Significance
Mosquito blood cells are central players of immunity against the vector-borne pathogens that devastate the lives of millions of people worldwide. However, their molecular identity and classification remain controversial. By applying single-cell RNA sequencing and high-content imaging flow cytometry, we defined the molecular fingerprint of a subset of mosquito blood cells and characterized two transcriptionally distinct blood cell populations that resemble previously described cell types. Surprisingly, cell population analyses at a single-cell level uncovered an active molecular transfer between the two cell types that may contribute to cellular diversity and plasticity seen across biological systems.
Keywords: entomology, insect immunity, malaria, blood cells, single-cell sequencing
Abstract
Mosquito blood cells are immune cells that help control infection by vector-borne pathogens. Despite their importance, little is known about mosquito blood cell biology beyond morphological and functional criteria used for their classification. Here, we combined the power of single-cell RNA sequencing, high-content imaging flow cytometry, and single-molecule RNA hybridization to analyze a subset of blood cells of the malaria mosquito Anopheles gambiae. By demonstrating that blood cells express nearly half of the mosquito transcriptome, our dataset represents an unprecedented view into their transcriptional program. Analyses of differentially expressed genes identified transcriptional signatures of two cell types and provide insights into the current classification of these cells. We further demonstrate the active transfer of a cellular marker between blood cells that may confound their identification. We propose that cell-to-cell exchange may contribute to cellular diversity and functional plasticity seen across biological systems.
The cell is the basic building block of all living organisms. Cells found in the blood, or hemolymph in invertebrates, have received numerous designations, such as hemocytes, amebocytes, phagocytes, coelomocytes, and immunocytes (1). Regardless of their names, these cells play key roles in shaping the extracellular environment and helping fight infection all throughout the animal kingdom. In insects, blood cells circulate in the hemolymph or associate with internal organs as sessile cells (2). They are functionally equivalent to mammalian leukocytes as they display functions similar to neutrophils, monocytes, and macrophages: e.g., phagocytic abilities, chemotaxis, production of antimicrobial peptides, free radicals, and cytokine-like molecules (3–7). Contrary to the well-established classification of human leukocytes, insect blood cell type classification is controversial, with the same terms being used for different cell morphologies even within the same insect order (2, 8–10). Most studies of insect blood cells have focused on the embryonic and larval stages of the Drosophila model (11–14), but these observations are not immediately applicable to other insects.
Mosquitoes are the deadliest animals on Earth, transmitting pathogens that cause a variety of diseases and infect millions of people every year (15). While feeding on blood to reproduce, adult females acquire pathogens from an infected host. Pathogen development and replication within the mosquito is an absolute requirement for transmission so the disease cycle depends on the mosquito’s capacity to counterattack these invaders. Blood cells represent the cellular arm of mosquito immunity and participate in humoral responses by secreting pathogen-killing factors, such as components of the melanization pathway (16, 17) and of the complement-like system that help eliminate malaria parasites (18, 19). Landmark studies have used ultrastructure, enzymatic activity, lectin binding, immunocytochemistry, and function to characterize hemocytes from different mosquito species (8, 9, 20–22). More recently, hemocytes have also been classified based on their DNA content into euploid and polypoid (23). Transcriptomics studies have explored the molecular basis of mosquito hemocyte immunity upon infection with bacteria and Plasmodium (24, 25). Smith et al. (26) used mass spectrometry to analyze the proteome of hemocytes isolated based on the uptake of magnetic beads (26). To date, mosquito hemocytes are divided into (i) granulocytes, phagocytic cells that exhibit granules in their cytoplasm and quickly spread onto glass; (ii) oenocytoids, spherical and poorly adhesive cells that produce phenoloxidases, enzymes involved in melanization defenses; and (iii) prohemocytes, small cells of reportedly 2 μm (27, 28) or 4 to 6 μm in size suggested to function as hemocyte progenitors (9), and/or represent small phagocytic cells arising from asymmetrical divisions (29). These cell types exhibit distinct ultrastructural features, with granulocytes containing numerous granules, vacuoles, and a dilated rough endoplasmic reticulum (RER), whereas oenocytoids exhibit a homogenous cytoplasm and an eccentric nucleus, and prohemocytes have a large nucleus and almost no organelles (8, 20, 21). Phenoloxidase activity has been described in oenocytoids (9, 22) but has been also observed in granulocytes poststimulation (9) and even in prohemocytes (30). Prohemocytes stain only weakly for reactive nitrogen and oxygen species, but all cell types can bind lectins (9). A few studies have also identified mosquito plasmatocytes as a cell type distinct from granulocytes due to their fibroblast-like shape, lack of granules, and a well-developed RER (8, 21). Although an increase in hemocyte numbers was reported upon blood feeding and infection (23, 29, 31), the pathways underlining their differentiation into these classes remain unknown. Whether the current classification represents true cell types or states, and if cell subpopulations exist, are also yet to be explored. In mammals, single-cell transcriptomics has recently begun to tackle similar questions. It is increasingly evident that cell populations long considered to be of the same type display significant functional differences and considerable variability in gene expression (32–34). The use of single-cell approaches to explore cellular heterogeneity in nonmodel organisms holds the promise to uncover unforeseen complexity and to identify cellular populations that would be undetectable in bulk measurements.
Here, we unravel the molecular fingerprint of a subset of mosquito blood cells at a single-cell level and show that naive hemocytes express nearly half of the mosquito transcriptome. By applying fluorescence-activated cell sorting (FACS), single-cell RNA sequencing (scRNA-seq), and high-content imaging flow cytometry, our unparalleled study identifies two distinct cell populations that resemble granulocytes and oenocytoids. Our findings further reveal active molecular exchange between blood cells and the presence of extracellular vesicles (EVs) in the mosquito hemolymph. Altogether, our study contributes molecular insights into the established classification of mosquito blood cells.
Results
scRNA-Seq of Blood Cells from PPO6::RFP Transgenic Mosquitoes.
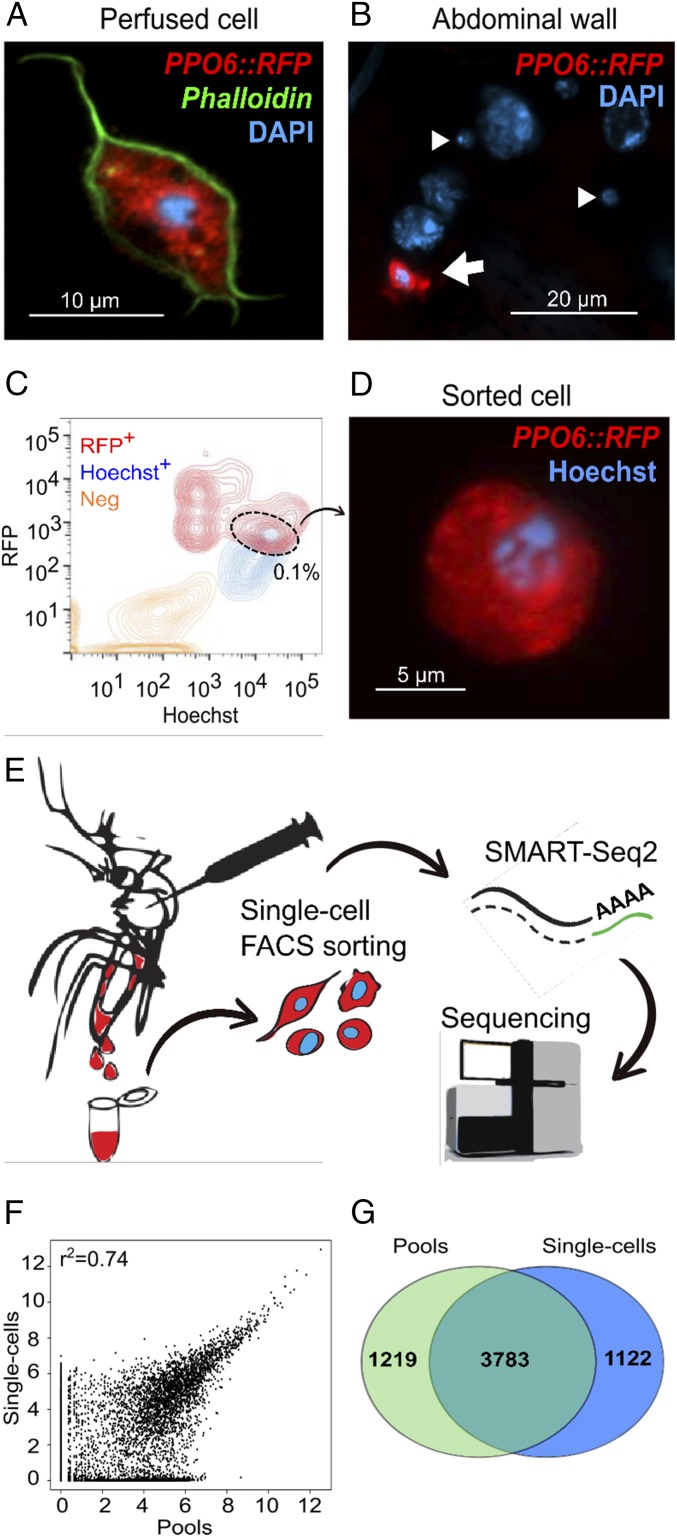
