death from pneumonia in frail elderly adults in nursing homes (15). This result suggests that methods to optimize the tongue microbiota composition might be beneficial in health maintenance of the elderly. However, no previous study has focused on the tongue microbiota composition of community-dwelling elderly adults, and therefore the tongue microbiota of normal elderly adults remains poorly understood. In this population-based study, we investigated the tongue microbiota and dental conditions of Japanese elderly adults aged 70 to 80 years inhabiting the town of Hisayama, Japan. This study aimed to understand the variations in tongue microbiota among community-dwelling elderly adults with various oral conditions and to identify oral health-related factors associated with the dysbiotic shift in the tongue microbiota.
RESULTS
This study investigated the tongue microbiota status of 506 elderly adults aged 70 to 80 years (231 males and 275 females) inhabiting the town of Hisayama who received a dental examination during a health examination of Hisayama residents performed in 2016. Institutionalized or hospitalized elderly adults and very frail elderly residents, such as bedridden people, did not participate in the health examination or this study. Nevertheless, this study population accounts for 50.6% of the total residents of this town in this age group, suggesting that our data largely cover the variations in tongue microbiota among the independent elderly. The tongue microbiota was collected from the center area of the tongue dorsum using a modified electric toothbrush as the sampling device (see Fig. S1 in the supplemental material). The bacterial and fungal density per 15-mm-diameter circular area at the center of the tongue dorsum and the bacterial composition of the microbiota of each individual were investigated using quantitative PCR and 16S rRNA gene next-generation sequencing approaches, respectively.
A tongue coating sampling device based on the Braun Oral-B Pro 500 electric toothbrush (Procter & Gamble, Cincinnati, OH) (A). A circular bonded-fiber fabric (15 mm in diameter) was attached to this brush head, from which the bristles were preliminarily removed (B). The head was placed on the center of the tongue dorsum, 3-s vibrations were applied, and the collected tongue coating samples adhered to the fabric. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tongue microbiota composition and predominant bacterial taxa.
The bacterial composition of tongue microbiota was determined by a 16S rRNA gene sequencing approach using a next-generation sequencer, Ion PGM (Thermo Fisher Scientific, Waltham, MA), which provided 9,544,681 quality-passed bacterial 16S rRNA gene sequences (V1-V2 regions). These sequences were assigned to 730 species-level operational taxonomic units (OTUs), using a cutoff distance of 0.03. Of these, 21 predominant OTUs with a mean relative abundance of >1% are listed in Table 1. The OTU corresponding to Streptococcocus salivarius indicated that this species was the most predominant in the tongue microbiota of the elderly, followed by other bacteria, such as Prevotella melaninogenica, Rothia mucilaginosa, and Veillonella atypica. The predominant taxa were mostly consistent with the Human Microbiome Project data, which recruited healthy younger people aged 18 to 40 years (16). These OTUs were mostly shared across this study population and constituted the majority of the microbiota in each individual (mean ± standard deviation [SD], 75.2% ± 9.5%).
TABLE 1 .
Twenty-one predominant OTUs with mean relative abundances of >1% in the tongue microbiota of 506 elderly adults
| OTU no. | Bacterial species corresponding to each OTU (taxon ID)a |
Mean relative abundance (%) |
Detection rate (%) |
|---|---|---|---|
| OTU2 | Streptococcus salivarius (755) | 9.5 ± 8.6 | 99.4 |
| OTU4 | Prevotella melaninogenica (469) | 9.2 ± 6.4 | 98.8 |
| OTU1 | Rothia mucilaginosa (681) | 8.8 ± 8.1 | 99.6 |
| OTU9 | Veillonella atypica (524) | 6.0 ± 3.2 | 100 |
| OTU6 | Neisseria flavescens (610) | 5.8 ± 8.3 | 89.9 |
| OTU3 | Prevotella histicola (298) | 4.7 ± 6.0 | 90.3 |
| OTU19 | Streptococcus parasanguinis II (411) | 4.3 ± 3.9 | 98.0 |
| OTU10 | Actinomyces odontolyticus (701) | 3.9 ± 3.4 | 99.6 |
| OTU7 | Granulicatella adiacens (534) | 3.3 ± 2.5 | 99.6 |
| OTU5 | Haemophilus parainfluenzae (718) | 3.2 ± 4.0 | 94.7 |
| OTU421 | Genus Neisseriab | 2.4 ± 5.1 | 58.3 |
| OTU25 | Actinomyces sp. (172) | 2.2 ± 2.9 | 93.5 |
| OTU8 | Fusobacterium periodonticum (201) | 1.8 ± 2.3 | 90.5 |
| OTU15 | Prevotella pallens (714) | 1.5 ± 1.5 | 91.3 |
| OTU14 | Porphyromonas pasteri (279) | 1.5 ± 2.7 | 75.5 |
| OTU11 | Gemella sanguinis (757) | 1.3 ± 1.3 | 97.2 |
| OTU23 | Actinomyces graevenitzii (866) | 1.3 ± 2.1 | 88.1 |
| OTU22 | Streptococcus sp. (074) | 1.3 ± 1.5 | 93.1 |
| OTU28 | Genus Streptococcusb | 1.3 ± 1.5 | 96.0 |
| OTU12 | Leptotrichia sp. (417) | 1.1 ± 2.8 | 86.8 |
| OTU17 | Alloprevotella sp. (308) | 1.0 ± 1.4 | 85.2 |
Oral taxon IDs in HOMD are given in parentheses following bacterial names.
No BLAST hit with ≥98.5% identity was found in the Human Oral Microbiome Database (HOMD).
