Significance
Finding the genes that control a complex trait is difficult because each gene may have only minor phenotypic effects. Quantitative trait loci mapping and genome-wide association study techniques have been developed for this purpose but are laborious and time-consuming. Here we developed a method combining pedigree analysis, whole-genome sequencing, and CRISPR-Cas9 technology. By sequencing the parents and descendants of IR8, the Green Revolution “miracle rice,” we identified many genes that had been retained in the pedigree by selection for high yield. Knockout and knockdown studies showed that a large proportion of the identified genes are essential or have phenotypic effects related to production. Our approach provides a powerful means for identifying genes involved in a complex trait.
Keywords: high-yield gene, pedigree analysis, Green Revolution, gene knockout
Abstract
Repeated artificial selection of a complex trait facilitates the identification of genes underlying the trait, especially if multiple selected descendant lines are available. Here we developed a pedigree-based approach to identify genes underlying the Green Revolution (GR) phenotype. From a pedigree analysis, we selected 30 cultivars including the “miracle rice” IR8, a GR landmark, its ancestors and descendants, and also other related cultivars for identifying high-yield genes. Through sequencing of these genomes, we identified 28 ancestral chromosomal blocks that were maintained in all the high-yield cultivars under study. In these blocks, we identified six genes of known function, including the GR gene sd1, and 123 loci with genes of unknown function. We randomly selected 57 genes from the 123 loci for knockout or knockdown studies and found that a high proportion of these genes are essential or have phenotypic effects related to rice production. Notably, knockout lines have significant changes in plant height (P < 0.003), a key GR trait, compared with wild-type lines. Some gene knockouts or knockdowns were especially interesting. For example, knockout of Os10g0555100, a putative glucosyltransferase gene, showed both reduced growth and altered panicle architecture. In addition, we found that in some retained chromosome blocks several GR-related genes were clustered, although they have unrelated sequences, suggesting clustering of genes with similar functions. In conclusion, we have identified many high-yield genes in rice. Our method provides a powerful means to identify genes associated with a specific trait.
Complex traits, which might be related to survival in natural environments or to crop productivity (1), are genetically difficult to dissect. This is in part because the effect of a single gene on a phenotype is usually small (2). To determine the genetic architecture of a complex trait (and the underlying gene networks), the most commonly employed methods are quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS). QTL mapping is suitable for relatively simple quantitative traits (3), while GWAS provides valuable insights into trait architecture or candidate loci (4). Both methods have limitations, however. In QTL, the effects detected may be sensitive to external environments (5), and the span of chromosomal regions detected is often too long [owing to limited recombination events (6)] to pinpoint the causative gene(s). Similarly, in GWAS, the effects detected are sensitive to population structure, leading to both false positives and false negatives (7, 8).
Recently, a pedigree from crosses between different founding genotypes was used to fine-map QTLs in Arabidopsis (1, 9). The pedigree-based analysis combines linkage and association study (6). A pedigree with a founding genotype (e.g., derived from a single cross of two ancestors) and with recombination events over many generations could overcome the disadvantages inherent in QTL and GWAS. To reduce the sensitivity to environmental effects, however, it is necessary to have a clear phenotypic difference between the two ancestors. Identification of chromosomal blocks preserved in all members of the pedigree under selection for a given trait will facilitate identification of candidate genes. The question, then, is whether these candidates are indeed associated with the trait. In principle, the CRISPR-cas9 system (10) can be used to knock out each candidate gene to get an insight into its function. Below we describe an application of this pedigree/knockout approach to the identification of high-yield genes in rice.
Our study takes advantage of the diploid rice pedigree in the Green Revolution. The Green Revolution has dramatically increased agriculture production worldwide since the 1960s, saving millions of lives from food shortage (11). The novel technologies allowed agronomists to breed high-yield varieties of maize, wheat, and rice. The yields were more than doubled in developing countries from 1961 to 1985 (12). Perhaps the most significant milestone of the Green Revolution was the introduction of semidwarfing genes into selected rice cultivars by hybridization.
The first semidwarf and high-yield modern rice variety of the Green Revolution, known as the “miracle rice” IR8, was created by crossing the Indonesian variety “Peta” with the Chinese variety “Dee-geo-woo-gen” (DGWG). It represented the first generation of the high-yielding plant type which provided a significantly higher yield potential for irrigated rice (13). In addition to the significant reduction in stem length, the high-yield rice cultivars have other important traits such as an early flowering time, improvement in photosynthetic allocation, and insensitivity to day length, directly or indirectly influencing the grain yield and yield stability (14, 15). These high-yield traits could be traced from the pedigree of miracle rice IR8 that consists of its parents and high-yield progenies.
We assume that the genes related to high yield were under strong artificial selection because yield was the major target trait of rice breeding since the 1960s. In this scenario, we note (i) if the multiple lineages descended from an original cross have all been placed under the same selection, the alleles responsible for the trait in question should be found in all the descendants but not in all control populations; (ii) in principle, these alleles can be traced back to their origination, and any variants inherited in all generations can be identified; (iii) a gene under strong artificial selection should be more commonly present in progeny than genes not under selection; and (iv) when knocking out a high-yield gene, a changed plant phenotype (e.g., an observable change in morphology or a physiological response such as sterility) should be observed. All these expectations can be tested by sequencing the cultivars at important nodes in the pedigree and then by a knockout study using the CRISPR-Cas9 system.