We chose to explore mosquito blood cells using a transgenic strain expressing a red fluorescence reporter (tdTomato, herein RFP) under the control of the prophenoloxidase 6 (PPO6) melanization-related gene (PPO6::RFP) (35). Melanization is a well-established immune response of invertebrates that controls infection against bacteria and parasites (36–39). Several reports suggest that melanization is mostly mediated by a specific cell population called oenocytoids, which represents ∼10% of the blood cells. We focused on cells obtained in the absence of infection or blood feeding as a baseline for analysis of cell-to-cell variation. We first confirmed that RFP-positive hemocytes were present in the circulation by the identification of cells displaying RFP fluorescence in hemolymph perfusate (Fig. 1A). RFP signal was also observed in the hemocytes attached to the inner abdominal wall of dissected adult female mosquitoes, where fat body cells are most prominent (Fig. 1B, arrow). As expected, the hemolymph perfusate was heavily contaminated with a mixture of cells and subcellular/tissue debris (9). To purify live RFP-positive cells, we developed a FACS approach based on RFP expression and Hoechst nuclear staining and validated our method by microscopic inspection of sorted cells. The sorted cell population corresponded to 0.1% of the total events measured (n = 100,000) in the perfusate of at least 10 mosquitoes (Fig. 1 C and D). This is in accordance with previous work indicating that only a small subset of adult mosquito hemocytes produces PPO (39). We FACS-sorted single blood cells and performed scRNA-seq to capture the transcriptome of single PPO-producing blood cells (Fig. 1E). Hemocytes were sorted into a 96-well plate and, after sample processing and quality assessment, we obtained successful cDNA amplification for 56 single cells in addition to two pools of 30 cells each, from which 28 high quality cDNA libraries were sequenced, representing 26 single hemocytes and the two pooled samples (SI Appendix, Fig. S1 A–D). As a single mosquito can contain as few as 500 blood cells in the circulation (23, 40), we believe the small number of cells analyzed reflects a combination of technical limitations inherent to our approach. Mosquito blood cells reportedly vary in size from as little as 2 to 20 μm (8–10, 41), and variability in cell size can affect RNA recovery, as small cells contain small amounts of RNA. The adaptation of the protocol to the study of an invertebrate system (e.g., the choice of lysis buffer and chemistry) may also have influenced our results, especially since different biological cell types show distinct technical quality features in scRNA-seq (42). A small number of cells have, nevertheless, been used in other scRNA-seq studies (43, 44), and this does not preclude identification of cellular types when an adequate sequencing depth is used. We therefore prioritized the deep sequencing of individually curated, very high-quality samples representing a small subset of cells obtained ex vivo.
Fig. 1.
Single-cell RNA sequencing of transgenic mosquitoes. A PPO6::RFP transgenic mosquito strain was used for isolation of blood cells. (A) An RFP-expressing hemocyte (red) obtained by perfusion of a female adult mosquito. (B) Only a proportion of adult mosquito blood cells display RFP expression (red, arrow) whereas other cells of sizes suggestive of hemocytes do not (arrowheads). (C) FACS sorting of RFP+ (R+) Hoechst+ (H+) blood cells from hemolymph of perfused PPO6::RFP females. (D) Representative image of a sorted cell. (E) The pipeline developed for the study of mosquito blood cells based on single-cell RNA sequencing. Cells were sorted into a 96-well plate, processed according to the SMART-Seq2 protocol, and sequenced in a HiSeq Illumina platform. Image courtesy of Suzana Zakovic (Max Planck Institute for Infection Biology, Berlin). (F) Scatter plot for the average normalized read counts from pools and single cells. r2 indicates Pearson correlation. (G) Venn diagram of genes detected in single cells and pools (normalized count ≥ 1). (Scale bars: A, 10 µm; B, 20 µm; D, 5 µm.) DNA is stained with DAPI (A and B) and Hoechst (D).
Our sequencing generated on average 4.5 million reads per sample, well-above the minimum of one million reads previously suggested as a requirement for adequate single-cell studies (45). Over 70% of the reads were successfully mapped, with exonic reads comprising of more than 40% (SI Appendix, Fig. S1E and Table S1). All samples achieved saturation at 2 million reads, comparable with that previously observed for mammalian cells (45). For further analyses, we discarded one cell as it showed gene expression suggestive of a doublet (SI Appendix, Fig. S1F). Doublets have been reported in the circulation of adult female mosquitoes (29) and may fall within a size range comparable with larger hemocytes. Around 3,800 genes were detected in each pool whereas single cells expressed between 450 and 1,400 genes. This large transcriptional variability likely results from the diverse functions hemocytes exhibit as they circulate in the open body cavity, exposed to biochemical changes and in close contact with internal organs. This is also in agreement with recent findings of cellular heterogeneity in subtypes of immune cells of other organisms, such as humans (46), mice (47), and fish (48). Similar expression profiles were obtained for pools and single cells with comparable numbers of detected genes (1,100 and 1,200 genes, respectively) (Fig. 1 F and G). The marker genes used for FACS sorting (PPO6 and tdTomato) were identified in both single cells and pools (Dataset S1), confirming the efficiency of our method. Altogether, our results showed that the transcriptome of mosquito hemocytes comprises over 6,000 genes, of which more than half (3,022) had not been identified in earlier studies (SI Appendix, Fig. S1G) (24, 25). As previous studies were based on pooled samples, their datasets were also compared with our pools and over 2,200 genes were identified only by our sequencing (SI Appendix, Fig. S1H). In addition, our approach revealed sequences for over 80% (n = 914) of the proteins reported by an earlier proteomics study based on magnetic beads isolation of Anopheles gambiae phagocytes (26) (SI Appendix, Fig. S1I). Most of the identified genes were present in more than one cell, but a few genes were detected in only one cell (SI Appendix, Fig. S1J). About 140 genes were found in 90% of the cells sequenced (Dataset S2), and gene ontology (GO) analyses indicated that this core transcriptome represents metabolism, biosynthesis, translation, and immune defenses (Datasets S5 and S6).
Mosquito hemocyte biology has been mostly studied in the context of immunity. We thus inspected our dataset for previously identified immune genes. Naive hemocytes expressed low levels of such immune genes as the transcription factors REL1 (AGAP009515) and REL2 (AGAP006747), Cactus (AGAP007938), IκBβ (AGAP009166) and IκBγ (AGAP005933), and the receptors PGRP-LC (AGAP005203) and PGRP-S1 (AGAP000536) (Dataset S1). Components of the complement cascade [e.g., TEP1 (AGAP010815), APL1C (AGAP007033), LRIM1 (AGAP006348), and HPX2 (AGAP009033)] were also detected in some cells, along with the LPS-induced TNFα transcription factor (LITAF)-like 3 (AGAP009053) described to control Plasmodium survival in the gut (28). The phagocytic and antibacterial activities of these cells can be illustrated by the expression of Eater (AGAP012386), Ninjurin (AGAP006745), and Nimrod (AGAP009762), alongside that of several fibrinogen-related proteins (FREPs/FBNs), such as FBN8 (AGAP011223), FBN9 (AGAP011197), FBN10 (AGAP011230), and FBN30 (AGAP006914) (49–51). Although no ortholog for a major Drosophila hemocyte marker, hemolectin, has been described in the A. gambiae genome, mosquito hemocytes expressed both Pannier (AGAP002235) and Serpent (AGAP002238) GATA factors, as well as misshapen (AGAP006340), the genes associated with blood cell differentiation, maturation, and activation in fruit fly larvae (52–54). Genes involved in cell adhesion and polarity, such as integrin β-1 (AGAP010233), laminin (AGAP010548), Notch1 (AGAP001015), and Armadillo (AGAP001043), and components of extracellular matrix like collagen type IV (AGAP009200) were also identified. Genes encoding other immune-related proteins previously observed in hemocytes by antibody staining, like Sp22D (AGAP005625), SRPN6 (AGAP009212), and SRPN10 (AGAP005246) (9), were also present. Out of two panhemocyte markers identified before (25), one gene (AGAP002267) is absent from the current genome annotation and could not be mapped to our sequences, and the other (AGAP007314) was not detected by our analysis. Altogether, our data suggested that, in addition to immunity, naive blood cells perform tissue maintenance and morphogenesis tasks. The processed gene expression data for visualization in single cells is accessible at https://scb.sanger.ac.uk/#/base/main.
Identification of Blood Cell Populations.