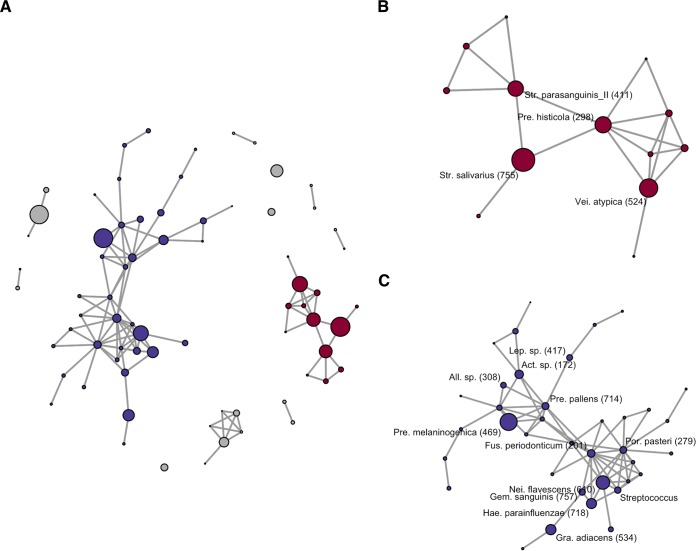
The co-occurrence network analysis using SparCC (17) suggested that these predominant members construct two major cohabiting groups (including multiple predominant OTUs) in the microbiota (Fig. 1A). One was primarily composed of a bacterial complex including Prevotella histicola, Veillonella atypica, Streptococcus salivarius, and Streptococcus parasanguinis II (commensal group I [Fig. 1B]). Another was mainly assembled from Neisseria flavescens, Haemophilus parainfluenzae, Fusobacterium periodonticum, Porphyromonas pasteri, Gemella sanguinis, Prevotella melaninogenica, and Prevotella pallens (commensal group II [Fig. 1C]). Negative associations were observed between the group I and group II bacteria, especially in subnetwork II (N. flavescens, F. periodonticum, P. pasteri, H. parainfluenzae, G. sanguinis, and Granulicatella adiacens [Fig. 1B and 2]). Principal-coordinate analysis (PCoA) plots based on unweighted UniFrac metrics showed that the individuals with microbiota containing greater relative abundances of group I commensals were localized in the negative direction of principal coordinate 1, whereas, in contrast, those with microbiota containing greater relative abundances of group II commensals were localized in its positive direction (see Fig. S2 in the supplemental material). It is suggested that the ratio of the two predominant commensal groups is strongly associated with the overall composition of the tongue microbiota.
FIG 1 .
Co-occurrence network in tongue microbiota of 506 elderly adults built from SparCC correlation coefficients between sequence abundances. Nodes corresponding to operational taxonomic units (OTUs) and connecting edges indicate the correlation between them. Only correlations with values greater than 0.40 are represented as edges. The node size indicates a mean relative abundance of each OTU. (A) All OTUs excluding isolated nodes corresponding to the OTUs with a mean relative abundance of <1% are shown as nodes in the diagram (72 of all 730 OTUs). Most of the predominant OTUs (with mean relative abundance of >1%) belonged to either of the two major networks including multiple predominant OTUs (cohabiting commensal groups I and II). The nodes belonging to commensal groups I and II are colored red and blue, respectively. (B) Only the co-occurrence network of OTUs belonging to commensal group I is shown. The bacterial taxa corresponding to the predominant OTUs are described. Oral taxon IDs in the Human Oral Microbiome database are given in parentheses following the bacterial names. Abbreviations: Pre., Prevotella; Vei., Veillonella; Str., Streptococcus. (C) Here, only the co-occurrence network of OTUs belonging to commensal group II is shown. The bacterial taxa corresponding to the predominant OTUs are described. Oral taxon IDs in the Human Oral Microbiome database are given in parentheses following bacterial names. Abbreviations: Nei., Neisseria; Hae., Haemophilus; Por., Porphyromonas; Gra., Granulicatella; Fus., Fusobacterium; Pre., Prevotella; Act., Actinomyces; Lep., Leptotorichia; All., Alloprevotella.
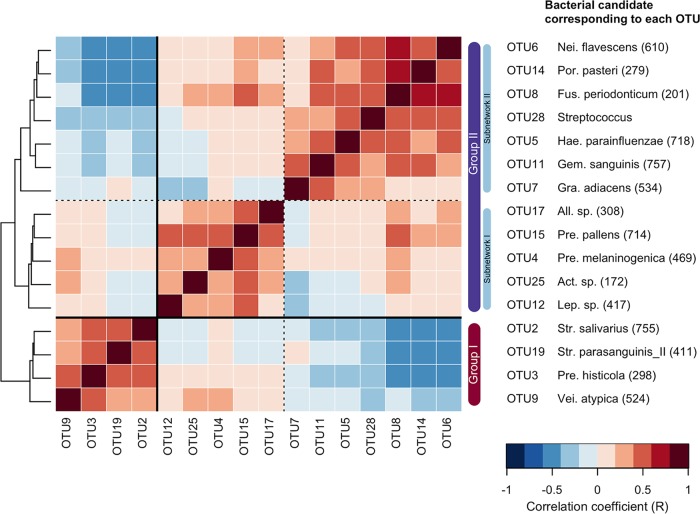
FIG 2 .
SparCC correlation coefficients between sequence abundances of predominant operational taxonomic units (OTUs) (with a mean relative abundance of >1%) belonging to cohabiting commensal groups I and II. The pairwise correlation coefficient is shown in each grid by the color intensity. The OTUs are ordered according to the result of a hierarchical cluster analysis using the Euclidean distance with average linkage (shown as a dendrogram on the left). Correlations with values greater than 0.40 are represented as edges in Fig. 1.
A principal-coordinate analysis plot showing similarity relationships among tongue microbiota samples from 506 community-dwelling elderly adults using an unweighted UniFrac distance metric. The points corresponding to the individuals with a higher relative abundance of each commensal group are depicted as bolder red. To correct for the unequal number of sequences, we evaluated 5,000 randomly selected sequences per sample. The two axes explained 12.9 and 9.7% of the variance, respectively. Download FIG S2, TIF file, 0.9 MB (945.3KB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Of the five predominant OTUs not belonging to the two commensal groups, the relative abundance of the OTU corresponding to Rothia mucilagiosa was significantly correlated with that of commensal group I, whereas those of the two OTUs corresponding to Streptococcus sp. strain OT-074 and Neisseria species were significantly correlated with commensal group II (see Table S1 in the supplemental material).
Spearman’s correlation coefficient between the relative abundances of group I and II commensals and five predominant operational taxonomic units (OTUs) that were not present in both cohabiting groups. Download TABLE S1, DOCX file, 0.1 MB (67.3KB, docx) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
OTUs corresponding to mutans streptococci (MS) and lactobacilli, which are typical dental caries-associated taxa (18), and 10 periodontitis-associated taxa validated by Oliveira et al. (19) from 18 candidate bacteria listed in a systematic review (20) were detected from some of the individuals as minority members (mean relative abundance of <0.1% [Table 2]). Furthermore, they were not part of the above-mentioned cohabiting commensal groups.
TABLE 2 .