Using the strategy described above, we studied the extended pedigree of the ancestors and descendants of IR8 and other related lines (Fig. 1A) to determine a set of genes that played a critical role in the rice Green Revolution. By resequencing 30 cultivars from the pedigree (Fig. 1), we identified 28 chromosomal blocks, including 129 candidate gene loci, that have been preserved by artificial selection (Fig. 2). Fifty-seven gene loci with unknown function were selected for knockout using the CRISPR-Cas9 technique. If the knockout failed, then a knockdown experiment was conducted. We found that 79% of the knocked out loci (15/19) and 62% of the knocked down loci (10/16) have phenotypic changes. These studies revealed a striking enrichment compared with control in yield/morphology-associated genes among the candidate genes. Thus, our pedigree-guided approach provides a simple, robust, and fast means to identify candidate genes under directional selection.
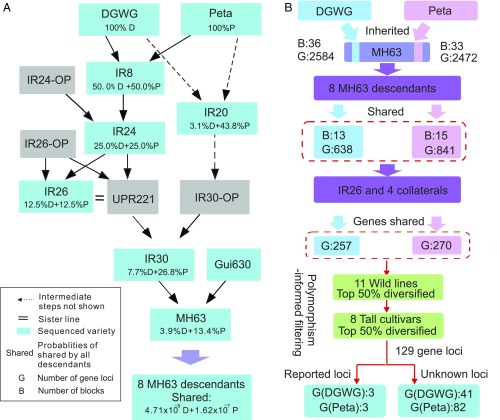
Fig. 1.
Pedigree and flowchart for the identification of gene loci under selection. (A) An abridged pedigree of the major rice cultivars used in this study. The cultivars shown in blue boxes were resequenced; cultivars shown in gray boxes were not. “OP” means “the other parent”; cultivars so identified were not sequenced. Percentages in boxes show the expected probability of a given locus being inherited from DGWG (D) or Peta (P) in that generation. The bottom box indicates the expected probabilities that a locus is shared by all eight MH63 descendants, which are extremely low (SI Appendix, Table S4). A solid arrow denotes a direct parent (i.e., IR24), and a dashed arrow indicates an indirect ancestor (i.e., IR20). (B) Flowchart of the approach used to identify candidate blocks and gene loci derived from DGWG or Peta. Numbers of blocks (B) and gene loci (G) within the high-confidence blocks are shown in each step of filtering. The six reported genes (three from DGWG and three from Peta) are the gene loci that have clear functions reported in literature. Most of the 129 gene loci contain only one gene, but 28 loci contain two or more overlapped genes (Methods).
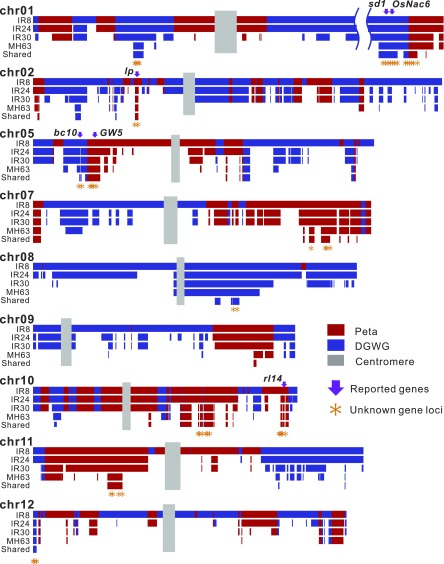
Fig. 2.
Blocks inherited from DGWG and Peta in IR8, IR24, IR30, MH63, and the eight descendants of MH63. Blue and red bars represent blocks derived from DGWG and Peta, respectively. “Shared” denotes the regions shared in all eight MH63 descendants. The purple arrows represent the six genes reported with functions related to plant type or high yield; asterisks represent the 123 gene loci with unknown functions; the six genes are shared by all eight MH63 descendants and five collateral series. Chromosomes 3, 4, and 6, which contain no regions shared by all eight MH63 descendants, are shown in SI Appendix, Fig. S7. The next-to-last block on chromosome 1 was shortened using breaks.
Results
Rice Cultivar Selection and SNP Identification.
The famous miracle rice IR8 is the key cultivar in our pedigree analysis (Fig. 1A). Its descendants and derivatives have been extensively used in the field, and its parents have been widely utilized to breed desired plant types (16). Another key cultivar is Minghui63 (MH63), which is a fourth-generation descendant of IR8 and was the restorer line for a number of rice hybrids. MH63 accounted for >20% of the total production area in China during the 1980s and 1990s (17). Because of its wide planting areas with a stable high yield, environmental or epigenetic effects could be excluded. IR8 and MH63 form the basis of our pedigree analysis. The pedigree further expands upward to the parents of IR8 (i.e., DGWG and Peta) and MH63 (IR30 and Gui630) and downward to the descendants of IR8 (i.e., IR24) and MH63. IR20, which has the same parents as IR8, and eight extensively used descendants of MH63 are also included in the analysis (Fig. 1A). All descendants of IR8 possessed the common feature of high yield. To enhance the resolution in identifying genes under selection, we also sequenced four IR8 collateral series, eight tall landraces, and a wild rice as the controls (SI Appendix, Fig. S1B and Table S1). The alleles present in the control groups were considered unlikely to contribute to high yield.