To account for the technical noise arising from the small amounts of RNA, we included in our samples External RNA Controls Consortium (ERCC) spike-ins before cDNA amplification (55). We analyzed the percentage of ERCC and mitochondrial counts as a proxy for sequencing efficiency, RNA degradation, or incomplete lysis and potential cell death. As anticipated, variation was observed (SI Appendix, Fig. S1 K–L), but caution was taken in applying these criteria and attributing them biological meaning because variability could have arisen from true cell type-related processes. As differences in the total number of expressed genes could also have stemmed from different morphologies and cell types, we manually curated their individual mappings to confirm that the samples corresponded to potentially true representations of blood cells. To estimate technical noise, we applied the variability threshold based on the square of the coefficient of variation (CV2) of the ERCCs (56) and identified 148 genes whose expression exceeded the threshold (Fig. 2A). These highly variable genes included a scavenger receptor, fibrinogen-related and leucine-rich repeat-containing proteins, as well as genes involved in vesicle transport, metabolism, and transcription (Dataset S4). No genes directly associated with cell cycle had high variability although several cyclin genes were detected in specific cells (Dataset S1), corroborating previous reports of the potential of mosquito hemocytes to undergo cellular division (23, 29, 41).
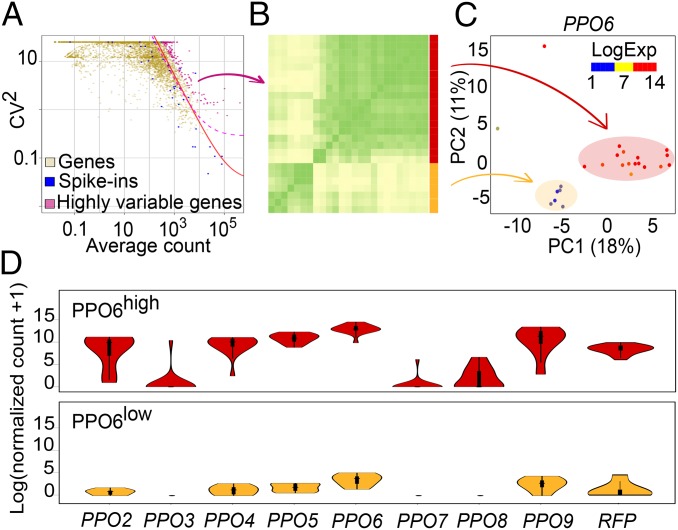
Fig. 2.
Identification of mosquito blood cell subpopulations. (A) The expression variability of individual genes measured by the squared coefficient of variation (CV2) is plotted against the mean expression level (normalized counts). Magenta points indicate mosquito genes showing higher than expected expression variability compared with ERCC spike-ins (blue) (adjusted P value of <0.1). The red line is the fitted line of the spike-ins, and the dashed line (pink) marks the margin for genes with 50% biological CV. (B) Pearson correlation heat map of single hemocytes based on the expression of the highly variable genes identified in A. Correlation suggests the presence of two groups of cells (red and yellow). (C) PCA plot based on the expression of highly variable genes. The first two principal components are shown, and each point represents one single hemocyte. Two clusters were identified and correspond to the subgroups in B. PPO6 expression, as log10 (normalized counts +1), is overlaid onto the PCA plot. (D) Violin plots of PPOs and RFP expression in the identified groups.
Hierarchical clustering of the variable genes based on pairwise Pearson correlation suggested the presence of at least two groups of cells (Fig. 2B). Principal component analysis (PCA) also yielded two cell populations, supporting our clustering (Fig. 2C). Interestingly, PPO6 showed high variability, and the overlay of PPO6 expression onto the PCA plot suggested that the two clusters were largely characterized by low and high expression of PPO6. PCA analysis in the absence of outlier cells confirmed the existence of the cell groups (SI Appendix, Fig. S2A). Comparable results were also found when the whole dataset was used for similar analyses (SI Appendix, Fig. S2 B and C). Differences in the PPO6 expression have been previously described by immunofluorescence microscopy (41) but have not been associated with cell types. Moreover, from the 10 PPO genes encoded in the A. gambiae genome, 8 were observed in our sequencing, and 6, as well as the RFP reporter, had variable expression between the groups (Fig. 2D and Dataset S4). PPO genes contributed significantly to the separation of the cells into the subpopulations, as suggested by multidimensional scaling (MDS) analysis (SI Appendix, Fig. S2D). Transcriptional heterogeneity was observed not only between the two groups, but also within cells of the same group, with several genes being highly expressed in individual or subsets of cells (SI Appendix, Fig. S2 E and F). Based on the abundance of PPO6 in the two cell groups, we designated them PPO6high and PPO6low.
PPO6high and PPO6low Cells Represent Transcriptionally Distinct Subpopulations.
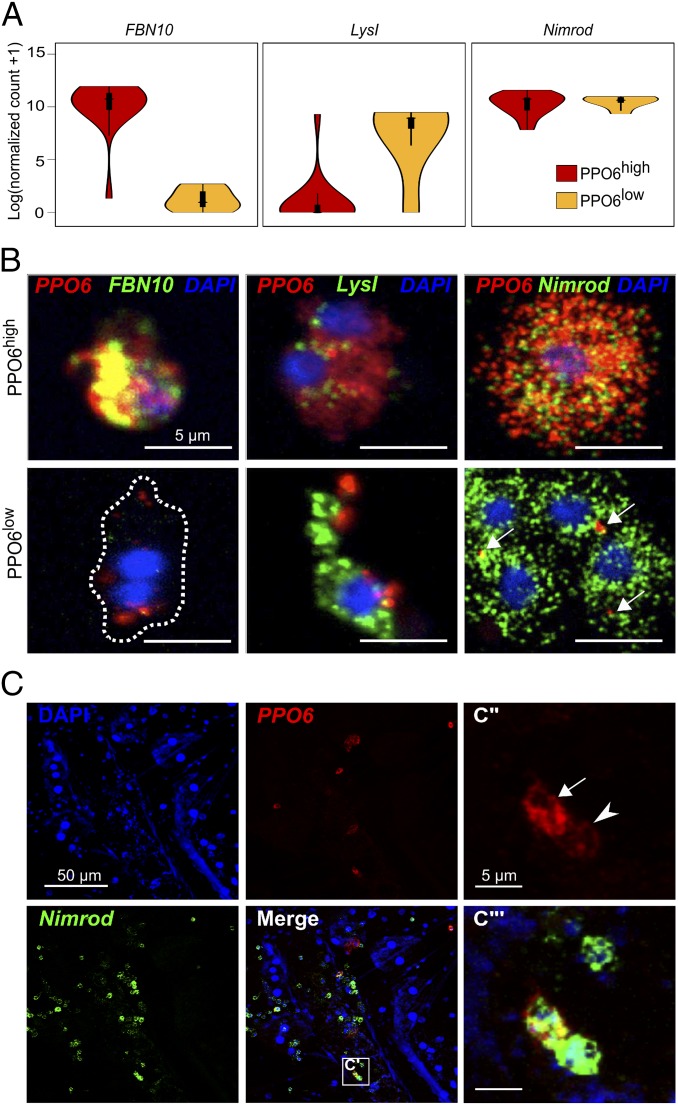
Among the highly variable genes, we detected several FBN sequences, such as FBN8 and -10. PPO6high cells showed high expression levels of FBN10 (Fig. 3 A, Left) whereas PPO6low cells exhibited weak or lack expression of FBN8, -10, and -30 (Fig. 3A, Dataset S4). Although below the ERCC-defined variability threshold, likely due to the small number of cells analyzed, expression of the antimicrobial peptide gene lysozyme type I (LysI) (AGAP011119) was more characteristic of PPO6low cells (Fig. 3 A, Middle). In the search for a panhemocyte marker, we also identified expression of phagocytic receptor Nimrod in both groups of cells (Fig. 3 A, Right). The in silico results were validated by single-molecule RNA fluorescence in situ hybridization (RNA-FISH), showing coexpression of tdTomato and PPO6 in all PPO6::RFP hemocytes, with no detection of tdTomato in blood cells isolated from WT mosquitoes (SI Appendix, Fig. S3 A and B). RNA-FISH accurately distinguished PPO6high and PPO6low hemocytes. Consistently, PPO6high cells showed high levels of FBN10, which were very low or absent in PPO6low cells. High levels of LysI were found in PPO6low cells, reinforcing the presence of PPO6low/FBN10low/LysIhigh cells; and Nimrod transcripts were observed in all perfused hemocytes (Fig. 3B). We took advantage of the high conservation of PPO6, LysI, and Nimrod genes in the closely related mosquito species Anopheles stephensi to examine the discovered blood cell groups in other anopheline mosquitoes. We detected PPO6high and PPO6low/LysIhigh cells, along with a low levels of Nimrod expression. No FBN10 was observed (SI Appendix, Fig. S3C), probably due to the specificity of the probe to A. gambiae and the large diversity of this gene family.
Fig. 3.
Characterization of PPO6high and PPO6low cell subpopulations. (A) Violin plots of the expression of putative population and panhemocyte markers. (B) RNA-FISH validation of identified PPO6high and PPO6low cell subpopulations in perfused cells based on markers shown in A. Cells were classified as PPO6high (Upper) or PPO6low (Lower) according to the expression of PPO6 (red). Arrows indicate lower PPO6 signal. (C–C‴) PPO6high and PPO6low cell subpopulations can also be seen as tissue-resident blood cells attached to the inner abdominal wall of female mosquitoes. Arrow and arrowhead indicate PPO6high and PPO6low cell subpopulations, respectively. C‴ shows higher amplification of the C′ boxed area. (Scale bars: B, 5 µm; C and C′, 50 µm; C″ and C‴, 5 µm.) DNA is stained with DAPI.