Relative abundances and detection rate of OTUs corresponding to mutans streptococci and lactobacilli and periodontitis-associated taxa in the tongue microbiota of 506 elderly adults
| OTU no. | Bacterial species corresponding to each OTU (taxon ID)a |
Mean relative abundance (%) |
Detection rate (%) |
|---|---|---|---|
| 14 OTUs corresponding to mutans streptococci and lactobacilli |
|||
| OTU73 | Streptococcus mutans (686) | 0.020 ± 0.131 | 17.8 |
| OTU75 | Streptococcus sobrinus (768) | 0.039 ± 0.412 | 11.1 |
| OTU157 | Lactobacillus iners (838) | 0.002 ± 0.055 | 0.2 |
| OTU230 | Lactobacillus paracasei (716) | 0.001 ± 0.008 | 2.2 |
| OTU235 | Lactobacillus fermentum (608) | 0.023 ± 0.222 | 4.9 |
| OTU238 | Lactobacillus vaginalis (051) | 0.008 ± 0.064 | 4.0 |
| OTU291 | Lactobacillus ultunensis (461) | 0.001 ± 0.011 | 3.0 |
| OTU293 | Lactobacillus reuteri (938) | 0.001 ± 0.020 | 0.6 |
| OTU338 | Lactobacillus oris (709) | 0.001 ± 0.007 | 1.2 |
| OTU375 | Lactobacillus pentosus (883) | 0.000 ± 0.004 | 0.6 |
| OTU40 | Lactobacillus gasseri (615) | 0.042 ± 0.437 | 8.3 |
| OTU41 | Lactobacillus crispatus (817) | 0.029 ± 0.426 | 4.7 |
| OTU47 | Lactobacillus salivarius (756) | 0.077 ± 0.722 | 8.5 |
| OTU599 | Lactobacillus sp. (052) | 0.000 ± 0.001 | 0.2 |
| 11 OTUs corresponding to periodontitis-associated taxab |
|||
| OTU60 | Porphyromonas gingivalis (619) | 0.066 ± 0.168 | 49.4 |
| OTU92 | Tannerella forsythia (613) | 0.008 ± 0.021 | 22.3 |
| OTU167 | Filifactor alocis (539) | 0.003 ± 0.012 | 8.3 |
| OTU194 | Selenomonas sputigena (151) | 0.001 ± 0.008 | 4.0 |
| OTU228 | Selenomonas sputigena (151) | 0.004 ± 0.023 | 10.9 |
| OTU239 | Fretibacterium sp. (360) | 0.002 ± 0.007 | 6.5 |
| OTU254 | Bacteroidales sp. (274) | 0.002 ± 0.011 | 7.1 |
| OTU534 | Bacteroidales sp. (274) | 0.000 ± 0.002 | 0.8 |
| OTU286 | Desulfobulbus sp. (041) | 0.000 ± 0.003 | 2.0 |
| OTU346 | Fretibacterium sp. (362) | 0.001 ± 0.006 | 2.8 |
| OTU81 | TM7 sp. (356) | 0.011 ± 0.031 | 21.7 |
Oral taxon IDs in HOMD are given in parentheses following bacterial names.
Bacterial taxa listed as periodontitis-associated bacteria by Oliveira et al. (19).
Correlation between parameters associated with tongue microbiota status.
To investigate the relationship between tongue microbiota status and dental conditions, this study focused on total bacterial density and richness, total fungal density, and the relative abundances of the OTUs corresponding to four bacterial groups: MS and lactobacilli as typical dental caries-associated taxa (14 OTUs [Table 2]) (18), the above-mentioned 10 periodontitis-associated taxa listed in a previous study (11 OTUs [Table 2]) (19), and the predominant bacteria in commensal group I (4 OTUs [Fig. 1B and 2]) and II (12 OTUs [Fig. 1C and 2]).
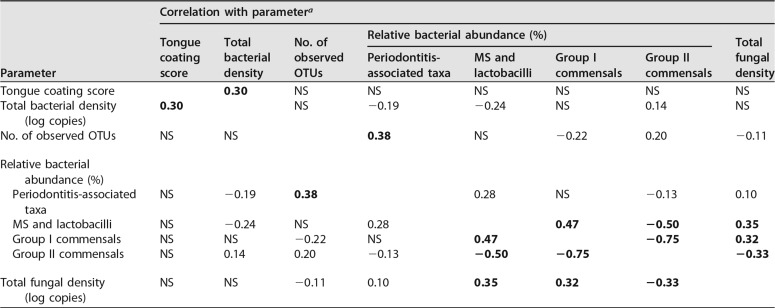
Prior to this analysis, we evaluated the relationships between eight parameters associated with the microbiota. Spearman’s correlation coefficients for these parameters are described in Table 3. A tongue coating score based on visual inspections of the tongue coating area and thickness reflected the total bacterial density rather than the bacterial composition. A higher relative abundance of periodontitis-associated taxa showed a significant association with a greater bacterial richness of the microbiota (number of observed OTUs). This relationship is reasonable, considering that deepening periodontal pockets resulting from periodontitis progression supply saliva to the subgingival site-specific bacteria, which are rarely present on the tongue. Of the two cohabiting commensal groups, the relative abundance of group I was positively correlated with the relative abundance of the dental caries-associated bacterial group (MS and lactobacilli) and the fungal density. On the other hand, their relative abundances exhibited inverse correlations with the relative abundances of the members of commensal group II.
TABLE 3 .
Spearman’s correlation coefficient between parameters associated with the tongue microbiota status
| Parameter | Correlation with parametera |
|||||||
|---|---|---|---|---|---|---|---|---|
| Tongue coating score |
Total bacterial density |
No. of observed OTUs |
Relative bacterial abundance (%) |
Total fungal density |
||||
| Periodontitis- associated taxa |
MS and lactobacilli |
Group I commensals |
Group II commensals |
|||||
| Tongue coating score | 0.30 | NS | NS | NS | NS | NS | NS | |
| Total bacterial density (log copies) |
0.30 | NS | −0.19 | −0.24 | NS | 0.14 | NS | |
| No. of observed OTUs | NS | NS | 0.38 | NS | −0.22 | 0.20 | −0.11 | |
| Relative bacterial abundance (%) |
||||||||
| Periodontitis-associated taxa |
NS | −0.19 | 0.38 | 0.28 | NS | −0.13 | 0.10 | |
| MS and lactobacilli | NS | −0.24 | NS | 0.28 | 0.47 | −0.50 | 0.35 | |
| Group I commensals | NS | NS | −0.22 | NS | 0.47 | −0.75 | 0.32 | |
| Group II commensals | NS | 0.14 | 0.20 | −0.13 | −0.50 | −0.75 | −0.33 | |
| Total fungal density (log copies) |
NS | NS | −0.11 | 0.10 | 0.35 | 0.32 | −0.33 | |
An absolute correlation coefficient of >0.3 is shown in boldface. NS, not significant.
Tongue microbiota status and oral health conditions.
We compared the above-mentioned tongue microbiota statuses among the elderly with various oral conditions, including the number of remaining teeth, dental hygiene (maximum plaque index), dental caries experience (percentage of caries-experienced teeth), active dental caries (number of decayed teeth), gingivitis (percentage of teeth with bleeding on probing), periodontitis (mean periodontal pocket depth), and the use of dentures (Fig. 3, 4, and 5).