The 30 diploid rice accessions selected above were resequenced with a reasonable coverage depth (>20×) in our study (SI Appendix, Fig. S2 and Table S1). Because pedigree information and independent resequencing of descendants from the same ancestor offer the unique advantage of discriminating against false markers, each inherited block of interest can be double-checked not only between successive generations but also between nodes independently by more than one generation and between lineages. Based on the linked markers in the majority of the successive generations, this approach can exclude false markers, infer correct SNPs, and improve the accuracy of SNP identification (SI Appendix, Fig. S3). In the two most important parent–offspring trios, DGWG–Peta–IR8 and IR30–Gui630–MH63, a total of 592,603 and 481,385 high-quality SNPs, respectively, were called to detect the chromosomal blocks inherited from IR8 and its parents (Methods and SI Appendix, Fig. S4).
Expected and Observed Proportions of Inherited Blocks.
With the pedigree information, the probability of a block or a gene being passed on to the next generation can be computed using classical genetic theory. One can then compare the computed probability with the observed proportion (SI Appendix, SI Materials and Methods). In the absence of selection, the probabilities of a gene locus in MH63 from DGWG and Peta are expected to be 3.9% and 13.4%, respectively (Fig. 1A and SI Appendix, Table S2). The probability of one or more DGWG or Peta blocks being present in all eight descendants of MH63 is extremely low (4.71 × 10−8 or 1.62 × 10−7) (Fig. 1A, Table 1, and SI Appendix, Tables S3 and S4). Therefore, every block retained in all the MH63 progenies is likely to have been targeted by artificial selection for the high-yield phenotype.
Table 1.
Numbers of blocks derived from DGWG and Peta in different descendants
| Ancestor | Descendant | Chromosome | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Total | ||
| DGWG | IR8 | 4 | 15 | 17 | 12 | 7 | 11 | 4 | 3 | 3 | 13 | 6 | 13 | 108 |
| IR24 | 4 | 12 | 9 | 8 | 6 | 11 | 4 | 3 | 3 | 13 | 3 | 9 | 85 | |
| IR30 | 4 | 11 | 7 | 7 | 6 | 7 | 4 | 2 | 3 | 12 | 2 | 8 | 73 | |
| MH63 | 2 | 5 | 3 | 2 | 4 | 2 | 2 | 1 | 3 | 7 | 2 | 3 | 36 | |
| Shared* | 2 | 2 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 3 | 1 | 1 | 13 | |
| Genes† | 442 | 75 | 0 | 0 | 136 | 0 | 0 | 64 | 0 | 28 | 6 | 34 | 785 | |
| Peta | IR8 | 4 | 15 | 17 | 12 | 6 | 11 | 5 | 2 | 2 | 12 | 5 | 12 | 103 |
| IR24 | 4 | 14 | 11 | 6 | 4 | 6 | 5 | 1 | 2 | 10 | 3 | 10 | 76 | |
| IR30 | 4 | 12 | 10 | 6 | 4 | 2 | 5 | 0 | 2 | 10 | 3 | 8 | 66 | |
| MH63 | 1 | 5 | 7 | 1 | 3 | 0 | 3 | 0 | 1 | 6 | 1 | 5 | 33 | |
| Shared* | 1 | 2 | 0 | 0 | 3 | 0 | 2 | 0 | 1 | 5 | 1 | 0 | 15 | |
| Genes† | 101 | 34 | 0 | 0 | 115 | 0 | 265 | 0 | 42 | 308 | 95 | 0 | 960 | |
Shared represents the blocks and enclosed genes observed in MH63 and in all its eight descendants.
Genes contained in the shared blocks.
Theoretically, the heterozygosity of the F1 hybrid will be reduced to half in its F2 progeny through selfing and will eventually be reduced to almost zero in an inbred line (e.g., IR8 or MH63). Therefore, the crossover events can be detected in both IR8 and MH63 to determine the origin of each block (SI Appendix, Tables S5 and S6). The block information in MH63 enabled us to exclude the genetic blocks from Gui630 and identify those from DGWG or Peta based on the pedigree in Fig. 1A. In MH63, we found 57 and 59 blocks that were derived from 36 DGWG and 33 Peta blocks in IR8, respectively (Fig. 2). Thus, many of the original blocks inherited from DGWG and Peta had been fragmented into smaller ones in MH63 by recombination. The average length is 483 kb for the 57 DGWG-derived blocks and 398 kb for the 59 Peta-derived blocks, which are 5.45- and 3.20-fold shorter, respectively, than the average lengths of the original blocks in IR8 (SI Appendix, Tables S7 and S8). Among those original blocks, only a total of 6.26 Mb DGWG and a total of 8.76 Mb Peta segments are inherited in all eight MH63 descendants. They were 2.39- and 1.55-fold shorter, respectively, than the inherited blocks observed in MH63. The sequences shared by all eight MH63 descendants contained 785 DGWG- and 960 Peta-specific genes (Figs. 1B and 2).
Identification of Candidate Genes for the High-Yield Phenotype.
When only a limited number of genes in a block are under selection, the ancestral block will become shorter and shorter over generations because of recombination events. Fig. 2 includes an example in which a block on Peta chromosome 5 became shorter and shorter by crossover events from IR8 to MH63. Interestingly, a candidate gene, GW5, which is responsible for rice grain width, shape, quality, and yield, is located near recombination hotspots (18) but has been retained. The pattern displays efficient selection on this block.