We next compared the overall gene expression between PPO6high and PPO6low cells. Based on differentially expressed genes, GO analyses uncovered that melanization characterized PPO6high cells whereas metabolism and RNA processing defined the PPO6low subset (Datasets S5 and S6). Although not significant, PPO6low cells appeared to express more genes in total, but mitochondrial counts did not differ between the groups (SI Appendix, Fig. S3D). These findings suggest that PPO6high cells are specialized for melanization responses, expressing genes involved in these processes at very high levels, whereas PPO6low cells execute a broader range of biological tasks. The identified differences between the cell groups may represent different cell lineages, mediate diverse melanization processes [e.g., metamorphosis and cuticle sclerotization (57–59)], or reflect localization patterns of the cells inside the mosquito body (29). To assess whether differences were related to tissue residency, we performed RNA-FISH in tissues and observed both cell populations in close contact with the fat body cells within the abdominal wall with no conspicuous cell clusters. The majority of the sessile cells were positive for Nimrod, independent of PPO expression (Fig. 3C), indicating that Nimrod is a potential marker for both circulating and tissue-resident blood cells. Altogether, these results demonstrate that both circulatory and tissue-resident hemocytes display transcriptional heterogeneity and that PPO6high and PPO6low cell populations are present in two mosquito species.
PPO6high and PPO6low Cells Share Functional and Morphological Features.
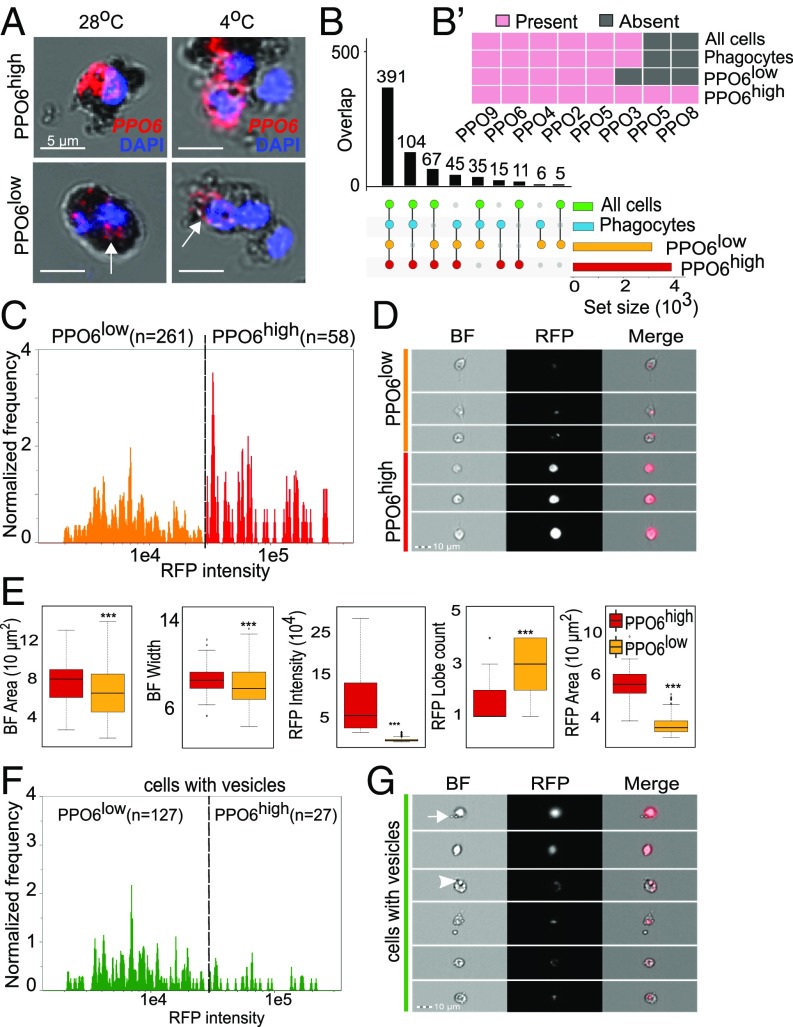
Mosquito blood cells are separated into three classes—granulocytes, oenocytoids, and prohemocytes. Our GO analyses suggested the presence of a PPO-specialized cell population and a second cell subset of a less specific nature. We reasoned that PPO6high and PPO6low cell groups could be representatives of oenocytoids and granulocytes, respectively. As phagocytosis is a hallmark of granulocytes, we first explored functional differences using magnetic bead uptake as a means for “phagocyte” isolation, as suggested before (26). To this end, we injected mosquitoes with magnetic beads and either allowed them to rest at 28 °C before perfusion or incubated the mosquitoes at 4 °C to inhibit phagocytosis. To our surprise, both PPO6high and PPO6low cells were identified among magnetically isolated cells and under both conditions, suggesting that, instead of phagocytosis, both cell types endocytosed the beads (Fig. 4A, arrows). We next compared the gene profiles of PPO-producing cells to the proteomics results obtained by Smith et al. (26) using magnetic bead isolation. Our analyses revealed that similarities were the strongest when profiles were compared across all samples—PPO6high, PPO6low, phagocytes, and all cells. PPO6high and PPO6low shared expression of more genes with phagocytes when considered together rather than alone, indicating that both PPOhigh and PPOlow cell types shared similarities with “phagocytes” at the gene/protein level (Fig. 4B). In agreement, nearly all PPOs were present in all samples (Fig. 4B). Based on this assay, we did not detect functional differences between the groups as both PPO6high and PPO6low performed endocytosis. This was in agreement with the identification of endocytosis-related genes in the transcriptome (Dataset S1) and with a previous observation that oenocytoids can internalize beads and bacteria (16).
Fig. 4.
PPO6high and PPO6low cell subpopulations share functional and morphological features. (A) Magnetic bead isolation assays followed by RNA-FISH for identification of cell populations. Arrows indicate lower PPO6 signal. (B) Intersection analyses between PPO6high and PPO6low cell populations and proteomics results obtained by Smith et al. (26) for phagocytes and all cells (unselected). The presence or absence of PPO genes/proteins in each of the samples is shown in B′. (C) Hemocytes were perfused from PPO6::RFP mosquitoes and analyzed using imaging flow cytometry. RFP-positive cells were identified by their RFP Median Pixel and further separated into PPO6high and PPO6low based on their level of RFP fluorescence. The number of cells analyzed per group is shown between parentheses, and the dotted line indicates the RFP threshold level used for separation of the two populations. (D) Image gallery containing representative images of PPO6high and PPO6low populations. Variation in cell shape and RFP intensity can be observed in the bright-field (BF), RFP, and merged images. (E) Box plots show the distribution of five morphological measurements according to the groups. Asterisks represent P < 0.01 based on Mann–Whitney–Wilcoxon test. (F) RFP-positive cells were interrogated for the presence of membrane protrusions or internal structures suggestive of vesicles. Similar to C, cells were grouped into PPO6high and PPO6low, and an RFP intensity histogram of cells displaying vesicles (green population) was generated to illustrate that vesicles are observed in cells from both groups. Representative images of cells in this population subset are shown in G. Arrow and arrowhead indicate representative membrane protrusions and internal vesicles, respectively. Box plots indicate the median, first and third quartile, and min and max values. (Magnification: D and G, 40×.)
Next, we sought to investigate the morphology of the cell populations using imaging flow cytometry and RFP fluorescence as a proxy for PPO6 expression. We measured a series of morphological features of 319 single RFP-positive cells, which were divided into RFPhigh (PPO6high, n = 58) and RFPlow (PPO6low, n = 261) based on their fluorescence intensity (Fig. 4 C and D and SI Appendix, Fig. S4). Overall, RFP-positive cells had a mean area of 67 µm2, ranging from 18 µm2 to nearly 140 µm2. These measurements are in accordance with the reported cell sizes (9, 23, 41). Similar to recent studies based on flow cytometry of fixed cells (23, 41), we did not detect the cells of 2 µm in size described by other research groups based on label-free light microscopy alone (27, 60). When comparing the cell groups in terms of bright-field measurements of their cytoplasm, PPO6low cells showed smaller area, width, and minor axis than PPO6high cells (Fig. 4E and SI Appendix, Fig. S4). Our expectation was to find that PPO6high comprised oenocytoids: i.e., spherical cells with weak or no granularity. To our surprise, no differences between the groups were detected in granularity or cell shape, and PPO6high and PPO6low cells were equally circular. (SI Appendix, Fig. S4 and Dataset S7). Cells from both groups also displayed an elongated shape, typical of the cytoplasmic extensions seen in fusiform or spindle-shaped cells. This shape is characteristic of plasmatocytes described in other insects and mosquito species (2, 8, 21). These results failed to assign RFP-positive cells to any of the groups previously characterized based on light microscopic morphological and ultrastructural analyses: granulocytes, plasmatocytes, or oenocytoids. In fact, the highest discriminating factors (Fisher’s linear discriminant) separating PPO6high and PPO6low subpopulations relied on RFP intensity alone, with bright-field parameters scoring poorly and failing to establish a morphological distinction between the cells (Dataset S7). Importantly, our imaging flow cytometry approach relied on morphological analyses of cells in suspension, which is unbiased and likely more relevant for the identification of the cellular types found in the hemolymph circulation. Fluorescence microscopy upon cell attachment reinforced our finding that elongated cells are found in both groups, with no particular cell shape being attributed to either group (SI Appendix, Fig. S5A). Round and oval cells were also observed as PPO6high and PPO6low using anti-PPO6 antibodies. PPO6 and RFP signals overlapped, but we occasionally identified cells expressing spotty and cytoplasmic PPO6 patterns that did not show any RFP signal (SI Appendix, Fig. S5B), suggesting differences in regulation or stability between the mRNAs and/or proteins.