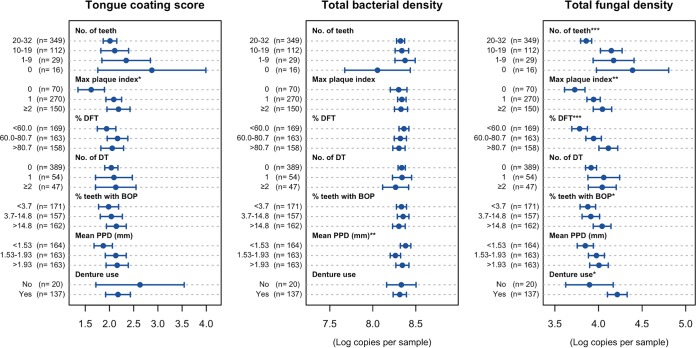
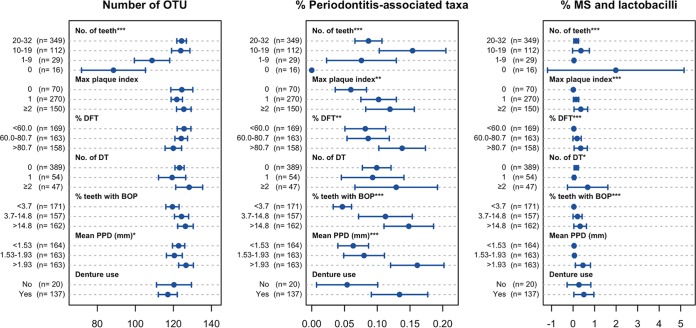
FIG 3 .
Tongue coating score and total bacterial and fungal density per 15-mm-diameter circular area at the center of the tongue dorsum of the elderly adults with various oral conditions. Of the 506 individuals, the 16 edentulous elderly were excluded from the maximum plaque index, percentage of decayed and filled teeth (DFT), number of decayed teeth (DT), percentage of teeth with bleeding on probing (BOP), and mean periodontal pocket depth (PPD), whereas 349 individuals with ≥20 teeth were excluded from denture use. The individuals were categorized into the tertile of percentage of DFT, percentage of teeth with BOP, and mean PPD, and then the parameters of the microbiota status were compared. Dots indicate the mean; the error bars indicate 95% confidence intervals. ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by Kruskal-Wallis analysis.
FIG 4 .
Alpha diversity and relative abundances of periodontitis- and dental caries-associated taxa in tongue microbiota of the elderly adults with various oral conditions. Of the 506 individuals, the 16 edentulous elderly were excluded from the maximum plaque index, percentage of decayed and filled teeth (DFT), number of decayed teeth (DT), percentage of teeth with bleeding on probing (BOP), and mean periodontal pocket depth (PPD), whereas 349 participants with ≥20 teeth were excluded from denture use. The individuals were categorized into the tertile of percentage of DFT, percentage of teeth with BOP, and mean PPD, and the parameters of the microbiota status were compared. Dots indicate the mean; the error bars indicate 95% confidence intervals. ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by Kruskal-Wallis analysis.
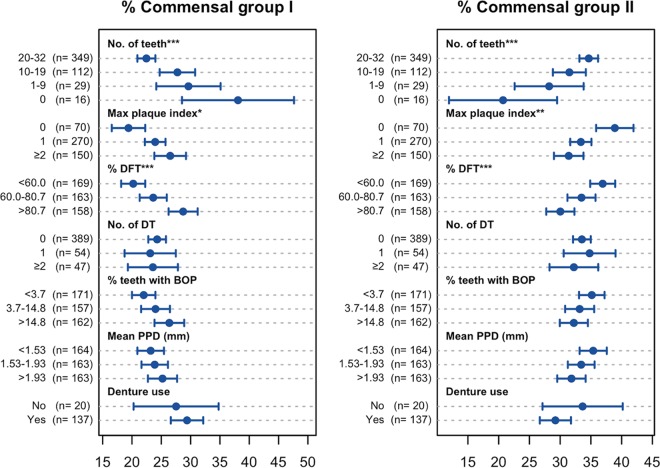
FIG 5 .
Relative abundances of two cohabiting commensal groups in tongue microbiota of elderly adults with various oral conditions. Of the 506 individuals, the 16 edentulous elderly were excluded from the maximum plaque index, percentage of decayed and filled teeth (DFT), number of decayed teeth (DT), percentage of teeth with bleeding on probing (BOP), and mean periodontal pocket depth (PPD), whereas 349 participants with ≥20 teeth were excluded from denture use. The individuals were categorized into the tertile of percentage of DFT, percentage of teeth with BOP, and mean PPD, and the parameters of the microbiota status were compared. Dots indicate the mean; the error bars indicate 95% confidence intervals. ***, P < 0.001, **, P < 0.01, and *, P < 0.05, in Kruskal-Wallis analysis.
A notable difference was not observed in the tongue coating scores based on visual inspections, except for a low score in the individuals without visible dental plaque (Fig. 3). The bacterial density of the tongue microbiota was also mostly independent of the oral conditions (Fig. 3). Although a significant difference in bacterial densities was observed among the individuals with different mean periodontal pocket depths, the density in the middle category of mean periodontal pocket depth was lower than in the other categories. The cause of this nonlinear relationship remains unclear.
A higher density of fungi was observed in the microbiota of individuals with less than 20 teeth, a higher plaque index, and more caries-experienced teeth (Fig. 3). In the individuals with less than 20 teeth, the use of dentures was significantly associated with a higher fungal density.
A drastic decrease in the bacterial richness (number of observed OTUs) was observed in the individuals with fewer teeth (Fig. 4). This reduction was mostly due to the dissipation of various minority taxa in the tongue microbiota, rather than the elimination of specific predominant members. It is likely to be caused by the supply decline of bacteria shed from tooth surfaces.
Periodontitis-associated taxa were enriched in the individuals with a greater amount of dental plaque and gingival and periodontal inflammation, as expected (Fig. 4). The lack of periodontitis-associated taxa in the edentulous individuals was not unexpected, as the periodontal niches disappeared from the oral cavity due to the loss of the tooth itself. A higher abundance of MS and lactobacilli was observed in the individuals with multiple decayed teeth, more caries-experienced teeth, and a larger amount of dental plaque, as well as those with more severe gingivitis and edentulous individuals (Fig. 4).
The ratio of the two predominant commensal groups varied drastically in accordance with tooth conditions and dental hygiene (Fig. 5). Commensal group I, including P. histicola, was observed in a higher proportion in the microbiota of individuals with less teeth, a higher plaque index, and more caries-experienced teeth. On the other hand, their microbiota contained a lower abundance of group II commensals such as N. flavescens.
The PCoA plot based on unweighted UniFrac metrics showed that the individuals with better oral health status tend to localize in the positive direction of principal coordinate 1 (see Fig. S3 in the supplemental material). The significant relationships between overall composition of the tongue microbiota and five oral health-related parameters (the number of remaining teeth, maximum plaque index, percentage of caries-experienced teeth, percentage of teeth with bleeding and on probing, and mean periodontal pocket depth) were confirmed by permutational analysis of variance (PERMANOVA) (Table 4).