In a block with many genes, some alleles that are not subjected to selection may be inherited due to linkage (i.e., hitchhiking). Several strategies were employed to exclude the hitchhiked genes and identify the genes that were most likely the target of selection, including those with unannotated functions (Fig. 1B). The π (polymorphic sites/informative sites) was calculated for each 10-kb window to compare the diversity values within and between different groups. First, we assumed that targeted alleles should also have been retained in the IR8 collateral series because those cultivars are also of high-yield plant types. With this assumption, we selected four cultivars of the IR8 collateral series (SI Appendix, Fig. S1 and Table S1) and calculated the nucleotide diversity of these candidate genes between MH63 and each of the four collateral cultivars together with IR26, a progeny of IR24 and a sister line of UPR221 (a parent of IR30 in Fig. 1A). Only the genes that had an average diversity <0.0001 and in which the compared pairs were identical in the majority (three or more) of the collateral series were kept. Second, we assumed that a gene with an extremely low diversity among wild rice lines and cultivars should be excluded because it is more likely to be essential for fundamental biological processes rather than being responsible for the high-yield phenotype. Therefore, we further filtered out the bottom 50% of genes in terms of the diversity between MH63 and the 11 wild rice varieties. Third, we filtered further by comparison with tall cultivars as follows. All the resequenced cultivars in this study were grown in the field, and their heights were measured. Because the semidwarfism trait was specifically selected for the Green Revolution, we expect that the alleles related to the Green Revolution would be divergent with tall cultivars and would be kept only in the genes showing a diversity higher than the median between MH63 and each of the eight tall cultivars (SI Appendix, Tables S1 and S9).
The above filtering procedure identified 129 gene loci, which can be divided into 101 single loci and 28 loci with overlapping genes (i.e., loci in which two or more genes overlap completely or partly within the same locus). As an example of overlapping genes, the coding sequence of Os01g0883850 is completely contained in the reported gene sd1 (Os01g0883800). These two genes are thus considered as a single entity in our analysis. Each locus is named by one gene it contains. Of the 129 gene loci, 44 are from DGWG- and 85 are from Peta-specific blocks (Fig. 1B and SI Appendix, Table S10). These 129 gene loci are located on 17 blocks which are inherited in all eight descendants of MH63. Six of the 129 gene loci contain genes with known functions, including the semidwarf gene sd1, known as the “green revolution gene.” This gene encodes gibberellin 20-oxidase, the key enzyme in the gibberellin biosynthesis pathway. Another gene, larger panicle (lp), which controls the panicle architecture (19), has recently been found to be a target of selection in Indica cultivars by a GWAS study of 1,479 rice accessions (20). The others are GW5, BC10, RL14, and OsNAC6, responsible for grain width, brittle culm, leaf rolling, and stress tolerance, respectively (18, 21–24). Interestingly, three of these six genes were identified from natural mutants, whereas most functional genes commonly were identified from transfer DNA insertion and mutagen-induced mutants (accounting for roughly 90% of reported genes with a known function). This suggests that the genes identified from a pedigree analysis could reflect the real targets of selection in plant breeding better than the genes identified from artificial mutants.
Knockout Phenotypes of Candidate Gene Loci.
To determine whether a gene locus with unknown function has a phenotypic effect when knocked out, 57 of the 123 loci with unknown function were randomly sampled for knockout by the CRISPR-cas9 system. Of these 57 loci, 19 had knockout mutants, which were confirmed by PCR and Sanger sequencing. However, in the other 38 gene loci, no knockout mutants were obtained even after at least two independent transformations. We suspected that many of these genes are essential in callus development, so that no transformant survived. This possibility is supported by the observation that most of these genes (91.2%) had medium or high expression levels in callus (SI Appendix, Table S11).
As positive controls, we also attempted to knock out the six genes with known functions. As expected, five knockout mutants exhibited phenotypic changes similar to or stronger than the changes reported in previous studies (SI Appendix, Table S12) (18, 19, 21, 23, 24). However, one, RL14, had no knockout mutant (SI Appendix, Table S12). In a previous report, rl14, which carries a single amino acid mutation, exhibited severe leaf rolling; therefore RL14 may have essential functions so that its knockout could not survive (22). In addition, as random controls, 10 genes were randomly sampled from the 1-kb to 300-kb regions (SI Appendix, Table S13) adjacent to the retained ancestor blocks (which were shared by all eight descendants of MH63). The near-neighbor controls may be considered as conservative random controls because, unlike true random controls, these controls in part allow possibly important position effects, such as the clustering of genes with similar expression profiles (25). In all 10 cases the knockout mutant showed no phenotypic changes (SI Appendix, Table S14), in contrast to 79% (15/19) of the unknown gene loci that showed observable phenotypic changes when the gene was knocked out (Table 2; detailed changes in phenotypes and genotypes are given in SI Appendix, Table S15).
Table 2.