In addition to intensity, the cell groups differed in their RFP area. PPO6high cells displayed an overall cytoplasmic distribution of the RFP signal whereas a more localized globular signal was detected in the cytoplasm of PPO6low cells (Fig. 4 D and E). Microscopic examination also revealed that nearly half of the cells from both groups displayed internal structures and “budding” extensions of the cytoplasm suggestive of vesicles (Fig. 4 F and G, arrowhead and arrow, respectively). To confirm that, we performed correlative scanning electron microscopy (SEM) and demonstrated the presence of membrane protrusions or “blebs” in RFP-positive cells (SI Appendix, Fig. S6A). Altogether, these results established that morphological plasticity of the mosquito blood cells is independent from their transcriptional profile and that mosquito blood cells have membrane vesicles and protrusions.
Mosquito Blood Cells Exchange Molecular Information.
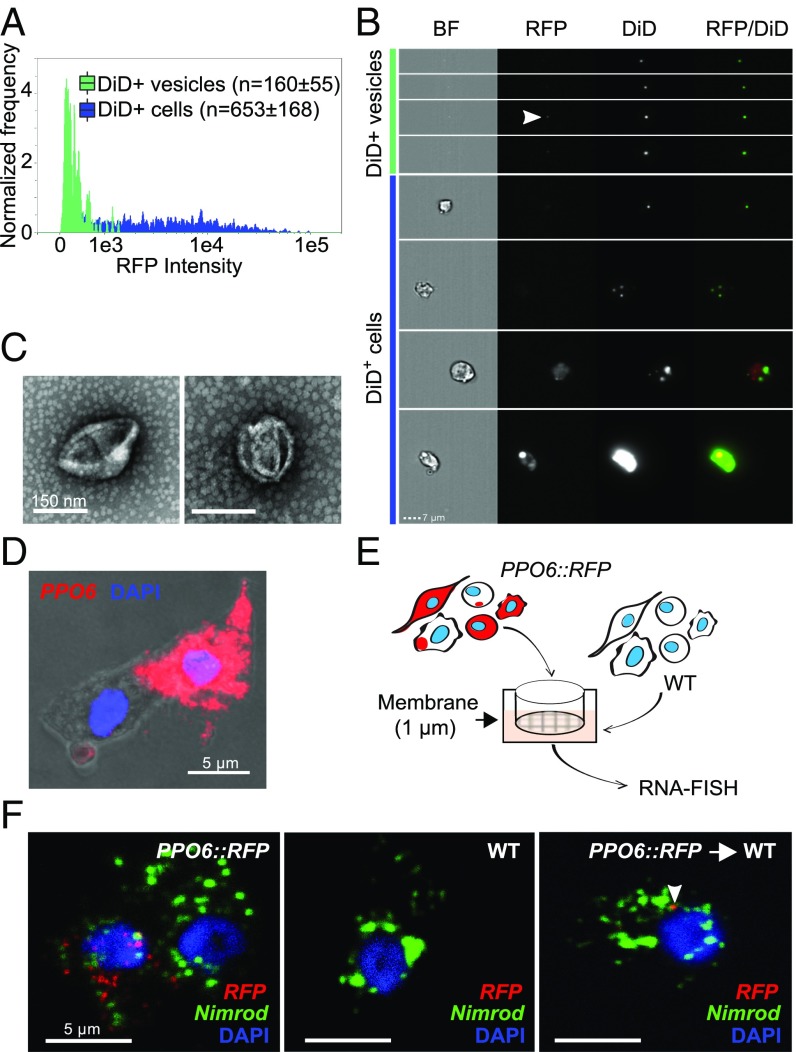
We were puzzled by the possibility that the RFP signal analyzed in our cell-sorting approach could have originated from RFP-positive vesicles. Earlier reports used DiD, a lipophilic cyanine dye, to label both mosquito hemocytes and hemocyte-derived vesicles (61, 62). To test whether the localized RFP signal seen in our imaging was associated with vesicles, we first stained PPO-producing cells with DiD and observed that RFP-positive cells indeed contained DiD-positive membrane-bound and internal vesicles that were both RFP-positive and negative (SI Appendix, Fig. S6 B and C). To identify EVs in the mosquito circulation, we performed imaging flow cytometry using DiD and a recently published approach (63). Both DiD-positive cells and EVs could be identified in hemolymph perfusate (Fig. 5 A and B). EVs were detected based on their small size, weak dark-field and positive DiD fluorescence, with a few EVs also displaying weak RFP signal (Fig. 5B, arrowhead). The degree of DiD intensity differed between cells and did not depend on RFP fluorescence. Differential centrifugation followed by EM confirmed the presence of EVs in hemolymph perfusate of naive female mosquitoes (Fig. 5C). SEM of perfused cells also revealed that vesicles of different sizes and shapes, corresponding to the different vesicle types described in the literature—exosomes, microvesicles, and apoptotic vesicles (64)—could be indeed observed in association with naive mosquito blood cells (SI Appendix, Fig. S6D). These findings suggested that EV production is a general phenomenon that is not limited to PPO-associated cells.
Fig. 5.
Vesicle identification and molecular exchange in mosquito blood cells. Hemocytes were perfused from PPO6::RFP mosquitoes, stained with the lipophilic DiD dye, and analyzed using imaging flow cytometry. DiD-positive cells and EVs were first identified according to their DiD and dark-field intensity. EVs were further separated from cells based on their small bright-field area. Cells were identified considering their area and aspect ratio. An RFP fluorescence histogram for both DiD-positive cells and EVs is shown in A. (B) Representative images of DiD-positive cells and EVs. The arrowhead indicates a representative of an RFP-positive vesicle. (C) Negative staining electron microscopy of vesicles obtained by differential centrifugation (10,000 × g) of hemolymph perfusate (representative images of two independent experiments are shown). (D) PPO6 mRNA detection by RNA-FISH within budding extensions of blood cells. (E) Schematics of a transwell assay developed to test the transfer of RFP between blood cells from PPO6::RFP transgenic mosquitoes and WT mosquitoes that do not express any reporter genes. Hemolymph perfusate from WT mosquitoes was pipetted onto a coverslip placed under a 1-µm membrane. Perfusate collected from PPO6::RFP mosquitoes was placed on top of the membrane. (F) RNA-FISH was performed on coverslips obtained from the transwell assay described in E using probes to detect RFP and Nimrod expression in WT acceptor cells (Right, arrowhead). Representative images of two independent experiments are shown. (Magnification: B, 63×.)
A growing body of evidence has demonstrated that RNA can be transferred between mammalian cells. As RNA can be found in EVs and our data showed that EVs are present in the mosquito hemolymph, we explored whether a potential exchange between PPO-positive and negative cells could be responsible for the identification of PPO6high and PPO6low cells. Strengthening this idea were the observations that (i) our scRNA-seq results uncovered cells with minute levels of PPO6 and RFP transcripts and (ii) expression of PPO6 by RNA-FISH was detected inside budding extensions associated with PPO6-positive cells (Fig. 5D). To test whether RFP mRNA can be transferred between naive transgenic and WT blood cells, we developed a transwell assay using blood cells from PPO6::RFP transgenic and WT mosquitoes (Fig. 5E). Remarkably, after exposure to hemolymph perfusate from transgenic mosquitoes, RFP transcripts were observed by RNA-FISH inside WT blood cells (Fig. 5F, arrowhead). This result indicated that RFP mRNAs can be shuttled between blood cells and might account for the PPO6low population identified by our scRNA-seq and imaging. Taken together, our findings demonstrated that molecular exchange between cells, likely via EVs, impacts their transcriptional profile. As EVs have been shown to carry lipids, proteins, and RNA and can be secreted by virtually all cells, our results revealed an unappreciated role of intercellular molecular exchange in defining cellular identity.