TABLE 4 .
PERMANOVA results for variance of the unweighted UniFrac distance among 490 dentulous elderly adults
| Variable | R2 value | P value |
|---|---|---|
| Gender | 0.002 | 0.631 |
| No. of teeth (3 categories)a,b | 0.011 | <0.001 |
| Max plaque index (3 categories)a | 0.009 | <0.001 |
| % DFT (3 categories)a | 0.009 | <0.001 |
| No. of DT (3 categories)a | 0.004 | 0.290 |
| % of teeth with BOP (3 categories)a | 0.008 | <0.001 |
| Mean PPD (mm [3 categories])a | 0.007 | <0.001 |
| Denture use | 0.002 | 0.197 |
| Residuals | 0.944 | |
| Total | 1.000 |
A principal-coordinate analysis plot showing similarity relationship among tongue microbiota samples from 506 community-dwelling elderly adults using an unweighted UniFrac distance metric. The points corresponding to the individuals with different categories in each oral health status are depicted in different colors. To correct for the unequal number of sequences, we evaluated 5,000 randomly selected sequences per sample. The two axes explained 12.9 and 9.7% of the variance, respectively. Download FIG S3, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
This population-based study revealed the tongue microbiota composition of 506 Japanese community-dwelling elderly adults aged 70 to 80 years. The predominant bacterial taxa, including S. salivarius, were mostly common among the individuals (Table 1). On the other hand, a co-occurrence network analysis suggested that these commensal members constructed two competitive cohabiting groups in the microbiota (Fig. 1 and 2), and the ratio of these two groups varied among individuals with different dental conditions (Fig. 5). One group assembled from group I commensals, including N. flavescens and P. pasteri, was observed in a lower proportion in the microbiota of individuals with fewer teeth, poorer dental hygiene, and more dental caries experience. Alternatively, the other group composed of group II commensals, including P. histicola, V. atypica, S. salivarius, and S. parasanguinis, was more predominant in their microbiota. These bacteria were specifically predominant in the microbiota on oral mucosal surfaces, including the tongue dorsum, and were only minor components of the tooth-associated microbiota (16, 21). Therefore, there was little doubt that the observed differences occurred in the tongue microbiota itself, rather than from the contamination of dental plaque shed from the teeth. Our result suggests that the global composition of the tongue microbiota was associated with the tooth conditions surrounding but separated from the tongue, although whether it is the cause or consequence of dental conditions remains unclear.
The tongue microbiota is a primary source of numerous bacterial populations ingested with saliva (6–8). The results of this study imply that the laryngopharynx of the elderly with fewer teeth, poorer dental hygiene, and more dental caries experience is constantly exposed to bacterial populations containing a higher relative abundance of group I commensals. Although Prevotella, Veillonella, and Streptococcus are not typical respiratory pathogens, they were identified as etiologic agents of pulmonary infections such as aspiration pneumonia (22) and lung abscesses (23). Our prospective cohort study demonstrated that a higher relative abundance of group I commensals on the tongue was implicated in an increased mortality risk from pneumonia in frail elderly adults in nursing homes (15). It is assumed that an aspiration of oral contents into the lower respiratory tract would be more health threatening for the elderly with poorer dental conditions. In addition, recent studies revealed that the lungs of healthy adults are colonized by bacteria mainly carried by microaspiration, which frequently contains Prevotella, Veillonella, and Streptococcus species (24). Segal et al. also demonstrated that the predominance of Prevotella and Veillonella species in the lung microbiota was associated with enhanced subclinical lung inflammation, such as enhanced expression of inflammatory cytokines and elevated Th17 lymphocytes (25). These results suggest that the tongue microbiota in which group I commensals are predominant would also be undesirable in the nondysphagic elderly.
The relative abundance of group I commensals was significantly correlated with that of MS and lactobacilli, which are well-known aciduric bacteria associated with dental caries (Table 3). The commensal group I itself also included the two predominant Streptococcus species on the tongue, i.e., S. salivarius and S. parasanguinis, which can survive in acidic conditions (26, 27), indicating an enrichment of the acid-tolerant bacteria in the tongue microbiota. In addition, elevated levels of the Veillonella species that metabolize lactate were associated with higher acid production within plaque (28). Therefore, it is reasonable to assume that the more acidified oral ecosystem is correlated with a tongue microbiota containing a predominance of group II commensals. The prolonged low-pH conditions might contribute to a higher susceptibility to dental caries, resulting in a larger number of dental caries-experienced teeth in the oral cavity.
A previous Dutch study demonstrated that a higher Candida load in the saliva of elderly adults occurred in aciduric bacterial communities with a greater prevalence of streptococci (29). Consistent with these results, a greater amount of fungi was contained in the tongue microbiota of the elderly with fewer teeth, poorer dental hygiene, and more dental caries experience, which contained higher relative abundances of Streptococcus species (Fig. 5). Synergistic interactions between streptococci and Candida via coaggregation, signaling, and metabolic processes (30) might promote their growth on the tongue. Furthermore, a previous study showed that the number of Candida species on the tongue dorsum tended to be higher in those subjects who died of aspiration pneumonia (31), although Candida pneumonia is a rare infection of the lung (32). The tongue dorsum should be treated with caution as a potential reservoir of oral fungi in the individuals who wore dentures (33), as well as in the dentate elderly with fewer teeth, poorer dental hygiene, and more dental caries experience.
No significant differences were observed in the relative abundance of both commensal groups I and II among individuals with different gingivitis (percentage of teeth with bleeding on probing) and periodontitis (mean periodontal pocket depth) statuses (Fig. 5), suggesting that the ratio of predominant commensals in the tongue microbiota was associated more strongly with tooth conditions compared with gingival conditions. Nevertheless, the tongue microbiota of elderly adults with poorer gingival or periodontal health contained a drastically higher relative abundance of the OTUs corresponding to periodontitis-associated taxa (Fig. 4), although they were only minor components even in the elderly with poor gingival health (up to 2.1%). Tongue microbiota analysis using a 16S rRNA gene deep sequencing approach would be useful for monitoring subgingival bacteria shed from the periodontal pockets.
The relative abundance of group I commensals varied with the tooth conditions of each elderly subject (Fig. 5); however, no significant relationship was observed with the total bacterial density on their tongue dorsum (Table 3). This result indicates that the predominance of group I commensals was unlikely to be a consequence of bacterial overgrowth on the tongue. Mechanical cleaning of the coated tongue is an effective approach to decrease the bacterial burden on the tongue dorsum (12, 34, 35). However, our data suggest that a future prospective intervention study is essential to develop another tongue care approach, which can systematically alter the ratio of the two commensal groups in the microbiota. Noteworthy is the inverse correlation between the group I and group II commensals in subnetwork II (N. flavescens, F. periodonticum, P. pasteri, H. parainfluenzae, G. sanguinis, and G. adiacens [Fig. 2]). Their growth promotion might be also helpful for shifting the microbiota to a healthier one.