Phenotype when a specific gene was knocked out
| Sampled ancestral block | Locus | Observed phenotypes |
| DGWG chr01:37602014–39226171 | Os01g0884200 | Dwarf, sterile |
| Os01g0884400* | Late heading, sterile | |
| Os01g0884450 | ||
| Os01g0885000 | Small, growth retarded, fewer tillers | |
| Os01g0886000 | Late heading, fewer tillers, sterile | |
| Peta chr01:40248759–40971796 | Os01g0925600* | Rolling leaves, shorter panicle, dwarf |
| Os01g0925700 | ||
| Os01g0930800 | Late heading, sterile | |
| Os01g0930900 | No phenotypic change | |
| Peta chr10:21769689–21922126 | Os10g0555600* | Dwarf |
| Os10g0555651 | ||
| Os10g0555900* | Dwarf, late heading | |
| Os10g0556000 | ||
| Os10g0556200 | Dwarf | |
| Os10g0556900 | No phenotypic change | |
| Os10g0555100 | Dwarf, spike shape change | |
| Os10g0555200 | Dwarf, sterile | |
| Os10g0555300 | Dwarf, sterile | |
| Os10g0555700 | Sterile | |
| Os10g0556100 | Small, growth retarded, leaf rolling | |
| Peta chr10:21992900–22072751 | Os10g0558850 | Rolling leaves, dwarf, weak |
| Os10g0559800* | No phenotypic change | |
| Peta chr11:6540176–7824094 | Os10g0559833 | |
| Os11g0242400 | No phenotypic change |
The 123 gene loci that passed our filtration came from 16 blocks, which ranged in size from 43 to 1,624 kb. In total, 19 gene loci from five blocks of different sizes (80 kb–1,624 kb) were successfully knocked out. For each gene, about 15 independently transgenic plants were obtained, and on average, in 79.5% of the cases, the gene was knocked out in both homologous chromosomes. The phenotypic change was based on the observation of the homozygous knockout plants. “No phenotypic change” means no significant change in phenotype; e.g., the knockout of the locus Os01g0930900 showed shorter plants and shorter awns, but the changes were not statistically significant. In total, 15 of the 19 knockouts exhibited phenotypes different from the WT, suggesting that a large portion of these unknown-function gene loci are involved in flowering, fertility, leaf morphology, and so forth. The genotype and phenotype of each gene studied are described in SI Appendix, Table S15. All the knockout plants in this table were in the Kasalath background.
In five pairs, the two genes in a pair overlapped partly or completely. For example, Os01g0884450 is completely contained in Os01g0884400.
High-yield plants are typically dwarf, because dwarfism reduces the investment in stalk, thereby potentially increasing investment into seeds. Therefore, we studied the growth difference between the mutated and unmutated versions. We compared plant heights in knockout and WT lines by the paired t test (Table 3). As expected, the random control genes showed no difference in height between mutant and nonmutant versions (P = 0.42, 95% CI: −1.15 to 2.54 cm), while the positive controls showed a significantly shorter height in mutants than in WT plants (P = 0.017, CI: −99.6 to −20.84 cm). Importantly, for the test group we also saw a strong dwarfism phenotype (P = 0.0013; 95% CI: −31.98 to −9.22 cm). As these were random samples from the 123 unknown gene loci, it implies that a high proportion of the 123 loci have a phenotype similar to that of the well-described positive control genes identified by the same method. However, the extent of the dwarfism is reduced in the test sample compared with the positive controls (t test on percentage difference comparing positive control and test samples, P = 0.029, 95% CI: −67.82 to 5.48). These genes may have weaker effects than the previously reported ones, and this may be why they have not been identified.
Table 3.
Comparison of plant height in Kasalath knockout mutants and WT plants
| Locus | Mutant height, cm | WT height, cm | % height change: (mutant height − WT height)/WT height | |
| Four positive controls* | Os01g0883800 | 62.8 | 132.4 | −52.6 |
| Os01g0884300 | 65.5 | 129.3 | −49.3 | |
| Os05g0170000 | 47.8 | 130.3 | −63.4 | |
| Os02g0260200 | 98.3 | 123.3 | −20.3 | |
| 18 target gene loci† | Os01g0884200 | 110.7 | 129.3 | −14.40 |
| Os01g0884400 | 125.3 | 127.3 | −1.6 | |
| Os01g0884450 | ||||
| Os01g0886000 | 127.7 | 129.6 | −1.5 | |
| Os01g0925600 | 93.3 | 125.6 | −25.7 | |
| Os01g0925700 | ||||
| Os01g0930800 | 125.3 | 131.2 | −4.5 | |
| Os01g0930900 | 130.2 | 129.1 | 0.9 | |
| Os10g0555100 | 99.6 | 130.1 | −23.4 | |
| Os10g0555200 | 99.7 | 130.3 | −23.5 | |
| Os10g0555300 | 105.2 | 129.2 | −18.6 | |
| Os10g0555600 | 95.2 | 130.2 | −26.9 | |
| Os10g0555651 | ||||
| Os10g0555700 | 127.1 | 130.6 | −2.7 | |
| Os10g0555900 | 64.6 | 132.4 | −51.2 | |
| Os10g0556000 | ||||
| Os10g0556100 | 65.2 | 127.3 | −48.8 | |
| Os10g0556200 | 73.7 | 122.1 | −39.6 | |
| Os10g0556900 | 131.2 | 129.1 | 1.6 | |
| Os10g0558850 | 117.2 | 130.2 | −10.0 | |
| Os10g0559800 | 130.1 | 129.3 | 0.6 | |
| Os10g0559833 | ||||
| Os11g0242400 | 129.4 | 128.6 | 0.6 | |
| 10 random controls‡ | Os01g0936100 | 130.3 | 131.5 | −0.9 |
| Os05g0375600 | 134.0 | 132.4 | 1.2 | |
| Os05g0571700 | 126.5 | 129.2 | −2.1 | |
| Os05g0573600 | 132.0 | 130.1 | 1.5 | |
| Os10g0341750 | 134.0 | 130.1 | 3.0 | |
| Os10g0342300 | 132.0 | 129.2 | 2.2 | |
| Os10g0341700 | 133.0 | 130.2 | 2.2 | |
| Os05g0571300 | 134.3 | 132.4 | 1.5 | |
| Os10g0558400 | 128.5 | 132.4 | −2.9 | |
| Os10g0342650 | 131.3 | 131.5 | −0.1 |
On average, positive controls showed a 46.4% reduction in plant height (P = 0.017, two-tailed t test, 95% CI: −99.6 to 0.84), while 18 target gene loci showed a 16.4% reduction in plant height (P = 0.0013, two-tailed t test, 95% CI: −31.98 to 9.22). Ten random controls only showed a slight difference (knockout effect) (average 0.5%, P = 0.42, two-tailed t test, 95% CI: −1.16 to 2.54).