Discussion
Understanding how transcriptional networks influence cell identity is a central problem in modern molecular biology. Our study describes mosquito blood cells as a source of key components of immunity, development, and tissue homeostasis and places them as a central hub coordinating mosquito biology at different levels. Using a combination of single-cell genomics and imaging, we reveal that hemocytes display an unexpected degree of complexity where two transcriptionally defined cellular “populations” suggestive of distinct cell types share morphological and functional features. We also demonstrate that mosquito blood cells exchange mRNA, leading to the detection, by RNA-FISH, of an “exogenous” gene in acceptor cells. Altogether, our results contribute insights into cellular cross-talk and cell type classification, in addition to illustrating the power of single cell-based approaches in discovering unappreciated events at the core of biological processes.
Using single-cell RNA sequencing, we describe the baseline expression of a mosquito blood cell in exceptional detail. An average mosquito blood cell under resting conditions expresses ∼1,000 genes, or 7% of the mosquito transcriptome. In total, about half of the genes currently annotated in the mosquito genome were detected by RNA sequencing of naive, unstimulated mosquito hemocytes. Our dataset represents a substantial gene expression resource for further studies of tissue-specific alternative splicing, RNA editing, and gene and transcript models. It also illustrates the importance of tissue-specific approaches and paves the road toward the detailed mapping of gene expression in cells and tissues of insects, with the ultimate goal of creating a comprehensive reference atlas of cellular diversity.
By successfully applying single-cell RNA sequencing to the study of mosquito blood cells, we demonstrated proof of the existence of at least two transcriptionally distinct cell groups that are similar to currently defined cell types. PPO6low cells have a rich transcriptional program suggestive of mosquito granulocytes. The second subpopulation, with a transcriptional profile specialized in melanization, is indicative of oenocytoids. Nonetheless, these two transcriptionally defined cell types could not be distinguished by the morphological tests used here. Recent studies have relied on bright-field microscopy alone to quantify hemocyte types in hemocytometer and analyze their role in immune responses (27, 28, 65). Our imaging flow and fluorescence microscopy findings clearly showed that cells with morphological features suggestive of granulocytes belong to both PPO6low and PPO6high populations. Studies incorporating the cell-type markers identified here would help elucidate the contribution of these cell types to mosquito biology. The spatial distribution of these blood cells, especially as no lymph glands or hematopoietic cell clusters have been described in mosquitoes, also warrants a thorough investigation. Tissue-resident blood cells likely contribute to local responses and help regulate tissue-specific events. This is exemplified by the recent discovery of macrophage subsets regulating electrical pulsing in the mouse heart (66) and of ovarian hemocytes that control germline stem cell niche maintenance in Drosophila (67). As macrophages, along with other immune cells, have their evolutionary roots in ancestral invertebrates (5, 68) and new cell types arise as a result of evolutionary processes (69), the study of insect blood cells can help elucidate the origins of the immune system.
Distinct levels and patterns of fluorescence of cellular markers are widely used in microscopy and flow cytometry as a means to mammalian cell type classification. Cellular cross-talk may, however, affect such approaches. Acquisition of macrophage-derived blebs by lymphocytes has been described, resulting in misrepresentation of lymphocytes as macrophages in flow cytometry studies and suggesting that these two cells may interact to control early responses in the lymph node (70). More importantly, translation of transferred mRNAs into functional proteins has been demonstrated before (71), along with the reprogramming of acceptor cells upon microvesicle-mediated exchange (72). It is, therefore, intriguing to consider that transcript detection of specific cellular markers might be influenced by EV uptake. This observation calls for a critical reassessment of cellular markers by the scientific community. It is also imperative to investigate whether certain protein-coding RNAs are preferentially exchanged compared with other RNAs found in the cells. How PPO6 and RFP transcripts, and potentially other PPO genes, contribute to the function of acceptor cells is another exciting question. PPO proteins lack the signal peptides required for their secretion, and it has been suggested that PPO6 is secreted by exocytosis as cell rupture has not been observed (41). It is plausible that PPO transcripts are shed by PPO6high cells and processed by PPO6low or negative cells that locally activate melanization only under specific conditions: e.g., upon infection with specific pathogens or during wounding and tissue repair. Molecular signals exchanged between cells can, thus, coordinate cellular plasticity and contribute to the diversity of functional subsets or “hybrid” cells that express markers of different or multiple cell types.
Although we cannot rule out that RFP mRNAs were transferred between mosquito cells by means other than EVs, the demonstration of mosquito blood-borne EVs indicates that different cells and tissues likely communicate through vesicles secreted into the insect open circulatory system. Several recent reports have suggested EV-mediated immune responses in dipteran insects. Exosome-like vesicles containing virus-derived siRNAs have been identified in Drosophila and contribute to systemic antiviral immunity (73). Apoptotic vesicles released by hemocytes in the proximity of invading parasites have been implicated in anti-Plasmodium responses by activating the complement pathway in A. gambiae mosquitoes (61). Interestingly, using a GFP reporter strain, Volohonsky et al. (74) reported that the antimalaria mosquito complement-like factor TEP1 is predominately expressed in the fat body as a transcript, but, at the protein level, it is found in hemocytes upon blood feeding and infection (74). The authors speculate that this is due to the uptake by the blood cells of TEP1 attached to bacterial cells. As in our sequencing only one cell contained low levels of TEP1, we suggest that EV-mediated delivery of TEP1 (mRNA or protein) may better explain these findings. We propose that vesicles found in the mosquito hemolymph contain proteins and transcripts that coordinate cell-to-cell and tissue communication not only in infection but also under physiological conditions. Disturbance of homeostasis, be it by infection, metabolic changes, tissue damage, or stress, may escalate secretion of vesicles containing an array of different cargo that can be targeted to specific tissues and complement systemic responses. We believe that, similar to how environment, microbiota, and genetic make-up influence phenotypic variation, cellular exchange can also drive cellular identity and represents an inventive and unexplored way through which nature coordinates who and what we are.
Materials and Methods
Mosquito Rearing, Fluorescence Microscopy, and Hemolymph Perfusion.
A. gambiae sensu lato PPO6::RFP transgenic and WT strains were reared at 28 °C under 80% humidity and at a 12/12-h day/night cycle. Larvae were fed with cat food, and adult mosquitoes were fed ad libitum with 10% sugar. For tissue microscopy, mosquitoes were dissected in 1× PBS, fixed in 4% paraformaldehyde (PFA), washed, and mounted using Vectashield mounting medium containing DAPI. For hemolymph perfusion, 3- to 5-d-old female mosquitoes were anesthetized on ice for 10 min, microinjected with 700 nL of a buffer containing 60% Schneider's medium, 10% FBS, and 30% citrate buffer (anticoagulant; 98 mM NaOH, 186 mM NaCl, 1.7 mM EDTA, 41 mM citric acid, pH 4.5), and allowed to rest for 10 min on ice. A small cut was made between the last two abdominal segments with the help of dissection scissors, and the flow through was collected after further injection of 10 µL of buffer. For microscopy analyses, mosquitoes were perfused directly onto glass slides or coverslips, and cells were allowed to attach for at least 15 min before fixation in PFA. Cells were stained with a 1:100 dilution of Alexa 488 Phalloidin (ThermoFisher) for 30 min at room temperature. For the DiD analyses, cells were stained with DiD (5 µM) for 20 min before PFA fixation. The anti-PPO6 immunofluorescence was performed as previously described (41). Following washes, cells were mounted as described above and analyzed on a Zeiss Axiovert microscope.
FACS and Single-Cell RNA Sequencing by SMART-Seq2.
Hemolymph from 10 to 12 mosquitoes was collected with the help of a pipette, transferred into a siliconized microtube, and diluted to a final volume of 500-µL buffer containing 2 µg/mL Hoechst 3342 (Molecular Probes). Cells were immediately analyzed in a BD ARIA II Cell Sorter equipped with lasers at 405 and 561 nm. Cells were first gated based on their RFP fluorescence, followed by positive Hoechst signal, with area vs. width being used for doublet discrimination. The FACS machine was standardized with fluorochrome-containing beads, and sorting purity was validated by visualization of cells sorted onto a coverslip. Cells were sorted into a 96-well PCR plate containing 5 µL of 0.2% Triton X-100 supplemented with 2 U/µL RNase inhibitor (Clontech), with two wells containing 30 cells each (pool samples) and one column (eight wells) containing only the lysis buffer as a negative control. We added ERCC spike-ins (Ambion) at a 1:2 billion dilution into the plate before cDNA synthesis, and all samples were processed according to the SMART-Seq2 protocol using up to 22 PCR cycles for cDNA synthesis (75). PCR products were purified with AMPure XP beads (Beckman Coulter). Quality control was performed for each sample individually both as cDNA input and sequencing library using a high sensitivity DNA kit (Agilent). A total of 125 pg of cDNA was used for library construction. Libraries were pooled at a 10-nM final concentration, and 100-bp paired-end sequencing was performed in one lane of a HiSeq2000 Sequencer (Illumina).
RNA-Seq Data Analysis.