Our previous population-based study demonstrated that saliva of both middle-aged and elderly adults often contains high relative abundances of group I commensals such as P. histicola, V. atypica, S. salivarius, and S. parasanguinis (36), suggesting that the dysbiotic pattern of the tongue microbiota is not unique to elderly adults. On the other hand, the study also indicated that the percentage of individuals with higher relative abundances of these taxa significantly increased according to age after adjustment for confounding factors (36). It is therefore assumed that dysbiotic tongue microbiota would occur with a higher frequency in older people.
Our large-scale population-based study revealed variations of the tongue microbiota among Japanese community-dwelling elderly adults aged 70 to 80 years. This result suggests that the elderly with less teeth, poorer dental hygiene, and more dental caries experience constantly ingest more dysbiotic microbiota, with a higher relative abundance of Prevotella, Veillonella, and Streptococcus species and a larger number of fungal species. Careful attention should be given to the tongue microbiota status in elderly adults with poorer dental conditions.
MATERIALS AND METHODS
Ethics approval and consent to participate.
All participants understood the nature of the study and provided informed consent. The Ethics Committee of Kyushu University, Fukuoka, Japan, approved the study design as well as the procedure for obtaining informed consent (reference no. 28-31). All experiments were performed in accordance with the approved guidelines.
Study population.
Dental examinations and the collection of tongue coating samples were conducted for elderly residents in the town of Hisayama, Fukuoka, Japan, which is a suburb of the Fukuoka metropolitan area in western Japan. Hisayama is recognized to be demographically representative of Japan in terms of its age and occupational distributions based on national consensus data (37). The population of Hisayama is approximately 8,000. A health examination of adult residents, including dental examinations, was conducted in 2016. Among all residents aged 70 to 80 years (1,000 individuals), 508 residents who received the dental examination consented to participate in the study.
Dental examination.
The details of the dental examination as a component of a health examination of Hisayama residents were described previously (36). In brief, the numbers of present, decayed, or filled teeth were examined. Periodontal conditions were evaluated based on periodontal pocket depth and bleeding on probing at two sites for all teeth (mesio- and mid-buccal sites) based on the NHANES III method, with the exception of the third molars. The tongue coating was visually scored by multiplying the area score (0 to 3) and thickness score (0 to 2) based on conventional criteria (38).
Sample collection.
Following the dental examination, tongue coating samples were collected by using a sampling device based on a modified Braun Oral-B Pro 500 electric toothbrush (Procter & Gamble, Cincinnati, OH [Fig. S1]). A circular bonded-fiber fabric (15 mm in diameter; Kao Corporation, Tokyo, Japan) was attached to this brush head, of which the bristles were preliminarily removed, as described previously (39). The head was placed on the center of the tongue dorsum, 3-s vibrations were applied, and the collected tongue coating samples adhered to the fabric. The fabric peeled from the brush head was immersed in 200 µl of lysis buffer containing 10 mM Tris-HCl, 1 mM EDTA, and 1% sodium dodecyl sulfate and then stored at −80°C until further analysis. DNA extraction from the tongue coating sample was performed using a bead-beating method with 0.3 g zirconia silica beads (0.1-mm diameter [BioSpec Products, Bartlesville, OK]) and a tungsten-carbide bead (3-mm diameter [Qiagen, Hilden, Germany]) as described previously (40). Two individuals were excluded from the latter analysis due to the poor quality of the extracted DNA.
Quantitative PCR analysis.
Quantitative PCR analysis of bacterial or fungal density in the collected sample was performed using a QuantiFast SYBR green PCR kit (Qiagen) according to the manufacturer’s instructions. The primers 806F (5′ TTA GAT ACC CYG GTA GTC C 3′) and 926R (5′ CCG TCA ATT YCT TTG AGT TT 3′), which target V5 regions of the 16S rRNA gene, were used for the quantification of bacterial density (41). The 16S rRNA gene of P. pasteri was inserted into the vector plasmid pBluescript SKII(+) (Stratagene, La Jolla, CA) and used as a real-time control. The fungal density was estimated using the primers 5.8SR (5′ TCG ATG AAG AAC GCA GC 3′) (42) and ITS4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) (43) for the amplification of a fungal internal transcribed spacer 2 region of the rRNA operon. This primer set covers a variety of fungal species, excluding several species of basidiomycetous fungi (the phylum Basidiomycota) (44). The internal transcribed spacer region of Candida albicans was inserted into the vector plasmid pBluescript SKII(+) (Stratagene) and used as a real-time control.
Ion Torrent 16S rRNA gene analysis.
The 16S rRNA gene sequencing analysis was performed using tongue coating samples collected from the participants. The V1-V2 region of 16S rRNA genes from each sample was amplified using the following primers: 8F (5′ AGA GTT TGA TYM TGG CTC AG 3′), with Ion Torrent adaptor A and the sample-specific 8-base tag sequence, and 338R (5′ TGC TGC CTC CCG TAG GAG T 3′>), with the Ion Torrent trP1 adaptor sequence. PCR amplification, purification, and quantification of each PCR adaptor were performed as previously described (36). Emulsion PCR and enrichment of template-positive particles were performed using an Ion PGM Hi-Q View OT2 kit (Thermo Fisher Scientific) with the Ion One Touch 2 system (Thermo Fisher Scientific), and sequencing was performed with the Ion PGM (Thermo Fisher Scientific) using an Ion PGM Hi-Q View sequencing kit (Thermo Fisher Scientific).
Data processing.
The raw sequence reads were quality filtered using a script written in R (version 3.3.1). The reads were excluded from the analysis if they exhibited ≤240 bases (not including the tag sequence), had an average quality score of ≤25, did not include the correct forward primer sequence, did not include the correct reverse primer sequence (one mismatch was allowed), or had a homopolymer run of >7 nucleotides (nt). The remaining reads were assigned to the appropriate sample by examining the tag sequence. Similar sequences were assigned into operational taxonomic units (OTUs) using UPARSE (45), with a minimum pairwise identity of 97%. The taxonomy of representative sequences was determined using BLAST against 889 oral bacterial 16S rRNA gene sequences (HOMD 16S rRNA RefSeq version 14.51) in the Human Oral Microbiome Database (46). Nearest-neighbor species with ≥98.5% identity to the representative sequence were selected as candidates for each OTU. The taxonomy of sequences without hits was further determined using the RDP classifier with a minimum support threshold of 80%. The number of observed OTUs and the relative abundance of each OTU were calculated following rarefaction with a depth of 5,000 reads per sample using R. We confirmed that the rarefaction curve for the number of OTUs per sample approached a plateau after 5,000 sequence reads (see Fig. S4 in the supplemental material).