Four of the six positive controls were knocked out in Kasalath. GW5 was knocked out in Wuyungeng, and rl14 was not successfully knocked out.
The knockout plant Os01g0885000 died before the tilling stage, and the plant height could not be compared with the others. Therefore, only 18 target mutants were measured.
Ten genes adjacent to the target blocks were randomly chosen as controls.
A gene of particular interest is Os10g0555100, as its knockout showed a different panicle architecture and a 23% reduction in height. Note that one of the reported genes, lp, showed an altered panicle architecture as well. The protein product may be a glycogenin glucosyltransferase (see ic4r.org), suggesting a possible role in controlling free glucose and glucose storage. However, this speculation requires further analysis. Among the other genes some, such as Os10g0558850, had rolled leaves (Fig. 3) but a relatively modest (∼10%) reduction in plant height. All the 15 unknown gene loci with knockout phenotypes have various protein-level motifs with unknown function, suggesting that the plant type and the high-yield phenotype are controlled by many types of genes.
Fig. 3.
Photographs of knockout mutants with changed phenotypes compared with WT Kasalath. These six examples show shorter plants (A and B), rolling leaves (C), a later heading date (D), changed panicles (E), and empty seeds (F) in the mutants compared with WT plants. The other nine knockout mutants with observable phenotypic changes and the controls are shown in SI Appendix, Fig. S8.
Interestingly, the physically proximal gene loci, although showing no sequence similarity, have similar functions. For example, knockouts of three of the six gene loci on chromosome 1 (Os01g0884400–Os01g0930900) resulted in late heading, and knockouts of 6 of the 11 loci on chromosome 10 (Table 2 and SI Appendix, Table S15) resulted in dwarf phenotypes relative to the background line. This clustering mirrors the previously observed clustering of QTL signals (26). The clustering may reflect selection for coordinated gene expression or may possibly be the result of epistatic effects. Importantly, this result also suggests a strategy for finding genes with similar functions: If you have found one, investigate its neighbors.
Knockdown Phenotypes of Gene Loci with No Knockout Transformant.
To investigate the 38 loci with no knockout mutants, we randomly selected 26 loci to knock down their expression level, using the dCas9 knockdown technique (27). Similar to the knockout results, in 10 of the 26 loci (38.5%) no knockdown mutants were obtained due to the death of the transformed callus after hygromycin selection. Most of the 26 loci also have medium or higher expression levels in callus (SI Appendix, Table S16). Moreover, even in the 16 loci with knockdown transgenic plants, 10 of the knockdown plants showed distinct negative phenotypic changes, and seven died during plant regeneration (Table 4 and SI Appendix, Fig. S5). As expected, the qRT-PCR study confirmed that the expression of these target loci in knockdown transformants was indeed down-regulated (SI Appendix, Fig. S6). These results suggest that most of the 38 candidate genes are essential genes in rice.
Table 4.
Phenotypic changes in knockdown mutants
| Locus | Abnormal phenotypes |
| Os01g0883900 | Curled leaves, retarded growth; died before maturity |
| Os01g0931600 | Retarded growth, multiple tillers |
| Os05g0170200 | Retarded growth, curled leaves |
| Os10g0556500 | Brown and curled leaves; died before maturity |
| Os10g0556700 | Normal |
| Os10g0559866 | Normal |
| Os02g0258900 | Retarded growth, brown and curled leaves |
| Os10g0391100 | Normal |
| Os10g0391200 | Normal |
| Os10g0392400 | Curled leaves; died before maturity |
| Os10g0554900 | Normal |
| Os12g0103000 | Brown leaves; died before maturity |
| Os12g0104250 | Normal |
| Os12g0104400 | Brown leaves; died before maturity |
| Os12g0104700 | Retarded growth, curled leaves; died before matuity |
| Os12g0104733* | Grew only roots, no seedling |
| Os12g0104766* | Curled leaves |
These genes are included in the same locus.
Discussion
Determining the genes that explain complex traits has never been easy. The two much used methods, QTL and GWAS, have both led to important discoveries, but such analyses are typically very labor intensive. Indeed, during the past decades much effort has gone into dissecting the genetic basis of high-yielding traits based on molecular linkage maps, e.g., the identification of many QTLs (28–31), but relatively few genes have been identified. The pedigree-based method that we expanded here has, in some cases, reduced much of the effort. It requires a good pedigree and consistent directional selection, however. Until recently, the confirmation of such results also would have been very time consuming, but CRISPR-Cas9 can greatly reduce the amount of work required. In this study, we not only have identified the three well-known loci for the Green Revolution (the green revolution gene sd1, the grain size-related gene GW5, and the domestication gene lp) but also have identified more than 100 candidates. Among the 57 candidate genes selected for knockout and knockdown studies, we found that many are essential genes or showed phenotypic effects. Thus, the pedigree approach seems to be highly efficient in identifying candidate genes that were subject to strong selection.