Sequencing reads were demultiplexed using bcl2fastq (version 1.8.4) and mapped to an A. gambiae genome (P4), ERCC92 (Ambion), and dTomato sequence (35) with the STAR aligner (version 2.4.2a) (76). The genome index was generated with an A. gambiae geneset file in gtf format (P4.4), and gene count tables were produced during mapping (–quantMode Genecounts). They were next normalized with size factors calculated from the ERCCs using DESeq2 (77). A gene was considered expressed if at least one normalized read was identified in at least one sample. Genes were annotated using Vectorbase (78) and manual curation. For comparisons with previous studies (24–26), IDs were converted using Vectorbase and BioMart. Intersection analyses were performed in R using the VennDiagram and upsetR packages. Technical noise estimation and identification of the highly variable genes were performed as reported before (56), using the 60-percentile as the mean cutoff to include more ERCC genes in the technical fit. PCAs were done with the prcomp function using the variable genes or the whole dataset: i.e., genes expressed in at least one cell. For the MDS analysis, only the expression of PPO genes was taken into consideration, and Euclidean distances and the cmdscale function were used. Differential expression analyses were based on DESeq2, using the ERCC size factors and PPO6low versus PPO6high as comparison. For GO analyzes, we used topGO (79), and GO terms were obtained from the org.Ag.eg.db package. Analyses were performed in R, and scripts are available at https://github.com/mssevero/hemo-scRNASeq. The sequencing results were deposited in the European Nucleotide Archive (accession no. PRJEB23372), and the expression data can be accessed at https://scb.sanger.ac.uk/#/base/main for single gene visualization.
Imaging Flow Cytometry.
Female mosquitoes (n = 10 to 12) were perfused into a final volume of 20 to 40 µL, and the samples were immediately analyzed on an Amnis ImageStreamX MKII (Merck). For PPO6::RFP analyses, WT mosquitoes were used to set background fluorescence, and cells were measured with a 40× objective. Comparisons between populations were performed using the “Object” mask and based on the built-in function that uses Fisher’s discriminant ratio (Rd) to determine the best statistical separation (largest Rd) between identified populations. For the DiD analyses, cells were collected into FBS-free buffer containing 1 µM DiD and analyzed at 60×. Single staining controls representing RFP, DiD, and buffer alone were used for calibration and manual compensation. Experiments were repeated at least twice. Cell gating was confirmed considering the images and manually curated to exclude debris and doublets that could not be excluded by the gating alone. Vesicle detection was performed as reported before (63). DiD-positive events were interrogated based on the level of DiD fluorescence and size scatter intensity. DiD-labeled vesicles showed a low scatter along with low to mid DiD fluorescence whereas cells displayed mid to high fluorescence and scatter measurements. Speed beads, used for instrument calibration and focusing, were easily gated out as a discrete population displaying very high levels of side-scatter intensity. The RFP intensity was measured based on the median pixel by means of histogram, and cellular debris and doublets were excluded according to their bright-field area and aspect ratio. The identified populations were compared using the “Feature Finder” function of the IDEAS software (MilliporeSigma). Statistical analyses were based on Mann–Whitney–Wilcoxon, and graphs were done in R.
RNA in Situ Hybridization Using RNAscope.
RNA in situ studies were performed according to the RNAscope Multiplex Fluorescence manual (Advanced Cell Diagnostics). Cells were perfused onto glass slides, allowed to attach, and fixed in PFA as described above. If needed, slides were dehydrated and kept in 100% ethanol at −20 °C until processing. Tissue samples were processed immediately after dissection in RNase-free PBS. All RNAscope probes were designed by Advanced Cell Diagnostics and are commercially available. Each probe was tested against a negative control before and during each analysis. Images were acquired using a Leica SP8 equipped with 405-, 488-, 561-, and 647-nm lasers and prepared for submission using the basic features of the LAS X software.
Bead Uptake Assay.
For magnetic isolation of hemocytes, we followed the protocol by Smith et al. (26). Briefly, 20 females were cold-anesthetized and injected with 300 µL of a 2 mg/mL suspension of MagnaBind Carboxyl Derivatized Beads (Thermo Scientific). Mosquitoes were kept at 28 °C or 4 °C for 2 h and perfused. Hemolymph was collected with a pipette tip and transferred into a 0.5-µL Eppendorf tube containing 100 µL of injection buffer. Samples were diluted to 200 µL and incubated in a magnetic stand for 20 min at 4 °C. Supernatant was removed by pipetting, and the pellet was ressuspended in RNase-free PBS and transferred to a microscopy slide. Cells were allowed to attach for 15 min and then processed for RNA-FISH.
Transwell Assay.
Hemolymph was collected onto the top of a glass coverslip placed inside a 24-well plate. A total of 100 µL of buffer was added to prevent dehydration. Cell inserts (Merck) were then placed over individual wells, and hemolymph from PPO6::RFP females was gently pipetted onto the 1-µm membrane. Diluted hemolymph from at least two WT and four transgenic mosquitoes was used per treatment. Plates were kept at room temperature for 1 h, fixed with PFA, washed, and immediately processed based on the RNAscope manual. Images were obtained by confocal microscopy as described above. Experiments were repeated at least twice.
Scanning Electron Microscopy.
Cells were perfused from at least two females directly onto coverslips and fixed with 4% PFA. To facilitate exosome imaging, poly-l-lysine–treated coverslips were used. For correlative SEM, cells were placed onto microscopic dishes with finder grids (ibidi) and imaged directly after fixation using a Zeiss Axiovert microscope, before SEM processing. Samples were postfixed in 2.5% glutaraldehyde, 0.5% osmium-tetroxide, tannic acid, and osmium-tetroxide again. The coverslips or optical membranes were then dehydrated in a graded ethanol series, dried in carbon dioxide at a critical point, and vacuum coated with 3 nm of Carbon-Platinum. Imaging was performed using a LEO 1550 (Zeiss) scanning electron microscope. Experiments were repeated at least twice.
Transmission Electron Microscopy.
EVs were isolated as described before (80). Hemolymph of at least 20 mosquitoes was differentially centrifuged at 10,000 × g at 4 °C, and pellets were processed for negative staining electron microscopy. Aliquots were applied to freshly glow discharged carbon- and pioloform film-coated copper grids and allowed to adsorb for 10 min. After washes, the grids were contrasted with 2% uranyl acetate, touched on filter paper, and air-dried. The grids were examined in a LEO 906 (Zeiss AG) electron microscope operated at 100 kV, and images were recorded with a Morada (SIS-Olympus) digital camera.
Supplementary Material
Acknowledgments
We thank S. Zakovic for sharing the drawing in Fig. 1 and all members of the Vector Biology Unit for intellectual input and technical support. We thank Dr. N. Regev-Rudzki for discussions and critical reading of the manuscript. We thank Dr. E. Marois (UPR9022 CNRS, U963 Inserm) for sharing the transgenic PPO6::RFP line and Dr. K. Müller (Humboldt University) for providing A. stephensi mosquitoes. We also acknowledge the support provided by the Flow Cytometry Core Facility [German Rheumatism Research Centre (DRFZ)/Max Planck Institut for Infection Biology (MPIIB)] and by I. Wagner (Microarrays Core Facility, MPIIB). We acknowledge funding by the CNRS Laboratoire International Associé “REL2 and resistance to malaria” project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB23372). Analyses were performed in R, and scripts are available on GitHub (https://github.com/mssevero/hemo-scRNASeq). The expression data can be accessed at https://scb.sanger.ac.uk/#/base/main for single gene visualization.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803062115/-/DCSupplemental.