Rarefaction curves for a number of observed operational taxonomic units (OTUs) per sample. The mean number of observed OTUs almost reached a plateau by 5,000 sequence reads. The error bars indicate the standard deviation. Download FIG S4, TIF file, 0.6 MB (643.5KB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis.
All statistical analyses were conducted using R (version 3.3.1). The co-occurrence network analysis was conducted using SparCC (17), which calculates correlations from compositional data using a log-ratio-transformed abundance with default parameters (10 inference iterations and 100 bootstraps). Co-occurrence networks with correlation values greater than 0.40 were represented as edges and were constructed using the gplot function in the sna library. The unweighted UniFrac metric (47) was used to determine the dissimilarity between any pair of bacterial communities. The similarity relationship, assessed using the UniFrac metric, was presented in a principal-coordinate analysis (PCoA) plot drawn by R. Permutational multivariate analysis of variance (PERMANOVA) was used to assess the effects of oral health-related factors on tongue microbiota using the Adonis function in the vegan library based on 9,999 permutations. The tongue microbiota statuses were compared among the elderly with different dental and gingival conditions, which included the number of present teeth, dental hygiene (maximum plaque index), dental caries (number of decayed teeth or caries-experienced teeth), gingivitis (percentage of teeth with bleeding on probing), periodontitis (mean periodontal pocket depth), and the use of dentures. The conditions of dental hygiene and dental caries, as well as gingival and periodontal inflammation, were compared among the individuals with ≥1 tooth, because such parameters were unable to be evaluated for the edentulous individuals. Kruskal-Wallis analysis was used to assess the differences in microbiota status between the categories.
Accession number(s).
The obtained sequence data obtained in this study have been deposited in the DDBJ Sequence Read Archive under accession no. DRA006979, DRA006980, DRA006981, DRA006982, and DRA006983.
ACKNOWLEDGMENTS
This study was supported in part by Grants-in-Aid by JSPS KAKENHI grant no. 16H05557 (T.T.), 16K15856 (T.T.), 16H02692 (Y.Y.), and 16H05850 (Y.Y.) from the Japan Society for the Promotion of Science (JP).
We thank Yukie Shibata, Rie Matsumi, Shino Suma, Akihiko Tanaka, Shigeyuki Yokayama, Naoko Yatabe, Koji Ogata, and Yukari Ihara for assistance with dental examination and sample collection and the staff of the Division of Health and Welfare of the Hisayama Town Office for their cooperation in this study.
REFERENCES
- 1.Yang I, Nell S, Suerbaum S. 2013. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev 37:736–761. doi: 10.1111/1574-6976.12027. [DOI] [PubMed] [Google Scholar]
- 2.Marik PE, Kaplan D. 2003. Aspiration pneumonia and dysphagia in the elderly. Chest 124:328–336. doi: 10.1378/chest.124.1.328. [DOI] [PubMed] [Google Scholar]
- 3.Teramoto S, Fukuchi Y, Sasaki H, Sato K, Sekizawa K, Matsuse T, Japanese Study Group on Aspiration Pulmonary Disease . 2008. High incidence of aspiration pneumonia in community- and hospital-acquired pneumonia in hospitalized patients: a multicenter, prospective study in Japan. J Am Geriatr Soc 56:577–579. doi: 10.1111/j.1532-5415.2008.01597.x. [DOI] [PubMed] [Google Scholar]
- 4.Cabre M, Serra-Prat M, Palomera E, Almirall J, Pallares R, Clavé P. 2010. Prevalence and prognostic implications of dysphagia in elderly patients with pneumonia. Age Ageing 39:39–45. doi: 10.1093/ageing/afp100. [DOI] [PubMed] [Google Scholar]
- 5.Sjögren P, Nilsson E, Forsell M, Johansson O, Hoogstraate J. 2008. A systematic review of the preventive effect of oral hygiene on pneumonia and respiratory tract infection in elderly people in hospitals and nursing homes: effect estimates and methodological quality of randomized controlled trials. J Am Geriatr Soc 56:2124–2130. doi: 10.1111/j.1532-5415.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 6.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. 2012. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. 2003. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644–654. doi: 10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, Fortenberry JD, Holland MJ, Burr SE, Shannon WD, Sodergren E, Weinstock GM. 2013. Biogeography of the ecosystems of the healthy human body. Genome Biol 14:R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loesche WJ. 2003. Microbiology and treatment of halitosis. Curr Infect Dis Rep 5:220–226. doi: 10.1007/s11908-003-0077-8. [DOI] [PubMed] [Google Scholar]
- 10.Spencer P, Greenman J, McKenzie C, Gafan G, Spratt D, Flanagan A. 2007. In vitro biofilm model for studying tongue flora and malodour. J Appl Microbiol 103:985–992. doi: 10.1111/j.1365-2672.2007.03344.x. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons RJ, Houte JV. 1975. Bacterial adherence in oral microbial ecology. Annu Rev Microbiol 29:19–44. doi: 10.1146/annurev.mi.29.100175.000315. [DOI] [PubMed] [Google Scholar]
- 12.Drinka PJ, El-Solh AA. 2010. The tongue, oral hygiene, and prevention of pneumonia in the institutionalized elderly. J Am Med Dir Assoc 11:465–467. doi: 10.1016/j.jamda.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Abe S, Ishihara K, Adachi M, Okuda K. 2008. Tongue-coating as risk indicator for aspiration pneumonia in edentate elderly. Arch Gerontol Geriatr 47:267–275. doi: 10.1016/j.archger.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Shimazaki Y, Tomioka M, Saito T, Nabeshima F, Ikematsu H, Koyano K, Yamashita Y. 2009. Influence of oral health on febrile status in long-term hospitalized elderly patients. Arch Gerontol Geriatr 48:411–414. doi: 10.1016/j.archger.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama S, Takeshita T, Furuta M, Tomioka M, Asakawa M, Suma S, Takeuchi K, Shibata Y, Iwasa Y, Yamashita Y. 2018. Relationships of variations in the tongue microbiota and pneumonia mortality in nursing home residents. J Gerontol A Biol Sci Med Sci 73:1097–1102. doi: 10.1093/gerona/glx205. [DOI] [PubMed] [Google Scholar]
- 16.Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A 111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caufield PW, Schön CN, Saraithong P, Li Y, Argimón S. 2015. Oral lactobacilli and dental caries: a model for niche adaptation in humans. J Dent Res 94:110S–118S. doi: 10.1177/0022034515576052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira RR, Fermiano D, Feres M, Figueiredo LC, Teles FR, Soares GM, Faveri M. 2016. Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res 95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, Duarte P, Feres M. 2014. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kageyama S, Takeshita T, Asakawa M, Shibata Y, Takeuchi K, Yamanaka W, Yamashita Y. 2017. Relative abundance of total subgingival plaque-specific bacteria in salivary microbiota reflects the overall periodontal condition in patients with periodontitis. PLoS One 12:e0174782. doi: 10.1371/journal.pone.0174782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akata K, Yatera K, Yamasaki K, Kawanami T, Naito K, Noguchi S, Fukuda K, Ishimoto H, Taniguchi H, Mukae H. 2016. The significance of oral streptococci in patients with pneumonia with risk factors for aspiration: the bacterial floral analysis of 16S ribosomal RNA gene using bronchoalveolar lavage fluid. BMC Pulm Med 16:79. doi: 10.1186/s12890-016-0235-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayanagi N, Kagiyama N, Ishiguro T, Tokunaga D, Sugita Y. 2010. Etiology and outcome of community-acquired lung abscess. Respiration 80:98–105. doi: 10.1159/000312404. [DOI] [PubMed] [Google Scholar]