While the knockout analysis suggested a low false-positive rate, the false-negative rate is unknown and probably is quite high, as our filters are quite stringent. Indeed, when we look at two genes that failed to pass the diversity cutoff, we find that one of them resulted in phenotypic change when knocked out. This suggests that slight relaxation of the stringent filtering will result in more candidates but potentially in a higher false-positive rate as well. More generally, we do not know how many genes are essential for the rice Green Revolution. As a consequence, the method should be considered a technique for greatly enriching relevant genes selectively rather than a method for an exhaustive search for relevant genes.
This study showed that rice is unusually well suited to this pedigree method. First, the well-documented pedigree information can be used to calculate the expected proportions of blocks (or loci) being transmitted from an ancestor to a descendant (e.g., Fig. 1A). By comparing the expected and observed proportions, the gene loci that were most likely to have been the target of artificial selection could be identified. For example, the probability of a DGWG block appearing in all eight MH63 descendants was estimated to be nearly zero. Thus, if a block is observed in the resequencing data, it was very likely subjected to strong artificial selection. Second, from the relationships in a pedigree, SNP markers can be verified and corrected by comparing the sequences of parents and offspring between generations (demonstrated in Fig. 2 and SI Appendix, Fig. S1). In rice we are fortunate to have access to the stocks of the prior generations. Third, pedigree analysis focuses on tracing relatively longer blocks from the parents to the offspring instead of single SNPs or genes. Therefore it is not difficult to identify selected targets. Finally, the CRISPR-Cas9 system provides an effective way of gene knockout to find a set of genes relevant to complex traits. In conclusion, our approach should be useful for many breeding projects.
Our choice of our model organism was motivated not only by its meeting the conditions for pedigree analysis but also by the enormous impact of the Green Revolution, as indicated by the generation of high-yielding plant types through breeding. The introduction of dwarfing genes has resulted in plants that possess short, strong stalks, which are less liable to lodging. The stability of shorter plants dramatically reduces the need for photosynthetic investment in the stem. Assimilates are then redirected to grain production, resulting in a better plant type and increased yield (32). The candidate genes identified in this study will be useful for understanding the underlying mechanism of this physiology.
Importantly, then, we have identified many genes responsible for high yield, an economically most important trait. Most of these gene loci have not yet been functionally annotated, although a few belong to the β-expansin family or contain a zinc finger domain, which are known to play an important role in plant height, flower development, and light-regulated morphogenesis (33–35). We highlighted Os10g0555100, the knockout of which showed a different panicle architecture and a 23% reduction in height. We also note that our results suggest that the genes identified from cultivated lines in a pedigree could reflect the real targets in plant breeding better than the genes identified from artificial mutants. Our catalog of 123 unannotated gene loci provides choices for downstream analysis. Our knockout and knockdown study of about half of these loci revealed that most of the genes in these loci are essential for rice phenotypes or for normal growth. Among the 159 genes we identified, at least 31 are yield-related genes, including 15 identified by knockout, 10 by knockdown, and six previously reported. This proportion (19.5%) is significantly higher than the expectation (2.33 in 159 = 1.5%) based on the reported yield-related genes in the rice genome (P < 0.001, χ2 = 334, df = 1, χ2 test with Yate’s correction; gene information is from Q-TARO, qtaro.abr.affrc.go.jp; see details in SI Appendix, SI Materials and Methods). However, the alleles contributing to the Green Revolution are not necessarily null alleles, so our knockout and knockdown studies did not directly test the contribution of allelic changes to the Green Revolution. Gene replacements in IR8 or MH63 would directly reveal the contributions of the alleles, but IR8 and MH63 are difficult to transform, and gene replacement is currently difficult in rice.
Our results also highlight the clustering of unrelated genes with similar yield-associated phenotypes in the genome. This observation is of relevance for those hunting for complex trait genes and for those interested in genome evolution. For the former, it suggests that looking at neighbors of functionally relevant genes would be an effective way to look for functionally related genes. The clustering may reflect epistasis between genes or selection for coexpression. Previous QTL analysis also suggested that genes of similar phenotypic effects tend to cluster together (26), but this could also reflect allelic versions of control elements for a single gene. The fact that the knockouts of the clustered genes tend to have similar phenotypes suggests allelic versions of control elements is not the case.
Methods
Detailed materials and methods are outlined in SI Appendix, SI Materials and Methods.
Plant Materials and Sequencing.
The seeds of all rice accessions were obtained from the International Rice Research Institute (IRRI) and China National Rice Research Institute (CNRRI) (Dataset S1). Pedigree information was obtained from the germplasm databases of the IRRI and CNRRI. All rice varieties were grown in the paddy field. DNA samples were prepared from fresh leaves of a single plant using the cetyltrimethylammonium bromide (CTAB) method and were sequenced at Beijing Genomics Institute (BGI)-Shenzhen. Briefly, paired-end sequencing libraries with an insert size of ∼500 bp were constructed for each plant, following the BGI-Shenzhen’s instructions, and 100-bp paired-end reads were generated on an Illumina HiSeq 2000 sequencer. The sequencing reads of the 30 rice accessions have been deposited in the National Center for Biotechnical Information (NCBI) Sequence Read Archive under accession numbers PRJNA271253 and SRR1060330. Indica cultivar 9311 callus RNA-sequencing data were downloaded from NCBI BioProject PRJNA117345, SRR037711–SRR037724.
Construction of CRISPR Genome-Editing Vectors and Knockout of Target Gene Loci.