References
- 1.Ottaviani E, Franceschi C. The invertebrate phagocytic immunocyte: Clues to a common evolution of immune and neuroendocrine systems. Immunol Today. 1997;18:169–174. doi: 10.1016/s0167-5699(97)84663-4. [DOI] [PubMed] [Google Scholar]
- 2.Ribeiro C, Brehélin M. Insect haemocytes: What type of cell is that? J Insect Physiol. 2006;52:417–429. doi: 10.1016/j.jinsphys.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. Superoxide production in Galleria mellonella hemocytes: Identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun. 2005;73:4161–4170. doi: 10.1128/IAI.73.7.4161-4170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne N, Heelan M, Kavanagh K. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence. 2013;4:597–603. doi: 10.4161/viru.25906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmann K. Evolution of innate Immunity: Clues from invertebrates via fish to mammals. Front Immunol. 2014;5:459. doi: 10.3389/fimmu.2014.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa SC, Ribeiro C, Girard PA, Zumbihl R, Brehélin M. Modes of phagocytosis of Gram-positive and Gram-negative bacteria by Spodoptera littoralis granular haemocytes. J Insect Physiol. 2005;51:39–46. doi: 10.1016/j.jinsphys.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 8.Brayner FA, Araújo HR, Cavalcanti MG, Alves LC, Peixoto CA. Ultrastructural characterization of the hemocytes of Culex quinquefasciatus (DIPTERA: Culicidae) Micron. 2005;36:359–367. doi: 10.1016/j.micron.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Castillo JC, Robertson AE, Strand MR. Characterization of hemocytes from the mosquitoes Anopheles gambiae and Aedes aegypti. Insect Biochem Mol Biol. 2006;36:891–903. doi: 10.1016/j.ibmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernández S, et al. Morphological and cytochemical characterization of female Anopheles albimanus (Diptera: Culicidae) hemocytes. J Med Entomol. 1999;36:426–434. doi: 10.1093/jmedent/36.4.426. [DOI] [PubMed] [Google Scholar]
- 11.Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics. 2008;180:1671–1678. doi: 10.1534/genetics.108.089748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlisidou I, Wood W. Drosophila blood cells and their role in immune responses. FEBS J. 2015;282:1368–1382. doi: 10.1111/febs.13235. [DOI] [PubMed] [Google Scholar]
- 13.Wood W, Jacinto A. Drosophila melanogaster embryonic haemocytes: Masters of multitasking. Nat Rev Mol Cell Biol. 2007;8:542–551. doi: 10.1038/nrm2202. [DOI] [PubMed] [Google Scholar]
- 14.Zdobnov EM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 15.WHO . A Global Brief on Vector-Borne Diseases. WHO Press; Geneva: 2014. [Google Scholar]
- 16.Hillyer JF, Schmidt SL, Christensen BM. Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J Parasitol. 2003;89:62–69. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Yassine H, Kamareddine L, Osta MA. The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 2012;8:e1003029. doi: 10.1371/journal.ppat.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blandin S, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 19.Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity. 2006;25:677–685. doi: 10.1016/j.immuni.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Hillyer JF, Christensen BM. Characterization of hemocytes from the yellow fever mosquito, Aedes aegypti. Histochem Cell Biol. 2002;117:431–440. doi: 10.1007/s00418-002-0408-0. [DOI] [PubMed] [Google Scholar]
- 21.Araújo HC, Cavalcanti MG, Santos SS, Alves LC, Brayner FA. Hemocytes ultrastructure of Aedes aegypti (Diptera: Culicidae) Micron. 2008;39:184–189. doi: 10.1016/j.micron.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Hillyer JF, Schmidt SL, Christensen BM. Hemocyte-mediated phagocytosis and melanization in the mosquito Armigeres subalbatus following immune challenge by bacteria. Cell Tissue Res. 2003;313:117–127. doi: 10.1007/s00441-003-0744-y. [DOI] [PubMed] [Google Scholar]
- 23.Bryant WB, Michel K. Blood feeding induces hemocyte proliferation and activation in the African malaria mosquito, Anopheles gambiae Giles. J Exp Biol. 2014;217:1238–1245. doi: 10.1242/jeb.094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baton LA, Robertson A, Warr E, Strand MR, Dimopoulos G. Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium berghei infection. BMC Genomics. 2009;10:257. doi: 10.1186/1471-2164-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto SB, et al. Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae. Proc Natl Acad Sci USA. 2009;106:21270–21275. doi: 10.1073/pnas.0909463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith RC, et al. Molecular profiling of phagocytic immune cells in Anopheles gambiae reveals integral roles for hemocytes in mosquito innate immunity. Mol Cell Proteomics. 2016;15:3373–3387. doi: 10.1074/mcp.M116.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353–1355. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith RC, Barillas-Mury C, Jacobs-Lorena M. Hemocyte differentiation mediates the mosquito late-phase immune response against Plasmodium in Anopheles gambiae. Proc Natl Acad Sci USA. 2015;112:E3412–E3420. doi: 10.1073/pnas.1420078112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King JG, Hillyer JF. Spatial and temporal in vivo analysis of circulating and sessile immune cells in mosquitoes: Hemocyte mitosis following infection. BMC Biol. 2013;11:55. doi: 10.1186/1741-7007-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. A systematic study on hemocyte identification and plasma prophenoloxidase from Culex pipiens quinquefasciatus at different developmental stages. Exp Parasitol. 2011;127:135–141. doi: 10.1016/j.exppara.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Castillo J, Brown MR, Strand MR. Blood feeding and insulin-like peptide 3 stimulate proliferation of hemocytes in the mosquito Aedes aegypti. PLoS Pathog. 2011;7:e1002274. doi: 10.1371/journal.ppat.1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaublomme JT, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 34.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volohonsky G, et al. Tools for Anopheles gambiae transgenesis. G3 (Bethesda) 2015;5:1151–1163. doi: 10.1534/g3.115.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham EG, et al. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc Natl Acad Sci USA. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen BM, Li J, Chen CC, Nappi AJ. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005;21:192–199. doi: 10.1016/j.pt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Hillyer JF, Strand MR. Mosquito hemocyte-mediated immune responses. Curr Opin Insect Sci. 2014;3:14–21. doi: 10.1016/j.cois.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hillyer JF. Mosquito immunity. Adv Exp Med Biol. 2010;708:218–238. doi: 10.1007/978-1-4419-8059-5_12. [DOI] [PubMed] [Google Scholar]
- 41.Bryant WB, Michel K. Anopheles gambiae hemocytes exhibit transient states of activation. Dev Comp Immunol. 2016;55:119–129. doi: 10.1016/j.dci.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Björklund AK, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 47.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmona SJ, et al. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017;27:451–461. doi: 10.1101/gr.207704.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284:9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Estévez-Lao TY, Hillyer JF. Involvement of the Anopheles gambiae Nimrod gene family in mosquito immune responses. Insect Biochem Mol Biol. 2014;44:12–22. doi: 10.1016/j.ibmb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 51.Lombardo F, Ghani Y, Kafatos FC, Christophides GK. Comprehensive genetic dissection of the hemocyte immune response in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9:e1003145. doi: 10.1371/journal.ppat.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun A, Lemaitre B, Lanot R, Zachary D, Meister M. Drosophila immunity: Analysis of larval hemocytes by P-element-mediated enhancer trap. Genetics. 1997;147:623–634. doi: 10.1093/genetics/147.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci USA. 2003;100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minakhina S, Tan W, Steward R. JAK/STAT and the GATA factor Pannier control hemocyte maturation and differentiation in Drosophila. Dev Biol. 2011;352:308–316. doi: 10.1016/j.ydbio.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker SC, et al. External RNA Controls Consortium The External RNA Controls Consortium: A progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 56.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 57.Dudzic JP, Kondo S, Ueda R, Bergman CM, Lemaitre B. Drosophila innate immunity: Regional and functional specialization of prophenoloxidases. BMC Biol. 2015;13:81. doi: 10.1186/s12915-015-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsao IY, Lin US, Christensen BM, Chen CC. Armigeres subalbatus prophenoloxidase. III. Cloning, characterization and potential role in morphogenesis. Insect Biochem Mol Biol. 2009;39:96–104. doi: 10.1016/j.ibmb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 59.Tsao IY, et al. The dual roles of Armigeres subalbatus prophenoloxidase V in parasite melanization and egg chorion melanization in the mosquito Ar. subalbatus. Insect Biochem Mol Biol. 2015;64:68–77. doi: 10.1016/j.ibmb.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 60.Smith RC, Jacobs-Lorena M. Malaria parasite Pfs47 disrupts JNK signaling to escape mosquito immunity. Proc Natl Acad Sci USA. 2015;112:1250–1251. doi: 10.1073/pnas.1424227112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castillo JC, Ferreira ABB, Trisnadi N, Barillas-Mury C. Activation of mosquito complement antiplasmodial response requires cellular immunity. Sci Immunol. 2017;2:eaal1505. doi: 10.1126/sciimmunol.aal1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.King JG, Hillyer JF. Infection-induced interaction between the mosquito circulatory and immune systems. PLoS Pathog. 2012;8:e1003058. doi: 10.1371/journal.ppat.1003058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Headland SE, Jones HR, D’Sa AS, Perretti M, Norling LV. Cutting-edge analysis of extracellular microparticles using ImageStream(X) imaging flow cytometry. Sci Rep. 2014;4:5237. doi: 10.1038/srep05237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 65.Ramirez JL, et al. The role of hemocytes in Anopheles gambiae antiplasmodial immunity. J Innate Immun. 2014;6:119–128. doi: 10.1159/000353765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hulsmans M, et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169:510-522.e520. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van De Bor V, et al. Companion blood cells control ovarian stem cell niche Microenvironment and homeostasis. Cell Reports. 2015;13:546–560. doi: 10.1016/j.celrep.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 68.Dzik JM. Evolutionary roots of arginase expression and regulation. Front Immunol. 2014;5:544. doi: 10.3389/fimmu.2014.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arendt D, et al. The origin and evolution of cell types. Nat Rev Genet. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 70.Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PLoS One. 2012;7:e38258. doi: 10.1371/journal.pone.0038258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 72.Ratajczak J, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 73.Tassetto M, Kunitomi M, Andino R. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell. 2017;169:314–325.e313. doi: 10.1016/j.cell.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Volohonsky G, et al. Transgenic expression of the anti-parasitic factor TEP1 in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2017;13:e1006113. doi: 10.1371/journal.ppat.1006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 76.Dobin A, et al. Star: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawson D, et al. VectorBase: A home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 80.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.