- 24.Huffnagle GB, Dickson RP, Lukacs NW. 2017. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol 10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, Diaz P, Huang L, Wikoff WR, Ubeda C, Artacho A, Rom WN, Sterman DH, Collman RG, Blaser MJ, Weiden MD. 2016. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paddick JS, Brailsford SR, Kidd EA, Beighton D. 2005. Phenotypic and genotypic selection of microbiota surviving under dental restorations. Appl Environ Microbiol 71:2467–2472. doi: 10.1128/AEM.71.5.2467-2472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensäter G, Larsson UB, Greif EC, Cvitkovitch DG, Hamilton IR. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol Immunol 12:266–273. doi: 10.1111/j.1399-302X.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 28.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJ, Crielaard W, Zaura E. 2012. The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One 7:e42770. doi: 10.1371/journal.pone.0042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol 29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awano S, Ansai T, Takata Y, Soh I, Akifusa S, Hamasaki T, Yoshida A, Sonoki K, Fujisawa K, Takehara T. 2008. Oral health and mortality risk from pneumonia in the elderly. J Dent Res 87:334–339. doi: 10.1177/154405910808700418. [DOI] [PubMed] [Google Scholar]
- 32.Shweihat Y, Perry J III, Shah D. 2015. Isolated Candida infection of the lung. Respir Med Case Rep 16:18–19. doi: 10.1016/j.rmcr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams DW, Kuriyama T, Silva S, Malic S, Lewis MA. 2011. Candida biofilms and oral candidosis: treatment and prevention. Periodontol 2000 55:250–265. doi: 10.1111/j.1600-0757.2009.00338.x. [DOI] [PubMed] [Google Scholar]
- 34.Matsui M, Chosa N, Shimoyama Y, Minami K, Kimura S, Kishi M. 2014. Effects of tongue cleaning on bacterial flora in tongue coating and dental plaque: a crossover study. BMC Oral Health 14:4. doi: 10.1186/1472-6831-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bordas A, McNab R, Staples AM, Bowman J, Kanapka J, Bosma MP. 2008. Impact of different tongue cleaning methods on the bacterial load of the tongue dorsum. Arch Oral Biol 53(Suppl 1):S13–S18. doi: 10.1016/S0003-9969(08)70004-9. [DOI] [PubMed] [Google Scholar]
- 36.Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, Shimazaki Y, Akifusa S, Ninomiya T, Kiyohara Y, Yamashita Y. 2016. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci Rep 6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hata J, Ninomiya T, Hirakawa Y, Nagata M, Mukai N, Gotoh S, Fukuhara M, Ikeda F, Shikata K, Yoshida D, Yonemoto K, Kamouchi M, Kitazono T, Kiyohara Y. 2013. Secular trends in cardiovascular disease and its risk factors in Japanese: half-century data from the Hisayama Study (1961–2009). Circulation 128:1198–1205. doi: 10.1161/CIRCULATIONAHA.113.002424. [DOI] [PubMed] [Google Scholar]
- 38.Oho T, Yoshida Y, Shimazaki Y, Yamashita Y, Koga T. 2001. Characteristics of patients complaining of halitosis and the usefulness of gas chromatography for diagnosing halitosis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 91:531–534. doi: 10.1067/moe.2001.112543. [DOI] [PubMed] [Google Scholar]
- 39.Yanou Y. 2011. Components of oral malodor caused by aging and its origin. Shikoku Dent Res 24 (In Japanese.) [Google Scholar]
- 40.Takeshita T, Nakano Y, Yamashita Y. 2007. Improved accuracy in terminal restriction fragment length polymorphism phylogenetic analysis using a novel internal size standard definition. Oral Microbiol Immunol 22:419–428. doi: 10.1111/j.1399-302X.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 41.Takeshita T, Nakano Y, Kumagai T, Yasui M, Kamio N, Shibata Y, Shiota S, Yamashita Y. 2009. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J 3:65–78. doi: 10.1038/ismej.2008.91. [DOI] [PubMed] [Google Scholar]
- 42.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 44.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. 2010. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. 2013. Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 46.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A tongue coating sampling device based on the Braun Oral-B Pro 500 electric toothbrush (Procter & Gamble, Cincinnati, OH) (A). A circular bonded-fiber fabric (15 mm in diameter) was attached to this brush head, from which the bristles were preliminarily removed (B). The head was placed on the center of the tongue dorsum, 3-s vibrations were applied, and the collected tongue coating samples adhered to the fabric. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A principal-coordinate analysis plot showing similarity relationships among tongue microbiota samples from 506 community-dwelling elderly adults using an unweighted UniFrac distance metric. The points corresponding to the individuals with a higher relative abundance of each commensal group are depicted as bolder red. To correct for the unequal number of sequences, we evaluated 5,000 randomly selected sequences per sample. The two axes explained 12.9 and 9.7% of the variance, respectively. Download FIG S2, TIF file, 0.9 MB (945.3KB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Spearman’s correlation coefficient between the relative abundances of group I and II commensals and five predominant operational taxonomic units (OTUs) that were not present in both cohabiting groups. Download TABLE S1, DOCX file, 0.1 MB (67.3KB, docx) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A principal-coordinate analysis plot showing similarity relationship among tongue microbiota samples from 506 community-dwelling elderly adults using an unweighted UniFrac distance metric. The points corresponding to the individuals with different categories in each oral health status are depicted in different colors. To correct for the unequal number of sequences, we evaluated 5,000 randomly selected sequences per sample. The two axes explained 12.9 and 9.7% of the variance, respectively. Download FIG S3, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rarefaction curves for a number of observed operational taxonomic units (OTUs) per sample. The mean number of observed OTUs almost reached a plateau by 5,000 sequence reads. The error bars indicate the standard deviation. Download FIG S4, TIF file, 0.6 MB (643.5KB, tif) .
Copyright © 2018 Asakawa et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.