For each target locus, gRNAs were designed to target specific sites at the beginning of exons to cause a frame shift mutation. For each target, a pair of DNA oligonucleotides with appropriate cloning linkers were synthesized (BGI, Inc.). Each pair of oligonucleotides was phosphorylated, annealed, and then ligated into BsaI-digested pRGEB31 vectors (Addgene no. 7722) (36). After transformation into Escherichia coli DH5-alpha, the resulting constructs were purified with the Plasmid Mini kit (Genebase, Inc.) for subsequent use in rice callus transformation. We selected the Kasalath and Wuyungeng24 varities to be the background because they have a high transformation success rate, while IR8 and MH63 are difficult to transform. Besides, Kasalath has a rather high stature, and it is easy to observe when it becomes dwarf. Each construct was transformed into calli of Kasalath (an Indica) or Wuyugeng24 (a Japonica) by the method reported in a previous study (37). About 10 transformed individuals were produced in two recipients for each vector (details are given in SI Appendix, Tables S13–S15).
Genotype Confirmation and Phenotype Observation.
The transgenic plants were examined under natural field conditions in the Nanjing University Experimental Station, Nanjing, China. For each plant, genomic DNA was extracted from fresh leaves by the CTAB method. To get double-knockout mutants, we amplified the target region by PCR and confirmed the genotypes by Sanger sequencing. Primers were designed to make PCR products of ∼1 kb that contain the target sites. The results showed that 82.1% of transgenic plants had a knockout allele, and 79.5% had double-knockout mutants. Phenotypes of the mutants were observed at different stages. Plant phenotypes were observed every 3 d to determine the changes in comparison with WT rice plants. Plant height was measured after the heading stage. Fertility and spike shape were observed when seeds were mature.
Supplementary Material
Acknowledgments
We thank the National Mid-term Genebank for Rice of the China National Rice Research Institute and the International Rice Research Institute for providing the rice germplasm collection. This work was supported by the National Major Special Project on New Varieties Cultivation for Transgenic Organisms Grant 2016ZX08009001-003 and National Natural Science Foundation of China Grants 91731308 and 31571267 (to D.T.). L.D.H. is funded by European Research Council Advanced Grant EvoGenMed 669207—EvoGenMed—ERC-2014-ADG/ERC-2014-ADG.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing reads reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession nos. PRJNA271253 and SRR1060330).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806110115/-/DCSupplemental.
References
- 1.Mitchell-Olds T. Complex-trait analysis in plants. Genome Biol. 2010;11:113. doi: 10.1186/gb-2010-11-4-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasaki T, Moore G, editors. Oryza: From Molecule to Plant. Springer; Dordrecht, The Netherlands: 1997. [Google Scholar]
- 3.Zhu M, Zhao S. Candidate gene identification approach: Progress and challenges. Int J Biol Sci. 2007;3:420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackay TFC. The genetic architecture of quantitative traits. Annu Rev Genet. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- 6.Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011;12:232. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kover PX, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 11.Hazell PBR. The Asian Green Revolution. International Food Policy Research Institute; Washington, DC: 2009. [Google Scholar]
- 12.Conway G. The Doubly Green Revolution: Food for All in the Twenty-First Century. Comstock Publishing Associates, Cornell Univ Press; Ithaca, NY: 1998. [Google Scholar]
- 13.Peng S, Khush GS, Virk P, Tang Q, Zou Y. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 2008;108:32–38. [Google Scholar]
- 14.Khush GS. Green revolution: Preparing for the 21st century. Genome. 1999;42:646–655. [PubMed] [Google Scholar]
- 15.Springer N. Shaping a better rice plant. Nat Genet. 2010;42:475–476. doi: 10.1038/ng0610-475. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki A, et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J. 2005;42:772–780. doi: 10.1111/j.1365-313X.2005.02408.x. [DOI] [PubMed] [Google Scholar]
- 18.Wan X, et al. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics. 2008;179:2239–2252. doi: 10.1534/genetics.108.089862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J. 2011;9:1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x. [DOI] [PubMed] [Google Scholar]
- 20.Xie W, et al. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc Natl Acad Sci USA. 2015;112:E5411–E5419. doi: 10.1073/pnas.1515919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monna L, et al. Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- 22.Fang L, et al. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol J. 2012;10:524–532. doi: 10.1111/j.1467-7652.2012.00679.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, et al. BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.) Plant J. 2009;57:446–462. doi: 10.1111/j.1365-313X.2008.03703.x. [DOI] [PubMed] [Google Scholar]
- 24.Nakashima K, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 25.Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 26.Cai W, Morishima H. QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet. 2002;104:1217–1228. doi: 10.1007/s00122-001-0819-7. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez-Vilar M, et al. A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods. 2016;12:10. doi: 10.1186/s13007-016-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JX, et al. Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor Appl Genet. 2000;101:248–254. [Google Scholar]
- 29.Xing Z, et al. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet. 2002;105:248–257. doi: 10.1007/s00122-002-0952-y. [DOI] [PubMed] [Google Scholar]
- 30.Yu SB, et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, et al. Gene discovery using mutagen-induced polymorphisms and deep sequencing: Application to plant disease resistance. Genetics. 2012;192:139–146. doi: 10.1534/genetics.112.141986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedden P. The genes of the green revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/s0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 33.Choi D, Lee Y, Cho H-T, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Kende H. Expression of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol. 2001;127:645–654. [PMC free article] [PubMed] [Google Scholar]
- 35.Takatsuji H. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6:1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 37.Yang S, et al. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci USA. 2013;110:18572–18577. doi: 10.1073/pnas.1318211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.